Abstract
The binary toxin Clostridium difficile transferase (CDT) is frequently observed in C. difficile strains and is associated with an increased severity of C. difficile infection. CDT-producing C. difficile infections cause higher fatality rates than infections with CDT negative isolates. Thus, the rapid and accurate identification of a CDT positive C. difficile infection is critical for effective treatment. The present study demonstrates how loop-mediated isothermal amplification (LAMP) can be used to detect CDT-producing C. difficile based on visual observation. This is a low complexity, rapid molecular method that has the potential to be used within a point of care setting. The specificity and sensitivity of the primers in the LAMP reactions for CDT detection were determined using two different methods, a real-time turbidity monitor and visual detection after the addition of calcein to the reaction tube. The results revealed that target DNA was amplified and visualized by these two detection methods within 60 min at a temperature of 60°C. The sensitivity of the LAMP assay was identified to be 10-fold greater than that of polymerase chain reaction analysis. When 25 alternative bacterial strains lacking CDT were tested, the results of the amplification were negative, confirming the specificity of the primers. In conclusion, the visual LAMP method established in the present study may be a rapid, reliable and cost-effective tool for detecting CDT-producing C. difficile strains at the point of care.
Keywords: Clostridium difficile, loop-mediated isothermal amplification, binary toxin C. difficile transferase, calcein, turbidity
Introduction
Clostridium difficile is a gram-positive toxin-producing bacillus that causes intestinal infections in humans and animals (1). C. difficile presents an emerging threat in hospital environments (2), causing a range of digestive disorders, including inflammation of the bowel, abdominal pain, fever and diarrhea (1). C. difficile infection has increased in prevalence and severity over the last decade (2,3). Its pathogenesis comes from its ability to release two different toxins; toxins A and B. These toxins glucosylate and inactivate the Rho factor proteins of host cells. In addition, certain C. difficile isolates produce the binary toxin C. difficile transferase (CDT) (1,4,5). CDT belongs to the family of binary adenosine diphosphate (ADP)-ribosylating toxins and is made up of two separate toxin components: i) The enzymatic ADP-ribosyltransferase that modifies actin (CDTa); and ii) CDTb that binds to host cells and translocates CDTa into the cytosol (6). These two independent, unlinked protein chains are encoded by two separate genes, designated cdtA and cdtB (1,5,7). CDT serves an important role in the immediate colonization of C. difficile; it can create thin microtubule protrusions on the surface of intestinal epithelial cells, which increase the adherence of C. difficile onto the surface of these cells (4). CDT positive strains were previously reported as an infrequent cause of C. difficile infection in human populations but have become increasingly prevalent over the past decade (6). CDT is an important contributing factor towards the pathogenesis of C. difficile, and CDT-producing C. difficile infections are known to be more severe and have higher fatality rates than those caused by CDT-negative isolates (4). This has lead to CDT being viewed as a serious health threat (1).
The accurate and rapid diagnostic testing for CDT-producing C. difficile is essential for patient management and the timely implementation of infection control measures; however, sensitive and specific diagnostic tests for CDT detection are lacking (3). Advances in molecular methods for the detection of pathogens may offer increased sensitivity and specificity. For example, the binary toxin CDT encoding genes cdtA and cdtB are potential biomarkers for diagnostic purposes (3).
In the present study, a low-complexity method for the detection of binary toxin-producing C. difficile, that makes use of loop-mediated isothermal amplification (LAMP) was used. LAMP is a nucleic acid detection technique first described in 2,000, which commands high specificity as the amplification of DNA is conducted by two to three pairs of primers recognizing six independent regions on a target gene (8). LAMP was developed to amplify target DNA without the temperature shifts normally required for denaturing, annealing, and extension during the polymerase chain reaction (PCR) (9). As this method proceeds at a constant temperature, requiring only a thermostat, it can be effectively performed in the field. In the present study, the CDT encoding genes cdtA and cdtB were used for the diagnosis of CDT-producing C. difficile using LAMP (10). Although LAMP has previously been applied to detect the C. difficile enterotoxin A encoding gene tcdA and the cytotoxin B encoding gene tcdB (2,3,11,12), electrophoresis or other sophisticated apparatus, such as a real-time turbidimeter, were still required for the determination of the amplified products. The present study describes a visual detection method based on a color change in the calcein/Mn2+ mixture, allowing immediate interpretation of the LAMP results by the naked eye. This newly developed visual LAMP assay is simple, fast and only requires thermostatic equipment.
Materials and methods
Collection of clinical samples
A total of 10 stool samples were collected from hospitalized patients in the Infection Control and Hospital Epidemiology (Beijing, China). Written informed consent was obtained from all patients prior to sample collection. Of the 10 C. difficile strains that tested positive for binary toxin genes, only 1 strain that was isolated from a patient with Crohn's disease was identified to be positive for the cdtA and cdtB genes by LAMP detection. This was consistent with the results of polymerase chain reaction (PCR) analysis.
Pathogen strains and DNA extraction
A total of 25 strains used in the present study are from our microorganism center (Institute of Disease Control and Prevention, Academy of Military Medical Sciences; Beijing, China). These strains included Bacillus megaterium, Vibrio piscium, Pseudomonas maltophilia, Mycobacterium tuberculosis 4368, Vibrio cholera O139, Bacillus anthracis, enterohemorrhagic Escherichia coli, Yersinia enterocolitica, Vibrio parahaemolyticus (5474), Enteropathic Escherichia coli, enteroadherent Escherichia coli, Enteroinvasive Escherichia coli, Enterotoxigenic Escherichia coli, Yersinia pestis, Streptococcus pneumoniae, Neisseria meningitides group B CMCC29022, Burkholderia pseudomallei, Methicillin resistant Staphylococcus aureus, Acinetobacter baumannii, Escherichia coli, Bordetella pertussis ATCC18530, Haemophilus influenzae, Corynebacterium diphtheriae CMCC38001, Mycobacterium tuberculosis 4368 and Neisseria meningitides (NM29019). In addition, C. difficile strains were cultured in a cycloserine cefoxitin fructose agar base (BD Biosciences, Franklin Lakes, NJ, USA); Mycobacterium tuberculosis, Vibrio parahaemolyticus (5474), Neisseria meningitides and Haemophilus influenzae were cultured in Chocolate Agar Plates (Beijing Land Bridge Biotechnology Co., Ltd., Beijing, China); and other strains were cultured in Brain Heart Infusion (BD Biosciences). The genomic DNA of all strains was extracted using the TIANamp Bacteria DNA kit (Tiangen Biotech Co., Ltd., Beijing, China) following the manufactureri's instructions.
Primer design
The sequences of the cdtA and cdtB genes, with the accession numbers HQ639673.1 and HQ639677.1 respectively, were retrieved from the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/genbank). Primer Explorer Version 4 (primerexplorer.jp/elamp4.0.0/index.html; Fujitsu Ltd., Tokyo, Japan) was used to analyze the sequences, and design 8 primers for cdtA and 6 for cdtB. The primers were synthesized commercially by Sangon Biotech Co., Ltd. (Shanghai, China) and their sequences are presented in Table I.
Table I.
Sequences of the primers used for the specific amplification of cdtA and cdtB.
| A, cdtA primers | ||
|---|---|---|
| Primer type | Reaction used in | Sequence (5′-3′) |
| F3 | LAMP | TCTGGTCCTCAAGAATTTGG |
| B3 | LAMP | AATAGCTGATAGATAAGCTCCA |
| FIP | LAMP | GCTTGTCCTTCCCATTTTGATTTAATTTTTAACTCTTACTTCCCCTGA |
| BIP | LAMP | ATTGGTAGTGTGAATATGAGTGCATTACCTTTAGGTATAGTTATACGTAGT |
| LF | LAMP | AAATCATATTCAGGGGAA |
| LB | LAMP | TTTGCTAAAAGAAAAATAGTACTAC |
| F | PCR | TGAACCTGGAAAAGGTGATG |
| B | PCR | AGGATTATTTACTGGACCATTTG |
| B, cdtB primers | ||
| Primer type | Reaction used in | Sequence (5′-3′) |
| F3 | LAMP | GAGTCAAATACTGCTGGAGA |
| B3 | LAMP | TAGTAGCTCTGGAAACAGTT |
| FIP | LAMP | CGGATCTCTTGCTTCAGTCTTTTTTCAGATTATGAAAAAGCTTCAGGTT |
| BIP | LAMP | AGTTGCAGCATATCCAATTGTTGGTTTCCTTGATCAGTAGAGGCATG |
| F | PCR | CTTAATGCAAGTAAATACTGAG |
| B | PCR | AACGGATCTCTTGCTTCAGTC |
cdt, Clostridium difficile transferase; LAMP, loop-mediated isothermal amplification; PCR, polymerase chain reaction; F, forward; B, backwards; 3, outer; L, loop; I, inner; P, primer.
LAMP assay
A LAMP assay was performed using a Loopamp DNA Amplification kit (Eiken Chemical Co., Ltd., Tochigi, Japan) according to the manufacturer's protocol.
LAMP assay product detection
The amplification products were detected using two methods; a real-time turbidimeter (La-320C; Eiken Chemical Co., Ltd.) at 650 nm and visual observation. Pyrophosphate ions are released during the LAMP reaction process and they form a white magnesium phosphate precipitate. This can be monitored using a real-time turbidimeter, drawing reaction curves every 6 sec with Mg2+ ions in the reaction buffer (13). For visual detection, 1 µl of Loopamp Fluorescent Detection reagent (Eiken Chemical Co., Ltd.), containing a metal indicator, was added into the reaction system prior to amplification. The reaction buffer initially turned orange because the calcein was quenched by Mn2+ ions. Then, during amplification the calcein was displaced by pyrophosphate ions from the calcein/Mn2+ complex, and the color changed from orange to green. By contrast, if no amplification occurred, no color change was observed.
PCR analysis
To assess the detection limit of LAMP compared with traditional PCR, genomic DNA extracted from C. difficile was serially diluted at a ratio of 1:10 (from 24.8 ng/µl to 0.000248 pg/µl) and tested using the real-time turbidity monitor, the visual method and using the GeneAmp® PCR system 9700 (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The reaction mixture for PCR contained 12.5 µl 2X Taq PCR Master Mix (Tiangen Biotech Co., Ltd.), 1 µl of each primer, 1 µl genomic DNA as a template and ddH2O to a final volume of 25 µl. The thermocycling conditions for PCR were as follows: 95°C for 5 min; followed by 30 cycles of 95°C for 30 sec 60°C for 5 min and 95°C for 5 min; and then a final extension step at 72°C for 7 min. For detection of the PCR products, a 1% agarose gel containing ethidium bromide was used. The marker used was DL2000 (Takara Biotechnology, Dalian, China). C. difficile was used as a positive control, ddH2O as the negative control and 25 different strains of pathogen (listed above) were used as test strains.
Results
Optimal conditions for the simultaneous detection of cdtA and cdtB
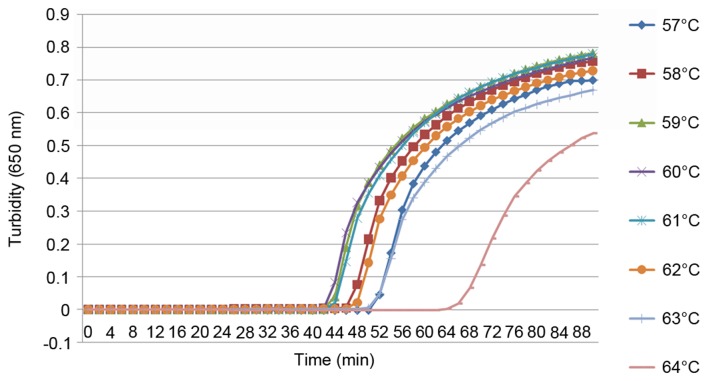
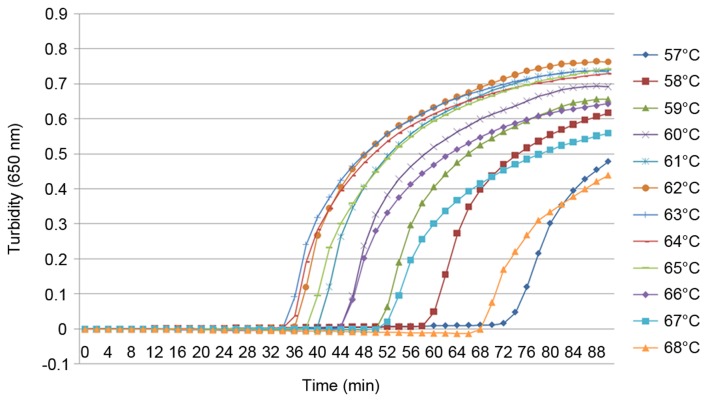
To the best of our knowledge, the present study was the first to use the cdtA and cdtB genes as the target sequences for a LAMP assay detecting C. difficile. For this reason it was necessary to establish the optimal conditions for the reaction. Six and four primers were designed for the detection of the cdtA and cdtB genes, respectively, using the same reaction conditions. Using a real-time turbidimeter, turbidity curves at 650 nm were constructed according to the amplification data. To optimize the reaction conditions for the LAMP assay, different reaction temperatures from 57–68°C were analyzed using the real-time tubidimeter (Figs. 1 and 2). When detecting cdtA, the optimal conditions were 59, 60 and 61°C for 60 min; however, for the detection of cdtB, the optimal conditions were 60°C for 60 min. Therefore, for the detection of cdtA and cdtB, the optimal conditions were set at 60°C for 60 min.
Figure 1.
LAMP specific to cdtA, performed at increasing temperatures. A LAMP reaction specific to cdtA was tested using a real time turbidimeter at 650 nm at increasing temperatures in order to determine the optimal conditions. LAMP, loop-mediated isothermal amplification; cdt, Clostridium difficile transferase.
Figure 2.
LAMP specific to cdtB, performed at increasing temperatures. A LAMP reaction specific to cdtB was tested using a real time turbidimeter at 650 nm at increasing temperatures in order to determine the optimal conditions. LAMP, loop-mediated isothermal amplification; cdt, Clostridium difficile transferase.
Specificity of the LAMP assay to C. difficile
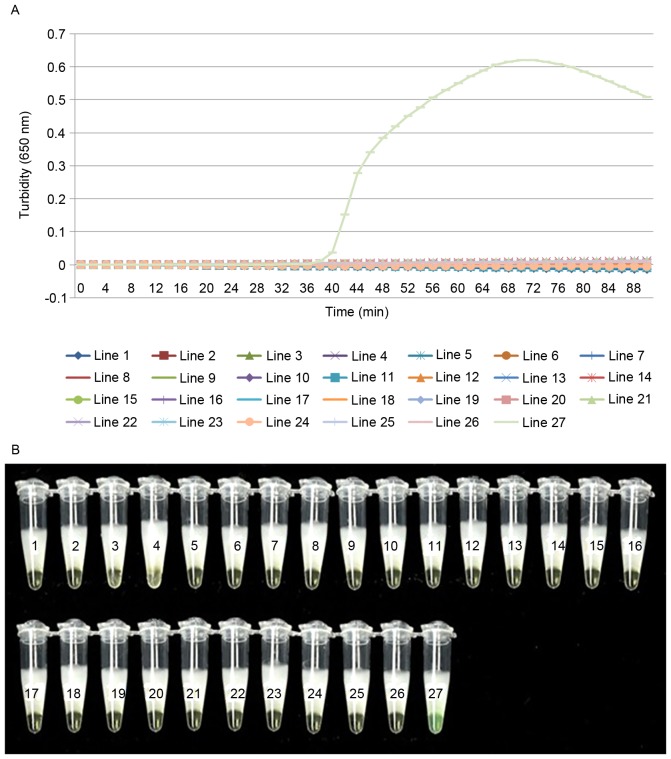
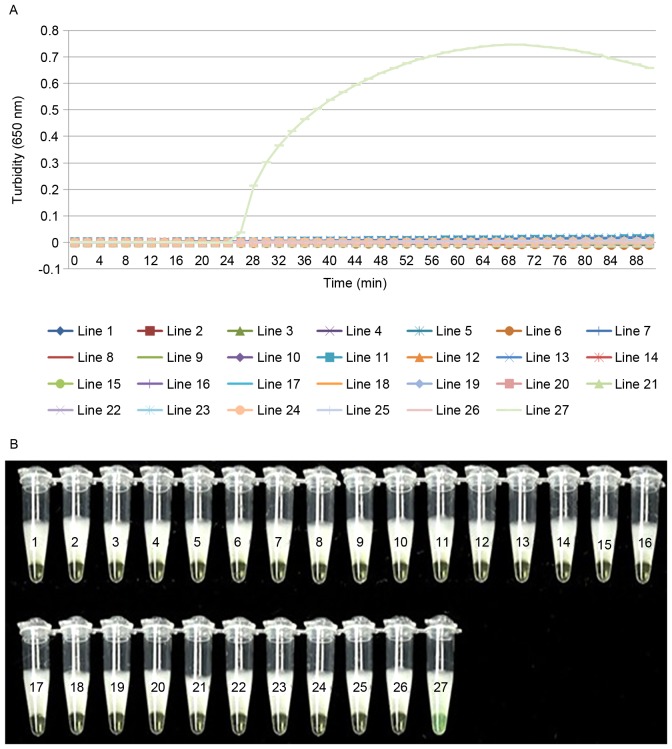
To evaluate the specificity of LAMP detection for cdtA and cdtB, genomic DNA extracted from C. difficile was used as a positive control, ddH2O as the negative control and 25 different strains of pathogen were used as test strains. The two detection methods gave the same results and no reactivity was recorded with any other pathogenic bacterial strains or the negative control (Figs. 3 and 4). This demonstrates that the LAMP assay used in the present study is specific to C. difficile.
Figure 3.
LAMP of cdtA has a high degree of specificity. Determination of the specificity of cdtA detection of the LAMP assay by (A) real-time turbidimeter or (B) visual detection after the addition of calcein to the reaction system. Amplification was performed at 60°C for 60 min. 1, Bacillus megaterium; 2, Vibrio piscium; 3, Pseudomonas maltophilia; 4, Mycobacterium tuberculosis 4368; 5, Vibrio cholera O139; 6, Bacillus anthracis; 7, enterohemorrhagic Escherichia coli; 8, Yersinia enterocolitica; 9, Vibrio parahaemolyticus (5474); 10, Enteropathic Escherichia coli; 11, enteroadherent Escherichia coli; 12, Enteroinvasive Escherichia coli; 13, Enterotoxigenic Escherichia coli; 14, Yersinia pestis; 15, Streptococcus pneumoniae; 16, Neisseria meningitides group B CMCC29022; 17, Burkholderia pseudomallei; 18, Methicillin resistant Staphylococcus aureus; 19, Acinetobacter baumannii; 20, Escherichia coli; 21, Bordetella pertussis ATCC18530; 22, Haemophilus influenzae; 23, Corynebacterium diphtheriae CMCC38001; 24, Mycobacterium tuberculosis 4368; 25, Neisseria meningitides (NM29019); 26, negative control; 27, positive control; LAMP, loop-mediated isothermal amplification; cdt, Clostridium difficile transferase.
Figure 4.
LAMP of cdtB has a high degree of specificity. Determination of the specificity of cdtB detection of the LAMP assay by (A) real-time turbidimeter or (B) visual detection after the addition of calcein to the reaction system. Amplification was performed at 60°C for 60 min. 1, Bacillus megaterium; 2, Vibrio piscium; 3, Pseudomonas maltophilia; 4, Mycobacterium tuberculosis4368; 5, Vibrio cholera O139; 6, Bacillus anthracis; 7, enterohemorrhagic Escherichia coli; 8, Yersinia enterocolitica; 9, Vibrio parahaemolyticus (5474); 10, enteropathic Escherichia. coli; 11, enteroadherent Escherichia coli; 12, enteroinvasive Escherichia coli; 13, enterotoxigenic Escherichia coli; 14, Yersinia pestis; 15, Streptococcus pneumoniae; 16, Neisseria meningitides group B CMCC29022; 17, Burkholderia pseudomallei; 18, Methicillin resistant Staphylococcus aureus; 19, Acinetobacter baumannii; 20, Escherichia coli; 21, Bordetella pertussis ATCC18530; 22, Haemophilus influenzae; 23, Corynebacterium diphtheriae CMCC38001; 24, Mycobacterium tuberculosis 4368; 25, Neisseria meningitides (NM29019); 26, negative control; 27, positive control; LAMP, loop-mediated isothermal amplification; cdt, Clostridium difficile transferase.
LAMP has a higher sensitivity in the detection of C. difficile compared with PCR
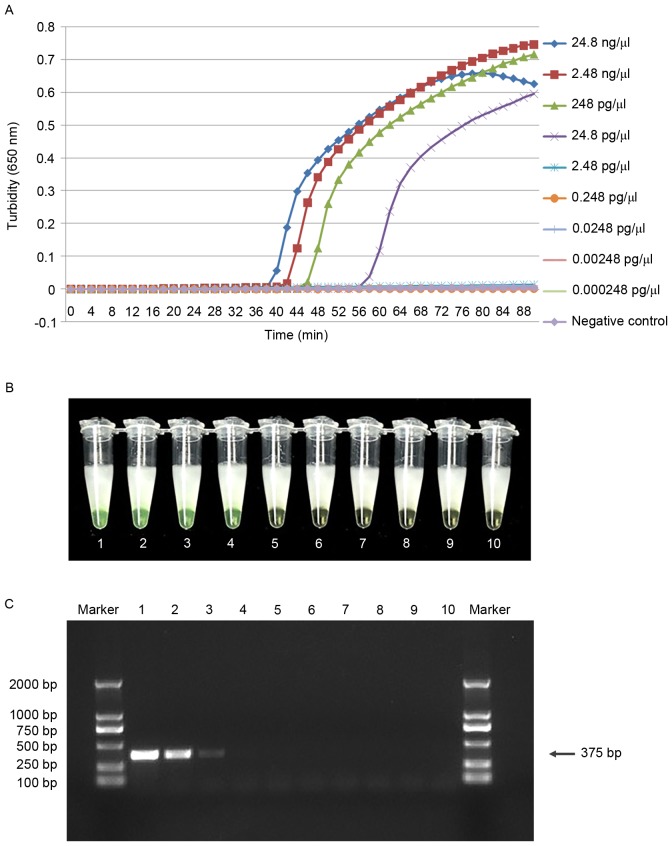
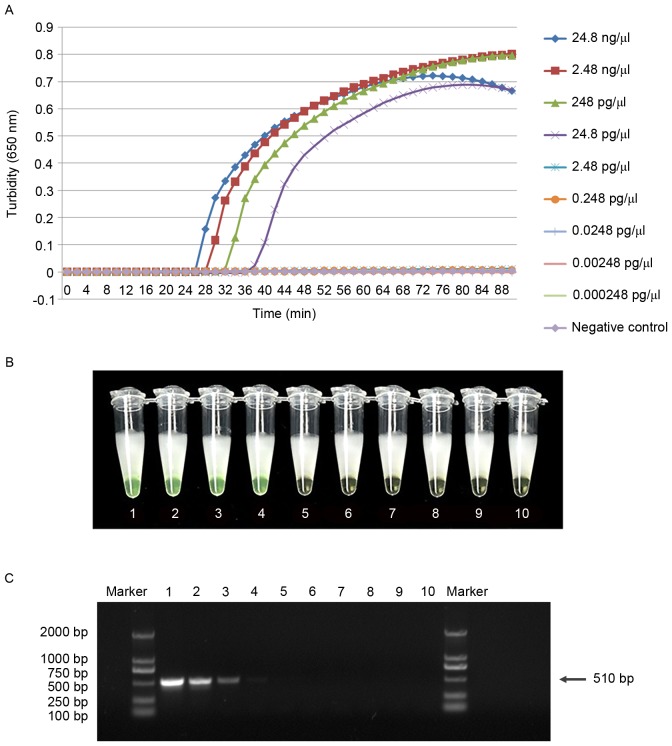
Serial dilutions of genomic DNA were tested using the real-time turbidity monitor, the visual method and PCR. The limit of detection for cdtA and cdtB was 24.8 pg/µl using the visual method and the real-time turbidimeter, which was 10-fold higher than that of the traditional PCR assay (Figs. 5 and 6). This suggests that the LAMP assay is more sensitive than traditional PCR for the detection of C. difficile. In addition, the minimum detectable concentration reaction time was within 60 min. To avoid non-specific amplification, the response time was limited to 60 min.
Figure 5.
LAMP is more sensitive in the detection of cdtA than PCR amplification. Comparative sensitivities of cdtA detection by (A) real-time turbidimeter observations of the LAMP assay, (B) visual observation of the LAMP assay and (C) traditional PCR. Tubes 1–9 and lanes 1–9 contain a 10-fold serial dilution of pure genomic DNA extracted from C. difficile from (24.8 ng/µl to 0.000248 pg/µl); tube 10 and lane 10 contain ddH2O as a negative control. Marker lane, DL-2000. LAMP, loop-mediated isothermal amplification; cdt, Clostridium difficile transferase; PCR, polymerase chain reaction.
Figure 6.
LAMP is more sensitive in the detection of cdtB than PCR amplification. Comparative sensitivities of cdtB detection by (A) real-time turbidimeter observations of the LAMP assay, (B) visual observation of the LAMP assay and (C) traditional PCR. Tubes 1–9 and lanes 1–9 contain a 10-fold serial dilution of pure genomic DNA extracted from C. difficile (from 24.8 ng/µl to 0.000248 pg/µl); tube 10 and lane 10 contain ddH2O as a negative control. Marker lane, DL-2000. LAMP, loop-mediated isothermal amplification; cdt, Clostridium difficile transferase; PCR, polymerase chain reaction.
Discussion
C. difficile is the most prevalent cause of healthcare-associated infectious diarrhea (14). Infection due to CDT-producing strains of C. difficile has been correlated with higher mortality rates compared to CDT negative strain infections (4). A previous study has suggested an association between the presence of CDT in infectious C. difficile strains and the increased mortality of patients (6). In infections due to CDT positive strains, abdominal pain and diarrhea were also reported to be more severe (4). As such, rapid and sensitive laboratory diagnostic testing for CDT positive strains of C. difficile is highly desirable. Diagnostic testing for toxigenic C. difficile has traditionally been accomplished by time-consuming culture methods and by immunoassays, which are faster but in general do not have sufficient sensitivity. Immunoassays that detect the glutamate dehydrogenase antigen display high sensitivity but poor specificity for C. difficile (15). By comparison, molecular tests have increased sensitivity and specificity (16). To the best of our knowledge, there have been no studies on the detection of CDT encoded genes. The present study investigated the efficacy of LAMP to directly detect the cdtA and cdtB genes in order to aid in the diagnosis of binary toxin-associated C. difficile infection. The cdtA and cdtB genes were targeted by specific primer sets in this molecular-based assay with rapid turnaround times. Although this method has been applied for the detection and identification of other microbial pathogens (17), to the best of our knowledge this is the first time this technique has been applied to detect CDT positive C. difficile. The LAMP assay does not require skilled operators or precise instruments, and is therefore practically applicable within point of care settings with poor resources. Since its introduction by Notomi et al (8) in 2000, it has been widely applied to a variety of fields, including clinical diagnosis, food safety, livestock breeding and determination of sex (18,19). This is largely due to its rapid and simple methodology. Numerous reports have demonstrated that LAMP offers high sensitivity and specificity for the detection of pathogens (20,21).
The present study investigated a visual LAMP method for detecting the cdtA and cdtB genes of C. difficile. The specificity of this assay was confirmed by testing strains of 26 bacterial species, including C. difficile, using a real-time turbidimeter and visual methods. A positive result was only obtained for C. difficile, all other species gave a negative result. This confirmed the specificity of the assay for detecting C. difficile, and indicated that the turbidimeter and visual detection methods exhibited the same level of specificity. The sensitivity of the LAMP assay for detecting the cdtA and cdtB genes was tested using a 10-fold serial dilution of pure genomic DNA extracted from C. difficile. Consistent with previous reports, LAMP was identified to be 10-fold more sensitive than PCR analysis, with a detection limit of 24.8 pg/µl for the real-time turbidimeter and visual methods. Furthermore, the LAMP assay was completed within 1.5 h using a simple thermostat, whereas PCR took 3 h and required a specialized thermocycler. The LAMP assay offers a simpler method and a faster result, and is therefore preferential for urgent diagnosis in the field.
In conclusion, the visual LAMP assay developed for detecting CDT positive C. difficile strains in the present study has a number of advantages over regular PCR amplification, such as the ability for results to be observed with the naked eye immediately after amplification is complete. The LAMP assay is also more sensitive than PCR in detecting CDT positive C. difficile. The results obtained with this visual method were consistent with the real-time turbidimeter observations. Consequently, this procedure offers a faster, simpler, more sensitive and lower cost method for diagnosing CDT-producing C. difficile, which may be particularly useful within point of care settings. No rapid diagnostic tool is available for detecting CDT-producing C. difficile at present, and this assay could be a useful and reliable diagnostic tool for infection control in clinical settings, even those lacking a clinical microbiology laboratory. Rapid diagnosis can be crucial during an infection outbreak and may potentially decrease the nosocomial transmission, morbidity and mortality associated with C. difficile infection.
Acknowledgements
The present study was supported by the Innovation and Development Fund of the Navy General Hospital (Beijing, China; grant no. KT-0821).
References
- 1.Doosti A, Mokhtari-Farsani A. Study of the frequency of Clostridium difficile tcdA, tcdB, cdtA and cdtB genes in feces of Calves in south west of Iran. Ann Clin Microbiol Antimicrob. 2014;13:21. doi: 10.1186/1476-0711-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norén T, Unemo M, Magnusson C, Eiserman M, Matussek A. Evaluation of the rapid loop-mediated isothermal amplification assay Illumigene for diagnosis of Clostridium difficile in an outbreak situation. APMIS. 2014;122:155–160. doi: 10.1111/apm.12121. [DOI] [PubMed] [Google Scholar]
- 3.Boyanton BL, Jr, Sural P, Loomis CR, Pesta C, Gonzalez-Krellwitz L, Robinson-Dunn B, Riska P. Loop-mediated isothermal amplification compared to real-time PCR and enzyme immunoassay for toxigenic Clostridium difficile detection. J Clin Microbiol. 2012;50:640–645. doi: 10.1128/JCM.01014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J, Seo MR, Kang JO, Choi TY, Pai H. Clinical and Microbiologic characteristics of Clostridium difficile infection caused by binary toxin producing strain in Korea. Infect Chemother. 2013;45:175–183. doi: 10.3947/ic.2013.45.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papatheodorou P, Hornuss D, Nölke T, Hemmasi S, Castonguay J, Picchianti M, Aktories K. Clostridium difficile binary toxin CDT induces clustering of the lipolysis-stimulated lipoprotein receptor into lipid rafts. MBio. 2013;4:e00244–e00213. doi: 10.1128/mBio.00244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerding DN, Johnson S, Rupnik M, Aktories K. Clostridium difficile binary toxin CDT: Mechanism, epidemiology, and potential clinical importance. Gut Microbes. 2014;5:15–27. doi: 10.4161/gmic.26854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silva RO, Santos RL, Pires PS, Pereira LC, Pereira ST, Duarte MC, de Assis RA, Lobato FC. Detection of toxins A/B and isolation of Clostridium difficile and Clostridium perfringens from dogs in Minas Gerais, Brazil. Braz J Microbiol. 2013;44:133–137. doi: 10.1590/S1517-83822013005000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JB, Han AR, Park EY, Kim JY, Cho W, Lee J, Seo EK, Lee KT. Inhibition of LPS-induced iNOS, COX-2 and cytokines expression by poncirin through the NF-kappaB inactivation in RAW 264.7 macrophage cells. Biol Pharm Bull. 2007;30:2345–2351. doi: 10.1248/bpb.30.2345. [DOI] [PubMed] [Google Scholar]
- 10.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 11.Gandelman O, Jackson R, Kiddle G, Tisi L. Loop-mediated amplification accelerated by stem primers. Int J Mol Sci. 2011;12:9108–9124. doi: 10.3390/ijms12129108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu C, Jiang DN, Xiang GM, Luo FK, Liu LL, Yu JC, Pu XY. DNA detection of Clostridium difficile infection based on real-time resistance measurement. Genet Mol Res. 2013;12:3296–3304. doi: 10.4238/2013.September.3.6. [DOI] [PubMed] [Google Scholar]
- 13.Mori Y, Nagamine K, Tomita N, Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. 2001;289:150–154. doi: 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- 14.Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, Rudensky AY. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 15.Pollock NR. Ultrasensitive detection and quantification of toxins for optimized diagnosis of Clostridium difficile infection. J Clin Microbiol. 2016;54:259–264. doi: 10.1128/JCM.02419-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hicke B, Pasko C, Groves B, Ager E, Corpuz M, Frech G, Munns D, Smith W, Warcup A, Denys G, et al. Automated detection of toxigenic Clostridium difficile in clinical samples: Isothermal tcdB amplification coupled to array-based detection. J Clin Microbiol. 2012;50:2681–2687. doi: 10.1128/JCM.00621-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansour SM, Ali H, Chase CC, Cepica A. Loop-mediated isothermal amplification for diagnosis of 18 World Organization for Animal Health (OIE) notifiable viral diseases of ruminants, swine and poultry. Anim Health Res Rev. 2015;16:89–106. doi: 10.1017/S1466252315000018. [DOI] [PubMed] [Google Scholar]
- 18.Yang L, Li J, Bi Y, Xu L, Liu W. Development and application of a reverse transcription loop-mediated isothermal amplification method for rapid detection of Duck hepatitis A virus type 1. Virus Genes. 2012;45:585–589. doi: 10.1007/s11262-012-0798-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirayama H, Kageyama S, Moriyasu S, Sawai K, Minamihashi A. Embryo sexing and sex chromosomal chimerism analysis by loop-mediated isothermal amplification in cattle and water buffaloes. J Reprod Dev. 2013;59:321–326. doi: 10.1262/jrd.2013-028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seki M, Yamashita Y, Torigoe H, Tsuda H, Sato S, Maeno M. Loop-mediated isothermal amplification method targeting the lytA gene for detection of Streptococcus pneumoniae. J Clin Microbiol. 2005;43:1581–1586. doi: 10.1128/JCM.43.4.1581-1586.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim DW, Kilgore PE, Kim EJ, Kim SA, Anh DD, Dong BQ, Kim JS, Seki M. The enhanced pneumococcal LAMP assay: A clinical tool for the diagnosis of meningitis due to Streptococcus pneumoniae. PLoS One. 2012;7:e42954. doi: 10.1371/journal.pone.0042954. [DOI] [PMC free article] [PubMed] [Google Scholar]