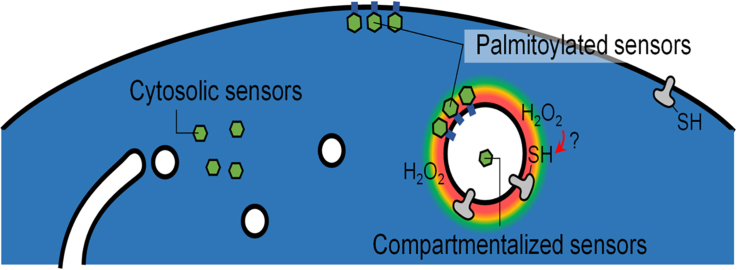
Abstract
Production of reactive oxygen species (ROS) and consequent glutathione oxidation are associated with various physiological processes and diseases, including cell differentiation, senescence, and inflammation. GFP-based redox sensors provide a straight-forward approach to monitor ROS levels and glutathione oxidation within a living cell at the subcellular resolution. We utilized palmitoylated versions of cytosolic glutathione and hydrogen peroxide sensors (Grx1-roGFP2 and roGFP2-Orp1, respectively) and demonstrated a unique redox environment near biological membranes. In HeLa cells, cytosolic glutathione was practically completely reduced (EGSH/GSSG = − 333 mV) and hydrogen peroxide level was under the detectable range. In contrast, the cytoplasmic milieu near membranes of intracellular vesicles exhibited significant glutathione oxidation (EGSH/GSSG > − 256 mV) and relatively high H2O2 production, which was not observed for the plasma membrane. These vesicles colocalized with internalized EGFR, suggesting that H2O2 production and glutathione oxidation are characteristics of cytoplasmic surfaces of the endocytosed vesicles. The results visually illustrate local redox heterogeneity within the cytosol for the first time.
Highlights
-
•
Palmitoylated versions of redox sensors were constructed.
-
•
The sensors successfully visualized local redox environment near membranes.
-
•
The cytosol is oxidative near endocytosed vesicles containing EGFR.
Graphical abstract
1. Introduction
Many biological processes involve reduction-oxidation (redox) reactions, which inevitably generate free radicals and other reactive oxygen species (ROS). Excess ROS potentially perturbs normal homeostasis through oxidative modifications of lipids, proteins, and nucleic acids and has been linked to human diseases and senescence [1], [2]. To protect the cell from ROS, biological systems utilize thiol-containing biomolecules, glutathione being the most abundant. ROS is catalytically detoxified at the expense of glutathione by glutathione peroxidase. Oxidatively modified protein thiols are reduced by glutaredoxins, which also depend on glutathione. These redox processes are coupled to glutathione oxidation (GSH to GSSG), making GSH/GSSG balance a useful measure of oxidative stress [3]. Oxidative shift of GSH/GSSG balance not only indicates an increase in ROS levels, but also causes modification of protein thiols, i.e., glutathionylation and disulfide formation. Therefore, maintaining the GSH/GSSG balance within a proper range is essential for normal homeostasis.
In physiological conditions, cells are supposed to intrinsically generate a certain level of ROS at least in part from respiratory chain in mitochondria [4], xanthine oxidase in peroxisomes [5], and NADPH oxidases at biological membranes of a cell and various vesicles [6], [7]. Many sensors have been developed to monitor ROS level and GSH/GSSG balance in a living cell and some sensors are targeted to specific subcellular compartments to monitor redox equilibrium in organelles [8], [9], [10], [11], [12]. The recently developed targeted sensors demonstrated that subcellular compartments contain different levels of ROS and maintain different balances of redox pairs [10]. While the lumen of some compartments are characterized by a relatively oxidized balance of glutathione (mitochondria and endoplasmic reticulum), the cytosol was shown to be particularly reduced (EGSH/GSSG is around − 330 mV [13]) at least partly because of the activities of cytosolic glutathione reductases.
While the average redox balance of the cytosolic glutathione pair is maintained at reductive level [10], it is possible that specific parts of the cytosol are locally oxidized, which has been potentially overlooked in previous studies using cytosolic redox sensors. The currently available fluorescent sensors such as roGFP derivatives and dichlorodihydrofluorescein diacetate (DCFDA) freely diffuse within the cytosol and local differences, if any, may not be detected. Based on previously determined diffusion coefficient of cytosolically expressed EGFP (1.3 × 10−11 m2/s [14]), the mean time for EGFP to traverse 1 µm square area is estimated to be in the order of 10−2 s which may not be sufficient for redox reactions in currently available redox sensors; even a rapidly-reacting glutathione sensor Grx1-roGFP2 [10] has t1/2 of ~ 10 s for full oxidation/reduction [10]. Thus, our current knowledge about the cytosolic redox status lacks local information. Considering that ROS is mainly generated within the organelle lumen or membranes [6], [7], the cytoplasmic sides of different organelle membranes and the plasma membrane (PM) may have unique redox states. In this study, we sought to compare the glutathione redox balances between the whole cytosol and the cytosolic surfaces of vesicular membranes (VM) and PM. To this end, we utilized palmitoylation of a roGFP-based glutathione sensor (Grx1-roGFP2 [10]), successfully targeted the sensor to PM and VM, and performed ratiometric analyses of local redox balance of cytosolic glutathione. The results demonstrated marked glutathione oxidation near VM. We further constructed a palmitoylated version of a hydrogen peroxide sensor (roGFP2-Orp1 [15]) and found relatively higher levels of H2O2 production near VM compared to PM and the whole cytosol. These results visually highlight a unique redox environment near biological membranes for the first time.
2. Material and methods
2.1. Construction of palmitoylated redox sensors
Plasmids pEIGW/Grx1-roGFP2 [10] and pEIGW/roGFP2-Orp1 [15] were gifts from Dr. Tobias Dick (Addgene plasmid # 64990 and # 64993, respectively). pPalmitoyl-mTurquoise2 [16] was a gift from Dr. Dorus Gadella (Addgene plasmid # 36209). Palmitoylated redox sensors were constructed from original plasmids pEIGW/Grx1-roGFP2 (Grx1-roGFP2Cyto), pEIGW/roGFP2-Orp1 (roGFP2-Orp1Cyto). Oligonucleotides used in the study are listed in Supplementary Data 1. Procedures of plasmid construction were fully documented in Supplementary Data 2. Briefly, DNA fragment coding for Grx1-roGFP2 was amplified by PCR using DNA primers XmaI_Grx1roGFP2_F and NotI_Grx1roGFP2_R and pEIGW/Grx1-roGFP2 as a template. The resulting fragment was inserted into pPalmitoyl-Turquoise2 plasmid in which Turquoise2 coding region was removed in advance. The resulting plasmid pPalmitoyl-Grx1-roGFP2 (Grx1-roGFP2Palm) was propagated in bacterial host XL10-Gold (Aggilent Technologies) and extracted using Plasmid Midi Kit (QIAGEN). Exactly the same strategy was adopted to construct pPalmitoyl-roGFP2-Orp1 (roGFP2-Orp1Palm) except that PCR was performed using primers XmaI_roGFP2Orp1_F and NotI_roGFP2Orp1_R and pEIGW/roGFP2-Orp1 as a template. The entire coding regions of pPalmitoyl-Grx1-roGFP2 and pPalmitoyl-roGFP2-Orp1 were sequenced for validation. DNA concentration was estimated by absorbance (λ=260 nm) using NanoDrop ND-1000 UV–Vis spectrophotometer.
2.2. Cell culture and transfection
HeLa cell line was kindly provided by Dr. Shigeru Sassa (The Rockefeller University, NY) and maintained in Dulbecco's minimal essential medium (DMEM) supplemented with 10% FBS (v/v), 100 U/ml Penicillin, and 100 µg/ml streptomycin. Cells were cultured on collagen-coated 35 mm Fluoro Dishes (World Precision Instruments, Inc., FD35-100). Plasmids were delivered into cells by transfecting 2 μg of plasmid DNA using Lipofectamine 3000 (Life Technologies). Culture medium was replenished 24 h after transfection and redox assay was performed typically 48 h after transfection. Prior to redox assay, culture medium was changed to 1 ml of roGFP assay medium (phenol-red free DMEM supplemented with 10% (v/v) FBS).
2.3. Confocal microscopy and immunostaining
Subcellular localizations of roGFP derivatives were analyzed using confocal microscope FV1000 (Olympus, Japan) equipped with a temperature- and CO2-controlled stage chamber. Images were processed using Image J [17] with Fiji package [18]. For immunostaining, cells were cultured on collagen-coated cover slips in a 12-well plate, fixed with 3.7% paraformaldehyde (PFA) in PBS for 20 min, permeabilized with 0.2% (w/v) Triton X-100 in PBS for 10 min, and blocked with 1% (w/v) BSA/1% (w/v) gelatin in PBS. Primary and secondary antibodies used in cell staining were listed in Supplementary Data 3. All antibodies were used at a dilution of 1:200. Stained samples were mounted with Fluoromount-G with DAPI (Southern Biotech, AL) and placed on slide glasses.
2.4. Ratiometric measurement of glutathione redox potential and hydrogen peroxide level
The degree of glutathione oxidation and H2O2 production was determined using previously documented method [8], [10]. Live cells expressing roGFP derivatives were analyzed by confocal microscopy as described above. For analyzing immunostained samples and taking z-section images, cells were treated with 10 mM N-ethylmaleimide (NEM) for 10 min at room temperature prior to PFA fixation. Each image was taken for two tracks with distinct excitation (488 nm and 405 nm) and a single emission range 540–560 nm. To avoid photobleaching and unwanted stress response, beam intensity was limited to as low as 0.5% and 0.2% for I405 and I488, respectively, and signals were detected using gallium arsenide phosphide (GaAsP) PMT high sensitivity detector unit (Olympus, Japan). The ratio of intensities for I405 and I488 (RI405/I488) represents the degree of sensor oxidation. For testing redox reactivity of the sensors, time-lapse images were taken during addition of exogenous oxidant and reductant; 0.1 mM H2O2 was added to culture medium for sensor oxidation and monitored for up to 30 min, followed by adding 5 mM DTT for sensor reduction. Percentage of sensor oxidation (OxD) was calculated from I405/I488 value using Hanson's equation. Redox potential was obtained by applying OxD and standard redox potential of glutathione pair (E°roGFP2, − 280 mV [19]) to Nernst equation. Calculations are fully described in Supplementary Data 4.
3. Results
3.1. Membrane targeting of redox sensors via palmitoylation
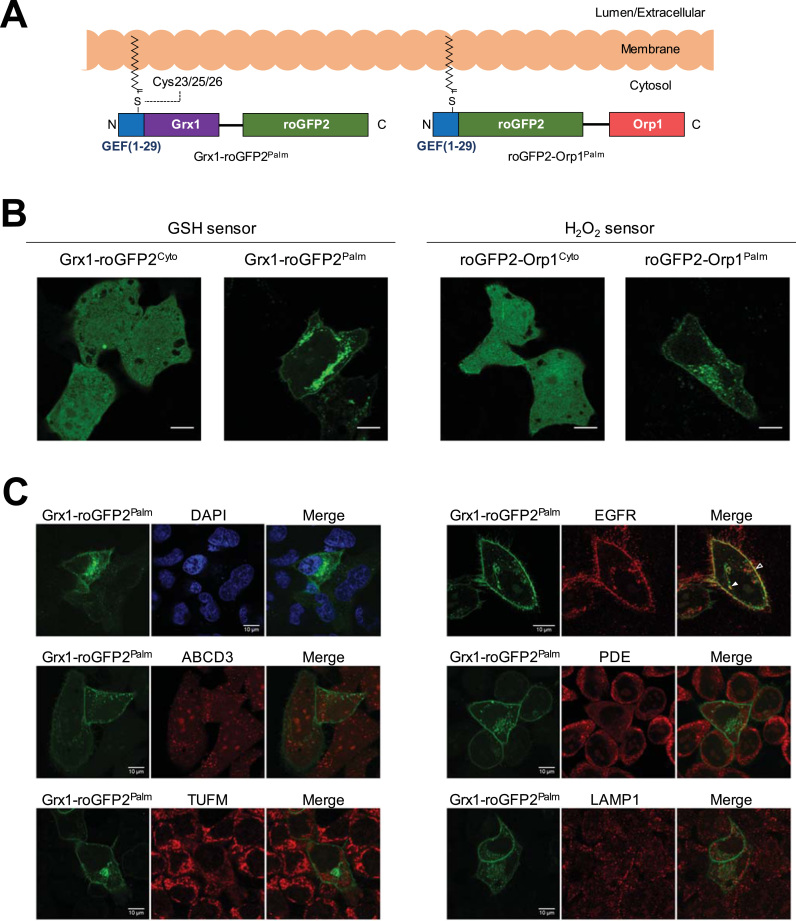
To target the Grx1-roGFP2 (glutathione) and roGFP2-Orp1 (H2O2) sensors to the cytoplasmic side of biological membranes, we fused them with an N-terminal palmitoylation motif, producing Grx1-roGFP2Palm (Fig. 1A) and roGFP2-Orp1Palm. We transfected the resulting plasmids into HeLa cells and observed specific fluorescent signals within membranous structures, including the plasma membrane (PM) and intracellular vesicles (Fig. 1B). The identity of these vesicles was assessed by immunostaining using various organelle markers (Fig. 1C). Grx1-roGFP2Palm colocalized with Epidermal Growth Factor Receptor (EGFR) not only at PM but also at vesicular membranes. EGFR is internalized by endocytosis and eventually sorted to late endosomes mainly located in the perinuclear region. Therefore, Grx1-roGFP2Palm is highly expected to reside on PM and late endosomal membranes. Vesicular patterns of Grx1-roGFP2Palm did not colocalize with neither ABCD3, phosphodiesterase (PDE), nor TUFM, indicating that the sensor was not localized in peroxisomes, endoplasmic reticulum, nor mitochondria. RoGFP2-Orp1Palm showed exactly the same localization pattern as observed for Grx1-roGFP2Palm (data not shown).
Fig. 1.
Membrane targeting of glutathione- and H2O2- specific redox sensors. A, Schematic representation of palmitoylated redox sensors, Grx1-roGFP2Palm and roGFP2-Orp1Palm. Grx1-roGFP2 and roGFP2-Orp1 were fused with the first 29 amino acid residues of human guanine nucleotide exchange factors (GEF) which contains S-palmitoylation motif (Cys23/25/26). Palmitoylated proteins can associate with biological membranes from the cytoplasmic side. B, Grx1-roGFP2Palm and roGFP2-Orp1Palm were transfected into HeLa cells and protein localization was analyzed using confocal microscopy. For comparison, the data for the original cytosolic constructs are shown. Scale bars, 10 µm. C, Localizations of Grx1-roGFP2Palm and roGFP2-Orp1Palm were assessed by immunostaining for organelle markers; EGFR at the plasma membrane and endocytic vesicles, ABCD3 at peroxisome, phosphodiesterase (PDE) at endoplasmic reticulum, TUFM in mitochondria, LAMP1 at lysosome. Scale bars, 10 µm. Co-localizations at the plasma membrane (PM; open arrow heads) and vesicular membranes (VM; filled arrow heads) are indicated.
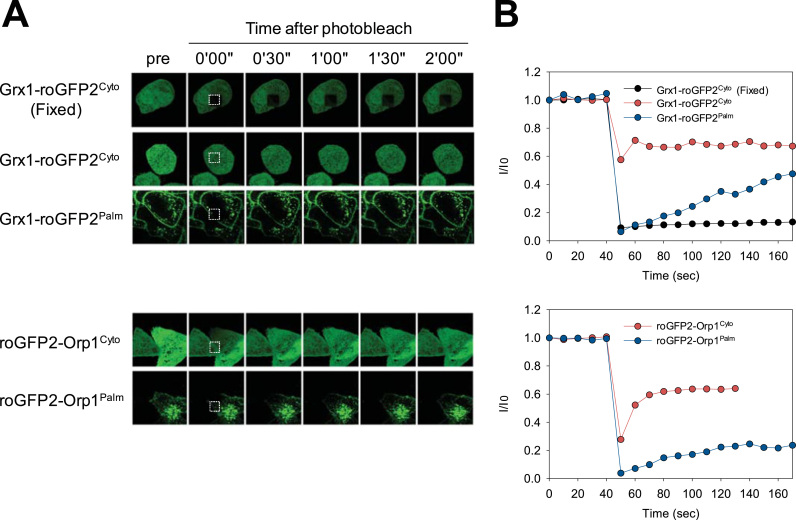
We further performed fluorescence recovery after photobleaching (FRAP) experiments to evaluate the association of the sensors to the biological membranes. Unlike soluble proteins in the cytosol, diffusion of membrane-bound proteins is limited within a lateral plane and exhibits slow FRAP kinetics [20]. As a static control, Grx1-roGFP2Cyto was overexpressed and fixed with paraformaldehyde (PFA) before FRAP assay, resulting in irreversible fluorescence quenching in a focused area (dashed box in Fig. 2A). Without PFA-fixation, photobleaching caused a significant decrease of overall fluorescence of Grx1-roGFP2Cyto within a cell, reflecting an immediate diffusion of fluorophore. In contrast, Grx1-roGFP2Palm was quenched only within a focused area and the fluorescence was slowly recovered with t1/2 of 300 s (Fig. 2B). Similarly, roGFP2-Orp1Cyto exhibited immediate diffusion, while roGFP2-Orp1Palm was characterized by slower recovery with t1/2 of 350 s. Thus, both Grx1-roGFP2Palm and roGFP2-Orp1Palm diffused in a much slower rate compared to their cytosolic forms. In conjunction with their localization patterns (Fig. 1B and C), we concluded that the majority of Grx1-roGFP2Palm and roGFP2-Orp1Palm associated with biological membranes.
Fig. 2.
Membrane association of Grx1-roGFP2Palm and roGFP2-Orp1Palm assessed by fluorescence recovery after photobleach (FRAP) experiments. A, Confocal images of HeLa cells expressing fluorescent redox sensors. Bleaching laser was shot on 10 µm × 10 µm area (dashed square) and images were taken after indicated time periods. As non-diffusible controls, cells were fixed with 3.7% paraformaldehyde prior to FRAP (Grx1-roGFP2 Fixed). B, FRAP kinetics for cells shown in A.
3.2. Palmitoylation does not affect the redox response of glutathione and H2O2 sensors
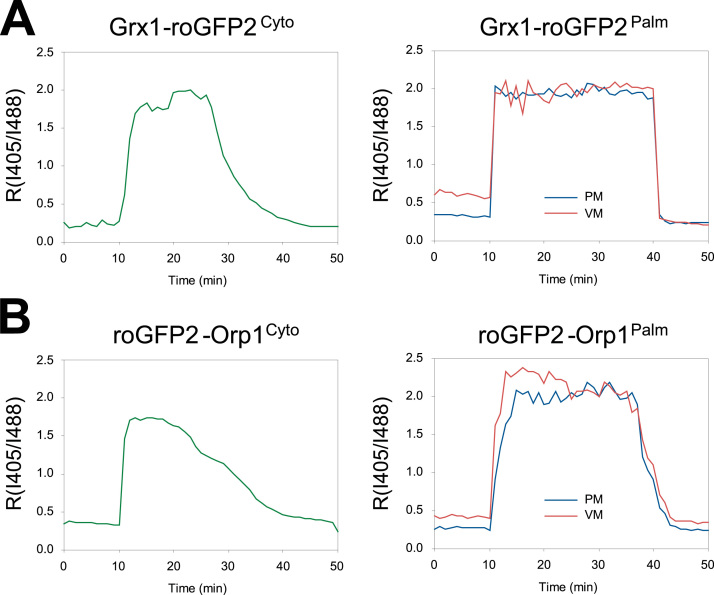
Introduction of an exogenous sequence into redox sensors can potentially disrupt redox reactivity or protein integrity. Therefore, we tested the reactivity of palmitoylated sensors. Glutathione oxidation was induced by adding H2O2 to the culture medium to a final concentration of 100 µM as previously described [10]. We transiently expressed Grx1-roGFP2Cyto and Grx1-roGFP2Palm in HeLa cells and treated the transfected cells with 100 µM H2O2 and 5 mM DTT to induce oxidation and reduction of the sensors. Grx1-roGFP2Cyto exhibited an immediate increase of RI405/I488, indicating H2O2-mediated oxidation of cytosolic glutathione and a rapid response of the sensor (Fig. 3A). As originally documented by Gutscher et al. [10], the RI405/I488 value gradually returned to the basal level, suggesting anti-oxidative processes within the cytosol. Interestingly, we observed prolonged response for Grx1-roGFP2Palm, which was sustained for at least 30 min after addition of 100 µM H2O2 (Fig. 3A, right). Signals from the PM and vesicles were separately analyzed and both signals produced a similar response against H2O2 and DTT. The maximal and minimal RI405/I488 values were comparable between Grx1-roGFP2Cyto and Grx1-roGFP2Palm, suggesting that the same parameters can be applied for estimating redox potentials. Similarly, roGFP2-Orp1Palm exhibited longer response compared to the original roGFP2-Orp1cyto construct, and maximal and minimal I405/I488 values were not changed by palmitoylation (Fig. 3B). These results confirmed intact redox reactivity of palmitoylated sensors and point to a unique redox equilibrium near biological membranes.
Fig. 3.
Palmitoylation did not affect reactivity of Grx1-roGFP2 and roGFP2-Orp1. Grx1-roGFP2Palm and roGFP2-Orp1Palm were transfected into HeLa cells. Redox reactivity was tested by sequential treatment with 100 µM H2O2 (oxidant, Time = 10 min) and 5 mM DTT (reductant, Time = 40 min in A and 35 min in B) during live cell imaging. I405 and I488 Images were taken by confocal microscopy and the ratio RI405/I488 was obtained for each cell. Time-dependent changes of RI405/I488 values for Grx1-roGFP2Palm (A) and roGFP2-Orp1Palm (B) are shown. PM, plasma membrane. VM, vesicular membrane.
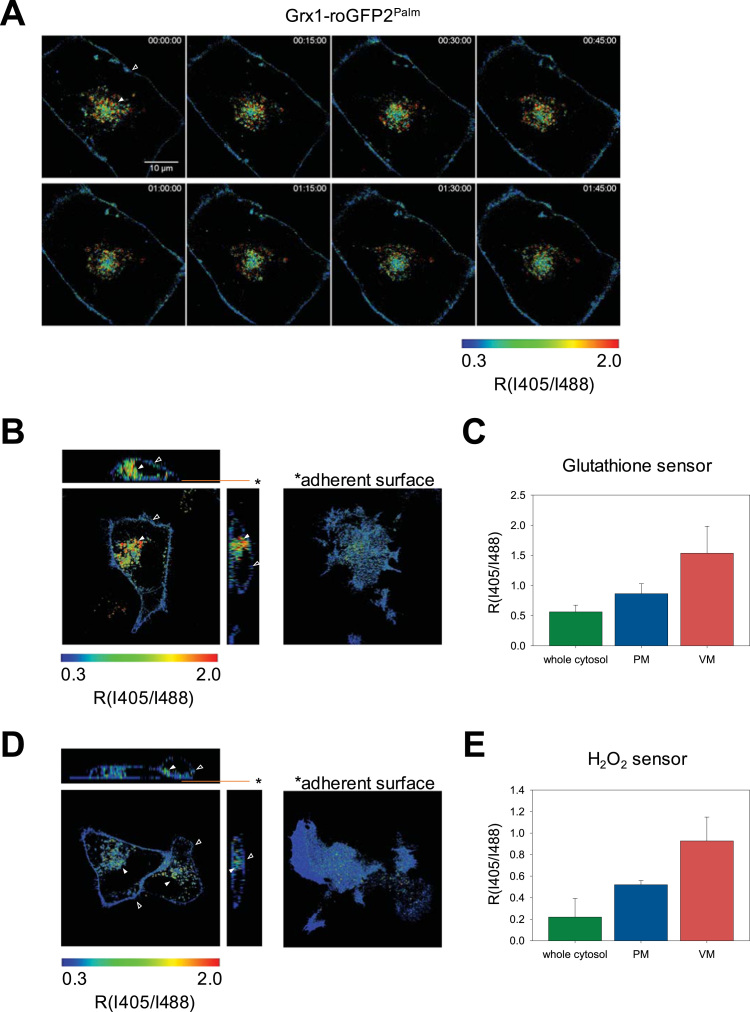
3.3. Constitutively high levels of H2O2 production and glutathione oxidation near vesicular membranes
The advantage of fluorophore-based redox sensors is the capability of redox imaging in living cells. As shown in Fig. 4A, using Grx1-roGFP2Palm, we visualized a unique spatial pattern of glutathione oxidation, which was far from being homogenously distributed (Time-lapse movie is available as Supplementary Data 5). Steady-state redox status of HeLa cells was monitored by time lapse imaging over 90 min, demonstrating that the analysis can be stably performed without apparent photoquenching or optical damage and that vesicular membrane (filled arrow head in Fig. 4A) was constitutively more oxidized than PM (open arrow head in Fig. 4A). Three-dimensional distribution of the sensor illustrates that perinuclear vesicles are specifically oxidized (filled arrow head in Fig. 4B). The average RI405/I488 values were 0.5 for the whole cytosol (determined by Grx1-roGFP2Cyto), 0.9 for PM, and 1.5 for vesicular membranes (Fig. 4C). These values correspond to redox potentials of − 333 mV, − 275 mV, and − 256 mV, respectively. Similarly, roGFP2-Orp1Palm was specifically oxidized on vesicular membranes (filled arrow head in Fig. 4D) while no oxidation was observed on PM (open arrow head in Fig. 4D), pointing to higher H2O2 level around perinuclear vesicles compared to PM (Fig. 4E).
Fig. 4.
Redox imaging demonstrated that the cytoplasmic surfaces the plasma membrane and vesicular membrane are characterized by different levels of glutathione oxidation and H2O2 production. A, Grx1-roGFP2Palm was transfected into HeLa cells and I405/I488 time-lapse imaging was performed by live-cell confocal microscopy. R value (I405/I488) represents the degree of sensor oxidation and shown in a false-color scale. Time is indicated in each image (h:m:s). Scale bar, 10 µm. Signals from the plasma membrane (PM; open arrow heads) and vesicular membranes (VM; filled arrow heads) are indicated. Time-lapse movie is available as Supplementary Data 5. B and D, Grx1-roGFP2Palm and roGFP2-Orp1Palm were transfected into HeLa cells and steady-state I405/I488 images were taken (z-sections). X-Y image in the right corresponds to the focal plane indicated by *. Signals from the plasma membrane (PM; open arrow heads) and vesicular membranes (VM; filled arrow heads) are indicated. C and E, Average RI405/I488 values from multiple cells (n = 5). PM, plasma membrane. VM, vesicular membrane.
Supplementary material related to this article can be found online at doi:10.1016/j.redox.2017.11.015.
The following is the Supplementary material related to this article Movie 1.
Time-lapse movie showing constitutively oxidized glutathione pair near the cytosolic vesicles. Grx1-roGFP2Palm was transfected into HeLa cells and I405/I488 time-lapse imaging was performed by live-cell confocal microscopy. R value (I405/I488) represents the degree of sensor oxidation and shown in a false-color scale. Representative images are shown in the main Fig. 4A. Time is indicated in each image (h:m:s).
4. Discussion
GFP-based redox sensors allow real-time analyses of various redox pairs in live cells. In this study, we utilized Grx1-roGFP2 [10] and roGFP2-Orp1 [15], designed to specifically report the redox status of glutathione and production of H2O2, respectively. Organelle-targeted versions of redox sensors have been previously constructed to assess glutathione oxidation in the mitochondria [10], [12], [21], nucleus [11], ER [9], and cytosol [10], [11], indicating that subcellular compartments have distinct redox equilibria and that the cytosolic glutathione is particularly reduced. Considering the presence of membrane-bound NADPH-oxidases [6], [7] and possible permeation of oxidants from the lumen [4], [22], the redox status of glutathione may not be completely homogenous within the cytosol. In this study, we tested this hypothesis and focused on the redox environment near the cytoplasmic side of the membrane surface. By attaching a palmitoylation motif, Grx1-roGFP2 [10] and roGFP2-Orp1 [15] were successfully targeted to biological membranes, including the PM and intracellular vesicles. The sensors sustained intact redox reactivity, allowing analyses of glutathione oxidation and H2O2 production near the membranes. The results demonstrated relatively high levels of glutathione oxidation and H2O2 production near vesicular membranes, which were clearly different from the cytosolic side of the PM or the rest of the cytosol (Fig. 4E). Based on a similarly high I405/I488 value for roGFP2-Orp1Palm at intracellular vesicles, glutathione oxidation around the vesicles is expected to be in part due to H2O2 production.
To target the redox sensors to biological membranes, we utilized a palmitoylation domain from Rho guanine nucleotide exchange factor (RhoGEF). Goedhart et al. successfully targeted the fluorescent protein mTurquoise2 to the PM by attaching the same sequence [16]. We swapped the fluorophore moiety in this original construct with either Grx1-roGFP2 or roGFP2-Orp1 to generate the palmitoylated redox sensors. The resulting proteins were efficiently targeted to the PM (Fig. 1B), but, seemingly, their localization to intracellular membranes was more prominent than that of the original palmitoyl-mTurquoise2 [16]. Protein aggregation was unlikely based on intact fluorescence, redox reactivity, and complete solubility after treatment with detergents (data not shown). Steady-state localization of palmitoylated proteins is not limited to the PM. Fyn is present in RhoD-positive endosomes [23] whereas p21rhoB is mainly targeted to early endosomes and a pre-lysosomal compartment [24]. Based on such endocytic-lysosomal pathway as well as colocalization with EGFR (Fig. 1C), vesicular signals of Grx1-roGFP2Palm and roGFP2-Orp1Palm is expected to reflect late endosomes.
Palmitoylation is an effective approach to target redox sensors to membranes of different intracellular structures, allowing direct comparison between distinct membranes. To analyze specific organelle membrane, the sensors need to be targeted using more specific approaches. The simplest and most reliable strategy for membrane targeting may be to attach the sensor to the cytoplasmic tail of a membrane protein destined to a specific organelle. To visualize intracellular structures in live cells, various fluorescent membrane proteins have been designed [16], [25], [26], [27], [28]. By replacing fluorophores in these organelle-targeted proteins with redox sensors, it will be possible to monitor local redox environment around specific intracellular structures.
5. Conclusions
We constructed palmitoylated versions of redox sensors that allow the analysis of glutathione oxidation and H2O2 production near the cytoplasmic sides of biological membranes. The results demonstrated clear differences from the rest of the cytosol; the cytosolic milieu was markedly oxidized near vesicular membranes. The degree of glutathione oxidation (EGSH/GSSG > − 256 mV) is in principle sufficiently high to induce thiol oxidation in some proteins, implicating the impact of the local redox environment within the cytosol. Such local redox environment is significantly different from the overall reduced state of the cytosol and may underlie pathologies related to redox perturbation.
Acknowledgements
The authors thank Dr. Tobias Dick (German Cancer Research Center, Heidelberg, Germany) for providing the Grx1-roGFP2 and roGFP2-Orp1 constructs, Dr. Dorus Gadella (University of Amsterdam, Amsterdam, Netherlands) for pPalmitoyl-mTurquoise2 construct, and Dr. Takeo Kitazawa (Yasuda Women's University, Hiroshima, Japan) for helpful discussions. Authors wish to thank Yasuda Women's University for grant support (to H.Y., I.S., A.R.), The Science Research Promotion Fund (to H.Y.), and JSPS KAKENHI Grant no. 17K18289 (to H.Y.)
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.11.015
Appendix A. Supplementary material
Supplementary material
References
- 1.Finkel T., Holbrook N.J. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 2.Nathan C., Ding A. Nonresolving inflammation. Cell. 2010;140:871–882. doi: 10.1016/j.cell.2010.02.029. [DOI] [PubMed] [Google Scholar]
- 3.Schafer F.Q., Buettner G.R. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 4.Chauvigné F., Boj M., Finn R.N., Cerdà J. Mitochondrial aquaporin-8-mediated hydrogen peroxide transport is essential for teleost spermatozoon motility. Sci. Rep. 2015;5:7789. doi: 10.1038/srep07789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonekamp N.A., Völkl A., Fahimi H.D., Schrader M. Reactive oxygen species and peroxisomes: struggling for balance. BioFactors. 2009;35:346–355. doi: 10.1002/biof.48. [DOI] [PubMed] [Google Scholar]
- 6.Brown D.I., Griendling K.K. Nox proteins in signal transduction. Free Radic. Biol. Med. 2009;47:1239–1253. doi: 10.1016/j.freeradbiomed.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupre-Crochet S., Erard M., Nusse O. ROS production in phagocytes: why, when, and where? J. Leukoc. Biol. 2013;94:657–670. doi: 10.1189/jlb.1012544. [DOI] [PubMed] [Google Scholar]
- 8.Morgan B., Sobotta M.C., Dick T.P., Measuring E(GSH) and H2O2 with roGFP2-based redox probes. Free Radic. Biol. Med. 2011;51:1943–1951. doi: 10.1016/j.freeradbiomed.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 9.Montero D., Tachibana C., Rahr Winther J., Appenzeller-Herzog C. Intracellular glutathione pools are heterogeneously concentrated. Redox Biol. 2013;1:508–513. doi: 10.1016/j.redox.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutscher M., Pauleau A.-L., Marty L., Brach T., Wabnitz G.H., Samstag Y., Meyer A.J., Dick T.P. Real-time imaging of the intracellular glutathione redox potential. Nat. Methods. 2008;5:553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 11.Dardalhon M., Kumar C., Iraqui I., Vernis L., Kienda G., Banach-Latapy A., He T., Chanet R., Faye G., Outten C.E., Huang M.-E. Redox-sensitive YFP sensors monitor dynamic nuclear and cytosolic glutathione redox changes. Free Radic. Biol. Med. 2012;52:2254–2265. doi: 10.1016/j.freeradbiomed.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhattacharjee A., Yang H., Duffy M., Robinson E., Conrad-Antoville A., Lu Y.-W., Capps T., Braiterman L., Wolfgang M., Murphy M.P., Yi L., Kaler S.G., Lutsenko S., Ralle M. The activity of Menkes disease protein ATP7A is essential for redox balance in mitochondria. J. Biol. Chem. 2016;291:16644–16658. doi: 10.1074/jbc.M116.727248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Go Y.-M., Jones D.P. Redox compartmentalization in eukaryotic cells. Biochim. Biophys. Acta. 2008;1780:1273–1290. doi: 10.1016/j.bbagen.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wachsmuth M., Waldeck W., Langowski J. Anomalous diffusion of fluorescent probes inside living cell nuclei investigated by spatially-resolved fluorescence correlation spectroscopy. J. Mol. Biol. 2000;298:677–689. doi: 10.1006/jmbi.2000.3692. [DOI] [PubMed] [Google Scholar]
- 15.Gutscher M., Sobotta M.C., Wabnitz G.H., Ballikaya S., Meyer A.J., Samstag Y., Dick T.P. Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J. Biol. Chem. 2009;284:31532–31540. doi: 10.1074/jbc.M109.059246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goedhart J., von Stetten D., Noirclerc-Savoye M., Lelimousin M., Joosen L., Hink M.A., van Weeren L., Gadella T.W.J., Royant A., Royant A. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93% Nat. Commun. 2012;3:751. doi: 10.1038/ncomms1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schindelin J., Rueden C.T., Hiner M.C., Eliceiri K.W. The ImageJ ecosystem: an open platform for biomedical image analysis. Mol. Reprod. Dev. 2015;82:518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dooley C.T., Dore T.M., Hanson G.T., Jackson W.C., Remington S.J., Tsien R.Y. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J. Biol. Chem. 2004;279:22284–22293. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- 20.Day C.A., Kraft L.J., Kang M., Kenworthy A.K. Analysis of protein and lipid dynamics using confocal fluorescence recovery after photobleaching (FRAP) Curr. Protoc. Cytom. 2012 doi: 10.1002/0471142956.cy0219s62. (Chapter 2, Unit 2.19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu J., Dong L., Outten C.E. The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix. J. Biol. Chem. 2008;283:29126–29134. doi: 10.1074/jbc.M803028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hara-Chikuma M., Satooka H., Watanabe S., Honda T., Miyachi Y., Watanabe T., Verkman A.S. Aquaporin-3-mediated hydrogen peroxide transport is required for NF-κB signalling in keratinocytes and development of psoriasis. Nat. Commun. 2015;6:7454. doi: 10.1038/ncomms8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandilands E., Brunton V.G., Frame M.C. The membrane targeting and spatial activation of Src, Yes and Fyn is influenced by palmitoylation and distinct RhoB/RhoD endosome requirements. J. Cell Sci. 2007;120 doi: 10.1242/jcs.003657. [DOI] [PubMed] [Google Scholar]
- 24.Adamson P., Paterson H.F., Hall A. Intracellular localization of the P21rho proteins. J. Cell Biol. 1992;119 doi: 10.1083/jcb.119.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez I., Chakrabarti S., Hellevik T., Morehead J., Fowler K., Andrews N.W. Synaptotagmin VII regulates Ca(2+)-dependent exocytosis of lysosomes in fibroblasts. J. Cell Biol. 2000;148:1141–1149. doi: 10.1083/jcb.148.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma D.K., Choudhury A., Singh R.D., Wheatley C.L., Marks D.L., Pagano R.E. Glycosphingolipids internalized via caveolar-related endocytosis rapidly merge with the clathrin pathway in early endosomes and form microdomains for recycling. J. Biol. Chem. 2003;278:7564–7572. doi: 10.1074/jbc.M210457200. [DOI] [PubMed] [Google Scholar]
- 27.Zurek N., Sparks L., Voeltz G. Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic. 2011;12:28–41. doi: 10.1111/j.1600-0854.2010.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., Voeltz G.K., Tubules Mark E.R. Sites of mitochondrial division. Science. 2011;80(334):358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time-lapse movie showing constitutively oxidized glutathione pair near the cytosolic vesicles. Grx1-roGFP2Palm was transfected into HeLa cells and I405/I488 time-lapse imaging was performed by live-cell confocal microscopy. R value (I405/I488) represents the degree of sensor oxidation and shown in a false-color scale. Representative images are shown in the main Fig. 4A. Time is indicated in each image (h:m:s).
Supplementary material