Abstract
AIM
To demonstrate that specific bacteria might release bacterial extracellular DNA (eDNA) to exert immunomodulatory functions in the mouse small intestine.
METHODS
Extracellular DNA was extracted using phosphate buffered saline with 0.5 mmol/L dithiothreitol combined with two phenol extractions. TOTO-1 iodide, a cell-impermeant and high-affinity nucleic acid stain, was used to confirm the existence of eDNA in the mucus layers of the small intestine and colon in healthy Male C57BL/6 mice. Composition difference of eDNA and intracellular DNA (iDNA) of the small intestinal mucus was studied by Illumina sequencing and terminal restriction fragment length polymorphism (T-RFLP). Stimulation of cytokine production by eDNA was studied in RAW264.7 cells in vitro.
RESULTS
TOTO-1 iodide staining confirmed existence of eDNA in loose mucus layer of the mouse colon and thin surface mucus layer of the small intestine. Illumina sequencing analysis and T-RFLP revealed that the composition of the eDNA in the small intestinal mucus was significantly different from that of the iDNA of the small intestinal mucus bacteria. Illumina Miseq sequencing showed that the eDNA sequences came mainly from Gram-negative bacteria of Bacteroidales S24-7. By contrast, predominant bacteria of the small intestinal flora comprised Gram-positive bacteria. Both eDNA and iDNA were added to native or lipopolysaccharide-stimulated Raw267.4 macrophages, respectively. The eDNA induced significantly lower tumor necrosis factor-α/interleukin-10 (IL-10) and IL-6/IL-10 ratios than iDNA, suggesting the predominance for maintaining immune homeostasis of the gut.
CONCLUSION
Our results indicated that degraded bacterial genomic DNA was mainly released by Gram-negative bacteria, especially Bacteroidales-S24-7 and Stenotrophomonas genus in gut mucus of mice. They decreased pro-inflammatory activity compared to total gut flora genomic DNA.
Keywords: Bacterial extracellular DNA, Flora, Immune-stimulatory property, Gut microbiota, Mouse, Small intestine
Core tip: Our results revealed that degraded bacterial genomic DNA was mainly released by Gram-negative bacteria, especially Bacteroidales-S24-7 and Stenotrophomonas genus in gut mucus of mice. They decreased pro-inflammatory activity compared to genomic DNA of total gut flora. Our study highlights that bacteria derived DNA plays an important role in modulating local immune response in mouse gut.
INTRODUCTION
The intestinal mucosal immune system of mammals evolved to coexist with densely populated microorganisms that reside in the intestinal mucus layer and lumen. The central physiological process for homeostatic immune response in the gut is induced by specific bacterial products. Unmethylated cytosine-guanine (CpG)-rich DNA is typical microbial products that are recognized by the vertebrate innate immune system[1]. Exposition to the TLR9 ligand CpG induces strong protective effects in different models of intestinal inflammation[2,3]. TLR9 activation by bacterial DNA has also been demonstrated to induce degranulation of Paneth cells and to induce increased resistance to Salmonella typhimurium infection[4]. However, the specific effect of the physiologic microbiota DNA on TLR9 pathway status within the intestine so far remains elusive. Because in the mucosal environment, dendritic cells (DCs) and enterocytes permanently monitor the bacterial burden and structure in the gut[5], it is conceivable that this physiologic interaction significantly contributes to gut homeostasis. It has been demonstrated that extracted DNA of gut lumen flora limited potently regulatory T cell (Treg) induction by DCs of the lamina propria, thus controlling the balance between Treg and effector T cell frequency and function[6].
Because of the large number of bacteria present in the gut, the amount of cell-free bacterial DNA is likely to be more significant. Small intestinal mucosa-associated bacteria might find it easier to release extracellular DNA (eDNA) because of the action of antimicrobial peptides[5], which would contact epithelial cells after penetration of the thin mucus layer. However, evidence is still lacking to support the existence of bacterial eDNA within the mucus layer.
It is worth to note that intestinal epithelial cells do not respond equally to bacterial DNA, and are capable of distinguishing between DNA from probiotic strains and DNA from pathogenic strains[7]. A bioinformatic analysis revealed that small intestine specific bacteria Enterococcus faecalis, Lactobacillus casei, Lactobacillus plantarum, and Lactobacillus rhamnosus, whose strains have been marketed as probiotics, had high counts of GTCGTT motifs, the optimal motif stimulating human TLR9[8]. The quantity and resource of CpG DNA can also be viewed as detrimental, depending on the host’s physiological status. Estimation of the load of bacterial released DNA by mucosa-associated bacteria could shed new light on host-microbe interactions across a range of diseases.
The present study aimed to demonstrate the existence of mucosa-associated bacteria released eDNA in the mucus layer in the mouse intestine. Furthermore, the major bacterial genera responsible for the release of eDNA in the small intestinal mucus layer were identified.
MATERIALS AND METHODS
Animals
Male C57BL/6 mice (four weeks old) were purchased from the Model Animal Research Center of Nanjing University (Nanjing, China). The animal protocol was designed to minimize pain or discomfort to the animals. The animals were acclimatized to laboratory conditions (23 °C, 12 h/12 h light/dark, 50% humidity, ad libitum access to normal chow and water) for one week prior to experimentation. All animals were euthanized by barbiturate overdose (intravenous injection, 150 mg/kg pentobarbital sodium) for tissue collection after being fasted overnight.
Staining of gut mucus and bacterial eDNA
The distal colon and small intestine were dissected longitudinally and placed in Carnoy’s solution (ethanol: chloroform: acetic acid = 6:3:1) for 3 h. The firm mucus layer of the colon was fixed using the same method except for scraping the surface slightly. The fixed tissues were then washed twice in absolute ethanol for 20 min each, followed by two washes in xylene for 15 min each, before paraffin embedding and sectioning as 4-μm-thin sections. The sections were placed on glass slides after floating on an air bath according to standard procedures[9].
An alcian blue-periodic acid Schiff (AB-PAS) staining kit was used to visualize the gut mucus. Visualization of eDNA in the intestinal biofilm was performed using the fluorescent dye TOTO-1 (Molecular Probes, Eugene, OR, United States).
Isolation of mucus bacterial eDNA
Ileums were opened longitudinally and food debris was removed carefully. The mucus was harvested with PBS containing 0.5 mmol/L dithiothreitol (PBS-DTT) and incubated for 3 min with gentle shaking. It was centrifuged for 10 min at 10000 rpm to harvest released DNA. This step was repeated twice and the supernatant was pooled. Then, 10% CTAB in 0.7 mol/L NaCl was added, and ethanol precipitation was used to concentrate DNA. Bead beating and the QIAamp DNA Stool Mini Kit (QIAGEN) were used to extract genomic DNA of mucoid bacteria.
Terminal restriction fragment length polymorphism (T-RFLP) analysis
Primers 334F/939R or 338F/806R[10] were labeled with 5′6-carboxyfluorescein (6-FAM) (forward) or 5′6-hexachlorofluorescein (HEX) (reverse). The 25-μL PCR reaction contained 1 × PCR buffer, 200 μmol/L of each deoxynucleoside triphosphate, 1.5 mmol/L MgCl2, 0.1 μmol/L of each primer, 100 ng of DNA template, and 0.5 U of Takara Taq DNA polymerase[11]. The PCR products were analyzed by 1.5% agarose gel electrophoresis.
After purification, the amplification products were digested with DdeI or AluI. The restriction enzyme digestion reaction mixture (20 μL), containing 2 U of DdeI or AluI, 2 μL of 1 × NEB buffer, and 500 ng of PCR product, was incubated at 37 °C overnight. Finally, the fluorescently labeled terminal fragments of sizes between 50 bp to 500 bp were analyzed by electrophoresis on an ABI PRISM 310 Genetic Analyzer in the GeneScan mode.
Bacterial 16S rRNA gene amplification and illumina MiSeq sequencing
The V4-V5 domains of 16S rRNA genes were amplified using primers 515F and 907R (see supplementary methods). The resulting amplicons were submitted to the Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China) for Illumina paired-end library preparation, cluster generation, and 300-bp paired-end sequencing on a MiSeq instrument in two separate runs. Details of the PCR amplification and sequencing are described in supplementary information. The raw reads were deposited in the NCBI Sequence Read Archive (SRA) database (Accession Number: SRP072153).
Stimulation of RAW264.7 cells for cytokine production
RAW264.7 cells (4 × 106 cells/mL) were treated with medium, lipopolysaccharide (LPS) (1 μg/mL), eDNA (1 ng/mL), iDNA (1 ng/mL), eDNA (1 ng/mL) + LPS (1 μg/mL), or iDNA (1 ng/mL) + LPS (1 μg/mL) for 12 h. Culture supernatants were analyzed by enzyme linked immunosorbent assays (ELISAs) for TNF-α, IL-6, IL-10, or IL-12p40. All recombinant murine cytokines and antibodies specific for murine cytokines were purchased from Xiamen Huijia Biotechnology Co., Ltd (Xiamen, China). Purified DNA was tested for contaminating endotoxins by using a Limulus amebocyte lysate kit (Xiamen Agent Company (Fujian, PR China). Only preparations with endotoxin levels not exceeding 0.05 endotoxin units/mL were used.
Statistical analysis
The statistical significance of the comparisons between multiple groups was carried out by ANOVA, followed by Tukey’s test. A 95% confidence interval was considered significant and was defined as P < 0.05. All values are expressed as the mean ± standard error of the mean (SEM). Each value is the mean of at least three separate experiments. Principal component analysis (PCA) and cluster analysis were used to analyze the terminal restriction fragment (TRF) profiles generated from the T-RFLP experiment, and were combined with the diversity index to study the bacterial communities. PCA plots were generated using the multivariate statistics software Canoco (version 4.5, Microcomputer Power, Ithaca, NY, United States). The biodiversity was measured using the Shannon-Wiener index (H = -∑pi•lnpi); the Simpson Index (D = 1-∑pi^2); and the Evenness Index (E = H/lnS) according to the relative height of each TRF (pi) and sum of the number of TRFs (S) in a sample.
RESULTS
Staining of gut mucus and eDNA
As shown in Figure 1, AB-PAS staining showed apparent differences in mucus thickness between the intact mucosa and post-suction mucosa. The loose layer and inner layer of the colon mucus were separated by gentle suction and scraping, which were stained blue after PAS staining. The impermeant nucleic acid dye TOTO-1 was used to visualize the eDNA. This fluorescent stain can bind DNA molecules via its positive charge and emits green fluorescence when excited at 514 nm. The mucus is reported to be composed of two layers with different characteristics and completely different distribution patterns of bacteria[12]. The loose layer contains bacteria, whereas the firm layer is reported to be free of bacteria[12]. We could see a clear deepening of the green fluorescent band on the colon surface, which proved the existence of specific microbial communities, which we attributed to the release of eDNA in the gut mucus loose layer. By contrast, the fluorescence intensity of the inner mucus was much weaker, which suggested no eDNA release because of the absence of bacteria in this firm layer. In the small intestine, the thin surface mucus layer was positive for TOTO-1 staining. Note that the bottom of the crypt lumen was positive and stained green under a confocal microscope, which might be explained by the accumulation of eDNA around such cells.
Figure 1.
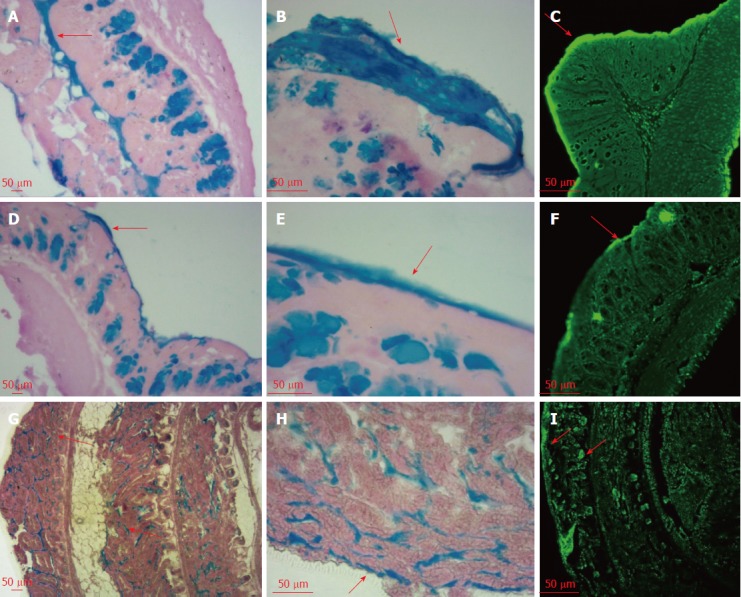
Mucus layer and eDNA in the mouse small intestine and colon. The first two columns are images obtained by optical microscopy with AB-PAS staining, and the third one are images obtained by laser scanning confocal microscopy with TOTO-1 staining. Pictures from the first two rows are the colon mucus layer, while those of the third row are the small intestinal mucus layer. A-F: Colon; G-I: Small intestine; A, D, G: AB-PAS 10×; B, E, H: AB-PAS 40×; C, F, I: TOTO-1 10 ×.
Results of T-RFLP
The T-RFLP data representing the gut microbial community profiles were analyzed using multivariate statistics for the intestinal mucus separately. First, the T-RFLP data from each individual were normalized and entered into a data matrix that comprised the TRFs as variables and individuals as objects. A consensus T-RFLP profile from each biological replicate was constructed by averaging the technical duplicates[13].
The PCA in Figure 2 clearly demonstrates remarkably different TRF profiles between eDNA and iDNA, using either AluI (Figure 2A) or DdeI (Figure 2B). Samples from eDNA or iDNA were found to gather in a concentrated area and were separate from each other. The results indicated that the components of eDNA varied greatly from those of iDNA, indicating that they were derived from two different bacterial communities.
Figure 2.
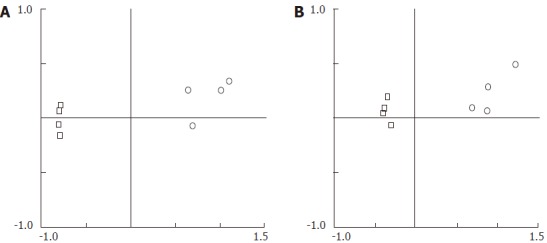
Principal component analysis plots for terminal restriction fragment length polymorphism profiles (including TRF size and relative abundance data). A: with AluI. B: with DdeI; Empty circle, eDNA; Empty square, iDNA.
The cluster analysis in supplementary figure 1 showed that eDNA was clustered separately from iDNA, which agreed with the conclusion of PCA. The unique TRFs were extracted that belonged to the iDNA or eDNA. Some unique TRFs from the same set could be filtered for further confirmation. The TRFs in supplementary figure 1A and C were specific for the iDNA using AluI and DdeI, respectively, while the TRFs in supplementary figure 1B and D belonged to eDNA using the two endonucleases.
By analyzing the diversity index (Table 1), the values of Shannon-wiener index, equitability index, and Simpson’s diversity index from the eDNA were observed to be smaller than those from the iDNA (P < 0.05). The lower index values indicate poorer abundance and stability of the eDNA. The special properties of eDNA microbiota could be the factor that distinguishes them from the iDNA resource. However, further studies are needed to provide more evidence to authenticate the particularity of eDNA.
Table 1.
The analysis of diversity index
| Shannon-wiener index | Equitability index | Simpson’s diversity index | |
| iDNA | 2.09 ± 0.04 | 0.87 ± 0.01 | 0.84 ± 0.00 |
| eDNA | 1.64 ± 0.09a | 0.77 ± 00.04a | 0.73 ± 0.04a |
Values are expressed as mean ± SEM (n = 6).
P < 0.05, significant differences between iDNA and eDNA.
Results of illumina MiSeq sequencing
We failed to amplify the 16S rRNA gene from the eDNA using primers 338F/806R, which should have generated a 468 bp amplicon (data not shown). Using primers 515F/907R, we produced a 392-bp product successfully. Consistent with our T-RLFP analysis, the sequencing results of this amplicon indicated a significant difference in proportions of major phyla between the eDNA and the iDNA (Figure 2). Firmicutes was the most abundant group in the iDNA (68%-77%), while 11% of Bacteroidetes occupied the second place. Whereas in the eDNA, we found that Bacteroidetes and Proteobacteria were more dominant at 54 % and 29%, respectively. Analysis at the genus level (Figure 3 and supplementary table 1) provided more detailed information. The results revealed that genera of two Gram-negative bacteria, Bacteroidales S24-7 and Stenotrophomonas, were the dominant genera in the eDNA resource, reaching a proportion of 77%-91%. While two Gram-positive genera, Staphylococcus and Allobaculum, were main components in iDNA. The iDNA also contained a small quantity of Bacteroidales S24-7. Gram-negative genera represented 83.4% of the genera in the eDNA and Gram-positive ones represented 86.1 % of the genera in the iDNA
Figure 3.
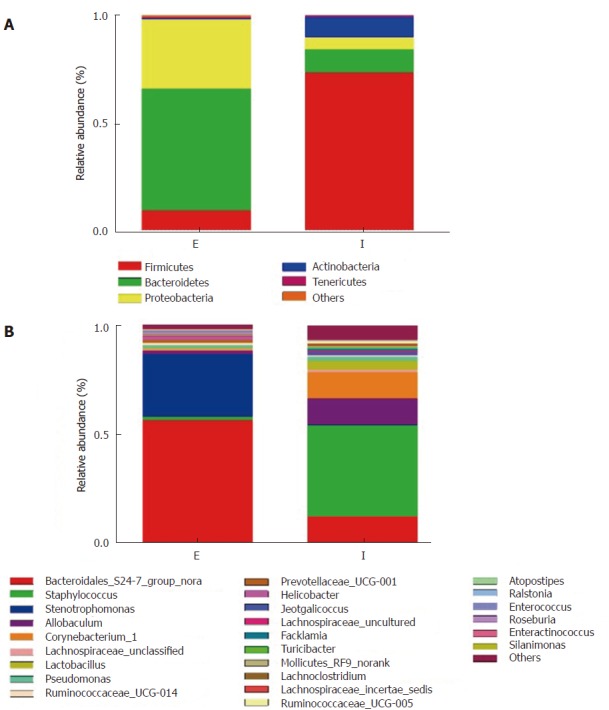
Community structure component diagram at the phylum (A) and genus (B) levels. Samples were from eDNA (E) and iDNA (I) in triplicate, respectively. eDNA: Extracellular bacterial DNA; iDNA: Intracellular bacterial DNA.
The results of fluorescence in situ hybridization with probes for Bacteroidales and Staphylococcus demonstrated different degrees of positive reaction in the crypt lumen (supplementary figure 2). These results indicated that eDNA of Gram-negative genera often migrated to the crypt.
Supplementary figure 3 showed that other detectable bacteria, such as Lactobacillus, Facklamia, and Ralstonia, were significantly different between eDNA and iDNA. All these results suggested that the different constituents and proportions of micro-community in the eDNA and iDNA might explain their specific functions.
Furthermore, the 16S rRNA marker gene sequences were used to establish the predictive functional profiles of the microbial communities using PICRUST (supplementary figure 4). Interestingly, the operational taxonomic unit (OTU) abundances representing certain specific functions in eDNA were generally higher than those of the iDNA, for example, cell motility (N), vesicular transport (U), and cell membrane biogenesis (M), which have been confirmed to be closely associated with the function of the eDNA.
Cytokine production from Raw264.7 cells is stimulated differently by eDNA and iDNA
As shown in Figure 4, the production of inflammatory cytokines TNF-α and IL-6 in Raw264.7 macrophages were stimulated significantly by both eDNA and iDNA (P < 0.05). Only eDNA promoted a very low, but significantly higher level of IL-12 compared with the iDNA (P < 0.05). However, iDNA suppressed the production of the anti- inflammatory cytokine IL-10 significantly (P < 0.05). LPS stimulated the production of all cytokines significantly (P < 0.01), which were greatly suppressed when cells were exposed to iDNA simultaneously (P < 0.05). Only the iDNA showed an inhibitory effect on LPS stimulated IL-12 production (P < 0.05). Considering the ratio of proinflammatory cytokine to IL-10, the iDNA showed a stronger proinflammatory effect than the eDNA and LPS, which was reflected by the significantly higher TNF-a/IL-10 and IL-6/IL-10 levels (P < 0.01).
Figure 4.
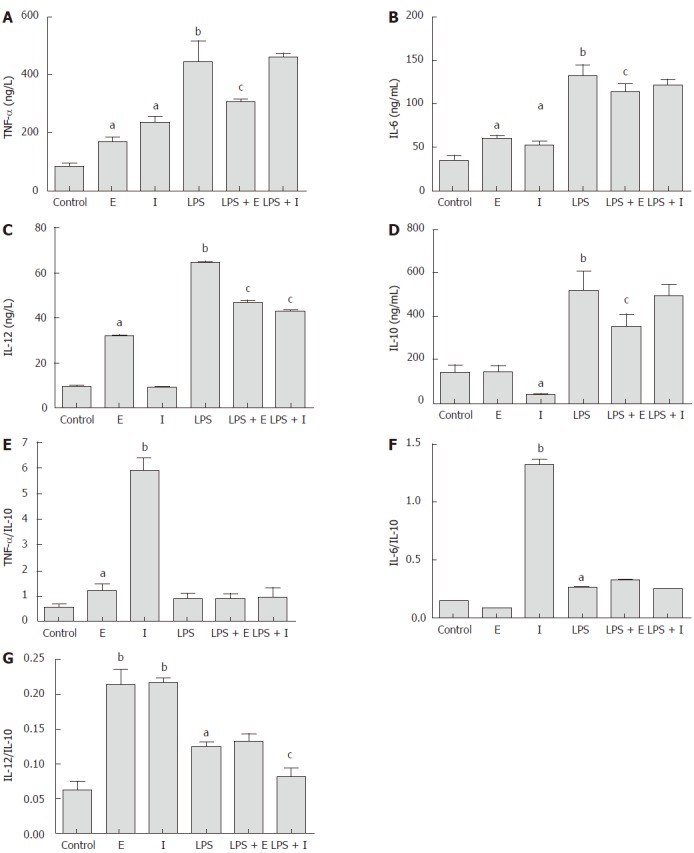
Effect of iDNA and eDNA on cytokine production by Raw267.4 cells, including TNF-α (A), IL6 (B), IL-12 (C), IL-10 (D), and ratio of TNF-α (E), IL6 (F) and IL-12 (G) to IL-10. Cells were treated for 12 h with medium, lipopolysaccharide (LPS) (1 μg/mL), eDNA (1 ng/mL), iDNA (1 ng/mL), eDNA(1 ng/mL) + LPS (1 μg/mL), or iDNA (1 ng/mL) + LPS (1 μg/mL) for 12 h. Values are expressed as mean ± SEM (n = 6). aP < 0.05, Significant differences between control and treatment; bP < 0.01, significant differences between control and treatment; cP < 0.05, significant differences between LPS treatment and treatment. E: Extracellular bacterial DNA; I: Intracellular bacterial DNA; LPS: Lipopolysaccharide.
DISCUSSION
In the present study, we extracted eDNA from the mouse small intestine and colon and established a suitable PCR-TRFLP protocol to distinguish the microorganism diversity, which consisted of eDNA and iDNA. Analysis of the Illumina MiSeq sequencing data demonstrated the significantly different constitutions and functions between the eDNA and iDNA. Our results provided a sound basis for research into the structure and function of eDNA from the aspect of microorganism diversity.
AB-PAS and TOTO-1 staining revealed that eDNA was enriched in the mucus layer of the colon and small intestine. Some of the eDNA would be derived from shed epithelial cells, and approximately 1400 cells are shed from each villus every 24 h[14]. Our study confirmed that some mucus DNA originated from intestinal bacteria. In contrast to pig small intestinal crypts[15], eDNA was observed frequently at the bottom of the crypts of Lieberkühn in mice. This might be the result of the accumulation of eDNA migrating from the mucus layer, which cannot prevent the diffusion of linear DNA[16]. The eDNA might also be released by bacteria killed by antimicrobial peptides secreted by Paneth cells residing at the base of the small intestinal crypts[17]. The sentinel role of Paneth cells requires the interaction between TLR9 and DNA containing CpG sequences[4,18]; therefore, crypt eDNA might play a significant role in maintaining small intestinal immune homeostasis.
We used DTT in eDNA extraction. It is a strong reducing agent that can break protein disulfide bonds to aid its dissolution. Using DTT allowed the efficient extraction of eDNA, demonstrating disruption of the interaction between eDNA and mucin. This is in agreement with the finding of Macierzanka et al that both large and small DNA particulates appeared to form a network or were held in place by the mucin network[15].
In consideration of lower integrity of eDNA compared with iDNA, primer selection was carried out before T-RFLP analysis, to recover a high percentage of bacterial species. Primers 27F/1492R and 530F/1492R, which have been used widely in previous studies, were found to be unsuitable for the amplification of 16S rRNA fragments from mucus eDNA (data not shown). The primers 334F/939R (V3-V5) produced a relatively small product that was highly specific for the spectrum of bacterial species in mucus eDNA, suggesting that eDNA was degraded genomic DNA. This is in agreement with the finding in pigs that degraded nuclei embedded in mucus was often fragmented[15]. This was confirmed by Illumina MiSeq sequencing analysis, where primers 515F/907R were more successful in PCR compared with primers 338F/806R.
PCA of T-RFLP and Illumina MiSeq sequencing data revealed significant differences between iDNA and eDNA, suggesting that the eDNA release characteristics varied among strains in the small intestinal microbiota. The eDNA was mainly released by Gram-negative bacteria of the Bacteroidales S24-7 and Stenotrophomonas genera. Gram negative-bacteria contain eDNA specifically associated with the outer membrane vesicle (OMV)[19,20]. The presence of OMV-associated DNA within Gram-negative bacteria biofilms has also been confirmed[21]. In the present study, the result of PICRUST analysis of eDNA and iDNA sequences showed that the functions of vesicular transport and cell membrane biogenesis were related to the bacterial source of the eDNA. This result might suggest the presence of OMV-associated DNA in the eDNA. In particular, the genus Stenotrophomonas released eDNA into the mucus, as demonstrated by a significant difference in their proportions in the eDNA and iDNA. Members of this genus are strict aerobic bacteria that belong to the γ-β subclass of Proteobacteria[22]. Stenotrophomonas is reported as a dominant member of the plant-associated bacterial community[23]. It has also been reported in the colonic mucosa-associated microbiota of healthy humans[24] and the mouse colonic crypt[25]. Notable features of this genus are their weak invasiveness, variety of colonization mechanisms, and strong ability to form biofilms, making them a successful colonizer of various hosts[26]. Further studies should be carried out to clarify the mechanism involved in the release of Stenotrophomonas eDNA into the mucus layer and the physiological significance. Our findings are also in agreement with those of Ou et al[27], who reported a predominance of Streptococci in the small intestinal mucus. Itzek et al[28] reported that certain oral Streptococci produce H2O2, which causes the release of eDNA to promote biofilm formation. However, in the present study, this genus released trace eDNA, indicating that habitat is a key factor in their eDNA releasing property. Streptococci have been reported to attach to Bacteroides-produced OMV[29], which might help their incorporation into the mucus biofilm without actively releasing eDNA.
Bacterial DNA stimulates not only potent pro-inflammatory activities, but also the interferon regulatory factor pathway that induces anti-inflammatory activities[30]. In the present study, iDNA and eDNA showed distinct cytokine stimulation patterns. The eDNA showed a lower pro-inflammatory effect, according to the low TNF-a/IL-10 and IL-6/IL-10 levels, and induced very low levels of IL-12. TLR-9 activation by bacterial DNA is dependent on the individual CpG content and intracellular delivery rate[31]. Thus, it is necessary to further study the difference in the CpG contents of eDNA and iDNA.
In conclusion, our results indicated that eDNA is located in the intestinal mucus layer and at the bottom of the crypt lumen in the small intestine. DTT promoted the release of bacterial eDNA from the small intestinal mucus layer. The eDNA was degraded bacterial genomic DNA mainly released by Gram-negative bacteria, especially by the Bacteroidales-S24-7 and Stenotrophomonas genera. The eDNA showed decreased pro-inflammatory activity compared with total gut flora genomic DNA. Further studies are needed to clarify the actual source of eDNA, and its relationship with the gut immune response, especially the production of AMPs in Paneth cells of the small intestinal crypt.
ARTICLE HIGHLIGHTS
Research background
Many studies strongly suggest that signals, including bacterial DNA, from colonizing microbes greatly alter host local immune system in the gut. Bacterial cells do not contact with enterocytes in normal physiological status. They might release DNA into the mucus layer to influence host innate immune cell through specific receptors, like Toll-like receptor 9. Evidence supporting this hypothesis is needed.
Research motivation
This research investigated the existence of extracellular bacterial DNA (eDNA) in the mouse gut mucus layer, their resource, and immune modulatory function. There were differences in DNA’s immuno-stimulatory properties among different bacteria as reported by other researchers. Therefore, host immune response would be modulated by targeted change of DNA releasing bacteria in the mucus through specific medicine or food components.
Research objectives
This study aimed to confirm the existence of bacterial eDNA in the mouse gut mucus layer, and to identify bacterial genera that release them. Immuno-stimulatory properties of eDNA were also studied in vitro. This provided basic knowledge about bacteria and host interaction through bacterial DNA and related signal pathways. This will also promote nutritional strategy development to modulate local immune response through changing DNA releasing microbiota.
Research methods
Bacterial eDNA in the mucus layer and crypts was visualized by TOTO-1 staining. Small intestinal mucosal microbiota and eDNA were analyzed using T-RFLP and Illumina MiSeq amplicon sequencing. Immuno-stimulatory effects of microbiota and eDNA were determined after incubation with mouse RAW264.7 macrophages.
Research results
TOTO-1 iodide staining confirmed existence of eDNA in the mucus layer. The composition of the eDNA was significantly different from that of the intracellular DNA (iDNA). The eDNA sequences came mainly from Gram-negative bacteria of Bacteroidales S24-7. The eDNA induced significantly lower TNF-α/IL-10 and IL-6/IL-10 ratios in LPS stimulated RAW264.7 cells than iDNA. This is the first report related to bacteria genus responsible for DNA release in the gut mucus layer.
Research conclusions
Our results indicated that eDNA was located in the intestinal mucus layer. The eDNA was degraded bacterial genomic DNA mainly released by Gram-negative bacteria especially Bacteroidales S24-7. They showed decreased pro-inflammatory activity compared with total gut flora genomic DNA.
Research perspectives
Further studies are needed to clarify the specific bacterial species/strains that release eDNA, and its relationship with the gut immune response, especially the production of antimicrobial peptides in Paneth cells of the small intestinal crypt.
Footnotes
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
Institutional animal care and use committee statement: All procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Committee of Jiangnan University (IACUC protocol number: No. 8/2014/JU).
Conflict-of-interest statement: No potential conflicts of interest.
Data sharing statement: No additional data are available.
Peer-review started: September 23, 2017
First decision: October 11, 2017
Article in press: October 27, 2017
P- Reviewer: Gassler N, Handra-Luca A, Ulasoglu C S- Editor: Gong ZM L- Editor: Wang TQ E- Editor: Lu YJ
Contributor Information
Ce Qi, The Key Laboratory of Carbohydrate Chemistry and Biotechnology, Ministry of Education, School of Biotechnology, Jiangnan University, Wuxi 214122, Jiangsu Province, China; Guo-wei Le, Jin Sun, School of Food Science and Technology, Jiangnan University, Wuxi 214122, Jiangsu Province, China.
Ya Li, Guo-wei Le, Jin Sun, School of Food Science and Technology, Jiangnan University, Wuxi 214122, Jiangsu Province, China. sunj@jiangnan.edu.cn.
Ren-Qiang Yu, Wuxi Maternal and Child Health Hospital, Wuxi 212422, Jiangsu Province, China.
Sheng-Li Zhou, Quality of Research and Development Department, COFCO Fortune Food Sales & Distribution Co., Ltd. Tianjin 300452, China.
Xing-Guo Wang, Guo-wei Le, Jin Sun, School of Food Science and Technology, Jiangnan University, Wuxi 214122, Jiangsu Province, China.
Hang Xiao, Department of Food Science, University of Massachusetts, Amherst, MA 01003, United States.
Jin Sun, The Key Laboratory of Carbohydrate Chemistry and Biotechnology, Ministry of Education, School of Biotechnology, Jiangnan University, Wuxi 214122, Jiangsu Province, China.
References
- 1.Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol. 2002;20:709–760. doi: 10.1146/annurev.immunol.20.100301.064842. [DOI] [PubMed] [Google Scholar]
- 2.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bleich A, Janus LM, Smoczek A, Westendorf AM, Strauch U, Mähler M, Hedrich HJ, Fichtner-Feigl S, Schölmerich J, Falk W, et al. CpG motifs of bacterial DNA exert protective effects in mouse models of IBD by antigen-independent tolerance induction. Gastroenterology. 2009;136:278–287. doi: 10.1053/j.gastro.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 4.Rumio C, Besusso D, Palazzo M, Selleri S, Sfondrini L, Dubini F, Ménard S, Balsari A. Degranulation of paneth cells via toll-like receptor 9. Am J Pathol. 2004;165:373–381. doi: 10.1016/S0002-9440(10)63304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 6.Hall JA, Bouladoux N, Sun CM, Wohlfert EA, Blank RB, Zhu Q, Grigg ME, Berzofsky JA, Belkaid Y. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouladoux N, Hall JA, Grainger JR, dos Santos LM, Kann MG, Nagarajan V, Verthelyi D, Belkaid Y. Regulatory role of suppressive motifs from commensal DNA. Mucosal Immunol. 2012;5:623–634. doi: 10.1038/mi.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kant R, de Vos WM, Palva A, Satokari R. Immunostimulatory CpG motifs in the genomes of gut bacteria and their role in human health and disease. J Med Microbiol. 2014;63:293–308. doi: 10.1099/jmm.0.064220-0. [DOI] [PubMed] [Google Scholar]
- 9.Johansson ME, Hansson GC. Preservation of mucus in histological sections, immunostaining of mucins in fixed tissue, and localization of bacteria with FISH. Mucins Methods Protoc. 2012:229–235. doi: 10.1007/978-1-61779-513-8_13. [DOI] [PubMed] [Google Scholar]
- 10.Armingohar Z, Jørgensen JJ, Kristoffersen AK, Abesha-Belay E, Olsen I. Bacteria and bacterial DNA in atherosclerotic plaque and aneurysmal wall biopsies from patients with and without periodontitis. J Oral Microbiol. 2014:6. doi: 10.3402/jom.v6.23408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M, Gong J, Cottrill M, Yu H, de Lange C, Burton J, Topp E. Evaluation of QIAamp DNA Stool Mini Kit for ecological studies of gut microbiota. J Microbiol Methods. 2003;54:13–20. doi: 10.1016/s0167-7012(02)00260-9. [DOI] [PubMed] [Google Scholar]
- 12.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagashima K, Hisada T, Sato M, Mochizuki J. Application of new primer-enzyme combinations to terminal restriction fragment length polymorphism profiling of bacterial populations in human feces. Appl Environ Microbiol. 2003;69:1251–1262. doi: 10.1128/AEM.69.2.1251-1262.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potten CS, Loeffler M. Stem cells: attributes, cycles, spirals, pitfalls and uncertainties. Lessons for and from the crypt. Development. 1990;110:1001–1020. doi: 10.1242/dev.110.4.1001. [DOI] [PubMed] [Google Scholar]
- 15.Macierzanka A, Mackie AR, Bajka BH, Rigby NM, Nau F, Dupont D. Transport of particles in intestinal mucus under simulated infant and adult physiological conditions: impact of mucus structure and extracellular DNA. PLoS One. 2014;9:e95274. doi: 10.1371/journal.pone.0095274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen H, Hu Y, Saltzman WM. DNA diffusion in mucus: effect of size, topology of DNAs, and transfection reagents. Biophys J. 2006;91:639–644. doi: 10.1529/biophysj.105.077404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer-Hoffert U, Hornef MW, Henriques-Normark B, Axelsson LG, Midtvedt T, Pütsep K, Andersson M. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut. 2008;57:764–771. doi: 10.1136/gut.2007.141481. [DOI] [PubMed] [Google Scholar]
- 18.Rumio C, Sommariva M, Sfondrini L, Palazzo M, Morelli D, Viganò L, De Cecco L, Tagliabue E, Balsari A. Induction of Paneth cell degranulation by orally administered Toll-like receptor ligands. J Cell Physiol. 2012;227:1107–1113. doi: 10.1002/jcp.22830. [DOI] [PubMed] [Google Scholar]
- 19.Dorward DW, Garon CF. DNA Is Packaged within Membrane-Derived Vesicles of Gram-Negative but Not Gram-Positive Bacteria. Appl Environ Microbiol. 1990;56:1960–1962. doi: 10.1128/aem.56.6.1960-1962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pérez-Cruz C, Delgado L, López-Iglesias C, Mercade E. Outer-inner membrane vesicles naturally secreted by gram-negative pathogenic bacteria. PLoS One. 2015;10:e0116896. doi: 10.1371/journal.pone.0116896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schooling SR, Hubley A, Beveridge TJ. Interactions of DNA with biofilm-derived membrane vesicles. J Bacteriol. 2009;191:4097–4102. doi: 10.1128/JB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anzai Y, Kim H, Park JY, Wakabayashi H, Oyaizu H. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int J Syst Evol Microbiol. 2000;50 Pt 4:1563–1589. doi: 10.1099/00207713-50-4-1563. [DOI] [PubMed] [Google Scholar]
- 23.Berg G, Egamberdieva D, Lugtenberg B, Hagemann M. Symbiotic plant-microbe interactions: stress protection, plant growth promotion, and biocontrol by Stenotrophomonas. In: Symbioses and Stress. Springer; 2010. pp. 445–460. [Google Scholar]
- 24.Rausch P, Rehman A, Künzel S, Häsler R, Ott SJ, Schreiber S, Rosenstiel P, Franke A, Baines JF. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci USA. 2011;108:19030–19035. doi: 10.1073/pnas.1106408108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pédron T, Mulet C, Dauga C, Frangeul L, Chervaux C, Grompone G, Sansonetti PJ. A crypt-specific core microbiota resides in the mouse colon. MBio. 2012;3:e00116–12. doi: 10.1128/mBio.00116-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryan RP, Monchy S, Cardinale M, Taghavi S, Crossman L, Avison MB, Berg G, van der Lelie D, Dow JM. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat Rev Microbiol. 2009;7:514–525. doi: 10.1038/nrmicro2163. [DOI] [PubMed] [Google Scholar]
- 27.Ou G, Hedberg M, Hörstedt P, Baranov V, Forsberg G, Drobni M, Sandström O, Wai SN, Johansson I, Hammarström ML, et al. Proximal small intestinal microbiota and identification of rod-shaped bacteria associated with childhood celiac disease. Am J Gastroenterol. 2009;104:3058–3067. doi: 10.1038/ajg.2009.524. [DOI] [PubMed] [Google Scholar]
- 28.Itzek A, Zheng L, Chen Z, Merritt J, Kreth J. Hydrogen peroxide-dependent DNA release and transfer of antibiotic resistance genes in Streptococcus gordonii. J Bacteriol. 2011;193:6912–6922. doi: 10.1128/JB.05791-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh U, Grenier D, McBride BC. Bacteroides gingivalis vesicles mediate attachment of streptococci to serum-coated hydroxyapatite. Oral Microbiol Immunol. 1989;4:199–203. doi: 10.1111/j.1399-302x.1989.tb00252.x. [DOI] [PubMed] [Google Scholar]
- 30.Kumagai Y, Takeuchi O, Akira S. TLR9 as a key receptor for the recognition of DNA. Adv Drug Deliv Rev. 2008;60:795–804. doi: 10.1016/j.addr.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Dalpke A, Frank J, Peter M, Heeg K. Activation of toll-like receptor 9 by DNA from different bacterial species. Infect Immun. 2006;74:940–946. doi: 10.1128/IAI.74.2.940-946.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


