ABSTRACT
The emergence and rapid dissemination of colistin-resistant Escherichia coli carrying the plasmid-mediated mcr-1 gene have created an urgent need to develop specific screening methods. In this study, we evaluated four assays based on the inhibition of MCR-1 activity by EDTA: (i) a combined-disk test (CDT) comparing the inhibition zones of colistin and colistin (10 μg) plus EDTA (100 mM); (ii) reduction of colistin MIC (CMR) in the presence of EDTA (80 μg/ml); (iii) a modified rapid polymyxin Nordmann/Poirel test (MPNP); and (iv) alteration of zeta potential (RZP = ZP+EDTA/ZP−EDTA). We obtained encouraging results for the detection of MCR-1 in E. coli isolates recovered from human, food, and animal samples, using the following assay parameters: ≥3 mm difference in the inhibition zones between colistin disks without and with EDTA; ≥4-fold colistin MIC decrease in the presence of EDTA; RZP of ≥2.5; and the absence of metabolic activity and proliferation, indicated by unchanged color of phenol red in the presence of colistin-EDTA, in the MPNP test. In this regard, the CDT, CMR, RZP, and MPNP assays exhibited sensitivities of 96.7, 96.7, 95.1, and 96.7% and specificities of 89.6, 83.3, 100, and 100%, respectively, for detecting MCR-1-positive E. coli. Our results demonstrate that inhibition by EDTA and zeta potential assays may provide simple and inexpensive methods for the presumptive detection of MCR-1-producing E. coli isolates in human and veterinary diagnostic laboratories.
KEYWORDS: polymyxins, colistin resistance, combined disk test, polymyxin NP test, charge modification
INTRODUCTION
Colistin (polymyxin E) and polymyxin B belong to a group of polypeptide antibiotics classified as polymyxins, which are considered one of the last lines of therapy for the treatment of lethal infections caused by multidrug-resistant Gram-negative pathogens (1, 2). Th antibacterial activity of polymyxins is based on an electrostatic interaction between cationic polypeptide antibiotics and negatively charged moieties present on the lipid A portion of the lipopolysaccharide (LPS) that form the outer membrane of Gram-negative bacteria (1–3). Consequently, the outer membrane is destabilized, increasing its permeability and leading to leakage of the cytoplasmic content, with subsequent lysis and bactericidal activity. Polymyxin resistance is usually caused by LPS modifications (3). In most resistant strains, 4-amino-4-deoxy-l-arabinose (l-Ara4N), phosphoethanolamine (PEtN), or galactosamine moieties are enzymatically added to the lipid A or the LPS core (1–3). These modifications result in a decrease in a net negative charge of phosphate residues, leading to a reduction in polymyxin affinity (2, 3). Some species are naturally resistant to polymyxins, including Proteus spp., Morganella morganii, Providencia spp., Serratia marcescens, and nonfermentative Burkholderia mallei, and Burkholderia cepacia (2, 4, 5), whereas in clinical isolates of Escherichia coli and Klebsiella pneumoniae, the two-component regulatory systems (TCSs) PmrA/PmrB and PhoP/PhoQ have been identified as regulatory systems involved in resistance to polymyxins, where PmrAB activates l-Ara4N synthesis leading to polymyxin resistance. Additionally, the insertional inactivation of the PhoQ/PhoP mgrB-encoding regulator has also been associated with colistin resistance (2, 3).
Recently, the plasmid-encoded polymyxin resistance determinant MCR-1 has been identified in clinically significant Enterobacteriaceae (particularly E. coli), and new plasmid-borne colistin resistance genes, mcr-2, mcr-3, and mcr-4, were further described (6–12). mcr-type genes encode phosphoethanolamine transferases that add PEtN to the phosphate group of the lipid A moiety (at the 4′ position) anchored on LPS, reduce negative charges that are present in LPS, and consequently confer resistance to polymyxins (4, 9–12). The mcr-1 gene has been identified as a plasmid-mediated resistance mechanism being widely disseminated among human, animal, food, and environmental E. coli isolates (6–8, 13–19).
Recent structural studies have revealed that the catalytic domain of the MCR-1 phosphoethanolamine transferase resembles a zinc metalloprotein, where zinc deprivation has reduced colistin MICs in MCR-1-producing E. coli isolated from different sources, revealing the importance of zinc to MCR-1 activity and supporting the notion that assays under zinc-limiting conditions could represent a strategy for phenotypic detection of MCR-1 (9, 20–23). Since the emergence of both intrinsic and transferable mechanisms of polymyxin resistance is becoming a critical issue worldwide, the development of rapid and reliable methods to determine the susceptibility and resistance to polymyxins is an urgent need for clinical laboratories. In addition, phenotypic tests for screening colistin-resistant Escherichia coli carrying the plasmid-mediated mcr-1 gene are highly desirable (24, 25). In this study, we evaluated four specific assays based on the inhibition of the MCR-1 activity by EDTA: (i) a combined-disk test (CDT) comparing the inhibition zones of colistin and colistin (10 μg) plus EDTA (100 mM); (ii) reduction of colistin MIC (CMR) in the presence of EDTA (80 mg/liter); (iii) a modified rapid polymyxin Nordmann/Poirel test (MPNP); and (iv) alteration of zeta potential (RZP = ZP+EDTA/ZP−EDTA).
RESULTS
MCR-1 detection by CDT.
From the different EDTA concentrations tested, 100 mM EDTA was chosen for inhibition activity of MCR-1 in the CDT, since this concentration showed no inhibitory activity across the bacterial growth of all screened isolates when sterile blank disks impregnated with 10 μl of 100 mM EDTA were tested. When all colistin-resistant (MIC, >2 μg/ml) MCR-1-positive E. coli isolates were analyzed, an increase of ≥3 mm in the size of inhibition zones around the 10 μg of colistin–100 mM EDTA in comparison to the inhibition zones of colistin without EDTA was observed. Table 1 summarizes the results of the CDT, which was performed three times on distinct dates. Under these conditions, the sensitivity (SN) and specificity (SP) of CDT were 96.7 and 89.6%, respectively. In this regard, a colistin-susceptible (MIC 1 μg/ml) MCR-1-positive E. coli isolate (strain ICBEC 146) and a colistin-resistant MCR-1-positive K. pneumoniae isolate (CCBH24080) were not identified by the CDT (Table 1). On the other hand, five colistin-resistant MCR-1-negative E. coli isolates displayed an increase of ≥3 mm in the size of inhibition zones around colistin-EDTA.
TABLE 1.
Evaluation of CDT using EDTA for detection of MCR-1-producing Escherichia coli
| Isolatea | Species | Source | Inhibition zone diam (mm)b |
Mechanism of colistin resistance | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Assay 1 |
Assay 2 |
Assay 3 |
||||||||||
| −E | +E | Increase | −E | +E | Increase | −E | +E | Increase | ||||
| Colistin-resistant isolates | ||||||||||||
| 50H | E. coli | Human | 9 | 13 | 4 | 10 | 13 | 3 | 10 | 14 | 4 | Plasmid-mediated mcr-1 gene |
| 51H | E. coli | Human | 10 | 14 | 4 | 10 | 13 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| 77H | E. coli | Human | 11 | 14 | 4 | 10 | 13 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| 200H | E. coli | Human | 10 | 13 | 3 | 9 | 13 | 4 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| ICBEC 72H | E. coli | Human | 10 | 13 | 3 | 10 | 13 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| ICBEC 79H | E. coli | Human | 10 | 13 | 3 | 9 | 12 | 3 | 9 | 12 | 3 | Plasmid-mediated mcr-1 gene |
| ICBEC 2.6 | E. coli | Chicken | 11 | 14 | 3 | 10 | 13 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| ICBEC 3.6 | E. coli | Chicken | 10 | 13 | 3 | 10 | 13 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| ICBEC 5.2.1 | E. coli | Chicken | 9 | 12 | 3 | 9 | 12 | 4 | 9 | 13 | 4 | Plasmid-mediated mcr-1 gene |
| ICBEC 5.3 | E. coli | Chicken | 9 | 12 | 3 | 8 | 12 | 4 | 8 | 12 | 4 | Plasmid-mediated mcr-1 gene |
| ICBEC 5.5 | E. coli | Chicken | 10 | 14 | 4 | 10 | 14 | 4 | 10 | 14 | 4 | Plasmid-mediated mcr-1 gene |
| ICBEC 6.3 | E. coli | Chicken | 10 | 13 | 3 | 10 | 13 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| ICBEC 9.3 | E. coli | Chicken | 11 | 14 | 3 | 11 | 14 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| ICBEC 9.6 | E. coli | Chicken | 10 | 14 | 4 | 10 | 13 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| ICBEC 11.3 | E. coli | Chicken | 11 | 14 | 3 | 10 | 13 | 3 | 11 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| ICBEC 11.8 | E. coli | Chicken | 11 | 14 | 3 | 10 | 13 | 3 | 11 | 14 | 3 | Plasmid-mediated mcr-1 gene |
| ICBEC 12.3 | E. coli | Chicken | 9 | 13 | 4 | 10 | 14 | 4 | 10 | 14 | 4 | Plasmid-mediated mcr-1 gene |
| ICBEC 12.6 | E. coli | Chicken | 10 | 13 | 3 | 11 | 14 | 3 | 11 | 14 | 3 | Plasmid-mediated mcr-1 gene |
| 96 | E. coli | Chicken | 10 | 13 | 3 | 11 | 14 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| 662 | E. coli | Chicken | 11 | 14 | 3 | 11 | 14 | 3 | 11 | 14 | 3 | Plasmid-mediated mcr-1 gene |
| 284 | E. coli | Bovine | 8 | 12 | 4 | 8 | 12 | 4 | 9 | 13 | 4 | Plasmid-mediated mcr-1 gene |
| 946 | E. coli | Bovine | 8 | 12 | 4 | 9 | 13 | 4 | 8 | 12 | 4 | Plasmid-mediated mcr-1 gene |
| CF 1.2 | E. coli | Chicken meat | 9 | 12 | 3 | 10 | 13 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| CF 101 | E. coli | Chicken meat | 10 | 13 | 3 | 10 | 13 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| CF 111 | E. coli | Chicken meat | 9 | 12 | 3 | 10 | 13 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| CF 131 | E. coli | Chicken meat | 9 | 12 | 3 | 9 | 13 | 4 | 9 | 12 | 3 | Plasmid-mediated mcr-1 gene |
| CF 132 | E. coli | Chicken meat | 10 | 13 | 3 | 9 | 13 | 4 | 9 | 12 | 3 | Plasmid-mediated mcr-1 gene |
| CF 341 | E. coli | Chicken meat | 9 | 13 | 4 | 10 | 13 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| CF 351 | E. coli | Chicken meat | 10 | 13 | 3 | 10 | 13 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| 28 | E. coli | Turkey | 9 | 12 | 3 | 9 | 12 | 3 | 10 | 14 | 4 | Plasmid-mediated mcr-1 gene |
| 69 | E. coli | Turkey | 9 | 12 | 3 | 11 | 14 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| 71 | E. coli | Turkey | 10 | 13 | 3 | 8 | 11 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| 73 | E. coli | Turkey | 11 | 14 | 3 | 10 | 13 | 3 | 9 | 12 | 3 | Plasmid-mediated mcr-1 gene |
| 75 | E. coli | Turkey | 10 | 13 | 3 | 10 | 13 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| 77 | E. coli | Turkey | 10 | 13 | 3 | 10 | 14 | 4 | 9 | 12 | 3 | Plasmid-mediated mcr-1 gene |
| 79 | E. coli | Turkey | 10 | 14 | 4 | 10 | 13 | 3 | 10 | 14 | 4 | Plasmid-mediated mcr-1 gene |
| 84 | E. coli | Turkey | 10 | 13 | 3 | 10 | 13 | 3 | 9 | 13 | 4 | Plasmid-mediated mcr-1 gene |
| 93 | E. coli | Turkey | 10 | 13 | 3 | 10 | 13 | 3 | 9 | 12 | 3 | Plasmid-mediated mcr-1 gene |
| 02 | E. coli | Swine | 10 | 13 | 3 | 10 | 14 | 4 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| 06 | E. coli | Swine | 9 | 12 | 3 | 9 | 13 | 4 | 9 | 12 | 3 | Plasmid-mediated mcr-1 gene |
| 08 | E. coli | Swine | 10 | 13 | 3 | 9 | 13 | 4 | 10 | 14 | 4 | Plasmid-mediated mcr-1 gene |
| 10 | E. coli | Swine | 9 | 12 | 3 | 10 | 14 | 4 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| 12 | E. coli | Swine | 8 | 11 | 3 | 10 | 13 | 3 | 9 | 12 | 3 | Plasmid-mediated mcr-1 gene |
| 14 | E. coli | Swine | 10 | 13 | 3 | 9 | 12 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| 18 | E. coli | Swine | 8 | 11 | 3 | 10 | 13 | 3 | 9 | 13 | 4 | Plasmid-mediated mcr-1 gene |
| 24 | E. coli | Swine | 10 | 13 | 3 | 10 | 14 | 4 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| 26 | E. coli | Swine | 10 | 13 | 3 | 10 | 13 | 3 | 9 | 12 | 3 | Plasmid-mediated mcr-1 gene |
| 29 | E. coli | Swine | 11 | 14 | 3 | 9 | 13 | 4 | 10 | 14 | 4 | Plasmid-mediated mcr-1 gene |
| 33 | E. coli | Swine | 10 | 13 | 3 | 11 | 14 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| 35 | E. coli | Swine | 9 | 13 | 4 | 8 | 13 | 5 | 8 | 12 | 4 | Plasmid-mediated mcr-1 gene |
| 53 | E. coli | Swine | 10 | 13 | 3 | 9 | 12 | 3 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| 55 | E. coli | Swine | 10 | 14 | 4 | 10 | 14 | 4 | 9 | 12 | 3 | Plasmid-mediated mcr-1 gene |
| 60 | E. coli | Swine | 11 | 15 | 4 | 10 | 14 | 4 | 12 | 15 | 3 | Plasmid-mediated mcr-1 gene |
| 86 | E. coli | Swine | 10 | 13 | 3 | 10 | 14 | 4 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| 88 | E. coli | Swine | 11 | 14 | 3 | 10 | 13 | 3 | 9 | 12 | 3 | Plasmid-mediated mcr-1 gene |
| 95 | E. coli | Swine | 9 | 12 | 3 | 8 | 11 | 3 | 8 | 12 | 4 | Plasmid-mediated mcr-1 gene |
| 98 | E. coli | Swine | 10 | 13 | 3 | 10 | 14 | 4 | 10 | 13 | 3 | Plasmid-mediated mcr-1 gene |
| ICBEC 171 | E. coli | Swine | 9 | 13 | 4 | 9 | 13 | 4 | 10 | 14 | 4 | Plasmid-mediated mcr-1 gene |
| 7P | E. coli | Penguin | 10 | 13 | 3 | 10 | 13 | 3 | 9 | 12 | 3 | Plasmid-mediated mcr-1 gene |
| HC113 | E. coli | Human | 8 | 11 | 3 | 9 | 12 | 3 | 8 | 11 | 3 | Unknown |
| HC629 | E. coli | Human | 12 | 17 | 5 | 10 | 13 | 3 | 10 | 13 | 3 | Unknown |
| M6 | E. coli | Wild bird | 9 | 11 | 2 | 10 | 12 | 2 | 9 | 11 | 2 | Unknown |
| M37A | E. coli | Wild bird | 8 | 12 | 4 | 8 | 12 | 4 | 8 | 12 | 4 | Unknown |
| M51 | E. coli | Wild bird | 8 | 12 | 4 | 8 | 12 | 4 | 8 | 12 | 4 | Unknown |
| M55 | E. coli | Wild bird | 8 | 12 | 4 | 9 | 13 | 4 | 10 | 13 | 3 | Unknown |
| Δ806 mutant | E. coli | Human | 10 | 12 | 2 | 10 | 11 | 1 | 10 | 12 | 2 | PmrB P92T |
| CCBH24080 | K. pneumoniae | Human | 9 | 10 | 1 | 10 | 10 | 0 | 9 | 11 | 2 | Plasmid-mediated mcr-1 gene |
| Alerta 06 | K. pneumoniae | Human | 13 | 14 | 1 | 13 | 13 | 0 | 13 | 13 | 0 | phoQ overexpressionc |
| Alerta 08 | K. pneumoniae | Human | 14 | 14 | 0 | 14 | 14 | 0 | 13 | 14 | 1 | Unknown |
| Alerta 09 | K. pneumoniae | Human | 14 | 14 | 0 | 13 | 14 | 1 | 13 | 13 | 0 | Decreased mgrB expressionc |
| Alerta 10 | K. pneumoniae | Human | 9 | 9 | 0 | 10 | 11 | 1 | 10 | 10 | 0 | phoP/phoQ overexpressionc |
| Alerta 12 | K. pneumoniae | Human | 12 | 12 | 0 | 10 | 10 | 0 | 12 | 12 | 0 | MgrB IS903-like |
| Alerta 13 | K. pneumoniae | Human | 9 | 9 | 0 | 9 | 9 | 0 | 9 | 9 | 0 | MgrB IS903b-like |
| Alerta 14 | K. pneumoniae | Human | 8 | 8 | 0 | 9 | 9 | 0 | 8 | 8 | 0 | MgrB truncated |
| Alerta 15 | K. pneumoniae | Human | 9 | 10 | 1 | 8 | 9 | 1 | 9 | 10 | 1 | MgrB ISKpn13 |
| Alerta 16 | K. pneumoniae | Human | 10 | 10 | 0 | 9 | 10 | 1 | 10 | 10 | 0 | mgrB promoter IS1 family |
| Alerta 17 | K. pneumoniae | Human | 10 | 12 | 2 | 10 | 10 | 0 | 11 | 11 | 0 | Unknown |
| Alerta 31 | K. pneumoniae | Human | 11 | 11 | 0 | 12 | 12 | 0 | 11 | 11 | 1 | PmrB T246C-R256G |
| Alerta 32 | K. pneumoniae | Human | 12 | 12 | 0 | 12 | 12 | 0 | 12 | 12 | 0 | PmrB T246A-R256G-A282T-V290G-E291K |
| Alerta 33 | K. pneumoniae | Human | 11 | 12 | 1 | 10 | 11 | 1 | 10 | 11 | 1 | Unknown |
| Alerta 35 | K. pneumoniae | Human | 10 | 11 | 1 | 10 | 11 | 1 | 10 | 11 | 1 | Unknown |
| Alerta 36 | K. pneumoniae | Human | 12 | 12 | 0 | 12 | 13 | 1 | 11 | 12 | 1 | Unknown |
| Alerta 37 | K. pneumoniae | Human | 11 | 12 | 1 | 10 | 11 | 1 | 10 | 11 | 1 | Unknown |
| Alerta 38 | K. pneumoniae | Human | 10 | 11 | 1 | 11 | 12 | 1 | 11 | 12 | 1 | PhoP W84C |
| Alerta 39 | K. pneumoniae | Human | 11 | 12 | 1 | 12 | 13 | 1 | 11 | 12 | 1 | MgrB IS903b-like |
| Kp 148 | K. pneumoniae | Human | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Unknown |
| BL-II-04(2) | M. morganii | Human | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Intrinsic |
| SM 26 | S. marcescens | Human | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Intrinsic |
| 25933 | P. mirabilis | ATCC | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | Intrinsic |
| Colistin-susceptible isolates | ||||||||||||
| 31 | E. coli | Turkey | 13 | 13 | 0 | 12 | 13 | 1 | 13 | 13 | 0 | |
| 51 | E. coli | Turkey | 12 | 13 | 1 | 11 | 13 | 2 | 11 | 11 | 0 | |
| 91 | E. coli | Turkey | 12 | 13 | 1 | 11 | 12 | 1 | 11 | 13 | 2 | |
| 04 | E. coli | Swine | 12 | 13 | 1 | 11 | 13 | 2 | 10 | 12 | 2 | |
| 49 | E. coli | Swine | 12 | 14 | 2 | 10 | 12 | 2 | 11 | 13 | 2 | |
| 58 | E. coli | Swine | 13 | 13 | 0 | 13 | 14 | 1 | 10 | 12 | 2 | |
| 62 | E. coli | Swine | 13 | 14 | 1 | 11 | 13 | 2 | 11 | 12 | 1 | |
| 64 | E. coli | Swine | 14 | 15 | 1 | 14 | 15 | 1 | 13 | 14 | 1 | |
| 65 | E. coli | Swine | 11 | 12 | 1 | 13 | 13 | 0 | 13 | 13 | 0 | |
| 89 | E. coli | Swine | 13 | 14 | 1 | 12 | 14 | 2 | 11 | 12 | 1 | |
| 100 | E. coli | Swine | 12 | 13 | 1 | 10 | 10 | 0 | 12 | 12 | 0 | |
| 60198 | E. coli | Human | 12 | 13 | 1 | 11 | 12 | 1 | 11 | 12 | 1 | |
| ICBEC 146 | E. coli | Swine | 9 | 10 | 1 | 10 | 10 | 0 | 10 | 10 | 1 | Plasmid-mediated mcr-1 gene |
| 25922 | E. coli | ATCC | 13 | 13 | 0 | 14 | 14 | 0 | 13 | 13 | 0 | |
| Alerta 26 | K. pneumoniae | Human | 10 | 11 | 1 | 10 | 12 | 2 | 11 | 12 | 1 | |
| Alerta 27 | K. pneumoniae | Human | 11 | 12 | 1 | 10 | 11 | 1 | 11 | 12 | 1 | |
| Alerta 28 | K. pneumoniae | Human | 12 | 12 | 0 | 12 | 12 | 0 | 10 | 11 | 1 | |
| Alerta 29 | K. pneumoniae | Human | 10 | 11 | 1 | 11 | 12 | 1 | 11 | 12 | 1 | |
| Alerta 30 | K. pneumoniae | Human | 10 | 11 | 1 | 11 | 12 | 1 | 10 | 12 | 2 | |
| 13883 | K. pneumoniae | ATCC | 13 | 13 | 0 | 14 | 14 | 0 | 13 | 13 | 0 | |
PFGE and/or MLST data obtained from earlier studies revealed that most mcr-1-positive E. coli isolates were clonally unrelated (8, 13–17, 33).
The combined disk test method was performed in triplicate. Two 10-μg colistin disks without (−E) and with (+E) EDTA (10 μl of a 100 mM solution [pH 8]) were used. An increase of ≥3 mm in the inhibition zone diameter in the presence of EDTA was considered a positive result.
Although a variation in the gene expression was verified compared to the K. pneumoniae MGH 78578 strain, no mutations in amino acid or nucleotide sequences were detected in any studied genes.
CMR in the presence of EDTA.
For CMR assays, the final concentration of EDTA was fixed at 80 μg/ml, since this concentration showed no antibacterial activity against all colistin-resistant screened isolates, allowing us to observe a ≥4-fold colistin MIC decrease among MCR-1-positive E. coli isolates in the presence of EDTA. In Table 2, the results of reproducible replicates, performed three times on 3 distinct occasions, are shown. Under these conditions, the SN and SP of CMR were 96.7 and 83.3%, respectively. Nevertheless, for both colistin-susceptible MCR-1-positive E. coli strain ICBEC 146 and colistin-resistant MCR-1-positive K. pneumoniae strain CCBH24080, only a 2-fold colistin MIC decrease was recorded (Table 2). Moreover, two MCR-1-negative colistin-susceptible (isolates 58 and 89) and six MCR-1-negative colistin-resistant E. coli strains (isolates HC113, HC629, M6, M37A, M51, and M55) exhibited a ≥4-fold colistin MIC decrease in the presence of EDTA.
TABLE 2.
Alteration of zeta potential, MPNP test results, and CMR induced by EDTA for detection of MCR-1-producing Escherichia coli
| Isolate | Species | Zeta potential (mean [SD]) (mV)a |
Colistin MIC (μg/ml)b |
MIC reduction (fold change)c | MPNP test resultd | mcr-1 confirmation | |||
|---|---|---|---|---|---|---|---|---|---|
| −E | +E | RZP | −E | +E | |||||
| Colistin-resistant isolates | |||||||||
| 50H | E. coli | −7.09 (0.54) | −38.21 (1.09) | 5.39 | 16 | 2 | 8 | + | + |
| 51H | E. coli | −4.23 (0.56) | −37.65 (1.78) | 8.90 | 8 | 1 | 8 | + | + |
| 77H | E. coli | −5.98 (0.45) | −38.09 (1.08) | 6.37 | 8 | 0.5 | 16 | + | + |
| 200H | E. coli | −5.32 (0.97) | −32.09 (1.65) | 6.03 | 16 | 1 | 16 | + | + |
| ICBEC 72H | E. coli | −4.91 (0.53) | −34.06 (1.02) | 6.94 | 8 | 1 | 8 | + | + |
| ICBEC 79H | E. coli | −4.88 (0.46) | −39.50 (0.39) | 8.09 | 8 | 1 | 8 | + | + |
| ICBEC 2.6 | E. coli | −4.88 (0.90) | −40.48 (0.29) | 8.30 | 8 | 1 | 8 | + | + |
| ICBEC 3.6 | E. coli | −4.23 (0.89) | −36.52 (0.91) | 8.63 | 8 | 1 | 8 | + | + |
| ICBEC 5.2.1 | E. coli | −5.73 (1.00) | −37.24 (0.41) | 6.50 | 8 | 2 | 4 | + | + |
| ICBEC 5.3 | E. coli | −7.19 (0.63) | −40.62 (0.57) | 5.65 | 8 | 0.5 | 16 | + | + |
| ICBEC 5.5 | E. coli | −8.19 (1.72) | −39.85 (1.61) | 4.87 | 8 | 0.5 | 16 | + | + |
| ICBEC 6.3 | E. coli | −5.28 (0.95) | −35.71 (0.87 | 6.76 | 8 | 1 | 8 | + | + |
| ICBEC 9.3 | E. coli | −6.05 (0.34) | −40.26 (0.98) | 6.65 | 8 | 0.5 | 16 | + | + |
| ICBEC 9.6 | E. coli | −7.20 (0.91) | −33.30 (0.95) | 4.63 | 8 | 1 | 8 | + | + |
| ICBEC 11.3 | E. coli | −8.92 (0.48) | −34.93 (2.57) | 3.92 | 8 | 2 | 4 | + | + |
| ICBEC 11.8 | E. coli | −7.24 (0.84) | −38.74 (1.16) | 5.35 | 8 | 0.5 | 16 | + | + |
| ICBEC 12.3 | E. coli | −5.19 (0.47) | −40.81 (1.20) | 7.86 | 16 | 2 | 8 | + | + |
| ICBEC 12.6 | E. coli | −7.66 (0.92) | −34.71 (2.26) | 4.53 | 8 | 2 | 4 | + | + |
| 96 | E. coli | −5.04 (0.81) | −35.26 (1.02) | 7.00 | 4 | 0.25 | 16 | + | + |
| 662 | E. coli | −4.37 (1.38) | −38.86 (0.81) | 8.89 | 4 | 1 | 4 | + | + |
| 284 | E. coli | −6.40 (0.52) | −36.42 (1.71) | 5.69 | 4 | 1 | 4 | + | + |
| 946 | E. coli | −4.39 (0.90) | −39.96 (0.76) | 9.10 | 4 | 0.5 | 8 | + | + |
| CF 1.2 | E. coli | −6.08 (1.87) | −31.65 (0.98) | 5.21 | 8 | 2 | 4 | + | + |
| CF 101 | E. coli | −6.31 (0.85) | −34.89 (0.35) | 5.53 | 8 | 2 | 4 | + | + |
| CF 111 | E. coli | −8.76 (0.74) | −31.89 (1.43) | 3.64 | 4 | 0.25 | 16 | + | + |
| CF 131 | E. coli | −9.43 (0.23) | −34.50 (0.29) | 3.66 | 8 | 1 | 8 | + | + |
| CF 132 | E. coli | −7.21 (0.83) | −33.03 (0.98) | 4.58 | 8 | 0.25 | 32 | + | + |
| CF 341 | E. coli | −4.99 (0.67) | −32.01 (0.39) | 6.41 | 4 | 1 | 4 | + | + |
| CF 351 | E. coli | −5.24 (0.87) | −38.67 (0.65) | 7.34 | 8 | 1 | 8 | + | + |
| 28 | E. coli | −6.86 (1.71) | −35.99 (1.02) | 5.25 | 4 | 0.5 | 8 | + | + |
| 69 | E. coli | −8.92 (0.65) | −31.09 (0.74) | 3.49 | 4 | 0.5 | 8 | + | + |
| 71 | E. coli | −4.22 (0.32) | −21.13 (1.02) | 5.01 | 4 | 1 | 4 | + | + |
| 73 | E. coli | −5.67 (0.98) | −37.27 (0.36) | 6.57 | 8 | 1 | 8 | + | + |
| 75 | E. coli | −9.03 (1.16) | −35.27 (0.57) | 3.91 | 4 | 0.5 | 8 | + | + |
| 77 | E. coli | −8.20 (0.99) | −38.19 (1.39) | 4.66 | 4 | 0.25 | 16 | + | + |
| 79 | E. coli | −9.87 (1.67) | −36.78 (0.67) | 3.73 | 4 | 1 | 4 | + | + |
| 84 | E. coli | −7.41 (1.08) | −34.47 (1.32) | 4.65 | 4 | 0.25 | 16 | + | + |
| 93 | E. coli | −6.25 (0.17) | −36.96 (0.96) | 5.91 | 4 | 0.5 | 8 | + | + |
| 02 | E. coli | −6.08 (0.54) | −37.65 (1.03) | 6.19 | 4 | 1 | 4 | + | + |
| 06 | E. coli | −4.75 (0.36) | −38.64 (1.31) | 8.13 | 4 | 0.5 | 8 | + | + |
| 08 | E. coli | −8.57 (1.96) | −37.57 (2.07) | 4.38 | 4 | 0.125 | 32 | + | + |
| 10 | E. coli | −5.68 (0.73) | −37.11 (1.74) | 6.53 | 8 | 1 | 8 | + | + |
| 12 | E. coli | −6.14 (0.20) | −37.97 (1.69) | 6.18 | 4 | 0.5 | 8 | + | + |
| 14 | E. coli | −4.28 (1.98) | −35.83 (0.54) | 8.37 | 4 | 0.125 | 32 | + | + |
| 18 | E. coli | −7.56 (0.84) | −38.51 (1.23) | 5.10 | 8 | 2 | 4 | + | + |
| 24 | E. coli | −6.13 (0.51) | −37.18 (0.84) | 6.10 | 4 | 0.25 | 16 | + | + |
| 26 | E. coli | −6.39 (0.79) | −36.58 (0.38) | 5.72 | 4 | 1 | 4 | + | + |
| 29 | E. coli | −6.54 (0.71) | −37.56 (1.47) | 5.74 | 4 | 1 | 4 | + | + |
| 33 | E. coli | −4.20 (0.87) | −39.20 (1.46) | 9.33 | 4 | 0.5 | 8 | + | + |
| 35 | E. coli | −7.38 (0.95) | −37.02 (0.82) | 5.01 | 4 | 0.125 | 32 | + | + |
| 53 | E. coli | −3.34 (1.78) | −36.79 (0.09) | 11.01 | 4 | 0.06 | 67 | + | + |
| 55 | E. coli | −8.71 (0.37) | −37.56 (1.63) | 4.31 | 4 | 0.5 | 8 | + | + |
| 60 | E. coli | −5.89 (1.19) | −35.73 (0.61) | 6.07 | 8 | 0.5 | 16 | + | + |
| 86 | E. coli | −6.14 (2.10) | −35.85 (0.38) | 5.78 | 4 | 0.5 | 8 | + | + |
| 88 | E. coli | −6.96 (0.33) | −38.11 (0.63) | 5.48 | 4 | 0.5 | 8 | + | + |
| 95 | E. coli | −5.21 (1.96) | −32.28 (0.66) | 6.20 | 4 | 1 | 4 | + | + |
| 98 | E. coli | −6.32 (0.81) | −30.65 (0.12) | 4.85 | 4 | 0.25 | 16 | + | + |
| ICBEC 171 | E. coli | −5.32 (0.64) | −32.41 (1.23) | 6.09 | 8 | 0.5 | 16 | + | + |
| 7P | E. coli | −19.34 (0.58) | −30.42 (0.52) | 1.57 | 8 | 2 | 4 | + | + |
| HC113 | E. coli | −15.03 (1.27) | −17.04 (1.78) | 1.13 | 8 | 1 | 8 | − | − |
| HC629 | E. coli | −24.07 (0.84) | −42.30 (0.88) | 1.75 | 4 | 1 | 4 | − | − |
| M6 | E. coli | −3.52 (1.10) | −7.08 (1.02) | 2.01 | 4 | 1 | 4 | − | − |
| M37A | E. coli | −23.34 (1.58) | −22.51 (1.10) | 0.96 | 4 | 0.5 | 8 | − | − |
| M51 | E. coli | −21.54 (0.24) | −16.02 (0.84) | 0.74 | 8 | 1 | 8 | − | − |
| M55 | E. coli | −28.32 (0.70) | −29.15 (0.20) | 1.03 | 8 | 1 | 8 | − | − |
| Δ806 mutant | E. coli | −23.34 (1.58) | −21.53 (1.05) | 0.92 | 4 | 4 | — | − | − |
| CCBH24080 | K. pneumoniae | −31.9 (0.48) | −28.51 (0.69) | 0.89 | 8 | 4 | 2 | − | + |
| Alerta 06 | K. pneumoniae | −34.85 (1.42) | −37.51 (0.28) | 1.07 | 16 | 16 | — | − | − |
| Alerta 08 | K. pneumoniae | −38.59 (1.78) | −39.83 (2.23) | 1.03 | 4 | 4 | — | − | − |
| Alerta 09 | K. pneumoniae | −35.45 (0.93) | −39.28 (1.49) | 1.11 | 8 | 4 | 2 | − | − |
| Alerta 10 | K. pneumoniae | −36.69 (2.38) | −39.25 (0.66) | 1.07 | 4 | 2 | 2 | − | − |
| Alerta 12 | K. pneumoniae | −29.23 (0.82) | −34.36 (3.22) | 1.18 | 4 | 4 | — | − | − |
| Alerta 13 | K. pneumoniae | −30.59 (0.77) | −34.19 (1.07) | 1.12 | 16 | 16 | — | − | − |
| Alerta 14 | K. pneumoniae | −34.28 (2.28) | −47.21 (1.13) | 1.38 | 8 | 8 | — | − | − |
| Alerta 15 | K. pneumoniae | −40.96 (0.52) | −35.83 (0.54) | 0.87 | 4 | 4 | — | − | − |
| Alerta 16 | K. pneumoniae | −30.56 (2.37) | −33.52 (1.23) | 1.10 | 8 | 8 | — | − | − |
| Alerta 17 | K. pneumoniae | −34.26 (0.88) | −35.52 (1.64) | 1.04 | >32 | >32 | — | − | − |
| Alerta 31 | K. pneumoniae | −31.09 (2.10) | −33.11 (1.24) | 1.06 | 8 | 8 | — | − | − |
| Alerta 32 | K. pneumoniae | −37.65 (1.09) | −38.08 (0.34) | 1.01 | 8 | 4 | 2 | − | − |
| Alerta 33 | K. pneumoniae | −36.03 (1.24) | −35.49 (1.93) | 0.99 | 8 | 8 | — | − | − |
| Alerta 35 | K. pneumoniae | −33.44 (2.47) | −32.10 (0.25) | 0.96 | 8 | 4 | 2 | − | − |
| Alerta 36 | K. pneumoniae | −38.09 (1.40) | −39.89 (1.05) | 1.05 | 4 | 4 | — | − | − |
| Alerta 37 | K. pneumoniae | −39.20 (0.56) | −35.98 (1.86) | 0.92 | 16 | 16 | — | − | − |
| Alerta 38 | K. pneumoniae | −38.76 (0.98) | −39.21 (1.90) | 1.01 | 8 | 8 | — | − | − |
| Alerta 39 | K. pneumoniae | −34.54 (1.03) | −33.23 (0.10) | 0.96 | 8 | 4 | 2 | − | − |
| Kp 148 | K. pneumoniae | −30.91 (0.61) | −33.91 (0.26) | 1.10 | 32 | 32 | — | − | − |
| BL-II-04(2) | M. morganii | −36.91 (1.05) | −36.99 (0.98) | 1.00 | 512 | 512 | — | − | − |
| SM 26 | S. marcescens | −36.85 (1.81) | −33.43 (1.40) | 0.91 | 512 | 512 | — | − | − |
| 25933 | P. mirabilis | −32.90 (1.42) | −35.06 (0.59) | 1.07 | 512 | 512 | — | − | − |
| Colistin-susceptible isolates | |||||||||
| 31 | E. coli | −36.57 (2.08) | −36.08 (1.26) | 0.99 | 2 | 1 | 2 | − | − |
| 51 | E. coli | −30.10 (3.10) | −35.93 (0.52) | 1.19 | 0.06 | 0.06 | — | − | − |
| 91 | E. coli | −33.13 (0.64) | −37.01 (0.57) | 1.12 | 0.25 | 0.25 | — | − | − |
| 04 | E. coli | −31.91 (0.51) | −37.33 (1.81) | 1.17 | 1 | 0.5 | 2 | − | − |
| 49 | E. coli | −29.60 (1.03) | −34.36 (2.71) | 1.16 | 0.06 | 0.06 | — | − | − |
| 58 | E. coli | −31.05 (2.48) | −36.38 (2.36) | 1.17 | 1 | 0.06 | 16 | − | − |
| 62 | E. coli | −32.14 (1.88) | −35.31 (1.14) | 1.10 | 1 | 1 | — | − | − |
| 64 | E. coli | −29.93 (2.63) | −36.48 (0.94) | 1.22 | 1 | 1 | — | − | − |
| 65 | E. coli | −30.23 (0.87) | −30.96 (1.03) | 1.02 | 1 | 0.5 | 2 | − | − |
| 89 | E. coli | −32.85 (1.04) | −37.37 (0.90) | 1.14 | 0.25 | 0.06 | 4 | − | − |
| 100 | E. coli | −35.08 (1.22) | −39.45 (0.93) | 1.12 | 0.25 | 0.25 | — | − | − |
| 60198 | E. coli | −44.21 (0.69) | −39.54 (2.70) | 1.11 | 2 | 1 | 2 | − | − |
| ICBEC 146 | E. coli | −28.21 (2.74) | −30.29 (2.78) | 1.08 | 1 | 0.5 | 2 | − | + |
| 25922 | E. coli | −32.01 (1.75) | −37.53 (0.45) | 1.17 | 1 | 1 | — | − | − |
| Alerta 26 | K. pneumoniae | −30.23 (1.32) | −31.14 (0.98) | 1.03 | 0.5 | 0.5 | — | − | − |
| Alerta 27 | K. pneumoniae | −38.90 (1.98) | −38.21 (0.35) | 0.98 | 0.25 | 0.25 | — | − | − |
| Alerta 28 | K. pneumoniae | −39.24 (1.47) | −38.98 (0.64) | 0.99 | 1 | 0.5 | 2 | − | − |
| Alerta 29 | K. pneumoniae | −32.35 (1.65) | −31.29 (0.87) | 0.97 | 0.5 | 0.5 | — | − | − |
| Alerta 30 | K. pneumoniae | −34.29 (2.09) | −35.60 (1.09) | 1.04 | 1 | 1 | — | − | − |
| 13883 | K. pneumoniae | −32.60 (1.03) | −32.04 (0.92) | 0.98 | 1 | 1 | — | − | − |
Bacterial surface charge (in millivolts) measures were determined for colistin-resistant and colistin-susceptible strains grown in Mueller-Hinton broth without (−E) and with (+E) EDTA (80 μg/ml). After 18 h of incubation at 37°C, cells were washed in 1.0 mM NaCl adjusted to 1 × 104 CFU/ml. Zeta potential (in millivolts) measures were determined using a Zeta Potential Analyzer (ZETAPALS; Brookhaven). The zeta potential ratio (RZP) is calculated as ZP+EDTA/ZP−EDTA, where ZP+EDTA and ZP−EDTA correspond to zeta potential values obtained for bacterial suspensions grown in the presence or absence of 80 μg/ml EDTA, respectively. Each value represents the mean of at least 5 individual measurements ± the standard deviation.
Colistin MICs were determined by microdilution broth method according to EUCAST guidelines (32). Colistin MIC reduction was evaluated in the presence of EDTA at a final concentration of 80 μg/ml, and the values represent the results of reproducible replicates, performed three times on 3 distinct occasions.
Dashes in empty cells indicate no MIC reduction (fold change) in the presence of of 80 µg/ml EDTA.
MPNP was performed in triplicate. The MPNP test was considered positive to MCR-1 production when the growth of colistin-resistant E. coli in wells containing colistin solution (3.75 μg/ml) was inhibited by the addition of EDTA (80 μg/ml), as indicated by the unchanged color of phenol red in the NP solution (i.e., absence of metabolic activity and proliferation).
MPNP.
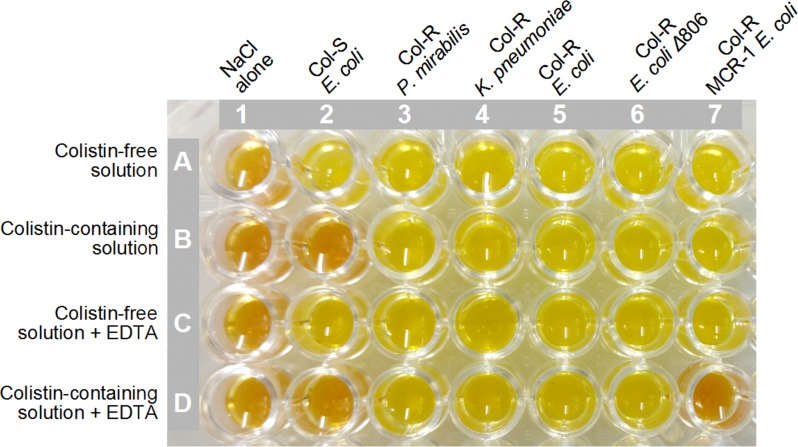
The MPNP was based on the original NP test proposed for the rapid identification of polymyxin-resistant and -susceptible Enterobacteriaceae (26, 27). Interestingly, while all colistin-resistant isolates showed a positive NP test result, only colistin-resistant MCR-1-positive E. coli isolates were inhibited by EDTA, as depicted in Fig. 1, where an absence of metabolic activity and bacterial proliferation (negative NP test indicated by unchanged color of phenol red in the well containing colistin-EDTA) was observed after a 2.5-h incubation at 37°C. The SN and SP were 96.7 and 100%, respectively. Indeed, the colistin-susceptible MCR-1-positive E. coli strain ICBEC 146 was not detected by the NP or the MPNP test. On the other hand, the colistin-resistant MCR-1-positive K. pneumoniae CCBH24080 strain showed positive and negative NP and MPNP results, respectively (Table 2).
FIG 1.
MCR-1 detection by the modified rapid polymyxin NP test (MPNP). Modification of the NP test was based on incorporation of two additional wells, which were filled with colistin-free solution plus EDTA (80 μg/ml) and colistin-containing (5 μg/ml) solution plus EDTA, respectively. Wells A1 to A7 were filled with 150 μl of a colistin-free NP solution. Wells B1 to B7 were completed with 150 μl of NP solution supplemented with 5 μg/ml colistin sulfate. Wells C1 to C7 were filled with 150 μl of colistin-free NP solution supplemented with 80 μg/ml EDTA. Wells D1 to D7 were added with 150 μl of NP solution containing 5 μg/ml colistin sulfate and 80 μg/ml EDTA. Wells in column 1 were filled with 50 μl of 0.85% NaCl (negative sterility control), whereas for each isolate, 50 μl of a 3.0 to 3.5 McFarland bacterial suspension (∼109 UFC/ml) was dispensed and mixed with 150 μl of reaction solution contained in each of the wells in columns 2 to 7. Columns 2 to 7 represent the MPNP test performed for E. coli ATCC 25922, P. mirabilis ATCC 25933, colistin-resistant (Col-R) K. pneumoniae Alerta 16 (mgrB promoter IS1 family), colistin-resistant mcr-1-negative E. coli strain HC113, colistin-resistant mcr-1-negative E. coli Δ806 mutant strain, and mcr-1-positive E. coli strain ICBEC72H, respectively. The plates were incubated at 35 ± 2°C under aerobic conditions for 4 h, and visual changes in the color of the wells were monitored each hour. In wells with added colistin (B1 t oB7), a color change from orange to yellow was considered positive to colistin resistance, whereas the MPNP test was considered positive to MCR-1 phosphoethanolamine transferase production when the colistin-containing solution supplemented with EDTA (wells D1 to D7) remained orange (i.e., absence of glucose metabolization); this shows that growth of the colistin-resistant E. coli (mcr-1-positive) in the well containing colistin solution (well D7) was inhibited by EDTA.
Alteration of zeta potential.
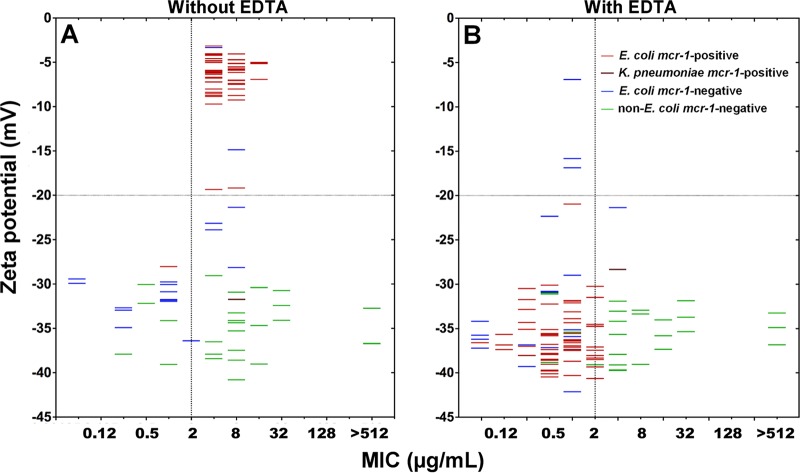
The zeta potential results for all strains evaluated in this study are summarized in Table 2. The replacement of lipid A with the PEtN-4′-lipid A, mediated by MCR-1, reduced the negative membrane charge of all colistin-resistant E. coli isolates to less than or equal to −20 mV (−4.20 to −19.34 mV), whereas for the K. pneumoniae strain CCBH24080, no charge reduction was observed. On the other hand, with the exception of two colistin-resistant MCR-1-negative E. coli strains (M6 and HC113), most colistin-susceptible and colistin-resistant (MCR-1-negative) Enterobacteriaceae presented zeta potential values between −21.54 and −44.21 mV (Fig. 2). For colistin-resistant MCR-1-positive E. coli isolates, bacterial growth in the presence of EDTA (80 μg/ml) resulted in an alteration of zeta potential ranging from −21.13 to −40.81 mV (Fig. 2), with an RZP value of ≥1.5. However, since two colistin-resistant MCR-1-negative E. coli strains (HC629 and M6) presented RZP values of 1.75 and 2.01, respectively, we have established an RZP value of ≥2.5 as the cutoff criterion for the presumptive identification of MCR-1-positive E. coli isolates. The SN and SP of RZP were 95.1 and 100%, respectively. For the colistin-susceptible MCR-1-positive E. coli strain ICBEC 146, an RZP of 1.08 was obtained, which was interpreted as a false-negative result.
FIG 2.
Bacterial surface charge distribution as a function of colistin MIC (A) and alteration of zeta potential induced by EDTA (B). Zeta potential was determined for colistin-resistant and colistin-susceptible strains grown in Mueller-Hinton broth without (A) and with (B) EDTA (80 μg/ml). After 18 h of incubation at 37°C, cells were washed in 1.0 mM NaCl adjusted to 1 × 104 CFU/ml. Zeta potential (millivolts) measures were determined using a Zeta Potential Analyzer (ZETAPALS; Brookhaven). mcr-1-positive E. coli, mcr-1-positive K. pneumoniae, mcr-1-negative E. coli, and non-E. coli strains are represented by red, brown, blue, and green dashes, respectively.
DISCUSSION
In this study, we have evaluated distinct phenotypic tests for the detection of colistin-resistant MCR-1-positive E. coli from human, food, and animal samples, based on the inhibition of the MCR-1 phosphoethanolamine transferase by EDTA. Microbiological assays relying on the synergy between EDTA and carbapenems have been previously developed and standardized for the screening of metallo-β-lactamase-producing Gram-negative bacteria, since it is well known that EDTA inhibits metallo-β-lactamase activity (28). Similarly, molecular and structure studies of the catalytic domain of MCR-type proteins have supported that phosphoethanolamine transferases can be assigned as a member of the alkaline phosphatase metalloenzyme superfamily, with zinc being required for MCR activity (9, 20–23). In fact, for MCR-1-positive strains, a clear reduction in colistin MIC was observed in the presence of EDTA, supporting the idea that zinc-limiting conditions induced by EDTA represent a good alternative for phenotypic identification of MCR-1-producing E. coli (20). Moreover, previous studies have demonstrated that the activity of putative Ca2+-induced PEtN transferases, known to modify the other Kdo (3-deoxy-d-manno-octulosonic acid) residue of E. coli LPS, can be strongly inhibited by EDTA (29, 30). More recently, the inhibition of MCR-1 by dipicolinic acid (another metalloenzyme chelator) was reported as a useful method (named the colistin-MAC test) for phenotypic screening of mcr-1-positive colistin-resistant E. coli strains (31).
The CDT method was based on the utilization of disks containing colistin (10 μg), which were impregnated with 10 μl of 100 mM EDTA. Although colistin-resistant E. coli strains could be accurately classified as MCR-1 using as an interpretative criterion an increase of ≥3 mm in the inhibition zone of colistin disks plus EDTA in comparison to those observed for colistin disks without EDTA, this method may fail to detect MCR-1-positive isolates with colistin MICs of ≤2 μg/ml. In fact, a single strain was not identified as MCR-1 positive or colistin resistant by CDT, CMR, or MPNP, suggesting that mcr-1 might not be expressed in this isolate. On the other hand, five colistin-resistant MCR-1-negative E. coli strains displayed a positive result, whereas a colistin-resistant MCR-1-positive K. pneumoniae strain was not identified by the CDT method. In this regard, in a previous study of MCR-1 inhibition induced by dipicolinic acid (DPA), the disk diffusion format presented no significant differences in inhibition zones between mcr-1-positive and mcr-1-negative colistin-resistant strains, probably due to the low and variable diffusibility of colistin from disks into the Mueller-Hinton agar, as previously reported by Coppi et al. (31).
Although the colistin MICs of MCR-1-positive E. coli strains in the presence of EDTA resulted in a ≥4-fold colistin MIC reduction, this reduction was more evident for E. coli exhibiting high-level resistance to colistin. On the other hand, under this experimental condition, the specificity of MCR-1 detection was affected by the effect of the chelating agent against two colistin-susceptible E. coli strains. Moreover, this colistin MIC reduction in the presence of EDTA was also observed among colistin-resistant MCR-1-negative E. coli strains, with exception of the mutant E. coli strain.
The rapid polymyxin NP test was originally proposed for the detection of polymyxin-resistant and -susceptible Enterobacteriaceae, regardless of the resistance mechanism exhibited (26, 27). This method has been reported as an easy-to-perform, rapid, sensitive, and specific test. Interestingly, in this study, modification of the NP test based on the incorporation of two wells containing colistin-free solution plus EDTA, and a colistin-containing solution plus EDTA, resulted in the specific detection of MCR-1-positive colistin-resistant E. coli isolates, enhancing the accuracy of this method.
In order to develop colistin resistance, Gram-negative pathogens have developed multiple mechanisms to modify the lipid A structure of the LPS (2–4). In this study, we confirm that the biochemical mechanism by which MCR-1-positive bacteria acquire resistance to colistin is dependent on the reduction of the net negative charge of the bacterial outer membrane, which consequently decreases the binding affinity of colistin to the bacterial surface (2–4). Furthermore, under limited conditions of zinc for the MCR-1 biochemical function, as caused by EDTA, an increase in the net negative charge for colistin-resistant MCR-1-positive E. coli strains was observed. On the other hand, colistin MIC values in the presence of EDTA were reverted to a susceptible category, as interpreted according to the EUCAST breakpoint (32). The increase in net negative charge in the presence of EDTA reached zeta potential values identical to those of colistin-susceptible bacterial isolates. Thus, alteration of the zeta potential allowed us to generate an RZP index, where a value of ≥2.5 was associated with the MCR-1 phenotype. Finally, the lack of inhibitory effect of EDTA on the mcr-1-positive K. pneumoniae strain was also reported for DPA, which indeed could be due to a reduced permeability and/or additional unknown mechanisms of resistance to polymyxins (31).
There are certain limitations of this study, such as the reduced number of MCR-1-positive K. pneumoniae isolates investigated and the lack of other Enterobacteriaceae species (i.e., Salmonella spp., Enterobacter spp., and Citrobacter spp.) encoding MCR-1 and any isolates carrying other variants of the MCR-like PEtN transferase (10–12). We also should be aware of EDTA chelator activity that could act to nonspecifically affect other bacterial processes. On the other hand, the coproduction of an additional mechanism of colistin resistance, as related to the activation of TCSs, could interfere in the assays of inhibition by EDTA. Therefore, the results should be cautiously interpreted and confirmed with a reference method. Finally, all screening methods evaluated here fail in detecting the colistin-susceptible E. coli strain carrying the mcr-1 gene, confirming that gene detection by molecular methods continues to be the gold standard.
In summary, our results demonstrate that assays of inhibition by EDTA may provide simple and inexpensive methods for detecting MCR-1-producing E. coli in human and veterinary diagnostic laboratories, mainly under the conditions described for the MPNP and zeta potential methods, which displayed the highest SN and SP values. However, additional studies are necessary to confirm the accuracy of these methodologies for phenotypically detecting MCR-producing isolates by testing other Enterobacteriaceae species or MCR variants isolated worldwide.
MATERIALS AND METHODS
Bacterial isolates.
A total of 109 isolates belonging to the Enterobacteriaceae family were tested in this study (Table 1). Colistin-resistant Escherichia coli (n = 66) and Klebsiella pneumoniae (n = 20) and colistin-susceptible E. coli (n = 13) and K. pneumoniae (n = 5) isolates were recovered from humans, food, and animals. Intrinsically colistin-resistant Morganella morganii and Serratia marcescens (which were isolated from clinical specimens) and Proteus mirabilis ATCC 25933 were included. Additionally, the E. coli ATCC 25922 and K. pneumoniae ATCC 13883 strains were also evaluated. Among colistin-resistant E. coli, a total of 59 isolates were mcr-1 positive, as confirmed by PCR and sequencing (6, 8, 10–17, 33), whereas colistin resistance in K. pneumoniae was related to MCR-1 production (8), activation of the two-component regulatory systems PhoPQ/PmrAB, or mgrB inactivation. In this regard, K. pneumoniae isolates were submitted to PCR amplification, followed by DNA sequencing (ABI 3500 genetic analyzers; Applied Biosystems, Foster City, CA) to the pmrA, pmrB, pmrD, phoP, phoQ, and mgrB genes (34, 35). Mutational analysis was performed by SeqManII version 5.0 and the BLASTn tool using K. pneumoniae MGH 78578 as the reference strain. Insertion sequences were confirmed by ISfinder (https://www-is.biotoul.fr). For those isolates that did not show any mutation, a reverse transcription-quantitative PCR (qRT-PCR) assay was performed to quantify the relative expression of the above-mentioned genes (35, 36). Additionally, a colistin-resistant mutant E. coli strain (Δ806) was developed under selective colistin pressure, as previously described (37).
CDT.
The combined-disk test was adapted from methods described for the detection of metallo-β-lactamase-producing isolates (38, 39). Initially, in order to select an EDTA concentration displaying no antibacterial activity against all screened isolates, different concentrations and volumes of EDTA solution (Sigma-Aldrich, St. Louis, MO, USA) were added to both blank disks and 10-μg colistin disks (Oxoid, Basingstoke, UK). In brief, we tested combinations of 5, 10, and 20 μl at 50, 80, 90, 100, 150, 200, 300, 400, and 500 mM EDTA (pH 8.0). In this regard, the use of 10 μl of a 100 mM EDTA solution was selected for further tests. In this way, for each screened bacterium, two 10-μg colistin disks without and a blank disk with EDTA and a blank disk were placed onto Mueller-Hinton agar plates (Becton Dickinson, Le Pont de Claix, France) inoculated with a 0.5 McFarland bacterial suspension. Inhibition zone diameters around the colistin disks (with and without EDTA) were measured and compared after 18 to 24 h of incubation at 37°C.
CMR in the presence of EDTA.
The MIC for colistin was performed by the broth microdilution method according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (32). However, for evaluation of the reduction in colistin MIC in the presence of EDTA, cation supplementation of Mueller-Hinton broth was not performed, since further addition of calcium and magnesium would impair the inhibitory activity of EDTA. Therefore, nonspecific binding of EDTA to excess of calcium and magnesium could reduce the concentration of free EDTA needed to chelate zinc ions required to MCR-1 activity (17). Moreover, it has been shown that calcium supplementation could favor the activity of putative PEtN transferases in E. coli (29, 30). In order to select an EDTA concentration exhibiting no antibacterial activity against all screened isolates, five different concentrations of EDTA (64, 80, 100, 128, and 256 μg/ml) were evaluated. The 80 μg/ml EDTA solution was selected for further tests. In brief, bacterial inocula were adjusted to a 0.5 McFarland turbidity standard and diluted to a ratio of 1:100 in Mueller-Hinton broth (Becton Dickinson, Le Pont de Claix, France). All isolates were tested in serial dilutions of colistin sulfate (Sigma-Aldrich, St. Louis, MO, USA) ranging from 0.06 to 32 μg/ml, except for the intrinsically colistin-resistant isolates, which were tested using serial dilutions of colistin sulfate ranging from 0.06 to 512 μg/ml. In this regard, E. coli ATCC 25922 and K. pneumoniae ATCC 13883 were used as susceptible controls, whereas P. mirabilis ATCC 25933 was used as a resistant control (Table 2). MIC interpretation was performed according to EUCAST breakpoints (32).
MPNP.
The MPNP rapid colorimetric test was based on the detection of bacterial growth in the presence of colistin sulfate at a final concentration of 3.75 μg/ml (26), where bacterial growth detection was supported by glucose metabolism with acid formation related to glucose metabolism in Enterobacteriaceae, resulting in a color change of a pH indicator (26, 27). In this study, modification was based on incorporation of two additional wells, which were filled with a colistin-free solution plus EDTA (80 μg/ml) and a colistin-containing (5 μg/ml) solution plus EDTA. Using sterile 96-well polystyrene plates (Sarstedt, Newton, NC, USA), the following experimental conditions were established in the MPNP test: (i) wells A1 to A12 were filled with 150 μl of colistin-free NP solution; (ii) 150 μl of NP solution supplemented with 5 μg/ml colistin sulfate was added to wells B1 to B12; (iii) wells C1 to C12 were filled with 150 μl of colistin-free NP solution supplemented with 80 μg/ml EDTA; (iv) 150 μl of NP solution supplemented with 5 μg/ml colistin sulfate and 80 μg/ml EDTA was deposited into wells D1 to D12; (v) except for the wells in column 1 (filled with 50 μl of 0.85% NaCl [negative sterility control]), for each isolate, 50 μl of a 3.0 to 3.5 McFarland bacterial suspension (∼109 CFU/ml) was dispensed and mixed with 150 μl of reaction solution contained in each of the wells in columns 2 to 12. Therefore, the plate was organized so that each column represented the MPNP test performed for each strain. Finally, the plates were incubated at 35 ± 2°C under aerobic conditions for 4 h, and visual changes in the color of the wells were monitored each hour (26, 27). In this regard, in wells with added colistin-containing solution (B1 to B12), a color change from orange to yellow was considered positive to colistin resistance, whereas the MPNP test was considered positive to MCR-1 phosphoethanolamine transferase production when the colistin-containing solution supplemented with EDTA (wells D1 to D12) remained orange (i.e., absence of glucose metabolization) (Fig. 1). E. coli ATCC 25922 and Proteus mirabilis ATCC 25933 strains were used as controls.
Zeta potential measurement.
Particle size (mean diameter in nanometers) and zeta potential (millivolts) of bacterial cells were measured with a ZetaPALS ZetaPotential Analyzer (Brookhaven Instruments Corporation, Holtsville, NY), which was equipped with a 677-nm laser and dynamic light-scattering (PCS) at 90° for particle sizing (40). Zeta potential was determined from electrophoretic mobility μ at 25°C in 1 mM NaCl and using Smoluchowski's equation ζ = μη/ε, where η is the medium viscosity and ε the medium dielectric constant (40–42). Prior to sample analysis, bacterial suspensions grown in the absence or presence of 80 μg/ml EDTA were centrifuged (5,000 rpm for 5 min at 5°C) and pellets were washed twice, being suspended in 2 ml of sterile 1 mM NaCl solution and adjusted to the turbidity of a 0.5 McFarland standard solution. For each bacterial sample, an additional 1:4 dilution was performed in 1 mM NaCl, and particle size and zeta potential were determined in 2-ml aliquots. Each value is shown as a mean of at least 5 individual measurements ± the standard deviation. Alterations of zeta potential induced by EDTA were calculated from the zeta potential ratio (RZP = ZP+EDTA/ZP−EDTA), where ZP+EDTA and ZP−EDTA correspond to zeta potential values obtained for bacterial suspensions grown in the presence or absence of 80 μg/ml EDTA, respectively.
Statistical analysis.
Sensitivity (SN) and specificity (SP) were calculated for CDT, CMR, MPNP, and RZP. PCR and direct sequencing for the mcr-1 gene were considered the gold standard. SN and SP were calculated with the formulas a/(a + b) and d/(c + d), respectively, where a is the number of isolates correctly identified as MCR-1 by the tested methods, b is the number of true MCR-1 positives that were incorrectly assigned non-MCR-1 by the tested methods, d is the number of true isolates that are non-MCR-1 producers that were correctly identified by the tested methods, and c is the number of isolates that were incorrectly identified as MCR-1 producers.
ACKNOWLEDGMENTS
We thank Jean-Yves Madec, Marisa Haenni, Carlos E. Levy, Flávia Rossi, Raquel Girardello, Ana Paula Carvalho-Assef, and Daniel F. Monte for kindly supplying bacterial control strains and Gabriel Gutkind (Faculty of Pharmacy and Biochemistry, University of Buenos Aires, Buenos Aires, Argentina) for comments and suggestions.
This work was supported by research grants from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2016/08593-9), and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 462042/2014-6) and by a fellowship to M.R.F. from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2015/13527-2). N.L. and A.M.M. are research grant fellows of CNPq. M.R.F. and M.P.C. are research grant fellows of FAPESP. We are also grateful to the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for providing grants to W.M.M. and R.C. (PNPD 20131991) and to the National Council for Science and Technological Development (CNPq) for providing a grant to A.C.G. (process no. 305535/2014-5).
We declare no conflicts of interest.
REFERENCES
- 1.Falagas ME, Kasiakou SK. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant Gram-negative bacterial infections. Clin Infect Dis 40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 2.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeannot K, Bolard A, Plésiat P. 2017. Resistance to polymyxins in Gram-negative organisms. Int J Antimicrob Agents 49:526–535. doi: 10.1016/j.ijantimicag.2016.11.029. [DOI] [PubMed] [Google Scholar]
- 4.Liu YY, Chandler CE, Leung LM, McElheny CL, Mettus RT, Shanks RM, Liu JH, Goodlett DR, Ernst RK, Doi Y. 2017. Structural modification of lipopolysaccharide conferred by mcr-1 in Gram-negative ESKAPE pathogens. Antimicrob Agents Chemother 61:e00580-17. doi: 10.1128/AAC.00580-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kieffer N, Nordmann P, Poirel L. 2017. Moraxella species as potential sources of MCR-like polymyxin-resistance determinants. Antimicrob Agents Chemother 61:e00129-17. doi: 10.1128/AAC.00129-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 7.Schwarz S, Johnson AP. 2016. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother 71:2066–2070. doi: 10.1093/jac/dkw274. [DOI] [PubMed] [Google Scholar]
- 8.Aires CAM, da Conceição-Neto OC, Oliveira TRTE, Dias CF, Montezzi LF, Picão RC, Albano RM, Asensi MD, Carvalho-Assef APD. 2017. Emergence of plasmid-mediated mcr-1 gene in clinical KPC-2-producing Klebsiella pneumoniae ST392 in Brazil. Antimicrob Agents Chemother 61:e00317-17. doi: 10.1128/AAC.00317-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun J, Xu Y, Gao R, Lin J, Wei W, Srinivas S, Li D, Yang RS, Li XP, Liao XP, Liu YH, Feng Y. 2017. Deciphering MCR-2 colistin resistance. mBio 8(3):e00625-17. doi: 10.1128/mBio.00625-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21:pii=30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 11.Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, Zhang R, Walsh TR, Shen J, Wang Y. 2017. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8(3):e00543-17. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, Pezzotti G, Magistrali CF. 2017. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill 22:30589. doi: 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandes MR, Sellera FP, Esposito F, Sabino CP, Cerdeira L, Lincopan N. 2017. Colistin-resistant mcr-1-positive Escherichia coli in public beaches, an infectious threat emerging in recreational waters. Antimicrob Agents Chemother 61:e00234-17. doi: 10.1128/AAC.00234-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandes MR, McCulloch JA, Vianello MA, Moura Q, Pérez-Chaparro PJ, Esposito F, Sartori L, Dropa M, Matté MH, Lira DP, Mamizuka EM, Lincopan N. 2016. First report of the globally disseminated IncX4 plasmid carrying the mcr-1 gene in a colistin-resistant Escherichia coli sequence type 101 isolate from a human infection in Brazil. Antimicrob Agents Chemother 60:6415–6417. doi: 10.1128/AAC.01325-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandes MR, Moura Q, Sartori L, Silva KC, Cunha MP, Esposito F, Lopes R, Otutumi LK, Gonçalves DD, Dropa M, Matté MH, Monte DF, Landgraf M, Francisco GR, Bueno MF, de Oliveira Garcia D, Knöbl T, Moreno AM, Lincopan N. 2016. Silent dissemination of colistin-resistant Escherichia coli in South America could contribute to the global spread of the mcr-1 gene. Euro Surveill 21:pii=30214. doi: 10.2807/1560-7917.ES.2016.21.17.30214. [DOI] [PubMed] [Google Scholar]
- 16.Sellera FP, Fernandes MR, Sartori L, Carvalho MP, Esposito F, Nascimento CL, Dutra GH, Mamizuka EM, Pérez-Chaparro PJ, McCulloch JA, Lincopan N. 2017. Escherichia coli carrying IncX4 plasmid-mediated mcr-1 and blaCTX-M genes in infected migratory Magellanic penguins (Spheniscus magellanicus). J Antimicrob Chemother 72:1255–1256. doi: 10.1093/jac/dkw543. [DOI] [PubMed] [Google Scholar]
- 17.Monte DF, Mem A, Fernandes MR, Cerdeira L, Esposito F, Galvão JA, Franco BDGM, Lincopan N, Landgraf M. 2017. Chicken meat as a reservoir of colistin-resistant Escherichia coli strains carrying mcr-1 genes in South America. Antimicrob Agents Chemother 61:e02718-16. doi: 10.1128/AAC.02718-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corbella M, Mariani B, Ferrari C, Comandatore F, Scaltriti E, Marone P, Cambieri P. Three cases of mcr-1-positive colistin-resistant Escherichia coli bloodstream infections in Italy, August 2016 to January 2017. Euro Surveill 22:pii=30517. doi: 10.2807/1560-7917.ES.2017.22.16.30517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trung NV, Matamoros S, Carrique-Mas JJ, Nghia NH, Nhung NT, Chieu TT, Mai HH, van Rooijen W, Campbell J, Wagenaar JA, Hardon A, Mai NT, Hieu TQ, Thwaites G, de Jong MD, Schultsz C, Hoa NT. 2017. Zoonotic transmission of mcr-1 colistin resistance gene from small-scale poultry farms, Vietnam. Emerg Infect Dis 23:529–532. doi: 10.3201/eid2303.161553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinchliffe P, Yang QE, Portal E, Young T, Li H, Tooke CL, Carvalho MJ, Paterson NG, Brem J, Niumsup PR, Tansawai U, Lei L, Li M, Shen Z, Wang Y, Schofield CJ, Mulholland AJ, Shen J, Fey N, Walsh TR, Spencer J. 2017. Insights into the mechanistic basis of plasmid-mediated colistin resistance from crystal structures of the catalytic domain of MCR-1. Sci Rep 7:39392. doi: 10.1038/srep39392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stojanoski V, Sankaran B, Prasad BV, Poirel L, Nordmann P, Palzkill T. 2016. Structure of the catalytic domain of the colistin resistance enzyme MCR-1. BMC Biol 14:81. doi: 10.1186/s12915-016-0303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma G, Zhu Y, Yu Z, Ahmad A, Zhang H. 2016. High resolution crystal structure of the catalytic domain of MCR-1. Sci Rep 6:39540. doi: 10.1038/srep39540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu M, Guo J, Cheng Q, Yang Z, Chan EW, Chen S, Hao Q. 2016. Crystal structure of Escherichia coli originated MCR-1, a phosphoethanolamine transferase for colistin resistance. Sci Rep 6:38793. doi: 10.1038/srep38793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caspar Y, Maillet M, Pavese P, Francony G, Brion JP, Mallaret MR, Bonnet R, Robin F, Beyrouthy R, Maurin M. 2017. mcr-1 colistin resistance in ESBL-producing Klebsiella pneumoniae, France. Emerg Infect Dis 23:874–876. doi: 10.3201/eid2305.161942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donà V, Bernasconi OJ, Kasraian S, Tinguely R, Endimiani A. 2017. A SYBR Green-based real-time PCR method for improved detection of mcr-1-mediated colistin resistance in human stool samples. J Glob Antimicrob Resist 9:57–60. doi: 10.1016/j.jgar.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Nordmann P, Jayol A, Poirel L. 2016. Rapid detection of polymyxin resistance in Enterobacteriaceae. Emerg Infect Dis 22:1038–1043. doi: 10.3201/eid2206.151840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jayol A, Dubois V, Poirel L, Nordmann P. 2016. Rapid detection of polymyxin-resistant Enterobacteriaceae from blood cultures. J Clin Microbiol 54:2273–2277. doi: 10.1128/JCM.00918-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hrabák J, Chudáčková E, Papagiannitsis CC. 2014. Detection of carbapenemases in Enterobacteriaceae: a challenge for diagnostic microbiological laboratories. Clin Microbiol Infect 20:839–853. doi: 10.1111/1469-0691.12678. [DOI] [PubMed] [Google Scholar]
- 29.Reynolds CM, Kalb SR, Cotter RJ, Raetz CR. 2005. A phosphoethanolamine transferase specific for the outer 3-deoxy-d-manno-octulosonic acid residue of Escherichia coli lipopolysaccharide. Identification of the eptB gene and Ca2+ hypersensitivity of an eptB deletion mutant. J Biol Chem 280:21202–21211. doi: 10.1074/jbc.M500964200. [DOI] [PubMed] [Google Scholar]
- 30.Kanipes MI, Lin S, Cotter RJ, Raetz CR. 2001. Ca2+-induced phosphoethanolamine transfer to the outer 3-deoxy-d-manno-octulosonic acid moiety of Escherichia coli lipopolysaccharide. A novel membrane enzyme dependent upon phosphatidylethanolamine. J Biol Chem 276:1156–1163. doi: 10.1074/jbc.M009019200. [DOI] [PubMed] [Google Scholar]
- 31.Coppi M, Cannatelli A, Antonelli A, Baccani I, Di Pilato V, Sennati S, Giani T, Rossolini GM. A simple phenotypic method for screening of MCR-1-mediated colistin resistance. Clin Microbiol Infect, in press. doi: 10.1016/j.cmi.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 32.European Committee on Antimicrobial Susceptibility Testing. 2016. Breakpoints tables for interpretation of MICs and zone diameters, version 6.0. European Committee on Antimicrobial Susceptibility Testing, Växjö, Sweden: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_6.0_Breakpoint_table.pdf. [Google Scholar]
- 33.Monte DF, Fernandes MR, Cerdeira L, de Souza TA, Mem A, Franco BDGM, Landgraf M, Lincopan N. 2017. Draft genome sequences of colistin-resistant MCR-1-producing Escherichia coli ST1850 and ST74 strains isolated from commercial chicken meat. Genome Announc 5(20):e00329-17. doi: 10.1128/genomeA.00329-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braun G, Cayô R, Barbosa AC, Gales AC. 2016. In-vivo emergence of polymyxin B-resistant Klebsiella pneumoniae in patients with bloodstream infections. J Hosp Infect 94:338–340. doi: 10.1016/j.jhin.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Cannatelli A, Giani T, D'Andrea MM, Di Pilato V, Arena F, Conte V, Tryfinopoulou K, Vatopoulos A, Rossolini GM, COLGRIT Study Group. 2014. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 58:5696–5703. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramos PI, Custódio MG, Quispe Saji GD, Cardoso T, da Silva GL, Braun G, Martins WM, Girardello R, de Vasconcelos AT, Fernández E, Gales AC, Nicolás MF. 2016. The polymyxin B-induced transcriptomic response of a clinical, multidrug-resistant Klebsiella pneumoniae involves multiple regulatory elements and intracellular targets. BMC Genomics 17:737. doi: 10.1186/s12864-016-3070-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phan M-D, Nhu NTK, Achard MES, Forde BM, Hong KW, Chong TM, Yin W-F, Chan K-G, West NP, Walker MJ, Paterson DL, Beatson SA, Schembri MA. 2017. Modifications in the pmrB gene are the primary mechanism for the development of chromosomally encoded resistance to polymyxins in uropathogenic Escherichia coli. J Antimicrob Chemother 72:2729–2736. doi: 10.1093/jac/dkx204. [DOI] [PubMed] [Google Scholar]
- 38.Picão RC, Andrade SS, Nicoletti AG, Campana EH, Moraes GC, Mendes RE, Gales AC. 2008. Metallo-beta-lactamase detection: comparative evaluation of double-disk synergy versus combined disk tests for IMP-, GIM-, SIM-, SPM-, or VIM-producing isolates. J Clin Microbiol 46:2028–2037. doi: 10.1128/JCM.00818-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andrade SS, Picão RC, Campana EH, Nicoletti AG, Pignatari AC, Gales AC. 2007. Influence of disk preparation on detection of metallo-beta-lactamase-producing isolates by the combined disk assay. J Clin Microbiol 45:2058–2060. doi: 10.1128/JCM.02467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lincopan N, Espíndola NM, Vaz AJ, da Costa MH, Faquim-Mauro E, Carmona-Ribeiro AM. 2009. Novel immunoadjuvants based on cationic lipid: preparation, characterization and activity in vivo. Vaccine 27:5760–5771. doi: 10.1016/j.vaccine.2009.07.066. [DOI] [PubMed] [Google Scholar]
- 41.Halder S, Yadav KK, Sarkar R, Mukherjee S, Saha P, Haldar S, Karmakar S, Sen T. 2015. Alteration of Zeta potential and membrane permeability in bacteria: a study with cationic agents. Springerplus 4:672. doi: 10.1186/s40064-015-1476-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soon RL, Nation RL, Cockram S, Moffatt JH, Harper M, Adler B, Boyce JD, Larson I, Li J. 2011. Different surface charge of colistin-susceptible and -resistant Acinetobacter baumannii cells measured with zeta potential as a function of growth phase and colistin treatment. J Antimicrob Chemother 66:126–133. doi: 10.1093/jac/dkq422. [DOI] [PMC free article] [PubMed] [Google Scholar]