Abstract
Acute myeloid leukaemia (AML) is a type of cancer affecting all ages but it is more common in adults, as compared to children. Recent advancements in proteomics and mass spectrometry tools, offer a comprehensive solution to study the molecular complexity of diseases, such as cancers. This study is focused on the proteomic profiling of AML in comparison to healthy control for which, a systematic 5D proteomic approach for the fractionation of pooled plasma samples was used. Methodology includes depletion of Top-7 abundant proteins, ZOOM-isoelectric focusing (ZOOM-IEF), two-dimensional gel electrophoresis (2-DGE), and matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) analysis followed by the validation of identified biomarker proteins using enzyme linked immunosorbent assay (ELISA). Up-/down-fold changes in concentration of proteins were observed in 2-DGE of AML in comparison with the healthy control and a total of 34 proteins were identified in fractioned plasma. Among them, fifteen proteins were significantly differentiated and five proteins; SAA1, complement factor C7, ApoE, plasminogen, and ApoA1 were later verified by ELISA in individual samples, which showed that SAA1 and plasminogen could be used as potential biomarker for AML.
Introduction
Acute myeloid leukaemia (AML) is an aggressive blood disease comprising of many subtypes with the dominant features of infiltration and accumulation of myeloid lineage hematopoietic blast cells in bone marrow and peripheral blood1,2. It is common in adults, and its incidence rate increases with increase in age however, also affects small percentage of children (10–15%)3. Though 40–50% of the older patients with AML achieve complete remission, but 5-years survival rate in patients above 65 years is 5% due to increased rate of relapse1. Sub-classification is based on cellular morphology, hematopoietic lineage as well as common translocations and mutations4. Among cytogenetic abnormalities, the most common abnormality is internal tandem repeats of the FLT-3 gene. Some cases show chromosomal translocation inv(16) and t(8; 21) generating fusion protein which involve the core binding factor (CBF) genes. These, together with t(15; 17) variant show good prognosis. Many cases of AML also carry mutations in the nucleoplasmin (NPM) gene. Though different AML subtypes are considered as separate genetic diseases, but they can be grouped together because usually their treatment and prognosis are similar. Clinical features include low platelets count and increased tendency to bleed along with disseminated intravascular coagulation. In advanced cases, metastasis to tissues, such as skin, gums, and central nervous system tissues also occurs. Prognosis of patients and planning of treatment largely depends on cytogenetic and molecular analysis. Treatment strategy for AML is both supportive and specific involving the use of intensive and selective chemotherapy. Another treatment option is stem cell transplant which minimizes the rate of relapse but adds further toxicity, because of the treatment regime5.
Proteomic profiling using various approaches including mass spectrometry are being effectively used as a powerful tool for the understanding of diseases and identification of biomarkers particularly in cancers6. However, identification of reliable and sensitive protein biomarker depends on many factors specially the proteomic fractionation strategy being applied. A number of studies based on mass spectrometry, combining proteomic technologies such as two-dimensional gel electrophoresis (2-DGE), difference gel electrophoresis (DIGE), immunoprecipitation, and affinity chromatography in combination with matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), surface-enhanced laser desorption/ionization time-of-flight (SELDI-TOF) mass spectrometry, liquid chromatography–mass spectrometry (LC-MS) and shotgun proteomics have been investigated for the proteomic profiling of AML to reveal new insights and diagnostic biomarkers7–12.
Our key objective in this study is to identify the proteomics based biomarkers for AML in comparison to healthy control using 5D protein strategy i.e. depletion of most abundant proteins using fast protein liquid chromatography (FPLC), ZOOM-IEF, 2-DGE, and MALDI-MS analysis followed by the validation of identified biomarker proteins through ELISA. In-solution isoelectric focusing (IEF) helps in the detection of low abundant proteins and improves the detection range because large amounts of proteins of specific pH can be applied to narrow pH range gel13. An early study has evaluated the technical use of microscale solution-phase IEF after immunodepletion of human proteome organization (HUPO) plasma, combined with narrow range 2-DGE, and stated to be effective and reproducible14. Therefore, it is expected that this developed strategy will be useful for the quest of proteomic biomarkers in other diseases as well. Moreover, similar fractionation strategy combining with MALDI-MS has already been used for the biomarker profiling of renal cancer patients15.
Experimental
Auxological data of subjects
A total of fasting 50 plasma samples of AML were included in this study. The subtypes of AML which were included in this study are listed in Table S1. A total of 50 healthy male and 50 female healthy volunteers were also selected for the study (Table S2). All healthy individuals were free of AML, and other haematological diseases, and no previous familial history of AML. At the time of sampling, the physical condition was good, with normal vital signs and no history of any other disease.
Sample collection and processing
The study was approved by the Independent Ethics Committees, National Institute of Blood Diseases & Bone Marrow Transplantation Hospital, Karachi, and the International Centre for Chemical and Biological Sciences (ICCBS), University of Karachi, Pakistan. All samples were collected according to approved protocol; a written informed consent and a thorough questionnaire was filled by every patient and healthy volunteer. Sample collection was carried out in accordance with relevant guidelines and regulations. Individual blood samples were collected, processed, and stored according to the human proteome organization (HUPO) standard protocol16. Human blood (5 mL) was transferred by venepuncture into evacuated blood collection tubes, containing K2-ethylenediaminetetra acetic acid (K2-EDTA). The plasma was separated by centrifugation at 2,200 × g for 10 min at 4 °C. Pool of individual samples were used in this study, because pooling is cost effective, include greater statistical power, improved ability to compare results and validation of models. Pooled plasma has already been applied in many studies17–20. To make healthy pool, equal volumes of each individual healthy plasma sample ware mixed to obtain the healthy Pakistani pooled plasma. Similarly, AML samples were also pooled by combining equal amounts of every AML specimen. Pooled plasma samples were then subjected to aliquoting, and stored at −80 °C until further processing.
1D SDS-PAGE analysis of samples
One-dimensional sodium dodecyl sulphate polyacrylamide gel electrophoresis (1D SDS-PAGE) was performed for comparative analysis of healthy and diseased samples on X Cell SureLock system (Invitrogen). Chemicals and reagents for 1D SDS-PAGE were purchased from Invitrogen (USA). β-mercaptoethanol, sucrose, and Tris HCL were purchased from Sigma Aldrich (USA).
Depletion of abundant proteins through MARS column
Depletion of top seven most abundant proteins was carried out using Multiple Affinity Removal Column (MARS) Hu-7 (4.6 × 50 mm), purchased from Agilent (USA) on ÄKTA™ FPLC system (GE Healthcare, Sweden). The column has an affinity for the albumin, IgG, IgA, transferrin, antitrypsin, haptoglobin (HPT), and fibrinogen in plasma. Protease inhibitors; ethylene diamine tetra-acetic acid (EDTA), leupeptin, pepstatin-A, and phenyl methanesulphonyl flouride (PMSF) were purchased from Sigma Chemicals (USA). 500 mL Vacuum Filter/Storage Bottle System, 0.22 µm was purchased from Corning (USA). The plasma sample (300 µL) was depleted according to the kit protocol. The plasma amount used was according to the column capacity, and required sample size for multiple replicate depletions. 1D SDS-PAGE analysis of all fractions, including bound and unbound fractions was performed to check the sample recovery after depletion. Unbound fractions were pooled and concentrated using 5 kDa molecular weight cut off (MWCO) tubes for both pools. For concentrating the pooled bound fractions, the sample was centrifuged several times at 3,500 rpm for 15 min at 4 °C to obtain an amount of 200 µL. Enrichment efficiency was checked by loading the equivalent to 0.1 µL plasma from the fractions before concentrating and after concentrating.
Reduction and alkylation
Urea was purchased from Invitrogen (USA), tris(hydroxymethyl)aminomethane (Tris) from Boehringer Mannheim (Germany), dichlorodiphenyltrichloroethane (DTT) and iodoacetamide (IAM) from SERVA (Germany). Acetone, and trichloroacetic acid (TCA) were purchased from Fisher Scientific (UK) and Scharlau (Spain), respectively. For denaturation and pH adjustment, concentrated depleted sample was adjusted to 8 M urea and 20 mM Tris. To reduce the proteins, the sample was adjusted to 20 mM DTT, and subsequently for alkylation, to 50 mM IAM. To quench the alkylation, the sample was adjusted to 1% DTT. Protein precipitation was carried out using TCA/acetone protocol with some modifications21. Sample, ice-cold acetone and 100% TCA solution, were mixed and placed at −20 °C for 1 hr. to precipitate the proteins. The tube was then centrifuged at 5,000 rpm for 30 min to have the proteins pelleted down. The pellet was washed two to three times with ice-cold acetone.
ZOOM-IEF
The ZOOM-IEF® Fractionator Combo Kit including ZOOM-IEF fractionator was purchased from Invitrogen (USA). The sample pellet was dissolved in ZOOM buffer by vortexing for 5 min, followed by the addition of 26 µL of ZOOM carrier ampholytes (pH 3–10) along with small amount of bromophenol blue. To remove the fine particles, the sample was centrifuged at 12,000 rpm for 1 min through 0.22 µm spin filter. Fractionation was performed according to established protocol22. 1D SDS-PAGE analysis of all five ZOOM fractions was performed to check the efficiency of IEF, and to compare the samples from healthy persons with AML patients.
2-D gel electrophoresis
2-DGE was performed on Bio-Rad PROTEAN IEF cell. The ReadyPrep 2-D Starter Kit, ReadyStrip IPG Strips, Ready Gel precast gel, mineral oil, 10X Tris/glycine/SDS Buffer, and paper wicks were purchased from Bio-Rad (USA). Buffers were prepared according to the kit protocol. 125 µL of freshly prepared rehydration/sample buffer for 7 cm IPG strip was added in a conical centrifuge tube containing the sample pellet, and vortexed to dissolve the pellet. The sample was centrifuged to settle down the fine particles. The whole procedure was performed according to the kit protocol. Gel images were taken through Gel DOC 800 system (Bio-Rad, USA).
Mass spectrometric analysis
Analysis was performed using MALDI-TOF-TOF MS (Ultraflex III, Broker Daltonics Germany). Mass spectrometric profile was obtained by flexAnalysis version 3.0. (Bruker Daltonics). The protein spots of interest were extracted from the stained gels using the manual cutting procedure and digested according to formerly mentioned protocol23,24.
The digested peptides were analysed using standard protocol25. Briefly, the samples were mixed with equal amounts of freshly prepared α-cyano-4-hydroxycinnamic acid in acetonitrile (ACN) in 1:1 ratio. Calibration of MALDI-TOF was carried out in the reflector positive mode using peptide calibrant standard I (Bruker Daltonics). A 337-nm nitrogen laser and a 2 GHz digitilizer were used. Mass spectra were obtained with 25 KV of ion acceleration, 6 KV lens potential, and high gating strength to deflect ions with a mass below 500 m/z values. Spectra were obtained in the mass range of 500–3000. Every spectrum was the sum of 2000 laser shots within the same spot (200 shots/position) and intensity of 20–40%.
ELISA analysis
ELISA was performed for five proteins, serum amyloid A (SAA1), plasminogen, apolipoprotein E (ApoE), complement factor C7 and apolipoprotein A1 (ApoA1) on Thermo Fischer Scientific™ Multiskan™ FC Microplate Photometer (USA). ELISA kits were purchased from Crystalchem (USA) and Assaypro (USA). Plasma samples of 15 healthy individuals and 18 AML patients were diluted according to the kit protocol, using 1X diluent. Standard and diluted samples were dispensed into wells. After one-hour incubation, the samples were aspirated and the wells were washed 4 times using 1X wash buffer. The antibody-HRP conjugate was then applied for 20 min. On completion of this step, 4 times washing was carried out again using 1X wash buffer. The substrate solution was then added and left for 10 min for reaction completion. Stop solution was added and readings were taken at 450 and 630 nm. This procedure with some modifications according to the manufacturer’s protocol was applied to validate our results for selected proteins; complement factor C7, plasminogen, and ApoE, for which ELISA kits were purchased from Assaypro (USA).
Statistics and data analysis
The analysis of 2-DGE images, detection of spots, spot matching, and semi-quantitative statistical analysis were performed using the Bio-Rad PDQuest version 8.0.1. Bio-Rad (USA). Master gel was used to compare the gels of healthy and AML pool. The analysis involved matching the gels, differences and similarities in spots pattern, background subtraction, and removal of artefacts (horizontal and vertical streaks). T-test was used to study the differential protein expression among protein spots from healthy and AML pool gels, with p < 0.05 and four-fold change in spot intensity was selected as threshold. After automated matching, the detected spots were manually edited for greater accuracy.
After MALDI-MS the protein identification was carried out through MASCOT database search – Matrix Science, based on mass fingerprinting (PMF) using Swiss-Prot and NCBInr databases. Peptide modification which we have chosen as a fixed modification during the search, was carbamidomethylation of cystein. The oxidation of methionine was used as variable modification. The maximum number of missed cleavages was set to 1, peptide tolerance 100 ppm/1 Da, and p < 0.05 were used to identify proteins.
Identified proteins were further subjected to Gene Ontology (GO) based analysis to know the function. The connections between differently expressed proteins with each other and their connections with the other proteins were assessed by the STRING: EMBL (European Molecular Biology Laboratory) software26. Minimum required interaction score of medium confidence 0.400 was used.
Results
Fractionation of plasma samples
Using a systematic strategy (as shown in Fig. 1) pool plasma samples of AML and healthy subjects were first depleted for the top-7 abundant proteins through a MARS column. The resultant FPLC spectrum showed a clear separation of unbound and bound fractions, and was in the agreement with the figure provided by the manufacturer (Fig. S1). The resultant low-abundant proteins in flow through were then analysed by 1D SDS-PAGE for depletion efficiency which showed an effective protein depletion (Fig. S2). After depletion, the unbound portion was concentrated using 5 kDa MWCO tubes, followed by enrichment efficiency checking by 1D SDS-PAGE and found to be acceptable (Fig. S3).
Figure 1.
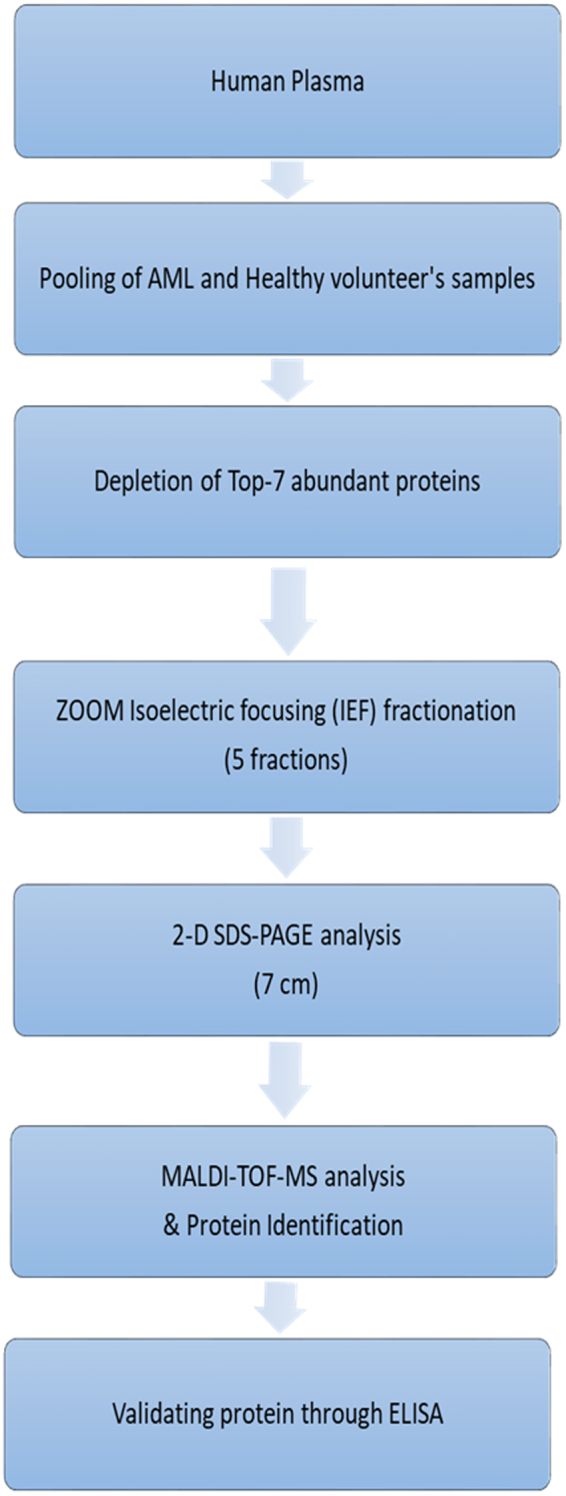
Scheme used for analysis of the human plasma sample.
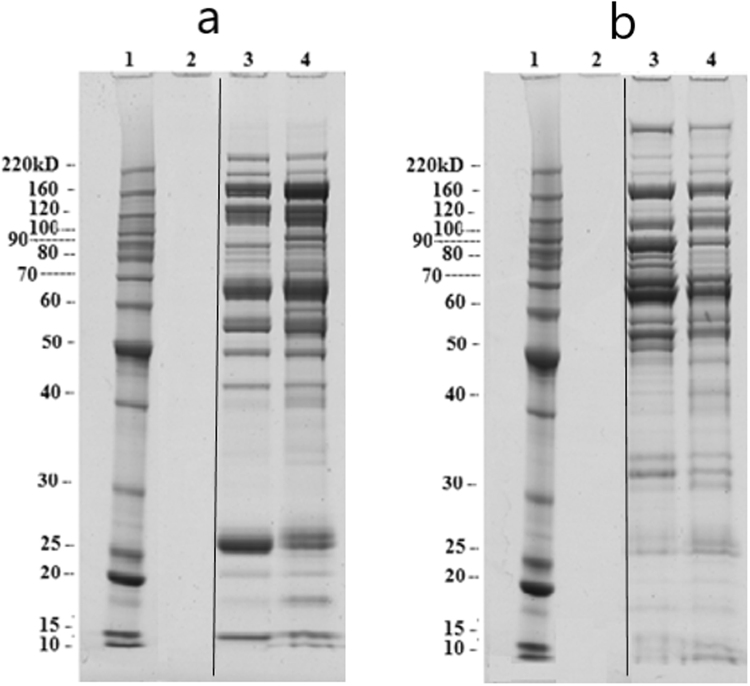
The unbound portion, after enrichment by 5 kDa MWCO and protein precipitation, was further resolved by ZOOM-IEF over a pH range of 3.0 to 10 into five fractions of different pH ranges; pH: 3.0–4.6, pH: 4.6–5.4, pH: 5.4–6.2, pH: 6.2–7.0, and pH: 7.0–10.0 (Fig. S4). 1D SDS-PAGE analysis of all fractions showed that among all fractions two of pH: 5.4–6.2 and pH: 6.2–7.0 from AML and healthy samples pool had many protein bands with some differential pattern in the molecular weight range of 10–266 kDa. Therefore, these two fractions were subjected to further analysis. A comparative 1D SDS-PAGE picture of these two fractions from AML and healthy samples pool is shown in Fig. 2.
Figure 2.
(a) Comparison of fraction-3 (pH: 5.4–6.2) after ZOOM_IEF. 1: Protein ladder, 2: Blank, 3: Healthy pool, 4: AML pool, (b) Comparison of fraction-4 (pH: 6.2–7.0) after ZOOM_IEF. 1: Protein ladder, 2: Blank, 3: Healthy pool, 4: AML pool. The amount of fractions loaded into gel was equivalent to 3 uL of original plasma.
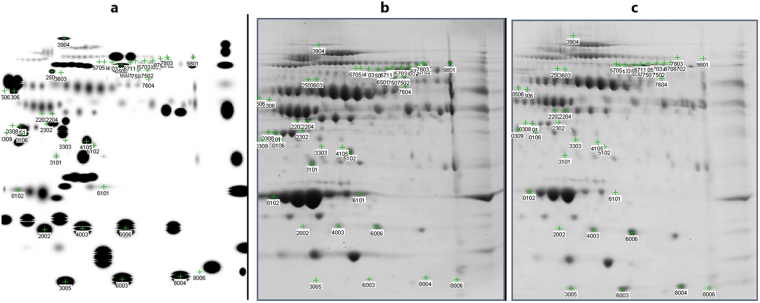
These two fractions were then mixed to make a single fraction of pH range 5.4 to 7.0. After protein precipitation, 2-DGE of this fraction was performed on IPG strip of pH range 4 to 7 followed by electrophoretic separation. Master gel image was created by combining the spots from both AML and healthy gels (Fig. 3a). That was used for comparison of AML gel with healthy gel image (Fig. 3a and b). The comparison between healthy and AML samples is shown in the scatter plot (Fig. S5). Out of 182 spots, 42 spots were having a 4X quantity difference, while 137 spots were with significance level ≥95%. The spots which were with 4X quantity difference and ≥95% significant both were 41. Altogether, 182 gel spots from the control and the AML samples, when analysed by MALDI-TOF MS, led to the identification of 34 distinct proteins and/or their respective isoforms and subunits (Fig. 4). The list of identified proteins with details such as theoretical and experimental pI and molecular weight, MASCOT score, sequence coverage, etc. is shown in Table 1. A sample Mascot Score Histogram of hemopexin protein is given in supplementary section (Fig. S6).
Figure 3.
Comparison of AML pool and healthy pool 2-DGE image. Highlighted spot numbers are those who are more than 95% significant and quantity changes more than 4-fold, which were further analysed. (a) Master gel, created my adding spots from AML pool and healthy pool gel images using PDQuest software. (b) 2-DGE map of healthy pool in the range of pH 5.4–7.0. (c) 2-DGE map of AML pool in the range of pH 5.4–7.0.
Figure 4.
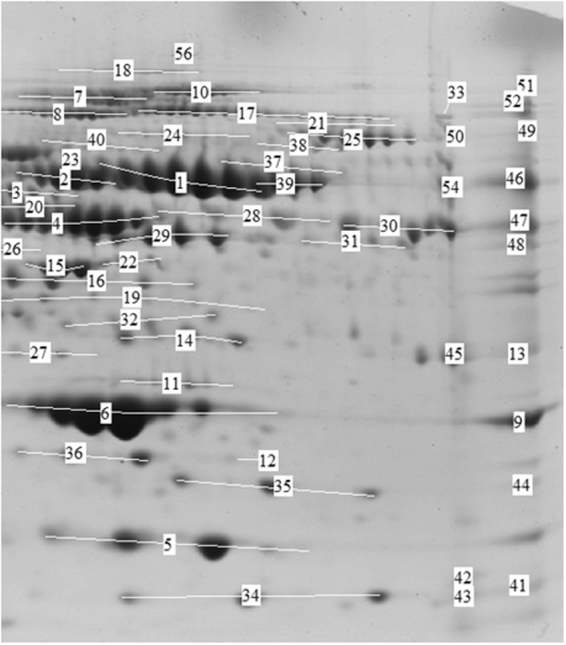
Identified variants and subunits of proteins in 2-DGE image through MALDI-MS and MASCOT database searching. [1, 39, 46: hemopexin, 2: alpha-1β-glycoprotein, 3, 20: kininogen-1, 4: Vit-D-binding protein, 5: transthyretin, 6, 9: apolipoprotein A-I, 7, 8, 52: ceruloplasmin, 10, 51, 55: complement factor H, 11: human serum amyloid-P component, 12, 13, 14, 32: apolipoprotein E, 15: apolipoprotein A-IV, 16, 35, 44: haptoglobin, 17, 53: alpha 2-macroglobulin, 18: fibronectin, 21: complement factor 7, 22: CD5 antigen-like, 23, 40: prothrombin, 24: complement C1r-subcomponent, 25: complement factor B, 26: complement C4-A, 27: alpha-1 microglobulin, 28, 29, 48: fibrinogen gamma chain, 30, 31, 47: fibrinogen beta chain, 33, 49, 50: plasminogen, 34, 41, 43: serum amyloid A-I, 36: retinol binding protein, 37, 39: C4b-binding protein alpha chain, 38: gelsolin, 39: human serum albumin, 42: serum amyloid A-IV, 43: haemoglobin β-component, 45: complement C4, 54: fibrinogen alpha chain].
Table 1.
List of 34 identified proteins through mass spectrometry and MASCOT database searching.
| Spot ID | Accession no. | Protein name | Score | Expect value | Matched peptides no. | Sequence coverage (%) | Molecular weight (Mr) | Isoelectric pH (pI) |
|---|---|---|---|---|---|---|---|---|
| 17, 53 | gi|224053 | Alpha-2-macroglobulin | 120 | 3.1e-07 | 38/146 | 29% | 162072 | 5.92 |
| 10, 51, 55 | CFAH_HUMAN | Complement factor H | 145 | 6.4e-11 | 45/136 | 35% | 143680 | 6.21 |
| 7, 8, 52 | CERU_HUMAN | Ceruloplasmin | 92 | 1.4e-05 | 19/107 | 26% | 122983 | 5.44 |
| 25 | CFAB_HUMAN | Complement factor B | 98 | 3.3e-06 | 20/90 | 30% | 86847 | 6.67 |
| 24 | C1R_HUMAN | Complement C1r subcomponent | 75 | 6.7e-04 | 16/64 | 21% | 81606 | 5.82 |
| 23, 40 | THRB_HUMAN | Prothrombin | 115 | 6.4e-08 | 18/88 | 34% | 71475 | 5.64 |
| 37, 39 | C4BPA_HUMAN | C4b-binding protein alpha chain | 109 | 2.5e-07 | 19/131 | 39% | 69042 | 7.15 |
| 26, 45 | gi|401871713 | Chain C, Complement C4 In Complex with Masp-2 | 80 | 0.003 | 12/104 | 38% | 33737 | 6.37 |
| 2 | A1BG_HUMAN | Alpha-1β-glycoprotein | 83 | 9.5e-05 | 18/125 | 35% | 54790 | 5.56 |
| 4 | VTDB_HUMAN | Vitamin D-binding protein | 79 | 2.5e-04 | 15/111 | 41% | 54526 | 5.40 |
| 1, 39, 46 | HEMO_HUMAN | Hemopexin | 130 | 2e-09 | 19/127 | 41% | 52385 | 6.55 |
| 28, 29, 48 | gi|223170 | Fibrinogen gamma chain | 76 | 7.5e-03 | 10/66 | 31% | 46823 | 5.54 |
| 3, 20 | gi|37748641 | Kininogen 1 | 112 | 2e-06 | 17/115 | 44% | 48954 | 6.29 |
| 16, 35, 44 | HPT_HUMAN | Haptoglobin | 114 | 8e-08 | 15/102 | 36% | 45861 | 6.13 |
| 18 | FINC_HUMAN | Fibronectin | 64 | 0.0079 | 43/179 | 18% | 266052 | 5.46 |
| 22 | CD5L_HUMAN | CD5 antigen-like | 112 | 1.3e-07 | 17/80 | 55% | 39603 | 5.28 |
| 6, 9 | gi|90108664 | Chain A, Apolipoprotein A-I | 135 | 9.9e-09 | 15/77 | 62% | 28061 | 5.27 |
| 11 | SAMP_HUMAN | Serum amyloid P-component | 70 | 2.e-03 | 9/76 | 29% | 25485 | 6.10 |
| 5 | gi|14719497 | Chain A, Transthyretin | 78 | 4.8e-03 | 6/81 | 66% | 12671 | 5.26 |
| 15 | APOA4_HUMAN | Apolipoprotein A-IV | 92 | 1.1e-05 | 15/72 | 38% | 45371 | 5.28 |
| 12, 13, 14, 32 | APOE_HUMAN | Apolipoprotein E | 112 | 1.3e-07 | 16/92 | 50% | 36246 | 5.65 |
| 5 | TTHY_HUMAN | Transthyretin | 57 | 0.04 | 4/95 | 48% | 15991 | 5.52 |
| 30, 31, 47 | FIBB_HUMAN | Fibrinogen beta chain | 184 | 8e-15 | 28/108 | 60% | 56577 | 8.54 |
| 21 | CO7_HUMAN | Complement component C7 | 68 | 0.0034 | 17/91 | 23% | 96550 | 9.06 |
| 54 | gi|11761629 | Fibrinogen alpha chain precursor | 83 | 0.0016 | 23/147 | 36% | 70227 | 8.23 |
| 43 | HBB_HUMAN | Haemoglobin subunit beta | 44 | 0.72 | 6/78 | 49% | 16102 | 6.75 |
| 42 | SAA4_HUMAN | Serum amyloid A-4 | 61 | 0.016 | 6/60 | 40% | 14851 | 9.17 |
| 33, 49, 50 | PLMN_HUMAN | Plasminogen | 183 | 1e-14 | 27/112 | 39% | 93247 | 7.04 |
| 38 | GELS_HUMAN | Gelsolin | 46 | 0.46 | 13/105 | 17% | 86043 | 5.90 |
| 34, 41, 43 | SAA1_HUMAN | Serum amyloid A-1 | 64 | 0.0079 | 5/70 | 46% | 13581 | 6.28 |
| 36 | RET4_HUMAN | Retinol-binding protein | 53 | 0.11 | 9/63 | 51% | 23337 | 5.76 |
| 24 | C1S_HUMAN | Complement C1s subcomponent | 48 | 0.29 | 18/113 | 25% | 78174 | 4.86 |
| 27 | gi|374977533 | Alpha-1-microglobulin | 79 | 0.0037 | 10/117 | 63% | 22030 | 6.25 |
| 39 | ALBU_HUMAN | Serum albumin | 94 | 7.7e-06 | 21/129 | 39% | 71317 | 5.92 |
Spot IDs are those mentioned in Fig. 4.
Using PDQuest, we further analysed the 2-DGE spots to identify most significant and consistently dysregulated 15 proteins in AML cases (showing ≥4-fold increase/decrease in spot intensity and with significance level more than 95% in t-test), in comparison to the control subject. The results unequivocally showed variability in the levels of the SAA1, HPT, complement factor B, CD5 antigen-like, kininogen-1, fibrinogen gamma chain, C4b-binding protein alpha chain, complement factor 7, ApoA1, ApoE, plasminogen, apolipoprotein A-IV, prothrombin, fibronectin, and gelsolin. The former 6 proteins were found to be up-regulated, while the latter 9 were down-regulated in the AML, as shown in column graphs (Fig. S7).
Gene ontology (GO) analysis
Based on the known or postulated biological functions of the proteins as found in the GO consortium using homo sapiens taxon, the functions of the identified 34 proteins could be categorized on the basis of their functions as binding (28%), enzyme regulation activity (7%), homeostasis (7%), structural (2%), receptor mediated activity (15%), catalytic (14%), biological process regulation (25%) while for 2% proteins the functions are not yet known (Fig. S8).
Validation through ELISA
ApoA1 was found to be down-regulated in AML pool (10.67 mg/dL), in agreement with the spot intensity in the 2-DGE image of AML, as compared to the healthy pool (18.15 mg/dL). However, when the individual samples of AML were analysed, the mean plasma concentration of ApoA1 in the healthy group was 10.55 ± 4.6 mg/dL, whereas in the AML subjects it was found to be 14.59 ± 6.2 mg/dL i.e., higher, quite contrary to the expected results (Fig. 5). We applied t-test with Welch’s correction (One-way ANOVA) to compare the variances. The mean concentrations were found to be significantly different with p < 0.0396, while the variances were not significantly different in individual samples.
Figure 5.
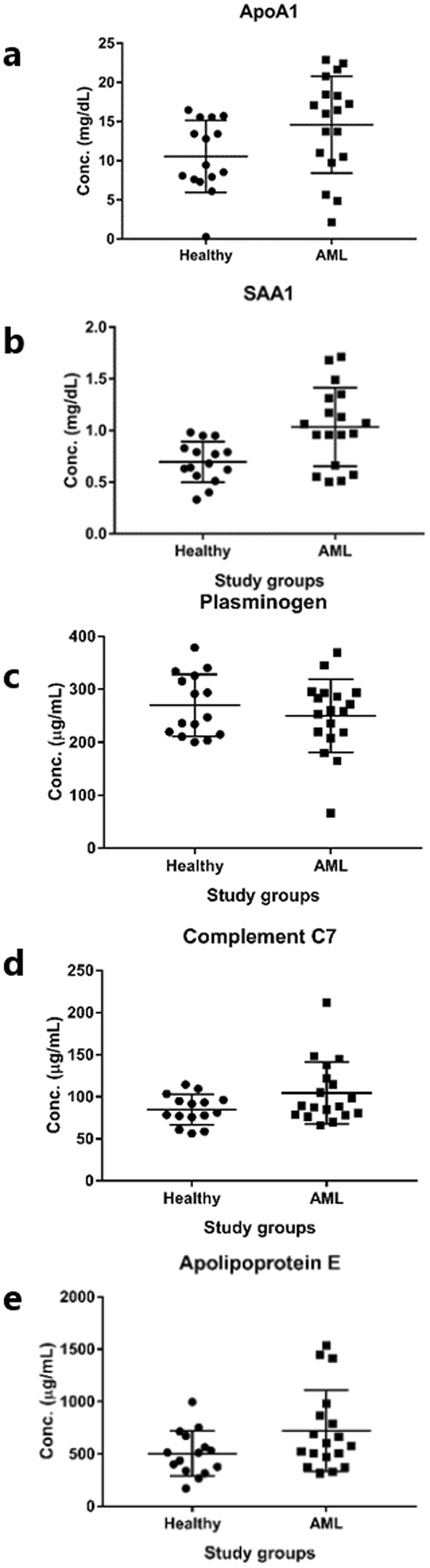
Scatter plot with standard deviation achieved through ELISA results of individual samples of healthy and AML subjects. (a) ApoA1 protein, (b) SAA1 protein, (c) plasminogen, (d) complement factor C7, (e) apolipoprotein E.
SAA1 was found to be up-regulated in AML pool sample (1.15 mg/dL), in comparison to the pool sample of healthy individuals (0.89 mg/dL). Validation results of individual samples also showed significant differences when validated through ELISA. The mean plasma concentration of SAA1 in the individual samples of control was 0.69 ± 0.19 mg/dL whereas it was found to be 1.04 ± 0.38 mg/dL i.e., high in the AML subjects (Fig. 5). Mean concentrations were found to be significantly different with p < 0.0029, and the variances were also significantly different in individual samples with p < 0.0162 by applying statistical analysis.
Plasminogen was found to be down-regulated in AML patients in comparison to the healthy individuals through ELISA validation and found to agree with 2-DGE results. When pool samples of AML and healthy individuals were compared, the value of plasminogen was found to be low in AML (216.42 µg/mL), in comparison to healthy pool (244.87 µg/mL). Similarly, the mean plasma concentration of plasminogen in healthy and AML individuals was 269.8 ± 58.74 µg/mL, and 250.1 ± 69.31 µg/mL, respectively (Fig. 5). Statistical analysis showed significant difference of mean concentrations with p < 0.3844, and the variances with p < 0.5372.
In AML complement factor 7 (C7) protein was found to be up-regulated in comparison to healthy individuals, contrary to 2-DGE results, where we found C7 was down-regulated in AML pool gel, in comparison to healthy pool gel. Although not a big difference, the complement C7 protein value in healthy pool was 94.68 µg/mL versus 98.09 µg/mL in AML pool. Plasma mean value was 84.62 ± 17.92 µg/mL in healthy, while 104.6 ± 36.96 µg/mL in AML patients (Fig. 5). T-test with Welch’s correction showed no significant difference between AML and healthy with p value 0.0537, while F-test to compare variances showed significant difference among study groups with p value 0.0090.
ApoE was found to be up-regulated in AML in contrast to 2-DGE results. ApoE value in healthy pool was 517.79 µg/mL versus AML pool was 564.82 µg/mL. Mean value in AML patients was found to be 504.4 ± 214.2 µg/mL, versus 720.2 ± 387.5 µg/mL in healthy group (Fig. 5). T-test with Welch’s correction gave no significant difference with p value 0.0532, and when variances were compared through F-test, a significant difference with p value 0.0300 was observed.
Discussion
The combination of 2-DGE and mass spectrometry offers a powerful tool to investigate the proteomic expression profiles to identify biomarkers which might serve as indicators of the disease27. Recently many potential protein biomarkers have been reported in the literature for the diagnosis of AML e.g. ApoE, complement factor H, HPT, apolipoprotein A-N, SAA1, and gelsolin using proteomics techniques28–31. Also various MS based studies have already been done on cell lines32–35. In this study, we have investigated differential proteomic profile pattern among AML and healthy samples in plasma by employing multi-dimensional fractionation strategy to identify biomarker proteins.
The major bottleneck in analysing plasma proteins is to analyse low abundant proteins in the presence of high abundant proteins36. Therefore, to overcome this difficulty, we first depleted the samples for high abundant proteins to unmask the proteins which are present in very low amount. The depletion column with antibodies for the top 7 abundant proteins, including albumin, fibrinogen and HPT were used. While doing depletion, we have noticed that 100% depletion of plasma samples was not achieved, because in flow-through (unbound fraction), we have also identified peaks of albumin, all three chains of fibrinogen (α, β and γ), and HPT (Fig. 4). In terms of specificity of immunodepletion, contaminants in bound and eluted portion have been already reported by manufacturers and researchers37. Albumin may be present due to non-specific interactions with other proteins in the plasma, which is called the “sponge-effect”37–40. HPT and three chains of fibrinogen were visible in unbound fraction possibly due to the narrow-range techniques applied, or the fragments may have low binding efficiency to the column. The presence of target proteins (to be depleted through column) as contaminants have already been reported22,41.
After depletion, many low abundant proteins were visualized by IEF. In-solution IEF eases the detection of low abundance proteins and increases the detection range as large amounts of proteins of specific pH can be loaded on the gel13. In 2-DGE maps, most of the identified proteins were represented by multiple spots in AML and control group (Fig. 4). Slight to moderate shifts in pI or mass were between theoretical and experimentally-calculated values has already been reported in most of the cases. Both observations i.e., representation of single protein by multiple spots and variations in theoretical and experimentally calculated pI/MW, seem to be the result of post-translational modifications, especially glycosylation, affecting the electrophoretic mobility of the proteins, as reported earlier42.
To gain an insight of the biological functions and interactive links that are known to be associated with the differentially expressed proteins in the dataset, STRING software program was used. Interactive links between 15 proteins of the dataset could be traced as presented in Fig. 6. Blue lines show phylogenetic co-occurrence between proteins, whereas light blue lines represent database evidence. The line thickness is a rough indicator of the power of the association. The visualizations among protein nodes show the predicted association between the proteins detected in the samples of AML and non-leukemic healthy patients. The pathways involved by these differentially regulated proteins are presented in Table S3.
Figure 6.
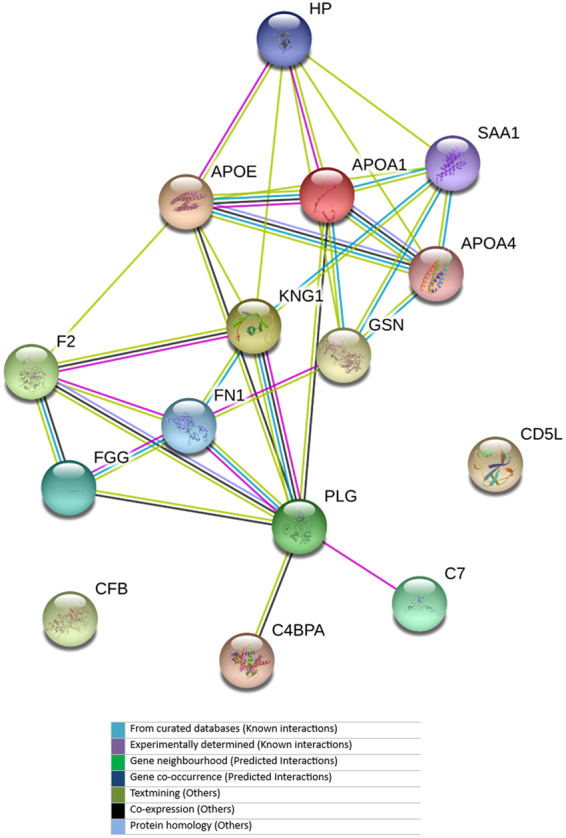
Curated pathway of fifteen differentially expressed proteins in acute myeloid leukaemia acquired from online STRING database. Balls with structures show that their 3D structures are also available in database.
We further validated some of the deregulated proteins through ELISA, as it has the potential to accelerate validation of protein biomarkers for clinical use43. HPT and all three chains of fibrinogen were not included for the validation because depletion column had affinity for HPT and fibrinogen, and most of them had depleted out of plasma sample in column, so these results seen in 2-DGE was not reliable.
SAA1 is a major acute phase reactant and is also found as apolipoprotein of the HDL complex. ELISA validation results of SAA1 show similar trend as 2-DGE results, and hence SAA1 may be used as a potential diagnostic biomarker for AML (Fig. 5). Plasminogen; Plasmin dissolves the fibrin of blood clots and acts as a proteolytic factor in a variety of other processes including, embryonic development, tissue remodelling, tumour invasion, and inflammation. Plasminogen was found to be down-regulated in ELISA validation.
ApoA1; participates in the reverse transport of cholesterol from tissues to the liver for excretion by promoting cholesterol efflux from tissues and by acting as a cofactor for the lecithin cholesterol acyltransferase (LCAT), and as part of the SPAP complex activates spermatozoa motility. The results from the analysis of ApoA1 protein from individual samples were in contradiction from the 2-DGE maps (Fig. 5). Therefore, these results of ApoA1 protein require further validation on large number of individual samples.
Complement component 7; constituent of the membrane attack complex (MAC) plays a key role in the innate and adaptive immune response by forming pores in the plasma membrane of target cells. ApoE mediates the binding, internalization, and catabolism of lipoprotein particles. It serves as a ligand for the LDL (ApoB/E) receptor, and for the specific ApoE receptor (chylomicron remnant) of hepatic tissues. Results of both complement C7 and ApoE were not in agreement with 2-DGE results.
During ELISA validation, we found that out of five differentiating proteins, only two proteins SAA1 and plasminogen showed potential of differentiation of AML from healthy group, during ELISA validation. These two proteins showed links to 4 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways including transcriptional misregulation in cancers, p53 signalling pathway, proteoglycans in cancers, and hypoxia-inducible factor 1 (HIF-1) signalling pathway (Fig. 7). These pathways regulate cell proliferation and apoptosis, inflammation, metabolism, angiogenesis, cell growth and migration, cell migration and metastasis, cell cycle arrest, cellular senescence, and DNA repair, which are properties of cancerous cell. Therefore, the relevant up-regulation of SAA1 and down-regulation of plasminogen may be due to the direct effect of these disturbed signalling pathways, and can be used for the diagnosis of acute myeloid leukaemia in future.
Figure 7.
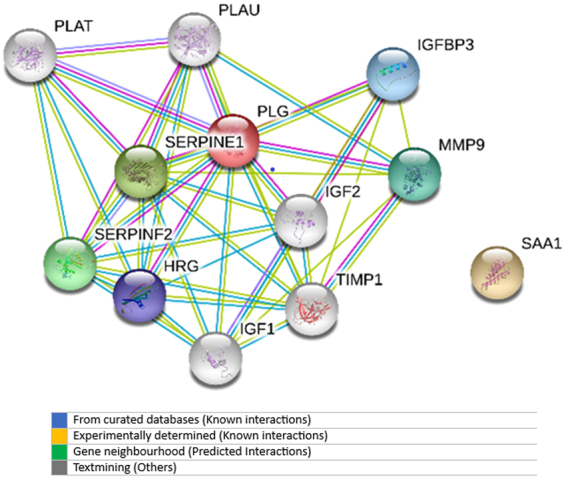
Pathways analysis of SAA1 and plasminogen.
Conclusion
In conclusion, 5D proteomic strategy using immunodepletion, 2-DGE, ZOOM-IEF and MALDI-MS, and ELISA analysis has shown a promising approach for the detection of differentiated proteins in AML in comparison with the control. Fifteen proteins were found to be deregulated in comparison to healthy control. Some of these deregulated proteins were further validated through ELISA technique and the results suggest that SAA1 and plasminogen can be used as biomarkers for the diagnosis of AML patients. However, as the validation was performed on a small number of proteins, therefore validation of these deregulated proteins on a larger number of individual subjects is needed.
Electronic supplementary material
Acknowledgements
The study was financially supported by the Higher Education Commission (No. 4493), Pakistan. Our special thanks to all AML patients and healthy participants, who voluntarily gave their blood samples for this research project.
Author Contributions
S.K.R. was involved in experimental work and manuscript writing. M.S. helped in the experiment. S.G.M. contributed to the study design, conceptual and technical guidance along with the laboratory equipment and expertise. T.S. provided the disease samples. A.R. and M.I.C. were involved in manuscript checking.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-16699-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Aasebo E, Forthun RB, Berven F, Selheim F, Hernandez-Valladares M. Global Cell Proteome Profiling, Phospho-signaling and Quantitative Proteomics for Identification of New Biomarkers in Acute Myeloid Leukemia Patients. Current Pharmaceutical Biotechnology. 2016;17:52–70. doi: 10.2174/1389201016666150826115626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marcucci G, Haferlach T, Dohner H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. Journal of Clinical Oncology. 2011;29:475–486. doi: 10.1200/JCO.2010.30.2554. [DOI] [PubMed] [Google Scholar]
- 3.Hoffbrand, V., Moss, P. & Pettit, J. Essential Haematology. (Wiley, 2006).
- 4.Vardiman, J. W. et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood 114, 937–951 (2009). [DOI] [PubMed]
- 5.Hoffbrand, M. Pettit. Essential Hematology. (2006).
- 6.Ray S, et al. Proteomic technologies for the identification of disease biomarkers in serum: advances and challenges ahead. Proteomics. 2011;11:2139–2161. doi: 10.1002/pmic.201000460. [DOI] [PubMed] [Google Scholar]
- 7.Ota J, et al. Proteomic analysis of hematopoietic stem cell-like fractions in leukemic disorders. Oncogene. 2003;22:5720. doi: 10.1038/sj.onc.1206855. [DOI] [PubMed] [Google Scholar]
- 8.Kwak J-Y, et al. The comparative analysis of serum proteomes for the discovery of biomarkers for acute myeloid leukemia. Experimental hematology. 2004;32:836–842. doi: 10.1016/j.exphem.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Balkhi M, et al. Proteomics of acute myeloid leukaemia: cytogenetic risk groups differ specifically in their proteome, interactome and post-translational protein modifications. Oncogene. 2006;25:7041. doi: 10.1038/sj.onc.1209689. [DOI] [PubMed] [Google Scholar]
- 10.Strassberger V, et al. A comprehensive surface proteome analysis of myeloid leukemia cell lines for therapeutic antibody development. Journal of proteomics. 2014;99:138–151. doi: 10.1016/j.jprot.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 11.Oellerich, T. et al. FLT3-ITD and TLR9 employ Bruton9s tyrosine kinase to activate distinct transcriptional programs mediating AML cell survival and proliferation. Blood, blood-2014-2006-585216 (2015). [DOI] [PubMed]
- 12.Gu T-L, et al. Survey of activated FLT3 signaling in leukemia. PLoS One. 2011;6:e19169. doi: 10.1371/journal.pone.0019169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuo X, Speicher DW. Comprehensive analysis of complex proteomes using microscale solution isoelectrofocusing prior to narrow pH range two-dimensional electrophoresis. Proteomics. 2002;2:58–68. doi: 10.1002/1615-9861(200201)2:1<58::AID-PROT58>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 14.Echan LA, Tang HY, Ali-Khan N, Lee K, Speicher DW. Depletion of multiple high-abundance proteins improves protein profiling capacities of human serum and plasma. Proteomics. 2005;5:3292–3303. doi: 10.1002/pmic.200401228. [DOI] [PubMed] [Google Scholar]
- 15.Vasudev NS, et al. Serum biomarker discovery in renal cancer using 2‐DE and prefractionation by immunodepletion and isoelectric focusing; increasing coverage or more of the same? Proteomics. 2008;8:5074–5085. doi: 10.1002/pmic.200800497. [DOI] [PubMed] [Google Scholar]
- 16.Rai AJ, et al. HUPO Plasma Proteome Project specimen collection and handling: towards the standardization of parameters for plasma proteome samples. Proteomics. 2005;5:3262–3277. doi: 10.1002/pmic.200401245. [DOI] [PubMed] [Google Scholar]
- 17.He P, et al. The human plasma proteome: analysis of Chinese serum using shotgun strategy. Proteomics. 2005;5:3442–3453. doi: 10.1002/pmic.200401301. [DOI] [PubMed] [Google Scholar]
- 18.Li X, et al. Comparison of alternative analytical techniques for the characterisation of the human serum proteome in HUPO Plasma Proteome Project. Proteomics. 2005;5:3423–3441. doi: 10.1002/pmic.200401226. [DOI] [PubMed] [Google Scholar]
- 19.Donahue MP, et al. Discovery of proteins related to coronary artery disease using industrial-scale proteomics analysis of pooled plasma. American heart journal. 2006;152:478–485. doi: 10.1016/j.ahj.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Gastwirth JL. The Efficiency of Pooling in the Detection of Rare Mutations. American Journal of Human Genetics. 2000;67:1036–1039. doi: 10.1086/303097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fic E, Kedracka-Krok S, Jankowska U, Pirog A, Dziedzicka-Wasylewska M. Comparison of protein precipitation methods for various rat brain structures prior to proteomic analysis. Electrophoresis. 2010;31:3573–3579. doi: 10.1002/elps.201000197. [DOI] [PubMed] [Google Scholar]
- 22.Vasudev NS, et al. Serum biomarker discovery in renal cancer using 2-DE and prefractionation by immunodepletion and isoelectric focusing; increasing coverage or more of the same? Proteomics. 2008;8:5074–5085. doi: 10.1002/pmic.200800497. [DOI] [PubMed] [Google Scholar]
- 23.de Roos B, et al. Proteomic methodological recommendations for studies involving human plasma, platelets, and peripheral blood mononuclear cells. Journal of Proteome Research. 2008;7:2280–2290. doi: 10.1021/pr700714x. [DOI] [PubMed] [Google Scholar]
- 24.Bouwman F, Suylen D, Renes J, Mariman E. Evaluation and improving the success rate of protein identification by peptide mass fingerprinting using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Communications in Mass Spectrometry. 2005;19:2465–2468. doi: 10.1002/rcm.2073. [DOI] [PubMed] [Google Scholar]
- 25.Musharraf SG, et al. Comparison of plasma from healthy nonsmokers, smokers, and lung cancer patients: Pattern-based differentiation profiling of low molecular weight proteins and peptides by magnetic bead technology with MALDI-TOF MS. Biomarkers. 2012;17:223–230. doi: 10.3109/1354750X.2012.657245. [DOI] [PubMed] [Google Scholar]
- 26.Jensen LJ, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Research. 2009;37:D412–416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong Y, et al. Different immunoaffinity fractionation strategies to characterize the human plasma proteome. Journal of Proteome Research. 2006;5:1379–1387. doi: 10.1021/pr0600024. [DOI] [PubMed] [Google Scholar]
- 28.Zheng RJ, Ma XD. Study on serum protein mass spectrometric characteristics of acute leukemia. Zhonghua Xue Ye Xue Za Zhi. 2013;34:426–429. doi: 10.3760/cma.j.issn.0253-2727.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 29.Braoudaki M, et al. Proteomic analysis of childhood de novo acute myeloid leukemia and myelodysplastic syndrome/AML: correlation to molecular and cytogenetic analyses. Amino Acids. 2011;40:943–951. doi: 10.1007/s00726-010-0718-9. [DOI] [PubMed] [Google Scholar]
- 30.Lee SW, et al. Use of MDLC-DIGE and LC-MS/MS to identify serum biomarkers for complete remission in patients with acute myeloid leukemia. Electrophoresis. 2012;33:1863–1872. doi: 10.1002/elps.201200047. [DOI] [PubMed] [Google Scholar]
- 31.Kwak JY, et al. The comparative analysis of serum proteomes for the discovery of biomarkers for acute myeloid leukemia. Experimental hematology. 2004;32:836–842. doi: 10.1016/j.exphem.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Weber C, Schreiber TB, Daub H. Dual phosphoproteomics and chemical proteomics analysis of erlotinib and gefitinib interference in acute myeloid leukemia cells. Journal of proteomics. 2012;75:1343–1356. doi: 10.1016/j.jprot.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Hahn CK, et al. Proteomic and genetic approaches identify Syk as an AML target. Cancer cell. 2009;16:281–294. doi: 10.1016/j.ccr.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walters DK, et al. Phosphoproteomic analysis of AML cell lines identifies leukemic oncogenes. Leukemia research. 2006;30:1097–1104. doi: 10.1016/j.leukres.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Casado P, et al. Kinase-substrate enrichment analysis provides insights into the heterogeneity of signaling pathway activation in leukemia cells. Science Signaling. 2013;6:rs6–rs6. doi: 10.1126/scisignal.2003573. [DOI] [PubMed] [Google Scholar]
- 36.Ebert MP, Korc M, Malfertheiner P, Rocken C. Advances, challenges, and limitations in serum-proteome-based cancer diagnosis. Journal of Proteome Research. 2006;5:19–25. doi: 10.1021/pr050271e. [DOI] [PubMed] [Google Scholar]
- 37.Zhou M, et al. An investigation into the human serum “interactome”. Electrophoresis. 2004;25:1289–1298. doi: 10.1002/elps.200405866. [DOI] [PubMed] [Google Scholar]
- 38.Lopez MF, et al. A novel, high-throughput workflow for discovery and identification of serum carrier protein-bound peptide biomarker candidates in ovarian cancer samples. Clinical Chemistry. 2007;53:1067–1074. doi: 10.1373/clinchem.2006.080721. [DOI] [PubMed] [Google Scholar]
- 39.Cho SY, et al. Efficient prefractionation of low-abundance proteins in human plasma and construction of a two-dimensional map. Proteomics. 2005;5:3386–3396. doi: 10.1002/pmic.200401310. [DOI] [PubMed] [Google Scholar]
- 40.Bjorhall K, Miliotis T, Davidsson P. Comparison of different depletion strategies for improved resolution in proteomic analysis of human serum samples. Proteomics. 2005;5:307–317. doi: 10.1002/pmic.200400900. [DOI] [PubMed] [Google Scholar]
- 41.Okano T, et al. Plasma proteomics of lung cancer by a linkage of multi-dimensional liquid chromatography and two-dimensional difference gel electrophoresis. Proteomics. 2006;6:3938–3948. doi: 10.1002/pmic.200500883. [DOI] [PubMed] [Google Scholar]
- 42.Barrabes S, et al. Effect of sialic acid content on glycoprotein pI analyzed by two-dimensional electrophoresis. Electrophoresis. 2010;31:2903–2912. doi: 10.1002/elps.200900764. [DOI] [PubMed] [Google Scholar]
- 43.Zangar RC, Daly DS, White AM. ELISA microarray technology as a high-throughput system for cancer biomarker validation. Expert review of proteomics. 2006;3:37–44. doi: 10.1586/14789450.3.1.37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.