Abstract
Aim:
Solithromycin is a new macrolide antibiotic for the potential treatment of bacterial pneumonia.
Materials & methods:
Solithromycin N-acetylation by human NAT1 and NAT2 was investigated following recombinant expression in yeast and in cryopreserved human hepatocytes from rapid, intermediate and slow acetylators.
Results:
Solithromycin exhibited over twofold higher affinity for recombinant human NAT2 than NAT1. Apparent maximum velocities for the N-acetylation of solithromycin catalyzed by the NAT2 allozyme associated with rapid acetylators were significantly (p < 0.01) higher than by the NAT2 allozymes associated with slow acetylators. Robust gene dose responses (rapid>intermediate>slow acetylators) were exhibited in cryopreserved human hepatocytes in situ following incubation with 100 μM solithromycin.
Conclusion:
Solithromycin is N-acetylated by human NAT1 and NAT2 and the role of the NAT2 acetylation polymorphism on solithromycin metabolism may be concentration dependent.
Keywords: : acetylation polymorphism, cryopreserved human hepatocytes, N-acetyltransferase 2, solithromycin
Lower respiratory infections, such as, community-acquired bacterial pneumonia frequently require physician intervention and are among the top infectious causes of death worldwide [1]. As recently reviewed [2], solithromycin is a novel fluoroketolide efficacious against the most common causes of community-acquired bacterial pneumonia, including macrolide-, penicillin- and fluoroquinolone-resistant isolates of Streptococcus pneumoniae as well as Haemophilus influenzae and atypical bacterial pathogens. Following completion of Phase I–III clinical trials, solithromycin clinical efficacy data have demonstrated the potential to improve treatment of community-acquired bacterial pneumonia in both hospital and community settings. General guidelines for pneumonia treatment recommend three broad categories (β-lactam antibiotic alone, β-lactam antibiotic and a macrolide, fluoroquinolone alone) and monotherapy with earlier-generation macrolides is not a preferred treatment option due to high pneumococcal resistance rates. Solithromycin, a fourth-generation macrolide antibiotic with activity against macrolide-resistant pneumonia pathogens is a potential drug for macrolide monotherapy [3].
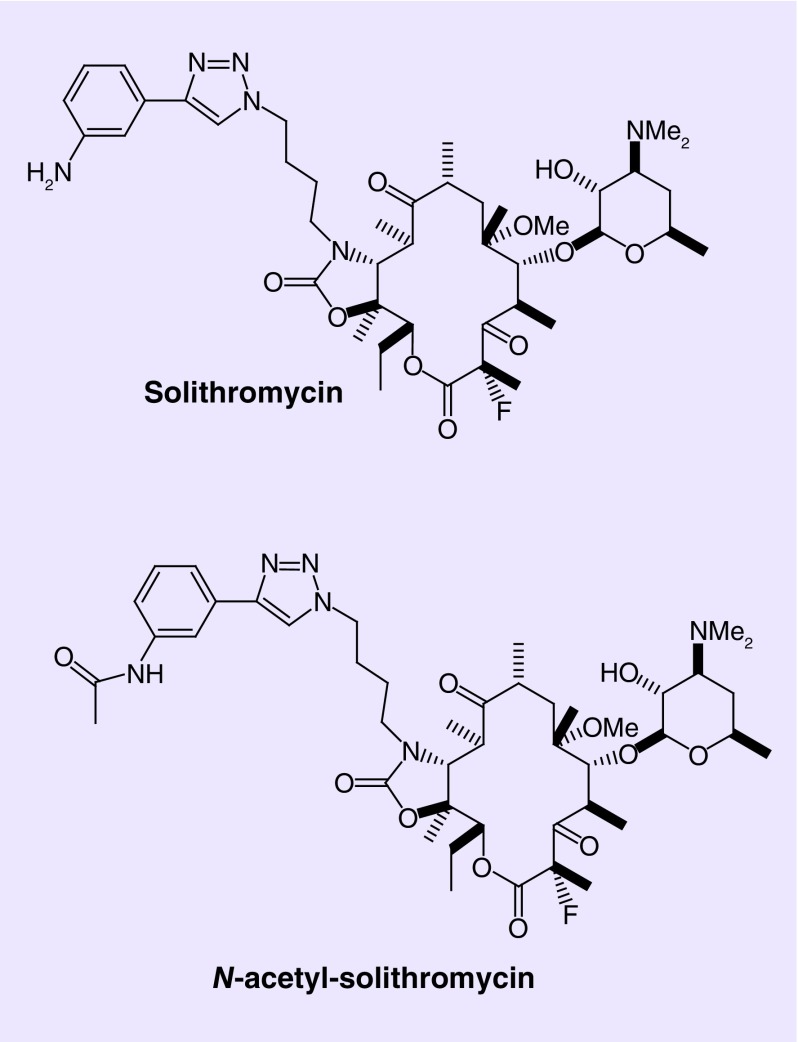
Human pharmacokinetic studies following solithromycin administration have identified an N-acetylated metabolite, N-acetyl-solithromycin [4]. N-acetyl-solithromycin presumably forms via hepatic and intestinal N-acetylation of solithromycin (Figure 1). Plasma concentrations of the active side-chain metabolites of solithromycin were low relative to the parent concentrations, since exposure (area under the curveτ) to N-acetyl-solithromycin was about 7% in healthy subjects increasing with degree of hepatic impairment [4]. Solithromycin exhibits potent antimicrobial activity toward Ureaplasma spp. isolates, a major cause of early preterm birth and N-acetyl-solithromycin, although active, exhibits reduced antimicrobial activity against this species [5].
Figure 1. . Chemical structures of solithromycin and N-acetyl-solithromycin.
Nonalcoholic steatohepatitis is a fatty liver disease in which increased liver fat is accompanied by cellular injury, inflammatory infiltrate and subsequently liver fibrosis, which can progress to cirrhosis and complications most often associated with liver-related mortality [6]. Solithromycin also has been proposed to treat noncirrhotic nonalcoholic steatohepatitis [7], and as a possible agent to target inflammation and fibrosis in nonalcoholic steatohepatitis-related kidney disease [8].
As previously reviewed [9], humans possess two functional N-acetyltransferase (NAT) isozymes identified as NAT1 and NAT2. The reference human NAT1 enzyme associated with high activity is NAT1 4 encoded by the NAT1*4 and the reference human NAT2 enzyme associated with high activity is NAT2 4 encoded by NAT2*4. Human NAT2 exhibits genetic polymorphisms resulting in rapid, intermediate and slow acetylator phenotypes resulting in differential efficacy and/or toxicity following administration of numerous drugs [10,11]. NAT2*4 is associated with rapid acetylator phenotype and the most common human NAT2 haplotypes associated with slow acetylator phenotype are NAT2*5B, NAT2*6A, NAT2*7B and NAT2*14B [9].
Human NAT1 and NAT2 allozymes have been useful to investigate N-acetylation of the antibiotic, sulfamethazine following recombinant expression in yeast (Schizosaccharomyces pombe) [12,13]. Cryopreserved human hepatocytes are an excellent model system for hepatic drug metabolism studies [14] and have been useful for investigation of hepatic N-acetylation of antibiotics, such as, sulfamethazine and isoniazid together with the effect of the acetylation polymorphism [15–17].
The purpose of this study was to investigate the N-acetylation of solithromycin by human NAT1 and NAT2 and their genetic variants following recombinant expression in yeast and in cryopreserved human hepatocytes from rapid, intermediate and slow acetylators.
Materials & methods
Chemicals
Solithromycin and N-acetyl-solithromycin were obtained from Cempra Pharmaceuticals, Inc., NC, USA.
Production of human recombinant NATs
The coding regions of NAT1*4 (reference NAT1), NAT2*4 (reference NAT2 associated with rapid acetylators), NAT2*5B, NAT2*6A and NAT2*14B (associated with slow acetylators) were amplified by PCR using previously constructed plasmids as previously described [12–13,18]. The yeast vector, pESP-3 (Stratagene, CA, USA) was digested with NdeI and AscI at 37°C overnight and gel purified in a similar manner to the PCR products. Purified PCR products and 80 ng of plasmid were ligated overnight at 16°C with T4 DNA ligase (New England Biolabs, Inc, MA, USA). Ligated plasmids were transformed into XL-10 Gold Ultracompetent Escherichia coli (Stratagene). Plasmids were isolated from cultures grown from selected colonies using the Qiagen Plasmid Midi kit (Qiagen, CA, USA) and sequenced using Thermo Sequenase (Amersham, IL, USA). Constructs were then transformed into competent S. pombe and expressed following the manufacturer's instructions (Stratagene). Mock-transformed yeast used pESP-3 vector with no NAT1 or NAT2 insert. Total cell lysates were prepared by vigorous agitation of yeast in ice-cold 20 mM NaPO4, 1 mM dithiothreitol, 1 mM EDTA, 0.2% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin A containing acid-washed glass beads (Stratagene) for 10 min at 4°C. Liquid fractions were collected from the lysed cells and centrifuged at 13,000 × g for 20 min. Supernatants were collected, aliquoted and stored at -70°C until utilized for enzymatic assays. Protein concentrations were determined using the Bio-Rad protein assay kit (Bio-Rad, CA, USA). To mitigate possible instability of human NAT1 and NAT2, supernatant aliquots were unthawed only one time on ice and used immediately to carry out the enzymatic reactions catalyzed by recombinant human NAT1 [12] and NAT2 [13,18], as previously described.
Source & processing of cryopreserved human hepatocytes
Cryopreserved human hepatocytes were obtained from BioreclamationIVT (MD, USA) and stored in liquid nitrogen until use. All hepatocytes were prepared from fresh human tissue with hepatocytes isolated and frozen within 24 h of organ removal. All hepatocytes were human transplant rejected. All hepatocytes were tested and were negative for hepatitis B and C, and HIV 1 and 2. Hepatocytes were treated as containing human-derived materials as potentially infectious, as no known test methods can offer assurance that products derived from human tissues will not transmit infectious agents. Upon removal from liquid nitrogen, hepatocytes were thawed according to the manufacturer's instructions by warming a vial of the hepatocytes at 37°C for 90 s and transferring to a 50 ml conical tube containing 45 ml of InVitroGRO HT medium (BioreclamationIVT). The cell suspension was centrifuged at 50 × g at room temperature for 5 min. The supernatant was discarded and cells washed once in ice-cold PBS before lysing the cells in ice-cold 20 mM NaPO4, 1 mM dithiothreitol, 1 mM EDTA, 0.2% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin A and 1 μg/ml aprotinin. The lysate was centrifuged at 15,000 × g for 20 min and the supernatant was aliquoted and stored at -70°C. Protein concentrations were determined using the Bio-Rad protein assay kit (Bio-Rad). To mitigate possible instability of human NAT1 and NAT2, supernatant aliquots were unthawed only one time on ice and used immediately to carry out the enzymatic reactions for human NAT1 and NAT2 as previously described [15,16].
NAT2 genotyping & assignment of acetylator phenotype
Genomic DNA was isolated from pelleted cells prepared from human cryopreserved hepatocyte samples as described above using the QIAamp DNA Mini Kit (Qiagen) according to the manufacturer's instructions. NAT2 genotypes and deduced phenotypes were determined as described previously [15,16,19]. Controls (no DNA template) were run to ensure that there was no amplification of contaminating DNA. Individuals possessing two NAT2 alleles associated with rapid acetylation activity (NAT2 *4, NAT2 *12 and NAT2 *13) were classified as rapid acetylators; individuals possessing one of these alleles and one allele associated with slow acetylation activity (NAT2 *5, NAT2 *6, NAT2 *7 and NAT2 *14) were classified as intermediate acetylators; and those individuals that possessed two slow acetylation alleles were classified as slow acetylators. Cryopreserved hepatocytes with rapid, intermediate and slow NAT2 acetylator genotype were selected at random for measurements of solithromycin N-acetylation as described below.
Measurement of solithromycin NAT activity in vitro
NAT assays containing yeast or hepatocyte lysate (<2 mg of protein/ml), 10–1000 μM solithromycin and 0.1 or 1 mM acetyl coenzyme A (AcCoA) were incubated at 37°C. Reactions were terminated by the addition of 1/10 volume of 1 M acetic acid and the reaction tubes were centrifuged to precipitate protein. The amount of N-acetyl-solithromycin produced was determined following separation and quantitation by HPLC. Separation of solithromycin and N-acetyl-solithromycin was accomplished with a 125 × 4 mm LiChrospher 100 RP-100 5μM C18 HPLC column using a gradient of 75% 20 mM sodium perchlorate pH 2.5/25% acetonitrile to 50% 20 mM sodium perchlorate pH 2.5/50% acetonitrile over 10 min. Solithromycin and N-acetyl-solithromycin were quantitated by measuring absorbance at 260 nm. Retention times were 11.7 and 13.4 min, respectively.
Determination of NAT kinetic constants
Solithromycin N-acetylation rates were determined with solithromycin concentrations ranging from 10 to 1000 μM in the presence of a fixed AcCoA concentration (1000 μM). Apparent Vmax and apparent Km were determined by nonlinear regression of the Michaelis–Menten equation (GraphPad Software, Inc, CA, USA).
Measurement of solithromycin N-acetylation in situ
Plateable cryopreserved human hepatocytes previously identified as rapid, intermediate or slow NAT2 acetylator genotypes were thawed as described above and contents of the vial were transferred into a 15 ml conical tube containing 12 ml of InVitroGRO CP (BioreclamationIVT) media prewarmed to 37°C. Cells (1.0 ml/well) were plated into Biocoat® collagen-coated 12-well plates (BD labware, MA, USA) and allowed to attach overnight. The next morning, media were removed and attached cells were washed with 1× PBS and replaced with fresh prewarmed InVitroGRO CP media containing 10 or 100 μM solithromycin. Hepatocytes were incubated for up to 48 h after which media were removed and protein precipitated by addition of 1/10 volume of 1 M acetic acid. Media were centrifuged at 15,000 × g for 10 min and supernatant was used to separate and quantitate solithromycin and N-acetyl-solithromycin by HPLC as described above.
Statistical analysis
Differences in N-acetylation rates among rapid, intermediate and slow NAT2 acetylator genotypes or recombinant human NAT2 allozymes were tested for significance by one-way analysis of variance followed by Tukey–Kramer multiple comparisons test (GraphPad Software, Inc).
Results & discussion
Human NAT1 and NAT2 were recombinantly expressed in yeast and tested for their capacity to N-acetylate solithromycin. Michaelis–Menten kinetic constants were generated for the N-acetylation of solithromycin (in the presence of 0.1 mM AcCoA) catalyzed by NAT1 4 and NAT2 4. Both human NAT1 and NAT2 catalyzed the N-acetylation of solithromycin. The apparent Vmax for recombinant human NAT2 4 (276 ± 45 nmoles/min/mg) catalysis of solithromycin N-acetylation was slightly higher than for recombinant human NAT1 4 (246 ± 28 nmoles/min/mg). By comparison, following identical recombinant expression in yeast, human NAT1 4 catalyzed the N-acetylation of p-aminobenzoic acid and 2-aminofluorene at rates over 12,000 and 30,000 nmoles/min/mg, respectively [12] whereas human NAT2 4 catalyzed the N-acetylation of sulfamethazine and 2-aminofluorene at rates less than 1500 nmoles/min/mg [13]. These results suggest that human NAT2 4 is at least as important as NAT1 4 in the N-acetylation of solithromycin but should be interpreted with caution as the absolute levels of catalytic activity most likely reflect differential recombinant expression of human NAT1 4 and NAT2 4 in yeast as opposed to actual expression of human NAT1 4 and NAT2 4 in a particular human tissue.
The mean ± standard error (n = 3) apparent Km was 1.26 ± 0.14 and 0.512 ± 0.078 mM for recombinant human NAT1 4 and NAT2 4, respectively. These findings suggest that solithromycin N-acetylation is selectively but not specifically catalyzed by human NAT2. Thus, human NAT2 genetic polymorphism may affect solithromycin metabolism most likely following oral administration.
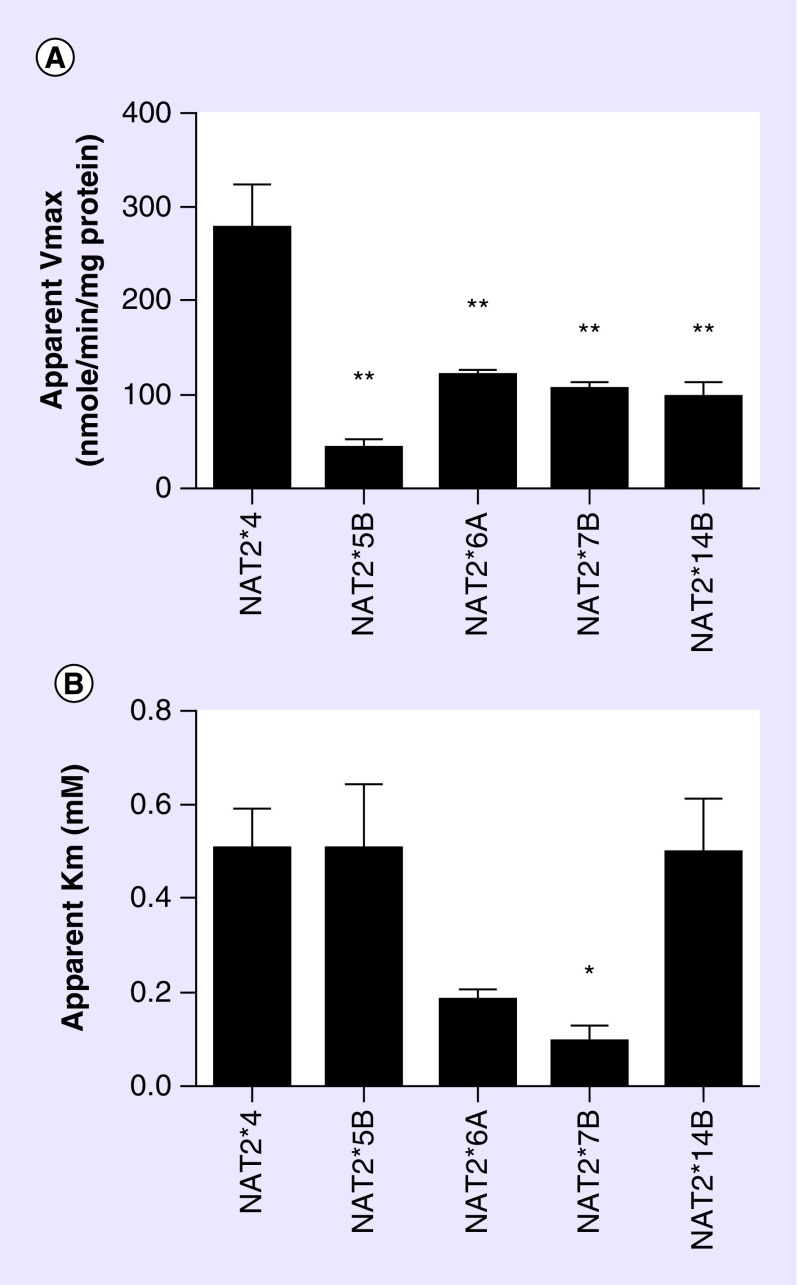
Because solithromycin exhibited over twofold higher affinity for recombinant human NAT2 than NAT1, Michaelis–Menten kinetic constants were generated for the N-acetylation of solithromycin (in the presence of 0.1 mM AcCoA) catalyzed by NAT2 4 (associated with rapid acetylators) and the most common slow acetylator human NAT2 allozymes. As shown in Figure 2, apparent maximum velocities for the N-acetylation of solithromycin were significantly (p < 0.01) higher when catalyzed by the allozyme associated with rapid acetylator haplotype (NAT2 4), than by the allozymes associated with slow acetylator haplotypes (NAT2 5B, NAT2 6A, NAT2 7B and NAT2 14B). These findings are consistent with N-acetylation, O-acetylation and N, O-acetylation rates for human NAT2 allozymes recombinantly expressed in Escherichia coli [20–23] and further support a role for NAT2 and the acetylation polymorphism in the metabolism of solithromycin. The apparent Km of solithromycin for NAT2 7B was significantly (p < 0.05) lower than for NAT2 4, NAT2 5B and NAT2 14B consistent with the apparent Km values of NAT2 allozymes following recombinant expression in E. coli [22,23].
Figure 2. . Michaelis–Menten kinetic constants of human N-acetyltransferase 2 following recombinant expression in yeast.
Each bar represents mean ± standard error for three separate determinations. Acetyl coenzyme A concentrations were 0.1 mM in all assays. Apparent Vmax values differed significantly (p = 0.0002) among the various NAT2 allozymes following one-way analysis of variance. *The apparent Km for NAT2 7B was significantly lower (p < 0.05) than NAT2 4, NAT2 5B and NAT2 14B following Tukey's multiple comparisons test.**The apparent Vmax for NAT2 5B, NAT2 6A, NAT2 7B and NAT2 14B were each significantly (p < 0.01) lower than NAT2 4 following Tukey's multiple comparisons test. Apparent Km values differed significantly (p < 0.05) among the various NAT2 allozymes following one-way analysis of variance.
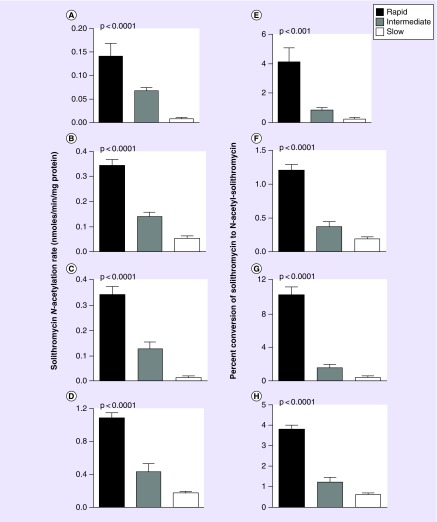
Rates of solithromycin N-acetylation in vitro were measured in cryopreserved human hepatocytes of rapid, intermediate and slow acetylator NAT2 genotype. As shown in Figure 3, a robust and highly significant (p < 0.0001) gene dose response was exhibited for solithromycin N-acetylation with the highest rates in rapid acetylators, intermediate rates in intermediate acetylators and the lowest rates in slow acetylators at multiple concentrations of solithromycin and AcCoA. Rates of N-acetylation increased as either the solithromycin or AcCoA concentrations were increased from 100 to 1000 μM reflecting the relatively high apparent Km for solithromycin determined for recombinant human NAT1 and NAT2 as described above. The solithromycin N-acetylation rates in rapid, intermediate and slow acetylator hepatocytes were slightly below those reported for sulfamethazine [15] but slightly above those reported for isoniazid [17], both are prototype drugs whose metabolism is modified by the NAT2 acetylation polymorphism in human populations [11].
Figure 3. . Solithromycin N-acetylation rates in vitro in cryopreserved human hepatocytes from rapid, intermediate and slow acetylators.
Solithromycin N-acetylation rates in nmoles/min/mg protein are shown in (A–D). Solithromycin N-acetylation rates as percent conversion of solithromycin to N-acetyl-solithromycin are shown in (E–H). In each case, differences between rapid, intermediate and slow acetylators were analyzed by one-way analysis of variance, and p-values for significance of the differences are shown. Assays were conducted at multiple substrate concentrations: (A & E; 100 μM solithromycin and 100 μM AcCoA), (B & F; 1 mM solithromycin and 100 μM AcCoA), (C & G; 100 μM solithromycin and 1 mM AcCoA), and (D & H; 1 mM solithromycin and 1 mM AcCoA). Each bar represents mean ± standard error measured in six to seven samples of cryopreserved human hepatocytes. Rapid acetylator hepatocytes (black bars) consisted of NAT2*4/*4 (five samples) and NAT2*4/*13 (one sample). Intermediate acetylator hepatocytes (gray bars) consisted of NAT2*4/*5B (three samples), NAT2*4/*6A (one sample), NAT2*4/*5A (one sample), NAT2*4/*7B (one sample) and NAT2*4/*14B (one sample). Slow acetylator hepatocytes (white bars) consisted of NAT2*5B/*5B (two samples), NAT2*5B/*6A (three samples) and NAT2*6A/*6A (one sample). The sex and ethnicity of the samples were six female Caucasians, four male Caucasians, two male Hispanics, two female Hispanics and one male of African descent.
AcCoA: Acetyl coenzyme A.
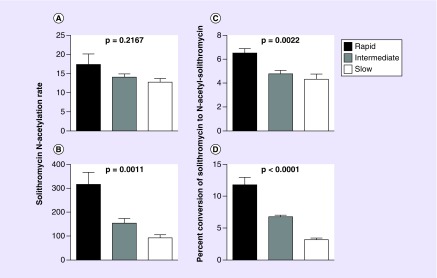
Rates of solithromycin N-acetylation were measured in rapid, intermediate and slow acetylator cryopreserved human hepatocytes in situ following incubation with solithromycin concentrations of 10 or 100 μM. Solithromycin N-acetylation exhibited a gene dose response in situ, with highest rates in hepatocytes from NAT2 rapid acetylator genotypes, intermediate rates in hepatocytes from NAT2 intermediate acetylator genotypes and lowest rates in hepatocytes from NAT2 slow acetylator genotypes, that was consistently significant following incubation with 100 μM but not 10 μM solithromycin (Figure 4). Following incubations with 100 μM solithromycin, N-acetyl-solithromycin levels were 315, 154 and 92 nmoles/24 h/million cells in the rapid, intermediate and slow acetylator hepatocytes which is 5- to 15-fold higher than for N-acetyl-isoniazid following incubations with 100 μM isoniazid in rapid, intermediate and slow acetylator hepatocytes [17].
Figure 4. . N-acetylation of solithromycin in situ in cryopreserved human hepatocytes from rapid, intermediate and slow acetylators following incubation with 10 μM (A & C) or 100 μM (B & D) solithromycin.
Solithromycin N-acetylation rates in nmoles N-acetyl-solithromycin/24 h/million cells are shown in A & B. Percent conversion of solithromycin to N-acetyl-solithromycin for these data are shown in C & D. In each case, differences between rapid, intermediate and slow acetylators were analyzed by one-way analysis of variance, and p values for significance of the differences are shown. The sex and ethnicity of the samples were four female Caucasians, five male Caucasians, three male Hispanics and three females of Hispanic descent. Each bar represents mean ± standard error measured in five samples of cryopreserved human hepatocytes. Rapid acetylator hepatocytes consisted of NAT2*4/*4 (four samples) and NAT2*4/*13 (one sample). Intermediate acetylator hepatocytes consisted of NAT2*4/*5B (two samples), NAT2*4/*6A (two samples) and NAT2*4/*5C (one sample). Slow acetylator hepatocytes consisted of NAT2*5B/*5B (two samples), NAT2*5B/*6A (two samples) and NAT2*6A/*6A (one sample).
The NAT2 genetic polymorphism was identified following administration of isoniazid for the treatment of tuberculosis [24]. Previous studies have shown an effect of NAT2 acetylator polymorphism on isoniazid efficacy [25,26], and pharmacogenetic-based therapy for tuberculosis has been proposed [27–31]. An NAT2 genotype-guided regime (isoniazid dose 7.5 mg/kg for rapid acetylators; 5.0 mg/kg for intermediate acetylators; 2.5 mg/kg for slow acetylators) reduced isoniazid-induced liver injury and early treatment failure in a randomized controlled trial [32].
A recent study evaluated the safety and pharmacokinetics of solithromycin in patients with chronic liver disease [4]. Subjects received one 800-mg dose on day 1 followed by once-daily doses of 400 mg on days 2–5. Maximum plasma concentrations of solithromycin measured under this dosing regimen ranged from 0.5 to 0.8 μg/ml, which is about tenfold lower than the 10 μM and about 100-fold lower than the 100 μM solithromycin concentrations utilized in the cryopreserved human hepatocytes. Significant differences in solithromycin N-acetylation between rapid, intermediate and slow acetylators were consistently observed at solithromycin concentrations fivefold (100 μM) but not 50-fold (10 μM) below the apparent Km observed for recombinant human NAT2 toward solithromycin (500 μM). These findings suggest it is likely that the role of the human NAT2 polymorphism on solithromycin metabolism is concentration dependent and may not be a factor with the dosing strategy employed in previous human pharmacokinetic studies of solithromycin metabolism [4].
Executive summary.
Aim
To investigate the N-acetylation of solithromycin by human NAT1 and NAT2 and their genetic variants following recombinant expression in yeast and in cryopreserved human hepatocytes from rapid, intermediate and slow acetylators.
Materials & methods
Michaelis–Menten kinetic constants were generated for the N-acetylation of solithromycin catalyzed by NAT1 and NAT2.
Plateable cryopreserved human hepatocytes previously identified as rapid, intermediate or slow NAT2 acetylator genotypes were incubated with 10 or 100 μM solithromycin.
Results
Solithromycin exhibited over twofold higher affinity for recombinant human NAT2 than NAT1.
Apparent maximum velocities for the N-acetylation of solithromycin were significantly (p < 0.01) higher catalyzed by the allozyme associated with rapid acetylators than by allozymes associated with slow acetylators.
Robust and significant (p < 0.0001) gene dose responses (rapid>intermediate>slow) were exhibited in cryopreserved human hepatocytes in vitro and in situ.
Conclusion
Solithromycin is N-acetylated by both human NAT1 and NAT2.
The role of the NAT2 acetylation polymorphism on solithromycin metabolism is likely dependent on concentration.
Footnotes
Financial & competing interests disclosure
Cempra Pharmaceuticals, Inc. (NC, USA), the developer of solithromycin, provided funding, solithromycin and N-acetyl-solithromycin, and suggestions for the manuscript. DW Hein serves as a consultant to Cempra Pharmaceuticals, Inc. and receives payment for services rendered. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Global Burden of Disease Study 2013 Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;386(9995):743–800. doi: 10.1016/S0140-6736(15)60692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhanel GG, Hartel E, Adam H, et al. Solithromycin: a novel fluoroketolide for the treatment of community-acquired bacterial pneumonia. Drugs. 2016;76(18):1737–1757. doi: 10.1007/s40265-016-0667-z. [DOI] [PubMed] [Google Scholar]
- 3.Barrera CM, Mykietiuk A, Metev H, et al. Efficacy and safety of oral solithromycin versus oral moxifloxacin for treatment of community-acquired bacterial pneumonia: a global, double-blind, multicentre, randomised, active-controlled, non-inferiority trial (SOLITAIRE-ORAL) Lancet Infect. Dis. 2016;16(4):421–430. doi: 10.1016/S1473-3099(16)00017-7. [DOI] [PubMed] [Google Scholar]
- 4.Jamieson BD, Ciric S, Fernandes P. Safety and pharmacokinetics of solithromycin in subjects with hepatic impairment. Antimicrob. Agents Chemother. 2015;59(8):4379–4386. doi: 10.1128/AAC.04652-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furfaro LL, Spiller OB, Keelan JA, Payne MS. In vitro activity of solithromycin and its metabolites, CEM-214 and N-acetyl-CEM-101, against 100 clinical Ureaplasma spp. isolates compared with azithromycin. Int. J. Antimicrob. Agents. 2015;46(3):319–324. doi: 10.1016/j.ijantimicag.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 6.Ekstedt M, Hagstrom H, Nasr P, et al. Fibrosis stage is the strongest predictor for disease-specific mortality in NAFLD after up to 33 years of follow-up. Hepatology. 2015;61(5):1547–1554. doi: 10.1002/hep.27368. [DOI] [PubMed] [Google Scholar]
- 7.Rotman Y, Sanyal AJ. Current and upcoming pharmacotherapy for non-alcoholic fatty liver disease. Gut. 2017;66(1):180–190. doi: 10.1136/gutjnl-2016-312431. [DOI] [PubMed] [Google Scholar]
- 8.Musso G, De Michieli F, Bongiovanni D, et al. New pharmacologic agents that target inflammation and fibrosis in nonalcoholic steatohepatitis-related kidney disease. Clin. Gastroenterol. Hepatol. 2016 doi: 10.1016/j.cgh.2016.08.002. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 9.Hein DW. N-acetyltransferase SNPs: emerging concepts serve as a paradigm for understanding complexities of personalized medicine. Expert Opin. Drug Metab. Toxicol. 2009;5(4):353–366. doi: 10.1517/17425250902877698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber WW, Hein DW. N-acetylation pharmacogenetics. Pharmacol. Rev. 1985;37(1):25–79. [PubMed] [Google Scholar]
- 11.McDonagh EM, Boukouvala S, Aklillu E, Hein DW, Altman RB, Klein TE. PharmGKB summary: very important pharmacogene information for N-acetyltransferase 2. Pharmacogenet. Genomics. 2014;24(8):409–425. doi: 10.1097/FPC.0000000000000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fretland AJ, Doll MA, Leff MA, Hein DW. Functional characterization of nucleotide polymorphisms in the coding region of N-acetyltransferase 1. Pharmacogenetics. 2001;11(6):511–520. doi: 10.1097/00008571-200108000-00006. [DOI] [PubMed] [Google Scholar]
- 13.Fretland AJ, Leff MA, Doll MA, Hein DW. Functional characterization of human N-acetyltransferase 2 (NAT2) single nucleotide polymorphisms. Pharmacogenetics. 2001;11(3):207–215. doi: 10.1097/00008571-200104000-00004. [DOI] [PubMed] [Google Scholar]
- 14.Gomez-Lechon MJ, Donato MT, Castell JV, Jover R. Human hepatocytes as a tool for studying toxicity and drug metabolism. Curr. Drug Metab. 2003;4(4):292–312. doi: 10.2174/1389200033489424. [DOI] [PubMed] [Google Scholar]
- 15.Doll MA, Zang Y, Moeller T, Hein DW. Codominant expression of N-acetylation and O-acetylation activities catalyzed by N-acetyltransferase 2 in human hepatocytes. J. Pharmacol. Exp. Ther. 2010;334(2):540–544. doi: 10.1124/jpet.110.168567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hein DW, Doll MA. Accuracy of various human NAT2 SNP genotyping panels to infer rapid, intermediate and slow acetylator phenotypes. Pharmacogenomics. 2012;13(1):31–41. doi: 10.2217/pgs.11.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doll MA, Salazar-González RA, Hein DW. The Sixth International Workshop on Arylamine N-acetyltransferases. Toronto, Canada: 4–6 October 2013. Acetylator genotype-dependent N-acetylation of isoniazid in human hepatocytes in situ . Presented at. [Google Scholar]
- 18.Leff MA, Fretland AJ, Doll MA, Hein DW. Novel human N-acetyltransferase 2 alleles that differ in mechanism for slow acetylator phenotype. J. Biol. Chem. 1999;274(49):34519–34522. doi: 10.1074/jbc.274.49.34519. [DOI] [PubMed] [Google Scholar]
- 19.Doll MA, Hein DW. Comprehensive human NAT2 genotype method using single nucleotide polymorphism-specific polymerase chain reaction primers and fluorogenic probes. Anal. Biochem. 2001;288(1):106–108. doi: 10.1006/abio.2000.4892. [DOI] [PubMed] [Google Scholar]
- 20.Hein DW, Ferguson RJ, Doll MA, Rustan TD, Gray K. Molecular genetics of human polymorphic N-acetyltransferase: enzymatic analysis of 15 recombinant wild-type, mutant, and chimeric NAT2 allozymes. Hum. Mol. Genet. 1994;3(5):729–734. doi: 10.1093/hmg/3.5.729. [DOI] [PubMed] [Google Scholar]
- 21.Hein DW, Doll MA, Rustan TD, Ferguson RJ. Metabolic activation of N-hydroxyarylamines and N-hydroxyarylamides by 16 recombinant human NAT2 allozymes: effects of 7 specific NAT2 nucleic acid substitutions. Cancer Res. 1995;55(16):3531–3536. [PubMed] [Google Scholar]
- 22.Grant DM, Hughes NC, Janezic SA, et al. Human acetyltransferase polymorphisms. Mutat. Res. 1997;376(1–2):61–70. doi: 10.1016/s0027-5107(97)00026-2. [DOI] [PubMed] [Google Scholar]
- 23.Hickman D, Palamanda JR, Unadkat JD, Sim E. Enzyme kinetic properties of human recombinant arylamine N-acetyltransferase 2 allotypic variants expressed in Escherichia coli . Biochem. Pharmacol. 1995;50(5):697–703. doi: 10.1016/0006-2952(95)00182-y. [DOI] [PubMed] [Google Scholar]
- 24.Evans DA, Manley KA, McKusick V. Genetic control of isoniazid metabolism in man. Br. Med. J. 1960;2(5197):485–491. doi: 10.1136/bmj.2.5197.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donald PR, Sirgel FA, Venter A, et al. The influence of human N-acetyltransferase genotype on the early bactericidal activity of isoniazid. Clin. Infect. Dis. 2004;39(10):1425–1430. doi: 10.1086/424999. [DOI] [PubMed] [Google Scholar]
- 26.Pasipanodya JG, Srivastava S, Gumbo T. Meta-analysis of clinical studies supports the pharmacokinetic variability hypothesis for acquired drug resistance and failure of antituberculosis therapy. Clin. Infect. Dis. 2012;55(2):169–177. doi: 10.1093/cid/cis353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinzig-Schippers M, Tomalik-Scharte D, Jetter A, et al. Should we use N-acetyltransferase type 2 genotyping to personalize isoniazid doses? Antimicrob. Agents Chemother. 2005;49(5):1733–1738. doi: 10.1128/AAC.49.5.1733-1738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zabost A, Brzezinska S, Kozinska M, et al. Correlation of N-acetyltransferase 2 genotype with isoniazid acetylation in Polish tuberculosis patients. BioMed Res. Int. 2013:853602. doi: 10.1155/2013/853602. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsumoto T, Ohno M, Azuma J. Future of pharmacogenetics-based therapy for tuberculosis. Pharmacogenomics. 2014;15(5):601–607. doi: 10.2217/pgs.14.38. [DOI] [PubMed] [Google Scholar]
- 30.Jung JA, Kim TE, Lee H, et al. A proposal for an individualized pharmacogenetic-guided isoniazid dosage regimen for patients with tuberculosis. Drug Des. Devel. Ther. 2015;9:5433–5438. doi: 10.2147/DDDT.S87131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi R, Jeong BH, Koh WJ, Lee SY. Recommendations for optimizing tuberculosis treatment: therapeutic drug monitoring, pharmacogenetics, and nutritional status considerations. Ann. Lab. Med. 2017;37(2):97–107. doi: 10.3343/alm.2017.37.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Azuma J, Ohno M, Kubota R, et al. NAT2 genotype guided regimen reduces isoniazid-induced liver injury and early treatment failure in the 6-month four-drug standard treatment of tuberculosis: a randomized controlled trial for pharmacogenetics-based therapy. Eur. J. Clin. Pharmacol. 2013;69(5):1091–1101. doi: 10.1007/s00228-012-1429-9. [DOI] [PMC free article] [PubMed] [Google Scholar]