Abstract
AIM
To identify chromosomal copy number aberrations (CNAs) in early-stage hepatocellular carcinoma (HCC) and analyze whether they are correlated with patient prognosis.
METHODS
One hundred and twenty patients with early-stage HCC were enrolled in our study, with the collection of formalin fixed, paraffin-embedded (FFPE) specimens and clinicopathological data. Tumor areas were marked by certified pathologists on a hematoxylin and eosin-stained slide, and cancer and adjacent non-cancerous tissues underwent extraction of DNA, which was analyzed with the Affymetrix OncoScan platform to assess CNAs and loss of heterozygosity (LOH). Ten individuals with nonmalignant disease were used as the control group. Another cohort consisting of 40 patients with stage I/II HCC were enrolled to analyze gene expression and to correlate findings with the OncoScan data.
RESULTS
Copy number amplifications occurred at chromosomes 1q21.1-q44 and 8q12.3-24.3 and deletions were found at 4q13.1-q35.2, 8p 23.2-21.1, 16q23.3-24.3, and 17p13.3-12, while LOH commonly occurred at 1p32.3, 3p21.31, 8p23.2-21.1, 16q22.1-24.3, and 17p 13.3-11 in early-stage HCC. Using Cox regression analysis, we also found that a higher percentage of genome change (≥ 60%) was an independent factor for worse prognosis in early-stage HCC (P = 0.031). Among the 875 genes in the OncoScan GeneChip, six were independent predictors of worse disease-free survival, of which three were amplified (MYC, ELAC2, and SYK) and three were deleted (GAK, MECOM, and WRN). Further, patients with HCC who exhibited ≥ 3 CNAs involving these six genes have worse outcomes compared to those who had < 3 CNAs (P < 0.001). Similarly, Asian patients with stage I HCC from The Cancer Genome Atlas harboring CNAs with these genes were also predicted to have poorer outcomes.
CONCLUSION
Patients with early-stage HCC and increased genome change or CNAs involving MYC, ELAC2, SYK, GAK, MECOM, or WRN are at risk for poorer outcome after resection.
Keywords: Early-stage hepatocellular carcinoma, Copy number aberration, Prognosis, OncoScan, Molecular inversion probe
Core tip: In this paper, we report that patients with early-stage hepatocellular carcinoma presenting a higher percentage of genome change or copy number aberrations affecting MYC, ELAC2, SYK, GAK, MECOM, or WRN are predicted to have worse outcomes, and they should be intensively followed after resection.
INTRODUCTION
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the second leading cause of cancer death worldwide[1,2]. Partial hepatectomy, ablation therapy, and liver transplantation are considered curative treatments for HCC; however, the high probability of recurrence frequently results in unsatisfactory outcomes, which has led to the increased importance of combined or multimodal treatments in recent years[3-6]. Among patients with HCC, most patients with early-stage (stage I/II ) cancer have a favorable outcome; nevertheless, a proportion of patients have poor prognosis after resection, which may arise from increased genomic instability[7]. Emergent efforts to resolve this dilemma include the integration of genomics, proteomics, metabolomics data, and clinical variants to predict outcomes for patients with early-stage HCC at both the research and clinical levels[8].
In particular, it appears that a differential gene expression profile in HCC arises from genetic instability or mutation[9,10]. Chromosome instability and copy number aberrations (CNAs) in HCC and other solid tumors could lead to the activation of oncogenes and the inactivation of tumor suppressor genes, which induce tumor invasiveness[11]. The common chromosome imbalances in HCC comprise of gains (amplification) at 1q, 8q, and 20q or losses (deletion) at 1p, 4q, 8p, 13q, 16q, and 17p across HCC specimens of different etiologies and cell lines using comparative genomic hybridization[12]. Some studies have also used formalin-fixed paraffin-embedded (FFPE) specimens for genome-wide copy number variation (CNV) analysis via high-density array, which disclosed common CNV regions, such as gains of 1q, 8q, 7q, 5p, 7p, Xq, 5q, and Xp and losses of 17p, 4q21.21-q26, 8p, 1p36.11-pter, and 9p[13,14]. In addition to these regions, chr12q13, 13q12, and 6p21-p24 may also contribute to the invasive phenotype of HCC[15,16]. However, little survival analysis has been performed in prior HCC FFPE studies owing to the limited number of cases or incomplete survival data[13,16].
Since FFPE specimens represent the most abundant bioresources in hospitals and the clinical outcome of some patients is already known, these factors allow scientists to integrate both complete clinical data and genomic information to reveal potential biomarkers either for cancer diagnosis or prognosis, especially for rare tumors or early-stage cancers. Therefore, an increasing number of studies have analyzed FFPE specimens using a global analysis of chromosome imbalance via the Affymetrix OncoScan FFPE Express 2.0 system with molecular inversion probes (MIPs) in ovary, breast, colon, and brain tumors[17-20].
In the current study, global chromosomal CNAs in early-stage (stage I/II) HCC FFPE samples were analyzed using the Affymetrix OncoScan platform to: (1) disclose genomic alterations; (2) determine their correlation with patient characteristics; (3) predict long-term outcomes with CNA percent change; and (4) identify the most significant altered genes.
MATERIALS AND METHODS
Patients
This study was approved by the Institutional Review Board of the Chang Gung Memorial Hospital (CGMH) in Linkou, Taiwan (#104-3511C). The inclusion criterion for participants was defined as having a resectable single HCC lesion (stage I or II as defined in the American Joint Committee on Cancer/International Union Against Cancer TNM system) and the exclusion criteria were the presence of distant metastasis or abnormal liver function tests[6]. One hundred and twenty patients with early-stage HCC were enrolled in this study, with the collection of FFPE specimens and clinicopathological data.
DNA extraction, FFPE sample gene chip analysis, and analysis of MIP data
FFPE samples were sliced into 10-μm sections and the tumor area was marked by certified pathologists on a hematoxylin and eosin-stained slide. Cancer and adjacent non-cancerous tissues underwent DNA extraction using the QIAamp DNA FFPE Tissue Kit (Qiagen, Sussex, United Kingdom) according to the manufacturer’s instructions. DNA concentration and purity were determined using the Qubit Fluorometer (Thermo Fisher Scientific UK Ltd., Paisley, United Kingdom). The extracted samples were further processed at the Genomic Medicine Core Laboratory at CGMH and analyzed with the Affymetrix OncoScan platform (Santa Clara, CA, United States) to assess CNAs and loss of heterozygosity (LOH)[21,22].
MIP data and the percentage of the genome that changed (percent genome change) were analyzed using the Nexus Copy Number software included in the Affymetrix OncoScan FFPE Express Service (Biodiscovery, El Segundo, CA, United States). The OncoScan GeneChip includes 875 gene targets representing tumor suppressor genes and oncogenes; each gene is represented by 20-40 probes depending on the length of the gene.
RNA extraction, microarray data processing, and Affymetrix GeneChip Human Genome U133 Plus 2.0 Array
Another cohort consisting of 40 patients with stage I/II HCC were enrolled to analyze gene expression and to correlate findings with the OncoScan data (IRB No.96-1371C, 99-1127B, 101-1186B, and 201600707B0). Total RNA was extracted with TRIzol as recommended by the manufacturer followed by RNA cleanup using the MinElute Kit (Thermo Fisher Scientific Inc.). RNA labeling, hybridization, washes, and processing were performed by the Genomic Medicine Core Laboratory of CGMH. To filter the lower variance genes, we used a standard deviation > 0.5 to filter 6522 probe sets from the original 22215. The differentially expressed genes between cancer and non-cancerous tissues were identified with paired t-tests, and P-values of gene expression were calculated as previously described[23].
TaqMan copy number assay
Validation of chromosome aberrations was performed with TaqMan copy number real-time polymerase chain reaction (PCR) using genomic DNA extracted from FFPE samples. Twenty nanograms of genomic DNA were mixed with 1 μL of target gene, RNASEP primer/probes (Thermo Fisher Scientific Inc.), and 1 × TaqPath ProAmp Master Mix (Thermo Fisher Scientific Inc.) to a final volume of 20 μL. Copy number PCR was performed in QuantStudio 3 (Thermo Fisher Scientific Inc.) and then further analyzed with CopyCaller v2.1 (Thermo Fisher Scientific Inc.) by normalization with RNASEP Ct values and calculating the log2 ratio of ΔCt between RNASEP and the target gene, which were further normalized to their normal counterpart tissues to show as 2-ΔΔCT (fold change).
Survival and long-term outcome analysis
Categorical data were analyzed using the chi-square test or Fisher’s exact test. Continuous variables were analyzed using Student’s t test. Survival rates in each group were determined by the Kaplan-Meier method and differences between groups were analyzed using log-rank test. Tumor recurrence was analyzed with area under the receiver operating characteristic curve (AUROC) comparisons using the percent genome change, α-fetoprotein (AFP), and tumor size for each patient. Long-term outcomes were determined using Cox regression analysis incorporating CNAs from the OncoScan data. All P-values calculated were two-tailed and significance was defined at the 95% level (P < 0.05). Statistical analyses were performed using SPSS statistical software version 17.0 (SPSS, Inc., Chicago, IL, United States).
RESULTS
Virtual karyotyping analysis and validation of OncoScan data via TaqMan copy number assay
Global genomic alterations in early-stage HCC FFPE were analyzed using the Affymetrix OncoScan platform. The most frequent CNAs identified by virtual karyotyping of chromosomes were amplifications of 1q21.1-q44 and 8q12.3-24.3 as well as deletions of 4q13.1-35.2, 8p23.2-21.1, 16q23.3-24.3, and 17p13.3-12. In addition, LOH was commonly identified at 1p32.3, 3p21.31, 8p23.2-21.1, 16q22.1-24.3, and 17p13.3-11 (Figure 1A). These CNAs were rarely found in patients with nonmalignant liver tumors or normal HCC counterpart tissue (Figure 1B). To confirm the OncoScan data, the same genomic DNA samples extracted from FFPE specimens were tested via TaqMan copy number assay using real-time PCR. Four target genes including MDM2 proto-oncogene (MDM2 at 12q15), KRAS proto-oncogene, GTPase (KRAS at 12p12.1), MYC at 8q24.21, and ribonucleotide reductase regulatory subunit M2 (RRM2 at 2q25.1) were chosen to validate the OncoScan CNV data. We found comparable results between the TaqMan copy number PCR results of these four genes and the corresponding OncoScan data, with robust Pearson correlation coefficients (r) of 0.684, 0.71, 0.67, and 0.77 for MDM2, KRAS, RRM2, and MYC, respectively (Figure 2).
Figure 1.
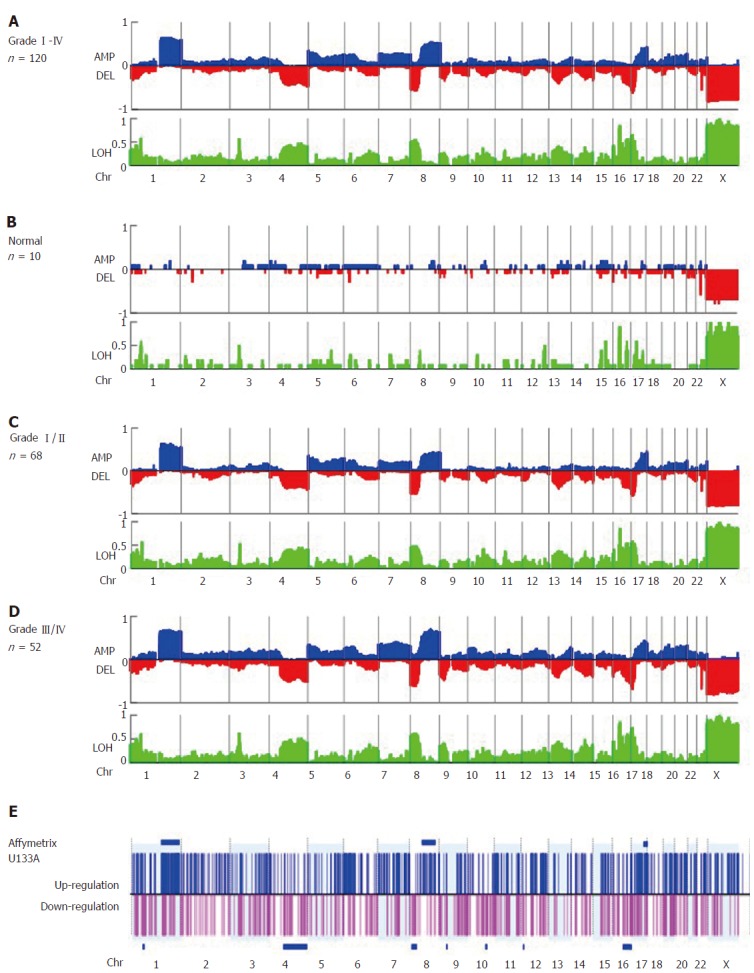
Virtual karyotyping analysis of amplification, deletion, and loss of heterozygosity in 120 formalin fixed, paraffin-embedded specimens of early-stage hepatocellular carcinoma. The frequency of copy number amplification (AMP), deletion (DEL), and loss of heterozygosity (LOH) in (A) specimens from all 120 patients with hepatocellular carcinoma (HCC), (B) patients with benign liver lesions (n = 10), (C) Edmonson grade I/II HCC specimens (n = 68), and (D) Edmonson grade III/IV HCC specimens (n = 52) are shown on the Y-axis of each panel. Relative chromosomal position is shown on the X-axis. Blue and red plots represent the frequency of copy number amplification and deletion, respectively (upper panel), and LOH (shown in green, lower panel). Note that +1, 0, and -1 represent a frequency of 100% amplification, no alteration, and 100% deletion, respectively. (E) The gene expression profiles of another set of early-stage HCC tumors (n = 40). The log ratio of each probe set was normalized by the normal counterpart of each HCC tumor [log(T-N)] and fold changes > 2 or ≤ 2 were considered up- (shown in blue) or down-regulated (shown in pink), respectively. The P-value for differential gene expression in tumor versus normal tissue was calculated and considered significant with P < 0.05. Clustered chromosome regions associated with genome change are shown as horizontal blue bars (P < 0.05).
Figure 2.
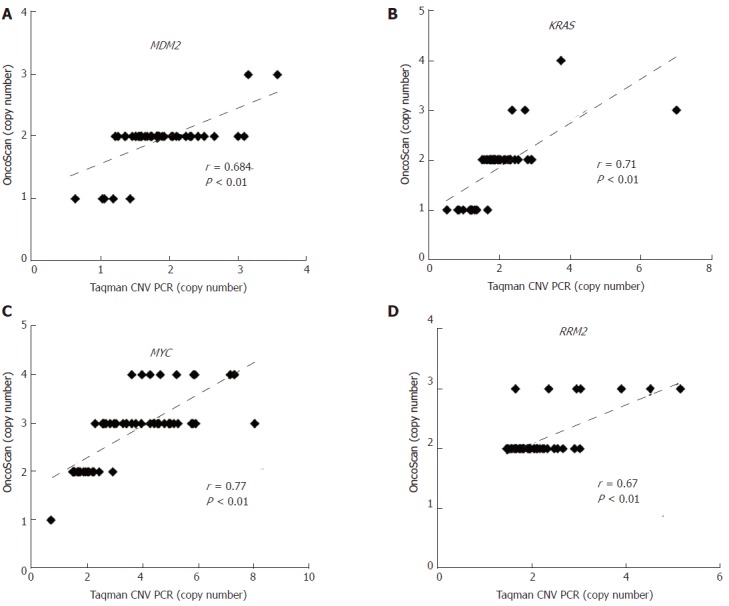
Validation of OncoScan copy number aberration data. The CNAs of (A) MDM2, (B) KRAS, (C) MYC, and (D) RRM2 in early-stage HCC were validated by TaqMan copy number assay. Correlations between OncoScan CNAs and TaqMan PCR-determined CNAs were calculated via Pearson’s correlation r and P-value. The X-axis shows the CNA results from TaqMan PCR and the Y-axis shows that of the OncoScan data. CNA: Copy number aberration.
Correlation of Affymetrix OncoScan results with gene expression microarray analysis
Next, gene expression of the global chromosome CNAs was investigated in the second cohort of 40 cases with stage I/II HCC, and six clustered regions were identified. Overexpressing genes in HCC tumors were found in 1q21.1-44, 8q12.3-24.3, and 17q22-25.3 with P-values of 7.72 × 10-25, 0.002, and 0.042, respectively. These regions were also found to be amplified based on OncoScan data (Figure 1E, blue bar). In addition, we found down-regulated genes in HCC tumors were clustered in 4q13.1-35.2, 8p23.2-21.1, and 16q23.3-24.3 with P-values of 0.002, 0.03, and 0.047, respectively, and these regions were deleted in tumors (Figure 1E, blue bar). These findings indicate that differential gene expression arises from CNVs in the HCC cancer genome.
Analysis of chromosome aberrations in early-stage HCC
Regarding the clinical characteristics of our patients with early-stage HCC, 60% were stage I and 40% were stage II ; 16.7% had grade I tumors, 40% had grade II tumors, and 43.3% had grade III/IV tumors. In our cohort, the mean tumor size was 4.2 cm, 7.5% presented with satellite lesions, 28.3% with vascular invasion, 0.8% with a microscopic margin, and all were Child-Pugh grade A (supplementary table 1). The mean disease-free survival (DFS) and overall survival were 43.8 ± 4.3 mo and 108.1 ± 10.5 mo, respectively. Tumor relapse occurred in 62.5% (75/120) of patients.
There were no correlations between CNAs and clinicopathological factors except for tumor grade. We found that amplification of chromosome 8q12.3-24.3 was associated with tumor differentiation and recurrence (%CNA = 39.99% and 62.01% in grade I/II vs III/IV , respectively, P = 0.006, Table 1, Figure 1C and D, Supplementary figure 1). Furthermore, we found from univariate analysis that factors conferring worse prognosis for early-stage HCC included older age (P = 0.004), larger tumor size (P = 0.001), higher tumor grading (P = 0.004), elevated AFP > 100 ng/mL (P = 0.006), elevated alkaline phosphatase level > 120 U/L (P = 0.016), and higher percent genome change ≥ 60% (P = 0.004), whereas Cox regression analysis showed that tumor size > 4.5 cm (P = 0.012), the presence of satellite lesions (P = 0.013), elevated serum alkaline phosphatase > 120 U/L (P = 0.042), and percent genome change (P = 0.031) were independent predictors (Table 2). Taken together, our findings show that HCC genomic alteration patterns are consistent with those of previous reports using fresh or FFPE specimens, and a higher percentage of genome change itself is a prognostic factor for early-stage HCC.
Table 1.
Summary of chromosome imbalances in 120 hepatocellular carcinoma specimens and normal paired tissues
| Chromosome region | Copy number | Cytoband | Grade I/II HCC | Grade III/IV HCC | P value |
| 1q | Gain | 1q21.1-44 | 57.86% | 60.63% | NS |
| 4q | Loss | 4q13.1- 35.2 | 39.56% | 47.51% | NS |
| 8p | Loss | 8p 23.2-21.1 | 48.56% | 56.28% | NS |
| 8q | Gain | 8q12.3-24.3 | 39.99% | 62.01% | 0.006a |
| 16q | Loss | 16q23.3-24.3 | 38.96% | 47.32% | NS |
| 17p | Loss | 17p13.3-12 | 52.22% | 53.73% | NS |
| 17q | Gain | 17q22-25.3 | 37.76% | 36.37% | NS |
P < 0.05, statistically significant (Grade I/II vs Grade III/IV). NS: Not significant; HCC: Hepatocellular carcinoma.
Table 2.
Disease-free survival analysis of clinicopathological data and percentage genome change of 120 patients with hepatocellular carcinoma
| Clinicopathological factor | Log-rank test, P value | Cox regression analysis, P value | HR (95%CI) |
| Age (yr), ≤ 49 (22.5%) vs > 49 (77.5%) | 0.004a | 0.169 | 0.674 (0.384-1.183) |
| Sex (M/F), M (81.7%) vs F (19.3%) | 0.146 | ||
| Tumor size (cm), ≤ 4.5 (69.2%) vs > 4.5 (30.8%) | 0.001a | 0.012a | 1.959 (1.160-3.309) |
| Satellite lesions (%), Yes (7.5%) vs No (92.5%) | 0.088 | 0.013a | 2.900 (1255-6.702) |
| Vascular invasion (%), Yes (28.3%) vs No (71.7%) | 0.240 | ||
| Grading I, II, III, IV (%), III & IV (43.3%) vs I & II (56.7%) | 0.004a | 0.063 | 1.626 (0.974-2.717) |
| Margin < 0.5 cm (%), ≤ 0.5 cm (47.5%) vs > 0.5 cm (52.5%) | 0.210 | ||
| Cirrhosis (%), Yes (54.2%) vs No (45.8%) | 0.258 | ||
| AFP (100 ng/mL), £100 (77.5%) vs > 100 (22.5%) | 0.006a | 0.462 | 1.244 (0.695-2.227) |
| Alkaline phosphatase (120 U/L), ≤ 120 (88.3%) vs > 120 (11.7%) | 0.016a | 0.042a | 1.976 (1.025-3.808) |
| Staging I/II, II (40.0%) vs I (60.0%) | 0.104 | ||
| Percentage genome changed (60%), ≥ 60% (7.5%) vs < 60% (92.5%) | 0.004a | 0.031a | 2.346 (1.080-5.097) |
P < 0.05, statistically significant. AFP: α-fetoprotein; HCC: Hepatocellular carcinoma; HR: Hazard ratio; CI: Confidence interval; M: Male; F: Female.
Area under the receiver operating characteristic curve (AUROC) analysis for percent genome change, AFP, and tumor size in early-stage HCC
We next compared percent genome change with current risk factors, such as AFP and tumor size, by performing AUROC analysis. We found that the AUROC of percent genome change was higher than that found with AFP and tumor size with values of 0.657, 0.598, and 0.633, respectively (Figure 3A). The sensitivity and specificity of percent genome change were 0.467 and 0.8 for HCC recurrence at a cutoff of 30% (Supplementary table 2). Additionally, patients with a higher percentage of genome change (≥ 60%) were associated with extremely poor outcomes as determined by Kaplan-Meier curve analysis (P = 0.004, Figure 3B).
Figure 3.
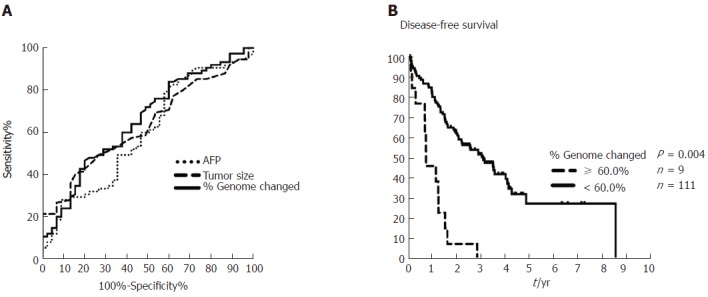
Comparisons of area under the receiver operating characteristic curve (AUROC) for percent genome change, α-fetoprotein, and tumor size in early-stage hepatocellular carcinoma, and Kaplan-Meier plot for 10-yr disease-free survival in patients with hepatocellular carcinoma with or without ≥ 60% genome change. A: AUROC analysis of AFP (dotted line, 0.598), tumor size (dashed line, 0.633), and percent genome change (solid line, 0.657) shows predictive ability for early-stage HCC recurrence. Cutoffs for AFP, tumor size, and percent genome change are 5.2 ng/mL, 4.25 cm, and 30%, respectively (Supplementary table 3). B: The Kaplan-Meier plot for 10-yr disease-free survival in patients with HCC with ≥ 60% (dashed line) or < 60% (solid line) genome change as determined from Affymetrix OncoScan CNA data. AFP: α-fetoprotein; HCC: Hepatocellular carcinoma.
Predictive genes with copy number alteration for early-stage HCC
Of the 875 oncogenes and tumor suppressor genes evaluated, we found that 83 amplified genes, 14 deleted genes, and 35 LOH genes were associated with recurrence in DFS analysis (Supplementary table 3). Cox regression analysis for DFS identified six independent genes: three of which were amplified, ELAC2 (P = 0.023), MYC (P = 0.025), and SYK (P = 0.001); and three deleted, GAK (P < 0.001), MECOM (P = 0.001), and WRN (P = 0.009) (Table 3). Patients with stage I/II HCC and any CNAs affecting these six genes have poor prognosis in Kaplan-Meier curve analysis (P = 0.0036, Figure 4A, dashed line). Furthermore, early-stage HCC with ≥ 3 CNAs affecting these six genes was associated with an extremely unfavorable outcome (P < 0.001, Figure 4B, dashed line). Subgroup analysis using tumor stage showed that CNAs of these six genes were associated with an unfavorable outcome in stage I HCC (n = 72, P = 0.0209) but not in stage II HCC (n = 48, P = 0.15) (Figure 4C and D, dashed line). Thus, genes within CNAs may predict poor outcome in early-stage HCC, especially patients with stage I cancer, using OncoScan GeneChips.
Table 3.
Cox regression analysis for disease-free survival in early-stage hepatocellular carcinoma
| Gene | Cytoband | CNA | HR | 95%CI | P value |
| ELAC2 | 17p12 | Amplification | 2.784 | 1.153-6.724 | 0.023a |
| MYC | 8q24.21 | Amplification | 1.772 | 1.074-2.924 | 0.025a |
| SYK | 9q22.2 | Amplification | 4.204 | 1.813-9.748 | 0.001a |
| GAK | 4p16.3 | Deletion | 2.916 | 1.635-5.199 | < 0.001a |
| MECOM | 3q26.2 | Deletion | 5.932 | 2.012-17.489 | 0.001a |
| WRN | 8p12 | Deletion | 1.9 | 1.172-3.080 | 0.009a |
P < 0.05, statistically significant. HCC: Hepatocellular carcinoma; CNA: Copy number aberration; HR: Hazard ratio; CI: Confidence interval.
Figure 4.

Disease-free survival analysis in patients with early-stage hepatocellular carcinoma with or without copy number aberrations involving six important genes identified using OncoScan data. A: Patients with stage I/II hepatocellular carcinoma (HCC) (n = 120) and copy number aberrations (CNAs) involving critical genes (dashed line) exhibited worse outcome compared to those without the predictive gene-containing CNAs (solid line) (P = 0.0036). B: Patients with early-stage HCC with three or more CNAs affecting these six identified genes had worse outcome (≥ 3 CNAs affecting these genes, dashed line, n = 14 vs < 3 CNAs affecting these genes, solid line, n = 106; P < 0.0001). C: Patients with stage I HCC (n = 72) and CNAs affecting any of the six identified genes (MYC, WRN, ELAC2, GAK, SYK, or MECOM) were associated with worse outcome after resection (dashed line, P = 0.0209). D: Patients with stage II HCC (n = 48) with/without any of the CNAs affecting these six genes (MYC, WRN, ELAC2, GAK, SYK, or ) showed no difference in DFS (P = 0.15). DFS: Disease-free survival.
Validation of CNAs as prognostic indicators for patients with early-stage HCC using The Cancer Genome Atlas Liver Hepatocellular Carcinoma database
CNAs involving these six putative prognostic genes for early-stage HCC were further analyzed using The Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC) database[24,25]. Of the 431 cases with HCC, we excluded those patients with liver dysfunction or lacking CNA or DFS follow-up data as well as patients with stage III/IV HCC, and selected 155 patients with early-stage HCC, of which 108 had stage I and 47 had stage II cancer. We found from this cohort those cases with a CNA that involved any of these six genes (MYC, WRN, ELAC2, MECOM, GAK, and SYK) have a trend towards worse prognosis (Figure 5C and D). Cases with stage I HCC and these CNAs showed a borderline difference in DFS (P = 0.0583, Figure 5A), while Asian cases with stage I (n = 73) and CNAs involving these genes have poorer outcome (P = 0.0203, Figure 5B). These findings from TCGA-LIHC patients were comparable to those found in our cohort.
Figure 5.
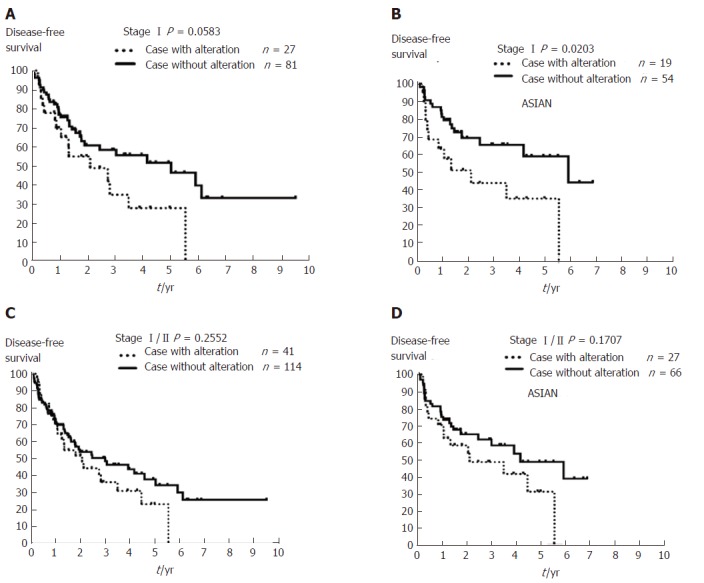
The Kaplan-Meier plot for 10-year disease-free survival in patients with stage I/II hepatocellular carcinoma with or without copy number aberrations involving MYC, WRN, ELAC2, GAK, SYK, or MECOM using The Cancer Genome Atlas Liver Hepatocellular Carcinoma data. A: Patients with stage I hepatocellular carcinoma (HCC) (n = 108) with any copy number aberrations (CNAs) affecting one of the six genes identified in this study (MYC, WRN, ELAC2, GAK, SYK, or MECOM) had a trend for worse outcome after resection (dashed line, P = 0.0583). B: Asian patients with stage I HCC (n = 73) and CNAs affecting these six genes (dashed line) exhibited poorer outcome compared to those cases without such CNAs (solid line, P = 0.0203). C: Patients with stage I/IIHCC (n = 155), and D: Asian patients with stage I/II HCC (n = 93) with/without any of these CNAs affecting MYC, WRN, ELAC2, GAK, SYK, or MECOM showed no change in DFS (P = 0.2552 and P= 0.1707, respectively). DFS: Disease-free survival.
DISCUSSION
Patients with early-stage HCC usually have a good survival outcome after curative treatment; however, the purpose of this study was to determine the underlying cause why some of those patients have a poor outcome[3,7]. In addition to driver gene mutations, and towards improvements in HCC treatment outcomes, we found that CNAs were also important in cancer genomics. In this study, we found that a higher percentage of genome change in CNAs identified via the Affymetrix OncoScan platform was of clinical significance, a finding which suggests that high-risk patients should be intensively followed after resection. The most important CNA region was gene amplification at chromosome 8q12.3-24.3, a region consistently reported in previous studies[13,26]. Within chromosome 8q12.3-24.3, MYC encodes a transcription factor with a basic helix-loop-helix leucine zipper domain that regulates various kinds of cellular processes. MYC has also been previously identified as a prognostic marker in HCC[27-30]. In our study, we found that MYC showed copy number amplification in 50.8% of patients with HCC and it was also an independent predictor of long-term survival for patients with early-stage HCC.
In contrast to MYC, the other five genes identified in our study have rarely been reported in either HCC carcinogenesis or prognosis[30]. WRN is associated with the autosomal recessive disorder Werner syndrome in which patients show premature aging and sarcomas because of chromosomal instability[31,32]. WRN’s functions are related to its DNA helicase and exonuclease activities, and therefore, it interacts with p53 in DNA replication, repair, and recombination pathways to influence genomic instability[33]. The functions of MECOM in HCC carcinogenesis are linked with hepatitis B X protein carcinogenesis and antagonized growth inhibition induced by transforming growth factor-β (TGF-β) signaling[34]. Another CNA-linked gene, ELAC2, encodes a protein that contributes to endoribonuclease activity for tRNA 3′ processing[35]. In addition, ELAC2 is associated with Smad2 and its nuclear partner, forkhead box H1 (also known as FAST-1), which suggests that ELAC2 may be involved in TGF-β/Smad signaling in prostate cancer cells[36]. SYK, a non-receptor tyrosine kinase, is widely expressed in hematopoietic cells[37]. Down-regulation of SYK has also been reported in epithelial malignancies implicated in tumor formation and progression[38,39]. The protein of another gene, GAK, regulates clathrin-mediated membrane trafficking and functions as a transcriptional repressor of the androgen receptor[40,41]. It has been reported that knockdown of GAK activates the spindle-assembly checkpoint to induce misaligned or abnormally condensed chromosomes, a finding which indicates that GAK has a role in the maintenance of chromosome stability[42]. These findings suggest that the roles of these five genes in HCC carcinogenesis may involve genome stability, although their specific functions require further exploration. It would be worthwhile in the future to evaluate the genetic signature of MYC, ELAC2, SYK, WRN, GAK, and MECOM with respect to the survival of patients with early-stage HCC using another large cohort.
This study demonstrated the value of using FFPE samples to investigate CNAs in patients. In fact, some patients in our cohort had samples taken over 14 years ago and there were no issues regarding their quality and quantity for use in our study. FFPE samples may be a vital resource for cancer genomic studies enabling the prediction of long-term outcomes in patients who had surgery years ago. Furthermore, the TCGA database has been used to validate findings for various cancer genomic studies[24,25]. Our finding of a trend towards survival outcome using the TCGA dataset conferred further validation of our finding that Asians have a worse outcome, especially regarding cases with stage I HCC. Although there was a different outcome based on the BRIDGE study, which may be because of ethical and/or regional differences in treatment choice, our use of the TCGA database consistently demonstrated that differences exist in the genomic background of patients with HCC[43].
In conclusion, our identification of percent genome change and six independently predictive genes from CNA data obtained using the Affymetrix OncoScan platform illustrates that chromosomal alterations are crucial regarding the outcome of patients with early-stage HCC after resection. Adoption of this platform could be useful in precision medicine for the prediction of early-stage HCC prognosis using FFPE specimens.
ARTICLE HIGHLIGHTS
Research background
Most patients with early-stage (stageI/II) hepatocellular carcinoma (HCC) have a favorable outcome; nevertheless, increased genomic instability possibly leads to postoperative recurrence.
Research motivation
Previous studies using frozen HCC tumor tissue with array comparative genomic hybridization were of limited clinical value because of the absence of patient survival data. Since formalin fixed, paraffin-embedded (FFPE) samples are the largest bio-resource with long-term patient survival data found in every hospital worldwide, the aim of this research was to determine whether FFPE specimens of early-stage HCC with long-term survival data may be used with OncoScan GeneChips towards prognostic analysis of patients.
Research objectives
The study enrolled 120 patients with early-stage HCC and ten nonmalignant liver tumors or normal HCC counterpart tissues to explore genome instability and copy number aberrations (CNAs) in early-stage HCC.
Research methods
Extracted DNA was processed at the Genomic Medicine Core Laboratory and analyzed with the Affymetrix OncoScan platform to assess CNAs and loss of heterozygosity (LOH). We reliably obtained global genome amplification/deletion and overall percentage genome change from all FFPE samples in our cohort.
Research results
CNA amplifications were clustered at chromosomes 1q21.1-q44 and 8q12.3-24.3 and deletions at 4q13.1-q35.2, 8p 23.2-21.1, 16q23.3-24.3, and 17p13.3-12 in patients with early-stage HCC. We found that percentage of genome change ≥ 60% was an independent factor for worse prognosis and MYC, ELAC2, and SYK (amplification) as well as GAK, MECOM, and WRN (deletion) were the most powerful predicting genes. Using Asian patients with stage I HCC from The Cancer Genome Atlas as an independent cohort, we found that patients harboring CNAs affecting these genes were also predicted to have poorer outcomes.
Research conclusions
The identification of percent genome change and six independently predictive genes from the Affymetrix OncoScan platform illustrates that chromosomal alterations are crucial for outcome of patients with early-stage HCC after resection, which may be further applied for clinical practice using OncoScan or a custom-designed chip covering these six genes regions.
Research perspectives
Genome instability was related to early-stage HCC clinical outcome and patients with CNAs affecting MYC, ELAC2, SYK, GAK, MECOM, or WRN are at risk for poorer outcome after resection. In the era of precision medicine, the identification of CNAs in these six genes could be further applied for clinical practice using a small custom-designed chip.
ACKNOWLEDGMENTS
We are grateful for the technological support of the Department of Cancer Center and Genomic Medicine Core Laboratory at the Linkou branch of the Chang Gung Memorial Hospital.
Footnotes
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Taiwan
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Institutional review board statement: This study was approved by the Medical Ethics Committee of the Chang Gung Memorial Hospital in Linkou, Taiwan (#104-3511C).
Conflict-of-interest statement: All authors declare no conflict of interest.
Data sharing statement: Clinical dataset available from Dr. Yu MC at mingchin2000@gmail.com.
Peer-review started: September 20, 2017
First decision: October 11, 2017
Article in press: November 7, 2017
P- Reviewer: Niu ZS, Zhu X S- Editor: Gong ZM L- Editor: Wang TQ E- Editor: Ma YJ
Contributor Information
Ming-Chin Yu, Department of Surgery, Chang Gung Memorial Hospital, Taoyuan 33305, Taiwan; Department of Surgery, Xiamen Chang Gung Hospital, Xiamen 361028, China.
Chao-Wei Lee, Department of Surgery, Chang Gung Memorial Hospital, Taoyuan 33305, Taiwan; Graduate Institute of Clinical Medical Sciences, Chang Gung University, Taoyuan 33302, Taiwan.
Yun-Shien Lee, Genomic Medicine Core Laboratory, Chang Gung Memorial Hospital, Taoyuan 33305, Taiwan; Department of Biotechnology, Ming-Chuan University, Taoyuan 33348, Taiwan.
Jang-Hau Lian, Genomic Medicine Core Laboratory, Chang Gung Memorial Hospital, Taoyuan 33305, Taiwan.
Chia-Lung Tsai, Genomic Medicine Core Laboratory, Chang Gung Memorial Hospital, Taoyuan 33305, Taiwan.
Yi-Ping Liu, Department of Surgery, Chang Gung Memorial Hospital, Taoyuan 33305, Taiwan.
Chun-Hsing Wu, Department of Surgery, Chang Gung Memorial Hospital, Taoyuan 33305, Taiwan.
Chi-Neu Tsai, Graduate Institute of Clinical Medical Sciences, Chang Gung University, Taoyuan 33302, Taiwan. pink7@mail.cgu.edu.tw; Department of Pediatrics, Chang Gung Memorial Hospital, Taoyuan 33305, Taiwan.
References
- 1.Sartorius K, Sartorius B, Aldous C, Govender PS, Madiba TE. Global and country underestimation of hepatocellular carcinoma (HCC) in 2012 and its implications. Cancer Epidemiol. 2015;39:284–290. doi: 10.1016/j.canep.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Cheng CH, Lee CF, Wu TH, Chan KM, Chou HS, Wu TJ, Yu MC, Chen TC, Lee WC, Chen MF. Evaluation of the new AJCC staging system for resectable hepatocellular carcinoma. World J Surg Oncol. 2011;9:114. doi: 10.1186/1477-7819-9-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi C, Choi GH, Kim TH, Tanaka M, Meng MB, Seong J. Multimodality Management for Barcelona Clinic Liver Cancer Stage C Hepatocellular Carcinoma. Liver Cancer. 2014;3:405–416. doi: 10.1159/000343861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fong ZV, Tanabe KK. The clinical management of hepatocellular carcinoma in the United States, Europe, and Asia: a comprehensive and evidence-based comparison and review. Cancer. 2014;120:2824–2838. doi: 10.1002/cncr.28730. [DOI] [PubMed] [Google Scholar]
- 6.Chan AC, Fan ST, Poon RT, Cheung TT, Chok KS, Chan SC, Lo CM. Evaluation of the seventh edition of the American Joint Committee on Cancer tumour-node-metastasis (TNM) staging system for patients undergoing curative resection of hepatocellular carcinoma: implications for the development of a refined staging system. HPB (Oxford) 2013;15:439–448. doi: 10.1111/j.1477-2574.2012.00617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah SA, Cleary SP, Wei AC, Yang I, Taylor BR, Hemming AW, Langer B, Grant DR, Greig PD, Gallinger S. Recurrence after liver resection for hepatocellular carcinoma: risk factors, treatment, and outcomes. Surgery. 2007;141:330–339. doi: 10.1016/j.surg.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 8.Satow R, Shitashige M, Kanai Y, Takeshita F, Ojima H, Jigami T, Honda K, Kosuge T, Ochiya T, Hirohashi S, et al. Combined functional genome survey of therapeutic targets for hepatocellular carcinoma. Clin Cancer Res. 2010;16:2518–2528. doi: 10.1158/1078-0432.CCR-09-2214. [DOI] [PubMed] [Google Scholar]
- 9.Chen T, Sun Y, Ji P, Kopetz S, Zhang W. Topoisomerase IIα in chromosome instability and personalized cancer therapy. Oncogene. 2015;34:4019–4031. doi: 10.1038/onc.2014.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Cho HJ, Kim SS, Wang HJ, Kim BW, Cho H, Jung J, Cho SS, Kim JK, Lee JH, Kim YB, et al. Detection of Novel Genomic Markers for Predicting Prognosis in Hepatocellular Carcinoma Patients by Integrative Analysis of Copy Number Aberrations and Gene Expression Profiles: Results from a Long-Term Follow-Up. DNA Cell Biol. 2016;35:71–80. doi: 10.1089/dna.2015.3026. [DOI] [PubMed] [Google Scholar]
- 12.Roessler S, Long EL, Budhu A, Chen Y, Zhao X, Ji J, Walker R, Jia HL, Ye QH, Qin LX, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142:957–966.e12. doi: 10.1053/j.gastro.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu YJ, Zhou Y, Yeh MM. Recurrent genetic alterations in hepatitis C-associated hepatocellular carcinoma detected by genomic microarray: a genetic, clinical and pathological correlation study. Mol Cytogenet. 2014;7:81. doi: 10.1186/s13039-014-0081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niu ZS, Niu XJ, Wang WH. Genetic alterations in hepatocellular carcinoma: An update. World J Gastroenterol. 2016;22:9069–9095. doi: 10.3748/wjg.v22.i41.9069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon SM, Kim DS, Won NH, Park SJ, Chwae YJ, Kang HC, Lee SH, Baik EJ, Thorgeirsson SS, Woo HG. Genomic copy number alterations with transcriptional deregulation at 6p identify an aggressive HCC phenotype. Carcinogenesis. 2013;34:1543–1550. doi: 10.1093/carcin/bgt095. [DOI] [PubMed] [Google Scholar]
- 16.Zain SM, Mohamed R, Cooper DN, Razali R, Rampal S, Mahadeva S, Chan WK, Anwar A, Rosli NS, Mahfudz AS, et al. Genome-wide analysis of copy number variation identifies candidate gene loci associated with the progression of non-alcoholic fatty liver disease. PLoS One. 2014;9:e95604. doi: 10.1371/journal.pone.0095604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bellido F, Pineda M, Sanz-Pamplona R, Navarro M, Nadal M, Lázaro C, Blanco I, Moreno V, Capellá G, Valle L. Comprehensive molecular characterisation of hereditary non-polyposis colorectal tumours with mismatch repair proficiency. Eur J Cancer. 2014;50:1964–1972. doi: 10.1016/j.ejca.2014.04.022. [DOI] [PubMed] [Google Scholar]
- 18.Geiersbach KB, Jarboe EA, Jahromi MS, Baker CL, Paxton CN, Tripp SR, Schiffman JD. FOXL2 mutation and large-scale genomic imbalances in adult granulosa cell tumors of the ovary. Cancer Genet. 2011;204:596–602. doi: 10.1016/j.cancergen.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Paxton CN, Rowe LR, South ST. Observations of the genomic landscape beyond 1p19q deletions and EGFR amplification in glioma. Mol Cytogenet. 2015;8:60. doi: 10.1186/s13039-015-0156-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan WJ, Lai JC, Thike AA, Lim JC, Tan SY, Koh VC, Lim TH, Bay BH, Tan MH, Tan PH. Novel genetic aberrations in breast phyllodes tumours: comparison between prognostically distinct groups. Breast Cancer Res Treat. 2014;145:635–645. doi: 10.1007/s10549-014-2982-y. [DOI] [PubMed] [Google Scholar]
- 21.Ji H, Kumm J, Zhang M, Farnam K, Salari K, Faham M, Ford JM, Davis RW. Molecular inversion probe analysis of gene copy alterations reveals distinct categories of colorectal carcinoma. Cancer Res. 2006;66:7910–7919. doi: 10.1158/0008-5472.CAN-06-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Carlton VE, Karlin-Neumann G, Sapolsky R, Zhang L, Moorhead M, Wang ZC, Richardson AL, Warren R, Walther A, et al. High quality copy number and genotype data from FFPE samples using Molecular Inversion Probe (MIP) microarrays. BMC Med Genomics. 2009;2:8. doi: 10.1186/1755-8794-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fundel K, Küffner R, Aigner T, Zimmer R. Normalization and gene p-value estimation: issues in microarray data processing. Bioinform Biol Insights. 2008;2:291–305. doi: 10.4137/bbi.s441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patil MA, Gütgemann I, Zhang J, Ho C, Cheung ST, Ginzinger D, Li R, Dykema KJ, So S, Fan ST, et al. Array-based comparative genomic hybridization reveals recurrent chromosomal aberrations and Jab1 as a potential target for 8q gain in hepatocellular carcinoma. Carcinogenesis. 2005;26:2050–2057. doi: 10.1093/carcin/bgi178. [DOI] [PubMed] [Google Scholar]
- 27.Abou-Elella A, Gramlich T, Fritsch C, Gansler T. c-myc amplification in hepatocellular carcinoma predicts unfavorable prognosis. Mod Pathol. 1996;9:95–98. [PubMed] [Google Scholar]
- 28.Guasch G, Ollendorff V, Borg JP, Birnbaum D, Pébusque MJ. 8p12 stem cell myeloproliferative disorder: the FOP-fibroblast growth factor receptor 1 fusion protein of the t(6;8) translocation induces cell survival mediated by mitogen-activated protein kinase and phosphatidylinositol 3-kinase/Akt/mTOR pathways. Mol Cell Biol. 2001;21:8129–8142. doi: 10.1128/MCB.21.23.8129-8142.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jang KY, Noh SJ, Lehwald N, Tao GZ, Bellovin DI, Park HS, Moon WS, Felsher DW, Sylvester KG. SIRT1 and c-Myc promote liver tumor cell survival and predict poor survival of human hepatocellular carcinomas. PLoS One. 2012;7:e45119. doi: 10.1371/journal.pone.0045119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedica F, Ruzzenente A, Bagante F, Capelli P, Cataldo I, Pedron S, Iacono C, Chilosi M, Scarpa A, Brunelli M, et al. A re-emerging marker for prognosis in hepatocellular carcinoma: the add-value of fishing c-myc gene for early relapse. PLoS One. 2013;8:e68203. doi: 10.1371/journal.pone.0068203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sommers JA, Sharma S, Doherty KM, Karmakar P, Yang Q, Kenny MK, Harris CC, Brosh RM Jr. p53 modulates RPA-dependent and RPA-independent WRN helicase activity. Cancer Res. 2005;65:1223–1233. doi: 10.1158/0008-5472.CAN-03-0231. [DOI] [PubMed] [Google Scholar]
- 32.Goto M, Miller RW, Ishikawa Y, Sugano H. Excess of rare cancers in Werner syndrome (adult progeria) Cancer Epidemiol Biomarkers Prev. 1996;5:239–246. [PubMed] [Google Scholar]
- 33.Blander G, Kipnis J, Leal JF, Yu CE, Schellenberg GD, Oren M. Physical and functional interaction between p53 and the Werner’s syndrome protein. J Biol Chem. 1999;274:29463–29469. doi: 10.1074/jbc.274.41.29463. [DOI] [PubMed] [Google Scholar]
- 34.Huang JF, Wang Y, Liu F, Liu Y, Zhao CX, Guo YJ, Sun SH. EVI1 promotes cell proliferation in HBx-induced hepatocarcinogenesis as a critical transcription factor regulating lncRNAs. Oncotarget. 2016;7:21887–21899. doi: 10.18632/oncotarget.7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takaku H, Minagawa A, Takagi M, Nashimoto M. A candidate prostate cancer susceptibility gene encodes tRNA 3’ processing endoribonuclease. Nucleic Acids Res. 2003;31:2272–2278. doi: 10.1093/nar/gkg337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noda D, Itoh S, Watanabe Y, Inamitsu M, Dennler S, Itoh F, Koike S, Danielpour D, ten Dijke P, Kato M. ELAC2, a putative prostate cancer susceptibility gene product, potentiates TGF-beta/Smad-induced growth arrest of prostate cells. Oncogene. 2006;25:5591–5600. doi: 10.1038/sj.onc.1209571. [DOI] [PubMed] [Google Scholar]
- 37.Kurosaki T, Takata M, Yamanashi Y, Inazu T, Taniguchi T, Yamamoto T, Yamamura H. Syk activation by the Src-family tyrosine kinase in the B cell receptor signaling. J Exp Med. 1994;179:1725–1729. doi: 10.1084/jem.179.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailet O, Fenouille N, Abbe P, Robert G, Rocchi S, Gonthier N, Denoyelle C, Ticchioni M, Ortonne JP, Ballotti R, et al. Spleen tyrosine kinase functions as a tumor suppressor in melanoma cells by inducing senescence-like growth arrest. Cancer Res. 2009;69:2748–2756. doi: 10.1158/0008-5472.CAN-08-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coopman PJ, Do MT, Barth M, Bowden ET, Hayes AJ, Basyuk E, Blancato JK, Vezza PR, McLeskey SW, Mangeat PH, et al. The Syk tyrosine kinase suppresses malignant growth of human breast cancer cells. Nature. 2000;406:742–747. doi: 10.1038/35021086. [DOI] [PubMed] [Google Scholar]
- 40.Eisenberg E, Greene LE. Multiple roles of auxilin and hsc70 in clathrin-mediated endocytosis. Traffic. 2007;8:640–646. doi: 10.1111/j.1600-0854.2007.00568.x. [DOI] [PubMed] [Google Scholar]
- 41.Sato J, Shimizu H, Kasama T, Yabuta N, Nojima H. GAK, a regulator of clathrin-mediated membrane trafficking, localizes not only in the cytoplasm but also in the nucleus. Genes Cells. 2009;14:627–641. doi: 10.1111/j.1365-2443.2009.01296.x. [DOI] [PubMed] [Google Scholar]
- 42.Shimizu H, Nagamori I, Yabuta N, Nojima H. GAK, a regulator of clathrin-mediated membrane traffic, also controls centrosome integrity and chromosome congression. J Cell Sci. 2009;122:3145–3152. doi: 10.1242/jcs.052795. [DOI] [PubMed] [Google Scholar]
- 43.Park JW, Chen M, Colombo M, Roberts LR, Schwartz M, Chen PJ, Kudo M, Johnson P, Wagner S, Orsini LS, et al. Global patterns of hepatocellular carcinoma management from diagnosis to death: the BRIDGE Study. Liver Int. 2015;35:2155–2166. doi: 10.1111/liv.12818. [DOI] [PMC free article] [PubMed] [Google Scholar]


