Heart failure is a progressive disease with significant morbidity and mortality, and can occur in response to the loss of cardiac muscle cells (cardiac myocytes) after cardiac injury. In mammals, the majority of cardiac myocytes exit the cell cycle soon after birth, hence the adult heart is unable to significantly regenerate after an insult. Reprogramming of pluripotent human embryonic stem cells (HESC) to functional cardiac myocytes represents an attractive approach to replace damaged cardiac myocytes to repair the heart. Therefore, scientists are using this model to study cardiac development in vivo by understanding key transcriptional changes for the specification of HESC towards the cardiac lineage. Transcriptional activity is regulated by protein coding and non-coding RNAs. Protein coding genes are the most well studied sequence, but only account for < 2% of the genome (Morris and Mattick, 2014). In the last decade, advances in sequencing technologies and computational methods have uncovered non-coding RNAs (ncRNAs), initially thought to be “junk”, being transcribed in many cell types and tissues. Among these ncRNAs are long non-coding RNAs (lncRNA; 200 nt in length, linear RNA) and circular RNAs (circRNA; ~ 100 nt, single stranded RNA forming a circle through covalent binding). Both lncRNAs and circRNAs have been shown to play roles in transcription regulation during cardiac development and disease; reviewed in (McMullen and Drew, 2016, Li et al., 2017a).
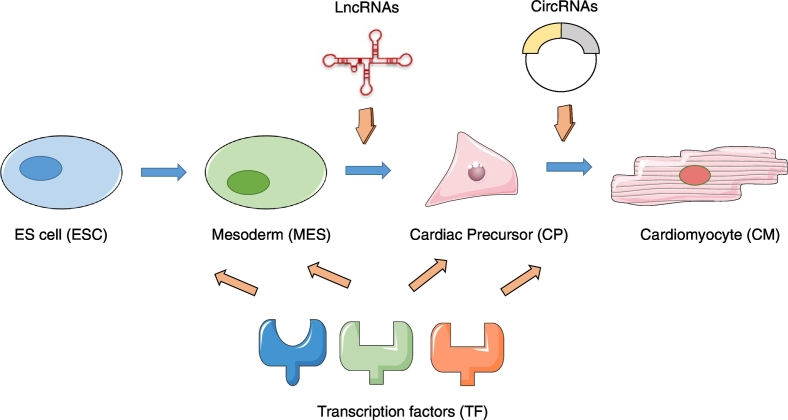
In a recent study published in EBioMedicine, Li and colleagues profiled protein-coding RNAs (transcriptome or mRNAs) and non-coding RNAs (lncRNA, circRNA) during the differentiation of HESC to cardiac myocytes (Li et al., 2017b). Using a previously published and publically available RNA-seq dataset, they reanalyzed the expression profiles in four stages of differentiation of HESC to cardiac myocytes- I) undifferentiated embryonic stem cells (ESC), II) the intermediate mesodermal stages (MES), III) cardiac precursor cells (CP) and lastly IV) cardiac myocytes (CM) (Fig. 1). The authors filtered for significant differential expression (FC > 2, FDR < 0.05; MES vs ESC, CP vs MES, CM vs CP) and identified 10,115 genes, 3488 lncRNAs and 198 circRNAs significantly regulated at each differentiation stage. LncRNAs were enriched during MES to CP differentiation while the number of significant circRNAs was higher during the CP to CM development, suggesting a role for lncRNAs in cardiac lineage commitment and circRNAs in cardiomyocyte differentiation/maturation (Li et al., 2017b) (Fig. 1).
Fig. 1.
The potential role of long non coding RNAs (lncRNAs), circular RNAs (circRNAs) and transcription factor regulation during cardiac differentiation stages.
To ascertain the potential function of the identified lncRNAs, circRNAs, and mRNAs, the authors performed pathway and transcription factor (TF) binding motif analyses at each differentiation stage. LncRNAs and circRNAs were enriched for pathways involved in cardiac development such as ESC differentiation, Wnt and calcium signaling. In addition, the authors identified stage specific TF binding sites on the promoters of lncRNAs and mRNAs (e.g., PAX7, NFAT), suggesting that lncRNAs and mRNAs may exhibit temporal expression under the regulation of TF (Fig. 1). Recent studies have suggested that the expression of circRNA does not always correlate with the expression of the linear transcript which it is derived from (Memczak et al., 2013). In this study, the authors identified a list of 23 differentially expressed host genes that could give rise to one or more circRNAs (positive or negative regulation). Finally, a weighted gene co-expression and transcription regulatory network analysis was performed to integrate TF-circRNA-lncRNA-mRNA interactions for each differentiation stage, showing that transcriptional regulation during cardiac differentiation is complex and involves the combinatorial interactions between TF, protein coding and non-coding RNAs (Li et al., 2017b). Previous work has identified complex interactions between other ncRNAs (microRNAs) and TFs in the heart (Ooi et al., 2017).
Overall, this study represents a comprehensive dataset of circRNAs, lncRNAs and mRNAs during each cardiac differentiation stage and is likely to be a valuable resource for scientists. In addition, the authors also identified novel interactions between protein coding and ncRNAs during stages of cardiomyocyte differentiation which may be important for therapeutic approaches for cardiac cell regeneration. However, it is important to be wary of false-positives generated with high throughput sequencing data. One method for limiting the follow-up of false positives is to focus on overlapping circRNAs/lncRNAs/mRNAs identified in this study with RNAs identified in independent profiling datasets using different bioinformatics algorithms. To determine which circRNAs or lncRNAs play a critical role in cardiac differentiation, validation and functional studies should be performed to confirm expression and interactions during heart development in vitro and in vivo using genetic loss-of-function approaches in mice. It may also be of interest to assess whether the identified circRNA/lncRNA play a role in cardiac reprogramming.
High throughput sequencing has enabled investigators to detect hundreds to thousands of lncRNAs and circRNAs but deciphering big datasets remains a huge challenge. Is one class of ncRNA superior compared to the other? As circRNAs are stable and have high conservation (vs lncRNA with low conservation), it could be that circRNAs play a more prominent role. However, additional studies will be required to better characterize the function and mechanisms of both lncRNA and circRNAs before this question can be answered. CircRNAs and lncRNAs have been shown to act as competing endogenous RNAs “microRNA sponges” for transcriptional regulation in the heart (Wang et al., 2016, Wang et al., 2014). Although lncRNAs and circRNAs have been classified as ncRNAs, recent studies have highlighted that some lncRNAs/circRNAs have coding potential (Anderson et al., 2015, Legnini et al., 2017). Subsequent studies are needed to fully understand the functional roles of proteins derived from these ncRNAs. In closing, the study by Li and colleagues mapping out the profiles of circRNA, lncRNA and mRNAs during cardiac development represents a good basis for future basic research in this field.
Disclosure
The authors declared no conflicts of interest.
Acknowledgments
Acknowledgements
JR McMullen is a National Health and Medical Research Council Senior Research Fellow (1078985).
References
- Anderson D.M., Anderson K.M., Chang C.L., Makarewich C.A., Nelson B.R., Mcanally J.R., Kasaragod P., Shelton J.M., Liou J., Bassel-Duby R., Olson E.N. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M., Laneve P., Rajewsky N., Bozzoni I. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66 doi: 10.1016/j.molcel.2017.02.017. (22-37.e9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Ding W., Sun T., Tariq M.A., Xu T., Li P., Wang J. Biogenesis of circular RNAs and their roles in cardiovascular development and pathology. FEBS J. 2017 doi: 10.1111/febs.14191. [DOI] [PubMed] [Google Scholar]
- Li Y., Zhang J., Huo C., Ding N., Li J., Xiao J., Lin X., Cai B., Zhang Y., Xu J. Dynamic organization of lncRNA and circular RNA regulators collectively controlled cardiac differentiation in humans. EBioMedicine. 2017 doi: 10.1016/j.ebiom.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen J.R., Drew B.G. Long non-coding RNAs (lncRNAs) in skeletal and cardiac muscle: potential therapeutic and diagnostic targets? Clin. Sci. (Lond.) 2016;130:2245–2256. doi: 10.1042/CS20160244. [DOI] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., Loewer A., Ziebold U., Landthaler M., Kocks C., Le Noble F., Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Morris K.V., Mattick J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi J.Y.Y., Bernardo B.C., Singla S., Patterson N.L., Lin R.C.Y., McMullen J.R. Identification of miR-34 regulatory networks in settings of disease and antimiR-therapy: implications for treating cardiac pathology and other diseases. RNA Biol. 2017;14:500–513. doi: 10.1080/15476286.2016.1181251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Liu F., Zhou L.Y., Long B., Yuan S.M., Wang Y., Liu C.Y., Sun T., Zhang X.J., Li P.F. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ. Res. 2014;114:1377–1388. doi: 10.1161/CIRCRESAHA.114.302476. [DOI] [PubMed] [Google Scholar]
- Wang K., Long B., Liu F., Wang J.X., Liu C.Y., Zhao B., Zhou L.Y., Sun T., Wang M., Yu T., Gong Y., Liu J., Dong Y.H., Li N., Li P.F. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]