ABSTRACT
Recent advances in nanopore sequencing technology have led to a substantial increase in throughput and sequence quality. Together, these improvements may permit real-time benchtop genomic sequencing and antimicrobial resistance gene detection in clinical isolates. In this study, we evaluated workflows and turnaround times for a benchtop long-read sequencing approach in the clinical microbiology laboratory using the Oxford Nanopore Technologies MinION sequencer. We performed genomic and plasmid sequencing of three clinical isolates with both MinION and Illumina MiSeq, using different library preparation methods (2D and rapid 1D) with the goal of antimicrobial resistance gene detection. We specifically evaluated the advantages of using plasmid DNA for sequencing and the value of supplementing MinION sequences with MiSeq reads for increasing assembly accuracy. Resequencing of three plasmids in a reference Klebsiella pneumoniae isolate demonstrated ∼99% accuracy of draft MinION-only assembly and >99.9% accuracy of assembly polished with MiSeq reads. Plasmid DNA sequencing of previously uncharacterized clinical extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and K. pneumoniae isolates using MinION allowed successful identification of antimicrobial resistance genes in the draft assembly corresponding to all classes of observed plasmid-based phenotypic resistance. Importantly, use of plasmid DNA enabled lower depth sequencing, and assemblies sufficient for full antimicrobial resistance gene annotation were obtained with as few as 2,000 to 5,000 reads, which could be acquired in 20 min of sequencing. With a MinION-only workflow that balances accuracy against turnaround time, full annotation of plasmid resistance gene content could be obtained in under 6 h from a subcultured isolate, less time than traditional phenotypic susceptibility testing.
KEYWORDS: MinION, nanopore, antibiotic resistance, genomics, plasmids
INTRODUCTION
Multidrug-resistant bacteria have been assessed as an urgent threat to global public health, and the sequencing of antimicrobial resistance (AMR) genes in these organisms is critical to understanding both the molecular mechanisms of resistance and the basis of their dissemination (1). As many resistance genes are located on plasmids, methods for plasmid sequencing are paramount to the success of these efforts. Sequencing of plasmids is also essential for accurate epidemiologic tracking in the settings of both hospital outbreaks and routine surveillance (2–4). However, the difficulty of achieving contiguous assemblies of plasmids using short-read sequencing technologies, such as those offered by the Illumina and Ion Torrent instruments, is well known (5). Sequence assemblies cannot unambiguously recover repeat-containing genomic regions if there are no reads that span the repeats. For short-read Illumina sequencing, fragment length is typically not longer than ∼400 to 500 bp, substantially shorter than the lengths of most mobile genetic elements. As a consequence, assemblies of plasmids that are enriched for these repeats are often extensively fragmented and frequently incomplete. Long-read sequencing with Pacific Biosystems (PacBio) instruments allows full assembly of plasmids to closure in many cases (3), but these instruments are expensive and large and remain inaccessible to most research and clinical labs.
Nanopore sequencers offer a potential solution to this problem for clinical microbiology labs through their long-read sequencing capabilities, low instrument cost (∼$1,000 U.S. dollars [USD]), and extremely small footprint (6). The Oxford Nanopore Technologies (ONT) MinION sequencer is a compact device that produces sequencing reads that are thousands to hundreds of thousands of nucleotides in length with real-time, read-by-read data availability (6). This rapid sequencing technology has many promising applications for the clinical microbiology laboratory and has been used successfully to sequence genomes of viruses, bacteria, and fungi (7–14), to identify viruses and bacteria in primary clinical specimens (15–21), and to characterize AMR genes in bacterial isolates and primary specimens (7, 9, 17, 20–24). Though MinION single-read error rates are higher than those for Illumina short reads, higher accuracy can be achieved by generating consensus sequences from multiple reads or by complementation with high-accuracy short reads (5, 6, 8, 9, 25, 26).
In addition to the remarkable capabilities of this evolving technology that led to its rapid adoption in the basic research lab, many studies have supported a potential role for nanopore sequencing for genome assembly, resolving plasmid structures, and identifying resistance genes in the context of diagnostics (8, 9, 17, 20–23, 27). In this study, we evaluated practical workflows and turnaround times for using the MinION device for real-time benchtop sequencing of plasmids and resistance gene detection in clinical isolates in the hospital laboratory setting. We performed genomic sequencing of three clinical isolates with both MinION and Illumina MiSeq platforms using different library preparation, sequencing, and base calling methods (2D versus rapid 1D) to compare sequencing workflows and specifically evaluated plasmid-enriched DNA extraction. We suggest an entirely benchtop workflow for plasmid sequencing in the clinical microbiology laboratory with tiered levels of turnaround time and assembly accuracy suited to different applications: (i) a rapid approach using only MinION sequencing for plasmid-based AMR gene detection and (ii) a higher-accuracy approach that uses a combination of long MinION reads and short MiSeq reads. With the rapid workflow based on plasmid DNA extraction, all steps from cultured isolate to full annotation of plasmid-based resistance genes could be achieved within 6 h, less time than traditional phenotypic susceptibility testing. Higher-accuracy plasmid assemblies can be completed in 72 h or less with combination MinION and MiSeq sequencing.
(This study was presented in part at the 2017 American Society for Microbiology Microbe Meeting, New Orleans, LA.)
RESULTS
MinION resequencing of KPNIH1 reference isolate.
To evaluate the accuracy of rapid MinION plasmid sequencing, and the efficiency with which AMR genes could be detected from MinION assemblies, we resequenced K. pneumoniae isolate KPNIH1, a blaKPC-positive, carbapenem-resistant isolate from the NIH Clinical Center (3, 4). The KPNIH1 isolate contains a 5.39-Mb genome and 15.1-kb, 113-kb, and 243-kb plasmids for which high-quality, closed reference genome sequences are available (3, 4). MinION sequencing was performed using plasmid DNA and ligation-based SQK-NSK007 R9 library prep followed by 2D base calling. In addition, we sequenced Nextera XT whole-genome libraries prepared from plasmid DNA and total DNA extracts using an Illumina MiSeq. Following a 48-h MinION run, 13,983 pass filter reads with a median length of 5.9 kb were obtained (Table 1), sufficient to provide at least 53× sequencing coverage depth of KPNIH1 plasmids. Importantly, MinION reads were distributed uniformly along each of the three plasmids individually, but the average depths of coverage were different between plasmids. Although all of the plasmids were well represented in the sequencing data, we observed a 5-fold difference in median coverage among them (Table 1), suggesting that plasmids might be isolated with different efficiencies during the DNA extraction, that other biases may exist during library preparation or sequencing, or that differences in plasmid copy number may be present. Therefore, although high depth of coverage is easily achieved by sequencing plasmid DNA extracts, our findings indicate that coverages will often be different among plasmids.
TABLE 1.
KPNIH1 reference genome coverage by MinION and Illumina readsa
| KPNIH1 chromosome and plasmids |
Plasmid DNA |
Total DNA Illumina |
|||||
|---|---|---|---|---|---|---|---|
| MinION |
Illumina |
||||||
| Genome component | Size (kb) | No. of reads (%) | Median fold coverage (IQR) | No. of reads (%) | Median fold coverage (IQR) | No. of reads (%) | Median fold coverage (IQR) |
| Chromosome | 5,394 | 3,266 (22.32) | 2.0 (1.0–3.0) | 137,068 (11.60) | 4.0 (3.0–6.0) | 2,195,476 (97.70) | 79.0 (67.0–89.0) |
| pKPN-498 | 243 | 3,240 (22.14) | 53.0 (44.0–61.0) | 153,196 (12.97) | 110 (89.0–129.0) | 30,754 (1.37) | 21.0 (17.0–26.0) |
| pKpQIL-6e6 | 113 | 7,566 (51.71) | 252 (215–284) | 493,226 (41.75) | 798 (692.0–884.0) | 10,492 (0.47) | 17.0 (14.0–21.0) |
| pAAC154-a50 | 15 | 401 (2.74) | 54.0 (46.0–63.0) | 393,641 (33.32) | 4633 (3731–5292) | 5,195 (0.23) | 63.0 (51.0–74.0) |
| Unmapped | 159 (1.09) | 4,234 (0.36) | 5,182 (0.23) | ||||
Note that some reads mapped ambiguously to multiple replicons due to shared elements. IQR, interquartile range. The accession numbers for KPNIH are NZ_CP008827 (chromosome), NZ_CP008829 (pKPN-498), NZ_CP008830 (pKpQIL-6e6), and NZ_CP008828 (pAAC154-a50).
Raw sequence reads were assembled into contigs with the ONT assemble-polish pipeline using both MinION and MiSeq data. The assemble-polish pipeline consecutively invokes canu 1.3 (26), racon (28), and pilon (29) and can generate both MinION-only and higher-accuracy Illumina-polished assemblies. All three plasmids were >98% covered by the generated assemblies (Table 2) without further refinement. No effort was made to close circular sequences or to eliminate redundancies in assemblies, as the goal in this case was to analyze direct output of the pipeline. The MinION-only assembly yielded >99% consensus accuracy (∼0.2% mismatches and 0.5 to 0.6% gaps), and when supplemented with MiSeq paired-end reads, the polished assembly accuracy increased to ∼99.9% (0.03% mismatches and ∼0.1% indels).
TABLE 2.
Reference genome coverage and alignment quality of different KPNIH1 assemblies
| Assembly | pKPN-498 |
pKpQIL-6e6 |
pAAC154-a50 |
|||
|---|---|---|---|---|---|---|
| Plasmid coverage (%) | No. of mismatches/indels per 100 kbp | Plasmid coverage (%) | No. of mismatches/indels per 100 kbp | Plasmid coverage (%) | No. of mismatches/indels per 100 kbp | |
| Draft MinION (canu) | 98.6 | 534.2/615.0 | 100 | 95.0/536.8 | 100 | 86.1/556.4 |
| Polished MinION (canu + racon) | 100 | 235.8/451.6 | 100 | 151.4/408.3 | 100 | 125.9/404.1 |
| Polished MinION + MiSeq (canu + racon + pilon) | 100 | 65.2/107.0 | 100 | 12.3/46.6 | 100 | 79.5/178.9 |
| Illumina WGS (SPAdes) | 96.9 | 73.7/4.7 | 97.8 | 468.7/7.2 | 100 | 13.3/0 |
We next performed AMR gene annotation on the assembled KPNIH1 plasmids using Resfinder (30) and compared detected AMR genes to those found in the reference genome annotation in GenBank. Resfinder AMR gene annotation was completed in as little as 30 min. As expected from the nearly complete coverage of all three plasmids, we identified all annotated plasmid-based resistance genes (see Table S1 in the supplemental material). Although both draft and polished assemblies allowed detection of all annotated AMR genes, the pilon-polished assemblies (using MiSeq reads) demonstrated closer sequence matches to reference AMR genes used by Resfinder. In contrast, polishing of the MinION-only assemblies with Racon was found to decrease the percent identity of the matches to the reference sequences in many cases (Table S1), for reasons not well understood.
Sequencing and assembly of a clinical ESBL-producing E. coli isolate (ECESBL-1).
Analysis of KPNIH1 resequencing data demonstrated that plasmid assemblies derived from our MinION workflow using plasmid DNA as input could be used for efficient AMR gene detection. We next asked whether we could similarly detect the expected set of AMR genes from isolates that had not been previously sequenced, again using plasmid DNA as input. To test this, we sequenced a clinical E. coli isolate (ECESBL-1) that was phenotypically characterized as an ESBL producer and was resistant to multiple classes of antimicrobials (Tables 3 and 4). As with KPNIH1, we employed ligation-based SQK-NSK007 library preparation using plasmid DNA, followed by 2D base calling. Following assembly and finishing, we obtained 5 contigs between 2.1 kb and 110 kb in length (Table 3). All five contigs were successfully circularized and are represented at a higher level in the MiSeq sequencing libraries prepared from plasmid DNA. For the 2.1-kb, 48.5-kb, and 110-kb contigs, we detected high-confidence hits with >95% identity to known plasmid replicons using PlasmidFinder (31) (Table 3). We noted that three incompatibility group loci (IncQ1, IncFIB, and IncFIA) were identified in the 110-kb contig 8, raising the possibility that two separate plasmids may have been recovered as a single contig 8 in the assembly. Manual inspection of raw read alignment to the finished assembly did not reveal evidence of discontinuities, and similarity searches identified a 97.6-kb E. coli plasmid, pEC316_3, of similar structure containing these three incompatibility loci (GenBank accession number CP018954.1; 99% identity and 83% coverage). Thus, we do not think that the presence of three incompatibility groups on contig 8 is a consequence of misassembly. Two remaining contigs of 3.2 kb and 6.6 kb have more distant (∼91%) matches to the ColRNAI probe. It was previously shown (31) that small, narrow-host-range ColE1-like plasmids represent the largest group of small plasmids in Enterobacteriaceae. We found multiple plasmid sequences closely related to the 3.2-kb contig in GenBank (for example, 97% match over the full length to E. coli plasmid p9705 [accession number AB040037.1]) and to the 6.6-kb contig (best match to plasmid pSH14-028_6; 99% identity and 100% coverage; accession number CP016513.1). We thus believe that these two contigs likely represent divergent members of ColE1-like narrow host range plasmid family.
TABLE 3.
Annotation of finished ECESBL-1 assembly and coverage by MinION reads
| Contig | Length (kb) | MinION readsa |
Predicted plasmid incompatibility group | AMR genes predicted | |
|---|---|---|---|---|---|
| No. (%) | Median fold coverage (IQR) | ||||
| 8 | 110 | 14,783 (50.7) | 472 (381–544) | IncQ/IncFIB/IncFIA | Yes |
| 91 | 48.5 | 7,527 (25.8) | 453 (353–539) | IncI | Yes |
| 94 | 6.64 | 150 (0.5) | 18.0 (14.0–22.0) | ∼ColRNAI (91%) | No |
| 41 | 3.17 | 193 (0.7) | 28.0 (24.0–33.0) | ∼ColRNAI (91%) | No |
| 83 | 2.10 | 175 (0.6) | 31.0 (26.0–36.0) | Col | No |
Closed, finished MinION assemblies were polished with Illumina reads.
TABLE 4.
ECESBL-1: phenotypic susceptibility and resistance gene annotation in MinION and whole-genome MiSeq assemblies
| Gene identified |
Targeted drug class | Antibiotic | MIC (μg/ml) or zone | Interpretationa | |
|---|---|---|---|---|---|
| MinION | Whole-genome SPAdes | ||||
| blaTEM-1B | blaTEM-1B | Penicillins | Ampicillin | >16 | R |
| blaCMY-42 | blaCMY-42 | Cephalosporins/cephamycins | Cefazolin | >4 | R |
| blaCTX-M-24 | Cefoxitin | >16 | R | ||
| Cefotaxime | >64 | R | |||
| Ceftriaxone | >32 | R | |||
| Ceftazidime | >128 | R | |||
| Cefepime | >16 | R | |||
| Monobactams | Aztreonam | >16 | R | ||
| Beta-lactams/beta-lactamase inhibitors | Amoxicillin-clavulanic acid | >16 | R | ||
| Piperacillin-tazobactam | 32 | I | |||
| strA | strA | Aminoglycosides | Amikacin | ≤8 | S |
| strB | strB | Gentamicin | >8 | R | |
| aac(3)-IId | aac(3)-IId | Tobramycin | ≤4 | S | |
| aadA5 | aadA5 | ||||
| sul1 | sul1 | Sulfonamides | Trimethoprim-sulfamethoxazole | >2 | R |
| sul2 | sul2 | ||||
| dfrA17 | dfrA17 | ||||
| tet(B) | tet(B) | Tetracyclines | Tetracycline | No zone | R |
| Doxycycline | No zone | R | |||
| erm(B) | mph(A) | Macrolides | Azithromycin | No zone | No CLSI interpretation |
| mph(A) | |||||
| None identified | None identified | Fluoroquinolones | Ciprofloxacin | >2 | R |
| Levofloxacin | >4 | R | |||
| None identified | None identified | Carbapenems | Doripenem | ≤1 | S |
| Ertapenem | ≤0.5 | S | |||
| Imipenem | ≤1 | S | |||
| Meropenem | ≤1 | S | |||
R, resistant; I, intermediate; S, susceptible.
We next used the ECESBL-1 assembled sequences for AMR gene detection. In this case, the whole-genome MiSeq short-read SPAdes assembly was used for comparison. This SPAdes assembly provides excellent coverage (see Table 2, for example) but is highly fragmented for the repeat-enriched plasmid sequences. Table 4 and Table S2 in the supplemental material list the AMR genes found in the two longest contigs and compare the resistance gene annotation with results of standard broth microdilution-based testing. With the exception of blaCTX-M-24, we found in the ONT assembly all of the AMR genes detected in the SPAdes assembly. Similarity searches using the 202-kb SPAdes contig on which blaCTX-M-24 is found detected exclusively chromosomal rather than plasmid sequences. Additionally, this contig has deeper coverage with whole-genome reads and is unusually long (SPAdes plasmid contigs were generally between 1.5 and 35 kb in length). These results support the conclusion that the blaCTX-M-24 is a chromosomal gene in the ECSBL-1 isolate.
Comparison with results of standard phenotypic susceptibility testing demonstrated that resistance genes were detected corresponding to all categories of phenotypic resistance, with the primary exception of fluoroquinolone resistance (Table 4). Though mechanisms conferring fluoroquinolone resistance may be plasmid based (qnr genes, for instance), resistance is commonly due to mutations in the chromosomal gyrase and topoisomerase IV genes (32), and thus, it is not surprising that genes were not located on the sequenced plasmids. Given that the details of mapping fluoroquinolone resistance mutations are beyond the scope of this paper, potential responsible chromosomal mutations were not further investigated. In addition to drug classes that were tested in the standard clinical phenotypic susceptibility panel, Resfinder also detected genes for macrolide resistance [erm(B) and mph(A)] and tetracycline resistance [tet(B)] in the MinION assemblies; resistance to these antimicrobials was subsequently confirmed by additional Kirby-Bauer disk testing (Table 4). We noted that the erm(B) gene was not found in the whole-genome SPAdes assembly, likely a reflection of the difficulty of assembling highly repetitive plasmids from short reads and relatively low representation of these plasmids in whole-genome DNA extracts. This finding highlights the value of MinION long-read sequences in resolving repetitive regions and improving assembly continuity and completeness.
Sequencing and assembly of a clinical ESBL-producing K. pneumoniae isolate (KPESBL-1).
Since the ligation-based library preparation is relatively time-consuming (2 to 4 h), we next evaluated the ultrarapid (10 to 15 min) transposon-based sequencing library preparation workflow available from ONT (rapid sequencing kit I, R9 version). To test this method, we sequenced a clinical multidrug-resistant ESBL-producing K. pneumoniae (KPESBL-1) isolate (Tables 5 and 6). Furthermore, rather than cloud-based 2D base calling, which was used for the previous two isolates, for this sequencing experiment, real-time 1D base calling was performed locally on a laptop personal computer (PC). The switch to local, real-time base calling resulted in a substantial reduction of raw data processing time. The finished and filtered MinION and MiSeq ONT pipeline assembly contains 3 circularized contigs 43.3 kb, 47.9 kb, and 207 kb in length (Table 5). Using PlasmidFinder we identified replicon sequences in each of the contigs.
TABLE 5.
Annotation of finished KPESBL-1 assembly and coverage by MinION reads
| Contig | Length (kb) | MinION readsa |
Predicted plasmid incompatibility group | AMR genes predicted | |
|---|---|---|---|---|---|
| No. (%) | Median fold coverage (IQR) | ||||
| 17 | 207 | 2,414 (5.0) | 18.51 (11.0–25.0) | IncFIB(K)/IncFII(K) | Yes |
| 14 | 47.9 | 15,053 (31.1) | 564 (366–781) | IncR | Yes |
| 8 | 43.3 | 10,659 (22.0) | 381 (248–533) | IncX3 | Yes |
Closed, finished MinION assemblies were polished with Illumina reads.
TABLE 6.
KPESBL-1: phenotypic susceptibility and resistance gene annotation in MinION and whole-genome MiSeq assemblies
| Gene identified |
Targeted drug class | Antibiotic | MIC (μg/ml) or zone (mm)a | Interpretation | |
|---|---|---|---|---|---|
| MinION | Whole-genome SPAdes | ||||
| blaSHV-12 | blaSHV-11 | Penicillins | Ampicillin | >16 | R |
| Cephalosporins/cephamycins | Cefazolin | >4 | R | ||
| Cefoxitin | ≤8 | S | |||
| Cefotaxime | 16 | R | |||
| Ceftriaxone | 16 | R | |||
| Ceftazidime | 128 | R | |||
| Cefepime | ≤2 | S | |||
| Monobactams | Aztreonam | >16 | R | ||
| Beta-lactams/beta-lactamase inhibitors | Amoxicillin-clavulanic acid | ≤8 | S | ||
| Piperacillin-tazobactam | ≤16 | S | |||
| aadA1 | aadA1 | Aminoglycosides | Amikacin | 16 | S |
| aadA2 | aadA2 | Gentamicin | >8 | R | |
| aac(3)-IVa | aac(3)-IVa | Tobramycin | >8 | R | |
| aac(6′)-Ib | aac(6′)-Ib | ||||
| aac(6′)Ib-cr | aac(6′)Ib-cr | ||||
| aph(4)-Ia | aph(4)-Ia | ||||
| sul1 | sul1 | Sulfonamides | Trimethoprim-sulfamethoxazole | >2 | R |
| sul3 | sul3 | ||||
| dfrA12 | |||||
| mph(A) | mph(A) | Macrolides | Azithromycin | 8* | No CLSI interpretation |
| catA1 | catA1 | Phenicols | Chloramphenicol | No zone | R |
| cmlA1 | cmlA1 | ||||
| aac(6′)Ib-cr | aac(6′)Ib-cr | Fluoroquinolones | Ciprofloxacin | >2 | R |
| oqxA | Levofloxacin | >4 | R | ||
| oqxB | |||||
| None identified | None identified | Carbapenems | Doripenem | ≤1 | S |
| Ertapenem | ≤0.5 | S | |||
| Imipenem | ≤1 | S | |||
| Meropenem | ≤1 | S | |||
| None identified | fosA | Fosfomycin | Fosfomycin | No test methods | No CLSI interpretation |
The zone (mm) value is indicated by an asterisk. All other values are MIC (μg/ml).
We annotated 17 distinct resistance genes in the KPESBL-1 contigs using Resfinder web software for both the whole-genome SPAdes assembly and polished ONT assembly (Table 6). With the exception of the oqxA, oqxB, and fosA genes, all of the AMR genes identified in the SPAdes whole-genome assembly were also identified in the MinION-only plasmid assemblies. Notably, the oqxA and oqxB genes are located in the middle of the longest (840 kb) SPAdes contig and the fosA gene is on a 272-kb contig. In contrast, and in agreement with the highly fragmented nature of short-read plasmid assemblies, 11 out of the 12 remaining AMR genes are located on short (2 to 10 kb) SPAdes contigs and only blaSHV-11 is on the 158-kb contig. We thus conclude that oqxA, oqxB, and fosA are located on the bacterial chromosome. A more detailed analysis of blaSHV genes revealed the presence of two distinct alleles: blaSHV-11 on a plasmid and blaSHV-12 on the chromosome. In the whole-genome sequence (WGS) SPAdes assembly, only one gene, blaSHV-12, was detected by Resfinder. In contrast, the draft MinION assembly has two genes, one chromosomal and one on a plasmid. Subsequent contig filtering retained only plasmid contigs, and only one copy of the blaSHV gene is present in the finished assembly. One gene, dfrA12, was detected only in the MinION plasmid assembly. Similar to the case with the erm(B) gene in the ECESBL-1 isolate, a likely explanation in the case of KPESBL-1 is a failure of SPAdes to assemble the repeat-enriched part of the plasmid, again highlighting the value of MinION long reads.
A comparison between phenotypic susceptibility testing for KPESBL-1 and the AMR genes annotated in the plasmid assemblies demonstrates that genes corresponding to all classes of plasmid-mediated phenotypic resistance were detected (Table 6; see also Table S3 in the supplemental material). In addition, resistance gene annotation demonstrated the presence of a cml gene, with the prediction of resistance to chloramphenicol, subsequently demonstrated by Kirby-Bauer testing. Good overall agreement between phenotypic testing results and AMR gene detection in MinION assemblies supports the conclusion that use of lower-accuracy 1D reads did not substantially affect our ability to detect AMR genes.
Determining the minimum amount of sequencing required to predict plasmid resistance genes.
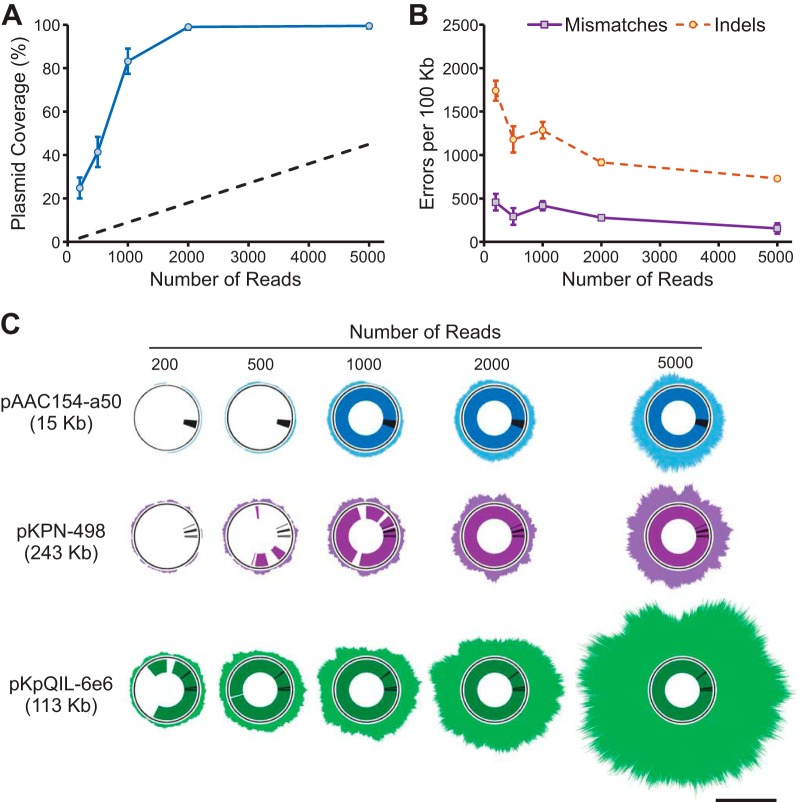
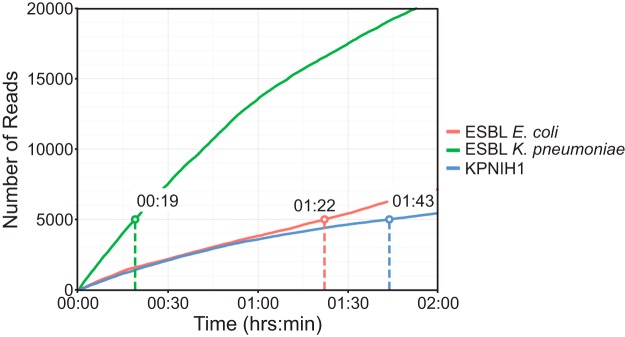
In the above-described experiments, we showed that we can detect all plasmid-borne AMR genes expected from phenotypic susceptibility testing using our MinION sequencing workflow. These experiments employed standard MinION sequencing protocols that ran for up to 48 h and generated data in substantial excess of the amount needed for complete plasmid assemblies and therefore for AMR gene detection. Indeed, in full data sets the median plasmid coverage reached 300- to 500-fold for better-represented plasmids (Table 1, 3, and 5), while assembly quality is generally not improved beyond ∼100× coverage and as little as 5× to 10× coverage might be sufficient to generate a low-depth assembly. To determine the minimum amount of time needed to acquire sufficient data for AMR gene annotation, we generated random subsets containing 200 to 5,000 MinION reads from the KPNIH1 sequencing run and used them for assembly and resistance gene annotation (Fig. 1). Since polishing and finishing require high-accuracy MiSeq reads that took us days to acquire, we used draft MinION-only assemblies generated with canu in this case. We found that all resistance genes were detected with 1,000 reads in KPNIH1, though the accuracy of assembly and resistance gene matches continued to improve with increasing coverage between 1,000 and 5,000 reads (Fig. 1). Importantly, our ability to assemble and subsequently detect AMR genes using read subsets was critically affected by the unequal representation of different plasmids in the sequencing data (see, for example, underrepresented pKPN-498 and pAAC154-a50 in Fig. 1 and Table 1). In other isolates, we observed similar differences in abundance, and three short plasmids in the ECESBL-1 isolate as well as the 207-kb plasmid in the ESBL-producing K. pneumoniae isolate were underrepresented. Even with unequal representation, we found that by 5,000 reads all of the plasmids were sufficiently covered to generate near-complete assemblies and for detection of all of the AMR genes (Fig. 1 and data not shown). In many cases, though, plasmids were not assembled as single contigs with this number of reads, and the number of reads required for plasmid assembly varied among isolates (Table S4 and Fig. S1). It is possible to generate 5,000 reads within 2 h of MinION runtime for older R7 sequencing chemistries and in as little as 20 min with the newer R9 sequencing kit (Fig. 1 and 2).
FIG 1.
Reference genome coverage and resistance gene detection in data subsets of KPNIH1. Random subsets of MinION reads (200, 500, 1,000, 2,000, and 5,000 reads) were selected to evaluate minimum requirements for plasmid assembly and resistance gene prediction. (A) Overall KPNIH1 plasmid coverage by assembled contigs (means ± SDs calculated over 10 random samples) at each sampled read count. Dashed lines indicates rough estimate of plasmid coverage achieved by sequencing of total DNA, calculated as straight-line projection from measured plasmid coverage data points and assuming ∼10% plasmid content. (B) Assembly accuracy (means ± SDs calculated over 10 random samples) for each data subset. (C) Reference genome coverage by raw MinION reads and assembled contigs for reach data subset. Outer rings represent raw read alignment to reference plasmid assemblies; inner rings represent percent reference sequence covered by assembled contigs. Black lines in inner rings indicate positions of resistance genes for reference. The scale bar indicates 100-fold sequencing depth.
FIG 2.
Acquisition of sequencing reads over time. The amount of time required to obtain 5,000 MinION reads was determined for all three sequencing runs performed. The KPNIH1 and ESBL-producing E. coli runs (red and blue lines) were significantly slower than the ESBL-producing K. pneumoniae run (green line) due to the older flow cell versions used. For the ESBL-producing K. pneumoniae isolate sequenced with the rapid 1D kit and the FLOW-MIN 105 (R9 SpotOn), 5,000 reads were obtained in as little as 19 min.
DISCUSSION
In this work, we assessed the feasibility of nanopore sequencing and AMR gene annotation in clinical isolates in the setting of the clinical microbiology laboratory. We found that plasmid DNA extraction followed by MinION sequencing permits efficient same-day detection of AMR genes in clinical isolates. For all of the isolates that were sequenced in this work, it was possible to assemble MinION reads into contigs covering plasmids and to detect the full complement of plasmid-borne AMR genes expected from phenotypic resistance testing, including those conferring beta-lactam, aminoglycoside, sulfonamide, tetracycline, macrolide, and phenicol resistance.
For the clinical ESBL-producing E. coli and K. pneumoniae isolates studied in this work, closed circular sequences were obtained for five and three distinct plasmids, respectively, using a combination of MinION and MiSeq reads. Based on comparison with KPNIH1 reference sequences, we estimate that consensus accuracy is ∼99.9% after polishing with MiSeq reads. However, for rapid turnaround time in the context of the clinical lab workflow, we favor a strategy that uses only MinION reads without MiSeq polishing, which can produce a draft assembly with an estimated consensus accuracy of ∼99%. Though such a strategy may not always produce single contig plasmid assemblies, and the ∼1% error rate may complicate exhaustive confirmation of functional open reading frames, we found that resistance gene annotation using MinION-only draft assemblies and MinION plus Illumina-polished assemblies produced essentially identical results. For certain applications, assembly at a higher level of accuracy may be required, including single nucleotide polymorphism (SNP)-based isolate tracking in the epidemiologic setting and categorization of resistance mechanisms determined by point mutations, such as those occurring in porin, gyrase, and topoisomerase genes.
To assess the minimal turnaround time for sequencing and comprehensive resistance gene identification, we generated plasmid assemblies and performed AMR prediction using data subsets of different sizes. Our analysis shows that the necessary amount of sequencing data can be acquired in less than 1 h, and a full antimicrobial gene detection workflow is possible to achieve in less than 6 h. Such a workflow would consist of 1- to 2-h plasmid DNA extraction followed by rapid transposase-based library preparation (15 min of hands-on time), MinION loading (∼30 min), and a 1-h MinION run with local 1D base calling and 1 to 2 h for sequence assembly and interpretation of results. These estimates for sequencing time are based on the use of a MIN105 (R9 SpotOn) flow cell. Newer flow cells, however, provide even higher throughput, and sequence acquisition time will likely decrease. For higher-quality assemblies, an output from a longer (24 to 48 h) MinION run is supplemented by Illumina MiSeq data. MiSeq sequencing takes from ∼16 h for a 150-cycle protocol to ∼60 h for a paired-end 600-cycle protocol. Therefore, taking into account the MiSeq run time and a longer assembly time, high-accuracy plasmid assemblies followed by annotation of AMR genes in these assemblies could be completed in approximately 3 days. We note that we performed plasmid purification from broth cultures, which deviates from standard procedures in a clinical microbiology laboratory. Plasmid preparation could also be performed from subcultures plated to solid media. We also note more generally that genomic sequencing itself is not standard in many clinical microbiology labs, and the assembly and analysis times mentioned above represent those that can be reasonably expected when appropriate bioinformatics expertise is available.
A further question that arises is whether it is necessary to assemble raw MinION reads at all for AMR gene detection. Indeed, the MinION can acquire sequences at extremely high speed—greater than 250 nucleotides (nt)/s—and from hundreds of pores simultaneously. Given this throughput, one can imagine that AMR gene detection could be achieved within minutes by read mapping without assembly. However, the 10% or higher single-read error rate results in sequences that may not distinguish between closely related alleles of AMR genes. Accuracy is improved critically by sequence assembly, allowing discrimination of closely related alleles with medium-depth assembly of MinION reads. It is conceivable that continued improvements in sequencing chemistry and base calling algorithms could improve raw read accuracy to the point that intermediate assembly is not needed prior to AMR gene identification in the future. It should be noted that the 2D library kit used in this study has been discontinued and a new 1D2 higher-accuracy chemistry has been introduced with a reported modal raw read accuracy of ∼97%. This single-read accuracy is still not high enough to skip consensus generation, but it is expected to increase with future improvement in computational tools.
A distinctive feature of our approach is the use of plasmid-enriched DNA. As plasmids often contain the bulk of resistance genes in Enterobacteriaceae isolates (33), much can be learned about an isolate's resistome by sequencing only the plasmid compartment, and this can be done at substantial savings in sequencing and assembly time. While the genome size in Enterobacteriaceae is often ∼5 to 6 Mb, the total aggregate length of the plasmids in an isolate usually does not exceed 0.5 Mb, and therefore, we estimate that no more than 1/10 of sequencing output is needed. Even though the plasmid DNA preparations in this study were not completely devoid of chromosomal DNA, its content did not exceed ∼10 to 20%, and plasmids were enriched approximately 10-fold compared to total DNA extracts. The reduced sequencing output results directly in reduced sequence acquisition time and much faster computational analysis. For our larger data sets and using a moderately powerful 20-core server, we observed runtime analyses of over 24 h, but for sequencing of plasmid content only, assembly time routinely does not exceed 1 h for 5,000-read data sets. Though many resistance genes in Enterobacteriaceae are located on plasmids as noted above, for complete characterization of AMR genes in these isolates (and in those non-Enterobacteriaceae isolates in which the bulk of resistance is not plasmid based), plasmid sequences should be supplemented by whole-genome assemblies. In addition, since plasmids may be underrepresented in genomic DNA preparations, we believe that supplementing whole-genome assemblies with plasmid sequences has substantial value for accurate determination of full plasmid content in isolates.
In our analysis, we noted unequal coverages of different plasmids by sequencing reads. Median coverage across plasmids varied between 5-fold (KPNIH1) and 20-fold (KPESBL-1). These differences may be explained by variable DNA extraction efficiencies, differences in plasmid copy number, or other biases introduced during library preparation or sequencing. With a plasmid DNA preparation, sufficient depth of coverage for all plasmids is more likely to be reached, and this strategy therefore avoids omissions from final assembly. Still, care should be taken to prevent loss of potentially underrepresented plasmids and to acquire data at sufficient depth. While we found that 5,000 reads provides efficient plasmid recovery for the three isolates analyzed in this study, this number might not be sufficient in all cases. Additional restrictions are imposed by desired assembly accuracy. Although canu and other genome assemblers can generate contigs from low-coverage data sets, consensus accuracy suffers, with the potential consequence of inability to detect shorter AMR genes or misassignment of related gene classes. Therefore, a strategy which requires acquisition of >10× minimal coverage for each contig might be justified if a comprehensive characterization of the plasmid-borne resistome is required.
As with any approach that predicts antimicrobial susceptibility from genomic sequence, translation from genomic annotation of AMR genes to quantitative phenotypic MIC is nontrivial. Some of the hurdles that remain for quantitative resistance class prediction in Enterobacteriaceae include prediction of exact aminoglycoside MICs from aminoglycoside-modifying enzymes, fluoroquinolone resistance prediction from single mutations in chromosome-based topoisomerase genes, and prediction of the complex changes in susceptibility that can result from mutations in outer membrane porin genes (1, 34). Recent research, however, shows great promise in defining both qualitative and quantitative antibiotic resistance phenotypes (35–37). Since the development of such sequence-to-phenotype prediction algorithms was outside the scope of the present work, we used a simple, previously established gene annotation-based approach (30). With this approach, we have found that it is possible to account for all of the phenotypically detected categories of plasmid-based resistance in these isolates. However, we do not suggest here that genomic sequencing can as yet be performed in place of phenotypic susceptibility testing, though we believe that information from such sequencing will complement phenotypic testing.
In conclusion, we found that rapid benchtop plasmid sequencing and annotation of plasmid-based AMR elements are feasible in the clinical microbiology laboratory. With the workflow presented here, plasmid DNA extraction, sequencing, assembly, and resistance gene annotation can be performed within 6 h, significantly less time than traditional phenotypic susceptibility testing requires. In the context of clinical lab implementation, it is important to note that this workflow requires strong experimental skills (plasmid extraction and sequencing) and substantial bioinformatics expertise. Though detection of resistance genes in clinical isolates does not yet replace traditional susceptibility testing, the reporting of individual genes has proven very helpful in guiding clinical management in many cases, including rapid testing for mecA in Staphylococcus aureus, vanA in enterococci, and blaKPC/blaNDM in Enterobacteriaceae. We anticipate that technologies, such as nanopore sequencing, that expand the scope of what is possible for rapid detection and reporting of resistance genes will likewise aid clinical management. We also expect that the roles of genomic analysis in susceptibility prediction will grow with improved resistance gene databases, better genotype-phenotype correlations, and user-friendly analysis tools. In addition to supporting clinical decision making, methods for rapid sequencing of bacterial plasmids will contribute substantially to the understanding of the mechanistic basis of resistance and its dissemination.
MATERIALS AND METHODS
Isolates.
Deidentified clinical bacterial isolates were subcultured from −80°C storage and incubated overnight in ambient atmosphere at 35°C. The previously sequenced clinical isolate KPNIH1 (3, 4) was plated on HardyCHROMCRE agar (Hardy Diagnostics, Santa Maria, CA). The clinical ESBL-producing E. coli (ECESBL-1) and ESBL-producing K. pneumoniae (KPESBL-1) isolates were plated on blood agar (Remel, Lenexa, KS) and passaged once. Single colonies of each isolate were picked from plates and inoculated into LB broth (Fisher Scientific, Hampton, NH). Liquid cultures (1 ml or 100 ml) were incubated at 37°C with ambient air and shaking overnight.
DNA purification.
Two different DNA extraction methods were used for plasmid and total DNA purification. Plasmid DNA was purified from 100 ml of liquid culture using the Qiafilter plasmid minikit (Qiagen, Valencia, CA) per the manufacturer's protocol for low-copy-number plasmids. Briefly, the bacterial cells were pelleted by centrifugation at 6,000 × g for 15 min at 4°C. Pelleted bacterial cells were resuspended in 6 ml of buffer P1 and lysed with the addition of 6 ml of buffer P2. Following 5 min of incubation at room temperature, proteins were precipitated with the addition of 6 ml of buffer P3. This precipitate was removed using a QIAFilter cartridge and the resulting supernatant was loaded onto a gravity flow DNA-binding column equilibrated with buffer QBT. The column was washed twice with 10 ml of buffer QC, and the purified DNA was eluted with 5 ml of buffer QF at 65°C. DNA was isopropanol precipitated and centrifuged at 20,800 × g for 30 min at 4°C. The resulting pellet was washed with 70% ethanol and resuspended in 1× Tris-EDTA (pH 8.0). Total genomic DNA was extracted from 1 ml of liquid culture with the Gentra Puregene Yeast/Bact. kit (Qiagen, Valencia, CA) per the manufacturer's instructions. For the KPESBL-1 isolate, a 10-μl loopful of the organism from a blood agar plate was resuspended in 1 ml of phosphate-buffered saline and then extracted as follows. Briefly, the bacterial culture was pelleted at 20,800 × g, resuspended in 600 μl of cell lysis solution, and incubated at 80°C for 5 min. RNase A solution (Qiagen) was added to the extract and incubated at 37°C for 15 to 60 min. The extract was briefly cooled on ice, and then 200 μl of protein precipitation solution was added and vigorously vortexed. The precipitate was separated by centrifugation at 20,800 × g for 3 min and the supernatant retained. DNA was precipitated by adding 600 μl of isopropanol to the supernatant. The DNA was pelleted by centrifugation at 20,800 × g for 1 min and washed once with 70% ethanol. The washed and dried DNA was rehydrated in DNA hydration solution and incubated at 65°C for 1 to 2 h, followed by overnight incubation at 30°C with gentle shaking for the KPNIH1 and ECESBL-1 isolates. DNA yields were quantified with a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA), and DNA quality was assessed by migration through a 0.8% agarose gel (Lonza, Rockland, ME).
Illumina MiSeq library preparation and sequencing.
Libraries for Illumina MiSeq sequencing were prepared using the Nextera XT library prep kit (Illumina, San Diego, CA). For each library, 1 ng of DNA was added to 10 μl of TD buffer and 5 μl of ATM. This reaction mixture was incubated at 55°C for 5 min to tagment the DNA and then neutralized with 5 μl of NT buffer. Tagmented DNA fragments were indexed with unique barcode primers and PCR amplified in a 50-μl reaction mixture as follows: 72°C for 3 min, 95°C for 30 s, and 12 cycles of 95°C for 10 s, 55°C for 30 s, and 72°C for 30 s, followed by 72°C for 5 min. PCR cleanup was performed by addition of 30 μl of AMPure XP beads (Beckman Coulter, Brea, CA), followed by 5 min of incubation and then two washings with 80% ethanol and elution into 50 μl of RSB. Plasmid and total DNA Nextera libraries for all isolates were multiplexed and sequenced in a paired-end 300-cycle mode on an Illumina MiSeq instrument using 600-cycle reagent kit v. 3 (Illumina).
MinION library preparation and sequencing.
MinION libraries of the KPNIH1 and ECESBL-1 clinical isolates were prepared from plasmid DNA preparations using the SQK-NSK007 nanopore sequencing kit, R9 version (Oxford Nanopore Technologies, Oxford, United Kingdom). Briefly, 1.5 μg (KPNIH1) or 2 μg (ECESBL-1) of plasmid DNA was sheared at 6,000 rpm in a g-TUBE (Covaris, Woburn, MA). The fragmented DNA was repaired using NEBNext FFPE repair mix (New England BioLabs [NEB], Ipswich, MA). After 15 min of incubation at 20°C with FFPE repair buffer and FFPE repair mix, the repaired DNA was purified using AMPure XP beads in a 1:1 ratio, washed with 70% ethanol, and eluted with nuclease-free water. The DNA fragments were end repaired and deoxyadenosine (dA) tailed using the NEBNext end repair/dA tailing module (NEB). DNA was mixed with DNA CS (positive-control strand) and Ultra II end prep reaction buffer and enzyme mix and incubated for 5 min at 20°C and 5 min at 65°C in a thermocycler. DNA was purified using AMPure XP beads in a 1:1 ratio, washed with 70% ethanol, and eluted with nuclease-free water. End-prepped DNA was incubated with hairpin adapter and NEB blunt/TA ligase master mix (NEB) for 10 min at room temperature, followed by addition of hairpin tether and 10 min of incubation at room temperature. The libraries were incubated with bead binding buffer-washed MyOne streptavidin C1 beads (Invitrogen, Carlsbad, CA) in a 1:1 ratio for 5 min at room temperature, washed with bead binding buffer, and eluted with elution buffer prior to loading.
For the KPESBL-1 clinical isolate, MinION libraries were prepared using DNA from both plasmid and total DNA extractions with the SQK-RAD001 rapid sequencing kit I, R9 version (Oxford Nanopore Technologies, Oxford, United Kingdom). Two hundred nanograms of DNA was tagmented by incubation with fragmentation mix (FRM) for 1 min at 30°C followed by 1 min at 75°C in a thermocycler. Tagmented DNA was then adapter ligated by incubation with rapid adapter mix (RAD) and NEB blunt/TA ligase master mix for 5 min at room temperature and directly loaded for sequencing.
KPNIH1 and ESBL-producing E. coli isolates were sequenced using FLOW-MIN 104 (R9) flow cells, and the KPESBL-1 isolate was sequenced using the FLOW-MIN 105 (R9 SpotOn) flow cell. Sequencing protocols appropriate to the flow cell version were used. At approximately 18 to 24 h (for KPNIH1 and ECESBL-1) and approximately 2 h (for KPESBL-1) into the sequencing protocol, additional library was added to the flow cell. For KPNIH1 and ECESBL-1, the second loading was with more of the initial plasmid library. For the KPESBL-1 isolate, the plasmid DNA library was loaded first and then the total DNA library was loaded second. Transient computer software interruptions (including an interruption due to a network-pushed update) were corrected by resets of sequencing protocol in certain instances, but total intended sequencing time was approximately maintained, though the four flow cell sequencing groups specified by the software were not necessarily completed in serial order. We did not find that this had significant impact on our analysis.
AST.
Phenotypic antimicrobial susceptibility testing (AST) was performed by broth microdilution with a Sensititre Aris 2x and custom gram-negative panels (Thermo Fisher Scientific, Waltham, MA) or by Kirby-Bauer disk diffusion (Becton, Dickinson and Company, Franklin Lakes, NJ). MIC data or zones of inhibition were interpreted per CLSI document M100-S27 (38).
Computational methods.
For the KPNIH1 and ECESBL-1 isolates, MinION 2D base calling was performed remotely using the Metrichor web-based service. 1D base calling was performed on a local computer in real time by MinKNOW v. 1.1.20. MinION data were further processed using poretools v. 0.6.0 (39) and converted to fasta/fastq files. Sequences were assembled using the ONT reference assemble-polish pipeline (https://github.com/nanoporetech/ont-assembly-polish), consisting of the programs canu (26), racon (28), and pilon (29), using only MinION reads longer than 1 kb. The ONT assemble-polish pipeline can be invoked to assemble MinION reads only with canu; alternatively, canu, racon, and pilon can be invoked consecutively to assemble and then polish assemblies with MiSeq reads. The ONT pipeline was run with canu assembler settings designed for better plasmid data recovery: “CANU_PARAMETERS = corMinCoverage = 0 corOutCoverage = 1000.” For assembly of the KPESBL-1 data set, we used only reads shorter than 30 kb to reduce assembly times. Whole-genome MiSeq data were assembled using SPAdes v. 3.9.1 (40). To produce finished and closed plasmid assemblies, contigs generated by the ONT assembly-polish pipeline were further processed. We first calculated the coverage of each contig by plasmid and whole-genome shotgun reads and retained only those contigs having much higher coverage by plasmid reads. Since the plasmid DNA extracts unavoidably contain a substantial proportion of chromosomal DNA, raw assembly may contain multiple chromosomal contigs, and this filtration step is necessary to separate plasmid-derived contigs from chromosomal contigs. The remaining contigs were subsequently circularized with the help of circlator v. 1.5.0 (41) and further polished with MiSeq data using pilon v. 1.20 (29). Assemblies were compared using quast v. 4.5 (42). For aligning raw MinION reads to generate coverage profiles, we used the BWA mem algorithm (bwa v. 0.7.12 [https://github.com/lh3/bwa]) with ont-optimized settings (bwa mem -x ont2d). AMR gene annotation on the assembled contigs was performed using Resfinder web software, using a 90.0% identity threshold and 60.0% minimum target gene coverage (30). Circular representations of coverage of KPNIH1 plasmids by raw reads and assembled contigs were plotted using Circos v. 0.69.3 (43).
Accession number(s).
The MiSeq and MinION data obtained in this study have been deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA392935 (https://www.ncbi.nlm.nih.gov/bioproject/392935).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the National Institutes of Health Clinical Center.
The opinions expressed in this presentation are those of the authors and do not necessarily reflect those of the Department of Health and Human Services or the National Institutes of Health.
This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01069-17.
REFERENCES
- 1.Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. 2015. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 2.Conlan S, Park M, Deming C, Thomas PJ, Young AC, Coleman H, Sison C, NISC Comparative Sequencing Program, Weingarten RA, Lau AF, Dekker JP, Palmore TN, Frank KM, Segre JA. 2016. Plasmid dynamics in KPC-positive Klebsiella pneumoniae during long-term patient colonization. mBio 7(3):e00742-16. doi: 10.1128/mBio.00742-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conlan S, Thomas PJ, Deming C, Park M, Lau AF, Dekker JP, Snitkin ES, Clark TA, Luong K, Song Y, Tsai YC, Boitano M, Dayal J, Brooks SY, Schmidt B, Young AC, Thomas JW, Bouffard GG, Blakesley RW, Program NCS, Mullikin JC, Korlach J, Henderson DK, Frank KM, Palmore TN, Segre JA. 2014. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 6:254ra126. doi: 10.1126/scitranslmed.3009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snitkin ES, Zelazny AM, Thomas PJ, Stock F, NISC Comparative Sequencing Program, Henderson DK, Palmore TN, Segre JA. 2012. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med 4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koren S, Phillippy AM. 2015. One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Curr Opin Microbiol 23:110–120. doi: 10.1016/j.mib.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 6.Jain M, Olsen HE, Paten B, Akeson M. 2016. The Oxford nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol 17:239. doi: 10.1186/s13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashton PM, Nair S, Dallman T, Rubino S, Rabsch W, Mwaigwisya S, Wain J, O'Grady J. 2015. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat Biotechnol 33:296–300. doi: 10.1038/nbt.3103. [DOI] [PubMed] [Google Scholar]
- 8.George S, Pankhurst L, Hubbard A, Votintseva A, Stoesser N, Sheppard AE, Mathers A, Norris R, Navickaite I, Eaton C, Iqbal Z, Crook DW, Phan HTT. 9 June 2017. Resolving plasmid structures in Enterobacteriaceae using the MinION nanopore sequencer: assessment of MinION and MinION/Illumina hybrid data assembly approaches. Microb Genom doi: 10.1099/mgen.0.000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Judge K, Hunt M, Reuter S, Tracey A, Quail MA, Parkhill J, Peacock SJ. 2016. Comparison of bacterial genome assembly software for MinION data and their applicability to medical microbiology. Microb Genom 2:e000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karamitros T, Harrison I, Piorkowska R, Katzourakis A, Magiorkinis G, Mbisa JL. 2016. De novo assembly of human herpes virus type 1 (HHV-1) genome, mining of non-canonical structures and detection of novel drug-resistance mutations using short- and long-read next generation sequencing technologies. PLoS One 11:e0157600. doi: 10.1371/journal.pone.0157600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loman NJ, Quick J, Simpson JT. 2015. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods 12:733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- 12.Quick J, Ashton P, Calus S, Chatt C, Gossain S, Hawker J, Nair S, Neal K, Nye K, Peters T, De Pinna E, Robinson E, Struthers K, Webber M, Catto A, Dallman TJ, Hawkey P, Loman NJ. 2015. Rapid draft sequencing and real-time nanopore sequencing in a hospital outbreak of Salmonella. Genome Biol 16:114. doi: 10.1186/s13059-015-0677-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szabó M, Nagy T, Wilk T, Farkas T, Hegyi A, Olasz F, Kiss J. 2016. Characterization of two multidrug-resistant IncA/C plasmids from the 1960s by using the MinION sequencer device. Antimicrob Agents Chemother 60:6780–6786. doi: 10.1128/AAC.01121-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Istace B, Friedrich A, d'Agata L, Faye S, Payen E, Beluche O, Caradec C, Davidas S, Cruaud C, Liti G, Lemainque A, Engelen S, Wincker P, Schacherer J, Aury JM. 2017. De novo assembly and population genomic survey of natural yeast isolates with the Oxford nanopore MinION sequencer. Gigascience 6:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faria NR, Quick J, Claro IM, Theze J, de Jesus JG, Giovanetti M, Kraemer MUG, Hill SC, Black A, da Costa AC, Franco LC, Silva SP, Wu CH, Raghwani J, Cauchemez S, du Plessis L, Verotti MP, de Oliveira WK, Carmo EH, Coelho GE, Santelli A, Vinhal LC, Henriques CM, Simpson JT, Loose M, Andersen KG, Grubaugh ND, Somasekar S, Chiu CY, Muñoz-Medina JE, Gonzalez-Bonilla CR, Arias CF, Lewis-Ximenez LL, Baylis SA, Chieppe AO, Aguiar SF, Fernandes CA, Lemos PS, Nascimento BLS, Monteiro HAO, Siqueira IC, de Queiroz MG, de Souza TR, Bezerra JF, Lemos MR, Pereira GF, Loudal D, Moura LC, Dhalia R, Franca RF, Magalhães T, Marques ET Jr, Jaenisch T, Wallau GL, de Lima MC, Nascimento V, de Cerqueira EM, de Lima MM, Mascarenhas DL, Neto JPM, Levin AS, Tozetto-Mendoza TR, Fonseca SN, Mendes-Correa MC, Milagres FP, Segurado A, Holmes EC, Rambaut A, Bedford T, Nunes MR, Sabino EC, Alcantara LCJ, Loman NJ, Pybus OG. 2017. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature 546:406–410. doi: 10.1038/nature22401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greninger AL, Naccache SN, Federman S, Yu G, Mbala P, Bres V, Stryke D, Bouquet J, Somasekar S, Linnen JM, Dodd R, Mulembakani P, Schneider BS, Muyembe-Tamfum JJ, Stramer SL, Chiu CY. 2015. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med 7:99. doi: 10.1186/s13073-015-0220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee RS, Pai M. 2017. Real-time sequencing of Mycobacterium tuberculosis: are we there yet? J Clin Microbiol 55:1249–1254. doi: 10.1128/JCM.00358-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quick J, Grubaugh ND, Pullan ST, Claro IM, Smith AD, Gangavarapu K, Oliveira G, Robles-Sikisaka R, Rogers TF, Beutler NA, Burton DR, Lewis-Ximenez LL, de Jesus JG, Giovanetti M, Hill SC, Black A, Bedford T, Carroll MW, Nunes M, Alcantara LC Jr, Sabino EC, Baylis SA, Faria NR, Loose M, Simpson JT, Pybus OG, Andersen KG, Loman NJ. 2017. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc 12:1261–1276. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quick J, Loman NJ, Duraffour S, Simpson JT, Severi E, Cowley L, Bore JA, Koundouno R, Dudas G, Mikhail A, Ouedraogo N, Afrough B, Bah A, Baum JH, Becker-Ziaja B, Boettcher JP, Cabeza-Cabrerizo M, Camino-Sanchez A, Carter LL, Doerrbecker J, Enkirch T, Garcia-Dorival I, Hetzelt N, Hinzmann J, Holm T, Kafetzopoulou LE, Koropogui M, Kosgey A, Kuisma E, Logue CH, Mazzarelli A, Meisel S, Mertens M, Michel J, Ngabo D, Nitzsche K, Pallasch E, Patrono LV, Portmann J, Repits JG, Rickett NY, Sachse A, Singethan K, Vitoriano I, Yemanaberhan RL, Zekeng EG, Racine T, Bello A, Sall AA, Faye O, Magassouba N, Williams CV, Amburgey V, Winona L, Davis E, Gerlach J, Washington F, Monteil V, Jourdain M, Bererd M, Camara A, Somlare H, Camara A, Gerard M, Bado G, Baillet B, Delaune D, Nebie KY, Diarra A, Savane Y, Pallawo RB, Gutierrez GJ, Milhano N, Roger I, Williams CJ, Yattara F, Lewandowski K, Taylor J, Rachwal P, Turner D, Pollakis G, Hiscox JA, Matthews DA, O'Shea MK, Johnston AM, Wilson D, Hutley E, Smit E, Di Caro A, Woelfel R, Stoecker K, Fleischmann E, Gabriel M, Weller SA, Koivogui L, Diallo B, Keita S, Rambaut A, Formenty P, Gunther S, Carroll MW. 2016. Real-time, portable genome sequencing for Ebola surveillance. Nature 530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt K, Mwaigwisya S, Crossman LC, Doumith M, Munroe D, Pires C, Khan AM, Woodford N, Saunders NJ, Wain J, O'Grady J, Livermore DM. 2017. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J Antimicrob Chemother 72:104–114. doi: 10.1093/jac/dkw397. [DOI] [PubMed] [Google Scholar]
- 21.Votintseva AA, Bradley P, Pankhurst L, Del Ojo Elias C, Loose M, Nilgiriwala K, Chatterjee A, Smith EG, Sanderson N, Walker TM, Morgan MR, Wyllie DH, Walker AS, Peto TEA, Crook DW, Iqbal Z. 2017. Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J Clin Microbiol 55:1285–1298. doi: 10.1128/JCM.02483-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao MD, Ganesamoorthy D, Elliott AG, Zhang H, Cooper MA, Coin LJ. 2016. Streaming algorithms for identification of pathogens and antibiotic resistance potential from real-time MinION(TM) sequencing. Gigascience 5:32. doi: 10.1186/s13742-016-0137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nair S, Ashton P, Doumith M, Connell S, Painset A, Mwaigwisya S, Langridge G, de Pinna E, Godbole G, Day M. 2016. WGS for surveillance of antimicrobial resistance: a pilot study to detect the prevalence and mechanism of resistance to azithromycin in a UK population of non-typhoidal Salmonella. J Antimicrob Chemother 71:3400–3408. doi: 10.1093/jac/dkw318. [DOI] [PubMed] [Google Scholar]
- 24.van der Helm E, Imamovic L, Hashim Ellabaan MM, van Schaik W, Koza A, Sommer MOA. 2017. Rapid resistome mapping using nanopore sequencing. Nucleic Acids Res 45:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao MD, Nguyen SH, Ganesamoorthy D, Elliott AG, Cooper MA, Coin LJ. 2017. Scaffolding and completing genome assemblies in real-time with nanopore sequencing. Nat Commun 8:14515. doi: 10.1038/ncomms14515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benítez-Páez A, Portune KJ, Sanz Y. 2016. Species-level resolution of 16S rRNA gene amplicons sequenced through the MinION portable nanopore sequencer. Gigascience 5:4. doi: 10.1186/s13742-016-0111-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaser R, Sovic I, Nagarajan N, Sikic M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27:737–746. doi: 10.1101/gr.214270.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yanat B, Rodriguez-Martinez JM, Touati A. 2017. Plasmid-mediated quinolone resistance in Enterobacteriaceae: a systematic review with a focus on Mediterranean countries. Eur J Clin Microbiol Infect Dis 36:421–435. doi: 10.1007/s10096-016-2847-x. [DOI] [PubMed] [Google Scholar]
- 33.Paterson DL. 2006. Resistance in gram-negative bacteria: Enterobacteriaceae. Am J Med 119:S20–S28; discussion, S62–S70. doi: 10.1016/j.amjmed.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 34.Partridge SR. 2015. Resistance mechanisms in Enterobacteriaceae. Pathology 47:276–284. doi: 10.1097/PAT.0000000000000237. [DOI] [PubMed] [Google Scholar]
- 35.Eyre DW, De Silva D, Cole K, Peters J, Cole MJ, Grad YH, Demczuk W, Martin I, Mulvey MR, Crook DW, Walker AS, Peto TE, Paul J. 2017. WGS to predict antibiotic MICs for Neisseria gonorrhoeae. J Antimicrob Chemother doi: 10.1093/jac/dkx067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metcalf BJ, Chochua S, Gertz RE Jr, Li Z, Walker H, Tran T, Hawkins PA, Glennen A, Lynfield R, Li Y, McGee L, Beall B, Active Bacterial Core surveillance team. 2016. Using whole genome sequencing to identify resistance determinants and predict antimicrobial resistance phenotypes for year 2015 invasive pneumococcal disease isolates recovered in the United States. Clin Microbiol Infect 22:1002.e1–1002.e8. doi: 10.1016/j.cmi.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 37.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. 2016. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother 60:5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.CLSI. 2017. Performance standards for antimicrobial susceptibility testing. M100-S27. Clinical and Laboratory Standards Institute, Wayne, PA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loman NJ, Quinlan AR. 2014. Poretools: a toolkit for analyzing nanopore sequence data. Bioinformatics 30:3399–3401. doi: 10.1093/bioinformatics/btu555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hunt M, Silva ND, Otto TD, Parkhill J, Keane JA, Harris SR. 2015. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol 16:294. doi: 10.1186/s13059-015-0849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.