Abstract
Neuroblastoma is one of the most commonly diagnosed solid cancers for children, and genetic factors may play a critical role in neuroblastoma development. Previous genome-wide association studies (GWASs) have identified nine genes associated with neuroblastoma susceptibility in Caucasians. To determine whether genetic variations in these genes are also associated with neuroblastoma susceptibility in Southern Chinese children, we genotyped 25 polymorphisms within these genes by the TaqMan method in 256 cases and 531 controls. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to evaluate the strength of the associations. We performed a meta-analysis to further evaluate the associations. Furthermore, we calculated the area under the receiver-operating characteristic curves (AUC) to assess which gene/genes may better predict neuroblastoma risk. We confirmed that CASC15 rs6939340 A > G, rs4712653 T > C, rs9295536 C > A, LIN28B rs221634 A > T, and LMO1 rs110419 A > G were associated with significantly altered neuroblastoma susceptibility. We also confirmed that rs6939340 A > G (G versus A: OR = 1.30, 95% CI = 1.13-1.50) and rs110419 G > A (A versus G: OR = 1.37, 95% CI = 1.19-1.58) were associated with increased neuroblastoma risk for all subjects. We also found that the combination of polymorphisms in CASC15, LIN28B, and LMO1 may be used to predict neuroblastoma risk (AUC = 0.63, 95% CI = 0.59-0.67). Overall, we verified five GWAS-identified polymorphisms that were associated with neuroblastoma susceptibility alteration for Southern Chinese population; however, these results need further validation in studies with larger sample sizes.
Introduction
Neuroblastoma is one of the most frequently occurring childhood tumors worldwide, affecting approximately 7.7 children per million in the Chinese population and accounting for approximately 9.8% of solid tumors in children [1]. Ethnic differences may influence the incidence of neuroblastoma. In the United States and most European countries, neuroblastoma accounts for approximately 7% to 10% of all childhood cancers with a standardized incidence rate of 8 to 14 neuroblastoma cases per million [2], [3]. In the Taiwan area, the incidence is approximately 7.8 children per million, which is quite similar to mainland China [4]. As for other countries, the incidence rate in children is approximately 9.6 per million for Australia [5], 4.5 per million for India [6], 9.1 per million for Uruguay, 4.7 per million for Chile, 3.8 per million for Mexico, 5.9 per million for Brazil, and 8.3 per million for Argentina [7]. To date, no environmental factors have been found to lead to the occurrence of neuroblastoma [8], [9], suggesting that genetic factors may play a crucial role in the occurrence of neuroblastoma [10], [11], [12], [13].
Because of the increased human genome knowledge and advancements in genotyping technology developed in the past decade, genome-wide association studies (GWASs) of human diseases became possible and have been widely utilized to study diseases such as cancer [14], [15]. In 2008, the first GWAS for neuroblastoma was conducted by Maris et al. [16], which included 1032 neuroblastoma patients and 2043 controls of European descent and was then confirmed with an additional 720 cases and 2128 controls. They confirmed that three polymorphisms (rs6939340 A > G, rs4712653 T > C, and rs9295536 C > A) within the CASC15 (also known as LINC00340) gene at the 6p22 chromosomal region were significantly associated with neuroblastoma susceptibility. When focusing on a high-risk subset, they found that common variations in the BARD1 gene at 2q35 were associated with high-risk neuroblastoma [17]. They also found that polymorphisms within DUSP12 at 1q23.3, DDX4 and IL31RA at 5q11.2, and HSD17B12 at 11p11.2 were associated with low-risk neuroblastoma [18]. In the fourth GWAS, by enlarging the sample size to 2251 cases and 6097 controls of European descent from four case series, Wang et al. [19] confirmed that four polymorphisms, especially the rs110419 A > G polymorphism within the LMO1 gene at 11p15.4 region, were significantly associated with altered susceptibility to neuroblastoma. In addition, Diskin et al. [20] analyzed data from 2817 neuroblastoma patients and 7473 controls and found that polymorphisms in the LIN28B and HACE1 genes at 6q16 were associated with neuroblastoma susceptibility.
The associations between polymorphisms within these GWAS-identified genes and neuroblastoma susceptibility have been validated in African-Americans [21], Italians [22], and Northern [23] and Southern Chinese children [24], [25], [26], [27], [28], [29]. Genetic background may differ among Europeans, African-Americans, and Chinese subjects, even among different regions of China. In the present study, we describe the relationship between genetic variations of the nine GWAS-identified genes and neuroblastoma susceptibility in Southern Chinese children including 256 cases and 531 controls. We also performed a meta-analysis to assess the association of the CASC15 rs6939340 A > G and LMO1 rs110419 G > A polymorphisms with neuroblastoma susceptibility for Southern Chinese children. We also calculated the area under the receiver-operating characteristic curves (AUC) to assess which gene/genes can best predict neuroblastoma susceptibility.
Materials and Methods
Study Subjects
This study consists of 256 neuroblastoma patients and 531 cancer-free controls that were matched by age, gender, and ethnicity as we described previously (Supplemental Table 1) [26], [30], [31]. Briefly, histopathologically confirmed neuroblastoma cases were recruited mainly between February 2010 and November 2015 with written, informed consent by their guardians. All the controls were collected in the same period from the Guangzhou Women and Children's Medical Center. This study was approved by the Institutional Review Board of Guangzhou Women and Children's Medical Center.
Genotyping and Quality Control
We genotyped the 25 polymorphisms within the nine GWAS-identified genes by TaqMan real-time PCR [32], [33]. To monitor quality control, eight negative controls (water) as well as eight replicate samples were included in each 384-well plate. Additionally, approximately 10% of the samples were randomly selected for further quality control, and the results were 100% concordant.
Meta-Analysis
We performed a meta-analysis by collecting data from all available publications on the CASC15 rs6939340 A > G and LMO1 rs110419 G > A polymorphisms. Crude odds ratios (ORs) and 95% confidence intervals (CIs) were used to investigate the strength of the associations under an allele-comparing model. Heterogeneity was measured by a χ2-based Q test. Random-effect modeling was used when Phet < .1 [34].
Statistical Analysis
We applied χ2 tests to compare categorical variables such as demographics and genotype frequencies. We used the goodness-of-fit χ2 test to assess the Hardy-Weinberg equilibrium for controls by using the observed genotypes for each polymorphism. Associations of the selected polymorphisms and the combined genotypes for the three most significant polymorphisms from each region with neuroblastoma susceptibility were estimated by ORs and 95% CIs were calculated using unconditional logistic regression with adjustment for age and gender. We adopted a nonparametric approach to compare the area under the receiver operating characteristic (ROC) curves (AUC) for the polymorphisms from the three most significant genes and the combined genes [35]. All statistical analyses were performed using SAS software (version 9.4; SAS Institute, Cary, NC, USA). All the P values were two sided, and P < .05 was considered statistically significant.
Results
Associations between Selected Polymorphisms and Neuroblastoma Susceptibility
As shown in Table 1, of the 25 selected polymorphisms, we confirmed that five were associated with neuroblastoma susceptibility: CASC15 gene polymorphisms rs6939340 G > A, rs4712653 C > T, and rs9295536 A > C; LIN28B gene polymorphism rs221634 A > T; and LMO1 gene polymorphism rs110419 A > G. No significant associations were observed for other polymorphisms.
Table 1.
Association between Polymorphisms in GWAS-Identified Genes and Neuroblastoma Risk in Southern Chinese Children
| Gene |
Polymorphism |
Allele |
Case (N = 256) |
Control (N = 531) |
Adjusted ORa |
Pa |
Adjusted ORb |
Pb |
HWE |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | AA | AB | BB | AA | AB | BB | (95% CI) | (95% CI) | |||||
| CASC15 | rs6939340 | G | A | 155 | 81 | 19 | 232 | 247 | 52 | 0.50 (0.37-0.68) | <.0001 | 0.74 (0.43-1.28) | .286 | 0.239 |
| CASC15 | rs4712653 | C | T | 171 | 69 | 15 | 285 | 209 | 37 | 0.57 (0.42-0.78) | .0004 | 0.84 (0.45-1.56) | .581 | 0.875 |
| CASC15 | rs9295536 | A | C | 168 | 76 | 11 | 282 | 212 | 37 | 0.59 (0.43-0.80) | .0008 | 0.61 (0.30-1.21) | .154 | 0.739 |
| BARD1 | rs7585356 | G | A | 120 | 114 | 21 | 235 | 237 | 59 | 0.88 (0.65-1.19) | .414 | 0.71 (0.42-1.20) | .199 | 0.948 |
| BARD1 | rs6435862 | T | G | 174 | 74 | 7 | 381 | 133 | 17 | 1.19 (0.86-1.65) | .291 | 0.85 (0.35-2.07) | .717 | 0.205 |
| BARD1 | rs3768716 | A | G | 166 | 81 | 8 | 364 | 148 | 19 | 1.18 (0.86-1.63) | .298 | 0.86 (0.37-1.99) | .723 | 0.415 |
| LIN28B[24] | rs221634 | A | T | 74 | 113 | 60 | 163 | 274 | 93 | 1.04 (0.75-1.45) | .798 | 1.50 (1.04-2.17) | .030 | 0.228 |
| LIN28B[24] | rs221635 | T | C | 176 | 64 | 7 | 345 | 168 | 17 | 0.74 (0.54-1.03) | .078 | 0.88 (0.36-2.14) | .771 | 0.527 |
| LIN28B[24] | rs314276 | C | A | 125 | 96 | 26 | 254 | 228 | 48 | 0.90 (0.67-1.22) | .497 | 1.19 (0.72-1.97) | .503 | 0.756 |
| LIN28B[24] | rs9404590 | T | G | 130 | 100 | 17 | 286 | 205 | 39 | 1.06 (0.78-1.43) | .723 | 0.93 (0.52-1.69) | .819 | 0.786 |
| LMO1[26] | rs110419 | A | G | 103 | 117 | 36 | 159 | 275 | 97 | 0.63 (0.46-0.86) | .004 | 0.74 (0.49-1.12) | .152 | 0.248 |
| LMO1[26] | rs4758051 | G | A | 95 | 126 | 35 | 194 | 242 | 95 | 0.99 (0.73-1.35) | .942 | 0.73 (0.48-1.11) | .144 | 0.199 |
| LMO1[26] | rs10840002 | A | G | 90 | 124 | 42 | 182 | 240 | 109 | 0.97 (0.71-1.33) | .863 | 0.76 (0.51-1.13) | .174 | 0.070 |
| LMO1[26] | rs204938 | A | G | 164 | 83 | 9 | 354 | 165 | 12 | 1.12 (0.82-1.54) | .470 | 1.55 (0.64-3.73) | .330 | 0.153 |
| DUSP12[28] | rs1027702 | T | C | 137 | 98 | 21 | 282 | 206 | 43 | 0.98 (0.73-1.33) | .915 | 1.02 (0.59-1.77) | .932 | 0.534 |
| IL31RA[28] | rs10055201 | A | G | 69 | 136 | 51 | 153 | 257 | 121 | 1.09 (0.78-1.53) | .607 | 0.83 (0.58-1.21) | .333 | 0.512 |
| DDX4[28] | rs2619046 | G | A | 57 | 132 | 67 | 151 | 257 | 123 | 1.39 (0.98-1.98) | .065 | 1.18 (0.84-1.67) | .345 | 0.499 |
| HSD17B12[28] | rs11037575 | C | T | 144 | 91 | 21 | 263 | 236 | 32 | 0.76 (0.57-1.03) | .077 | 1.38 (0.78-2.45) | .270 | 0.026 |
| HACE1 | rs6571212 | A | T | 137 | 102 | 17 | 310 | 185 | 36 | 1.22 (0.90-1.64) | .204 | 1.00 (0.55-1.82) | .995 | 0.246 |
| HACE1 | rs1316908 | C | T | 195 | 58 | 3 | 374 | 145 | 12 | 0.74 (0.52-1.04) | .080 | 0.51 (0.14-1.82) | .299 | 0.639 |
| HACE1[29] | rs2499667 | A | G | 90 | 118 | 41 | 181 | 248 | 101 | 0.91 (0.66-1.24) | .546 | 0.84 (0.56-1.25) | .394 | 0.330 |
| HACE1[29] | rs9404576 | T | G | 134 | 97 | 18 | 303 | 189 | 38 | 1.15 (0.85-1.55) | .380 | 1.03 (0.57-1.85) | .921 | 0.259 |
| HACE1[29] | rs2499663 | T | C | 93 | 115 | 41 | 189 | 243 | 98 | 0.92 (0.68-1.26) | .614 | 0.87 (0.59-1.30) | .508 | 0.204 |
| HACE1[29] | rs4336470 | C | T | 130 | 99 | 20 | 303 | 188 | 39 | 1.22 (0.90-1.65) | .197 | 1.13 (0.64-1.98) | .681 | 0.194 |
| HACE1[29] | rs4079063 | A | G | 92 | 116 | 41 | 189 | 242 | 99 | 0.94 (0.69-1.28) | .690 | 0.86 (0.58-1.29) | .466 | 0.169 |
HWE, Hardy-Weinberg equilibrium.
Adjusted for age and gender for dominant model.
Adjusted for age and gender for recessive model.
Estimates of Neuroblastoma Risk by Genotype
As shown in Table 2, we chose one of the most significant polymorphisms from each of the three regions (rs6939340, rs221634, and rs110419) to assess the joint impact on neuroblastoma risk. When the rs6939340 AG/AA, rs221634 AA/AT, and rs110419 GG/AG carriers were used as a reference, we found that risk genotype carriers may have increased neuroblastoma risk, particularly carriers of the rs6939340 GG, rs221634 TT, and rs110419 AA polymorphisms (adjusted OR = 4.11, 95% CI = 1.95-9.66).
Table 2.
Estimates of Neuroblastoma Risk by Genotypes at CASC15 (rs6939340), LIN28B (rs221634), and LMO1 (rs110419)
| Genotypes |
Case (N = 256) |
Control (N = 531) |
OR (95% CI) | P | Adjusted OR (95% CI)a | Pa | ||
|---|---|---|---|---|---|---|---|---|
| rs6939340 | rs221634 | rs110419 | N (%) | N (%) | ||||
| AG/AA | AA/AT | GG/AG | 45 (17.58) | 167 (31.45) | 1.00 | 1.00 | ||
| AG/AA | AA/AT | AA | 33 (12.89) | 76 (14.31) | 1.61 (0.95-2.72) | .075 | 1.59 (0.94-2.69) | .082 |
| AG/AA | TT | GG/AG | 11 (4.30) | 42 (7.91) | 0.97 (0.46-2.04) | .940 | 0.96 (0.46-2.02) | .913 |
| AG/AA | TT | AA | 11 (4.30) | 14 (2.64) | 2.92 (1.24-6.86) | .014 | 2.88 (1.22-6.79) | .016 |
| GG | AA/AT | GG/AG | 72 (28.13) | 138 (25.99) | 1.94 (1.25-2.99) | .003 | 1.92 (1.24-2.97) | .003 |
| GG | AA/AT | AA | 46 (17.97) | 57 (10.73) | 3.00 (1.80-4.98) | <.0001 | 3.01 (1.81-5.01) | <.0001 |
| GG | TT | GG/AG | 25 (9.77) | 25 (4.71) | 3.71 (1.95-7.07) | <.0001 | 3.66 (1.92-6.97) | <.0001 |
| GG | TT | AA | 13 (5.08) | 12 (2.26) | 4.02 (1.72-9.41) | .001 | 4.11 (1.75-9.66) | .001 |
Adjusted for age and gender.
Meta-Analysis Results
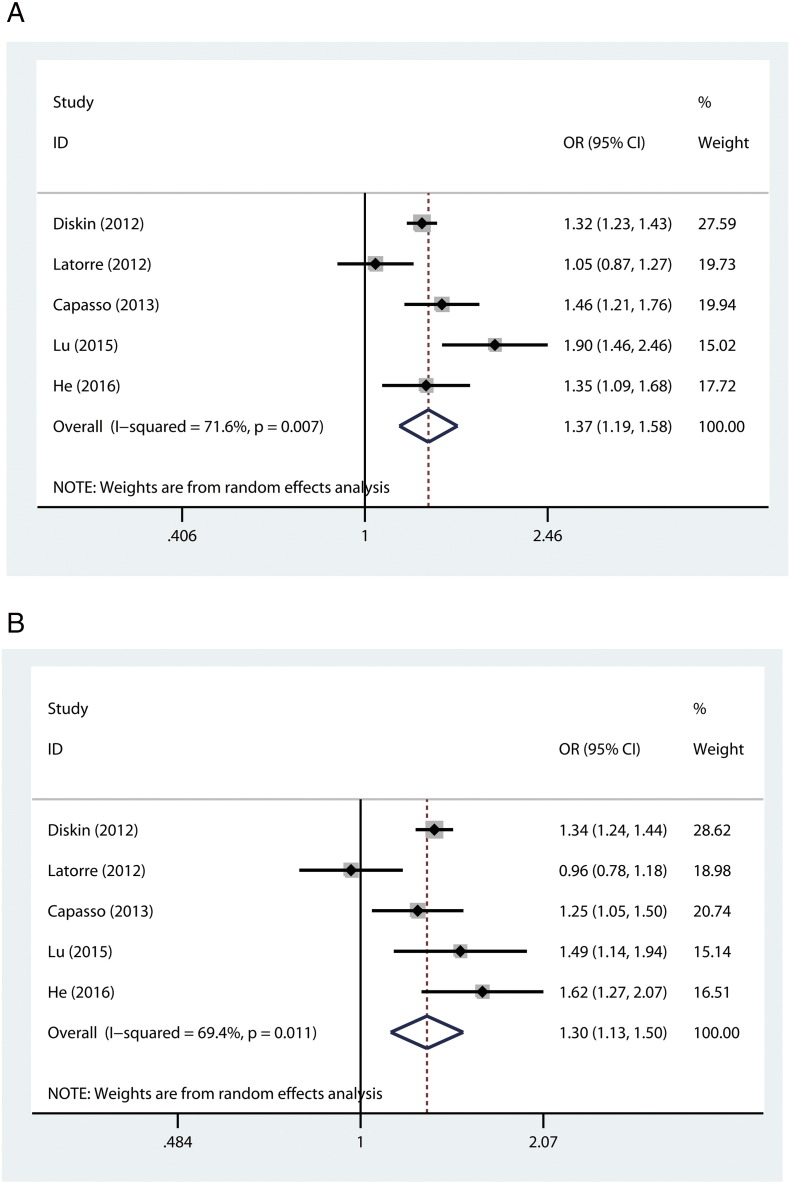
As shown in Table 3 and Figure 1, analysis of the rs6939340 G > A polymorphism in 3302 neuroblastoma cases and 8279 controls found that carrying the rs6939340 G allele is associated with increased neuroblastoma risk (G versus A: OR = 1.37, 95% CI = 1.19-1.58, P = 1.97*10−5). Similarly, for the rs110419 A > G polymorphism, a total of 3289 cases and 8303 controls were analyzed, and the combined results indicated that this polymorphism was significantly associated with neuroblastoma susceptibility (A versus G: OR = 1.30, 95% CI = 1.13-1.50, P = 3.15*10−4) (Figure 1).
Table 3.
Characteristics of Studies Included in This Meta-Analysis for CASC15 rs6939340 A > G and LMO1 rs110419 G > A Polymorphisms
| Surname | Year | Race | Case | Control | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CASC15 rs6939340 A > G | All | AA | AG | GG | A | G | G Freq | All | AA | AG | GG | A | G | G Freq | ||
| Diskin | 2012 | Caucasians | 2101 | / | / | / | 1895 | 2307 | 0.549 | 4202 | / | / | / | 4404 | 4000 | 0.476 |
| Latorre | 2012 | Africans | 363 | 12 | 103 | 248 | 127 | 599 | 0.825 | 2480 | 82 | 677 | 1721 | 841 | 4119 | 0.830 |
| Capasso | 2013 | Caucasians | 339 | 74 | 162 | 103 | 310 | 368 | 0.543 | 761 | 196 | 390 | 175 | 782 | 740 | 0.486 |
| Lu | 2015 | Asians | 244 | / | / | / | 124 | 364 | 0.746 | 305 | / | / | / | 205 | 405 | 0.660 |
| He | 2016 | Asians | 255 | 19 | 81 | 155 | 119 | 391 | 0.767 | 531 | 52 | 247 | 232 | 351 | 711 | 0.669 |
| Total | 3302 | 8279 | ||||||||||||||
| LMO1 rs110419 G > A | All | GG | AG | AA | G | A | A Freq | All | GG | AG | AA | G | A | A Freq | ||
| Diskin | 2012 | Caucasians | 2101 | / | / | / | 1853 | 2349 | 0.559 | 4202 | / | / | / | 4294 | 4110 | 0.489 |
| Latorre | 2012 | Africans | 365 | 18 | 124 | 223 | 160 | 570 | 0.781 | 2491 | 137 | 863 | 1491 | 1137 | 3845 | 0.772 |
| Capasso | 2013 | Caucasians | 323 | 84 | 152 | 87 | 320 | 326 | 0.505 | 774 | 271 | 370 | 133 | 912 | 636 | 0.411 |
| Lu | 2015 | Asians | 244 | / | / | / | 125 | 363 | 0.744 | 305 | / | / | / | 241 | 369 | 0.605 |
| He | 2016 | Asians | 256 | 36 | 117 | 103 | 189 | 323 | 0.631 | 531 | 97 | 275 | 159 | 469 | 593 | 0.558 |
| Total | 3289 | 8303 | ||||||||||||||
Figure 1.
Forest plots for the correlation of the (A) CASC15 rs6939340 G > A and (B) LMO1 rs110419 A > G polymorphisms with neuroblastoma susceptibility under the allele-comparing model. The horizontal line represents the OR and 95% CI for each investigation. The diamond represents the pooled OR and 95% CI.
AUC for GWAS-Identified Genes
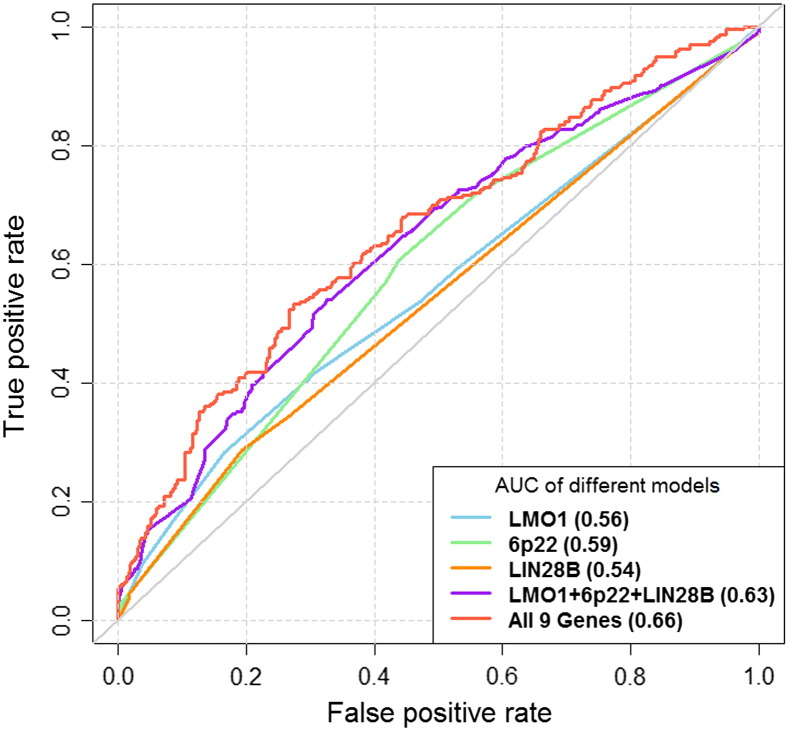
As shown in Figure 2, when all the polymorphisms for each gene are compared, the CASC15 gene (AUC = 0.59, 95% CI = 0.55-0.63) is a better predictor of neuroblastoma risk than the LMO1 gene (AUC = 0.56, 95% CI = 0.52-0.60) or LIN28B gene (AUC = 0.54, 95% CI = 0.51-0.58). However, these three genes combined have an AUC of 0.63 (95% CI = 0.59-0.67). When all the polymorphisms from the nine genes were combined, the AUC was further improved to 0.66 (95% CI = 0.61-0.70).
Figure 2.
ROC analysis for single and combined genes identified from GWAS for neuroblastoma. The areas under the ROC curves (AUCs) were calculated to measure the predictive power of risk-assessment models based on polymorphisms within gene/genes.
Discussion
In the described hospital-based case-control study with 256 neuroblastoma cases and 531 cancer-free controls from south China, we systematically evaluated the associations between polymorphisms derived from nine GWAS-identified genes and confirmed the role of five polymorphisms in predicting neuroblastoma susceptibility. We also found that risk genotype carriers have a significantly increased neuroblastoma risk, as high as 4.11-fold. By analyzing data from all available publications, we further confirmed that the CASC15 rs6939340 G > A and LMO1 rs110419 A > G polymorphisms were significantly associated with neuroblastoma risk.
In addition to environmental factors, genetic factors may also play a crucial role in the occurrence of neuroblastoma [13]. GWAS is a powerful tool in identifying disease-related loci. It has significantly improved our understanding of the genetic basis of cancer, providing the basis for discovering new options for targeted prevention and therapy [14]. To date, nine susceptibility genes have been discovered [16], [17], [18], [19], [20], and among them, polymorphisms within the CASC15, LMO1, LIN28B, and HCAE1 genes are significantly associated with neuroblastoma risk, including but not limited to high-risk and low-risk subtypes. The first identified and most prominent polymorphism associated with neuroblastoma was CASC15 rs6939340 G > A (P = 9.33 × 10−15) at 6p22 region. Two additional CASC15 gene polymorphisms (rs4712653 with P = 5.50*10−13 and rs9295536 with P = 1.24*10−11) were also associated with neuroblastoma susceptibility [16]. Following this discovery, using data from 1627 cases and 3254 controls in the discovery stage and 624 cases and 2843 controls in the replication stage, Wang et al. [19] discovered four LMO1 gene polymorphisms (rs110419 A > G, rs4758051 G > A, rs10840002 A > G, and rs204938 A > G) that were associated with neuroblastoma susceptibility. Among these polymorphisms, the rs110419 A > G was the most noteworthy one. In 2012, Diskin et al. [20] found that five polymorphisms in the HACE1 gene and one polymorphism in the LIN28B gene were associated with neuroblastoma susceptibility including a total of 10,290 subjects. It is also worth noting that BARD1 gene polymorphisms have been reported to be associated with high-risk neuroblastoma [17].
In their replication study consisting of African-Americans with 391 cases and 2500 controls, Latorre et al. [21] analyzed a total of 12 polymorphisms from the CASC15, BARD1, and LMO1 genes and confirmed that all of the five polymorphisms in the BARD1 gene were associated with neuroblastoma risk. However, they failed to confirm the effects of the CASC15 and LMO1 genes. In a replicated study in an Italian population with 370 cases and 809 controls, Capasso et al. [22] investigated 16 polymorphisms from the nine GWAS-identified genes and successfully confirmed the association of the CASC15, BARD1, LMO1, and HSD17B12 genes. As for Northern Chinese subjects, Lu et al. [23] analyzed a total of 244 cases and 305 controls and found that polymorphisms in the CASC15, LMO1, and HSD17B12 genes were associated with neuroblastoma susceptibility. In this study of Southern Chinese children, we confirmed that five polymorphisms within the nine GWAS-identified genes were associated with neuroblastoma susceptibility. Our meta-analysis also confirmed that the CASC15 rs6939340 G > A and LMO1 rs110419 A > G polymorphisms were significantly associated with increased neuroblastoma risk. Our failure to confirm an association with the additional polymorphisms may be due to the weak effect of SNPs, limited sample size, and ethnicity differences.
Several limitations should be mentioned. First, the sample size (256 neuroblastoma cases) is relatively small despite us including all the samples available. More samples from other regions of China should be investigated and combined in future multicenter studies. Second, we only included 25 polymorphisms in these nine genes and nearly none of them was potential functional according to SNPinfo (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html); inclusion of more polymorphisms, in particular, the potential functional ones [33] as well as low-frequency variants [36], needs to be considered. Third, we only investigated nine genes by previous GWAS; the latest ones such as MLF1 and CPZ [37] were not included in the current study. Fourth, relatively limited information was collected due to the nature of retrospective investigations. Other factors such as paternal exposures, living environment, and dietary intake were not available.
In summary, we provide an overview of the genetic variations within the GWAS-identified genes associated with neuroblastoma susceptibility in Southern Chinese children. Further investigations with larger samples and different ethnicities are needed to validate and confirm the effect of GWAS-identified genes for neuroblastoma susceptibility.
The following are the supplementary data related to this article.
Distribution of Demographic and Clinical-Pathologic Characteristics for Neuroblastoma Patients and Cancer-Free Controls
Footnotes
Novelty: In this study of 256 neuroblastoma cases and 531 controls, we evaluated the association of polymorphisms in nine GWAS-identified genes with neuroblastoma susceptibility and confirmed associations with five polymorphisms. We also found that risk genotype carriers have a significantly increased neuroblastoma risk of 4.11-fold. By analyzing data from all available publications, we further confirmed that the CASC15 rs6939340 G>A and LMO1 rs110419 A>G polymorphisms are significantly associated with neuroblastoma risk.
Conflict of Interest: None.
This work was supported by grants from the Pearl River S&T Nova Program of Guangzhou (No. 201710010086), State Clinical Key Specialty Construction Project (Pediatric Surgery) 2013 (No: GJLCZD1301), and the National Natural Science Foundation of China (Grant No. 81502046).
Contributor Information
Jing He, Email: hejing198374@gmail.com.
Huimin Xia, Email: xia-huimin@foxmail.com.
References
- 1.Bao PP, Li K, Wu CX, Huang ZZ, Wang CF, Xiang YM, Peng P, Gong YM, Xiao XM, Zheng Y. Recent incidences and trends of childhood malignant solid tumors in Shanghai, 2002-2010. Zhonghua Er Ke Za Zhi. 2013;51:288–294. [PubMed] [Google Scholar]
- 2.Gatta G, Capocaccia R, Coleman MP, Ries LA, Berrino F. Childhood cancer survival in Europe and the United States. Cancer. 2002;95:1767–1772. doi: 10.1002/cncr.10833. [DOI] [PubMed] [Google Scholar]
- 3.Spix C, Pastore G, Sankila R, Stiller CA, Steliarova-Foucher E. Neuroblastoma incidence and survival in European children (1978-1997): report from the Automated Childhood Cancer Information System project. Eur J Cancer. 2006;42:2081–2091. doi: 10.1016/j.ejca.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Liu YL, Lo WC, Chiang CJ, Yang YW, Lu MY, Hsu WM, Ho WL, Li MJ, Miser JS, Lin DT. Incidence of cancer in children aged 0-14 years in Taiwan, 1996-2010. Cancer Epidemiol. 2015;39:21–28. doi: 10.1016/j.canep.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Baade PD, Youlden DR, Valery PC, Hassall T, Ward L, Green AC, Aitken JF. Trends in incidence of childhood cancer in Australia, 1983-2006. Br J Cancer. 2010;102:620–626. doi: 10.1038/sj.bjc.6605503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swaminathan R, Rama R, Shanta V. Childhood cancers in Chennai, India, 1990-2001: incidence and survival. Int J Cancer. 2008;122:2607–2611. doi: 10.1002/ijc.23428. [DOI] [PubMed] [Google Scholar]
- 7.Moreno F, Lopez Marti J, Palladino M, Lobos P, Gualtieri A, Cacciavillano W. Childhood neuroblastoma: incidence and survival in Argentina. Report from the National Pediatric Cancer Registry, ROHA Network 2000-2012. Pediatr Blood Cancer. 2016;63:1362–1367. doi: 10.1002/pbc.25987. [DOI] [PubMed] [Google Scholar]
- 8.De Roos AJ, Olshan AF, Teschke K, Poole C, Savitz DA, Blatt J, Bondy ML, Pollock BH. Parental occupational exposures to chemicals and incidence of neuroblastoma in offspring. Am J Epidemiol. 2001;154:106–114. doi: 10.1093/aje/154.2.106. [DOI] [PubMed] [Google Scholar]
- 9.De Roos AJ, Teschke K, Savitz DA, Poole C, Grufferman S, Pollock BH, Olshan AF. Parental occupational exposures to electromagnetic fields and radiation and the incidence of neuroblastoma in offspring. Epidemiology. 2001;12:508–517. doi: 10.1097/00001648-200109000-00008. [DOI] [PubMed] [Google Scholar]
- 10.Capasso M, Diskin SJ. Genetics and genomics of neuroblastoma. Cancer Treat Res. 2010;155:65–84. doi: 10.1007/978-1-4419-6033-7_4. [DOI] [PubMed] [Google Scholar]
- 11.Deyell RJ, Attiyeh EF. Advances in the understanding of constitutional and somatic genomic alterations in neuroblastoma. Cancer Genet. 2011;204:113–121. doi: 10.1016/j.cancergen.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Capasso M, Diskin S, Cimmino F, Acierno G, Totaro F, Petrosino G, Pezone L, Diamond M, McDaniel L, Hakonarson H. Common genetic variants in NEFL influence gene expression and neuroblastoma risk. Cancer Res. 2014;74:6913–6924. doi: 10.1158/0008-5472.CAN-14-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oldridge DA, Wood AC, Weichert-Leahey N, Crimmins I, Sussman R, Winter C, McDaniel LD, Diamond M, Hart LS, Zhu S. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature. 2015;528:418–421. doi: 10.1038/nature15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stadler ZK, Thom P, Robson ME, Weitzel JN, Kauff ND, Hurley KE, Devlin V, Gold B, Klein RJ, Offit K. Genome-wide association studies of cancer. J Clin Oncol. 2010;28:4255–4267. doi: 10.1200/JCO.2009.25.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 16.Maris JM, Mosse YP, Bradfield JP, Hou C, Monni S, Scott RH, Asgharzadeh S, Attiyeh EF, Diskin SJ, Laudenslager M. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med. 2008;358:2585–2593. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Capasso M, Devoto M, Hou C, Asgharzadeh S, Glessner JT, Attiyeh EF, Mosse YP, Kim C, Diskin SJ, Cole KA. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet. 2009;41:718–723. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen le B, Diskin SJ, Capasso M, Wang K, Diamond MA, Glessner J, Kim C, Attiyeh EF, Mosse YP, Cole K. Phenotype restricted genome-wide association study using a gene-centric approach identifies three low-risk neuroblastoma susceptibility Loci. PLoS Genet. 2011;7:e1002026. doi: 10.1371/journal.pgen.1002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K, Diskin SJ, Zhang H, Attiyeh EF, Winter C, Hou C, Schnepp RW, Diamond M, Bosse K, Mayes PA. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469:216–220. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diskin SJ, Capasso M, Schnepp RW, Cole KA, Attiyeh EF, Hou C, Diamond M, Carpenter EL, Winter C, Lee H. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat Genet. 2012;44:1126–1130. doi: 10.1038/ng.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latorre V, Diskin SJ, Diamond MA, Zhang H, Hakonarson H, Maris JM, Devoto M. Replication of neuroblastoma SNP association at the BARD1 locus in African-Americans. Cancer Epidemiol Biomarkers Prev. 2012;21:658–663. doi: 10.1158/1055-9965.EPI-11-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capasso M, Diskin SJ, Totaro F, Longo L, De Mariano M, Russo R, Cimmino F, Hakonarson H, Tonini GP, Devoto M. Replication of GWAS-identified neuroblastoma risk loci strengthens the role of BARD1 and affirms the cumulative effect of genetic variations on disease susceptibility. Carcinogenesis. 2013;34:605–611. doi: 10.1093/carcin/bgs380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu J, Chu P, Wang H, Jin Y, Han S, Han W, Tai J, Guo Y, Ni X. Candidate gene association analysis of neuroblastoma in Chinese children strengthens the role of LMO1. PLoS One. 2015;10:e0127856. doi: 10.1371/journal.pone.0127856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He J, Yang T, Zhang R, Zhu J, Wang F, Zou Y, Xia H. Potentially functional polymorphisms in the LIN28B gene contribute to neuroblastoma susceptibility in Chinese children. J Cell Mol Med. 2016;20:1534–1541. doi: 10.1111/jcmm.12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He J, Zhang R, Zou Y, Zhu J, Yang T, Wang F, Xia H. Evaluation of GWAS-identified SNPs at 6p22 with neuroblastoma susceptibility in a Chinese population. Tumour Biol. 2016;37:1635–1639. doi: 10.1007/s13277-015-3936-7. [DOI] [PubMed] [Google Scholar]
- 26.He J, Zhong W, Zeng J, Zhu J, Zhang R, Wang F, Yang T, Zou Y, Xia H. LMO1 gene polymorphisms contribute to decreased neuroblastoma susceptibility in a Southern Chinese population. Oncotarget. 2016;7:22770–22778. doi: 10.18632/oncotarget.8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang R, Zou Y, Zhu J, Zeng X, Yang T, Wang F, He J, Xia H. The association between GWAS-identified BARD1 gene SNPs and neuroblastoma susceptibility in a Southern Chinese population. Int J Med Sci. 2016;13:133–138. doi: 10.7150/ijms.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, Zou Y, Zhu J, Zhang R, Yang T, Wang F, Xia H, He J, Feng Z. HSD17B12 gene rs11037575 C>T polymorphism confers neuroblastoma susceptibility in a Southern Chinese population. Onco Targets Ther. 2017;10:1969–1975. doi: 10.2147/OTT.S136006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, Zhang R, Zhu J, Wang F, Yang T, Zou Y, He J, Xia H. Common variations within HACE1 gene and neuroblastoma susceptibility in a Southern Chinese population. Onco Targets Ther. 2017;10:703–709. doi: 10.2147/OTT.S129042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He J, Wang F, Zhu J, Zhang R, Yang T, Zou Y, Xia H. Association of potentially functional variants in the XPG gene with neuroblastoma risk in a Chinese population. J Cell Mol Med. 2016;20:1481–1490. doi: 10.1111/jcmm.12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He J, Wang F, Zhu J, Zhang Z, Zou Y, Zhang R, Yang T, Xia H. The TP53 gene rs1042522 C>G polymorphism and neuroblastoma risk in Chinese children. Aging (Albany NY) 2017;9:852–859. doi: 10.18632/aging.101196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He J, Qiu LX, Wang MY, Hua RX, Zhang RX, Yu HP, Wang YN, Sun MH, Zhou XY, Yang YJ. Polymorphisms in the XPG gene and risk of gastric cancer in Chinese populations. Hum Genet. 2012;131:1235–1244. doi: 10.1007/s00439-012-1152-8. [DOI] [PubMed] [Google Scholar]
- 33.Lou J, Gong J, Ke J, Tian J, Zhang Y, Li J, Yang Y, Zhu Y, Gong Y, Li L. A functional polymorphism located at transcription factor binding sites, rs6695837 near LAMC1 gene, confers risk of colorectal cancer in Chinese populations. Carcinogenesis. 2017;38:177–183. doi: 10.1093/carcin/bgw204. [DOI] [PubMed] [Google Scholar]
- 34.He J, Shi TY, Zhu ML, Wang MY, Li QX, Wei QY. Associations of Lys939Gln and Ala499Val polymorphisms of the XPC gene with cancer susceptibility: a meta-analysis. Int J Cancer. 2013;133:1765–1775. doi: 10.1002/ijc.28089. [DOI] [PubMed] [Google Scholar]
- 35.Ruan HL, Qin HD, Shugart YY, Bei JX, Luo FT, Zeng YX, Jia WH. Developing genetic epidemiological models to predict risk for nasopharyngeal carcinoma in high-risk population of China. PLoS One. 2013;8:e56128. doi: 10.1371/journal.pone.0056128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Zou L, Zhou Y, Li L, Zhu Y, Yang Y, Gong Y, Lou J, Ke J, Zhang Y. A low-frequency variant in SMAD7 modulates TGF-beta signaling and confers risk for colorectal cancer in Chinese population. Mol Carcinog. 2017;56:1798–1807. doi: 10.1002/mc.22637. [DOI] [PubMed] [Google Scholar]
- 37.McDaniel LD, Conkrite KL, Chang X, Capasso M, Vaksman Z, Oldridge DA, Zachariou A, Horn M, Diamond M, Hou C. Common variants upstream of MLF1 at 3q25 and within CPZ at 4p16 associated with neuroblastoma. PLoS Genet. 2017;13:e1006787. doi: 10.1371/journal.pgen.1006787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of Demographic and Clinical-Pathologic Characteristics for Neuroblastoma Patients and Cancer-Free Controls