Abstract
Objective
Current non-invasive early detection of colorectal cancer (CRC) requires improvement. We aimed to identified a fecal Clostridium symbiosum-based biomarker for early and advanced colorectal cancer detection.
Design
In the test stage, the relative abundance of Clostridium symbiosum (C. symbiosum) was measured by qPCR in 781 cases including 242 controls, 212 colorectal adenoma (CRA) patients, 109 early CRC (tumor restricted to the submucosa) patients, 218 advanced CRC patients. The prediction accuracy was compared to Fusobacterium nucleatum (F. nucleatum), fecal immunochemical test (FIT) and CEA (carcinoembryonic antigen) and validated in an independent cohort of 256 subjects. Current status of the trial:ongoing/still enrolling. Primary endpoint:June, 2017 (Clinicaltrials.gov Identifier NCT02845973).
Results
Significant stepwise increase of C. symbiosum abundance was found in CRA, early CRC and advanced CRC (P < 0.01). C. symbiosum outperformed all the other markers in early CRC prediction performance. The combination of C. symbiosum and FIT achieved better performance (0.803 for test cohort and 0.707 for validation cohort). For overall discrimination of CRCs, the combination of all above markers achieved the performance of 0.876.
Conclusions
Fecal C. symbiosum is a promising biomarker for early and noninvasive detection of colorectal cancer, being more effective than F. nucleatum, FIT and CEA. Combining C. symbiosum and FIT or CEA may improve the diagnosis power.
Keywords: Colorectal cancer, Early diagnosis, Clostridium symbiosum, Quantitative PCR
Highlights
-
•
The fecal abundance of Clostridium symbiosum was found increased in patients with colorectal neoplasia and it may serve as a potiential biomarker in non-invasive early differentiation of colorectal cancer from healthy controls.
-
•
The fecal abundance of Clostridium symbiosum was even more sensitive and efficient in diagnosis of both early and advanced colorectal cancer than reported markers like fecal immunochemical test, carcinoembryonic antigen and the abundance of Fusobacterium nucleatum.
-
•
Combining the abundance of Clostridium symbiosum and the other markers above may further enhance its predictive performance.
1. Introduction
Colorectal adenocarcinoma (CRC) ranks one of the 5 most lethal malignant tumors both in China and worldwide (Siegel et al., 2016, Chen et al., 2016). Early identification and treatment of CRC and the precancerous lesions, colorectal adenomas by screening may help reducing cancer mortality (Atkin et al., 2010, Schoen et al., 2012). Screening strategies including fecal immunochemical test (FIT) or serum carcinoembryonic antigen (CEA) test by several international guidelines were proved fairly efficient (Lieberman et al., 2012, Hassan et al., 2013, Moore and Aulet, 2017, Sung et al., 2015), however, large group of people may be still lack of accessible and affordable screening methods due to limitations of existing strategies and health care policies. Fecal occult blood test shares a large range of sensitivity of 24–86% according to population based reports and yet can be ambiguous towards adenomas and very early stage of CRC at which serum CEA test may work weakly as well (Raginel et al., 2013, Lieberman and Weiss, 2001, Nicholson et al., 2015, Fakih and Padmanabhan, 2006, Stiksma et al., 2014, Hewitson et al., 2007, Lee et al., 2014). Thus, a non-invasive economic biomarker sensitive to both early stage of CRC and high risk adenomas yields to be explored.
Quantities of previous researches revealed that shifts of gut microbiota may be an important part in the initiation and progression of CRC and further findings implicated the microbial stability can only be influenced by his or her genetic status, feeding pattern, age, gender, dietary habit, health situation, drug intake and wealth level, making gut microbiota a highly potential biomarker (Ding and Schloss, 2014, Costello et al., 2009, Quigley, 2017, Sun and Kato, 2016, Peters et al., 2016, Yatsunenko et al., 2012). Recent studies, including ours, have suggested that microbiota profiles determined by high-throughput sequencing may be effective in predicting CRCs (Zeller et al., 2014, Ai et al., 2017). However, sequencing-based methods are more resource-consuming and affectable by many factors in library construction, sequencing platform, etc. Therefore, mining the crucial factors/microbes in fecal microbiota and developing cost-effective, easy-to-apply methods are essential for translating this concept into clinical application.
F. nucleatum (Bacteria > Fusobacteria > Fusobacteriia > Fusobacteriales > Fusobacteriaceae > Fusobacterium > Fusobacterium nucleatum) has been suggested by a considerable number of studies as a potential marker for CRC detection (Yu et al., 2015). However, the effectiveness of F. nucleatum for detecting early CRC remains unclear due to limited numbers of cases in previous studies (n < 100 for each class) (Suehiro et al., 2016, Flanagan et al., 2014, Wong et al., 2016). Our previous work systematically evaluated the performance of different machine-learning models in microbiota-based CRC prediction, and selected significantly altered bacterial species in a French cohort and our own Chinese cohort (Ai et al., 2017, Zeller et al., 2014). Interestingly, we found significantly increasing level of C. symbiosum (Bacteria > Terrabacteria group > Firmicutes > Clostridia > Clostridiales > Lachnospiraceae > Lachnoclostridium > Clostridium symbiosum) and decreasing level of S. salivarius (Bacteria > Terrabacteria group > Firmicutes > Bacilli > Lactobacillales > Streptococcaceae > Streptococcus > streptococcus salivarius) in the CRC group (especially in early CRC), which were not reported elsewhere.
In our study, we developed qPCR-based assays to detect C. symbiosum and S. salivarius, and evaluated the performance of above-mentioned noninvasive markers (C. symbiosum, S. salivarius, F. nucleatum, FIT and serum CEA) in two independent large cohorts. By these efforts, we aim to identify an effective, noninvasive marker for detection of CRC, especially early CRC and advanced adenoma.
2. Materials and methods
2.1. Study design and participants
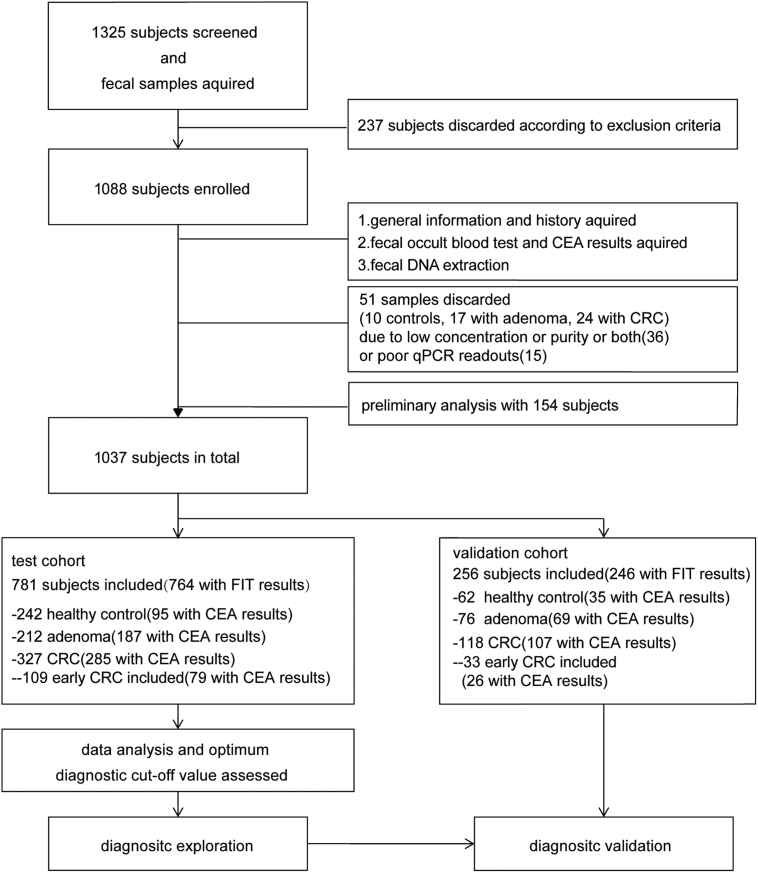
After the informed consent and general information questionnaire obtained, a total number of 1325 voluntary patients aging over 40 years old was recruited. The test and validation cohorts were recruited independently from different sites: (1) The test cohort was from Fudan University Shanghai Cancer Center (August 2016 to December 2016) and ECRJ-East Campus of Renji hospital (January 2012 to March 2017); (2) The validation cohort was from Shanghai Tenth People's Hospital (October 2015 to November 2016) and WCRJ-West Campus of Renji hospital (July 2016 to March 2017). To avoid potential alternation of the gut microbiota, excluding criteria were established as showed in workflow charts in Fig. 1. The exclusion criteria included: 1) with history of uninterested gastrointestinal tract neoplasia; 2) with history of upper GI tracts surgery; 3) with a history of FAP, HNPCC or P-J syndromes or uncontrolled diabetes, hypertension or other chronic metabolic disorder; 4) with eating habits changes in recent 4 weeks; 5) with active GI tracts bleeding in recent 6 months; 6) using any of the following medicine: NSAIDs (nonsteroidal anti-inflammatory drugs), immunosuppressor, antibiotics or probiotics at least 1 month prior to enrollment. Primary endpoint is June, 2017. The protocol had the approval of the Ethics Committee of the Shanghai Jiao-Tong University School of Medicine, Renji Hospital (Clinicaltrials.gov Identifier NCT02845973) and the research was carried out according to the provisions of the Helsinki Declaration of 1975.
Fig. 1.
Workflow Charts. CRC = colorectal cancer. CEA = carcinoembryonic antigen.
2.2. Definitions
The clinical phenotype was set by the endoscopic and pathological diagnosis. Tumors in the caecum, ascending colon, hepatic flexure, transverse colon or splenic flexure were considered to be proximal ones, while distal tumors included those in the rest of colon or rectum. The T stage was assessed by the AJCC (American Joint Committee on Cancer) TNM system. The early stage CRC was defined as cancers confined in submucosa including high-grade intraepithelial neoplasia. The advanced adenoma referred to adenomas with size over 10 mm, or with tubulovillous or villous component (Those advanced adenoma with high-grade intraepithelial neoplasia were considered to be early stage CRC). Healthy controls were those with normal or chronic inflammated colorectal mucosa.
2.3. Sample collection
All patients were asked to keep a steady dietary and life style and leave fecal sample over 0.5 g in the special germ-free containment before bowel preparation for any surgery or endoscopy. All samples were moved to − 20 °C for temporary preservation and transferred to − 80 °C for long-term storage within 48 h. Samples from different centers were gathered and preserved at the sample bank in the division of gastroenterology and hepatology, Renji Hospital waiting for further test. All positive blood test or fecal test results 6 months before the surgery or colonoscopy were recorded.
2.4. Fecal occult blood test (FIT) and CEA test
All enrolled subjects were asked to offer a valid fecal occult blood test and CEA test report from either a community hospital or a general hospital in recent 6 months. Stool samples with blank FIT result would have to be examined using Fecal Occult Blood Gold Gel Stripe kit (W.H.P.M. Bioresearch & technology co., LTD, Beijing) which has been approved by the Chinese Food and Drug Administration Bureau. The operator for FIT test had experience for at least 1000 samples test and kept blind to other study related results. The cut-off value for positive FIT is 200 ng/ml according to manufacturer's instructions.
2.5. Colonoscopy
All colonoscopies have been done in endoscopic department for each center by experienced endoscopists and have went through the entire colon reaching ileocecal valve with enough retreating time. Any neoplasia shall be biopsied and recorded right into report. Pathologists and endoscopists were blinded to the all study contents.
2.6. DNA extraction
QIAamp DNA Stool Mini Kit was used according to manufacturer's instructions (Qiagen, Hilden, Germany). Examined using NANO DROP 2000, those with a concentration lower than 10 ng/μl or OD260/OD280 ratio not in the range of 1.8–2.2 were discarded (36 samples). All extracts were preserved at − 20 °C before subsequent PCR amplification.
2.7. Primers and PCR amplification
The microbial primers of C. symbiosum, S. salivarius were designed in PRIMER3 (Rozen and Skaletsky, 2000) according to specific gene sequences, 2-hydroxyglutaryl-CoA dehydratase gene for C. symbiosum, Citrate synthase (gltA) gene for S. salivarius. The primers sequences were presented as follows: C. symbiosum. For-GTGAGATGATGTGCCAGGC; C. symbiosum. Rev-TACCGGTTGCTTCGTCGATT; S. salivarius. For-TTCGCTTCCCAGAGTCAAGT; S. salivarius. Rev-AAACGACCAGCCAGCAATTC; Internal reference primer for total bacterial DNA was determined by 16s rRNA (Kostic et al., 2013) using the following primers: 16s rRNA For-GGTGAATACGTTCCCGG; 16s rRNA Rev-TACGGCTACCTTGTTACGACTT; Positive control was used from F. nucleatum of which the primer was reported by well validated paper (Wong et al., 2016), with the following sequence: F. nucleatum. For-CAACCATTACTTTAACTCTACCATGTTCA; F. nucleatum. Rev.-GTTGACTTTACAGAA GGAGATTATGTAAAAATC.
All primers were synthesized and purified by Sangon Biotech Shanghai. 10 μl SYBR Green II was used as qPCR reaction system by TAKARA cooperation with SYBR®Premix Ex TaqTMII (TliRNaseH Plus). Stepone®plus by ABI company was used in qPCR with all the operation and configuration according to the manufacture's instruction with 40 cycles of 95 °C denaturation for 5 s, and 60 °C annealing and extension for 30s in total after a 30s of pre-denaturation at 95 °C.
2.8. Statistical analyses
FIT test results were recorded as positive or negative. Our data resulted in a cut-off value of 3.3 ng/ml which was similar with the clinical standard of 4.8 ng/ml so CEA test results were recorded as positive or negative according to the cut-off value of 4.8 ng/ml. All PCR samples were in triplicates and Ct value ranging over 5 (Ctmax − Ctmin) or a readout of underdetermined was excluded (15 samples). Average Ct value from the triplicates was calculated and the relative abundance of the target gut microbiota was based on the ΔCt value defined as the target Ct value subtracted Ct value for 16s rRNA, which means the all the relative abundance was normalized by the abundance of 16 s rRNA (Wong et al., 2016). As for statistical analysis, values were all expressed as mean ± SD. Wilcoxon signed-rank test, Mann-Whitney U test and Chi-square test were used in comparison for continuous, nonparametric and categorical variables analyses respectively. Factors independently associated with CRC diagnosis were estimated using univariate and multivariate logistic regression. Receiver Operating Characteristic (ROC) curve was used to evaluate the diagnostic value of bacterial candidates in distinguishing CRC and controls and to determine the best cut-off values that maximized the Youden index (J = Sensitivity + Specificity − 1). Pairwise comparison of areas under ROC (AUCs) was performed using the Delong's test. Logistic regression model was applied to obtain independent models concerning various markers. (The regression equations for all comparisons are provided in the Supplementary data). All tests were done by SPS.salivarius software v22.0 (SPS.salivarius, Chicago, IL) or Medcalc 15.2.2 (MedCalc Software). p < 0.05 was taken as statistical significance.
2.9. Role of the funding source
The funding sources did not contribute to the study design, data analysis, data interpretation, or writing of the report. The corresponding authors had full access to all the data and had final responsibility for the decision to submit for publication.
3. Results
3.1. Patient cohorts and quality control
A total number of 781 patients in the test cohort was consisted with 242 healthy controls, 212 adenomas patients, 327 CRC patients with 109 early CRC included. The independent validation cohort including 256 subjects with 62 healthy controls, 76 adenoma patients and 118 CRC patients included with 33 early CRCs (basic demographic characteristics see Supplementary Table S1; Study flowchart see Fig. 1). Single peak and band in expected size were observed in melt curves for primers of bacterial markers from a random selected sample group of 10 and further electrophoresis in agarose gel for qPCR amplified products (Supplementary Figs. S1 and S2). Primer efficiency for each primer was determined by qPCR standard curves from a series of diluted DNA samples as follows: 100.46% (16sRNA), 103.01% (Fn), 104.66% (Cs), 96.63% (Ss) (Supplementary Fig. S3).
3.2. Selection of candidate microbial markers
Based on our previous study using supervised models for the prediction of CRC (including 49 healthy controls, 37 adenoma patients, 34 early stage CRC patients and 24 advanced CRC patients), we observed an increase in the relative abundance of F. nucleatum and C. symbiosum in CRC patients over healthy controls. As for S. salivarius, given potential differences in subject races, population size and measuring means from sequencing data, no significant changes have been revealed among groups in current study (P = 0.398). In our specific cohort, C. symbiosum increased by 268.72 folds (P < 0.001), and F. nucleatum increased by 5.12 folds. Only C. symbiosum showed a statistical difference for early stage CRC over controls, which is 86.8 folds more (P < 0.001). Therefore, our present focuses on C. symbiosum as a candidate marker for detection of early CRC (Supplementary Fig. S4).
3.3. Stepwise increase of C. symbiosum in healthy control, CRA and CRC
In our test cohort (Fig. 2), the relative abundance of C. symbiosum increased in patients with all colorectal neoplasms (CRA, early CRC and advanced CRC) as compared to controls. While compared to healthy controls, the relative abundance of C. symbiosum increased stepwise in CRA (6.28 folds, p < 0.001), early CRC (12.38 folds, P < 0.001) and advanced CRC (21.25 folds, P < 0.001). However, the relative abundance of F. nucleatum exhibited mild increase in early CRC (2.46 folds, P = 0.006), but substantial increase in advanced CRC (10.70 folds, P < 0.001) as compared to healthy controls.
Fig. 2.
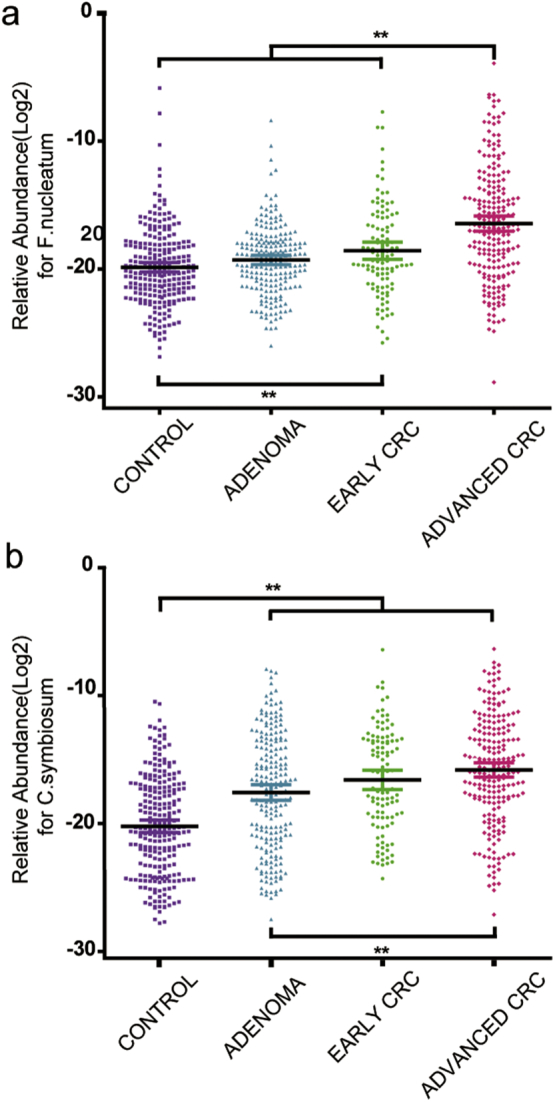
The fecal relative abundance of C. symbiosum and F. nucleatum among groups. (a) F. nucleatum for test cohort. (b) C. symbiosum for test cohort. CRC = colorectal cancer. C. symbiosum = clostridium symbiosum. F. nucleatum = fusobacteria nucleatum. **P < 0.01.
We further focused more on the bacterial distribution. No discrepancy for the F. nucleatum and C. symbiosum relative abundance has been revealed among distal and proximal colon for adenoma (PC. symbiosum = 0.55,PF. nucleatum = 0.21) or cancers (PC. symbiosum = 0.85,PF. nucleatum = 0.09). The depth of malignancy brought about different staging in pathology and thus leading to totally different clinical decision and prognosis. We thus compared F. nucleatum and C. symbiosum abundance in terms of T stages reported by pathologist. Though deeper infiltrated cancer (T4) patients seemed to have a higher level of fecal bacteria abundance (PC. symbiosum = 0.056, PF. nucleatum = 0.004), T staging may still not be a major factor to influence F. nucleatum nor C. symbiosum statistically especially for T2 and T3 cancers (Supplementary Fig. S5).
3.4. Performance of C. symbiosum in predicting advanced CRA and early CRC
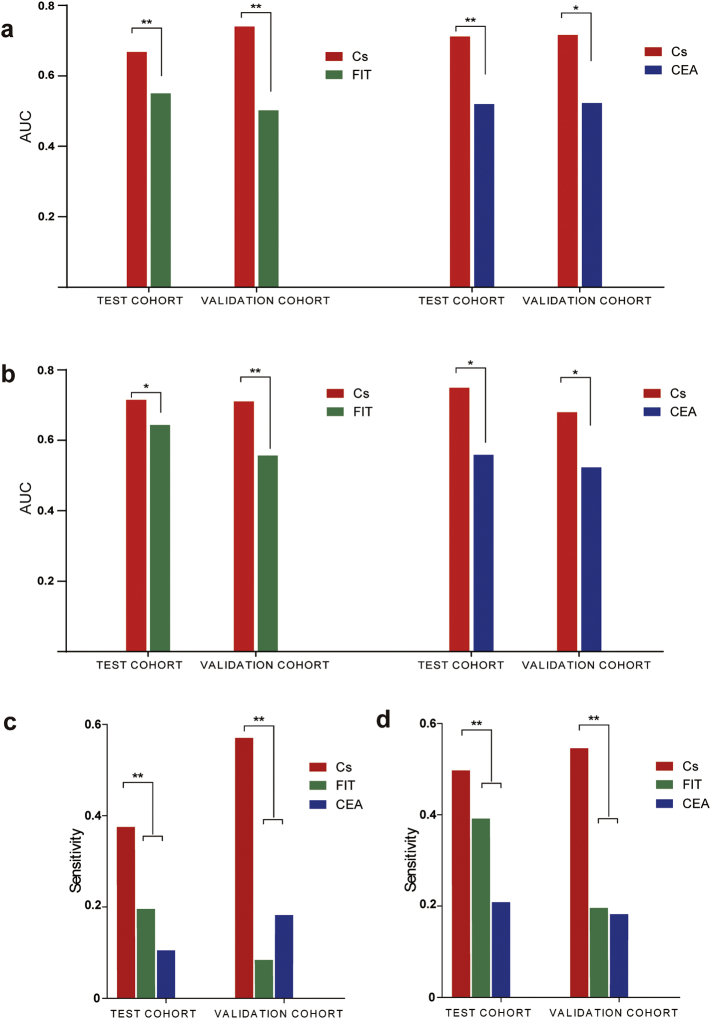
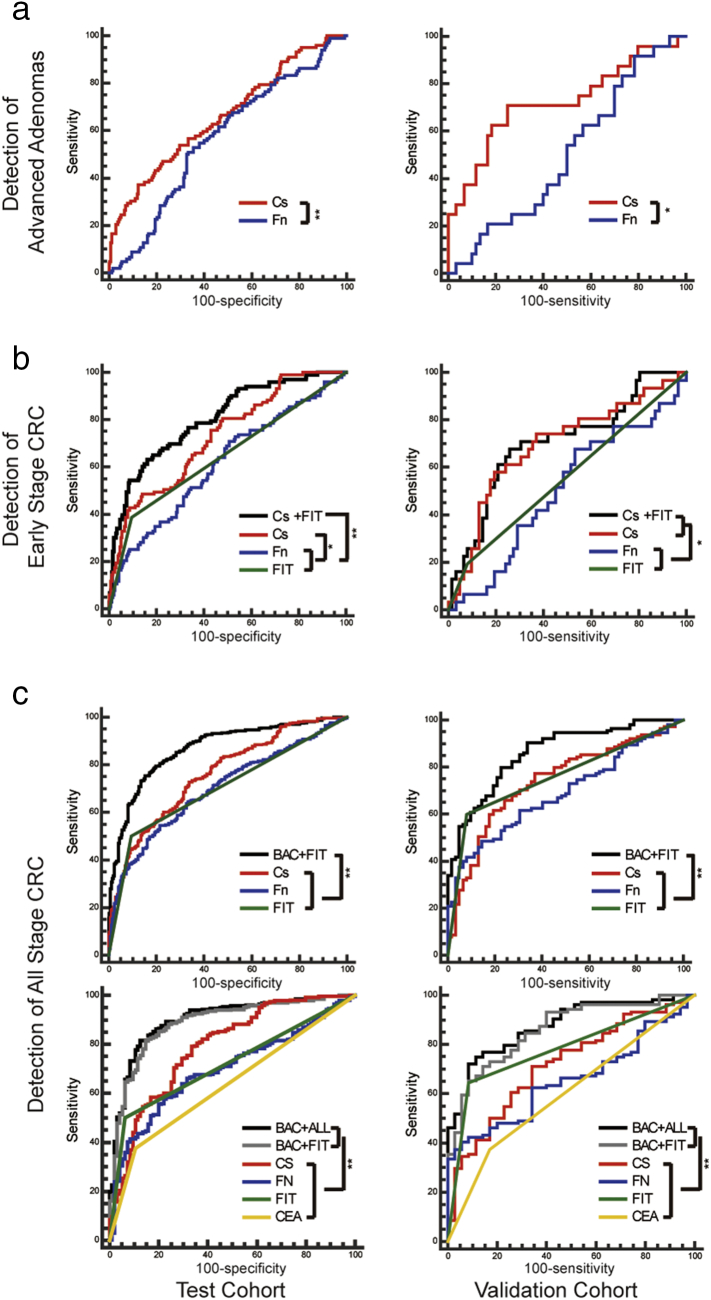
The relative abundance of C. symbiosum and F. nucleatum showed a increasing trend in the adenoma patients' feces as compared to healthy controls as described above (Fig. 1). In comparison with conventional markers like FIT and CEA, C. symbiosum showed higher sensitivity (P < 0.001) and similar specificity (P = 0.35 for FIT and P = 0.13 for CEA) so to reach stronger diagnostic performance in test cohorts (P < 0.001). With cut-off value set to match specificity among markers, the sensitivity from C. symbiosum marker may gain 6%–29% for FIT or 16%–20% for CEA. (Fig. 3 and Supplementary Table S2). Through AUCs from ROC curves, we also found C. symbiosum stronger than F. nucleatum (P < 0.001) (Fig. 4) in diagnosis of advanced adenomas. All such trend has also been observed in the validation cohort (Fig. 3, Fig. 4). To be more specific, further exploration emphasized similar changes between the non-advanced and advanced adenomas. Previous researches stated no substantial microbial changes from healthy people to adenoma people, but difference of 5.90 folds existed in our study for C. symbiosum in the advanced adenoma patients over the controls (P < 0.001). As for F. nucleatum, its relative abundance changed insignificantly for non-advanced adenoma (P = 0.076) and advanced adenomas (P = 0.126) compared to controls, making C. symbiosum more promising to predict advanced colorectal adenomas than F. nucleatum.
Fig. 3.
The diagnostic power of C. symbiosum in advanced adenoma and early stage CRC (cancers confined in submucosa including high-grade intraepithelial neoplasia). (a) AUC comparison for C. symbiosum abundance with FIT or with CEA in advanced adenoma. (b) AUC comparison for C. symbiosum abundance with FIT or with CEA in early stage CRC. (c) Sensitivity for C. symbiosum abundance, CEA and FIT test in advanced adenoma. (d) Sensitivity for C. symbiosum abundance, CEA and FIT test in early stage CRC. CRC = colorectal cancer. Cs = Clostridium symbiosum. Fn = Fusobacteria nucleatum. CEA = carcinoembryonic antigen. FIT = fecal immunochemical test. AUC = Area under curve for receiver operating characteristic curve. *P < 0.05. **P < 0.01.
Fig. 4.
The comparison of ROC for different markers in advanced adenoma, early stage CRC (cancers confined in submucosa including high-grade intraepithelial neoplasia) and all stage CRC. (a) AUC for C. symbiosum and F. nucleatum abundance in test and validation cohort in advanced adenoma. (b) AUC for Cs + FIT, FIT, C. symbiosum and F. nucleatum abundance in test and validation cohort in early stage CRC. (c) AUC for BAC + FIT, FIT, C. symbiosum and F. nucleatum abundance in test cohort and validation in all stage CRC for subjects with FIT results (upper figures) and for subjects with both FIT and CEA tests (lower figures). CRC = colorectal cancer. Cs = Clostridium symbiosum. Fn = Fusobacteria nucleatum. CEA = carcinoembryonic antigen. FIT = fecal immunochemical test. Cs + FIT = marker combined C. symbiosum and FIT. BAC + ALL = marker combined C. symbiosum and F. nucleatum abundance with FIT and CEA test. BAC + FIT = marker combined C. symbiosum and F. nucleatum abundance with FIT test. AUC = Area under curve for receiver operating characteristic curve. *P < 0.05. **P < 0.01.
As for the 109 early stage CRCs patients out of 327 CRC patients in test cohort, increasing C. symbiosum level reached an higher AUC over F. nucleatum (P < 0.001) and FIT (P < 0.001) as well as for CEA which was also observed in validation cohort (Fig. 3, Fig. 4). According to the cut-off value set by ROC curves, the fecal relative abundance of C. symbiosum was proven significantly more sensitive to early stage CRC compared to FIT (P < 0.001) and CEA test (P < 0.001). With further specificity balanced, C. symbiosum may contribute to a sensitivity increase of 4%–24% for FIT and 23%–27% for CEA.(Fig. 3 and Supplementary Table S3).
3.5. Fecal microbial markers with higher sensitivity for CRC
In feces from 327 CRC patients, higher level of F. nucleatum and C. symbiosum has been easily detected in CRC patients over controls. As AUCs for all these markers were closed to each other. C. symbiosum seemed to have greater advantages over conventional methods as well as F. nucleatum in terms of sensitivity (0.73 for C. symbiosum over 0.38–0.54 for the rest markers, P < 0.01, same trend for validation cohort) (Supplementary Table.S4). In further comparison, positive rate of serum CEA and FIT test rose about 3.61 folds and 5.28 folds from healthy controls to CRC patients.
3.6. Combination of fecal microbial markers with CEA and FIT
Based on the results above, we further divided enrolled people into F. nucleatum and C. symbiosum positive group and negative group according to the cut-off value identified previously. From univariate analysis, history of HTN or DM seemed futile in early stage CRC diagnosis while the abundance of C. symbiosum and F. nucleatum, FIT test and CEA level as well as gender and age may be of help. Further multivariate analysis indicated FIT test (OR = 8.557, P < 0.001), age (OR = 1.079, P < 0.01), CEA level (OR = 3.650, P = 0.014) and the relative abundance of C. symbiosum (OR = 4.354, P < 0.01) contributed to the diagnosis of early stage CRC and F. nucleatum showed no further statistical significance (OR = 1.804, P = 0.145) (Supplementary Table S5). We then tried to unite these contributing markers as an independent model. Combination of C. symbiosum level and FIT may raise AUC to 0.803 in test cohort and 0.707 in validation cohort that showed statistically stronger diagnostic power over single FIT test (P < 0.05) and F. nucleatum (P < 0.05) (Supplementary Table S6), further combining CEA level did not enhance such trend (P = 0.09 for test cohort and P = 0.56 for validation cohort) but still reached stronger ability than single CEA marker or FIT alone (P < 0.001) in both cohort (Table 1 and Fig. 4).
Table 1.
AUC for different markers in patients with CEA and FIT results for predicting early CRC (cancers confined in submucosa including high-grade intraepithelial neoplasia).
| Test cohort |
Validation cohort |
Overall |
||||
|---|---|---|---|---|---|---|
| AUC | 95% CI | AUC | 95% CI | AUC | 95% CI | |
| In patients with both valid CEA and FIT results. Difference in comparison with Cs + FIT | ||||||
| CEA | 0.551⁎⁎ | 0.474–0.627 | 0.544⁎⁎ | 0.409–0.674 | 0.527⁎⁎ | 0.461–0.593 |
| FIT | 0.646⁎⁎ | 0.569–0.718 | 0.582⁎⁎ | 0.446–0.709 | 0.630⁎⁎ | 0.564–0.693 |
| Cs + FIT | 0.836 | 0.772–0.888 | 0.743 | 0.612–0.848 | 0.809 | 0.752–0.857 |
| Cs + ALL | 0.856 | 0.794–0.905 | 0.726 | 0.594–0.834 | 0.822 | 0.766–0.869 |
Cs + ALL = marker combined C. symbiosum and F. nucleatum abundance with FIT and CEA test. Cs + FIT = marker combined C. symbiosum abundance with FIT test. Cs = Clostridium symbiosum. Fn = Fusobacteria nucleatum. CEA = carcinoembryonic antigen. FIT = fecal immunochemical test. CI = confidence interval.
P < 0.01.
Univariate and multivariate analysis for all stage CRC patients indicated the relative abundance of F. nucleatum (OR = 4.282, P < 0.001) and C. symbiosum (OR = 4.237, P < 0.001), serum CEA concentration (OR = 3.897, P < 0.01) and FIT test (OR = 12.024, P < 0.001) as well as age (OR = 1.054, P < 0.01) were contributing to diagnosis yet gender, history of HTN and DM showed no statistical difference (Supplementary Table S5). Combination of F. nucleatum, C. symbiosum and FIT may set one step further to differentiate more CRC with an AUC of 0.861 (Supplementary Table S6). Taking CEA into account, the independent model of F. nucleatum, C. symbiosum, FIT and CEA may have an AUC of 0.876. Both models above were stronger than any of the markers alone (P < 0.05), which also has been matched quite well in both cohorts (Table 2 & Fig. 4).
Table 2.
AUC for different markers in patients with CEA and FIT results for predicting CRC of all stages.
| Development cohort |
Validation cohort |
Overall |
||||
|---|---|---|---|---|---|---|
| AUC | 95% CI | AUC | 95% CI | AUC | 95% CI | |
| In patients with both valid CEA and FIT results. Difference in comparison with BAC + FIT | ||||||
| CEA | 0.636⁎⁎ | 0.585–0.684 | 0.602⁎⁎ | 0.515–0.684 | 0.627⁎⁎ | 0.583–0.668 |
| FIT | 0.718⁎⁎ | 0.670–0.763 | 0.779⁎ | 0.701–0.845 | 0.735⁎⁎ | 0.694–0.772 |
| BAC + FIT | 0.888 | 0.852–0.918 | 0.859 | 0.790–0.912 | 0.875 | 0.844–0.902 |
| BAC + ALL | 0.900⁎ | 0.866–0.929 | 0.876 | 0.810–0.926 | 0.888⁎ | 0.857–0.913 |
BAC + ALL = marker combined C. symbiosum and F. nucleatum abundance with FIT and CEA test. BAC + FIT = marker combined C. symbiosum and F. nucleatum abundance with FIT test. Cs = Clostridium symbiosum. Fn = Fusobacteria nucleatum. CEA = carcinoembryonic antigen. FIT = fecal immunochemical test. CI = confidence interval.
P < 0.05.
P < 0.01.
4. Discussion
In this study, we identified a potential microbial marker Clostridium symbiosum for early detection of CRC and validated its performance in differentiating colorectal neoplasia including early stage cancer and advanced adenoma from controls with superior performance to conventional FIT and CEA test as well as the recently reported marker of F. nucleatum.
To draw such a conclusion firmly, we introduced the so far largest multi-center based clinical cohorts of 1037 subjects in total. The most vital finding in our study was the significant stepwise increase for the abundance of C. symbiosum in CRA, early CRC and advanced CRC, which also maintained to be the a rare-reported specie among various CRC-associated gut microbiota, making it a promising marker in colon neoplasia especially for early stage CRC.
Existing screening methods exert fairly good sensitivity and specificity for advanced CRC. However, 5-year survival rate of advanced CRC (< 10%) is drastically lower than that of early CRC (> 80%) (Siegel et al., 2016, Brenner et al., 2014), thus early screening of CRC represents a major factor in developing biomarkers. For population-wide screen of early CRC, non-invasive and cost-effective methods are highly desired, because these features are obviously beneficial to the compliance of patients.
Although FIT and CEA are widely applied markers for CRC screening, our data based on two large cohorts suggest that neither method can be sufficiently accurate for detecting early CRC. The true positive rates of FIT were low for early CRC (18–33%) and advanced CRA (9–19%), being consistent with its reported weakness in predicting advanced adenoma (Hundt et al., 2009, van Doorn et al., 2015), FIT also exhibits a wide range of sensitivity for all stages CRC as low as 36.4% or even lower as 25% while reaching as high as 79% according to several solid population-based researches (Lee et al., 2014, Lieberman and Weiss, 2001). In our work, though sharing similar low false positive rate of 91%–92% as reported (94%) (Lee et al., 2014), FIT differentiated 50–60% CRC patients reflected its limited performance. Gastrointestinal bleeding of any reasons may occasionally leave a positive FIT result and it has been highly related to the size and vessel distribution of the lesions while less connected to the tumor malignancy. CEA, instead, may report CRC with a sensitivity of 43–46%, but also displayed very limited sensitivity of 38%–39% in our study, which also has been considered of greater value in the CRC prognosis (Wang et al., 2014). Such low positive rate for CEA has also been reported by other studies concerning early stage CRC (Nicholson et al., 2015, Fakih and Padmanabhan, 2006).
In contrast to above-mentioned conventional markers, the fecal abundance of C. symbiosum displayed considerably higher accuracy in predicting early CRC. The true positive rate of C. symbiosum remained high in all gut tumors including both advanced adenomas and early or late stage of CRC, reaching 37–58% for advanced adenoma and 50%–56% for early stage CRC respectively in both cohorts, was approximately 2-folds higher than FIT test (see above) and CEA test (11%–18% and 17%–20%). Given such differences in sensitivity, none significant changes in the high negative rate (82%–93%) in controls have been observed between C. symbiosum and FIT (0.08–0.35 for P value) or CEA (0.13–0.70 for P value) in both cohorts (Supplementary Fig. S6).
As for early dectection of colorectal neoplasia, C. symbiosum even showed stronger diagnostic value in microbial markers. F. nucleatum is one of the most widely studied bacteria associated with CRC. By detecting the fecal abundance of F. nucleatum alone, qPCR assays have been reported with diagnostic potential for CRCs by several papers (Suehiro et al., 2016, Flanagan et al., 2014, Wong et al., 2016). However, due to limitations in the population enrolled for these studies, F. nucleatum has not been sufficiently verified for its predictive significance for early CRC and advanced CRA. Our data provided support to previous findings that F. nucleatum was a promising marker for predicting CRC (especially advanced CRC), but seemed to propose a weakness of F. nucleatum in identifying CRC in its early stage. The increase of F. nucleatum was subtle from healthy control to CRA and to early CRC, and for discriminating early CRC F. nucleatum exhibited a rather low AUC of 0.611 in the test cohort or 0.521 in the validation cohort. In contrast, the abundance of C. symbiosum displayed significant difference between healthy control and early CRC, and C. symbiosum achieved higher AUC for predicting early CRC (P < 0.05) and advanced adenoma (P < 0.05) than F. nucleatum. In the multivariate analysis, F. nucleatum exhibited no significant predictive value for early CRC. Thus making C. symbiosum as a better marker for discriminating CRC. These findings all suggest that C. symbiosum may be a stronger microbial marker than F. nucleatum in early colorectal tumor detection.
Another interesting finding was the improved predictive power for combining C. symbiosum and other existing markers such as FIT and CEA. Such combination increased sensitivity by 12–15% and recognized nearly a third more patients for early stage cancer, but having no obvious loss in specificity. For predicting all CRC patients, F. nucleatum and C. symbiosum combined with FIT and CEA also exhibited improved specificity up to 82%–86% with 69%–87% specificity.
Furthermore, the wide applying of this marker of C. symbiosum in CRC screening as we introduced, could be rather feasible since qPCR detection of bacterial DNA in stool is technically more reliable and cost-effective than other promising methods. For example, the multi-target stool DNA test based on free or shedding tumor cells in feces or blood has been recommended by recent CRC screening guidelines (Imperiale et al., 2014). Since each detection requires nearly 600 USD (Ladabaum and Mannalithara, 2016), well-trained technician and carefully installed devices, these resource-consuming features may limits its use for population-wide screening. Similarly, high-throughput metagenomic markers also have concerns in the high dependency on equipment and platforms, and difficulties in technical standardization and quality control. Here we have demonstrated that qPCR-based detection of C. symbiosum, in combination with conventional FIT or CEA, can achieve considerable predictive accuracy with acceptable cost. To facilitate its clinical application in early CRC screening, a strict cost-utility analysis should be further explored in future studies (Knudsen et al., 2016).
Is the increase of C. symbiosum stool a cause or consequence of CRC development? While there is no clear answer yet, it certainly deserves in-depth mechanistic studies. Clostridium symbiosum is a strict anaerobic colonizing bacteria with flagellum which is gram-negative non-toxin producing (Elsayed and Zhang, 2004). It shares similar micro-forms as F. nucleatum that has once been confused with the family of Fusobacterium. Differentiation have made in recent years thank to more precise sequencing methods. Study showed transplanting C. symbiosum specifically to germ-free nutrition-deficient mice may result in increasing level of acylcarnitines in gut epithelium which may contribute to decreasing level of protein synthesis and overoxidation of amino acids in liver (Blanton et al., 2016). Therefore, we postulate that C. symbiosum may promote protein synthesis in local gut epithelium which acted as potential supporter to development of carcinogenesis.
One limitation for our study is the absence of data from other diseases which may cause positive FIT or CEA, like IBD or gastric cancer. Very few relating researches have been done so far even for F. nucleatum. Another concern is lack of data from patients with curative surgery since several years of following up more should have been adequate for further solid judgement. Studies in larger cohorts enrolling more patients need to be carried out in future.
In conclusion, fecal Clostridium Symbiosum is a promising biomarker for early and noninvasive detection of colorectal neoplasia, being more effective than reported markers such as F. nucleatum, FIT and CEA. Combining the abundance of fecal C. symbiosum and fecal immunochemical test may further improve the noninvasive diagnosis of early CRC.
Acknowledgments
Acknowledgments
This project was supported by grants from the National Natural Science Foundation of China (81421001, 81320108024, 81530072, 81572303 and 81001070), the National Key Technology R&D Program (2014BAI09B05). The project was also partially supported by National Key Technology R&D Program (2016YFC0906002), Top-Notch project of China and Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20152514). The funding sources had no role in writing, data collection, analysis, or interpretation, or any aspect pertinent to the study. The decision to submit this manuscript has been made by the corresponding authors. The authors thank Dr. Jing-Xian Chen and Dr. Zhong Ming for supporting the study.
Declaration of Interests
All authors declare no commercial nor associative interest that represents a conflict of interest in connection with the work submitted.
Contributors
Study concept and design: JX, JYF, YHX; Acquisition of data: YHX, THZ, HMC, SYY, YWQ; Analysis and interpretation of data: YHX, SYY, YWQ, JX, YXC, JYF; Writing the manuscript: YHX, JX, JYF, YXC; Critical revision of the manuscript for important intellectual content: QYG, GXC, XMS, WQG, XYC, YC, DFS, ZJL, SJC, JX, YXC, JYF; Statistical analysis: YHX, JX.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2017.10.005.
Contributor Information
San-Jun Cai, Email: csj@shca.org.cn.
Jie Xu, Email: jiexu@sjtu.edu.cn.
Ying-Xuan Chen, Email: yingxuanchen71@sjtu.edu.cn.
Jing-Yuan Fang, Email: jingyuanfang@sjtu.edu.cn.
Appendix A. Supplementary data
Supplementary material
References
- Ai L., Tian H., Chen Z., Chen H., Xu J., Fang J.Y. Systematic evaluation of supervised classifiers for fecal microbiota-based prediction of colorectal cancer. Oncotarget. 2017;8(6):9546–9556. doi: 10.18632/oncotarget.14488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin W.S., Edwards R., Kralj-Hans I., Wooldrage K., Hart A.R., Northover J.M., Parkin D.M., Wardle J., Duffy S.W., Cuzick J. Once-only flexible sigmoidoscopy screening in prevention of colorectal cancer: a multicentre randomised controlled trial. Lancet. 2010;375(9726):1624–1633. doi: 10.1016/S0140-6736(10)60551-X. [DOI] [PubMed] [Google Scholar]
- Blanton L.V., Charbonneau M.R., Salih T., Barratt M.J., Venkatesh S., Ilkaveya O., Subramanian S., Manary M.J., Trehan I., Jorgensen J.M., Fan Y.M., Henrissat B., Leyn S.A., Rodionov D.A., Osterman A.L., Maleta K.M., Newgard C.B., Ashorn P., Dewey K.G., Gordon J.I. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016;351(6275) doi: 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner H., Kloor M., Pox C.P. Colorectal cancer. Lancet. 2014;383(9927):1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- Costello E.K., Lauber C.L., Hamady M., Fierer N., Gordon J.I., Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding T., Schloss P.D. Dynamics and associations of microbial community types across the human body. Nature. 2014;509(7500):357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsayed S., Zhang k. Bacteremia caused by Clostridium symbiosum. J. Clin. Microbiol. 2004;42(9):4390–4392. doi: 10.1128/JCM.42.9.4390-4392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakih M.G., Padmanabhan A. CEA monitoring in colorectal cancer. What you should know. Oncology (Williston Park) 2006;20(6):579–587. (discussion 588, 594, 596 passim) [PubMed] [Google Scholar]
- Flanagan L., Schmid J., Ebert M., Soucek P., Kunicka T., Liska V., Bruha J., Neary P., Dezeeuw N., Tommasino M., Jenab M., Prehn J.H., Hughes D.J. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33(8):1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- Hassan C., Quintero E., Dumonceau J.M., Regula J., Brandao C., Chaussade S., Dekker E., Dinis-Ribeiro M., Ferlitsch M., Gimeno-Garcia A., Hazewinkel Y., Jover R., Kalager M., Loberg M., Pox C., Rembacken B., Lieberman D. Post-polypectomy colonoscopy surveillance: European Society of Gastrointestinal Endoscopy (ESGE) Guideline. Endoscopy. 2013;45(10):842–851. doi: 10.1055/s-0033-1344548. [DOI] [PubMed] [Google Scholar]
- Hewitson P., Glasziou P., Irwig L., Towler B., Watson E. Screening for colorectal cancer using the faecal occult blood test, Hemoccult. Cochrane Database Syst. Rev. 2007;1:CD001216. doi: 10.1002/14651858.CD001216.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundt S., Haug U., Brenner H. Comparative evaluation of immunochemical fecal occult blood tests for colorectal adenoma detection. Ann. Intern. Med. 2009;150(3):162–169. doi: 10.7326/0003-4819-150-3-200902030-00005. [DOI] [PubMed] [Google Scholar]
- Imperiale T.F., Ransohoff D.F., Itzkowitz S.H., Levin T.R., Lavin P., Lidgard G.P., Ahlquist D.A., Berger B.M. Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 2014;370(14):1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- Knudsen A.B., Zauber A.G., Rutter C.M., Naber S.K., Doria-Rose V.P., Pabiniak C., Johanson C., Fischer S.E., Lansdorp-Vogelaar I., Kuntz K.M. Estimation of benefits, burden, and harms of colorectal cancer screening strategies: modeling study for the US preventive services task force. JAMA. 2016;315(23):2595–2609. doi: 10.1001/jama.2016.6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic A.D., Chun E., Robertson L., Glickman J.N., Gallini C.A., Michaud M., Clancy T.E., Chung D.C., Lochhead P., Hold G.L., El-Omar E.M., Brenner D., Fuchs C.S., Meyerson M., Garrett W.S. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladabaum U., Mannalithara A. Comparative effectiveness and cost effectiveness of a multitarget stool DNA test to screen for colorectal neoplasia. Gastroenterology. 2016;151(3):427–439.e6. doi: 10.1053/j.gastro.2016.06.003. [DOI] [PubMed] [Google Scholar]
- Lee J.K., Liles E.G., Bent S., Levin T.R., Corley D.A. Accuracy of fecal immunochemical tests for colorectal cancer: systematic review and meta-analysis. Ann. Intern. Med. 2014;160(3):171. doi: 10.7326/M13-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman D.A., Weiss D.G. One-time screening for colorectal cancer with combined fecal occult-blood testing and examination of the distal colon. N. Engl. J. Med. 2001;345(8):555–560. doi: 10.1056/NEJMoa010328. [DOI] [PubMed] [Google Scholar]
- Lieberman D.A., Rex D.K., Winawer S.J., Giardiello F.M., Johnson D.A., Levin T.R. Guidelines for colonoscopy surveillance after screening and polypectomy: a consensus update by the US multi-society task force on colorectal cancer. Gastroenterology. 2012;143(3):844–857. doi: 10.1053/j.gastro.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Moore J.S., Aulet T.H. Colorectal cancer screening. Surg. Clin. North Am. 2017;97(3):487–502. doi: 10.1016/j.suc.2017.01.001. [DOI] [PubMed] [Google Scholar]
- Nicholson B.D., Shinkins B., Pathiraja I., Roberts N.W., James T.J., Mallett S., Perera R., Primrose J.N., Mant D. Blood CEA levels for detecting recurrent colorectal cancer. Cochrane Database Syst. Rev. 2015;12:CD011134. doi: 10.1002/14651858.CD011134.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B.A., Dominianni C., Shapiro J.A., Church T.R., Wu J., Miller G., Yuen E., Freiman H., Lustbader I., Salik J., Friedlander C., Hayes R.B., Ahn J. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome. 2016;4(1):69. doi: 10.1186/s40168-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley E.M. Basic definitions and concepts: organization of the gut microbiome. Gastroenterol. Clin. N. Am. 2017;46(1):1–8. doi: 10.1016/j.gtc.2016.09.002. [DOI] [PubMed] [Google Scholar]
- Raginel T., Puvinel J., Ferrand O., Bouvier V., Levillain R., Ruiz A., Lantieri O., Launoy G., Guittet L. A population-based comparison of immunochemical fecal occult blood tests for colorectal cancer screening. Gastroenterology. 2013;144(5):918–925. doi: 10.1053/j.gastro.2013.01.042. [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000:132365–132386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Schoen R.E., Pinsky P.F., Weissfeld J.L., Yokochi L.A., Church T., Laiyemo A.O., Bresalier R., Andriole G.L., Buys S.S., Crawford E.D., Fouad M.N., Isaacs C., Johnson C.C., Reding D.J., O'Brien B., Carrick D.M., Wright P., Riley T.L., Purdue M.P., Izmirlian G., Kramer B.S., Miller A.B., Gohagan J.K., Prorok P.C., Berg C.D. Colorectal-cancer incidence and mortality with screening flexible sigmoidoscopy. N. Engl. J. Med. 2012;366(25):2345–2357. doi: 10.1056/NEJMoa1114635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- Stiksma J., Grootendorst D.C., van der Linden P.W. CA 19-9 as a marker in addition to CEA to monitor colorectal cancer. Clin. Colorectal Cancer. 2014;13(4):239–244. doi: 10.1016/j.clcc.2014.09.004. [DOI] [PubMed] [Google Scholar]
- Suehiro Y., Sakai K., Nishioka M., Hashimoto S., Takami T., Higaki S., shindo Y., Hazama S., Oka M., Nagano H., Sakaida I., Yamasaki T. Highly sensitive stool DNA testing of Fusobacterium nucleatum as a marker for detection of colorectal tumours in a Japanese population. Ann. Clin. Biochem. 2016;54(1):86–91. doi: 10.1177/0004563216643970. [DOI] [PubMed] [Google Scholar]
- Sun J., Kato I. Gut microbiota, inflammation and colorectal cancer. Genes Dis. 2016;3(2):130–143. doi: 10.1016/j.gendis.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung J.J., Ng S.C., Chan F.K., Chiu H.M., Kim H.S., Matsuda T., Ng S.S., Lau J.Y., Zheng S., Adler S., Reddy N., Yeoh K.G., Tsoi K.K., Ching J.Y., Kuipers E.J., Rabeneck L., Young G.P., Steele R.J., Lieberman D., Goh K.L. An updated Asia Pacific consensus recommendations on colorectal cancer screening. Gut. 2015;64(1):121–132. doi: 10.1136/gutjnl-2013-306503. [DOI] [PubMed] [Google Scholar]
- van Doorn S.C., Stegeman I., Stroobants A.K., Mundt M.W., de Wijkerslooth T.R., Fockens P., Kuipers E.J., Bossuyt P.M., Dekker E. Fecal immunochemical testing results and characteristics of colonic lesions. Endoscopy. 2015;47(11):1011–1017. doi: 10.1055/s-0034-1392412. [DOI] [PubMed] [Google Scholar]
- Wang R.F., Song B.R., Peng J.J., Cai G.X., Liu F.Q., Wang M.H., Cai S.J., Ye X. The prognostic value of preoperative serum CEA and CA19-9 values in stage I-III colorectal cancer. Hepato-Gastroenterology. 2014;61(132):994–999. [PubMed] [Google Scholar]
- Wong S.H., Kwong T.N., Chow T.C., Luk A.K., Dai R.Z., Nakatsu G., Lam T.Y., Zhang L., Wu J.C., Chan F.K., Ng S.S., Wong M.C., Ng S.C., Wu W.K., Yu J., Sung J.J. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. 2016;66(8):1441–1448. doi: 10.1136/gutjnl-2016-312766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M., Magris M., Hidalgo G., Baldassano R.N., Anokhin A.P., Heath A.C., Warner B., Reeder J., Kuczynski J., Caporaso J.G., Lozupone C.A., Lauber C., Clemente J.C., Knights D., Knight R., Gordon J.I. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Feng Q., Wong S.H., Zhang D., Liang Q.Y., Qin Y., Tang L., Zhao H., Stenvang J., Li Y., Wang X., Xu X., Chen N., Wu W.K., Al-Aama J., Nielsen H.J., Kiilerich P., Jensen B.A., Yau T.O., Lan Z., Jia H., Li J., Xiao L., Lam T.Y., Ng S.C., Cheng A.S., Wong V.W., Chan F.K., Xu X., Yang H., Madsen L., Datz C., Tilg H., Wang J., Brunner N., Kristiansen K., Arumugam M., Sung J.J., Wang J. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2015;66(1):70–78. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- Zeller G., Tap J., Voigt A.Y., Sunagawa S., Kultima J.R., Costea P.I., Amiot A., Bohm J., Brunetti F., Habermann N., Hercog R., Koch M., Luciani A., Mende D.R., Schneider M.A., Schrotz-King P., Tournigand C., Tran V.N.J., Yamada T., Zimmermann J., Benes V., Kloor M., Ulrich C.M., von Knebel D.M., Sobhani I., Bork P. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014;10766 doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material