Abstract
Because of differences in the downstream signaling patterns of its pathways, the role of the human epidermal growth factor family of receptors (HER) in promoting cell growth and survival is cell line and context dependent. Using two model cell lines, we have studied how the regulatory interaction network among the key proteins of HER signaling pathways may be rewired upon normal to cancerous transformation. We in particular investigated how the transcription factor STAT3 and several key kinases’ involvement in cancer-related signaling processes differ between normal 184A1L5 human mammary epithelial (HME) and MDA-MB-231 breast cancer epithelial cells. Comparison of the responses in these cells showed that normal-to-cancerous cellular transformation causes a major re-wiring of the growth factor initiated signaling. In particular, we found that: i) regulatory interactions between Erk, p38, JNK and STAT3 are triangulated and tightly coupled in 184A1L5 HME cells, and ii) STAT3 is only weakly associated with the Erk-p38-JNK pathway in MDA-MB-231 cells. Utilizing the concept of pathway substitution, we predicted how the observed differences in the regulatory interactions may affect the proliferation/survival and motility responses of the 184A1L5 and MDA-MB-231 cells when exposed to various inhibitors. We then validated our predictions experimentally to complete the experiment-computation-experiment iteration loop. Validated differences in the regulatory interactions of the 184A1L5 and MDA-MB-231 cells indicated that instead of inhibiting STAT3, which has severe toxic side effects, simultaneous inhibition of JNK together with Erk or p38 could be a more effective strategy to impose cell death selectively to MDA-MB-231 cancer cells while considerably lowering the side effects to normal epithelial cells. Presented analysis establishes a framework with examples that would enable cell signaling researchers to identify the signaling network structures which can be used to predict the phenotypic responses in particular cell lines of interest.
Electronic supplementary material
The online version of this article (doi:10.1007/s12079-017-0389-3) contains supplementary material, which is available to authorized users.
Keywords: EGFR signaling, Receptor tyrosine kinase, Signaling network, Signaling pathway crosstalk, Proliferation, Pro-survival, STAT3, MAPK, Mathematical modeling, Network inference, Modular response analysis
Introduction
Cells constantly receive and process signals through interactions with their microenvironment. Variations in the signaling patterns is a common mechanism for cells to regulate their phenotypic responses. Human epidermal growth factor family of receptors (HER) belong to the receptor tyrosine kinase (RTK) superfamily. HER receptors (also known as ErbB receptors) play a critical regulatory role in fundamental physiological processes through the activation of proliferation, pro-survival and stress-response pathways of cells (Schlessinger 2000; Yarden and Sliwkowski 2001; Prenzel et al. 2000; Arteaga 2003a; Hynes and MacDonald 2009; Volinsky and Kholodenko 2013). There is a significant positive correlation between the expression of HER receptors in cancers with a poor prognosis (Normanno et al. 2006; Holbro et al. 2003; Britten 2004; Arteaga 2003b; Klinger et al. 2013; Libermann et al. 1984). Although HER signaling promotes growth and survival in almost all normal (untransformed) and transformed cell types, the role of HER receptors is cell line and context dependent (Joslin et al. 2010; Alexander et al. 2011). This mainly stems from differences in the downstream signaling among multiple pathways: Activation of HER family of receptors, in particular its first member EGFR/HER1/ErbB1, leads to robust stimulation of major kinases and transcriptional regulators downstream of the receptors (Naegle et al. 2012; Rodland et al. 2008; Avraham and Yarden 2011; Aksamitiene et al. 2012). This provides a useful context to study the crosstalk between oncogenic proteins in breast cancer cells (Rodland et al. 2008; Avraham and Yarden 2011; Aksamitiene et al. 2012; Zhang et al. 2005; Jones et al. 2006; Kumar et al. 2008).
Stimulation of epithelial cells through HER receptors leads to the activation of Erk, p38, JNK (SAPK), and Akt kinases, as well as various transcription regulators such as STAT3 (Yarden and Sliwkowski 2001; Aksamitiene et al. 2012; Katz et al. 2007; Pawson and Warner 2007; Neve et al. 2002). As we have discussed earlier (Gong et al. 2015), proteins Erk, p38, JNK, Akt and STAT3 form a group of sentinels representative of the HER signaling pathways associated with cell proliferation, pro-survival, stress-responses, and enhanced cell migration. Erk, p38 and JNK kinases, which are the three major subgroups of the MAPK family, are activated by a large set of growth factors, cytokines, transforming agents, and carcinogens (Cobb 1999). Signaling through the Erk pathway can have significant functional consequences such as speedy cell migration, enhanced cell transformation and resistance to apoptosis (Joslin et al. 2007; Wu et al. 1999; Raman et al. 2007). JNK and p38 are stress-activated kinases involved in apoptosis/survival. The p38 kinase regulates EGFR down-regulation, survival, and cellular migration (Wagner and Nebreda 2009; Mueller et al. 2012; Frey et al. 2006), and it can suppress cell proliferation by antagonizing the JNK/c-Jun pathway (Hui et al. 2007). Activity of the Erk-p38-JNK axis plays a universal role in balancing autophagy and apoptosis related signals and this role can be context dependent (Wagner and Nebreda 2009; Frey et al. 2006; Chiacchiera et al. 2012). The serine/threonine kinase Akt promotes cellular survival (Song et al. 2005), and it has been implicated in many types of cancers because of its possible role in angiogenesis and tumor development (Chen et al. 2005).
The activation of MAPKs by RTKs leads to the activation of the signal transducer and activator of transcription (STAT) family of transcription factors (Katz et al. 2007). STATs are stimulated through a vast set of cytokine and growth factors. STATs, in particular STAT3, transcriptionally regulate a diverse array of cellular processes, including the transcription of genes related to cell proliferation (Katz et al. 2007; Rawlings et al. 2004; Berclaz et al. 2001; Levy and Darnell 2002; Yue and Turkson 2009; Banerjee and Resat 2015; Sen et al. 2012).
Because of these properties, Erk, p38, JNK, Akt, and STAT3 form a sentinel set whose activation patterns provide a good representation of the integrated signal flow downstream of HER receptors which is directly related to cell survival and proliferation (Gong et al. 2015). We have earlier shown that STAT3 occupies a central role in the crosstalk between cell proliferation, survival, and migration related signaling pathways when normal (untransformed) 184A1L5 human mammary epithelial (HME) cells were stimulated through HER receptors, and that wiring of the regulatory interactions was dependent on the expression of the members of HER family (Gong et al. 2015). In contrast to our initial study which compared the interactions in the 184A1L5 HME cell library, in this study we have investigated whether STAT3 has a similar role in the triple negative breast cancer cell line MDA-MB-231 and normal 184A1L5 HME cells when stimulated through EGFR. Our hypothesis was that transformation from normal to cancerous cells involves a considerable rewiring of the regulatory interaction network of HER signaling pathways. Since differences in signaling networks can impact cellular signal propagation and regulation capabilities, rewiring of the HER regulatory interaction network can lead to significant physiological response differences between the compared cell lines. Deriving the regulatory interaction network among studied sentinels in MDA-MB-231 breast cancer cells and comparing it to the network in normal 184A1L5 cells has allowed us to address this hypothesis and to investigate differential involvement of STAT3 and several key kinases in cancer-related signaling processes in normal mammary and MDA-MB-231 breast cancer epithelial cells. Our results have validated our hypothesis and established that: i) regulatory interactions between Erk, p38, JNK and STAT3 are triangulated and tightly coupled in 184A1L5 cells and ii) STAT3 is only weakly associated with the Erk-p38-JNK pathway in MDA-MB-231 cells. Combining these findings with the concept of pathway substitution enabled by the crosstalk between related signaling pathways, we predicted the differences in the proliferation/survival and motility responses of 184A1L5 and MDA-MB-231 cells when these cells were exposed to various inhibitors. We then tested our predictions to validate the inferred network models, and argue that our validated predictions can provide clues to developing new therapeutic strategies. Examples to possible strategies based on our results are discussed in the Discussion and Conclusions section below. Most importantly, this study presents a conceptual framework to identify the network structures involved in cellular signaling. We show with examples that the new framework can be used to predict cell phenotypic responses.
Materials and methods
General considerations
Establishing the relationships between signaling proteins requires multi-factorial perturbation experiments where signaling patterns are altered such that the possible effects of crosstalk between pathways are sampled. We have selectively inhibited the studied proteins one at a time and measured how a particular inhibition affected the activities of the other studied sentinels. We then analyzed the collected data separately for the 184A1L5 and MDA-MB-231 cell lines using inference methods to identify the regulatory interactions between investigated signaling molecules. This analysis enabled us to establish that EGFR signaling pathways are wired differently in 184A1L5 and MDA-MB-231 cells and that this differential wiring can lead to significant differences in cellular phenotype responses.
Cell lines
Our study compared the normal human mammary epithelial (HME) and MDA-MB-231 breast cancer cells. As the cancer cell line we utilized MDA-MB-231 cells (ATCC listing: HTB-26), which are a human EGFR+ (Davidson et al. 1987) and ER−/PR−/HER2− (i.e., triple negative) basal breast cancer cell type (Neve et al. 2006). HME cell line 184A1L5 (Stampfer et al. 1993) expresses approximately 200,000 molecules of EGFR/HER1 and relatively low copy numbers of HER2 (~30,000) and HER3 (~1000), two other members of the EGFR (HER) family of receptor tyrosine kinases (Hendriks et al. 2003; Zhang et al. 2009). We note that, unlike many model cell lines with limited physiological responses to growth factors, 184A1L5 HME cells are an excellent model system for studying the properties of the HER signaling. This is because, like many epithelium derived cell types, HME cells require EGFR activation for proper proliferation and migration responses (Stampfer et al. 1993; Dong et al. 1999). Because they express EGFR at comparable levels and have low levels of HER2 and HER3 (Shankaran et al. 2013), MDA-MB-231 cells was the most logical choice for comparison studies with 184A1L5 cells (which was labeled as the “parental” or “HER2−/3−” cell line in our earlier studies with this HME cell library (Gong et al. 2015; Zhang et al. 2009; Shankaran et al. 2013)). This close similarity eliminated the HER receptor expression difference concern and made cell line comparisons more realistic. We note that our earlier studies compared the signaling patterns between the cell lines of our HME library while this current study compares the 184A1L5 cells to MDA-MB-231 cancer cells.
The MDA-MB-231 cells were maintained at 37 °C in atmospheric air in L-15 media supplemented with 10% FBS and 1% penicillin-streptomycin. At approximately 70–80% confluency, the growth medium was replaced with medium lacking FBS and cells were brought to quiescence for 16–18 h prior to treatment in Western blot, proliferation and imaging experiments. These experiments were run in triplicate for every treatment condition. The MDA-MB-231 cells were activated by stimulating them using L-15 media containing 1% FBS, 12 ng/ml EGF and 40 ng/ml HRG-β. The 184A1L5 cells were maintained at 37 °C with 5% CO2 in DFCl-1 media supplemented with 10% FBS and 12.5 ng/ml EGF (Gong et al. 2015). A DFHB minimal media containing 0.1% bovine serum albumin and deficient of EGF was used to bring the cells to quiescence. DFCl-1 media supplemented with ligands was used to stimulate the 184A1L5 cells and data was collected in duplicate (Gong et al. 2015). Thus, the data for the normal and cancer epithelial cells were collected under equivalent conditions.
Western blot analysis of sentinel proteins - inhibition studies
We selectively inhibited the studied proteins one at a time and measured how a particular inhibition affected the activities of all sentinels. Inhibitors were added to starvation medium 2 h prior to ligand-induced stimulation. They were also included in the treatment medium, i.e., cells were exposed to respective inhibitors continuously before and after the stimulation with ligands. The following inhibitors and doses were used: U0126 (Erk1/2 inhibitor; 20 μM), JNK Inhibitor VIII (JNK inhibitor; 20 μM), Stattic (STAT3 inhibitor; 10 μM), Akt Inhibitor VIII (Akt inhibitor; 10 μM), and Birb-796 (p38 MAPK inhibitor; 5 μM). No-ligand measurements where cells were treated with minimal media provided the negative control.
Lysates for Western blotting studies were collected 15 min after activation. Data for the 184A1L5 cells was collected earlier (Gong et al. 2015). To collect the data anew for the MDA-MB-231 cells, plates were washed with ice-cold PBS and cells were then immediately lysed using RIPA buffer prepared with protease and phosphatase inhibitors. Plates were lysed over a period of 30 min, on ice, prior to cell scraping and transfer of the lysate to microtubes. Lysates were centrifuged at 12,000 rpm for 10 min. The supernatant was aspirated into fresh microfuge tubes for analysis and the pellet was discarded. Lysates were stored at −80 °C between analyses. Total protein concentration of cell lysates was determined by the Bicinchoninic Acid assay technique (reagents from Sigma). Total protein amount was matched to 20 μg protein/well and loaded to SDS-PAGE gels. TGX precast 10% gels from Bio-Rad with 10 wells were used. Electrophoresis was done using the Bio-Rad Mini-Protean Tetra System and proteins were transferred from the gel to PVDF membranes using the Bio-Rad Trans-blot Turbo transfer system. PVDF membranes were developed using ECL. Western blot images were captured by a LI-COR C-DiGit scanner. Images were analyzed using the Image Studio Digits v3.1 software.
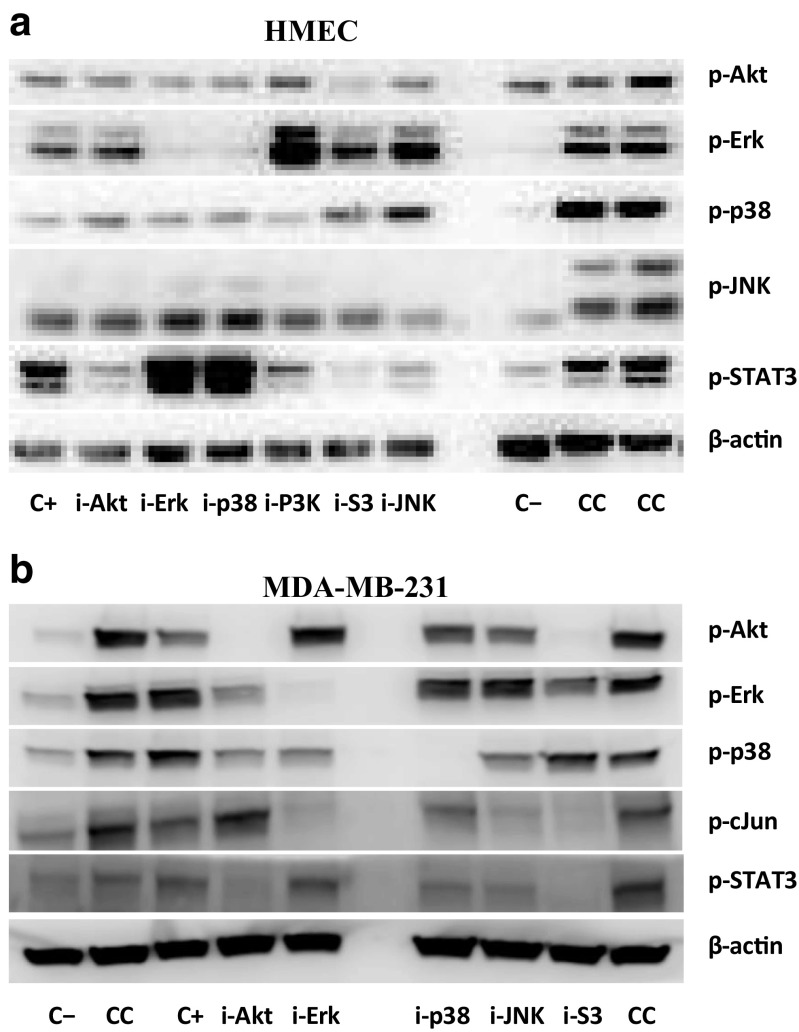
The Western blots (Fig. 1) illustrate that the chosen inhibition conditions effectively block phosphorylation of their designated targets, thus meeting the perturbation requirements necessary to infer regulatory interactions with MRA and BVSA. We note that our studies require comparison between cell lines and stimulation conditions; i.e., data from multiple plates or gels need to be combined during analysis. To increase the quality of merging data from multiple gels, we used “common positive controls” (CC; cf. Fig. 1), which are measurements for a strong signal condition that are performed on every gel in one or more wells. The lysate material used for CC is prepared in large batches at once and same lysate is later used on every gel. Measured CC values are then used for quality checks and normalization of gels against each other.
Fig. 1.
Representative Western blots used in the cell line comparison study for the (a, top) HME and (b, bottom) MDA-MB-231 cells. Cells were activated for 15 min with 12 ng/ml EGF and 40 ng/ml HRG. Labels on the right indicate the measured phospho-protein. Labels below blots indicate the treatment conditions used in the study: C− (negative control), C+ (positive control), CC (common control, used for normalization between gels, analogous to the positive control condition. Loading material used for the two CC lanes were from different batches.), i-Akt (Akt inhibition), i-Erk (Erk inhibition), i-p38 (p38 inhibition), i-JNK (JNK inhibition), and i-S3 (STAT3 inhibition). Data for the PI3K inhibition (i-P3K) was not used in the analysis. Note that the layout of the gels is slightly different for the two cell types. Loading control measurements using β-actin are shown for illustration purposes
Reagents
Epidermal growth factor (EGF, human recombinant) and heregulin-β1 (HRG-β, human recombinant) were purchased from Peprotech (Rocky Hill, NJ). L-15 and FBS were purchased from HyClone. Primary Western blot antibodies for phosphorylated p38 MAPK (Thr180/Tyr182, #9215), SAPK/JNK (Thr183/Tyr185, #9255), c-Jun (Ser73, #9164), Akt (Ser473, #4060) and STAT3 (Tyr705, #9145) were purchased from Cell Signaling Technology (Danvers, MA). As it is a direct target of JNK activity, c-Jun was used as a measure of output of integrated JNK signaling by assuming that c-Jun phosphorylation is directly proportional to JNK activation. Primary antibody for β-actin (mouse monoclonal, #A2228) was purchased from Sigma. WesternSure HRP goat-anti rabbit IgG (926–80,011) and HRP goat-anti mouse IgG (926–80,010) were purchased from LI-COR. U0126 (Erk1/2 inhibitor), JNK Inhibitor VIII, and Akt Inhibitor VIII were purchased from Calbiochem (EMD Millipore, Billerica, MA). Birb-796 (p38 MAPK inhibitor) was purchased from Selleckchem, and Stattic (STAT3 inhibitor) was purchased from Enzo Life Sciences. Protease and phosphatase inhibitors were obtained from Fisher. SDS-PAGE gels, PVDF membranes, SDS buffer and transfer buffer were purchased from Bio-Rad. Bicinchoninic acid assay reagents were obtained from Sigma. The peroxidase-based electrochemiluminescence (ECL) immunoassay substrate (femto maximum sensitivity) for Western blot was purchased from Thermo Scientific (Pittsburg, PA). All the other reagents were purchased from Sigma (St. Louis, MO) unless otherwise indicated.
Cell proliferation studies
We constructed GFP-expressing versions of 184A1L5 and MDA-MB-231 cells by transfecting them with GFP-Bsd lentiviral particles (GenTarget, Inc.). GFP-tagged cells were then seeded at a count of 10,000 cells/well using opaque black 96-well plates with transparent ultra-thin clear bottom (Corning, special optics plate). Cells were grown overnight and fluorescence from wells were measured daily to track growth. Fluorescence intensity was used as a measure of relative cell counts. All fluorescence readings were taken using a Cytation5 (BioTek) multi-well plate reader at an excitation wavelength of 485 nm and emission wavelength of 528 nm. Cell maintenance medium was changed every 12 h during the measurement period. Medium contained the respective inhibitors when used. Protein inhibitors and their doses matched those used in the Western blotting studies described above.
Cell motility studies
Motilities of the 184A1L5 and MDA-MB-231 cells were measured in duplicate using 12 well, flat bottom plates. Plates were coated with 300 μg/ml collagen-I and incubated overnight at room temperature. Plates were rinsed with nano-pure water before cells were plated. After plating, cells were allowed to attach to the collagen coated surface for at least 5 h. Cells were then treated with inhibitors for 2 h by adding the inhibitors at concentrations that were used in the Western blot experiments. Afterwards, media (L-15 media supplemented with 10% FBS for MDA-MB-231 cells and DFCl-1 media supplemented with 10% FBS and 12.5 ng/ml EGF for 184A1L5 cells) were replaced with fresh inhibitor containing media and cell positions were recorded at 4 min intervals for 2 h using time lapse imaging with a Zeiss Axio Vert.1 microscope with AxioCan MRm camera using the Zen software. Collected images were then analyzed using the Volocity image analysis software for cell tracking and in house Matlab codes for additional quantification and statistical analysis.
Motion of the cells were tracked at the individual cell level and average instantaneous velocity of cells (vins) and the root-mean-squared distance (RMSD) of cellular trajectories were computed. Average instantaneous velocity vins of a cell is computed from its measured locations as where k is the frame index of the image time-series data collected at intervals ∆t. This average instantaneous velocity is equal to the ratio of the total trajectory length of a cell to the total elapsed time. RMSD is a measure of the compactness of a trajectory. It was computed as <r k 2 > 1/2 where r k is the radial distance of the cell from the start of the trajectory at the k th time point, and the mean was taken over all the N measurement time points. Computed vins and RMSD for individual cells were then averaged over cells to obtain the reported values.
Network inference and analysis
We have combined the modular response analysis (MRA) and its Bayesian variable selection algorithm (BVSA) variant to identify the interactions among the sentinels. MRA reverse engineers a system to infer the existing regulatory interactions and their causality in a quantitative manner from measurements of how perturbing one system element affects the responses of the other elements in the system (Rkl below) (Bruggeman et al. 2002; Kholodenko et al. 2002; Andrec et al. 2005). MRA derives the local response matrix elements rkl = ∂xk/∂xl, which quantify the sensitivity of element k to changes in element l provided that the activities of all other nodes are kept constant, where xk denotes the activation of protein k in our case (Kholodenko et al. 2002; Andrec et al. 2005). For example, rerk,jnk corresponds to the Erk ← JNK interaction describing how JNK regulates Erk activation. When multiple interactions could be involved, measured changes in the activities are characterized with a global response matrix with elements Rkl = ∂lnxk/∂lnPl, which corresponds to the change in xk in response to a perturbation in component l, Pl. Rkl may be approximated as Rkl ≅ 2 (xk (l) - xk (0)) / (xk (l) + xk (0)) for small variations, where xk (0) and xk (l) are the activity levels of component k before and after the perturbation of component l, respectively (Kholodenko et al. 2002). MRA then computes the strengths of the interactions between module elements rkl from the measured Rkl by solving Σn rkn Rnl = Rkl where k ≠ l and the sum is over the nodes n (n ≠ k) that were perturbed in the experiments and rnn = −1 [51, 52]. Typically the total least squares estimation is used in Monte Carlo based simulations to estimate the local response coefficients (Gong et al. 2015; Santos et al. 2007). Utilizing this approach, we have computed the estimated probability distributions of the local response coefficients by forming random realizations of the node activities that were drawn from a normal distribution with a mean equal to those of the measured values. Standard deviation in the distribution was assumed to be 20% of the mean for each of the measured xk activity values, which is a typical deviation for Western blot experiments. Response matrices were calculated for 5 × 105 realizations of the randomly sampled data sets, and statistical distribution data obtained for the resulting local response coefficients were used to calculate the mean (μkl = <rkl>) and standard deviation (σkl) of the computed response coefficients. When tested, increasing the sampling 10-fold to 5 × 106 runs have hardly changed the results. So obtained distributions were well converged. MRA simulations were performed using the Matlab codes written in-house (Gong et al. 2015).
When the data are noisy, which usually is the case in biological experiments, global responses Rkl contain uncertainties and obtaining the local response coefficients rkl can be problematic. BVSA integrates Bayesian selection into MRA to infer regulatory interactions in a network and, instead of estimating rkl as in the MRA algorithm, it estimates the probability of interaction between nodes (Santra et al. 2013). The constitutive equation above is changed to Σn Akn rkn Rnl = 0, where the added variable Akn represents the probability of direct interaction between k and n (with Akn = 1 present to Akn = 0 absent scale). As a statistical inference algorithm, BVSA uses Bayesian statistics: An initial prior distribution is updated based on the experimental data using Bayes’ theorem to obtain the posterior distribution of the probabilities of connections between network modules (Santra et al. 2013). Our simulations utilized the Gibbs sampler Matlab program provided as Supplementary Material by Sandra et al. (Santra et al. 2013). Simulations were run for 1000 Gibbs scans (parameter: noit) and 5000 iterations (parameter: times). 50% of the early samples were assumed to be burn-ins and were not considered in the calculation of posterior edge probabilities. Prior and posterior distributions were computed using the default values for the other parameters of the program.
As further discussed in the Results section, two different strategies were used in our analysis. First, we utilized the combined BVSA-MRA approach that we have developed earlier (Gong et al. 2015). Combined BVSA-MRA approach integrates the predictions of the BVSA and MRA algorithms in two-stages:
-
i)
Perform BVSA analysis separately for 184A1L5 and MDA-MB-231 cells (cf., Table 1). Identify which interactions are likely to be present, i.e., find which Akl’s are not zero. The threshold to decide the presence of interactions was chosen to be equal to the average posterior edge probability, as recommended (Santra et al. 2013). Interaction set selection was done in two different ways; for each individual cell line by itself (BVSA-cell) or forming a collective (union) set by combining the interactions for both cell lines (BVSA-both).
-
ii)
In the MRA stage, set the local response matrix rkl elements corresponding to the interactions that were found to be not-likely to exist in the BVSA analysis to zero and compute the rkl values for the remaining interactions (cf., Tables 2, 3, 4, 5, and 6) and their interaction type. It should be noted that reactions with r > 0 are activating and with r < 0 are suppressing interactions.
Table 1.
Bayesian variable selection algorithm (BVSA) results
| Regulation of Erk by | ||||
| Cell line | Akt | p38 | STAT3 | JNK |
| HME | 0.154 | 0.250 | 0.661 | 0.165 |
| MDA-MB-231 | 0.283 | 0.494 | 0.194 | 0.196 |
| Regulation of Akt by | ||||
| Cell line | Erk | p38 | STAT3 | JNK |
| HME | 0.189 | 0.161 | 0.166 | 0.165 |
| MDA-MB-231 | 0.140 | 0.138 | 0.908 | 0.160 |
| Regulation of p38 by | ||||
| Cell line | Akt | Erk | STAT3 | JNK |
| HME | 0.151 | 0.207 | 0.750 | 0.185 |
| MDA-MB-231 | 0.122 | 0.425 | 0.126 | 0.485 |
| Regulation of JNK by | ||||
| Cell line | Akt | Erk | p38 | STAT3 |
| HME | 0.169 | 0.200 | 0.331 | 0.274 |
| MDA-MB-231 | 0.127 | 0.823 | 0.171 | 0.132 |
| Regulation of STAT3 by | ||||
| Cell line | Akt | Erk | p38 | JNK |
| HME | 0.189 | 0.759 | 0.429 | 0.204 |
| MDA-MB-231 | 0.338 | 0.201 | 0.231 | 0.181 |
Response matrix elements are classified into 3 classes: Elements with interaction probabilities A that are i) A < μr, ii) μr < A < μr + σr, and iii) A > μr + σr where μr and σr are the mean and standard deviation of the derived probabilities, respectively. In our analysis, they were μr = 0.288 and σr = 0.195 for the normal HME cells and μr = 0.294 and σr = 0.223 for the MDA-MB-231 breast cancer cells. Interactions in the first category are unlikely to be present. The ones in the last category are highly likely to be present in our system and they are marked in bold. Interactions with probabilities in the second category are likely to be present and they are marked in bold italics
Table 2.
Modular response analysis (MRA) results
| Regulation of Erk by | ||||
| Analysis set | Akt | p38 | STAT3 | JNK |
| HME cells | ||||
| BVSA-cell | N/I | N/I | -0.541 ± 0.049 | N/I |
| BVSA-both | N/I | −1.326 ± 0.701 | −1.223 ± 0.386 | N/I |
| MRA-0 | 0.500 ± 0.841 | −0.759 ± 1.521* | −1.287 ± 0.694 | 0.772 ± 1.216* |
| MRA-1 | N/I | -1.332 ± 0.725 | −1.261 ± 0.411 | 0.121 ± 0.296 |
| MRA-2 | N/I | -1.326 ± 0.701 | −1.223 ± 0.386 | N/I |
| MDA-MB-231 cells | ||||
| BVSA-cell | N/I | 0.318 ± 0.053 | N/I | N/I |
| BVSA-both | N/I | 0.294 ± 0.053 | 0.176 ± 0.058 | N/I |
| MRA-0 | 0.204 ± 0.114 | 0.251 ± 0.093 | −0.009 ± 0.130 | 0.099 ± 0.222 |
| MRA-1 | 0.195 ± 0.054 | 0.244 ± 0.080 | N/I | 0.130 ± 0.191 |
| MRA-2 | 0.199 ± 0.053 | 0.282 ± 0.053 | N/I | N/I |
| MRA-3 | N/I | 0.318 ± 0.053 | N/I | N/I |
Interaction strengths, rkl, are expressed as mean prediction +/− standard deviation. Interactions with r > 0 are activating and with r < 0 are suppressing interactions. Interaction are classified into 3 classes: Elements with interaction strengths that are i) |r| < μr, ii) μr < |r| < μr + σr, and iii) |r| > μr + σr where μr and σr are the mean and standard deviation of the absolute values of the derived interaction strengths (|r|), respectively. In our analysis, they were μr = 0.727 and σr = 0.461 for the normal HME cells and μr = 0.527 and σr = 0.567 for the MDA-MB-231 breast cancer cells. Interactions in the first category are unlikely to be present. The ones in the last category are highly likely to be present in our system and they are marked in bold. Interactions in the second category are likely to be present and they are marked in bold italics
N/I Not included in the analysis. These interactions were assumed to have r = 0
*The strength of these interactions had a large coefficient of variation (cv = σr/μr), and therefore, they were not well identified
Table 3.
Modular response analysis (MRA) results
| Regulation of Akt by | ||||
| Analysis set | Erk | p38 | STAT3 | JNK |
| HME | ||||
| BVSA-cell | N/I | N/I | N/I | N/I |
| BVSA-both | N/I | N/I | 0.087 ± 0.075 | N/I |
| MRA-0 (33.2 vs 66.8%)a |
−2.448 ± 0.813 1.655 ± 0.839 |
−1.772 ± 0.874 2.263 ± 0.771 |
−2.180 ± 0.879 2.440 ± 0.830 |
−0.526 ± 0.517 |
| MRA-1 (58.5 vs 41.5%)a |
−2.546 ± 0.792 2.157 ± 0.896 |
−2.079 ± 0.796 2.627 ± 0.811 |
−2.436 ± 0.762 2.581 ± 0.835 |
N/I |
| MRA-3 | N/I | N/I | N/I | -0.100 ± 0.175 |
| MDA-MB-231 | ||||
| BVSA-cell | N/I | N/I | 0.754 ± 0.050 | N/I |
| BVSA-both | N/I | N/I | 0.754 ± 0.050 | N/I |
| MRA-0 | 0.298 ± 0.464 | 0.141 ± 0.190 | 0.897 ± 0.112 | −0.941 ± 0.729 |
| MRA-1 | 0.557 ± 0.819 (*) | N/I | 0.857 ± 0.133 | −1.093 ± 1.092 (*) |
| MRA-2a | N/I | N/I | 0.832 ± 0.061 | −0.435 ± 0.151 |
| MRA-2b | −0.242 ± 0.103 | N/I | 0.793 ± 0.055 | N/I |
| MRA-3 | N/I | N/I | 0.754 ± 0.050 | N/I |
Notation used in this table is the same as in Table 2
aAs discussed in the main text, estimated interaction strengths for Akt ← Erk, Akt ← p38 and Akt ← STAT3 interactions had bimodal distributions in some of the analyzed cases. These values report the percentage of the runs that resulted in the indicated modes, respectively
Table 4.
Modular response analysis (MRA) results
| Regulation of p38 by | ||||
| Analysis set | Akt | Erk | STAT3 | JNK |
| HME | ||||
| BVSA-cell | N/I | N/I | -0.559 ± 0.050 | N/I |
| BVSA-both | N/I | −0.487 ± 0.575 | −0.860 ± 0.188 | 0.202 ± 0.360 |
| MRA-0 | 0.341 ± 0.243 | −0.253 ± 0.634 | −0.986 ± 0.192 | 0.591 ± 0.484 |
| MRA-1 | 0.318 ± 0.142 | N/I | -0.882 ± 0.132 | 0.622 ± 0.223 |
| MRA-2 | N/I | N/I | -0.721 ± 0.091 | 0.449 ± 0.182 |
| MRA-3 | N/I | N/I | -0.559 ± 0.050 | N/I |
| MDA-MB-231 | ||||
| BVSA-cell | N/I | −0.047 ± 0.687 | N/I | 0.990 ± 0.922 (*) |
| BVSA-both | N/I | 0.074 ± 0.722 | −0.082 ± 0.118 | 0.871 ± 0.935 (*) |
| MRA-0 | 0.494 ± 0.270 | 0.057 ± 0.409 | −0.558 ± 0.271 | 1.127 ± 0.550 |
| MRA-1 | 0.464 ± 0.286 | N/I | -0.497 ± 0.292 | 1.233 ± 0.247 |
| MRA-2a | N/I | N/I | -0.083 ± 0.079 | 1.011 ± 0.149 |
| MRA-2b | 0.044 ± 0.078 | N/I | N/I | 0.985 ± 0.140 |
| MRA-3 | N/I | N/I | N/I | 0.978 ± 0.138 |
Notation used in this table is the same as in Table 2
Table 5.
Modular response analysis (MRA) results
| Regulation of JNK by | ||||
| Dataset | Akt | Erk | p38 | STAT3 |
| HME | ||||
| BVSA-cell | N/I | N/I | 0.817 ± 0.282 | N/I |
| BVSA-both | N/I | −0.475 ± 0.144 | 0.847 ± 0.223 | N/I |
| MRA-0 | -0.085 ± 0.262 | 0.955 ± 0.498 | 1.945 ± 0.560 | 1.403 ± 0.435 |
| MRA-1 | N/I | 0.783 ± 0.399 | 1.810 ± 0.419 | 1.186 ± 0.341 |
| MDA-MB-231 | ||||
| BVSA-cell | N/I | 0.663 ± 0.092 | N/I | N/I |
| BVSA-both | N/I | 0.680 ± 0.115 | −0.022 ± 0.115 | N/I |
| MRA-0 | 0.034 ± 0.311 | 0.753 ± 0.261 | −0.063 ± 0.198 | −0.159 ± 0.390 |
| MRA-1 | N/I | 0.767 ± 0.132 | −0.069 ± 0.124 | −0.153 ± 0.084 |
| MRA-2 | N/I | 0.707 ± 0.067 | N/I | -0.136 ± 0.073 |
| MRA-3 | N/I | 0.663 ± 0.092 | N/I | N/I |
Notation used in this table is the same as in Table 2
Table 6.
Modular response analysis (MRA) results
| Regulation of STAT3 by | ||||
| Dataset | Akt | Erk | p38 | JNK |
| HME | ||||
| BVSA-cell | N/I | −0.887 ± 0.139 | −0.802 ± 0.174 | N/I |
| BVSA-both | 0.049 ± 0.106 | −0.872 ± 0.139 | −0.778 ± 0.180 | N/I |
| MRA-0 | 0.260 ± 0.136 | −0.579 ± 0.174 | −0.715 ± 0.145 | 0.408 ± 0.158 |
| MRA-1 | N/I | -0.796 ± 0.174 | −0.811 ± 0.169 | 0.156 ± 0.161 |
| MRA-2 | N/I | -0.887 ± 0.139 | −0.802 ± 0.174 | N/I |
| MDA-MB-231 | ||||
| BVSA-cell | 0.600 ± 0.074 | N/I | N/I | N/I |
| BVSA-both | 0.487 ± 0.127 | 0.360 ± 0.458 | 0.004 ± 0.392 | N/I |
| MRA-0 (90.8 vs 9.2%)a |
0.767 ± 0.333 | −1.305 ± 0.464 1.634 ± 0.483 |
−0.210 ± 0.372 | 2.954 ± 0.771 −2.623 ± 0.848 |
| MRA-1 (90.2 vs 9.8%)a |
0.760 ± 0.354 | −1.651 ± 0.543 2.176 ± 0.545 |
N/I | 3.170 ± 0.853 −2.994 ± 0.898 |
| MRA-2 (17.7 vs 82.3%)a |
N/I | −1.985 ± 0.547 2.288 ± 0.420 |
N/I | 3.753 ± 0.846 −3.212 ± 0.689 |
| MRA-3 | 0.600 ± 0.074 | N/I | N/I | N/I |
Notation used in this table is the same as in Table 2
aAs discussed in the main text, estimated interaction strengths for STAT3 ← Erk and STAT3 ← JNK interactions had bimodal distributions in some of the analyzed cases. These values report the percentage of the runs that resulted in the indicated modes, respectively
Second, we analyzed the perturbation data using MRA repeatedly such that interactions with the weakest strength were eliminated one at a time until convergent results were obtained (cf., Tables 2, 3, 4, 5, and 6) as follows:
-
i)
Assume that all interactions are possible and include all the interactions in the MRA study (i.e., no interaction is excluded; cf., MRA-0). For our study set with 5 proteins, for a particular protein, this corresponds to including all of the 4 interactions with other proteins. Then, identify which of the 4 interactions have the weakest strength.
-
ii)
Discard the weakest interaction, i.e., set the corresponding rkl to be zero, and repeat the MRA with the reduced set (cf., MRA-1). Repeat step ii iteratively to further reduce the network (cf., MRA-1, MRA-2, and MRA-3) until the remaining links have converged to a subset of significant interactions. In this notation MRA-N corresponds to a results set where N interactions were excluded from the computation.
Note that, in MRA studies, error with the predicted set of rkl values can be computed (Gong et al. 2015; Andrec et al. 2005). MRA were run for 5 × 105 random realizations of the perturbation data. We filtered the MRA-0 runs based on errors before computing the statistical distributions of the rkl values and their averages. If the error of estimation for how a protein X (X = Erk, Akt, p38, JNK, STAT3) was impacted by the inhibition perturbations for a particular run was εx, we defined the total error for that run as εtot 2 = Σx εx 2 where the sum is over all 5 proteins. We also defined an error value for each protein εx,low by computing the mean of the errors of the 1000 runs with the smallest εx. A total error value εtot,low was defined similarly. We then selected which parameter estimation tries were included in the mean and statistical distribution computations as the runs that satisfied both of the following two criteria: i) εx < 5 εx,low for every protein, i.e., for X = Erk, Akt, p38, JNK, STAT3. This criterion ensured that not a single protein dominated the error computation and each protein was considered equally. ii) εtot < 1.5 εtot,low, which allowed for the selection of runs with reasonable errors. We then computed the maximum error for each protein separately among the kept MRA-0 runs, and defined them as the error cutoff for selection in the subsequent MRA runs when certain interactions were excluded from the analysis (i.e., MRA-N with N > 0; Tables 2, 3, 4, 5, and 6). This ensured that a consistent error selection was applied as the number of interactions (i.e., model parameters) were changed in the analysis. It should also be noted that error cutoff determination was done separately for the 184A1L5 and MDA-MB-231 cells.
Results
Our comparative analysis revealed that there are considerable differences in how the investigated sentinel proteins regulate each other in normal 184A1L5 and MDA-MB-231 breast cancer epithelial cells. As explained above, how the investigated proteins respond to inhibition perturbations were measured using Western blots. Fig. 1 reports the results for a typical blot from multiple replicates. We first explain our results individually for each protein and then provide a description of the overall network for each cell line.
Regulation of sentinel proteins in HME and MDA-MB-231 epithelial cells
Regulatory interactions between sentinels were inferred using two different approaches that utilized the modular response analysis (MRA) and its Bayesian variable selection algorithm (BVSA) variant (cf., Methods section). The first method used a combined BVSA-MRA approach, which integrates the predictions of the BVSA and MRA algorithms in two-stages. The second method analyzed the perturbation data using MRA repeatedly by eliminating the weakest interaction one at a time until convergent results were obtained. Details of these approaches can be found in the Methods section.
Regulation of Erk
Erk and Akt are relatively upstream of p38, JNK and STAT3 in the HER signaling pathways. Therefore, interactions of the latter proteins with Erk or Akt can be considered as feedbacks. In the combined BVSA-MRA analysis (cf., Methods section), BVSA identified interactions of Erk with STAT3 in normal 184A1L5 cells and with p38 in MDA-MB-231 breast cancer cells (Table 1). Interestingly, MRA utilizing only these interactions resulted in conflicting outcomes (Table 2). In 184A1L5 cells, interaction with STAT3 was predicted only when Erk ← p38 interaction was also included. As discussed below, this outcome was later confirmed in the second approach. In cancer cells, none of the BVSA predicted interactions were found to have a strong strength.
In the second approach, for 184A1L5 cells, the potential Erk ← Akt regulatory interaction was the weakest, and its inclusion in the analysis actually increased the variances in other Erk interactions considerably (Table 2, MRA-0). This was most likely due to over-fitting of the system using unsupported degrees of freedom. Its negligence (MRA-1) decreased the variances of the computed parameters and illustrated that the Erk ← JNK interaction was insignificant (MRA-1) and that Erk ← STAT3 and Erk ← p38 interactions (MRA-2 and BVSA-both) were strong. As their r < 0, both of these interactions are suppressive, i.e., feedback from p38 and STAT3 both repress Erk activation. Compared to BVSA predictions (Table 1), MRA results confirmed the Erk ← STAT3 interaction. BVSA analysis showed a weak Erk ← p38 interaction but it was still second to STAT3 in strength (Table 1). We therefore conclude that Erk is regulated by both STAT3 and p38 in normal 184A1L5 HME cells.
MRA analysis indicated that there are no significant regulatory interactions between Erk and other studied sentinel proteins in MDA-MB-231 cancer cells (Table 2). Even the prediction for the strongest interaction has a strength that is less than 60% of the average interaction strength in the signaling network. Since BVSA prediction for the likelihood of Erk interaction with p38 was only moderate, we conclude that feedback to Erk is insignificant in MDA-MB-231 cells when EGFR pathways are stimulated.
Regulation of Akt
BVSA identified no interactions between Akt and other proteins in normal 184A1L5 cells but a very strong Akt ← STAT3 interaction was identified in MDA-MB-231 breast cancer cells (Table 1). Follow up analysis with MRA has fully supported these predictions that Akt was activated by STAT3 in MDA-MB-231 cells (Table 3).
Full MRA using the second approach has led to several interesting observations (Table 3). In 184A1L5 cells, distribution of the strengths of the Akt regulation by Erk, p38 and STAT3 were found to be bi-modal (Table 3 and Supplementary Fig. 1) and derived strengths of the three interactions were highly correlated (with correlation coefficients >0.95; Supplementary Fig. 2). This meant that Akt could be regulated by Erk/p38/STAT3 collectively. However, MRA analysis was not able to capture this unambiguously. Interestingly, Akt was predicted to be regulated by Erk, p38 and STAT in the same manner, i.e., all three either repress or activate Akt.
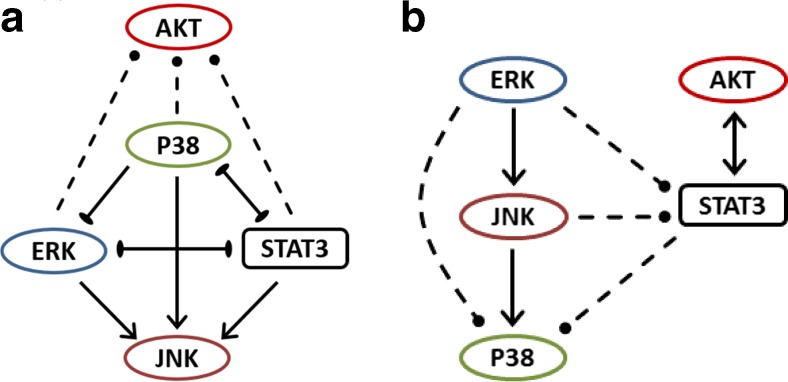
Fig. 2.
Inferred regulatory interaction networks in a 184A1L5 human mammary epithelial cells, and b MDA-MB-231 breast cancer cells. Solid lines indicate the regulatory interactions that were predicted by the used inference methods as robust interactions (cf. Tables 1, 2, 3, 4, 5, and 6). Arrows with a pointed end indicate activation interaction and with a blunt end indicate repression interaction. Arrows with dashed lines indicate the regulatory interactions which were either not reliably predicted by the used inference methods or predicted by only one of the two utilized methods. So their presence may not be as reliable as the interactions shown with solid lines. To identify these latter interactions, results of BVSA-MRA and repeated MRA analyses were contrasted for consensus and an inclusive selection was used. If an interaction was predicted as highly likely by either method, it was included in the model. When an interaction was predicted as likely by one of the methods and as unlikely by the other, consensus decision was weighted towards the repeated MRA method because pursuing MRA in different ways by leaving various interactions out (cf., Tables 2, 3, 4, 5, and 6) also provided information about the robustness of the derived strengths of the interactions. If the strength of the likely interaction under question was not significantly affected by the deletion of other interactions, the interaction was evaluated to be possible and included in the model. If it showed sensitivity to deletions, then it was excluded
Full MRA investigation of MDA-MB-231 cells confirmed the Akt ← STAT3 interaction which was predicted by the BVSA. Stepwise MRA analysis established the Akt ← p38 interaction to be the weakest (MRA-0, Table 3). Although the Akt ← JNK interaction seemed to be significant, its predicted interaction strength varied considerably and it was closely correlated with the Akt ← Erk interaction strength (MRA-1, Table 3). For example, the correlation coefficient between the predicted strengths of Akt ← JNK and Akt ← Erk interactions were 0.92 and 0.99 in MRA-0 and MRA-1 runs, respectively. This indeterminate condition was more obvious in MRA-2 runs, which showed that the strengths of the Akt ← JNK or Akt ← Erk interactions were weak when one of these interactions was excluded from the analysis (Table 3). In all cases, Akt ← STAT3 interaction was unambiguously predicted to be a strong activation reaction (Table 3). We therefore conclude that Akt ← STAT3 in MDA-MB-231 cells is the only Akt interaction supported by our data when EGFR pathways are stimulated with growth factors.
Regulation of p38
BVSA identified different interactions for p38 in normal 184A1L5 and MDA-MB-231 breast cancer epithelial cells (Table 1): p38 interacts with STAT3 in 184A1L5 cells and with Erk and JNK in MDA-MB-231 cells. The p38 ← STAT3 interaction in 184A1L5 cells was also supported by the MRA analysis (Table 4). However, although its distribution had a small variance, the strength of the p38 ← STAT3 interaction was found to depend on the inclusion of other interactions in MRA analysis. This dependence was obvious in particular for the p38 ← JNK interaction, which had an insignificant but sizable strength (Table 4). However, because of the low likelihood estimate for this interaction in BVSA analysis (Table 1), we conclude that the only significant regulatory interaction for p38 in 184A1L5 cells is its repression by STAT3 (Table 4).
For the MDA-MB-231 cells, BVSA analysis predicted p38 interactions with Erk and JNK. However, the likelihood estimate for both of the interactions were relatively low (Table 1), and only the p38 ← JNK interaction was clearly supported by the MRA analysis (Table 4). It should be noted that distribution of the interaction strength between p38 and JNK had an asymmetrical distribution with a long tail (Supplementary Fig. 3). Although its strength was very low (Table 4), obtained estimates for the p38 ← Erk interaction strength was highly correlated with the p38 ← JNK interaction strength (correlation coefficient was 0.99) and p38 ← Erk interaction strength had a relatively large variance. This correlation was also evident in the asymmetric distribution of the p38 ← Erk interaction strength that mimicked the distribution for p38 ← JNK interaction (Supplementary Fig. 3). Therefore, although our analysis predicted the p38 ← JNK activation interaction to be highly likely, p38 ← Erk activation interaction may also be present in the MDA-MB-231 cells because of the significant BVSA prediction. The MRA analysis indicated that the p38 ← STAT3 interaction may also be present in MDA-MB-231 cells (Table 4) but the strength of the interaction depended on the inclusion of other interactions in the analysis.
Fig. 3.
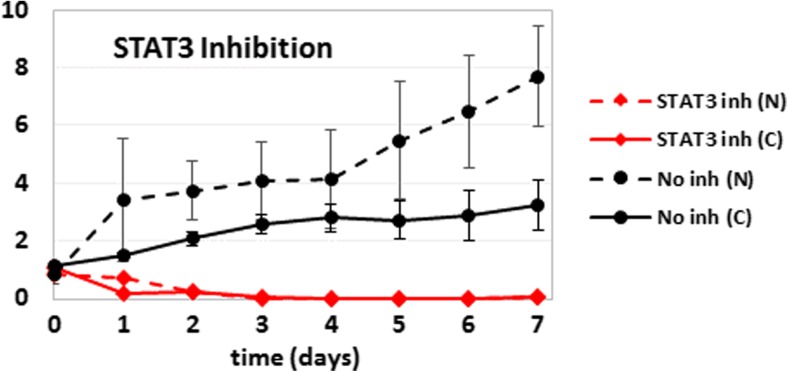
Cellular proliferation of HME (dashed lines; N) and MDA-MB-231 (solid lines; C) cells with (red) and without (black) inhibition of STAT3 with chemical inhibitors. The vertical axis shows the cell counts normalized with respect to the starting seeding. Error bars (one standard deviation) are shown for no inhibition cases only to illustrate the typical error sizes for this type of experiments
Regulation of JNK
Based on BVSA, JNK activation is regulated by p38 in normal 184A1L5 cells and by Erk in MDA-MB-231 breast cancer cells (Table 1). MRA analysis supported these predictions to a large degree but also identified the JNK ← Erk and JNK ← STAT3 activation as possible interactions in 184A1L5 cells (Table 5).
In the stepwise MRA approach, analysis for 184A1L5 cells has indicated that the only insignificant interaction between JNK and other sentinels was with Akt. The remaining three interactions were detected to activate JNK strongly (Table 5). It should be noted that, unlike the regulation of Akt case discussed above, distributions of the strengths of these three interactions were unimodal and had reasonably low correlations: pairwise correlations were only 0.59, 0.85, and 0.78. In the MDA-MB-231 cells, linkage of JNK to the rest of the network was more limited and only the JNK ← Erk activation interaction was detected (Table 5). This interaction was detected uniformly among different run sets (Table 5) and this MRA prediction agreed well with BVSA results (Tables 1 and 5).
Regulation of STAT3
STAT3 has been detected to be regulated by Erk and p38 in normal 184A1L5 cells and by Akt in MDA-MB-231 breast cancer cells in the BVSA analysis (Table 1). These predictions were supported by the MRA results (Table 6). In 184A1L5 cells, stepwise MRA approach established that both of the STAT3 ← Erk and STAT3 ← p38 interactions suppress the STAT3 activation strongly (Table 6).
For MDA-MB-231 cells, stepwise MRA has clearly predicted the STAT3 ← Akt activation interaction. Distribution of the predicted interaction strengths for STAT3 ← Erk and STAT3 ← JNK interactions was bi-modal (Supplementary Fig. 4) with opposite signs for the two interactions (Table 6; e.g., if STAT3 ← Erk is an activation interaction then STAT3 ← JNK is a repression or vice versa). Bi-modal distribution was observed regardless of the inclusion of the other two, i.e., STAT3 ← Akt and STAT3 ← p38, interactions in the analysis. However, the population of the “activation/repression” interaction prediction was dependent on the included interaction set (Table 6). Latter observation implies that both of STAT3 ← Erk and STAT3 ← JNK regulatory interactions may exist in the MDA-MB-231 cells. We therefore conclude that, as it is clearly predicted by both BVSA and MRA, STAT3 is regulated by Akt in MDA-MB-231 cells and it may also be regulated by Erk and JNK, where the latter two act in opposing manner when activating or repressing STAT3.
Fig. 4.
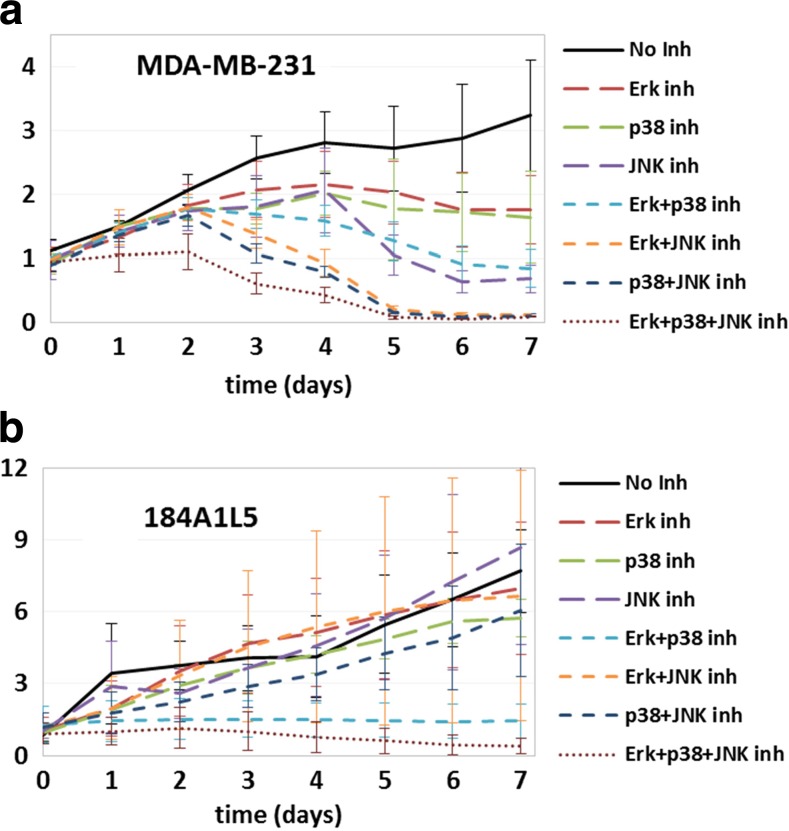
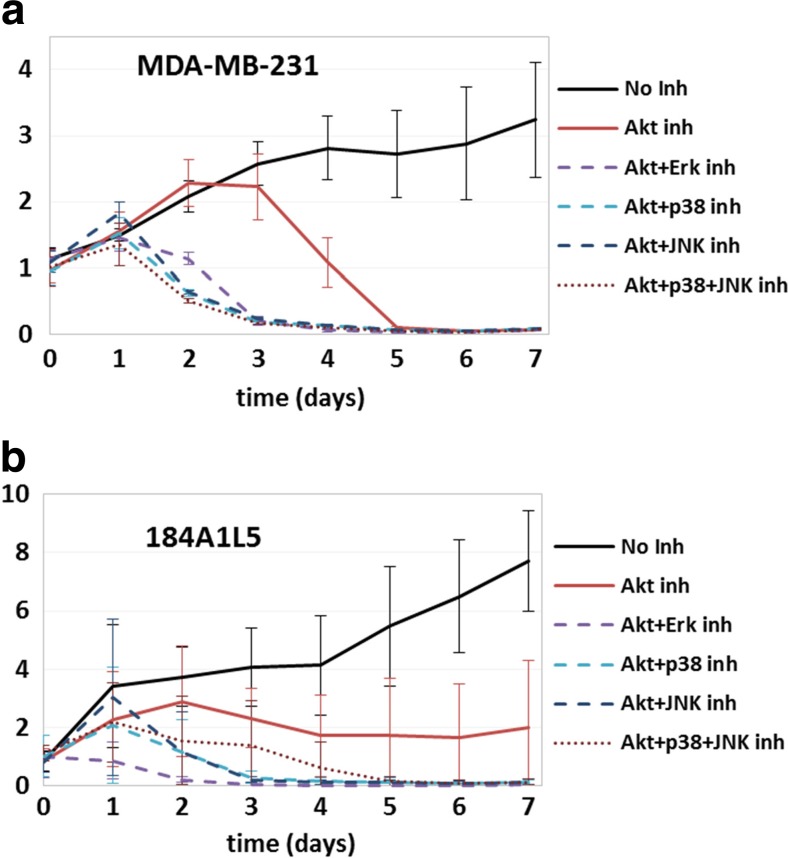
Cellular proliferation of a MDA-MB-231 and b 184A1L5 cells over 7-day duration for the inhibition conditions listed in the legends. Plots show the proliferation profiles when no protein was inhibited (solid line), single proteins were inhibited (long dashed lines), two proteins were inhibited simultaneously (short dashed lines), and three proteins were inhibited (dotted line). The vertical axis shows cell counts normalized with respect to the starting seeding. Typical error bars for this type of experiments can be seen in Fig. 3
Interaction networks in 184A1L5 and MDA-MB-231 epithelial cells
Inferred regulatory interactions between sentinel proteins can be combined into a network model. Figure 2 reports the signaling networks for normal 184A1L5 HME and MDA-MB-231 breast cancer epithelial cells. Comparison of the reconstructed network models show that, although there are similarities, EGFR signaling pathways are wired considerably differently in these two cell lines. Most notably:
Erk-p38-JNK network is common in both cell lines
JNK and p38 kinases are known to be functionally associated with the Erk pathway. Although observed interactions and feedbacks between these three kinases were somewhat different (Fig. 2), the Erk-p38-JNK pathway is largely conserved between 184A1L5 and MDA-MB-231 cells.
STAT3 is coupled to the Erk-p38-JNK pathway differently in normal and breast cancer cells
In 184A1L5 cells, STAT3 is tightly integrated to the Erk-p38-JNK pathway (Fig. 2a). In contrast, in MDA-MB-231 cells, it is directly coupled to Akt and only weakly associated with the Erk-p38-JNK pathway (Fig. 2b). This loose crosstalk could be an indicator of decoupled regulation of Erk and Akt pathways in cancer cells.
Interactions are triangulated in 184A1L5 cells
Interactions between Erk-p38-STAT3, between Erk-p38-JNK, and between p38-JNK-STAT3 are triangulated in 184A1L5 cells (Fig. 2a). This implies that occurrence of pathway substitution for signaling under different conditions would be much more likely in normal 184A1L5 HME cells. Lack of such permutations among interactions in MDA-MB-231 breast cancer cells implies that normal-to-cancer cell transformation causes a reduction in the inter-connectedness of regulatory interactions between Akt/STAT3 and key MAP kinases involved in proliferation and survival related responses.
Phenotypic cell response prediction based on inferred signaling networks
The ultimate aim of computational analysis and modeling studies is to generate predictive knowledge which can be used to determine the outcome of a system in response to various internal/external perturbations or stimuli. Querying biological data with model-based computational analysis can be a powerful approach to both eliminating invalid hypotheses and discovering the most physiologically relevant interactions (Shankaran et al. 2013; Shankaran et al. 2007a; Shankaran et al. 2006; Shankaran et al. 2007b; Shankaran et al. 2012; Shankaran et al. 2008). As described above, we have used the BVSA and MRA methods to infer network models from experimental perturbation data for EGFR signaling networks that regulate increased cell proliferation/viability and motility processes. Since the predictions of any inference method may contain false positives and false negatives, we have used the derived network models to make predictions and develop new hypotheses related to the proliferation and motility of 184A1L5 and MDA-MB-231 cells to show the utility and predictive power of the inferred models. We have then further validated the derived networks by experimentally testing the model predictions.
Predictions for cell proliferation
We have used the derived network models to make predictions and develop new hypotheses related to the proliferation of 184A1L5 and MDA-MB-231 cells. It is clear from the obtained network topologies (Fig. 2) that the Akt-STAT3 pathway is loosely connected to the Erk-p38-JNK pathway in MDA-MB-231 cells while the two pathways are coupled at multiple levels in 184A1L5 cells. JNK and p38 kinases are both implicated in the regulation of stress responses and cell survival. The Erk pathway plays a major role in the proliferation and motility of many cell lines, and STAT3 is an important transcriptional regulator of cell survival and proliferation. Crosstalk between related signaling pathways can enable pathway substitution when cells respond to external stimuli. Based on this expectation, we have hypothesized that tighter linkage between STAT3 and Erk-JNK-p38 pathway would make possible pathway substitution much more likely thus lowering the effects of STAT3 blocking in 184A1L5 cells. Therefore, inhibition of STAT3 is expected to decrease the proliferation and viability of the MDA-MB-231 cells more than the 184A1L5 cells. Although the effects of STAT3 inhibition on cell proliferation/death are very strong in both MDA-MB-231 and 184A1L5 cells, which is expected because of STAT3’s severe toxicity, we have found that STAT3 inhibition causes cell death about one day earlier in MDA-MB-231 cells than 184A1L5 cells (Fig. 3; an expanded and simplified version of the figure can be seen in Supplementary Fig. 5), which supported our expectation based on the inferred models.
Fig. 5.
Cellular proliferation of a MDA-MB-231 and b 184A1L5 cells over 7-day duration when Akt was inhibited alone or together with other proteins. Plots show the proliferation profiles when no protein was inhibited (solid black line), Akt was inhibited alone (solid red line), Akt and another protein (listed in the legends) were inhibited (dashed lines), and Akt, p38 and JNK were inhibited simultaneously (dotted line). The vertical axis shows cell counts normalized with respect to the starting seeding. Typical error bars for this type of experiments can be seen in Fig. 3
We have also hypothesized that pathway substitution due to the triangulated interactions between Erk-p38-JNK would lower the impact of inhibition of these proteins in normal 184A1L5 cells compared to MDA-MB-231 breast cancer cells. Comparison of the results for the two cell lines have validated this prediction (Fig. 4): In MDA-MB-231 cells, individual inhibition of Erk, JNK or p38 lowered the proliferation rate (Fig. 4a) but individual inhibition of Erk, JNK or p38 had only a minimal impact on the proliferation of 184A1L5 cells (Fig. 4b) where the impact of p38 inhibition was largest of the three. Implications of our hypothesis were much more evident in multi-inhibition cases. Since the interactions along the Erk-p38-JNK pathway has a linear branch structure in MDA-MB-231 cells (Fig. 2b), inhibition of two or all three of these proteins are expected to have a larger impact than inhibition of individual proteins. Our results supported this expectation fully in the MDA-MB-231 cells where cell survival was reduced for dual inhibition cases and cells started to die after a few days (Fig. 4a). For the 184A1L5 HME cells, we found that dual inhibition of Erk + JNK or p38 + JNK had a minimal impact on cellular growth while dual inhibition of Erk and p38 had a more noticeable impact (Fig. 4b). The latter was most likely due to p38 and Erk being upstream of JNK and activating JNK (Fig. 2b). This amplifies their regulatory role, and their simultaneous inhibition lowers JNK activation as well thus significantly reducing the total signaling through the Erk-p38-JNK pathway. As expected simultaneous inhibition of all three proteins led to cellular death and corresponded to the lowest cell count among the studied combinations in both cell lines (Fig. 4a, b).
Akt is a regulator of cell survival, and therefore, controls cellular growth and apoptosis. Our network reconstruction analysis (Fig. 2) have shown that Akt interacts strongly with STAT3 and the Akt-STAT3 pathway is weakly coupled to Erk-p38-JNK in MDA-MB-231 cells while Akt crosstalks with STAT3-Erk-p38-JNK at multiple levels in 184A1L5 cells. We therefore hypothesized that, because of its direct role, the impact of Akt inhibition would be stronger in MDA-MB-231 cells. This is because Akt inhibition would reduce the activation of STAT3 and hence simultaneously lower proliferation and induce cell death. In contrast, although it would have some impact, Akt inhibition would affect the proliferation of 184A1L5 cells to a lesser extent. Our results have validated this hypothesis: We have found that while it reduced the proliferation of 184A1L5 cells (Fig. 5a), Akt inhibition led to the death of MDA-MB-231 cells (Fig. 5b). We have further tested our hypothesis by blocking both Akt and Erk-p38-JNK pathways in cells. One implication of our hypothesis is that simultaneous blocking of Akt and some of the elements of the Erk-p38-JNK pathway should lower the proliferation below levels observed for Akt inhibition alone by shutting down both signaling pathways. Our results validated this expectation for both MDA-MB-231 and 184A1L5 cells (Fig. 5).
Predictions for cell motility
Like proliferation, increased motility is another hallmark of cancer related cellular processes. We have therefore also investigated how the observed differences in the coupling between Akt-STAT3 and Erk-p38-JNK pathways may impact the motilities of the MDA-MB-231 breast cancer and normal 184A1L5 HME cells. We measured the cellular motilities under 12 different treatment conditions by inhibiting one or more of the investigated sentinels (cf., Methods section). Trajectories of individual cells were analyzed to compute the average instantaneous velocity vins of the cells and the root-mean-squared distance RMSD of cellular trajectories (Table 7). Computed vins and RMSD were highly correlated for the studied conditions: Correlation coefficient between vins and RMSD was 0.97 for 184A1L5 and 0.94 for MDA-MB-231 cells. Therefore, derived conclusions were equally applicable to both properties.
Table 7.
Motility of MDA-MB-231 breast cancer and human mammary epithelial cells
| Inhibitor treatment | Normal | Cancer | ||
|---|---|---|---|---|
| vins (μm/min) | RMSD (μm) | vins (μm/min) | RMSD (μm) | |
| No inhibitor | 0.086 ± 0.038 | 3.99 ± 4.72 | 0.228 ± 0.155 | 8.25 ± 9.98 |
| Erk | 0.100 ± 0.050 | 4.31 ± 4.99 | 0.119 ± 0.113 | 5.80 ± 8.75 |
| p38 | 0.089 ± 0.043 | 3.82 ± 4.53 | 0.197 ± 0.155 | 8.67 ± 11.43 |
| JNK | 0.187 ± 0.093 | 11.76 ± 12.95 | 0.216 ± 0.126 | 10.16 ± 12.85 |
| Akt | 0.134 ± 0.057 | 8.74 ± 9.61 | 0.205 ± 0.135 | 8.52 ± 11.92 |
| STAT3 | 0.057 ± 0.051 | 2.15 ± 3.52 | 0.105 ± 0.097 | 4.40 ± 6.94 |
| STAT3 + p38 | 0.051 ± 0.071 | 1.72 ± 3.20 | 0.123 ± 0.131 | 4.11 ± 6.70 |
| STAT3 + JNK | 0.046 ± 0.035 | 1.79 ± 2.28 | 0.115 ± 0.109 | 3.66 ± 5.98 |
| p38 + JNK | 0.080 ± 0.061 | 3.61 ± 5.07 | 0.200 ± 0.161 | 7.50 ± 10.01 |
| Erk + p38 | 0.087 ± 0.099 | 2.78 ± 5.07 | 0.103 ± 0.094 | 4.63 ± 6.36 |
| Erk + JNK | 0.087 ± 0.069 | 4.56 ± 6.26 | 0.129 ± 0.125 | 5.10 ± 7.16 |
| Erk + Akt | 0.098 ± 0.110 | 4.55 ± 5.05 | 0.093 ± 0.081 | 3.55 ± 5.26 |
How vins and RMSD were computed is explained in the Methods section. They were computed for each cell individually and then averaged over the cells to obtain the tabulated values. Cases that show significant differences from the no inhibitor control case are highlighted in bold. Plain bold marks significantly different than control where motility decreased upon inhibition. Italicized bold marks significantly different from control where motility increased upon inhibition
Our most notable findings were that: i) MDA-MB-231 cells had a considerably higher motility than 184A1L5 cells (Table 7). ii) STAT3 inhibition lowered the motilities of both MDA-MB-231 and 184A1L5 cell lines significantly (Table 7). This observation was valid when STAT3 was inhibited alone or together with other proteins. Iii) Akt or JNK inhibition increased the motility of normal 184A1L5 HME cells while not affecting the motility of the MDA-MB-231 breast cancer cells. Akt has been shown to function by either enhancing or reducing the cell migration (Chin and Toker 2009). Its opposing dual functionality depends on the cell type. Our results show that Akt is differentially involved in the motility of normal and cancer epithelial cells, which supports its possible cell type dependent multiple functional role. Observed increase in cell motility upon Akt or JNK inhibition was negated when either Erk or p38 was also inhibited (compare Erk + Akt, Erk + JNK and p38 + JNK dual inhibition cases to Akt or JNK inhibition cases in Table 7). Dual inhibition of Erk and p38 also decreased the motility of 184A1L5 HME cells (Table 7). iv) Like STAT3, Erk strongly decreased the motility of MDA-MB-231 cells when it was inhibited by itself or together with other proteins (Table 7).
These results parallel the results of the proliferation studies discussed earlier. In particular, as in the proliferation case: i) STAT3 is a key regulator of cell motility in both cells, and ii) pathway substitution which results from alternative interaction pathways between signaling elements has a more pronounced effect on the motility of 184A1L5 cells compared to MDA-MB-231 cells. The dominant role of Erk in regulating the motility of MDA-MB-231 cells is likely due to its position as the most upstream element of the almost linear Erk-p38-JNK pathway (Fig. 2b) while the complex topology of the regulatory network in 184A1L5 cells (Fig. 2a) requires simultaneous inhibition of multiple elements to negate the effects of pathway substitution.
Discussion and conclusion
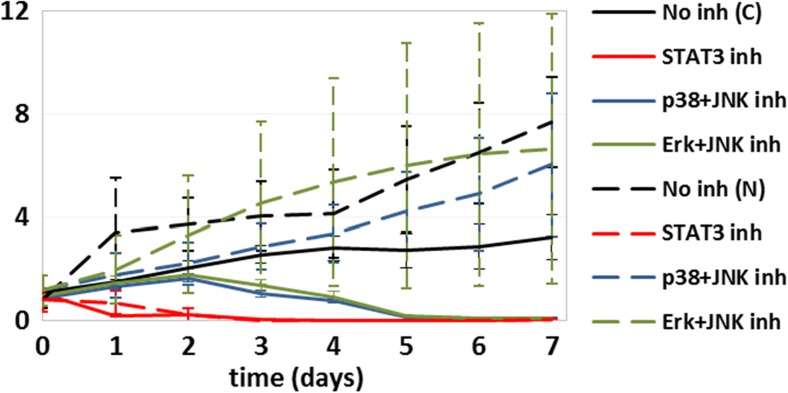
We have used an integrated reverse engineering approach to infer regulatory interactions from perturbation datasets. It is anticipated that the use of such integrated approaches to model-based computational analysis of the biological data will lead to more rapid advancement in the understanding of cellular signaling networks involved in oncogenesis and other diseases. We have combined the BVSA and MRA methods to infer network models from experimental perturbation data for EGFR signaling networks that regulate increased cell proliferation/viability and motility processes, which are hallmarks of cancer. Based on the inferred networks, we have developed new hypotheses about the differential role of pathway crosstalk and substitution in normal 184A1L5 HME and MDA-MB-231 breast cancer cells and then successfully validated these hypotheses in in vitro experiments. This enabled us to establish the major differences in the regulatory interactions between key kinases and STAT3 in 184A1L5 and MDA-MB-231 cells and the effects of identified differences on the cellular proliferation and motility responses. We have found that there are substantial differences in how the Akt-STAT3 and Erk-JNK-p38 pathways are cross-wired in MDA-MB-231 breast cancer and normal 184A1L5 epithelial cells. The observed differences and their in vitro validation have provided new ideas that may lead to improved therapeutic strategies: For example, instead of inhibiting STAT3, which has severe toxic side effects (Banerjee and Resat 2015), simultaneous inhibition of JNK together with Erk or p38 could be equally effective in imposing cell death in MDA-MB-231 (and possibly other triple-negative breast cancer) cells while only reducing the survival of the normal epithelial cells but not killing them (Fig. 6). Identification of such new therapy ideas that use multiple targets to significantly reduce toxicity and other side effects could have a wide impact in oncology and other diseases. More detailed and targeted studies that will further extend and validate our results in vivo can allow the establishment of carefully designed inhibition conditions which minimize toxicity to normal cells while effectively causing the death of cancer cells by taking advantage of the network models such as the one derived in this study.
Fig. 6.
Cellular proliferation of MDA-MB-231 (solid lines; C) and 184A1L5 (dashed lines; N) cells when no protein was inhibited (black), STAT3 was inhibited (red), p38 and JNK were inhibited (blue), and Erk and JNK were inhibited (green). Dual inhibition of p38 and JNK or Erk and JNK cause death of MDA-MB-231 cells but not of 184A1L5 HME cells. The vertical axis shows cell counts normalized with respect to the starting seeding. Typical error bars for this type of experiments can be seen in Fig. 3
It should be noted that this study utilized a defined small set of molecules and signaling pathways. Although this may bias the outcomes, there were two main reasons for this selection: i) Use of a larger protein set could be beneficial but the required effort increases steeply as N2 with increasing set size because all combinations of inhibition (perturbation) and protein activities need to be measured. ii) Ability to predict the phenotypic response of cells with the minimal amount of required information is more desirable. Therefore, in addition to keeping the involved effort manageable, investigating a small set of proteins was intentional. As our results show, selected group of proteins were suitable to successfully predict the response differences between 184A1L5 HME and MDA-MB-231 breast cancer cells.
Electronic supplementary material
(DOCX 189 kb)
Acknowledgements
The research described in this paper was funded by the National Institutes of Health Grant 5R01GM072821 and by the WSU start-up funds to H.R.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s12079-017-0389-3) contains supplementary material, which is available to authorized users.
References
- Aksamitiene E, Kiyatkin A, Kholodenko BN. Cross-talk between mitogenic Ras/MAPK and survival PI3K/Akt pathways: a fine balance. Biochem Soc Trans. 2012;40:139–146. doi: 10.1042/BST20110609. [DOI] [PubMed] [Google Scholar]
- Alexander J, Lim D, Joughin BA, Hegemann B, Hutchins JRA, Ehrenberger T et al (2011) Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci Signal 4(179). doi:10.1126/scisignal.2001796 [DOI] [PMC free article] [PubMed]
- Andrec M, Kholodenko BN, Levy RM, Sontag E. Inference of signaling and gene regulatory networks by steady-state perturbation experiments: structure and accuracy. J Theor Biol. 2005;232(3):427–441. doi: 10.1016/j.jtbi.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Arteaga C. Targeting HERI/EGFR: a molecular approach to cancer therapy. Semin Oncol. 2003;30(3):3–14. doi: 10.1016/S0093-7754(03)70010-4. [DOI] [PubMed] [Google Scholar]
- Arteaga CL. ErbB-targeted therapeutic approaches in human cancer. Exp Cell Res. 2003;284(1):122–130. doi: 10.1016/S0014-4827(02)00104-0. [DOI] [PubMed] [Google Scholar]
- Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol. 2011;12(2):104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- Banerjee K, Resat H. Constitutive activation of STAT3 in breast cancer cells: a review. Int J Cancer. 2015 doi: 10.1002/ijc.29923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berclaz G, Altermatt HJ, Rohrbach V, Siragusa A, Dreher E, Smith PD. EGFR dependent expression of STAT3 (but not STAT1) in breast cancer. Int J Oncol. 2001;19(6):1155–1160. doi: 10.3892/ijo.19.6.1155. [DOI] [PubMed] [Google Scholar]
- Britten CD. Targeting ErbB receptor signaling: a pan-ErbB approach to cancer. Mol Cancer Ther. 2004;3(10):1335–1342. [PubMed] [Google Scholar]
- Bruggeman FJ, Westerhoff HV, Hoek JB, Kholodenko BN. Modular response analysis of cellular regulatory networks. J Theor Biol. 2002;218(4):507–520. doi: 10.1016/S0022-5193(02)93096-1. [DOI] [PubMed] [Google Scholar]
- Chen J, Somanath PR, Razorenova O, Chen WS, Hay N, Bornstein P, et al. Akt1 regulates pathological angiogenesis, vascular maturation and permeability in vivo. Nat Med. 2005;11(11):1188–1196. doi: 10.1038/nm1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiacchiera F, Grossi V, Cappellari M, Peserico A, Simonatto M, Germani A, et al. Blocking p38/ERK crosstalk affects colorectal cancer growth by inducing apoptosis in vitro and in preclinical mouse models. Cancer Lett. 2012;324(1):98–108. doi: 10.1016/j.canlet.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Chin YR, Toker A. Function of Akt/PKB signaling to cell motility, invasion and the tumor stroma in cancer. Cell Signal. 2009;21(4):470–476. doi: 10.1016/j.cellsig.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb MH. MAP kinase pathways. Prog Biophys Mol Biol. 1999;71(3–4):479–500. doi: 10.1016/S0079-6107(98)00056-X. [DOI] [PubMed] [Google Scholar]
- Davidson NE, Gelmann EP, Lippman ME, Dickson RB. Epidermal growth factor receptor Gene expression in estrogen receptor-positive and negative human breast cancer cell lines. Mol Endocrinol. 1987;1(3):216–223. doi: 10.1210/mend-1-3-216. [DOI] [PubMed] [Google Scholar]
- Dong J, Opresko LK, Dempsey PJ, Lauffenburger DA, Coffey RJ, Wiley HS. Metalloprotease-mediated ligand release regulates autocrine signaling through the epidermal growth factor receptor. Proc Natl Acad Sci U S A. 1999;96(11):6235–6240. doi: 10.1073/pnas.96.11.6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Dise RS, Edelblum KL, Polk DB. p38 kinase regulates epidermal growth factor receptor downregulation and cellular migration. EMBO J. 2006;25(24):5683–5692. doi: 10.1038/sj.emboj.7601457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Zhang Y, Shankaran H, Resat H. Integrated analysis reveals that STAT3 is central to the crosstalk between HER/ErbB receptor signaling pathways in human mammary epithelial cells. Mol BioSyst. 2015;11:146–158. doi: 10.1039/C4MB00471J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks BS, Opresko LK, Wiley HS, Lauffenburger D. Coregulation of epidermal growth factor receptor/human epidermal growth factor receptor 2 (HER2) levels and locations: quantitative analysis of HER2 overexpression effects. Cancer Res. 2003;63(5):1130–1137. [PubMed] [Google Scholar]
- Holbro T, Civenni G, Hynes NE. The ErbB receptors and their role in cancer progression. Exp Cell Res. 2003;284(1):99–110. doi: 10.1016/S0014-4827(02)00099-X. [DOI] [PubMed] [Google Scholar]
- Hui LJ, Bakiri L, Stepniak E, Wagner EF. p38 alpha - a suppressor of cell proliferation and tumorigenesis. Cell Cycle. 2007;6(20):2429–2433. doi: 10.4161/cc.6.20.4774. [DOI] [PubMed] [Google Scholar]
- Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol. 2009;21(2):177–184. doi: 10.1016/j.ceb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439(7073):168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- Joslin EJ, Opresko LK, Wells A, Wiley HS, Lauffenburger DA. EGF-receptor-mediated mammary epithelial cell migration is driven by sustained ERK signaling from autocrine stimulation. J Cell Sci. 2007;120(20):3688–3699. doi: 10.1242/jcs.010488. [DOI] [PubMed] [Google Scholar]
- Joslin EJ, Shankaran H, Opresko LK, Bollinger N, Lauffenburger DA, Wiley HS. Structure of the EGF receptor transactivation circuit integrates multiple signals with cell context. Mol BioSyst. 2010;6(7):1293–1306. doi: 10.1039/c003921g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz M, Amit I, Yarden Y. Regulation of MAPKs by growth factors and receptor tyrosine kinases. Biochimica Et Biophysica Acta-Molecular Cell Research. 2007;1773(8):1161–1176. doi: 10.1016/j.bbamcr.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholodenko BN, Kiyatkin A, Bruggeman FJ, Sontag E, Westerhoff HV, Hoek JB. Untangling the wires: a strategy to trace functional interactions in signaling and gene networks (vol 99, pg 12841, 2002) Proc Natl Acad Sci U S A. 2002;99(23):15245. doi: 10.1073/pnas.192442699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger B, Sieber A, Fritsche-Guenther R, Witzel F, Berry L, Schumacher D et al (2013) Network quantification of EGFR signaling unveils potential for targeted combination therapy. Mol Syst Biol 9. doi:10.1038/msb.2013.29 [DOI] [PMC free article] [PubMed]
- Kumar N, Afeyan R, Kim HD, Lauffenburger DA. Multipathway model enables prediction of kinase inhibitor cross-talk effects on migration of Her2-overexpressing mammary epithelial cells. Mol Pharmacol. 2008;73(6):1668–1678. doi: 10.1124/mol.107.043794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- Libermann TA, Razon N, Bartal AD, Yarden Y, Schlessinger J, Soreq H. Expression of epidermal growth-factor receptors in human-brain tumors. Cancer Res. 1984;44(2):753–760. [PubMed] [Google Scholar]
- Mueller KL, Powell K, Madden JM, Eblen ST, Boerner JL. EGFR tyrosine 845 phosphorylation-dependent proliferation and transformation of breast cancer cells require activation of p38 MAPK. Transl Oncol. 2012;5(5):327–334. doi: 10.1593/tlo.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naegle KM, White FM, Lauffenburger DA, Yaffe MB. Robust co-regulation of tyrosine phosphorylation sites on proteins reveals novel protein interactions. Mol BioSyst. 2012;8(10):2771–2782. doi: 10.1039/c2mb25200g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RM, Holbro T, Hynes NE. Distinct roles for phosphoinositide 3-kinase, mitogen-activated protein kinase and p38 MAPK in mediating cell cycle progression of breast cancer cells. Oncogene. 2002;21(29):4567–4576. doi: 10.1038/sj.onc.1205555. [DOI] [PubMed] [Google Scholar]
- Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366(1):2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Pawson T, Warner N. Oncogenic re-wiring of cellular signaling pathways. Oncogene. 2007;26(9):1268–1275. doi: 10.1038/sj.onc.1210255. [DOI] [PubMed] [Google Scholar]
- Prenzel N, Zwick E, Leserer M, Ullrich A. Tyrosine kinase signalling in breast cancer. Epidermal growth factor receptor: convergence point for signal integration and diversification. Breast Cancer Res. 2000;2(3):184–190. doi: 10.1186/bcr52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26(22):3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004;117:1281–1283. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- Rodland KD, Bollinger N, Ippolito D, Opresko LK, Coffey RJ, Zangar R, et al. Multiple mechanisms are responsible for transactivation of the epidermal growth factor receptor in mammary epithelial cells. J Biol Chem. 2008;283(46):31477–31487. doi: 10.1074/jbc.M800456200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos SDM, Verveer PJ, Bastiaens PIH. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat Cell Biol. 2007;9(3):324–U139. doi: 10.1038/ncb1543. [DOI] [PubMed] [Google Scholar]
- Santra T, Kolch W, Kholodenko BN (2013) Integrating Bayesian variable selection with modular response analysis to infer biochemical network topology. BMC Syst Biol 7. doi:10.1186/1752-0509-7-57 [DOI] [PMC free article] [PubMed]
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103(2):211–225. doi: 10.1016/S0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Sen M, Joyce S, Panahandeh M, Li CY, Thomas SM, Maxwell J, et al. Targeting Stat3 abrogates EGFR inhibitor resistance in cancer. Clin Cancer Res. 2012;18(18):4986–4996. doi: 10.1158/1078-0432.CCR-12-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaran H, Resat H, Wiley HS. Cell surface receptors for signal transduction and ligand transport: a design principles study. PLoS Comput Biol. 2007;3(6):986–999. doi: 10.1371/journal.pcbi.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaran H, Wiley HS, Resat H. Modeling the effects of HER/ErbB1-3 coexpression on receptor dimerization and biological response. Biophys J. 2006;90(11):3993–4009. doi: 10.1529/biophysj.105.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaran H, Wiley HS, Resat H. Receptor downregulation and desensitization enhance the information processing ability of signalling receptors. BMC Syst Biol. 2007;1:48. doi: 10.1186/1752-0509-1-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaran H, Zhang Y, Chrisler WB, Ewald JA, Wiley HS, Resat H. Integrated experimental and model-based analysis reveals the spatial aspects of EGFR activation dynamics. Mol BioSyst. 2012;8(11):2868–2882. doi: 10.1039/c2mb25190f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaran H, Zhang Y, Opresko L, Resat H. Quantifying the effects of co-expressing EGFR and HER2 on HER activation and trafficking. Biochem Biophys Res Commun. 2008;371(2):220–224. doi: 10.1016/j.bbrc.2008.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankaran H, Zhang Y, Tan YB, Resat H (2013) Model-based analysis of HER activation in cells co-expressing EGFR, HER2 and HER3. Plos Computational Biology 9(8). doi:10.1371/journal.pcbi.1003201 [DOI] [PMC free article] [PubMed]
- Song G, Ouyang G, Bao S. The activation of Akt/PKB signaling pathway and cell survival. J Cell Mol Med. 2005;9(1):59–71. doi: 10.1111/j.1582-4934.2005.tb00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stampfer MR, Pan CH, Hosoda J, Bartholomew J, Mendelsohn J, Yaswen P. Blockage of EGF receptor signal transduction causes reversible arrest of normal and immortal human mammary epithelial cells with synchronous reentry into the cell cycle. Exp Cell Res. 1993;208:175–188. doi: 10.1006/excr.1993.1236. [DOI] [PubMed] [Google Scholar]
- Volinsky N, Kholodenko BN (2013) Complexity of receptor tyrosine kinase signal processing. Cold Spring Harb Perspect Biol 5(8). doi:10.1101/cshperspect.a009043 [DOI] [PMC free article] [PubMed]
- Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9(8):537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- Wu CJ, Qian XL, O'Rourke DM. Sustained mitogen-activated protein kinase activation is induced by transforming erbB receptor complexes. DNA Cell Biol. 1999;18(10):731–741. doi: 10.1089/104454999314872. [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2(2):127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Yue P, Turkson J. Targeting STAT3 in cancer: how successful are we? Expert Opin Investig Drugs. 2009;18(1):45–56. doi: 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Opresko L, Shankaran H, Chrisler WB, Wiley HS, Resat H. HER/ErbB receptor interactions and signaling patterns in human mammary epithelial cells. BMC Cell Biol. 2009;10:78. doi: 10.1186/1471-2121-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, et al. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol Cell Proteomics. 2005;4(9):1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 189 kb)