Abstract
Selecting informative genes, including individually discriminant genes and synergic genes, from expression data has been useful for medical diagnosis and prognosis. Detecting synergic genes is more difficult than selecting individually discriminant genes. Several efforts have recently been made to detect gene-gene synergies, such as dendrogram-based I(X 1; X 2; Y) (mutual information), doublets (gene pairs) and MIC(X 1; X 2; Y) based on the maximal information coefficient. It is unclear whether dendrogram-based I(X 1; X 2; Y) and doublets can capture synergies efficiently. Although MIC(X 1; X 2; Y) can capture a wide range of interaction, it has a high computational cost triggered by its 3-D search. In this paper, we developed a simple and fast approach based on abs conversion type (i.e. Z = |X 1 − X 2|) and t-test, to detect interactions in simulation and real-world datasets. Our results showed that dendrogram-based I(X 1; X 2; Y) and doublets are helpless for discovering pair-wise gene interactions, our approach can discover typical pair-wise synergic genes efficiently. These synergic genes can reach comparable accuracy to the individually discriminant genes using the same number of genes. Classifier cannot learn well if synergic genes have not been converted properly. Combining individually discriminant and synergic genes can improve the prediction performance.
Introduction
Selection of informative genes, including individually discriminant genes and synergic genes, from expression data has been useful for medical diagnosis and prognosis. Individual gene ranking techniques such as t-test1 etc. can typically produce a “list of genes” that are correlated with disease2. However, they cannot provide insights into the interaction of these genes. According to information theory, the pair-wise interactions I (X 1; X 2; Y)3 is defined as
| 1 |
where I is the symbol for mutual information, I (X 1; Y) is the individual effect of gene X 1 relative to phenotype Y, I (X 2; Y) is the individual effect of gene X 2 relative to Y, and I (X 1, X 2; Y) is the joint effect of X 1 and X 2 relative to Y. A positive value of I (X 1; X 2; Y) indicates synergy, while a negative value of I (X 1; X 2; Y) indicates redundancy.
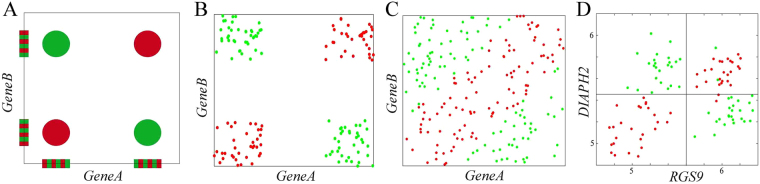
Figure 1 illustrates four typical pair-wise synergies examples from Watkinson et al.4 (Fig. 1A,B) and Chen et al.5 (Fig. 1C,D). Figure 1A–C are generated by simulated data, and Fig. 1D is generated by real-world data. As an example, when the RSG9 or DIAPH2 is evaluated individually and separately, neither of these two genes is correlated with cancer. Therefore, genes RGS9 and DIAPH2 would not be present in the output of any “individual gene ranking” techniques. However, when the pair-wise interactions is evaluated, the genes RGS9 -DIAPH2 are sufficient to distinguish cancer from normal samples (Fig. 1D).
Figure 1.
Four typical pair-wise synergies examples. Red and green dots represent cancer and normal samples, respectively.
Detecting synergic genes is more difficult than selecting individually discriminant genes. Several efforts have recently been made to detect gene–gene synergies. These efforts often fall into one of the two strategies. One is the non-conversion strategy, which uses formula (1) directly to measure I(X 1; X 2; Y)4 or uses the maximal information coefficient directly to measure MIC(X 1; X 2; Y)5. The way to discretize continuous variable is the key to estimate the value of mutual information. Binarization, such as the dendrogram-based4 technique, simplifies the estimation, and provides simple logical functions in the connection of the genes. However, it may result in information loss and estimation error. Although MIC(X 1; X 2; Y)5 can capture a wide range of interactions, it has a high computational cost triggered by its 3-D search. The other is the conversion strategy, such as doublets 6 and top scoring pair (TSP)7. They employ a new variable Z derived from the combinations between X 1 and X 2 (e.g. for the sum type of doublets, Z = X 1 + X 2) to measure I (Z; Y) instead of I(X 1; X 2; Y). This strategy is low computational cost, due to the search space reduced from 3-D to 2-D. However, it is unclear whether this conversion strategy can capture synergies8 efficiently.
Inspecting Fig. 1A–C, we found that they share the same pattern and can be characterized by the same function, Y = |X 1 − X 2|. The only difference between them is the value ranges of independent variables. Although Doublets 6 included sum, diff, mul and sign conversion types (TSP is similar to sign), it, unfortunately, ignored abs conversion type.
In this work, we developed a simple and fast approach based on abs conversion type and t-test, to discover pair-wise synergic genes that are related to cancer. Furthermore, we validated these synergic genes by using classification performance with simulation and real-world datasets. Our results show that these synergic genes can enhance the individually discriminant model and improve the prediction performance. We also demonstrated that these synergic genes should be converted into new variables (Z) prior to be used as input features for classifiers, especially for many pairs of synergistic genes.
Datasets and Methods
Datasets
Four binary class datasets are involved in this work. The reference, sample size, number of genes in each dataset, and the number of samples in each class are summarized in Table 1. All gene expression data have been normalized by using the RMA method9.
Table 1.
Four binary class gene expression datasets.
Conversion types and pair-wise gene rank
Suppose that a dataset has n samples and m genes, and can be denoted as {Y i, X ij}, i = 1,2,…,n; j = 1,2,…,m. X ij represents the expression value of the j th gene (G j) in the i th sample; and Y i represents the class label of i th sample. Y i ∈ {0, 1}, 0 denotes cancerous and 1 denotes normal tissue samples. Rank-based methods7 are robust to quantization effects and to overcome background differences between gene pairs. Therefore, let R ij denote the rank of the i th sample in the j th gene, we replace the expression values X ij by their ranks R ij and get a new data matrix {Y i, R ij}.
For two genes G p and G q, Doublets 6 lists four conversion types.
| 2 |
| 3 |
| 4 |
| 5 |
We add a new conversion type:
| 6 |
Here, i = 1,2,…,n; p = 1,2,…, m; q = 1,2,…, m; p ≠ q; s = 1,2,…, m(m−1)/2. Again, we get a new data matrix {Y i, Z is}. For each converted feature Z s, we use the t-score, instead of I (Z; Y), to rank the association between Z and Y, since Y ∈ {0, 1}.
The individually discriminant genes are also ranked by t- score.
Support Vector Machine Classifier and performance evaluation
Each gene pairs and each individually discriminant genes are ranked by t- score based on all samples. The Top N gene pairs and/or the Top N individually discriminant genes are selected as input features. Support Vector Machine (SVM) Classifier is available at http://www.csie.ntu.edu.tw/~cjlin/libsvm/ 10. We simply use the average accuracy of five-fold cross-validation (CV) to evaluate the classifier performance as the datasets involved in this paper have balanced numbers of positive and negative samples.
| 7 |
Here TP, TN, FP, FN denote true positives, true negatives, false positives and false negatives respectively.
Results and Discussion
Comparing gene pairs selected by different methods
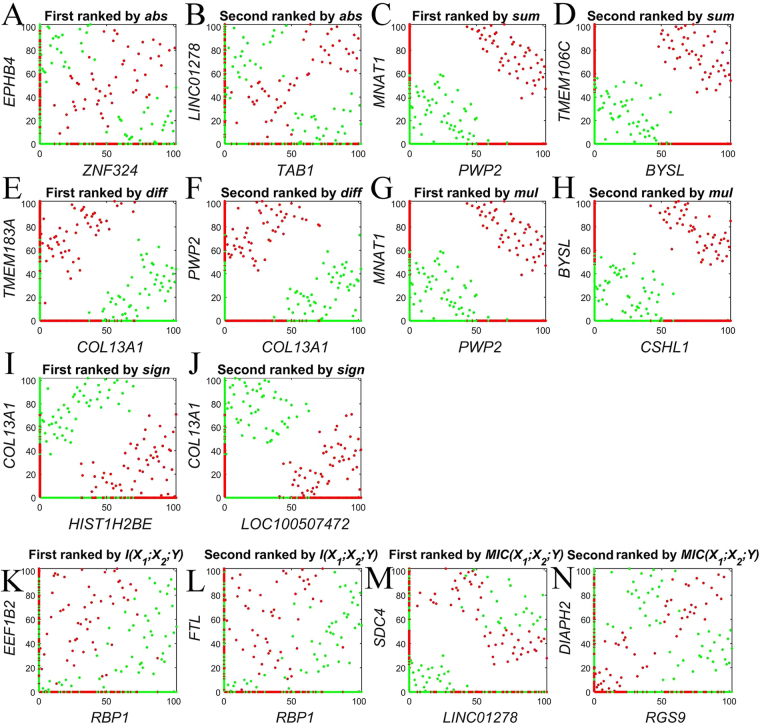
Figure 2 illustrates the scatterplot of the top-two gene pairs selected by abs conversion type and six reference methods in Prostate1 dataset11. In Fig. 2A,B,M and N, although the top-two synergic genes selected by abs conversion type and MIC(X 1; X 2; Y) are different, they share the same pattern: each individual gene is unrelated to cancer by individual gene evaluation, but the pair-wise genes are sufficient to distinguish the cancer from normal samples. Figure 2C–L are the top-two gene pairs selected from sum, diff, mul, sign and dendrogram-based I(X 1; X2; Y) methods. As an example (Fig. 2C), the higher the gene PWP2 expression level, the more likely to suffer cancer. The gene MNAT1 showed similar pattern as PWP2. Thus, these two genes (PWP2 and MNAT1) are related with cancer directly. However, they are individually discriminant rather than synergic genes. In a word, only abs conversion type and MIC(X 1; X 2; Y) can capture typical pair-wise synergies, dendrogram-based I(X 1; X 2; Y) and doublets are helpless for discovering pair-wise gene interactions.
Figure 2.
Top2 gene pairs selected by different methods in Prostate1 dataset. Red and green dots represent cancer and control, respectively. Gene expression levels are represented by the ranked values. K and L are from dendrogram-based I(X 1; X 2; Y)4, M and N are from MIC(X 1; X 2; Y)5.
We then compared the overlaps among the informative genes selected by Ind, Sum, Diff, Mul, Sign and Abs methods (Table 2). Clearly, a considerable number of similar informative genes can be detected by the first five methods. On the contrary, the informative genes selected by Abs method have little overlap with the informative genes selected by the others.
Table 2.
Overlaps among the informative genes selected by different methods in the Prostate1 dataset.
| Ind(100) | Sum(98) | Diff(94) | Mul(70) | Sign(128) | Abs(132) | |
|---|---|---|---|---|---|---|
| Ind(100) | ||||||
| Sum(98) | 35 | |||||
| Diff(94) | 36 | 41 | ||||
| Mul(70) | 23 | 20 | 21 | |||
| Sign(128) | 25 | 28 | 30 | 18 | ||
| Abs(132) | 1 | 0 | 0 | 0 | 0 |
Ind(100): The Top 100 individually discriminant genes selected by t-test. Sum (98): The Top 100 gene pairs selected by Sum conversion type and t-test, 98 genes reserved after removing repeated genes; the others as well.
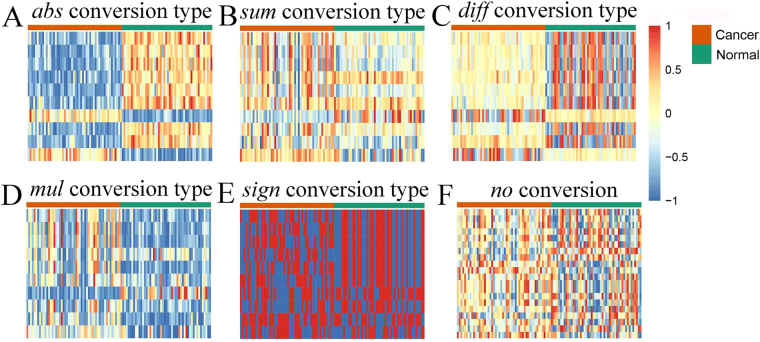
Given the top10 pair-wise synergic genes (16 genes) selected by abs conversion type, Fig. 3 contains the heat maps generated by these genes with different conversion type. Only the heat maps with abs conversion type (Fig. 3A) and diff conversion type (Fig. 3C) can distinguish cancer from normal samples. In diff conversion type, the Z values are medium in cancer samples, but they are either low or high in normal samples, and vice versa. Therefore, the pair-wise synergic genes converted by diff will receive low t-scores and cannot be highlighted.
Figure 3.
The heat maps generated by the same top10 synergic genes which were selected by abs conversion type. Each row corresponds to a pair of genes (A–E) or a gene (F), and each column corresponds to a sample. Gene expression levels are represented by the ranked values, and normalized to [−1, 1].
To answer whether the synergic genes selected by abs conversion type have any biological relevance to cancer, we further validated the top10 gene pairs (16 genes) according to UniHI12 database (http://www.unihi.org/) and PubMed (Table 3). UniHI is an enhanced database for retrieval and interactive analysis of human molecular interaction networks. In Top10 gene pairs, so far we have found two gene pairs (PARP1-HMGB1 and CCHCR1-GRAP) that are associated with interaction in UniHI. The interaction between PARP1 and HMGB1 has been verified by Dara et al. (2007)13, the activation of PARP1 induces release of the pro-inflammatory mediator HMGB1 from the nucleus13–15. Of the 16 genes, 15 of them have been reported to relate to cancer. Four of them have been reported to relate to prostate cancer directly. Although LINC01278 has not yet been reported to relate to cancer, abs conversion type suggests that it is an important informative gene. LINC01278 occurred three times in the top 10 gene pairs (Table 3), and should be given proper attention.
Table 3.
The top10 synergic genes selected by abs conversion type in Prostate1 dataset.
| Pair-wise synergic Genes | Related carcinoma and Ref. |
|---|---|
| ZNF324–EPHB4 | Breast cancer20 – Prostate cancer21 |
| TAB1–LINC01278 | Breast cancer22 – Unreported |
| CDH22–LINC01278 | Colorectal cancer23 – Unreported |
| KLF7–EXT1 | Oral carcinoma24 – Cartilage-capped tumor25 |
| SIPA1L3–LINC01278 | Breast cancer26 – Unreported |
| KLF7–DDR2 | Oral carcinoma24 – Lung cancer27 |
| MMP23A–DIP2C | Bladder cancer28 – Breast and lung cancer29 |
| CARM1–EPHB4 | Prostate cancers30 – Prostate cancer21 |
| CCHCR1–GRAP | Skin cancer31 – Medullary thyroid carcinoma32 |
| PARP1–HMGB1 | Prostate cancer33 – Prostate cancer13 |
Classifier cannot learn well if synergic genes have not been converted properly
Although we get the pair-wise synergic genes based on abs conversion type, Fig. 3F suggests that the no conversion feature (X or R) cannot distinguish cancer from normal samples. It also indicates that the input features for classifiers should be conversion feature Z (Fig. 3A). Therefore, we conducted an experiment to further validate this hypothesis. Ten simulation datasets were generated according to Table 4; their prediction accuracy of 5 fold cross-validation is listed in Table 5.
Table 4.
Ten simulation datasets and their input features.
| Dataset | Function | No converted input features | Converted input features |
|---|---|---|---|
| 1 | Y = |X 1 − X 2| = Z 1 | {X 1, X 2} | {Z 1} |
| 2 | Y = |X 1 − X 2| + |X 3 − X 4| = Z 1 + Z 2 | {X 1, X 2, X 3, X 4} | {Z 1, Z 2} |
| … | … | … | … |
| 10 | Y = |X 1 − X 2| + |X 3 − X 4| + … + |X 19 − X 20| = Z 1 + Z 2 + … + Z 10 | {X 1, X 2, X 3, X 4,…, X 19, X 20} | {Z 1, Z 2,…, Z 10} |
Here, X is assigned with random values between 0 and 1, and Y is binarized with the median. Sample size for each dataset is 200.
Table 5.
Prediction accuracy with converted and not converted input features.
| Dataset | SVM-RBFa | SVM-linearb | SVM-polyc | SVM-sigd | RF | ANNs | DT | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Con. | No con. | Con. | No con. | Con. | No con. | Con. | No con. | Con. | No con. | Con. | No con. | Con. | No con. | |
| 1 | 0.985 | 0.985 | 0.990 | 0.605 | 1.00 | 0.56 | 0.990 | 0.540 | 1.00 | 0.865 | 1.00 | 0.975 | 0.995 | 0.895 |
| 2 | 0.970 | 0.905 | 0.975 | 0.600 | 0.985 | 0.640 | 0.995 | 0.455 | 0.960 | 0.795 | 0.990 | 0.930 | 0.965 | 0.785 |
| 3 | 0.985 | 0.860 | 0.975 | 0.465 | 0.980 | 0.575 | 0.975 | 0.500 | 0.860 | 0.780 | 0.995 | 0.910 | 0.900 | 0.705 |
| 4 | 0.960 | 0.810 | 0.925 | 0.515 | 0.985 | 0.400 | 0.980 | 0.420 | 0.850 | 0.655 | 0.985 | 0.825 | 0.865 | 0.695 |
| 5 | 0.970 | 0.790 | 0.910 | 0.535 | 0.965 | 0.550 | 0.980 | 0.460 | 0.810 | 0.615 | 0.995 | 0.780 | 0.840 | 0.600 |
| 6 | 0.945 | 0.815 | 0.860 | 0.500 | 0.985 | 0.475 | 0980 | 0.485 | 0.770 | 0.620 | 0.990 | 0.770 | 0.795 | 0.615 |
| 7 | 0.940 | 0.715 | 0.905 | 0.530 | 0.980 | 0.500 | 0.980 | 0.535 | 0.865 | 0.610 | 0.985 | 0.670 | 0.795 | 0.585 |
| 8 | 0.970 | 0.675 | 0.955 | 0.410 | 0.970 | 0.455 | 0.955 | 0.455 | 0.760 | 0.545 | 0.995 | 0.695 | 0.760 | 0.610 |
| 9 | 0.955 | 0.660 | 0.885 | 0.515 | 0.960 | 0.460 | 0.955 | 0.435 | 0.790 | 0.510 | 0.990 | 0.665 | 0.770 | 0.580 |
| 10 | 0.955 | 0.655 | 0.860 | 0.480 | 0.955 | 0.525 | 0.975 | 0.525 | 0.735 | 0.520 | 0.960 | 0.600 | 0.750 | 0.625 |
Here, a: SVM with radial basis function (RBF) kernel; b: SVM with linear kernel; c: SVM with polynomial kernel; d: SVM with sigmoid kernel. RF: Random Forest; ANNs: artificial neuron network; DT: Decision Tree; Con: the converted input features; No con: the not converted input features.
For the less input features (e.g dataset1 and dataset2) (Table 5), all of the seven models perform well by applying with the converted features, whereas only two models (SVM-RBF and ANNs) perform well by applying with the not- converted features. For the larger input features (e.g dataset9 and dataset10) (Table 5), although four models (SVM-RBF, SVM-poly, SVM-sig and ANNs) still perform well by applying with the converted features, none of these seven models perform well by applying with the not converted features. Thus, we can conclude that pair-wise synergic genes should be converted into new variables (Z) prior to be used as input features for classifiers, especially for many pairs of synergistic genes.
This is a surprising and important discovery. Suppose phenotype Y is determined by individually discriminant genes X 1 and X 2, and pair-wise synergic genes X 3–X 4 and X 5–X 6. In other words, the true genetic model is , and the true optimal subset is {X 1, X 2, X 3, X 4, X 5, X 6}, X 7–X 1000 are genes unrelated to Y. Now we get the dataset {Y, X 1, X 2,…, X 1000} and want to construct a genomic prediction model16 based on machine learning, but don’t know the true genetic model. Even the individual discriminant genes X 1 and X 2 can be highlighted by t-test, and the synergic genes X 3, X 4, X 5 and X 6 can be highlighted by Abs conversion type or MIC(X 1; X 2; Y), classifier cannot learn well when the input features space is {X 1, X 2, X 3, X 4, X 5, X 6}. It means that learning machine can never tell us the true optimal subset, if synergic genes have not been converted properly. This indicates the complexity of genomic prediction, also provides a new explain for “missing heritability” in GWAS study.
Combining individually discriminant and synergic genes can improve prediction performance
To further validate the reliability of synergic genes selected by abs conversion type, we also evaluated the prediction performance of individually discriminant and synergic genes with three more recent and larger publicly available datasets (Lung, Prostate2 and Cardiovascular) (see Table 1). Meantime, the label randomization tests were performed. The top individually discriminant genes are selected by t-test, the top synergic genes are selected by abs conversion type + t-test. Here, we take the individually discriminant genes and/or converted synergic genes as the input features for the SVM-RBF classifier.
Table 6 illustrates the prediction of accuracy in different schemes of input features. The results show that: 1) By using the individually discriminant genes as input features alone, the average accuracies for Top10_Ind, Top20_Ind and Top40_Ind are 77.30%, 78.74% and 80.36%, respectively. By using the synergic genes as input features alone, the average accuracies for Top5_Syn, Top10_Syn and Top20_Syn are 75.58%, 81.67% and 84.63%, respectively. These indicate that the synergic genes receive comparable accuracy to the individually discriminant genes using the same number of genes. 2) When the input features involves 20 genes, the average accuracies for Top20_Ind, Top10_Syn and Top10_Ind + Top5_Syn are 78.74%, 81.67%, and 83.74%, respectively. When the input features involves 40 genes, the average accuracies for Top40_Ind, Top20_Syn and Top20_Ind + Top10_Syn are 80.36%, 84.63%, and 85.75%, respectively. These indicate that combining individually discriminant and synergic genes, rather than only using the individually discriminant genes or the synergic genes, can receive better prediction accuracies. 3) The classification performances of the label randomization tests drop to random, it validate the reliability of synergic genes selected by abs conversion type.
Table 6.
Prediction accuracies of 5-fold CV in different schemes of input features (%).
| Input features | Lung | Prostate2 | Cardiovascular | Average |
|---|---|---|---|---|
| Top10_Ind | 74.41 (43.81) | 84.20 (64.39) | 73.29 (63.22) | 77.30 (57.14) |
| Top20_Ind | 76.49 (43.31) | 85.13 (61.08) | 74.59 (61.65) | 78.74 (55.35) |
| Top40_Ind | 75.93 (46.02) | 84.20 (61.09) | 80.96 (62.95) | 80.36 (56.69) |
| Top5_Syn | 76.54 (47.03) | 74.52 (62.25) | 75.67 (62.99) | 75.58 (57.42) |
| Top10_Syn | 84.44 (50.28) | 76.18 (55.90) | 84.40 (61.38) | 81.67 (55.85) |
| Top20_Syn | 83.98 (47.06) | 80.20 (62.96) | 89.70 (62.17) | 84.63 (57.40) |
| Top10_Ind + Top5_Syn | 82.33 (48.17) | 86.34 (62.27) | 82.55 (63.22) | 83.74 (57.89) |
| Top20_Ind + Top10_Syn | 83.91 (40.11) | 86.31 (57.54) | 87.04 (62.44) | 85.75 (53.36) |
Ind represents the individually discriminant genes, Syn represents the synergic genes. A number in parentheses indicates the result of label randomization test.
The minimum number of individually discriminant and synergic genes required in the optimal subset remains to be determined by the further research.
We also compared the prediction performance of the 5 conversion types (Table 7). The results show that the genes selected by Abs conversion type have more powerful ability to improve prediction performance for the individually discriminant model than the genes selected by the other conversion types.
Table 7.
Prediction accuracies of 5-fold CV in different conversion types (%).
| Features | Lung | Prostate2 | Cardiovascular | Average |
|---|---|---|---|---|
| Top20_Ind | 76.49 | 85.13 | 74.59 | 78.73 |
| Top10_Sum | 80.68 | 81.61 | 78.83 | 80.37 |
| Top10_Diff | 83.37 | 85.84 | 76.97 | 82.06 |
| Top10_Mul | 80.81 | 81.61 | 79.09 | 80.50 |
| Top10_Sign | 78.08 | 84.68 | 79.38 | 80.71 |
| Top10_Abs | 84.44 | 76.18 | 84.40 | 81.67 |
| Top10_Sum + Top20_Ind | 79.70 | 85.14 | 80.42 | 81.75 |
| Top10_Diff + Top20_Ind | 82.33 | 84.44 | 83.33 | 83.37 |
| Top10_Mul + Top20_Ind | 78.11 | 86.55 | 79.64 | 81.43 |
| Top10_Sign + Top20_Ind | 81.35 | 84.43 | 76.21 | 80.66 |
| Top10_Abs + Top20_Ind | 83.91 | 86.31 | 87.04 | 85.75 |
Top20_Ind: The Top20 individually discriminant genes selected by t-test. Top10_Sum: the Top10 gene pairs selected by Sum conversion types + t-test, the others as well.
Conclusion
In this paper, we propose a fast approach based on the combination of abs conversion type and t-test, to detect gene–gene synergy. We find that dendrogram-based I(X 1; X 2; Y) and doublets are helpless for discovering pair-wise gene interactions, and the synergic genes selected by our method and the MIC(X 1; X 2; Y) method are consistent with the typical pair-wise synergy. However, MIC(X 1; X 2; Y) has a higher computational cost. For example, the running time of the entire process on Prostate1 dataset (12,600 × 12,599/2 gene pairs) by MIC(X 1; X 2; Y) method is approximately 20 hours (Intel Core i5-4590@3.3 GHz), whereas it is only 47 minutes by our method. Experiments on simulated and real-world data showed that combining the individually discriminant genes selected by t-test and the synergic genes selected by our methods can improve prediction performance. These synergic genes should be converted into new variables (Z) prior to be used as input features for classifiers.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (61701177 to Y.C.), the Science Research Projects of Hunan Provincial Department of Education (1071 to Z.Y.). We thank Dr. Alicia K. Byrd for helpful suggestions.
Author Contributions
P.X., Y.C., L.B and Z.Y. conceived and designed the experiments. P.X. and Y.C performed the experiments. P.X., Y.C., J.G., L.B and Z.Y. analyzed the data. P.X., J.G. and Z.Y. wrote the paper. All the authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Pengwei Xing and Yuan Chen contributed equally to this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lianyang Bai, Email: bailianyang2005@aliyun.com.
Zheming Yuan, Email: zhmyuan@sina.com.
References
- 1.Jafari P, Azuaje F. An assessment of recently published gene expression data analyses: reporting experimental design and statistical factors. BMC Medical Informatics and Decision Making. 2006;6:27. doi: 10.1186/1472-6947-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neumann U, Genze N, Heider D. EFS: an ensemble feature selection tool implemented as R-package and web-application. Biodata Mining. 2017;10:21. doi: 10.1186/s13040-017-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anastassiou D. Computational analysis of the synergy among multiple interacting genes. Molecular Systems Biology. 2007;3:83. doi: 10.1038/msb4100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watkinson J, Wang X, Tian Z. & Anastassiou, Dimitris. Identification of gene interactions associated with disease from gene expression data using synergy networks. BMC Systems Biology. 2008;2:1–16. doi: 10.1186/1752-0509-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, et al. Discovering Pair-wise Synergies in Microarray Data. Scientific Reports. 2016;6:30672. doi: 10.1038/srep30672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chopra P, Lee J, Kang J, Lee S. Improving Cancer Classification Accuracy Using Gene Pairs. PloS One. 2010;5:e14305. doi: 10.1371/journal.pone.0014305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geman D, et al. Classifying gene expression profiles from pairwise mRNA comparisons. Statistical Applications in Genetics & Molecular Biology. 2004;3:Article19. doi: 10.2202/1544-6115.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, et al. Informative gene selection and the direct classification of tumors based on relative simplicity. BMC Bioinformatics. 2016;17:1–16. doi: 10.1186/s12859-015-0844-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 10.Chang C, Lin C. LIBSVM: A library for support vector machines. Acm Transactions on Intelligent Systems & Technology. 2011;2:389–96. doi: 10.1145/1961189.1961199. [DOI] [Google Scholar]
- 11.Singh D, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203. doi: 10.1016/S1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 12.Kalathur, R. K. R. et al. UniHI 7: an enhanced database for retrieval and interactive analysis of human molecular interaction networks. Nucleic Acids Research42(Database issue), D408 (2014). [DOI] [PMC free article] [PubMed]
- 13.Dara DW-XZ, Craig B. Thompson. Activation of Poly(ADP)-ribose Polymerase (PARP-1) Induces Release of the Pro-inflammatory Mediator HMGB1 from the Nucleus. Journal of Biological Chemistry. 2007;282:17845. doi: 10.1074/jbc.M701465200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma A, et al. Overexpression of high mobility group (HMG) B1 and B2 proteins directly correlates with the progression of squamous cell carcinoma in skin. Cancer Investigation. 2008;26:43–51. doi: 10.1080/07357900801954210. [DOI] [PubMed] [Google Scholar]
- 15.Gnanasekar M, et al. HMGB1: A Promising Therapeutic Target for Prostate Cancer. Prostate Cancer. 2013;10:157103. doi: 10.1155/2013/157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bermingham ML, et al. Application of high-dimensional feature selection: evaluation for genomic prediction in man. Scientific Reports. 2015;5:10312. doi: 10.1038/srep10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spira A, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nature Medicine. 2007;13:361–366. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 18.Penney KL, et al. Association of Prostate Cancer Risk Variants with Gene Expression in Normal and Tumor Tissue. Cancer Epidemiology, Biomarkers & Prevention. 2015;24:255–260. doi: 10.1158/1055-9965.EPI-14-0694-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellsworth DL, et al. Intensive Cardiovascular Risk Reduction Induces Sustainable Changes in Expression of Genes and Pathways Important to Vascular Function. Circulation-cardiovascular Genetics. 2014;7:151–160. doi: 10.1161/CIRCGENETICS.113.000121. [DOI] [PubMed] [Google Scholar]
- 20.Lacroix M. Significance, detection and markers of disseminated breast cancer cells. Endocrine Related Cancer. 2006;13:1033. doi: 10.1677/ERC-06-0001. [DOI] [PubMed] [Google Scholar]
- 21.Xia G, et al. EphB4 expression and biological significance in prostate cancer. Cancer Research. 2005;65:4623–32. doi: 10.1158/0008-5472.CAN-04-2667. [DOI] [PubMed] [Google Scholar]
- 22.Neil JR, et al. TAB1:IκB Kinase Interaction Promotes Transforming Growth Factor β–Mediated Nuclear Factor-κB Activation during Breast Cancer Progression. Cancer Research. 2008;68:1462–70. doi: 10.1158/0008-5472.CAN-07-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou J, et al. Over-Expression of CDH22 Is Associated with Tumor Progression in Colorectal Cancer. Tumor Biology. 2009;30:130–40. doi: 10.1159/000225242. [DOI] [PubMed] [Google Scholar]
- 24.Ding X, et al. KLF7 overexpression in human oral squamous cell carcinoma promotes migration and epithelial-mesenchymal transition. Oncology Letters. 2017;13:2281–2289. doi: 10.3892/ol.2017.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mccormick C, et al. The putative tumour suppressor EXT1 alters the expression of cell-surfaceheparan sulfate. Nature Genetics. 1998;19:158. doi: 10.1038/514. [DOI] [PubMed] [Google Scholar]
- 26.Jönsson G, et al. Genomic subtypes of breast cancer identified by array-comparative genomic hybridization display distinct molecular and clinical characteristics. Breast Cancer Research. 2010;12:R42. doi: 10.1186/bcr2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammerman PS, et al. Mutations in the DDR2 Kinase Gene Identify a Novel Therapeutic Target in Squamous Cell Lung Cancer. Cancer Discovery. 2011;1:78. doi: 10.1158/2159-8274.CD-11-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matullo, G. et al. Abstract 778: DNA repair capacity, chromosomal damage, methylation and gene expression levels in bladder cancer: An integrated analysis 76, 778–778 (2016).
- 29.Larsson. et al. DIP2C regulates expression of the tumor suppressor gene CDKN2A. Genomics (2014).
- 30.Kim YR, et al. Differential CARM1 expression in prostate and colorectal cancers. BMC cancer. 2010;10:1–13. doi: 10.1186/1471-2407-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suomela S, et al. CCHCR1 Is Up-Regulated in Skin Cancer and Associated with EGFR Expression. PloS one. 2009;4:e6030. doi: 10.1371/journal.pone.0006030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludwig L, et al. Expression of the Grb2-related RET adapter protein Grap-2 in human medullary thyroid carcinoma. Cancer Letters. 2009;275:194–7. doi: 10.1016/j.canlet.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Schiewer MJ, et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discovery. 2012;2:1134. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]