Abstract
Lactobacillus (L. casei) Zhang is a koumiss-originated probiotic strain, which was used as a model in a long-term antibiotics-driven evolution experiment to reveal bacterial evolutionary dynamics; and we isolated gentamycin-resistant L. casei Zhang descendents. To decipher the gentamycin resistance mechanism, here we cultivated the parental L. casei Zhang and its descendent cells in an antibiotics-containing environment to compare their global protein expression profiles using the iTRAQ-based proteomic approach. A total of 72 proteins were significantly up-regulated (>2.0-fold, P < 0.05), whilst 32 proteins were significantly down-regulated <−2.0-fold, P < 0.05) in the descendent line. The gentamycin-resistant descendent line showed elevated expression in some carbohydrates, amino acids, and purine metabolic pathways. Several stress-related proteins were also differentially expressed. Among them, one alkaline shock protein, asp23, was up-regulated most in the gentamycin-resistant strain (21.9-fold increase compared with the parental strain). The asp23 gene disruption mutant was significantly more sensitive to gentamycin compared with the wild type, suggesting an important role of this gene in developing the gentamycin-resistant phenotype in L. casei. Our report has described the adaptation of a probiotic strain that has acquired antibiotics resistance through long-term antibiotics exposure at the proteome level, and we revealed a novel mechanism of gentamycin resistance.
Keywords: Lactobacillus casei Zhang, proteomic analysis, gentamycin, alkaline shock protein, asp23
Introduction
Lactobacillus (L. casei) is the dominant species within the genus Lactobacillus found in koumiss, a naturally fermented dairy product (Wu et al., 2009a). Besides, it has been detected in corn silage, wine, pickle, human gastrointestinal tracts, blood, yogurt, and cheese (Cai et al., 2007; Bao et al., 2016). The wide ecological distribution of L. casei reflects its flexibility in metabolizing carbohydrates (Zhang et al., 2010), and thus it is commonly used in food industry. The species Lactobacillus casei has been used both as starter cultures and as food additives for improving food texture properties (Dantas et al., 2016). Some strains are considered as probiotics bacteria due to their beneficial effects, such as antibacterial, antioxidative, and immunomodulatory properties (Ya et al., 2008; Zhang et al., 2014b; Wang et al., 2016). Meanwhile, the increasing availability of the whole genome sequences of representative L. casei strains and genetic tools for creating recombinant Lactobacillus has largely facilitated genetic and functional studies, leading to remarkable progress in our understanding of the cell biology of these bacteria (Xu and Kong, 2013; Lu et al., 2016).
Antibiotic resistance of bacteria is an increasingly serious public health threat (Normark and Normark, 2002). This is especially critical for pathogenic bacteria that can rapidly become antibiotic-resistant in response to clinical application of antibiotics (Arias and Murray, 2015). It is unlikely that the probiotics used in food industry face a similar situation due to low exposure to antibiotics within the food matrix. Moreover, strict regulations must be followed by the food industry to avoid unnecessary use of antibiotics (Lara et al., 2006). The dairy industry even routinely monitors raw milk against antibiotics contamination because such contaminants would suppress or even kill lactic acid bacteria (LAB) and subsequently affect milk fermentation. The addition of bacteriocins or bacteriocin-producing bacteria has been explored as a method of food preservation, but it is not a wide-spread practice (Galvez et al., 2007). However, owing to the imminent global health concern of bacterial antibiotic resistance, the use of probiotics has been proposed as a valuable adjunct or even alternative to antibiotic therapy in clinical practice due to both their health-promoting properties and intrinsic bacteriocidal effects (Boyanova and Mitov, 2012; Reid, 2017). Several clinical trials supported the use of probiotics in the management of acute gastroenteritis and antibiotic-associated diarrhea (Katz, 2010). In these situations, probiotics may be directly exposed to antibiotics, sometimes even at a high concentration. Thus, it would be crucial to characterize the adaptation of probiotics to antibiotics stress.
Lactobacillus casei Zhang is a probiotic strain isolated from koumiss (Zhang et al., 2014a, 2015). We previously performed a long-term evolution experiment using L. casei Zhang as a model to reveal bacterial evolutionary dynamics under antibiotic stress; and we generated gentamycin-resistant L. casei Zhang descendents (Wang et al., 2017). During the experiment, the bacteria developed resistance to gentamycin gradually, and the accumulation of genome point mutation stopped shortly after the descendent bacteria reached the maximum bacterial fitness. To decipher the mechanism of the resistance phenotype, here we compared the global protein expression profiles between the parental L. casei Zhang and its descendent cells grown under antibiotic selection force using the iTRAQ-based proteomic approach. Furthermore, we validated the expression of one selected differentially expressed protein using parallel reaction monitoring (PRM), followed by disrupting the corresponding gene to verify its function in the development of gentamycin-resistant phenotype of L. casei Zhang.
Materials and methods
Bacterial isolates and culture conditions
Lactobacillus casei Zhang-G-1200 was isolated from a long-term laboratory-based evolution experiment performed in our laboratory (Wang et al., 2017). The strain Zhang-G-1200 exhibited a higher resistance to gentamycin compared with the parental cells (minimum inhibitory concentration, MIC, of 32 μg/mL for Zhang-G-1200 vs. 2 μg/mL for the parental strain). The acquisition of resistance was a result of multigenerational and stepwise increase during the prolonged cultivation under gentamycin stress. Growth curves of the 2 strains were constructed based on optical density (OD) measurement every 2 h along 30-h fermentation in LSM supplemented with 1 μg/mL gentamycin. Meanwhile, changes in pH and viable counts were determined. All analyses were performed in triplicate.
The bacterial strain, Escherichia (E. coli) DH5α, was used as a host for standard cloning procedures. It was propagated aerobically in Luria Bertani broth at 37°C. Chloramphenicol (10 μg/ml for both E. coli and L. casei Zhang-G-1200) and erythromycin (250 and 5 μg/ml for E. coli and L. casei Zhang-G-1200, respectively) were used for selecting genetically modified bacterial clones.
Sample preparation, protein digestion and iTRAQ labeling
Both the parental and L. casei Zhang-G-1200 cells were collected after 24 h of cultivation in LSM supplemented with 1 μg/mL gentamycin. Four biological replicates were prepared for each of the L. casei Zhang and the Zhang-G-1200 strains. Cells were harvested by centrifugation, followed by washing with phosphate buffered saline for 4 times. One milliliter of lysis buffer was added to each sample, followed by sonication on ice and centrifugation at 13,000 rpm for 10 min at 4°C. The protein concentration of sample supernatants was determined by using the bicinchoninic acid protein assay.
Hundred microgram protein was transferred to a new tube and adjusted to a protein concentration of 1 μg/μL with 100 mM triethylammonium bicarbonate (TEAB). Five microliters of 200 mM DTT were added and incubated at 55°C for 1 h, then 5 μL of 375 mM iodoacetamide was added to the sample and incubated for 30 min at room temperature in the dark. For each sample, proteins were precipitated with ice-cold acetone before re-dissolving in 20 μL TEAB. Proteins were then tryptically digested with sequencing grade modified trypsin (Promega, Madison, WI), and the resultant peptide mixture was labeled using reagents from the iTRAQ reagents kit.
High pH reverse phase separation by UPLC
The peptide mixture was redissolved in buffer A (buffer A: 10 mM ammonium formate in water, pH 10, adjusted with ammonium hydroxide) and then fractionated by high pH separation using an Aquity UPLC system (Waters Corporation, Milford, MA) connected to a reverse phase column (BEH C18 column, 2.1 × 150 mm, 1.7 μm, 300 Å, Waters Corporation, Milford, MA). High pH separation was done using a linear gradient, starting from 0% B to 45% B in 35 min (B: 10 mM ammonium formate in 90% acetonitrile, pH 10.0, adjusted with ammonium hydroxide). The column flow rate and temperature were maintained at 250 μL/min and 45°C, respectively. Sixteen fractions were collected, and each fraction was dried in a vacuum concentrator prior to the next step.
Low pH nano-HPLC-MS/MS analysis
The dried fractions were re-suspended in a solution made of solvent C and D (C: water with 0.1% formic acid; D: acetonitrile with 0.1% formic acid), separated by nano LC, and analyzed by on-line electrospray tandem mass spectrometry. The experiments were performed on a Nano Aquity UPLC system (Waters Corporation, Milford, MA) connected to a quadrupole-Orbitrap mass spectrometer (Q-Exactive) (Thermo Fisher Scientific, Bremen, Germany) equipped with an online nano-electrospray ion source. Eight microliters of peptide sample were loaded onto the trap column (Thermo Scientific Acclaim PepMap C18, 100 μm × 2 cm), with a flow of 10 μl/min for 3 min, and subsequently separated on an analytical column (Acclaim PepMap C18, 75 μm × 25 cm) with a linear gradient, from 5% D to 30% D in 95 min. The column was re-equilibrated to the initial conditions for 15 min. The column flow rate and temperature were maintained at 300 nL/min and 45°C, respectively. An electrospray voltage of 2.0 kV was used against the inlet of the mass spectrometer.
The Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer was operated in the data-dependent mode to switch automatically between MS and MS/MS acquisition. Survey full-scan MS spectra (m/z 350–1,600) were acquired with a mass resolution of 70 K, followed by 15 sequential high-energy-collisional-dissociation (HCD) MS/MS scans with a resolution of 17.5 K. In all cases, one micro-scan was recorded using dynamic exclusion of 30 s, with an MS/MS fixed first mass of 100.
Database searching and data analysis
Tandem mass spectra were extracted by the Proteome Discoverer software (Thermo Fisher Scientific, version 1.4.0.288). The mass profiles generated from all samples were searched with Mascot (Matrix Science, London, UK; version 2.3) against the NCBI database (Taxonomy: L. casei Zhang). We used the percolator algorithm <1% to control peptide level false discovery rates. Only unique peptides were used for protein quantification and the normalization on protein median was applied to correct any experimental bias. The minimum number of proteins that must be observed was set to 1,000. Students't-tests were performed with the software package R, with p < 0.05 considered statistically significant. A 2.0-fold change was used as the threshold for selection of differentially regulated proteins. All regulated proteins were distributed over clusters of orthologous genes (COGs) and searched against the Kyoto Encyclopedia of Genes and Genomes database.
Validation of expression level of an alkaline shock protein by PRM
Based on the results of proteomic analysis, 1 protein (the alkaline shock protein, coded by the gene LCAZH_0227) was selected for PRM analysis. This protein was most up-regulated among all differentially expressed proteins.
Experiments were performed on a Q Exactive mass spectrometer coupled with Easy-nLC1200. Peptide mixtures were separated by C18-reversed phase chromatography on an Easy column (75 μm × 25 cm), and the analytical separation was run for 90 min using a linear gradient of ACN/FA 2%/0.1% (Solvent A) and ACN/FA 80%/0.1% (Solvent B) at a flow rate of 300 nL/min. The gradient programme was run as follows: 5% B at 1 min, ramping to 23% B at 41 min, 29% B at 51 min, rapid ramping to 100% B over 8 min and holding 100% B for 6 min before returning to initial condition of 5% B. The column was re-equilibrated to 5% B for 30 min after each run. All samples were analyzed using a multiplexed PRM method based on a scheduled inclusion list containing the target precursor ions representing the standard peptides. The full scan event was collected at m/z 300–1300, an Orbitrap resolution of 70,000, the automatic gain control target at 3e6, and the maximum fill time at 20 ms. Every full scan was followed by 10 PRM scans at a resolution of 17,500 with an isolation window: 2.0 m/z, an AGC value of 5e5, the maximum fill time of 100 ms, and a normalized collision energy of 28 in a higher-energy c-trap dissociation (HCD) cell.
PRM data analysis and data integration were performed with the Skyline Software. Routine assessment of instrument and chromatographic performance was done with a quality control (QC) standard consisting of all synthetic peptides, which was prepared at a concentration of 20 fmol/μl in 0.1% formic acid. Every sample was injected for three times, the peak areas of the target peptides were extracted using Skyline, peak peaking was manually checked and corrected in accordance to the retention time, transitions, mass accuracy, and MS/MS spectra. At least 3 transitions for each peptide were extracted from the PRM data. Peptides were quantified by summing the peak areas under curve (AUC) of each transition. Peptide abundance was normalized based on the total ion current (TIC) extracted from the full scan acquisition for each run. Proteins were quantified by summing the abundances of the selected peptides, and the accurate protein quantities had to match those of the synthetic standard peptides.
Construction and analysis of an alkaline shock protein gene disruption mutant
The gene LCAZH_0227 (asp23) was selected for target disruption. Plasmids and primers used for gene disruption are listed in Table 1. The mutant line was constructed by using a cre-lox-based system originally developed by Lambert et al. (2007). Briefly, the upstream (amplified with the primers 0227upF and 0227upR,) and downstream (amplified with 0227downF and 0227downR) fragments of the LCAZH_0227 gene were PCR amplified from the genomic DNA of L. casei Zhang-G-1200. The fragments were cloned between the SalI-HF or PmeI and Ecol 53KI or BglII restriction sites of the suicide vector pNZ5319 to form the recombinant mutagenesis vector, pNZ5319-0227 Up-Down. To inactivate the LCAZH_0227 gene, pNZ5319-0227 Up-Down was introduced into L. casei Zhang-G-1200 by electroporation. Chloramphenicol-resistant transformants were selected and replica plated to check for an erythromycin-sensitive phenotype. Candidate double-crossover mutant clones were identified by PCR, and correct integration of the lox66-P32-cat-lox71 cassette into the genome was further verified by PCR using the primers 0227upF or catR and catF or 0227downR. To excise the P32-cat selectable marker cassette, the cre expression plasmid pMSPcre was transformed into the 0227::lox66-P32-cat-lox71 gene replacement mutant. The Cre-mediated recombination and correct excision of the P32-cat cassette were checked by PCR using primers spanning the recombination loci (0227upF and 0227downR). The pMSPcre vector was cured from L. casei Zhang-G-1200 Δ0227 colonies by growth without erythromycin selection pressure. Additionally, PCR products were confirmed by sequencing when necessary.
Table 1.
Strains, primers, and plasmids used for constructing the gene disruption mutant.
| Strains, plasmids, and primers | Description or primer sequencea | Reference or source |
|---|---|---|
| STRAINS | ||
| E. coli DH5α | Cloning host | This study |
| L. casei Zhang | Isolated from home-made koumiss in Inner Mongolia, China | Wu et al., 2009a |
| L. casei Zhang-G-1200 | L. casei Zhang propagated in LSM broth containing gentamycin 1μg/mLfor 6 months | Wang et al., 2017 |
| L. casei Zhang-G-1200-0227::lox66-P32-cat-lox71 | Derivative of L. casei Zhang-G-1200 containing a lox66-P32-cat-lox71 replacement of LCAZH_0227 | This study |
| L. casei Zhang-G-1200-Δ0227 | Derivative of L. casei Zhang-G-1200-0227::lox66-P32-cat-lox71 containing a lox72 replacement of LCAZH_0227 | This study |
| PLASMIDS | ||
| pNZ5319 | CmrEmr; containing lox66-P32-cat-lox71 cassette for multiple gene replacement in Gram-positive bacteria | Lambert et al., 2007 |
| pNZ5319-0227Up-Down | CmrEmr; pNZ5319 derivative containing homologous regions upstream and downstream of LCAZH_0227 | This study |
| pMSPcre | Emr; expression of cre | Unpublished |
| PRIMERS | ||
| 0227upF | 5′-ACGCGTCGACGTCGCCTGTCTCGGTATTCCTGTG-3′ | This study |
| 0227upR | 5′-AGCTTTGTTTAAACGGCGCGCCGGTATTGATGCCAGCGTTT-3′ | This study |
| 0227downF | 5′-GGGTTTGAGCTCTTGCCAGTTCAGTCGTT-3′ | This study |
| 0227downR | 5′-GAAGATCTTCTCATTTGCCTCCCTTAT-3′ | This study |
| 85 | 5′-GTTTTTTTCTAGTCCAAGCTCACA-3′ | Lambert et al., 2007 |
| 87 | 5′-GCCGACTGTACTTTCGGATCCT-3′ | Lambert et al., 2007 |
| CatF | 5′-TCAAATACAGCTTTTAGAACTGG-3′ | Lambert et al., 2007 |
| CatR | 5′-ACCATCAAAAATTGTATAAAGTGGC-3′ | Lambert et al., 2007 |
| EryintF | 5′-CGATACCGTTTACGAAATTGG-3′ | Lambert et al., 2007 |
| EryintR | 5′-CTTGCTCATAAGTAACGGTAC-3′ | Lambert et al., 2007 |
The restriction sites in the primer sequences are underlined.
The growth performance and gentamycin-resistant phenotype of the mutant strain was evaluated by viable counts, OD measurements, and MIC of gentamycin (Guo et al., 2017). Phenotypic differences between the wild-type and mutant strains were determined statistically by Student's t-test.
Results
Growth performance of the parental and L. casei Zhang-G-1200 strains
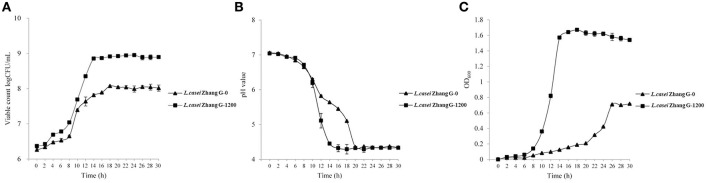
The growth of the two bacterial lines in the presence of gentamycin was monitored. Growth curves were plotted based on viable counts, pH values, and OD values. As shown in Figures 1A–C, the growth performance of L. casei Zhang-G-1200 was completely different from that of the parental line. The viable counts of L. casei Zhang-G-1200 increased more rapidly than that of the parental line, reaching a maximum cell density of 9.05 × 107 cfu/mL after 20 h (Figure 1A). Meanwhile, the pH of the LSM medium inoculated with L. casei Zhang-G-1200 dropped much faster than that of the parental line, indicating a higher fermentation rate (Figure 1B).
Figure 1.
Growth curves of the parental and L. casei Zhang-G-1200 strains based on viable counts (A), pH values (B) and OD values (C) in gentamycin-containing LSM.
Up-regulated L. casei Zhang-G-1200 proteins grown in gentamycin-containing medium
A total of 72 proteins were significantly up-regulated (>2.0-folds, P < 0.05) in L. casei Zhang-G-1200 compared with the parental strain (Table 2). Most of these proteins could be assigned to one of the functional COGs categories (Figure 2), with 26.3% of these proteins involved in carbohydrate transport and metabolism [G] and another 25.3% of them involved in amino acid transport and metabolism [E].
Table 2.
Up-regulated proteins of L. casei Zhang-G-1200 compared with its original strain in the presence of the gentamycin.
| Locus | Function | Fold change |
|---|---|---|
| CARBOHYDRATE TRANSPORT AND METABOLISM | ||
| LCAZH_0394 | Hypothetical protein | 2.07 |
| LCAZH_0395 | Mannose-6-phosphate isomerase | 3.06 |
| LCAZH_0403 | PTS system mannose/fructose/N-acetylgalactosamine-specific transporter subunit IIB | 2.23 |
| LCAZH_0503 | Sugar phosphate isomerase/epimerase | 2.19 |
| LCAZH_0604 | PTS system galactitol-specific transporter subunit IIB | 2.1 |
| LCAZH_0605 | PTS system galacitol transporter subunit EIIC | 2.22 |
| LCAZH_1336 | Tagatose-6-phosphate kinase | 2.41 |
| LCAZH_1768 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase | 2.13 |
| LCAZH_1771 | PTS system cellobiose-specific transporter subunit IIA | 2.2 |
| LCAZH_1772 | PTS system cellobiose-specific transporter subunit IIB | 2.61 |
| LCAZH_2151 | beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase | 2.14 |
| LCAZH_2624 | PTS system fructose-specific transporter subunit IIB | 2.36 |
| LCAZH_2633 | PTS system galactitol transporter subunit IIB | 2.97 |
| LCAZH_2645 | Hypothetical protein | 2.53 |
| LCAZH_2648 | PTS system galacitol transporter subunit EIIB | 5.79 |
| LCAZH_2649 | PTS system galacitol transporter subunit EIIA | 3.96 |
| LCAZH_2653 | Trehalose-6-phosphate hydrolase | 4.66 |
| LCAZH_2725 | Transaldolase | 4.56 |
| LCAZH_2895 | PTS system mannitol-specific transporter subunit IIBC | 3.96 |
| AMINO ACID TRANSPORT AND METABOLISM | ||
| LCAZH_0084 | Tryptophan synthase subunit alpha | 2.25 |
| LCAZH_0107 | Tetrahydrodipicolinate N-succinyltransferase | 2.25 |
| LCAZH_0201 | Oligopeptide ABC transporter periplasmic protein | 2.04 |
| LCAZH_0418 | Amino acid ABC transporter ATP-binding protein | 2.14 |
| LCAZH_0500 | Amino acid transporter | 2.59 |
| LCAZH_0506 | Shikimate 5-dehydrogenase | 2.08 |
| LCAZH_1596 | Oligopeptide ABC transporter periplasmic protein | 2.47 |
| LCAZH_1682 | Lactoylglutathione lyase-like lyase | 2.22 |
| LCAZH_1886 | Oligopeptide ABC transporter periplasmic protein | 2.92 |
| LCAZH_1980 | Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase | 2.04 |
| LCAZH_2023 | Dipeptide/oligopeptide/nickel ABC transporter ATPase | 2.21 |
| LCAZH_2024 | Dipeptide/oligopeptide/nickel ABC transporter permease | 2.45 |
| LCAZH_2025 | Dipeptide/oligopeptide/nickel ABC transporter permease | 3.27 |
| LCAZH_2026 | Oligopeptide ABC transporter periplasmic protein | 2.68 |
| LCAZH_2111 | Homoserine dehydrogenase | 2.36 |
| LCAZH_2302 | Aminopeptidase | 6.77 |
| LCAZH_2518 | NADPH-dependent glutamate synthase subunit beta-like oxidoreductase | 3.52 |
| LCAZH_2519 | Glutamate synthase domain 3 | 2.54 |
| LCAZH_2851 | Polar amino acid ABC transporter ATPase | 2.23 |
| ENERGY PRODUCTION AND CONVERSION | ||
| LCAZH_0188 | Acetate kinase | 2.17 |
| LCAZH_1301 | Acetoin/pyruvate dehydrogenase complex, E2 component, dihydrolipoamide succinyltransferase | 2.05 |
| LCAZH_1302 | Acetoin/pyruvate dehydrogenase complex, E3 component, dihydrolipoamide dehydrogenase | 2.13 |
| LCAZH_1396 | Pyruvate-formate lyase | 3.75 |
| LCAZH_2375 | Fumarase | 2.1 |
| NUCLEOTIDE TRANSPORT AND METABOLISM | ||
| LCAZH_1739 | Folate-dependent phosphoribosylglycinamide formyltransferase PurN | 2.5 |
| LCAZH_1740 | Phosphoribosylaminoimidazole (AIR) synthetase | 2.23 |
| LCAZH_1743 | Phosphoribosylformylglycinamidine (FGAM) synthase, glutamine amidotransferase domain | 2.21 |
| CELL WALL/MEMBRANE/ENVELOPE BIOGENESIS | ||
| LCAZH_0447 | Conjugated bile salt hydrolase-like amidase | 5.04 |
| TRANSCRIPTION | ||
| LCAZH_1410 | GNAT family acetyltransferase | 2.42 |
| LCAZH_2210 | Transcriptional regulator | 3.18 |
| COENZYME TRANSPORT AND METABOLISM | ||
| LCAZH_1463 | Lipoate-protein ligase A | 2.05 |
| TRANSLATION, RIBOSOMAL STRUCTURE AND BIOGENESIS | ||
| LCAZH_1880 | Acetyltransferase | 3.78 |
| POSTTRANSLATIONAL MODIFICATION, PROTEIN TURNOVER, CHAPERONES | ||
| LCAZH_0279 | ADP-ribosylglycohydrolase | 2.51 |
| LCAZH_1344 | Chaperone ClpB | 3.68 |
| LCAZH_1380 | Peptide methionine sulfoxide reductase | 2.23 |
| LCAZH_1398 | Pyruvate-formate lyase-activating enzyme | 2.88 |
| DEFENSE MECHANISMS | ||
| LCAZH_1217 | Multidrug ABC transporter ATPase | 6.01 |
| GENERAL FUNCTION PREDICTION ONLY | ||
| LCAZH_0294 | Alpha/beta hydrolase | 2.64 |
| LCAZH_0305 | NAD(FAD)-dependent dehydrogenase | 2.14 |
| LCAZH_0638 | ABC transporter periplasmic protein | 2.06 |
| LCAZH_0641 | ABC transporter permease | 2.3 |
| LCAZH_0642 | ABC transporter ATPase | 2.15 |
| LCAZH_1865 | Dinucleotide-binding enzyme | 2.05 |
| LCAZH_2372 | Oxidoreductase | 2.53 |
| LCAZH_2373 | Short-chain alcohol dehydrogenase | 3.71 |
| FUNCTION UNKNOWN | ||
| LCAZH_0227 | Alkaline shock protein | 21.93 |
| LCAZH_2030 | Hypothetical protein | 3.57 |
| LCAZH_2301 | Putative integral membrane protein | 3.32 |
| LCAZH_2056 | Hypothetical protein | 2.3 |
| LCAZH_2222 | Hypothetical protein | 3.29 |
| LCAZH_1464 | Hypothetical protein | 3.26 |
| LCAZH_1898 | Hypothetical protein | 2.42 |
| LCAZH_0186 | Hypothetical protein | 2.34 |
Figure 2.
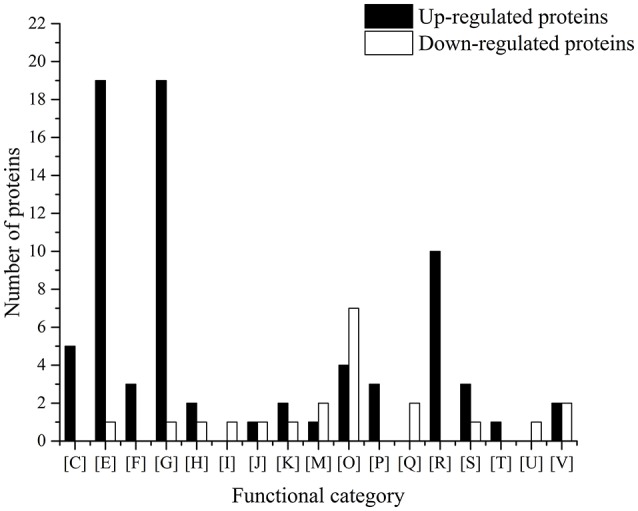
Proteomic profiles of differentially expressed proteins of the parental L. casei Zhang G-0 and gentamycin-resistant L. casei Zhang-G-1200 strains. Clusters of orthologous groups (COG) functional categories: [C], Energy production and conversion; [E], Amino acid transport and metabolism; [F] Nucleotide transport and metabolism; [G], Carbohydrate transport and metabolism; [H], Coenzyme transport and metabolism; [I], Lipid transport and metabolism; [J], Translation, ribosomal structure and biogenesis; [K], Transcription; [L], Replication, recombination and repair; [M], Cell wall/membrane/envelope biogenesis; [O], Posttranslational modification, protein turnover, chaperones; [P], Inorganic ion transport and metabolism; [Q], Secondary metabolites biosynthesis, transport and catabolism; [R], General function prediction only; [S], Function unknown; [T], Signal transduction mechanisms; [V], Defense mechanisms.
Over half (6 out of 10) of the up-regulated proteins were associated with the COGs category of carbohydrate transport and metabolism [G], which were the phosphotransferase system (PTS)-related components located within 3 different operons. According to the Transporter Classification Database (TCDB), 4 of these differentially regulated proteins belonged to the PTS Galactitol family, including the galacitol transporter subunits EIIA (LCAZH_2649), EIIB (LCAZH_2648), IIB (LCAZH_0604), and EIIC (LCAZH_0605), while the other 2 belonged to the PTS Lactose-N, N'-Diacetylchitobiose-β-glucoside family, namely the cellobiose-specific transporter subunits IIA (LCAZH_1771) and IIB (LCAZH_1772).
Another group of significantly up-regulated proteins was responsible for the uptake of oligopeptides (LCAZH_2023-LCAZH_2026). This genomic cluster coded for the ATPase and 2 permease components of a dipeptide/oligopeptide/nickel ABC transporter, as well as an oligopeptide ABC transporter periplasmic protein. Similar to the Opp system identified in other LAB genomes (Chen et al., 2015), another gene that coded for the ATP-binding subunit of the oligopeptide ABC transporter was found immediately downstream of the oligopeptide ABC transporter periplasmic protein of the genome of L. casei Zhang (Zhang et al., 2010). Besides, several other oligopeptide (LCAZH_0201, LCAZH_1596, LCAZH_1886, and LCAZH_2851) and amino acid (LCAZH_0500 and LCAZH_0418) transporter components, as well as subunits for de novo syntheses of tryptophan (LCAZH_0084) and glutamate (LCAZH_2518 and LCAZH_2519), were identified.
Other up-regulated proteins included those relating to purine biosynthesis (LCAZH_1739, LCAZH_1740, and LCAZH_1743) and stress response (LCAZH_1344 and LCAZH_0227). Notably, the alkaline shock protein (LCAZH_0227) was the most significantly up-regulated protein with a 21.9-fold increase in its expression compared with the parental line, suggesting its important role in gentamycin adaptation.
Down-regulated L. casei Zhang-G-1200 proteins grown in gentamycin-containing medium
A total of 36 proteins were significantly down-regulated (<−2.0-folds, P < 0.05) in L. casei Zhang-G-1200 compared with the parental strain (Table 3). According to the COGs functional classification, 33.3% of these proteins were involved in posttranslational modification, protein turnover, chaperones metabolism [G] (Figure 2). Indeed, most of them were stress-responsive proteins, including the protease subunit of ATP-dependent Clp protease (LCAZH_0907), the molecular chaperones GrpE, GroEL, and HSP20-2 (LCAZH_1553, LCAZH_2207, and LCAZH_2811), the co-chaperonin GroES (LCAZH_2208), and the Clp protease/DnaK/DnaJ chaperone ATP-binding subunit (LCAZH_1753), while the others were mainly hypothetical proteins of unknown functions.
Table 3.
Down-regulated proteins of L. casei Zhang-G-1200 compared with its original strain in the presence of the gentamycin.
| Locus | Function | Fold change |
|---|---|---|
| CARBOHYDRATE TRANSPORT AND METABOLISM | ||
| LCAZH_2698 | Fructose/tagatose bisphosphate aldolase | −3.3 |
| AMINO ACID TRANSPORT AND METABOLISM | ||
| LCAZH_1424 | Histidinol-phosphate/aromatic aminotransferase and cobyric acid decarboxylase | −2.51 |
| CELL WALL/MEMBRANE/ENVELOPE BIOGENESIS | ||
| LCAZH_0738 | D-alanyl transfer protein | −2.1 |
| LCAZH_2870 | Glycosyltransferase | −2.22 |
| TRANSCRIPTION | ||
| LCAZH_1554 | Transcriptional regulator | −2.06 |
| POSTTRANSLATIONAL MODIFICATION, PROTEIN TURNOVER, CHAPERONES | ||
| LCAZH_0497 | Membrane associated subtilisin-like serine protease | −2.77 |
| LCAZH_0907 | Protease subunit of ATP-dependent Clp protease | −2.04 |
| LCAZH_1553 | Molecular chaperone GrpE | −2.09 |
| LCAZH_1753 | Clp protease/DnaK/DnaJ chaperone ATP-binding subunit | −3.28 |
| LCAZH_2207 | Molecular chaperone GroEL | −2.14 |
| LCAZH_2208 | Co-chaperonin GroES (HSP10) | −2.77 |
| LCAZH_2811 | Molecular chaperone | −5.1 |
| COENZYME TRANSPORT AND METABOLISM | ||
| LCAZH_0348 | Thiamine monophosphate synthase | −2.21 |
| LIPID TRANSPORT AND METABOLISM | ||
| LCAZH_0739 | D-alanyl carrier protein | −3.1 |
| REPLICATION | ||
| LCAZH_0852 | 50S ribosomal protein L14 | −2.21 |
| SECONDARY METABOLITES BIOSYNTHESIS, TRANSPORT AND CATABOLISM | ||
| LCAZH_2835 | Amidase | −2.23 |
| DEFENSE MECHANISMS | ||
| LCAZH_1927 | Antimicrobial peptide ABC transporter permease | −2.34 |
| LCAZH_1928 | Antimicrobial peptide ABC transporter ATPase | −2.76 |
| LCAZH_1179 | XRE family transcriptional regulator | −4.68 |
| FUNCTION UNKNOWN | ||
| LCAZH_1052 | Hypothetical protein | −2.04 |
| LCAZH_1126 | Hypothetical protein | −2.01 |
| LCAZH_1754 | Hypothetical protein | −2.03 |
| LCAZH_0994 | Hypothetical protein | −2.06 |
| LCAZH_2341 | Prebacteriocin | −2.07 |
| LCAZH_0543 | Hypothetical protein | −2.1 |
| LCAZH_0824 | Hypothetical protein | −2.22 |
| LCAZH_1530 | Hypothetical protein | −2.23 |
| LCAZH_2528 | Hypothetical protein | −2.23 |
| LCAZH_1498 | Hypothetical protein | −2.24 |
| LCAZH_2472 | Hypothetical protein | −2.6 |
| LCAZH_1584 | Hypothetical protein | −2.85 |
| LCAZH_0616 | Hypothetical protein | −2.97 |
| LCAZH_0580 | Hypothetical protein | −3.06 |
| LCAZH_0521 | Hypothetical protein | −3.97 |
| LCAZH_2689 | Hypothetical protein | −4.46 |
| LCAZH_0113 | Hypothetical protein | −4.71 |
Validation of expression level of an alkaline shock protein by PRM
The expression of the alkaline shock protein (LCAZH_0227) was confirmed by PRM analysis. Fortunately, 1 unique peptide (FDDAVIAK) corresponding to this protein was found, which enabled the downstream quantification. As revealed by the analysis, the quantity of this alkaline shock protein was significantly higher in L. casei Zhang-G-1200 than the parental cells (2.938 ± 0.144 fmol/μg vs. 0.789 ± 0.057 fmol/μg). This result was consistent with the findings of the proteomic analysis.
Partial reversion of gentamycin-resistant phenotype of asp23 disruption mutant
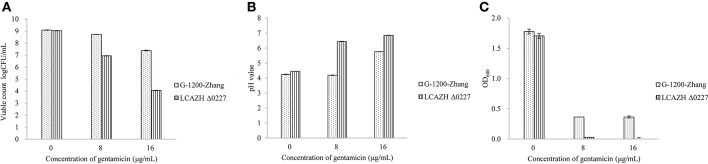
The asp23 gene (LCAZH_0227) was selected as a target for mutational analysis because it was the most up-regulated protein in L. casei Zhang-G-1200 when the cells were grown in the presence of gentamycin. As shown in Figure 3, asp23 inactivation did not affect the growth of L. casei Zhang-G-1200 in LSM without antibiotics. However, the L. casei Zhang-G-1200 asp23 disruption mutant was significantly more sensitive to gentamycin with a relatively low MIC value of 8 μg/mL though still higher than that of the parental strain (vs. 2 μg/mL and 32 μg/mL for the parental strain and L. casei Zhang-G-1200, respectively). When the concentration of gentamycin increased to 16 μg/mL, the wild type L. casei Zhang-G-1200 cells survived significantly better than the asp23 disruption mutant.
Figure 3.
The viable counts (A), pH values (B) and OD values (C) of the wild-type and mutant strains at the time of observing the minimum inhibitory concentration (MIC) by witness.
Discussion
Probiotics that are used in food industry have low exposure to antibiotics normally. However, owing to their intrinsic antimicrobial property and desirable health-promoting effects, some scientists and clinicians have suggested using them to manage gastrointestinal disorders in clinical cases, when they may be directly exposed to antibiotics, sometimes even at a high concentration. Thus, it would be crucial to understand the evolutionary adaptation mechanisms of probiotics bacteria toward antibiotics as part of the risk assessment. In a previous long-term antibiotic-driven evolution experiment, we isolated the L. casei Zhang-G-1200 strain that exhibited elevated resistance to gentamycin compared with the parental line. The present study further characterized the mechanism of gentamycin resistance of this isolate using the iTRAQ-based proteomic approach.
Lactobacillus casei is a highly adaptable bacterium that can live on a wide range of niches; and it has a great capacity for choosing specific nutritional elements that enable its growth within any complex environments (Wang et al., 2012b). Like many other bacteria, it has developed sophisticated cellular mechanisms to regulate its nutritional responses. Carbon is one of the most important macronutrients for the bacterial cells; thus, its carbon metabolism and regulation have been studied in detail (Titgemeyer and Hillen, 2002). The strong ability L. casei in carbohydrate utilization relies very much on a rich array of PTSs present in its genome, ranking highest among all members within the Lactobacillus genus (Zhang et al., 2010). The current proteomic analysis revealed an apparent up-regulated expression of some PTS-related components in the gentamycin-resistant L. casei Zhang-G-1200 strain. The main function of PTSs is catalyzing sugar transport and phosphorylation (Zhang et al., 2013). The enhanced expression in L. casei Zhang-G-1200 may suggest an elevated cellular demand for utilizing different carbon substrates due to the presence of gentamycin.
Amino acid regulation is another important aspect required for sustaining growth of L. casei, especially for the late growth stages (Wang et al., 2012a). L. casei Zhang is able to synthesize most but the branch-chained amino acids; it is thus necessary for the bacterial cells to acquire the missing amino acids from the direct growth environment by proteolysis. L. casei Zhang possesses a well-developed proteolytic system (Wang et al., 2012b). In the gentamycin-containing environment, some of the key proteins involving in tryptophan and glutamate synthesis were found to be up-regulated, e.g., the tryptophan synthase alpha subunit and the glutamate synthase subunits. The tryptophan synthase alpha subunit functions to convert indole-3-glycerolphosphate into indole, the terminal step of tryptophan biosynthesis (Lim et al., 1991), while the glutamate synthase (large subunit) and NADPH-dependent glutamate synthase (small subunit) together catalyze the transamidation of the amide group from glutamine to 2-oxoglutarate to form glutamate (Stannek et al., 2015). Moreover, there were several up-regulated amino acid transporters and transporter-associated components, which might facilitate the intake of dipeptide/oligopeptides from the growth medium. The simultaneous up-regulation of multiple transporters for dipeptide/oligopeptides may suggest a need for L. casei Zhang-G-1200 to assimilate amino acids more efficiently under gentamycin stress.
Purine nucleotides are substrates for RNA and DNA synthesis; and they are essential for the growth of some LAB species (Wang et al., 2012a). Here, we also observed the up-regulation of some purine biosynthesis-related proteins. Among these proteins, the folate-dependent phosphoribosylglycinamide formyltransferase catalyzes the steps whereby the formyl derivatives of tetrahydrofolic acid are donated to the precursors of inosinic acid during its biosynthesis (Hartman and Buchanan, 1959); and the phosphoribosylformylglycinamidine synthase converts glycinamide ribotide into glycinamidine ribotide (Melnick and Buchanan, 1957). As in some other bacteria, the genome of L. casei Zhang contains a typical gene cluster for de novo purine biosynthesis, namely PurCDFHKLM, consisting of 12 distinct genes. The bacterial purine nucleotide synthesis is a 10-step pathway that produces inosinic acid from 5-phosphoribosyl 1-pyrophosphate (Ebbole and Zalkin, 1987). Since the culture medium used in the present study lacked purine compounds, the purine nucleotides must be obtained from the bacterial de novo biosynthesis, which was reflected by the up-regulation of these proteins.
Another group of differentially expressed proteins was the stress-related proteins. The molecular chaperones, such as GroEL and GroES, play a central role in the control of general stress responses (Lemos and Burne, 2002). The GroEL and several other chaperones of L. casei have been shown to be up-regulated in response to acid and bile stress (Wu et al., 2009b, 2010). Here, we observed a general reduction in expression of these proteins in the L. casei Zhang-G-1200 strain compared with its parental line under gentamycin stress except for the Clp protein and an alkaline shock protein (encoded by LCAZH_0227, the asp2 gene). These simply indicate that Zhang-G-1200 coped better in the antibiotics-containing environment due to its long-term adaptation to the drug. This was also reflected by a better growth performance of the adapted strain than its parental line in the presence of gentamycin.
It is interesting to note the high expression of asp23 in L. casei Zhang-G-1200 (21.9-fold increase compared with the parental strain). The activation of this gene is normally associated with alkaline shock conditions (Kuroda et al., 1995). To investigate its role in the gentamycin-resistant phenotype in the adapted strain, we created an asp23 disruption mutant. The gene disruption mutant appeared to be significantly more sensitive to gentamycin compared with the wild type L. casei Zhang-G-1200, though it was still more resistant than the parental L. casei Zhang, suggesting that this alkaline shock protein was partially responsible for the resistant phenotype. The bactericidal action of gentamycin resembles that of a ribosome modulation factor via the irreversible binding of the bacterial 30S ribosomal subunit and hence interrupting protein synthesis. Some stress-responsive proteins are also known to serve as ribosome modulation factors (Niven and EI-Sharoud, 2008). For example, the E. coli RbfA is a cold shock protein that regulates cold shock response by binding to the 30S ribosomal binding factor; its absence promotes cold shock responses (Jones and Inouye, 1996). Although the exact cellular role of asp23 remains to be determined, it is tempting to speculate that it acts as a modulator of similar nature and competes with gentamycin. However, the facts that the gentamycin-resistant phenotype of Zhang-G-1200 was developed stepwise along the long-term cultivation and only partial reversion of gentamycin-resistant phenotype was observed in the asp23 disruption mutant together suggest that the resistant phenotype was not solely caused by the asp23 gene but possibly along with certain secondary pleiotrophic mutations accumulated in the bacterial genome. To our knowledge, this work reports for the first time the involvement of alkaline shock proteins in gentamycin resistance.
Conclusion
Several clinical trials supported the use of probiotics in the management of acute gastroenteritis and antibiotic-associated diarrhea, which elevates the chance of probiotics exposure to antibiotics. The present study characterized the mechanism of adaptive gentamycin resistance in a probiotic L. casei strain. We observed up-regulation in some carbohydrate, amino acid, and purine metabolic pathways in the resistant strain, which may function to support the bacterial growth under gentamycin stress. Meanwhile, some stress-related proteins were differentially expressed, particularly an alkaline shock protein encoded by the asp23 gene. The disruption of asp23 gene partially reversed the gentamycin-resistant phenotype, suggesting that this gene was involved in the resistance development although other accumulated mutations might play a role too. Normally, comparative proteomics should be performed in conditions, where the growth rates, cell densities and other culture parameters are as identical as possible between the strains that are compared. As the two strains studied here showed different growth performance in the presence of gentamycin, some changes in the proteome could result from the different physiological status between cultures and not directly from a specific differential response to gentamycin.
Author contributions
WZ, ZS, and HZ designed the study. WZ, L-YK, and ZS wrote the manuscript. ZS, HG, CC, LL, and L-YK performed experiments. WZ and ZS analyzed data. All authors reviewed the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (Grant Nos. 31571815 and 31622043) and the Program for Young Talents of Science and Technology in Universities of Inner Mongolia Autonomous Region (NJYT-17-B05). We would like to thank Prof. Jian Kong for providing the plasmid pMSPcre.
References
- Arias C. A., Murray B. E. (2015). A new antibiotic and the evolution of resistance. New Engl. J. Med. 372, 1168–1170. 10.1056/NEJMcibr1500292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Q., Song Y., Xu H., Yu J., Zhang W., Menghe B., et al. (2016). Multilocus sequence typing of Lactobacillus casei isolates from naturally fermented foods in China and Mongolia. J. Dairy Sci. 99, 5202–5213. 10.3168/jds.2016-10857 [DOI] [PubMed] [Google Scholar]
- Boyanova L., Mitov I. (2012). Coadministration of probiotics with antibiotics: why, when and for how long? Expert Rev. Anti Infect. Ther. 10, 407–409. 10.1586/eri.12.24 [DOI] [PubMed] [Google Scholar]
- Cai H., Rodriguez B. T., Zhang W., Broadbent J. R., Steele J. L. (2007). Genotypic and phenotypic characterization of Lactobacillus casei strains isolated from different ecological niches suggests frequent recombination and niche specificity. Microbiology 153(Pt 8), 2655–2665. 10.1099/mic.0.2007/006452-0 [DOI] [PubMed] [Google Scholar]
- Chen Y., Zhang W., Sun Z., Meng B., Zhang H. (2015). Complete genome sequence of Lactobacillus helveticus H9, a probiotic strain originated from kurut. J. Biotechnol. 194, 37–38. 10.1016/j.jbiotec.2014.11.038 [DOI] [PubMed] [Google Scholar]
- Dantas A. B., Jesus V. F., Silva R., Almada C. N., Esmerino E. A., Cappato L. P., et al. (2016). Manufacture of probiotic minas frescal cheese with Lactobacillus casei Zhang. J. Dairy Sci. 99, 18–30. 10.3168/jds.2015-9880 [DOI] [PubMed] [Google Scholar]
- Ebbole D. J., Zalkin H. (1987). Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis. J. Biol. Chem. 262, 8274–8287. [PubMed] [Google Scholar]
- Galvez A., Abriouel H., Lopez R. L., Ben Omar N. (2007). Bacteriocin-based strategies for food biopreservation. Int. J. Food Microbiol. 120, 51–70. 10.1016/j.ijfoodmicro.2007.06.001 [DOI] [PubMed] [Google Scholar]
- Guo H., Pan L., Li L., Lu J., Kwok L., Menghe B., et al. (2017). Characterization of antibiotic resistance genes from Lactobacillus isolated from traditional dairy products. J. Food Sci. 82, 724–730. 10.1111/1750-3841.13645 [DOI] [PubMed] [Google Scholar]
- Hartman S. C., Buchanan J. M. (1959). Biosynthesis of the purines. XXVI. The identification of the formyl donors of the transformylation reactions. J. Biol. Chem. 234, 1812–1816. [PubMed] [Google Scholar]
- Jones P. G., Inouye M. (1996). RbfA, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol. Microbiol. 21, 1207–1218. 10.1111/j.1365-2958.1996.tb02582.x [DOI] [PubMed] [Google Scholar]
- Katz J. (2010). Should probiotics be routine therapy for the prevention of antibiotic-associated diarrhea? J. Clin. Gastroenterol. 44, 83–84. 10.1097/MCG.0b013e3181bdf010 [DOI] [PubMed] [Google Scholar]
- Kuroda M., Ohta T., Hayashi H. (1995). Isolation and the gene cloning of an alkaline shock protein in methicillin resistant Staphylococcus aureus. Biochem. Biophys. Res. Commun. 207, 978–984. 10.1006/bbrc.1995.1281 [DOI] [PubMed] [Google Scholar]
- Lambert J. M., Bongers R. S., Kleerebezem M. (2007). Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl. Environ. Microbiol. 73, 1126–1135. 10.1128/AEM.01473-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara F. J., Garcia-Campana A. M., Ales-Barrero F., Bosque-Sendra J. M., Garcia-Ayuso L. E. (2006). Multiresidue method for the determination of quinolone antibiotics in bovine raw milk by capillary electrophoresis-tandem mass spectrometry. Anal. Chem. 78, 7665–7673. 10.1021/ac061006v [DOI] [PubMed] [Google Scholar]
- Lemos J. A., Burne R. A. (2002). Regulation and physiological significance of ClpC and ClpP in Streptococcus mutans. J. Bacteriol. 184, 6357–6366. 10.1128/JB.184.22.6357-6366.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim W. K., Shin H. J., Milton D. L., Hardman J. K. (1991). Relative activities and stabilities of mutant Escherichia coli tryptophan synthase alpha subunits. J. Bacteriol. 173, 1886–1893. 10.1128/jb.173.6.1886-1893.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W. W., Wang T., Wang Y., Xin M., Kong J. (2016). A food-grade fimbrial adhesin FaeG expression system in Lactococcus lactis and Lactobacillus casei. Can. J. Microbiol. 62, 241–248. 10.1139/cjm-2015-0596 [DOI] [PubMed] [Google Scholar]
- Melnick I., Buchanan J. M. (1957). Biosynthesis of the purines. XIV. Conversion of (alpha-N-formyl) glycinamide ribotide to (alpha-N-formyl) glycinamidine ribotide; purification and requirements of the enzyme system. J. Biol. Chem. 225, 157–162. [PubMed] [Google Scholar]
- Niven G. W., EI-Sharoud W. M. (2008). Ribosome modulation factor, in Bacterial Physiology, ed EI-Sharoud W. M. (Berlin; Heidelberg: Springer; ), 293–311. [Google Scholar]
- Normark B. H., Normark S. (2002). Evolution and spread of antibiotic resistance. J. Intern. Med. 252, 91–106. 10.1046/j.1365-2796.2002.01026.x [DOI] [PubMed] [Google Scholar]
- Reid G. (2017). Probiotic use in an infectious disease setting. Expert Rev. Anti Infect. Ther. 15, 449–455. 10.1080/14787210.2017.1300061 [DOI] [PubMed] [Google Scholar]
- Stannek L., Thiele M. J., Ischebeck T., Gunka K., Hammer E., Volker U., et al. (2015). Evidence for synergistic control of glutamate biosynthesis by glutamate dehydrogenases and glutamate in Bacillus subtilis. Environ. Microbiol. 17, 3379–3390. 10.1111/1462-2920.12813 [DOI] [PubMed] [Google Scholar]
- Titgemeyer F., Hillen W. (2002). Global control of sugar metabolism: a gram-positive solution. Antonie Van Leeuwenhoek 82, 59–71. 10.1023/A:1020628909429 [DOI] [PubMed] [Google Scholar]
- Wang J. C., Zhang W. Y., Zhong Z., Wei A. B., Bao Q. H., Zhang Y., et al. (2012a). Gene expression profile of probiotic Lactobacillus casei Zhang during the late stage of milk fermentation. Food Control 25, 321–327. 10.1016/j.foodcont.2011.10.036 [DOI] [Google Scholar]
- Wang J. C., Zhang W. Y., Zhong Z., Wei A. B., Bao Q. H., Zhang Y., et al. (2012b). Transcriptome analysis of probiotic Lactobacillus casei Zhang during fermentation in soymilk. J. Ind. Microbiol. Biotechnol. 39, 191–206. 10.1007/s10295-011-1015-7 [DOI] [PubMed] [Google Scholar]
- Wang J., Dong X., Shao Y., Guo H., Pan L., Hui W., et al. (2017). Genome adaptive evolution of Lactobacillus casei under long-term antibiotic selection pressures. BMC Genomics 18:320. 10.1186/s12864-017-3710-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xie J., Li Y., Dong S., Liu H., Chen J., et al. (2016). Probiotic Lactobacillus casei Zhang reduces pro-inflammatory cytokine production and hepatic inflammation in a rat model of acute liver failure. Eur. J. Nutr. 55, 821–831. 10.1007/s00394-015-0904-3 [DOI] [PubMed] [Google Scholar]
- Wu R., Sun Z., Wu J., Meng H., Zhang H. (2010). Effect of bile salts stress on protein synthesis of Lactobacillus casei Zhang revealed by 2-dimensional gel electrophoresis. J. Dairy Sci. 93, 3858–3868. 10.3168/jds.2009-2967 [DOI] [PubMed] [Google Scholar]
- Wu R., Wang L., Wang J., Li H., Menghe B., Wu J., et al. (2009a). Isolation and preliminary probiotic selection of lactobacilli from koumiss in Inner Mongolia. J. Basic Microbiol. 49, 318–326. 10.1002/jobm.200800047 [DOI] [PubMed] [Google Scholar]
- Wu R., Wang W., Yu D., Zhang W., Li Y., Sun Z., et al. (2009b). Proteomics analysis of Lactobacillus casei Zhang, a new probiotic bacterium isolated from traditional home-made koumiss in Inner Mongolia of China. Mol. Cell Proteomics 8, 2321–2338. 10.1074/mcp.M800483-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Kong J. (2013). Construction and potential application of controlled autolytic systems for Lactobacillus casei in cheese manufacture. J. Food Prot. 76, 1187–1193. 10.4315/0362-028X.JFP-12-307 [DOI] [PubMed] [Google Scholar]
- Ya T., Zhang Q., Chu F., Merritt J., Bilige M., Sun T., et al. (2008). Immunological evaluation of Lactobacillus casei Zhang: a newly isolated strain from koumiss in Inner Mongolia, China. BMC Immunol. 9:68. 10.1186/1471-2172-9-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wang L., Guo Z., Sun Z., Gesudu Q., Kwok L., et al. (2014a). 454 pyrosequencing reveals changes in the faecal microbiota of adults consuming Lactobacillus casei Zhang. FEMS Microbiol. Ecol. 88, 612–622. 10.1111/1574-6941.12328 [DOI] [PubMed] [Google Scholar]
- Zhang W., Sun Z., Menghe B., Zhang H. (2015). Short communication: single molecule, real-time sequencing technology revealed species- and strain-specific methylation patterns of 2 Lactobacillus strains. J. Dairy Sci. 98, 3020–3024. 10.3168/jds.2014-9272 [DOI] [PubMed] [Google Scholar]
- Zhang W. Y., Sun Z. H., Wu R. N., Meng H., Zhang H. P. (2013). Comparative genome analysis of a new probiotic strain Lactobacillus casei Zhang, Genomics II-Bacteria, Viruses and Metabolic Pathways (Brisbane: Iconcept Press; ), 276–296. [Google Scholar]
- Zhang W., Yu D., Sun Z., Wu R., Chen X., Chen W., et al. (2010). Complete genome sequence of Lactobacillus casei Zhang, a new probiotic strain isolated from traditional homemade koumiss in inner Mongolia, China. J. Bacteriol. 192, 5268–5269. 10.1128/JB.00802-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Guo X., Guo J., He Q., Li H., Song Y., et al. (2014b). Lactobacillus casei reduces susceptibility to type 2 diabetes via microbiota-mediated body chloride ion influx. Sci. Rep. 4:5654. 10.1038/srep05654 [DOI] [PMC free article] [PubMed] [Google Scholar]