Abstract
Ubiquinol cytochrome c reductase binding protein (UQCRB) is important for mitochondrial complex III stability, electron transport, cellular oxygen sensing and angiogenesis. However, its potential as a prognostic marker in colorectal cancer (CRC) remains unclear. The aim of this study was to determine whether UQCRB can be used as a diagnostic molecular marker for CRC. The correlation between the expression of three genes (UQCRB, UQCRFS1 and MT-CYB) in the mitochondrial respiratory chain complex III and clinico-pathological features was determined. Compared to non-tumor tissues, UQCRB gene expression was upregulated in CRC tissues. Gene and protein expression of the genes were positively correlated. Copy number variation (CNV) differences in UQCRB were observed in CRC tissues (1.32-fold) compared to non-tumor tissues. The CNV of UQCRB in CRC tissues increased proportionally with gene expression and clinical stage. Single-nucleotide polymorphisms in the 3′-untranslated region of UQCRB (rs7836698 and rs10504961) were investigated, and the rs7836698 polymorphism was associated with CRC clinical stage. DNA methylation of the UQCRB promoter revealed that most CRC patients had high methylation levels (12/15 patients) in CRC tissues compared to non-tumor tissues. UQCRB overexpression and CNV gain were correlated with specific CRC clinico-pathological features, indicating clinical significance as a prognostic predictor in CRC. Gene structural factors may be more important than gene transcription repression factors with respect to DNA methylation in UQCRB overexpression. Our results provide novel insights into the critical role of UQCRB in regulating CRC, supporting UQCRB as a new candidate for the development of diagnostics for CRC patients.
Introduction
Colorectal cancer (CRC) is a common malignancy worldwide and the leading cause of cancer-related death.1 Moreover, CRC is the fourth leading cause of cancer-related deaths in South Korea. Although the prevalence of CRC is lower in South Korea than in Western industrialized nations, the number of patients with CRC has increased rapidly over the past several decades.2, 3, 4 Despite recent advances in the early diagnosis and treatment of CRC, the overall prognosis for CRC patients remains poor. Thus, novel and effective targets must be identified to improve therapeutic efficacy and clinical outcomes.
The mitochondrion is the main energy-producing organelle of the cell, bearing five oxidative phosphorylation complexes that regulate ATP synthesis.5, 6 Of the five oxidative phosphorylation complexes, complex III carries out electron transport, ubisemiquinone radical stabilization and cellular oxygen sensing.7 Mitochondrial complex III (cytochrome bc1 complex) transfers electrons from ubiquinol to cytochrome c and contributes to the generation of an electrochemical proton gradient.8, 9, 10 Notably, mitochondrial complex III deficiency is associated with lactic acidosis, hypoglycemia, encephalopathy, among other disorders in humans.11
The UQCRB, UQCRFS1 (encoding Rieske iron–sulfur protein) and MT-CYB (encodingcytochrome b) genes are components of complex III. Ubiquinol cytochrome c reductase binding (UQCRB) is the homolog of the yeast Qcr7 subunit, which is located in the matrix-inner mitochondrial membrane interphase and is thought to interact with cytochrome b in the early stages of the assembly pathway.12 Qcr7 incorporation is essential for the stabilization of hemylated cytochrome b.13 UQCRB plays an important role in MT-CYB stability, explaining the low amounts of b-type cytochrome found in the mitochondria of CRC patients.
UQCRB is a functional gene with nuclear encoding of a 13.3-kDa subunit of mitochondrial complex III; UQCRB is located at the human chromosome location 8q22 and participates in stabilization of the ubisemiquinone radical through hydrophobic interactions with ubiquinone.14, 15 In addition, UQCRB is found in the respiratory chains of all aerobic organisms, as well as in the electron transfer systems of chloroplasts and photosynthetic bacteria.16, 17 Deletion of the gene encoding UQCRB causes a defect in complex III function, resulting in hypoglycemia and lactic acidosis. Disorders of complex III are comparatively rare, but they present as a clinically heterogeneous group of diseases.18
We previously demonstrated that loss of function of UQCRB by either genetic and pharmacological means inhibited angiogenesis, indicating that UQCRB plays a key role in this process and is a prognostic marker of angiogenesis and mitochondria-related diseases.19, 20 However, few studies have examined its clinical role in CRC. Thus, we investigated the clinical roles of UQCRB in CRC using genetic/proteomic analysis of human CRC tissues.
Materials and methods
Study participant data and tissue samples
This study was conducted on a total of 15 human CRC tissues and matched adjacent non-tumor tissues, which were histopathologically and clinically diagnosed at the Severance Hospital, Seoul, Korea, from November 2012 to December 2012. Disease-free tissue samples served as control groups. Tissues were flash-frozen in liquid nitrogen immediately after resection. Colorectal tissues were classified according to the World Health Organization tumor classification system. We obtained Institutional Review Board approval for the use of the CRC tissues from the Yonsei University Institutional Review Board (IRB No. 1040917-201408-BR-211-02E). The demographic and pathologic characteristics of the CRC patients are summarized in Table 1.
Table 1. Clinical characteristics of 15 CRC patients at the time of diagnosis.
| Parameter | Number of patients | % of patients | Range |
|---|---|---|---|
| Gender | |||
| Male | 10 | 66.7 | |
| Female | 5 | 33.3 | |
| Age at diagnosis (year) | |||
| <65 | 9 | 60.0 | 39–64 |
| ⩾65 | 6 | 40.0 | 65–79 |
| Localization | |||
| Rectum | 5 | 33.3 | |
| Colon | 9 | 60.0 | |
| Cecum | 1 | 6.7 | |
| Tumor size (cm) | |||
| <3 | 1 | 6.7 | 2.7–3 |
| 3–5 | 9 | 60.0 | 3–5 |
| >5 | 5 | 33.3 | 5–8.45 |
| Clinical (TNM) stage | |||
| I | 0 | 0 | |
| II | 6 | 40.0 | |
| III | 8 | 53.3 | |
| IV | 1 | 6.7 | |
Abbreviations: CRC, colorectal cancer; TNM, tumor-node metastasis.
Analysis of gene expression (quantitative real-time RT-PCR)
Total RNA from 15 pairs of CRC tissues and non-tumor tissues was extracted using aRNeasy Mini Kit from Qiagen (Hilden, Germany) according to the manufacturer’s instructions. RNA integrity was spectrophotometrically confirmed by electrophoresis on a 2% agarose gel. First-strand cDNA was synthesized from 0.5 μg of DNase-treated total RNA using the SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA) with random hexamers.
Real-time PCR was carried out using an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The RNA expression levels of UQCRB, UQCRFS1 and MT-CYB in CRC tissues with respect to their normal adjacent tissues were investigated by TaqMan Gene Expression Assay with a commercial primer/probe product (Applied Biosystems). The TaqMan Gene Expression Assay primers used for UQCRB, UQCRFS1 and MTCYB were Hs01890823_s1, Hs00705563_s1 and Hs02596867_s1, respectively. Human GAPDH was used as an internal control. PCR was carried out using 10 μl of 2 × TaqMan Universal PCR Master Mix, 1 μl of TaqMan Gene Expression Assay, 8 μl of RNase-free water and 1 μl of cDNA template. All reactions were conducted in duplicate along with no-template controls. The thermal cycling conditions comprised an initial step at 95 °C for 10 min, followed by 40 cycles at 95 °C for 10 s and 60 °C for 40 s. Relative expression was calculated using the 2−ΔΔCt method.21
Protein expression
Immunohistochemistry was performed to investigate alterations in protein expression in CRC tissues. The tissue sections, which were 4-μm thick from paraffin-embedded blocks, were mounted on coated slides and placed in drying racks. Subsequently, these samples were deparaffinized and rehydrated using xylene andgraded alcohol washes. The sections were heated in 10 mM citrate buffer (pH 6.0) in a pressure boiler for 10 min to achieve antigen recovery. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide. The primary antibody was rabbit polyclonal UQCRB antibody(1:200, HPA043060; Sigma-Aldrich, St Louis, MO, USA), which was applied overnight at 4 °C. After washing, tissue sections were treated with anti-rabbit secondary antibody (Sigma-Aldrich) for 30 min, followed by further incubation using the horseradish peroxidase complex method following the manufacturer’s instructions. The sections were developed with 3,3′-diaminobenzidine as the chromogen (DAKO, Carpenteria, CA, USA), and the microscopic images were captured digitally. As negative controls, we replaced the primary antibody with 5% fetal bovine serum.
The immunohistochemistry results were semiquantitatively examined by two independent observers who were blinded to the patient data. Staining intensity and the percentage of the stained cells were used as criteria for evaluation. A measure of scoring according to staining intensity (0, negative; I, weak; II, moderate; III, strong) and the percentage of stained cells (0, none; 1, <10% 2, 10–50% 3, >50%) was carried out to divide the samples into two groups: a high-expression group (moderate/strong, stain rated 2 and 3) and B low-expression group (negative/weak, stain rated 0 and 1).22
Copy number variation
Total genomic DNA from 15 pairs of CRC tissues and non-tumor tissues was extracted using aQIAamp DNA Mini Kit from Qiagen according to the manufacturer’s instructions. The relative changes in UQCRB and BRUNOL4 (CELF4)23 copy numbers between CRC tissues and non-tumor tissues were determined by real-time PCR and the TaqMan Copy Number Assay (Applied Biosystems). The TaqMan assay primers used for UQCRB and BRUNOL4 were Hs02791612_cn and Hs06439304_cn, respectively. PCR was performed in a total volume of 25 μl in each well, which contained 12.5 μl of TaqMan Universal MasterMix (Applied Biosystems), 1 μl of each primer (12.5 pM) and 25 ng of genomic DNA. The thermal cycling conditions included an initial step at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 60 s.
For all PCR assays, cycle threshold (Ct) numbers were established using 7500 Software v2.0.6 (Applied Biosystems), and the copy numbers, which were normalized against a reference gene (RNase P, two copies in a diploid genome) and the calibrator (normal sample), were determined using the 2−ΔΔCt method. A two-fold increase or decrease in the copy number of UQCRB and BRUNOL4 in CRC tissues compared to the corresponding normal sample within the pair was considered an amplification or a deletion, respectively. Copy number variations (CNVs) were calculated using Applied Biosystems CopyCaller software (Applied Biosystems).
Single-nucleotide polymorphism
Single-nucleotide polymorphisms (SNPs) were verified by high-throughput bi-directional Sanger DNA sequencing using the 3130 × l genetic analyzer (Applied Biosystems) for high-quality resolution of polymorphisms, insertions and deletions in the coding sequence (333 base pairs, NG_008237), marker rs7836698 and marker rs10504961.24 Major/minor alleles, the location and minor allele frequency of two SNPs (rs7836698 and rs10504961) were C/T, the 5′-untranslated region (UTR), and T-0.42 and T-0.50, respectively.
DNA methylation
Genomic DNA isolated from CRC tissues and non-tumor tissues was analyzed for DNA methylation in the promoter region of the human UQCRB gene using the EpiTect Methyl II PCR Array System according to the supplier’s instructions (Qiagen). Genomic DNA (250 ng) from tissues was digested using aEpitTect methyl II DNA restriction Kit (Qiagen) according to the manufacturer’s instructions. The UQCRB gene EpiTect methyl II Custom PCR array (Qiagen) was applied. Real-time PCR was performed using RT2 SYBR Green ROX qPCR Mastermix (Qiagen) according to the manufacturer’s recommendations in the ABI 7500 Fast Real-Time PCR System (Applied Biosystems). For this analysis, the relative concentrations of differentially methylated DNA (specifically, hypermethylated, unmethylated and intermediary methylated DNA) were determined by comparing the amount of each digest with that of a mock digest. For each sample, data were expressed as the sum of the percent of hypermediary and intermediary methylated DNA. Ct values were used to calculate the percentages of methylated and unmethylated DNA according to the recommendations of the manufacturer (Qiagen).
Statistical analysis
UQCRB expression and CNV profiles in the different groups are shown as the mean and standard error of the mean. Statistical calculations were conducted using the GraphPad Prism 5 (GraphPad, La Jolla, CA, USA) and MedCalc software packages (https://www.medcalc.org/).25 To compute P-values, Tukey’s multiple comparison test and one-way analysis of variance tests were used. A P-value of <0.05 was considered statistically significant. Each experiment was repeated at least twice.
Results
UQCRB gene expression
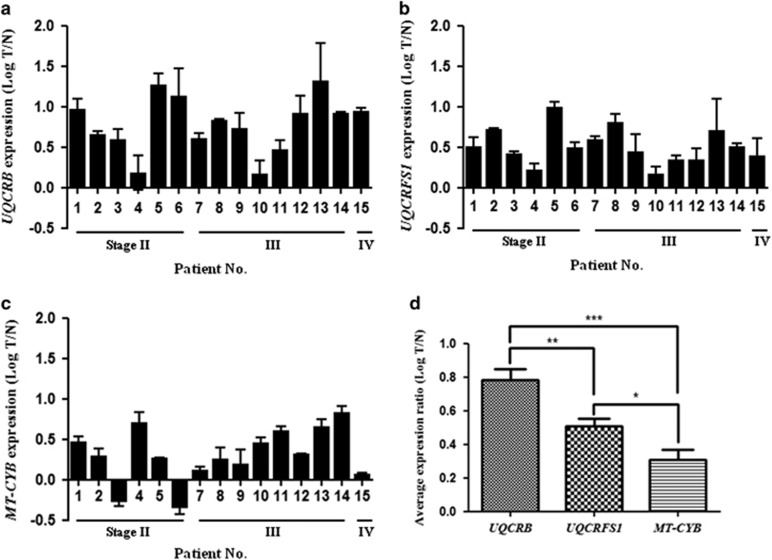
The UQCRB, UQCRFS1 and MT-CYB genes are important components in mitochondrial complex III.12, 13 Real-time PCR analysis showed that UQCRB, UQCRFS1 and MT-CYB mRNA expression levels were relatively higher in CRC tissues (T) than in the matched adjacent non-tumor tissues (N) (Figure 1a–c). Expression levels were normalized to GAPDH. UQCRB and UQCRFS1 mRNA expression levels were significantly upregulated in all 15 pairs (100%, 15/15 patients) of CRC tissues compared to non-tumor tissues (Figure 1a and b). UQCRB showed the highest mRNA expression level among the three target genes. The CRC tissues/non-tumor tissues (T/N) ratio of UQCRB mRNA expression was >1.3-fold in all samples, and the highest ratio was 20.61-fold (Figure 1a). By contrast, MT-CYB mRNA expression was upregulated in 13 pairs (86.6%, 13/15 patients) of CRC tissues compared to non-tumor tissues and was downregulated only in two pairs (13.3%, 2/15 patients) of CRC patients (Figure 1c). However, no significant difference was observed in the expression of other genes (UQCRB and UQCRFS1). UQCRB gene expression showed the highest expression (average log T/N, 0.78) of complex III-related three genes. On average, the three complex III-related genes (UQCRB, UQCRFS1 and MT-CYB) were confirmed to show high gene expression. The average log T/N expression of UQCRFS1 and MT-CYB were 0.51 and 0.31, respectively (Figure 1d).
Figure 1.
Logarithm of the ratio of colorectal cancer (CRC) tissues (T)/matched adjacent non-tumor tissues (N) for RNA levels of UQCRB (a), UQCRFS1 (b) and MT-CYB (c). (d) Expression level of complex III genes in CRC. Gene expression levels were normalized to that of GAPDH. Relative expression was calculated using the 2−ΔΔCt method. Data are presented as the mean±s.e.m. P-values are represented as *P<0.05, **P<0.01 and ***P<0.001.
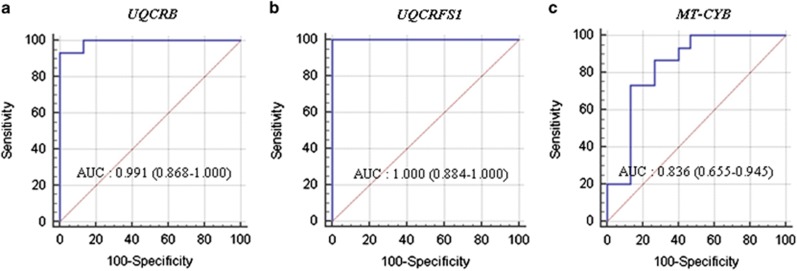
We used the area under the receiver operative characteristic curve (AUC) to evaluate the predictive efficacy of RNA expression. To evaluate the prediction power of the three genes in complex III, we compared the AUC of models developed with UQCRB and other genes. The AUC of UQCRB expression was 0.991 (95% CI, 0.868–1.000; P<0.0001). The Youden index, sensitivity and specificity of UQCRB expression were 0.933%, 93.3% and 100.0%, respectively. The AUC values of UQCRFS1 and MT-CYB were 1.000 (95% CI, 0.884–1.000; P<0.0001) and 0.836 (95% CI, 0.655–0.945; P<0.0001), respectively. The Youden index, sensitivity and specificity of UQCRFS1 expression were 1.000%, 100.0% and 100.0%, respectively. However, the Youden index, sensitivity and specificity for MT-CYB expression were 0.600%, 86.7% and 73.3%, respectively (Figure 2). From here forward, we decided to focus on UQCRB in the following study because UQCRB was significantly overexpressed among the complex III-related three genes (UQCRB, UQCRFS1 and MT-CYB).
Figure 2.
Analysis of ROC curve of UQCRB (a), UQCRFS1 (b) and MT-CYB (c) expression. The AUC of individual candidate genes was calculated to diagnose colorectal cancer (CRC). AUC was calculated using the MedCalc analysis program.
UQCRB protein expression
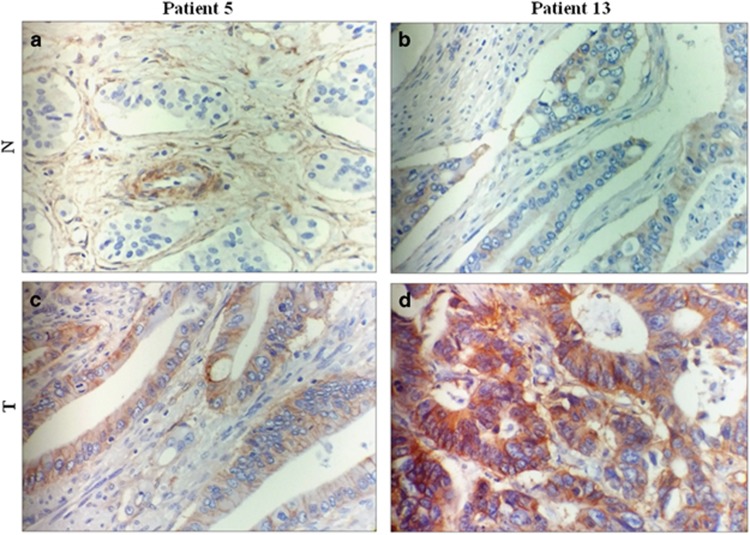
UQCRB protein was overexpressed within the tumor tissues examined in this study, with UQCRB staining mainly found in the cytoplasm. According to our definition, the rate of high UQCRB expression in CRC tissues was 13.3% (2/15 patients), whereas the rate of high UQCRB expression in non-tumor tissues was 0% (0/15 patients). As shown in Figure 3, the UQCRB protein of two patients (patient no. 5 and no. 13) was overexpressed in CRC tissues compared to its expression in non-tumor tissues. CRC tissues (Figure 3c and d) showed moderate to strong cytoplasmic staining of UQCRB, whereas non-tumor tissues (Figure 3a and b) showed either weak staining or no staining. Compared to non-tumor tissues, UQCRB protein of CRC tissues was highly expressed. Furthermore, two high-expression tissues in immunohistochemistry analysis were consistent with high gene expression (Figures 1 and 3). A positive correlation was observed between gene expression and protein expression. These results indicate that genetic and proteomic expression of UQCRB is upregulated in CRC tissues compared to that in non-tumor tissues.
Figure 3.
Detection of UQCRB protein expression in representative (patients 5 and13) colorectal cancer (CRC) tissues (T) and non-tumor tissues (N) as assessed by immunohistochemical staining with UQCRB-specific antibodies. CRC tissues (c, d) showed moderate to strong cytoplasmic staining of UQCRB, whereas non-tumor tissues (a, b) demonstrated either weak staining or no staining. Compared to non-tumor tissues, UQCRB protein of CRC tissues was highly expressed (original magnification; × 400).
Interestingly, there was no association between protein expression and other factors, such as gender, age, tumor localization, tumor size and clinical stage.
UQCRB gene CNV
CNV varies individually in human genomes at the genetic level. To determine the clinical stage-related changes in the genetic variations of UQCRB, we investigated the level of CNV in CRC tissues. UQCRB and BRUNOL4 copy numbers were calculated and normalized to the endogenous reference gene RNase P, which is known to be present in two copies in a diploid genome. TaqMan assay Copy Number Assays designed to specifically target UQCRB exon 4 and BRUNOL4 intron 2 sequences were tested on genomic DNA from 15 CRC tissues paired with non-tumor tissues.
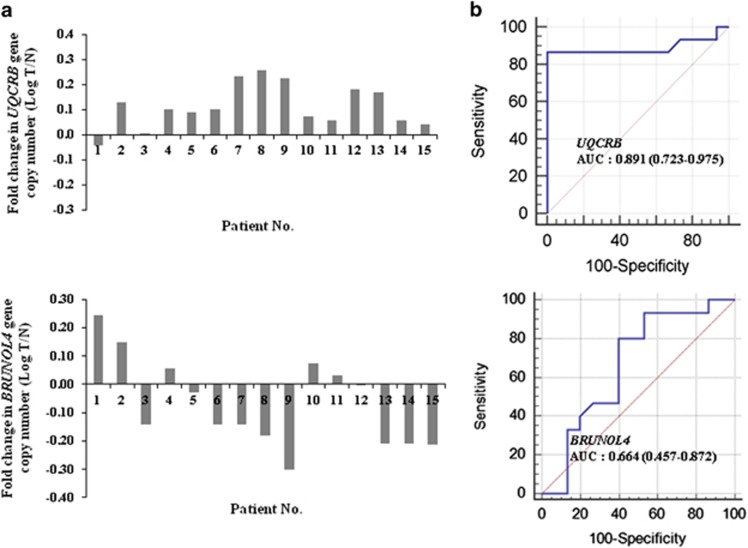
Figure 4 shows the results of the calculated UQCRB copy numbers in the tested CRC tissues/non-tumor tissues. The UQCRB copy number was significantly upregulated in 14 (14/15 patients, 93.3%) pairs of CRC tissues compared to its copy number in non-tumor tissues (Figure 4a). In addition, the BRUNOL4 copy number, representing a negative control gene in CRC, was upregulated in only five (5/15 patients, 33.3%) pairs of CRC tissues compared to that in non-tumor tissues (Figure 4a). Comparative analysis indicated that the average log T/N of the UQCRB and BRUNOL4 copy number levels were 0.11 and −0.07, respectively. The AUC of UQCRB CNV was 0.891 (95% CI, 0.723–0.975; P<0.0001). The Youden index, sensitivity and specificity of UQCRB CNV were 0.867%, 86.7% and 100.0%, respectively, whereas the AUC of BRUNOL4 was 0.664 (95% CI, 0.457–0.872; P<0.0001; Figure 4b).
Figure 4.
Gene copy number variation. (a) UQCRB and BRUNOL4 gene copy number alterations in patients with colorectal cancer (CRC). UQCRB copy number; logarithm T/N of patient No. 1 shows deletion, whereas the others show amplification. (b) Analysis of ROC curve of UQCRB, and BRUNOL4 CNV. The AUC of genes was calculated to diagnose CRC. AUC was calculated using the MedCalc analysis program.
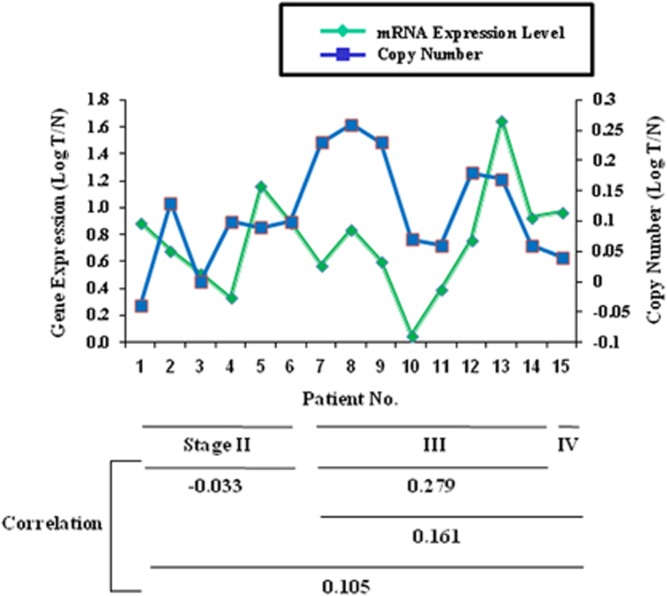
Relationship between gene expression and CNV
To explore whether this change in UQCRB gene expression was associated with a corresponding change in UQCRB copy number, the amount of mRNA of UQCRB and the corresponding UQCRB copy number were compared in CRC tissues/non-tumor tissues. As shown in Figure 5, the relationships between UQCRB gene expression, CNV and clinical stages were analyzed in 15 CRC patients. Gene expression was proportionately correlated with CNV and clinical stages. The samples included six cases of clinical stage II (40%, 6/15 patients), eight cases of clinical stage III (53.3%, 8/15 patients) and one case of clinical stage IV (6.6%, 1/15 patients). The numerical correlation of all samples was 0.105, but it was −0.033 in clinical stage II, 0.279 in clinical stage III and 0.161 in clinical stage III–IV. Although a limited number of samples were available for this analysis, we predict a high correlation for UQCRB gene expression with its CNV in a low clinical stage (stage II) for early diagnosis. However, the highest correlation was observed for a high clinical stage (stage III). These data confirm a high numerical correlation with a high clinical stage rather than a low clinical stage.
Figure 5.
Correlation coefficient was utilized to describe the association between gene expression and copy number in colorectal cancer (CRC) tissues per non-tumor tissues for UQCRB. Gene expression is shown on the left y axis; copy number is shown on the right y axis.
SNP analysis of UQCRB gene
SNP analysis results of theUQCRB CDS (NG_008237.1; 105..123, 2408..2479, 3695..3861, 4503..4580) were not noticed. We analyzed the two polymorphisms in the 3′-UTR region of UQCRB (rs7836698 and rs10504961). Two SNPs (rs7836698 and rs10504961) were already known as prognostic markers for CRC.24 As shown in Table 2, the marker rs7836698 in UQCRB showed an increased TT genotype in CRC tissues compared to non-tumor tissues. The TT genotype-to-CC genotype ratios of rs7836698 in CRC tissues and non-tumor tissues were 83.3% and 7.7%, respectively. In particular, the marker rs7836698 was significantly associated with CRC tumor cases compared to non-tumor tissues. The rs7836698 polymorphism was associated with clinical stage in CRC. However, the marker rs10504961 showed no difference between CRC tissues and non-tumor tissues. Interestingly, carriers of the two UQCRB polymorphisms, the TT carriers, were ~2-fold more likely to develop tumors in the colon than in the rectum.
Table 2. Association of two polymorphisms in UQCRB gene.
|
Clinical (TNM) stage |
||||||
|---|---|---|---|---|---|---|
| II | III | IV | ||||
| SNP | Tissue case | Genotype | No. | Total no. (%) | ||
| rs7836698 | CRC tissues | CC | 3 | 2 | 1 | 6 (40) |
| CT | 1 | 3 | 4 (26.6) | |||
| TT | 2 | 3 | 5 (33.3) | |||
| Non-tumor tissues | CC | 5 | 7 | 1 | 13 (86.6) | |
| CT | 1 | 1 (6.6) | ||||
| TT | 1 | 1 (6.6) | ||||
| rs10504961 | CRC tissues | CC | 4 | 4 | 1 | 9 (60) |
| CT | 1 | 3 | 4 (26.6) | |||
| TT | 1 | 1 | 2 (13.3) | |||
| Non-tumor tissues | CC | 4 | 4 | 1 | 9 (60) | |
| CT | 1 | 4 | 5 (33.3) | |||
| TT | 1 | 1 (6.6) | ||||
Abbreviations: CRC, colorectal cancer; TNM, tumor-node metastasis.
Major/minor allele and location of two SNPs (rs7836698 and rs10504961) were C/T and 5′-UTR, respectively.
DNA methylation analysis of UQCRB
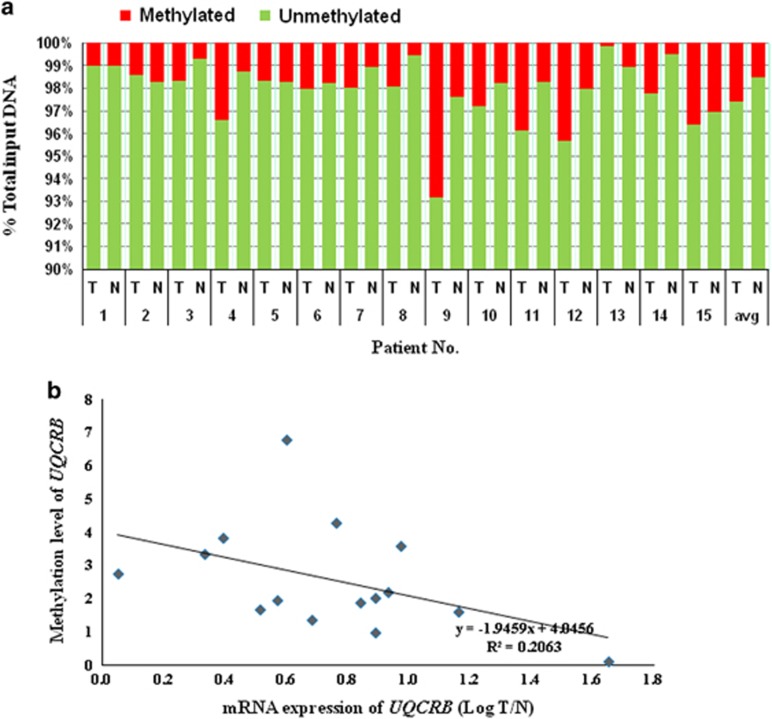
Cancer initiation and progression is regulated by both genetic and epigenetic modifications. Accordingly, we assessed epigenetic modifications of UQCRB and analyzed the classical dependence of DNA methylation in the gene promoter regions with that of gene expression. In a total of 30 samples, we determined UQCRB differential methylation in CRC tissues and non-tumor tissues from 15 patients with CRC. Through quantitative PCR-based DNA methylation analysis, we found that UQCRB was significantly more methylated in CRC tissues than in non-tumor tissues. Notably, CRC tissues in 12 of 15 patients showed a higher degree of methylation than non-tumor tissues (Figure 6a). The average degree of methylation in CRC tissues was 2.59%, whereas this value was 1.49% in non-tumor tissues. The methylation rate in the UQCRB promoter region was 1.7-fold higher in CRC tissues than in non-tumor tissues. Accordingly, there was a significant correlation between gene expression and methylation level of the UQCRB promoter (Figure 6b). To verify the result, however, alternative methods, such as bisulfite-pyrosequencing analysis, will be conducted in a following study.
Figure 6.
DNA methylation of UQCRB promoter. (a) DNA methylation level of UQCRB promoter region from patients. (b) The correlation was utilized to describe the association between the gene expression and methylation level of theUQCRB promoter region.
Discussion
Mitochondrial complex III deficiency is one of the least common oxidative phosphorylation defects associated with mitochondrial disease.26 However, complex III deficiency is a genetic condition that can affect several parts of the body, including the brain, kidneys, liver, heart and skeletal muscles. Signs and symptoms of complex III deficiency typically begin in infancy, but they can appear later.27, 28 Many studies have suggested that hereditary defects in UQCRB of complex III cause several mitochondrial diseases, such as hypoglycemia, lactic acidosis, myopathy and cardiomyopahty.18, 29
Few studies have examined UQCRB in CRC. The primary purpose of this study was to explore the relationship between the expression of UQCRB and the clinico-pathologic features of CRC. In the present study, the expression of three representative genes (UQCRB, UQCRFS1 and MT-CYB) associated with complex III in CRC was examined and assessed for its potential clinical value in CRC development and progression. The expression of the three genes was increased significantly in CRC tissues. Furthermore, we demonstrated that UQCRB expression was upregulated at both the gene and protein levels in CRC tissues compared to matched adjacent non-tumor tissues. High levels of UQCRB expression were more frequently observed in CRC patients with poor pathologic differentiation or in those with advanced stages. We demonstrated that high UQCRB expression was correlated with CRC tissues, whereas low UQCRB expression was correlated with non-tumor tissues.
Owens et al.30 quantified the mitochondrial oxidative phosphorylation activities of all five complexes and confirmed the activity and gene expression of complex III subunits in human breast cancer cell lines and primary tumors. The activity of complex III of breast cancer cell lines showed large differences compared to normal activity. In addition, UQCRFS1 overexpression was detected in breast tumors compared to matched normal subjects. These studies directly demonstrated the contribution of impaired mitochondrial oxidative phosphorylation complex III to breast tumorigenesis.30 In agreement with the results of previous studies, our results directly demonstrate the contribution of mitochondrial oxidative phosphorylation complex III to colon tumorigenesis. In particular, we found that unlike other genes, MY-CYB showed negative logarithm figures of the T/N ratio in clinical stage II, indicating that among the three genes, MT-CYB showed differences in clinical stages II and III. Moreover, patients with high gene expression generally showed an increased risk of distant metastasis and advanced clinical stage. These data demonstrate that complex III plays an important role in CRC progression and metastasis.
A previous study showed that UQCRFS1 was overexpressed (14/40 patients, 35%) in breast cancer.30 The expression of mitochondrial-encoded genes, including cytochrome B (complex III), generally decreased in myelodysplastic syndromes (MDS) and acute myeloid leukemia-MDS, whereas the mitochondrial DNA copy number increased rather than being reduced. Overall, MDS patients showing a decrease in mitochondrial gene expression displayed a stochiometric imbalance in mitochondrial-encoded genes. The authors of that study concluded that dysregulated mitochondrial gene expression extends beyond a simple age-related effect and is compatible with the putative role of mitochondrial dysfunction in MDS pathophysiology.31
Notably, there were significant correlations between UQCRB expression and tumor types, although we detected no relationship between UQCRB expression and tumor size. The expression of the three genes may also be associated with the localization (rectum, colon and cecum) of CRC. We found that gene expression was high in most of the rectum samples of CRC (5/5 patients, 100%). Accordingly, UQCRB could potentially be used as a biomarker of rectal cancer.
The AUC is a standard analytical tool for evaluating diagnostic tests.32 Although we were unable to accurately determine the AUC because of our limited sample number, we analyzed AUC with our tested CRC tissues (n=15). Genetic and proteomic expression of UQCRB was upregulated. The concordance rate between high expression of the UQCRB gene (100%, 15/15 patients) and UQCRB protein was 13.3% (2/15 patients).
UQCRFS1 expression in additional matched normal and breast tumors revealed overexpression (35%, 14/40 patients) in breast tumors. UQCRFS1 knockdown in breast tumor led to decreased mitochondrial membrane potential, and a reduction in Matrigel invasion mediated by reduced reactive oxygen species levels, coinciding with decreased expression of NADPH oxidase involved in reactive oxygen species production.30
Cancer biomarkers in mitochondrial genetic/proteomic studies have not been extensively studied. Deregulation of the mitochondrial tumor suppressor genes (SIRT3, SIRT4 and MTUS1) was detected in head and neck squamous cell carcinoma.33 Sirtuins, a family of orthologs of yeast silent information regulator 3 (Sirt3) and 4 (Sirt4), are important tumor suppressor genes located in the mitochondria.34, 35, 36, 37 However, UQCRB was reported to be frequently downregulated in endometrial stromal cells.38 In addition, the expression level of UQCRB was significantly decreased in Pallister–Killian syndrome samples, and an inverse linear correlation was observed between the level of miR-1224 and the UQCRB expression level.39
We observed that the UQCRB copy number was upregulated in CRC. Gene expression was proportionately correlated with CNV and clinical stage. In particular, a high numerical correlation was detected in clinical stage III. Similarly, UQCRB CNV appears to be more suitable as a candidate marker in CRC. A high correlation between expression and CNV has been reported in ovarian cancer.40 The BRUNOL4 copy number was downregulated, as expected. Poulogiannis et al.23 demonstrated that BRUNOL4 (encoding Bruno-like 4 splicing factor) copy number loss is an independent prognostic indicator in CRC. Lu et al.41 evaluated CNV and gene expression to identify genes reproducibly associated with tumorigenesis. CNV regions identified 475 genes differentially expressed between tumor and normal tissues and showed concordance between CNV and gene expression changes. Based on combined gene expression profiles and CNV, the identified genes/pathways may serve as prognostic biomarkers for lung tumorigenesis. Wrzeszczynski et al.42 evaluated gene expression data and predicted 156 genes with CNV and then correlated changes in expression. Among these genes, UQCRB was identified within The Cancer Genome Atlas (http://tcga.cancer.gov) data set and Memorial Sloan-Kettering Cancer Center data set in ovarian cancer. UQCRB is overexpressed in liver cancer cells, and CNV was found in cervical cancer and may be used as a cellular marker for cervical cancer diagnosis.42 Moreover, altered copy number and changes in UQCRB gene expression were identified in specimens of hepatocellular carcinoma, and pancreatic ductal adenocarcinoma from microarray studies indicated that UQCRB plays a significant role in tumorigenesis.43, 44
In the present study, we also demonstrated that for the UQCRB rs7836698 polymorphism, the TT genotype was associated with tumorigenesis in CRC tissues, but not in non-tumor tissues. There was no significant correlation between rs7836698 polymorphisms and gender, age, tumor localization, or tumor size. However, the TT genotype and CT genotype were more likely to be increased in stage III than in stage II. The genotype change at rs7836698 between CRC tissues and non-tumor tissues was proportional to clinical stage. Thus, rs7836698 polymorphisms are closely associated with biomarkers of CRC. Two SNPs (rs7836698 and rs10504961) in the 3′-UTR of UQCRB were associated with overall survival of CRC patients, suggesting that SNPs identified by multivariate analysis can be used as independent prognostic factors for CRC.24 As small numbers of analyzed SNPs strongly influence statistical analysis, a larger number of patient samples will be collected in the following study to obtain statistically significant results.
To determine whether the clinical stage-related increase in DNA methylation of the UQCRB promoter is a universal aging phenomenon, we measured the level of DNA methylation in the promoter of UQCRB. Clinical stage was not found to affect DNA methylation in UQCRB. In addition, there was no difference in the level of DNA methylation of the UQCRB promoter between elderly (⩾65 years) and young subjects (<65 years). However, the methylation rate in CRC tissues was higher than in non-tumor tissues. DNA methylation is a potential mechanism for regulating the expression of a gene. Cytosine residues in CG dinucleotides are targets for DNA methylation, and gene expression is typically reduced when DNA methylation occurs at a promoter. Accordingly, the level of DNA methylation was related to gene expression. Our results demonstrated that methylation level is highly correlated with mRNA expression, which is inversely proportional in CRC (R2=0.1926). However, the statistical correlation between the expression level and the methylation level was lower than the correlation between the expression level and the CNV level. Moarii et al.45 suggested that the high correlation between promoter DNA methylation and gene expression differentiates tumor tissues from non-tumor tissues. The gene expression of a few specific genes, particularly transcription factors, is associated with DNA methylation in a tissue-dependent manner. In addition, both methylation and CNV are important requisite predictors of gene expression variation.45 Ling et al.46 suggested that the relative mRNA level of UQCRB was also reduced in muscle in the elderly compared to in young twins. The level of DNA methylation was also found to be related to gene expression. UQCRB was among the genes showing reduced expression in diabetic muscle. Age-related DNA methylation of UQCRB increases, whereas expression of UQCRB decreases with aging. DNA methylation is associated with an age-related decrease in gene expression in human muscle in some, but not all, genes. Changes in the DNA methylation and expression of all genes associated with SNPs (amplified and deleted genes) were observed in ovarian cancer tumors. In addition, tumor suppressor and oncogenic features of these modalities and correlation analysis with expression were determined. Genes showing a strong correlation for methylation-dependent expression changes exhibited varying copy number aberrations.46
A previous study demonstrated the extent of genomic and epigenetic alterations for known tumor suppressors and oncogenes and used these defined features to identify potential ovarian cancer gene candidates.42 DNA methylation alterations are a significant feature in the cancer genome.47, 48 SNP amplification and promoter hypomethylation may play a role in the overexpression of oncogenes. By contrast, a particularly well-known oncogene may be deleted in a specific cancer, decreasing its pathogenic role within that cancer or an individual sample. Interestingly, for highly amplified genes, a high level of methylation accompanied by low expression indicates altered tumor suppressor function in the cancer cell. In highly amplified genes, low-level methylation and high expression indicate oncogenic features in a cancer cell. As one of the largest epigenetic studies of CRC, as well as the first investigation of UQCRB methylation in CRC, our findings highlight the potential for UQCRB methylation as a biomarker of CRC.
Recent reports have revealed genetic variations in UQCRB in several cancers, including hepatocellular carcinoma, ovarian cancer, pancreatic ductal adenocarcinoma and CRC. By contrast, our UQCRB study, which used multiple tools, was the first study of CRC. In addition, numerous studies detected UQCRB expression at the gene level, whereas our study focused on its expression at the gene and protein levels. In addition, we detected UQCRB SNPs and conducted epigenetic analysis at the gene level. Our results may be useful for predictive diagnosis in which UQCRB expression correlates with CRC patients’ clinical features. However, the precise mechanism by which UQCRB predicts CRC development remains unclear. Thus, further studies, including overexpression and knockdown of UQCRB expression in CRC cells, are needed to explore the mechanism by which UQCRB is involved in the development and progression of CRC and its exact regulating pathway in vitro and in vivo.
We previously reported that UQCRB plays a role in angiogenesis and that the developed cell-permeable PTD-UQCRB can be utilized as a pro-angiogenic agent.49 In addition, we provided a molecular basis for UQCRB-related biological processes and revealed the potential key roles of UQCRB in angiogenesis and mitochondria-mediated metabolic disorders.50 In this study, we found that UQCRB expression and CNV were significantly upregulated in CRC tissues compared to non-tumor tissues and were associated with clinical stage and pathologic differentiation. Representative CRC biomarkers discovered during the recent years continue to be closely examined: MSI, chromosome 18q loss of heterozygosity (18qLOH), p53, KRAS, BRAF, NRAS, PIK3CA mutations, PTEN expression, UGT1A1 gene polymorphism and ezrin protein. Mutations in the tumor suppressor gene p53 (chromosome region 17p13) occur in 50–70% of all CRC.51 Together with these known CRC biomarkers, multivariate analyses of this study demonstrated that UQCRB can be used as a novel prognostic biomarker of human CRC.
Acknowledgments
This work was partly supported by grants from the National Research Foundation of Korea, which is funded by the Korean government (MSIP; 2012M3A9D1054520, 2015K1A1A2028365, 2015M3A9B6027818, 2015M3A9C4076321); the Brain Korea 21 Plus Project, Republic of Korea; and the Seoul Medical Science Institute in 2015. The transcriptome analysis data of UQCRB overexpressing cells are available at Korean BioInformation Center (KBRS20171018_0000001∼KBRS20171018_0000336).
Footnotes
The authors declare no conflict of interest.
References
- Smith RA, Andrews K, Brooks D, DeSantis CE, Fedewa SA, Lortet-Tieulent J et al. Cancer screening in the United States, 2016: a review of current American Cancer Society Guidelines and current issues in cancer screening. CA Cancer J Clin 2016; 66: 96–114. [DOI] [PubMed] [Google Scholar]
- Kim DH. Risk factors of colorectal cancer. J Korean Soc Coloproctol 2009; 25: 356–362. [Google Scholar]
- Yang SK, Yun S, Kim JH, Park JY, Kim HY, Kim YH et al. Epidemiology of inflammatory bowel disease in the Songpa-Kangdong district, Seoul, Korea, 1986-2005: a KASID study. Inflamm Bowel Dis 2008; 14: 542–549. [DOI] [PubMed] [Google Scholar]
- Park HC, Shin A, Kim BW, Jung KW, Won YJ, Oh JH et al. Data on the characteristics and the survival of Korean patients with colorectal cancer from the Korea central cancer registry. Ann Coloproctol 2013; 29: 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med 2003; 348: 2656–2668. [DOI] [PubMed] [Google Scholar]
- Heo HJ, Kim HK, Youm JB, Cho SW, Song IS, Lee SY et al. Mitochondrial pyruvate dehydrogenase phosphatase 1 regulates the early differentiation of cardiomyocytes from mouse embryonic stem cells. Exp Mol Med 2016; 48: e254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crivellone MD, Wu M, Tzagoloff A. Assembly of the mitochondrial membrane system. Analysis of structural mutants of the yeast coenzyme QH2-cytochrome C reductase complex. J Biol Chem 1988; 263: 14323–14333. [PubMed] [Google Scholar]
- Berry EA, Guergova-Kuras M, Huang LS, Crofts AR. Structure and function of cytochrome bc complexes. Annu Rev Biochem 2000; 69: 1005–1075. [DOI] [PubMed] [Google Scholar]
- Darrouzet E, Moser CC, Dutton PL, Daldal F. Large scale domain movement in cytochrome bc1: a new device for electron transfer in proteins. Trends Biochem Sci 2001; 26: 445–451. [DOI] [PubMed] [Google Scholar]
- Trumpower BL. The protonmotive Q cycle. Energy transduction by coupling of proton translation by the cytochrome bc1 complex. J Biol Chem 1990; 265: 11409–11412. [PubMed] [Google Scholar]
- Lonlay PD, Valnot I, Barrientos A, Gorbatyuk M, Tzagoloff A, Taanman JW et al. A mutant mitochondrial respiratory chain assembly protein causes complex III deficiency in patients with tubulopathy, encephalopathy and liver failure. Nat Genet 2001; 29: 57–60. [DOI] [PubMed] [Google Scholar]
- Zara V, Conte L, Trumpower BL. Biogenesis of the yeast cytochrome bc1complex. Biochim Biophys Acta 2009; 1793: 89–96. [DOI] [PubMed] [Google Scholar]
- Hildenbeutel M, Hegg EL, Stephan K, Gruschke S, Meunier B, Ott M. Assembly factors monitor sequential hemylation of cytochrome b to regulate mitochondrial translation. J Cell Biol 2014; 205: 511–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Hosokawa Y, Toda H, Nishikimi M, Ozawa T. Cloning and sequencing of a cDNA for humanmitochondrialubiquinone-bindingprotein of complexIII. Biochem Biophys Res Commun 1988; 156: 987–994. [DOI] [PubMed] [Google Scholar]
- Malaney S, Heng HH, Tsui LC, Shi XM, Robinson BH. Localization of the human gene encoding the 13.3-kDa subunit of mitochondrial complex III (UQCRB) to 8q22 by in situ hybridization. J Cytogenet Genome Res 1996; 73: 297–299. [DOI] [PubMed] [Google Scholar]
- Mulder W, Scholten IH, van Roon H, Grivell LA. Isolation and characterisation of the linked genes APA2 and QCR7, coding for Ap4A phosphorylase II and the 14 kDa subunit VII of the mitochondrial bc1-complex in the yeast Kluyveromyces lactis. Biochim Biophys Acta 1994; 1219: 719–723. [DOI] [PubMed] [Google Scholar]
- Hemrika W, Jong MD, Berden JA, Grivell LA. The C-terminus of the 14-kDa subunit of ubiquinol-cytochrome-c oxidoreductase of the yeast Saccharomyces cerevisiae is involved in the assembly of a functional enzyme. Eur J Biochem 1994; 220: 569–576. [DOI] [PubMed] [Google Scholar]
- Haut S, Brivet M, Touati G, Rustin P, Lebon S, Garcia-Cazorla A et al. A deletion in the human QP-C gene causes a complex III deficiency resulting in hypoglycemia and lactic acidosis. Hum Genet 2003; 113: 118–122. [DOI] [PubMed] [Google Scholar]
- Cho YS, Jung HJ, Seok SH, Payumo AY, Chen JK, Kwon HJ. Functional inhibition of UQCRB suppresses angiogenesis in zebrafish. Biochem Biophys Res Commun 2013; 433: 396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HJ, Shim JS, Lee J, Song YM, Park KC, Choi SH et al. Terpestacin inhibits tumor angiogenesis by targeting UQCRB of mitochondrial complex III and suppressing hypoxia- induced reactive oxygen species production and cellular oxygen sensing. J Biol Chem 2010; 285: 11584–11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Huang Y, Yu H, Lei H, Xie C, Zhong Y. Matrix metalloproteinase 7 is a useful marker for 5-fluorouracil-based adjuvant chemotherapy in stage II and stage III colorectal cancer patients. Med Oncol 2014; 31: 824. [DOI] [PubMed] [Google Scholar]
- Poulogiannis G, Ichimura K, Hamoudi RA, Luo F, Leung SY, Yuen ST et al. Prognostic relevance of DNA copy number changes in colorectal cancer. J Pathol 2010; 220: 338–347. [DOI] [PubMed] [Google Scholar]
- Lascorz J, Bevier M, Schonfels WV, Kalthoff H, Aselmann H, Beckmann J et al. Polymorphisms in the mitochondrial oxidative phosphorylation chain genes as prognostic markers for colorectal cancer. BMC Med Genet 2012; 13: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Kim E, Lee S, Kim D, Chun J, Park KH et al. TFAP2C-mediated upregulation of TGFBR1 promotes lung tumorigenesis and epithelial–mesenchymal transition. Exp Mol Med 2016; 48: e273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Vizarra E, Zeviani E. Nuclear gene mutations as the cause of mitochondrial complex III deficiency. Front Genet 2015; 6: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez A, Gil-Borlado MC, Moran M, Verdu A, Cazorla-Calleja MR, Martin MA et al. Infantile mitochondrial encephalomyopathy with unusual phenotype caused by a novel BCS1L mutation in an isolated complex III-deficient patient. Neuromuscul Disord 2009; 19: 143–146. [DOI] [PubMed] [Google Scholar]
- De Meirleir L, Seneca S, Damis E, Sepulchre B, Hoorens A, Gerlo E et al. Clinical and diagnostic characteristics of complex III deficiency due to mutations in the BCS1L gene. Am J Med Genet 2003; 121A: 126–131. [DOI] [PubMed] [Google Scholar]
- Borisov VB. Mutations in respiratory chain complexes and human diseases. Ital J Biochem 2004; 53: 34–40. [PubMed] [Google Scholar]
- Owens KM, Kulawiec M, Desouki MM, Vanniarajan A, Singh KK. Impaired OXPHOS complex III in breast cancer. PLoS ONE 2011; 11: e23846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildgen V, Wulfert M, Gattermann N. Impaired mitochondrial gene transcription in myelodysplastic syndromes and acute myeloid leukemia with myelodysplasia-related changes. Exp Hematol 2011; 39: 666–675. [DOI] [PubMed] [Google Scholar]
- Zou KH, O’Malley AJ, Mauri L. Receiver-operating characteristic analysis for evaluating diagnostic tests and predictive models. Circulation 2007; 115: 654–657. [DOI] [PubMed] [Google Scholar]
- Mahjabeen I, Kayani MA. Loss of mitochondrial tumor suppressor genes expression is associated with unfavorable clinical outcome in head and neck squamous cell carcinoma: Data from retrospective study. PLoS ONE 2016; 11: e0146948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 2010; 17: 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 2010; 464: 121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, Pierce K et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell 2013; 23: 450–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibold S, Rudroff C, Weber M, Galle J, Wanner C, Marx M. Identification of a new tumor suppressor gene located at chromosome 8p21.3e-22. FASEB J 2003; 17: 1180–1182. [DOI] [PubMed] [Google Scholar]
- Chen J, Gu L, Ni J, Hu P, Hu K, Shi YL. MiR-183 regulates ITGB1P expression and promotes invasion of endometrial stromal cells. Biomed Res Int 2015; 2015: 340218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi K, Zhang Z, Kaur M, Krantz ID. 12p microRNA expression in fibroblast cell lines from probands with Pallister-Killian syndrome. Chromosome Res 2014; 22: 453–461. [DOI] [PubMed] [Google Scholar]
- Bowtell DD. The genesis and evolution of high-grade serous ovarian cancer. Nat Rev Cancer 2010; 10: 803–808. [DOI] [PubMed] [Google Scholar]
- Lu TP, Lai LC, Tsai MH, Chen PC, Hsu CP, Lee JM. Integrated analyses of copy number variations and gene expression in lung adenocarcinoma. PLoS ONE 2011; 6: e24829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzeszczynski KO, Varadan V, Byrnes J, Lum E, Kamalakaran S, Levine DA et al. Identification of tumor suppressors and oncogenes from genomic and epigenetic features in ovarian cancer. PLoS ONE 2011; 6: e28503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia HL, Ye QH, Qin LX, Budhu A, Forgues M, Chen Y et al. Gene expression profiling reveals potential biomarkers of human hepatocellular carcinoma. Clin Cancer Res 2007; 13: 1133–1139. [DOI] [PubMed] [Google Scholar]
- Harada T, Chelala C, Crnogorac-Jurcevic T, Lemoine NR. Genome-wide analysis of pancreatic cancer using microarray-based techniques. Pancreatology 2009; 9: 13–24. [DOI] [PubMed] [Google Scholar]
- Moarii1 M, Boeva1 V, Vert JP, Reyal F. Changes in correlation between promoter methylation and gene expression in cancer. BMC Genomics 2015; 16: 873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, Poulsen P, Simonsson S, Rönn T, Holmkvist J, Almgren P. Genetic and epigenetic factors are associated with expression of respiratory chain component NDUFB6 in human skeletal muscle. J Clin Invest 2007; 117: 3427–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting AH, McGarvey KM, Baylin SB. The cancer epigenome–components and functional correlates. Genes Dev 2006; 20: 3215–3231. [DOI] [PubMed] [Google Scholar]
- Sadikovic B, Al-Romaih K, Squire JA, Zielenska M. Cause and consequences of genetic and epigenetic alterations in human cancer. Curr Genomics 2008; 9: 394–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JH, Jung HJ, Park HJ, Cho SW, Lee SK, Kwon HJ. Cell-permeable mitochondrial ubiquinol–cytochrome c reductase binding protein induces angiogenesis in vitro and in vivo. Cancer Lett 2015; 366: 52–60. [DOI] [PubMed] [Google Scholar]
- Chang JH, Jung HJ, Jeong SH, Kim HK, Han J, Kwon HJ. A mutation in the mitochondrial protein UQCRB promotes angiogenesis through the generation of mitochondrial reactive oxygen species. Biochem Biophys Res Commun 2014; 433: 290–297. [DOI] [PubMed] [Google Scholar]
- Lech G, Słotwiński R, Słodkowski M, Krasnodębski IW. Colorectal cancer tumour markers and biomarkers: recent therapeutic advances. World J Gastroenterol 2016; 22: 1745–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]