Abstract
Background
Classically, Crotalus durissus terrificus (Cdt) venom can be described, according to chromatographic criteria, as a simple venom, composed of four major toxins, namely: gyroxin, crotamine, crotoxin and convulxin. Crotoxin is a non-covalent heterodimeric neurotoxin constituted of two subunits: an active phospholipase A2 and a chaperone protein, termed crotapotin. This molecule is composed of three peptide chains connected by seven disulfide bridges. Naturally occurring variants/isoforms of either crotoxin or crotapotin itself have already been reported.
Methods
The crude Cdt venom was separated by using RP-HPLC and the toxins were identified by mass spectrometry (MS). Crotapotin was purified, reduced and alkylated in order to separate the peptide chains that were further analyzed by mass spectrometry and de novo peptide sequencing.
Results
The RP-HPLC profile of the isolated crotapotin chains already indicated that the α chain would present isoforms, which was corroborated by the MS and tandem mass spectrometry analyses.
Conclusion
It was possible to observe that the Cdt crotapotin displays a preferred amino acid substitution pattern present in the α chain, at positions 31 and 40. Moreover, substitutions could also be observed in β and γ chains (one for each). The combinations of these four different peptides, with the already described chains, would produce ten different crotapotins, which is compatible to our previous observations for the Cdt venom.
Keywords: Crotalus durissus terrificus, Venom, Crotoxin, Crotapotin, Isoforms
Background
Snake venoms are complex mixtures rich in proteins and peptides, in which such molecules can comprise up to 95% of the venom dry weight [1, 2]. Such molecules do aid the animal survival, once they may be used either as a hunting tool or as a defense mechanism [3]. Moreover, these toxins are also involved in ophidian accidents [4]. Crotoxin, the major Crotalus durissus terrificus (Cdt) venom toxin, is also the most toxic [5, 6]. It is a heterodimeric neurotoxin comprised of a basic phospholipase A2 (PLA2) and an acidic protein, also known as crotapotin [7, 8].
Crotapotin, a 9.6-kDa peptide displaying a pI of 3.4, was initially characterized as a chaperone since the PLA2 would increase its toxicity and inhibit the PLA2 activity [9–12]. However, this peptide has also been described as presenting anti-inflammatory activity and being able to modulate the humoral immunity, including in some neurodegenerative autoimmune disorders [13–17].
Structurally, crotapotin is composed of three peptide chains, connected by seven disulfide bonds [16, 18]. These chains, called α, β and γ, were first sequenced in 1985 and determined to be composed of 40 (α-chain), 35 (β-chain) and 14 (γ-chain) amino acids [19, 20]. However, some authors have observed the occurrence of natural variations of crotapotin [17, 21].
In the present study, we have developed a method for the isolation and biochemical characterization of crotapotin from crude Cdt venom, including the chromatographic separation of the peptide chains after reduction and alkylation, and de novo mass spectrometry peptide sequencing.
Methods
Venoms and animals
Pooled Cdt venom was obtained from snakes kept in the Center for the Studies of Venoms and Venomous Animals (CEVAP) of UNESP, in Botucatu (Brazil). All procedures involving snake specimens were in accordance with the ethical standards of the institutional and/or national research committee. The study was approved by the responsible Ethics Committee on Animal Use of Botucatu Medical School (protocol no 1145/2015 – CEUA).
RP-HPCL
A 10 mg.mL−1 crude Cdt venom solution (0.1% trifluoroacetic acid – TFA) was centrifuged (3800 x g) and separated by RP-HPLC using a Luna C8 column (100 A, 250 × 10 mm, Phenomenex) coupled to a Shimadzu Proeminence binary HPLC system. A 20–40% linear gradient of B (90% acetonitrile – ACN, containing 0.1% TFA) over A (0.1% TFA) was used for 40 min after initial isocratic elution for 5 min, under a constant flow of 5 mL.min−1. UV monitoring was performed at 214 nm and fractions were manually collected. The reduced and alkylated crotapotin chains were separated by a Shimpack C18 column (100 A, 10 × 4.6 mm, Shimadzu), using a 0–50% linear gradient of B, for 20 min, under constant flow of 1 mL.min−1. UV monitoring was performed at 225 nm.
Chemical processing
Isolated crotapotin was reduced with 500 mM DTT in 50 mM NH4HCOOH for 25 min at 56 °C and alkylated with 500 mM IAA for 30 min, at 25 °C, protected from light. The isolated α and β chains were chemically hydrolyzed with 70% formic acid for 48 h, at 37 °C. Reaction was stopped by water addition followed by lyophilization.
Mass spectrometry and de novo peptide sequencing
A Bruker ESI-Q-TOF instrument, coupled to a Prominence Shimadzu binary HPLC, was employed for MS and MS/MS experiments. Samples were placed in the autosampler holder and submitted to a 10–80% linear gradient of B for 15 min, under constant flow of 0.2 mL.min−1, using a Shimpack C18 column (100 A, 10 × 2 mm). CID fragmentation for MS/MS experiments was performed with N2. Data were acquired under a 50–2000 m/z window and processed by Peaks Studio Suite.
Results
Crotapotin isolation
Figure 1 presents the C8-RP-HPLC profile of the crude Cdt venom separated according to the Methods section. Six fractions (F1-F6) were manually collected and submitted to MS analyses in order to identify the known toxins. F1 and F2 are crotamins, F3 corresponds to crotapotin and F4, F5 and F6 are PLA2s. The minor peaks were not collected or analyzed by MS. F3 was then submitted to another chromatographic step (Fig. 1, inset) in order to assess its homogeneity and the molecular mass was determined (Fig. 2). Figure 2 already points out to the presence of more than one molecule; however, the charge states are only indicated for the major ions.
Fig. 1.
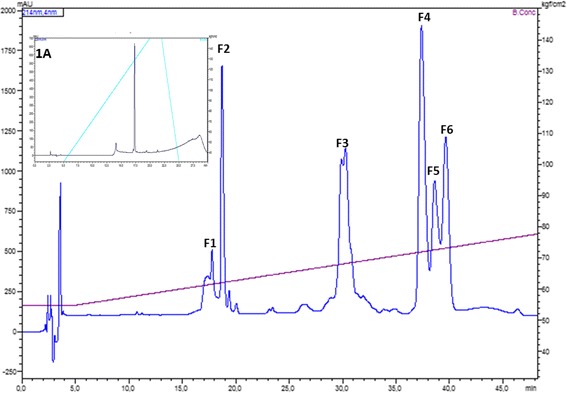
RP-HPLC profile of the crude Cdt venom. F1 to F6 correspond to the manually collected fractions. F1 and F2: crotamin; F3: crotapotin; F4, F5 and F6: PLA2. UV monitoring 214 nm. Inset: F3 analytical RP-HPLC demonstrating the proper molecule isolation. Chromatographic conditions are described in Methods section
Fig. 2.
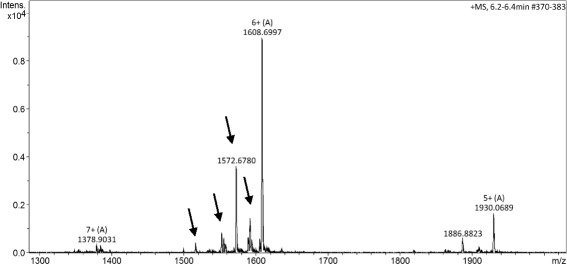
F3 ESI+ MS spectrum. The charge states of the major ions are presented above the m/z value. The presence of isoforms is indicated by the arrows for the [M + 6H]6+ ion
Crotapotin chains isolation
Following reduction and alkylation, as described in Methods, the processed crotapotin was subjected to RP-HPLC chromatographic separation in order to obtain the isolated α, β and γ chains. Figure 3a and b (zoomed chromatogram) present the chain separation. Major peaks in the chromatogram correspond to the reagents (data not shown). The chains were identified based on their molecular masses, as presented in Fig. 4a–c.
Fig. 3.
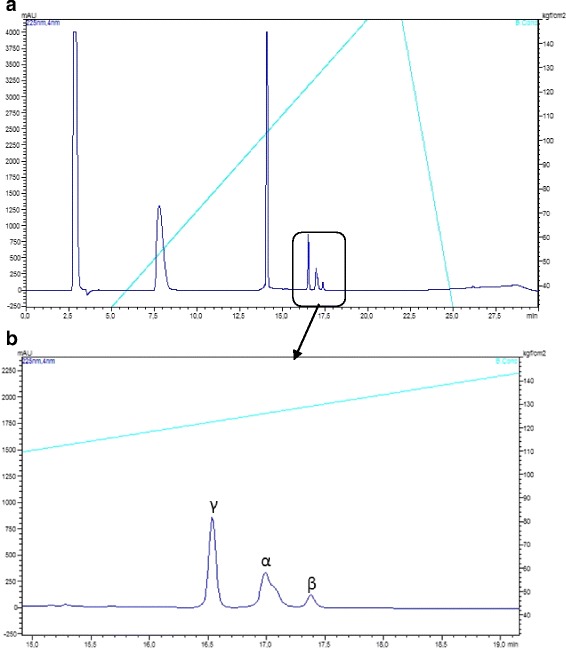
a Reduced and alkylated crotapotin (F3) RP-HPLC separation chromatographic profile. b Zoomed region with the identification of the individual chains. UV monitoring 225 nm. The major peaks in A correspond to the alkylation reagents
Fig. 4.
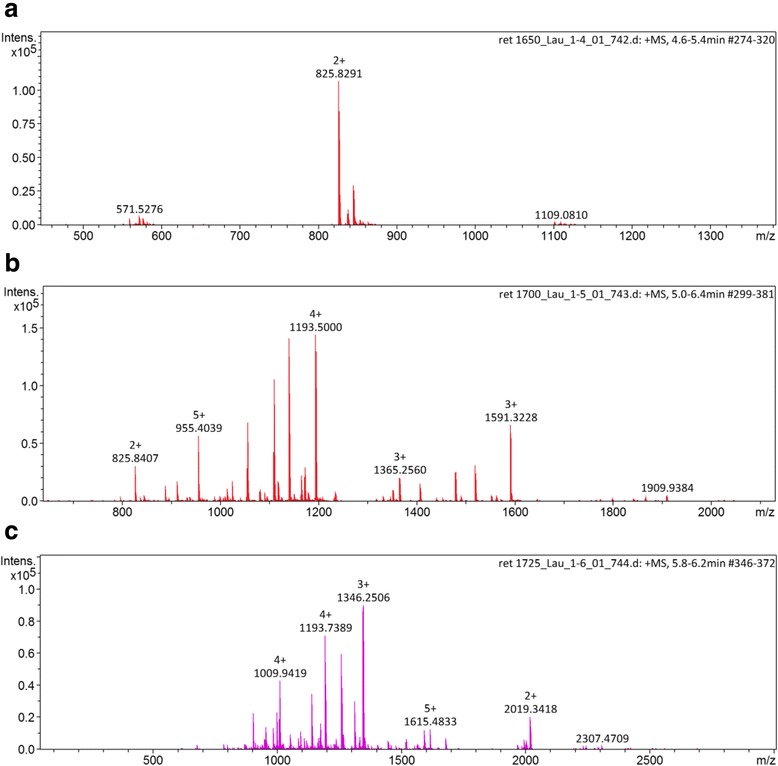
MS spectrum of the (a) γ, (b) β and (c) α chains. The charge states of the major ions are presented above the m/z value. The lack of homogeneity indicates the presence of isoforms
De novo peptide sequencing
The isolated formic acid hydrolyzed crotapotin chains were analyzed by LC-MS/MS (ESI-Q-TOF) and the fragmentation spectra are presented in Fig. 5a, b and c. Only the spectra of the isoforms are presented. Several spectra, corresponding to the known/deposited sequences, were obtained, but are not presented.
Fig. 5.
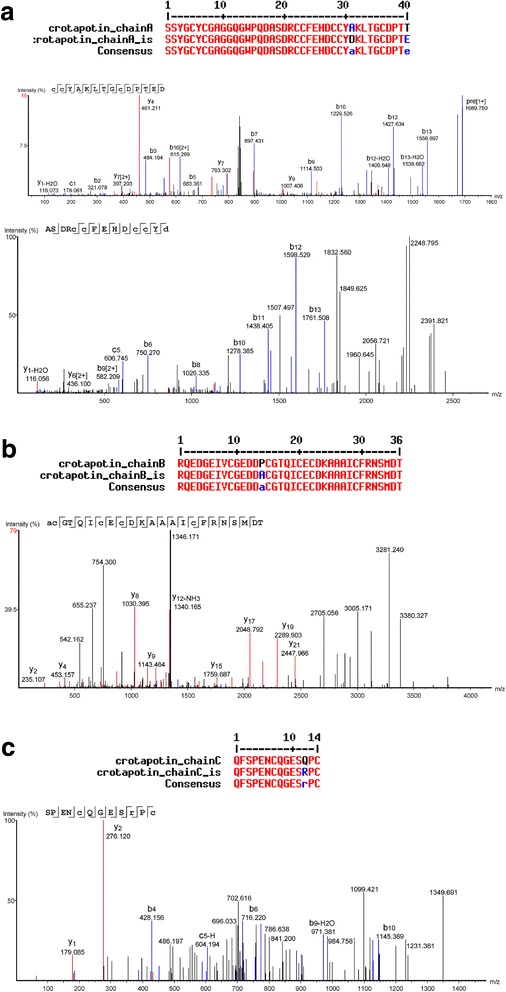
Representative annotated interpreted CID fragmentation spectra of the de novo sequenced isoforms of crotapotin (a) α, (b) β and (c) γ chains. Above each chain, the aligned sequences presenting the amino acid substitution are shown
Discussion
Crotoxin is a β neurotoxin, composed of two subunits: an active PLA2 and the catalytic inactive crotapotin [22, 23]. Since most venoms and toxins present isoforms, a consequence of an evolutionary strategy, we have chosen to evaluate whether there would be a preferred substitution site in a given crotapotin subunit that would give rise to the previously observed isoforms [21].
In order to achieve this goal, we developed a chromatographic method for the separation of crotapotin from the crude Cdt venom, and another method for the obtainment of the reduced and alkylated subunits. Moreover, due to the particular amino acid composition of the subunits, classical proteomics approaches (based on trypsin hydrolysis) could not be performed. Conversely, we have opted for the formic acid hydrolysis to adjust the peptide size to the CID fragmentation requirements. Interestingly, such strategy helped in the identification of a particular α-chain isoform, in which the deposited Ala31 residue was replaced by an Asp, yielding a new formic acid cleavage site that was successfully used in the de novo sequencing process (Fig. 5a).
According to our analyses, what happens in the venom gland is merely amino acid substitutions and not alternate processing, i.e., we were not able to detecte longer or shorter chains, only isoforms regarding residue changes [9]. Most of the isoforms did present molecular masses close to the already known molecule [16].
During the course of the work, we successfully covered 97% of crotapotin using the proteomics/de novo sequencing (data not shown). However, few spectra did not match the deposited sequence and, by using a combination of the Spider algorithm of Peaks Studio and manually checking the spectra for correction, we were able to identify four amino acid substitutions. Interestingly, the α chain bears more amino acid substitutions, as the asymmetrical HPLC peak already indicated (Fig. 3b). Since the α and β chains are homologous to the PLA2, such preference for mutation in these chains may have a counter part in the isoforms also observed for the Cdt PLA2 itself [21, 24].
Our de novo data specifically indicates the Thr → Glu77, Ala → Asp68 substitutions in the α chain. Although the Thr → Glu77 substitution would retain the hydrogen bond capabilities (but with the addition of a true charge), the Ala → Asp68 substitution seems more disturbing. Such mutation is located in an α helix and the introduction of a charged residue may alter such structure. Moreover, this particular region is mostly involved in the PLA2 interaction [23].
In the β chain, the Pro → Ala98 substitution would evoke the same type of alteration. Proline is a rigid, structurally relevant amino acid, typically present in protein ‘turn’ regions. On the other hand, alanine is a much more flexible amino acid. Such substitution would relax this molecule region, allowing for more flexibility and, therefore, different types of intermolecular interactions.
The γ chain (also known as crotalphine [25]) presents a Gln → Arg136 substitution. This modification has already been reported by Konno et al. [25] and is described as not capable of altering the analgesic properties of this peptide. This is in agreement with our proposal based on the conservation of the electrostatic characteristics of the residue that would retain the hydrogen bond-forming capabilities, in spite of the addition of a charge.
Unfortunately, the absence of genomic (or transcriptomic) data makes it more difficult to characterize other amino acid substitutions that are certainly occurring but, due to the lower relative concentration levels, have not yielded high quality spectra, suitable for the de novo sequencing (data not shown). Few studies report other amino acid substitutions, such as residue 84 of the β chain [26].
Evolution has long been ‘experimenting’ with amino acids substitutions in proteins and peptides in order to increase venom efficiency and efficacy as well as avoid prey evasive strategies [27, 28]. Not only that, but also synergism aroused by the presence of several isoforms of a given molecule also increases toxicity [29]. If one takes into account the biotechnological appeal of the clinical use of toxins, such isoforms may hint to specific targets when given residues in the toxins are substituted; the Conus toxins are a good example of this concept [30].
Conclusion
The mere existence of venoms and toxic animal secretions is itself a demonstration of an unbalanced (or biased) evolutionary strategy happening in a given organism. Toxins are molecules that must act in another organism, and not against the producing animal. Therefore, regulation mechanisms must exist to prevent local damage. They may include, but are not limited to, the presence of concentrated low affinity inhibitors; the absence of ionic co-factors; the lack of catalytic activity due to conformation restrains; the pH of the media; the timing of the activation of the precursor, among others. Certainly, one of those strategies targeted to increase the efficiency and efficacy of the venom is the presence of isoforms of a given toxin, as herein reported.
Acknowledgments
Special thanks are due to Mass Spectrometry Laboratory group of the Center for the Study of Venoms and Venomous Animals (CEVAP) of UNESP for their support on LC-MS/MS analysis.
Funding
This study was partly supported by grants from the São Paulo Research Foundation (FAPESP – proc. no 2012/08101–8) to RSFJ and from the Coordination for the Improvement of Higher Education Personnel (CAPES – Edital Toxinologia no 063/2010, proc. no 23038.006285/2011–21, AUXPE Toxinologia 1219) to BB. RSFJr is a CNPq DTI fellow researcher (310,395/2014–3). DCP is a fellow CNPq researcher (303,792/2016–7).
Abbreviations
- ACN
Acetonitrile
- Cdt
Crotalus durissus terrificus
- MS
Mass spectrometry
- MS/MS
Tandem mass spectrometry
- PLA2
Phospholipase A2
- TFA
Trifluoroacetic acid
Authors’ contributions
LAO, DCP and LDS designed the study, collected and analyzed data, and wrote the manuscript. FCTC and LCB collected and analyzed data. RSFJ, BB and DCP reviewed the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval
All procedures involving snake specimens were in accordance with the ethical standards of the institutional and/or national research committee. The present study was approved by the responsible Ethics Committee on Animal Use of Botucatu Medical School (protocol no 1145/2015 – CEUA).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests. One of the authors of this article, Dr. Benedito Barraviera, is the Editor-in-Chief of Journal of Venomous Animals and Toxins including Tropical Diseases. He did not get involved in the peer review process of this manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Laudicéia Alves de Oliveira, Email: laudiceia.biomed@gmail.com.
Rui Seabra Ferreira, Jr, Email: rseabra@cevap.unesp.br.
Benedito Barraviera, Email: bbviera@gnosis.com.br.
Francilene Capel Tavares de Carvalho, Email: francilene_capel@hotmail.com.
Luciana Curtolo de Barros, Email: luciana@cevap.unesp.br.
Lucilene Delazari dos Santos, Email: lucilenebio@gmail.com.
Daniel Carvalho Pimenta, Phone: +55 11 2627 9746, Email: dcpimenta@butantan.gov.br.
References
- 1.de la Rosa G, Pastor N, Alagón A, Corzo G. Synthetic peptide antigens derived from long-chain alpha-neurotoxins: Immunogenicity effect against elapid venoms. Peptides. 2017;88:80–86. doi: 10.1016/j.peptides.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Kerkis I, Hayashi MA, Prieto da Silva AR, Pereira A, De Sá Júnior PL, Zaharenko AJ, et al. State of the art in the studies on crotamine, a cell penetrating peptide from South American rattlesnake. Biomed Res Inter. 2014;2014:Article ID 675985. doi:10.1155/2014/675985. [DOI] [PMC free article] [PubMed]
- 3.Warrell DA. Snake bite. Lancet. 2010;375(9708):77–88. doi: 10.1016/S0140-6736(09)61754-2. [DOI] [PubMed] [Google Scholar]
- 4.Oliveira LMF, Ullah A, Masood R, Zelanis A, Spencer PJ, Serrano SMT, et al. Rapid purification of serine proteinases from Bothrops alternatus and Bothrops moojeni venoms. Toxicon. 2013;76:282–290. doi: 10.1016/j.toxicon.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Rangel-Santos A, Dos-Santos EC, Lopes-Ferreira M, Lima C, Cardoso DF, Mota I. A comparative study of biological activities of crotoxin and CB fraction of venoms from Crotalus durissus terrificus, Crotalus durissus cascavella and Crotalus durissus collilineatus. Toxicon. 2004;43(7):801–810. doi: 10.1016/j.toxicon.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Madey JJ, Price AB, Dobson JV, Stickler DE, McSwain SD. Facial diplegia, pharyngeal paralysis, and ophthalmoplegia after a timber rattlesnake envenomation. Pediatr Emerg Care. 2013;29(11):1213–1216. doi: 10.1097/PEC.0b013e3182aa472f. [DOI] [PubMed] [Google Scholar]
- 7.Vieira LF, Magro AJ, Fernandes CA, de Souza BM, Cavalcante WL, Palma MS, et al. Biochemical, functional, structural and phylogenetic studies on Intercro, a new isoform phospholipase A2 from Crotalus durissus terrificus snake venom. Biochimie. 2013;95(12):2365–2375. doi: 10.1016/j.biochi.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 8.Lomeo RS, Gonçalves AP, da Silva CN, de Paula AT, Costa Santos DO, Fortes-Dias CL, et al. Crotoxin from Crotalus durissus terrificus snake venom induces the release of glutamate from cerebrocortical synaptosomes via N and P/Q calcium channels. Toxicon. 2014;85:5–16. doi: 10.1016/j.toxicon.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 9.Bouchier C, Boulain JC, Bon C, Ménez A. Analysis of cDNAs encoding the two subunits of crotoxin, a phospholipase A2 neurotoxin from rattlesnake venom: the acidic non enzymatic subunit derives from a phospholipase A2-like precursor. Biochim Biophys Acta. 1991;1088(3):401–408. doi: 10.1016/0167-4781(91)90132-6. [DOI] [PubMed] [Google Scholar]
- 10.Ponce-Soto LA, Toyama MH, Hyslop S, Novello JC, Marangoni S. Isolation and preliminary enzymatic characterization of a novel PLA2 from Crotalus durissus collilineatus venom. J Protein Chem. 2002;21(3):131–136. doi: 10.1023/A:1015332314389. [DOI] [PubMed] [Google Scholar]
- 11.Bon C, Changeux JP, Jeng TW, Fraenkel-Conrat H. Postsynaptic effects of crotoxin and of its isolated subunits. Eur J Biochem. 1979;99(3):471–481. doi: 10.1111/j.1432-1033.1979.tb13278.x. [DOI] [PubMed] [Google Scholar]
- 12.Soares AM, Mancin AC, Cecchini AL, Arantes EC, França SC, Gutiérrez JM, et al. Effects of chemical modifications of crotoxin B, the phospholipase A2 subunit of crotoxin from Crotalus durissus terrificus snake venom, on its enzymatic and pharmacological activities. Int J Biochem Cell Biol. 2001;33(9):877–888. doi: 10.1016/S1357-2725(01)00065-6. [DOI] [PubMed] [Google Scholar]
- 13.Landucci EC, Toyama M, Marangoni S, Oliveira B, Cirino G, Antunes E, et al. Effect of crotapotin and heparin on the rat paw oedema induced by different secretory phospholipase A2. Toxicon. 2000;38(2):199–208. doi: 10.1016/S0041-0101(99)00143-9. [DOI] [PubMed] [Google Scholar]
- 14.Yonamine CM, Kondo MY, Juliano MA, Icimoto MY, Baptista GR, Yamane T, et al. Kinetic characterization of gyroxin, a serine protease from Crotalus durissus terrificus venom. Biochimie. 2012;94(12):2791–2793. doi: 10.1016/j.biochi.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 15.Nunes FP, Zychar BC, Della-Casa MS, Sampaio SC, Gonçalves LR, Cirillo MC. Crotoxin is responsible for the long-lasting anti-inflammatory effect of Crotalus durissus terrificus snake venom: involvement of formyl peptide receptors. Toxicon. 2010;55(6):1100–1106. doi: 10.1016/j.toxicon.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 16.Dos-Santos MC. Crotoxina e Crotoxina-símile isoladas de venenos de subespécies de Crotalus durrisus e suas múltiplas atividades biológicas. Sci Amazon. 2014;3(1):102–105. [Google Scholar]
- 17.Castro FR, Farias AS, Proença PL, de La Hoz C, Langone F, Oliveira EC. The effect of treatment with crotapotin on the evolution of experimental autoimmune neuritis induced in Lewis rats. Toxicon. 2007;49(3):299–305. doi: 10.1016/j.toxicon.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 18.Bon C. Synergism of the two subunits of crotoxin. Toxicon. 1982;20(1):105–109. doi: 10.1016/0041-0101(82)90173-8. [DOI] [PubMed] [Google Scholar]
- 19.Aird SD, Kaiser II, Bieber AL. Nomenclature of biological toxins. Toxicon. 1985;23(2):179–187. doi: 10.1016/0041-0101(85)90133-3. [DOI] [PubMed] [Google Scholar]
- 20.Aird SD, Kaiser II, Lewis RV, Kruggel WG. A complete amino acid sequence for the basic subunit of crotoxin. Arch Biochem Biophys. 1986;249(2):296–300. doi: 10.1016/0003-9861(86)90005-6. [DOI] [PubMed] [Google Scholar]
- 21.Lourenço A, Jr, Zorzella Creste CF, de Barros LC, Delazari dos Santos L, Pimenta DC, Barraviera B, et al. Individual venom profiling ofrotalus durissus terrifics specimens from a geographically limited region: crotamine assessment and captivity evaluation on the biological activities. Toxicon. 2013;69:75–81. doi: 10.1016/j.toxicon.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Fusco LS, Rodríguez JP, Torres-Huaco F, Huancahuire-Vega S, Teibler P, Acosta O, et al. P9a(Cdt-PLA2) from Crotalus durissus terrificus as good immunogen to be employed in the production of crotalic anti-PLA2 IgG. Toxicol Lett. 2015;238(1):7–16. doi: 10.1016/j.toxlet.2015.06.528. [DOI] [PubMed] [Google Scholar]
- 23.Fernandes CAH, Pazin WM, Dreyer TR, Bicev RN, Cavalcante WL, Fortes-Dias CL, et al. Biophysical studies suggest a new structural arrangement of crotoxin and provide insights into its toxic mechanism. Sci Rep. 2017;7:43885. doi: 10.1038/srep43885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Oliveira DG, Toyama MH, Martins AM, Havt A, Nobre AC, Marangoni S, et al. Structural and biological characterization of a crotapotin isoform isolated from Crotalus durissus cascavella venom. Toxicon. 2003;42(1):53–62. doi: 10.1016/S0041-0101(03)00100-4. [DOI] [PubMed] [Google Scholar]
- 25.Konno K, Picolo G, Gutierrez VP, Brigatte P, Zambelli VO, Camargo AC, et al. Crotalphine, a novel potent analgesic peptide from the venom of the south American rattlesnake Crotalus durissus terrificus. Peptides. 2008;29(8):1293–1304. doi: 10.1016/j.peptides.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Faure G, Guillaume JL, Camoin L, Saliou B, Bon C. Multiplicity of acidic subunit isoforms of crotoxin, the phospholipase A2 neurotoxin from Crotalus durissus terrificus venom, results from posttranslational modifications. Biochemistry. 1991;30(32):8074–8083. doi: 10.1021/bi00246a028. [DOI] [PubMed] [Google Scholar]
- 27.Blanchet G, Alili D, Protte A, Upert G, Gilles N, Tepshi L, et al. Ancentral protein ressurection and engineering opportunities of the mamba aminergic toxins. Sci Rep. 2017;7(1):2701. doi: 10.1038/s41598-017-02953-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang ZM, Yu H, Liu ZZ, Pei JZ, Yang YE, Yan SX, et al. Serine protease isoforms in Gloydius intermedius venom: full sequences, molecular phylogeny and evolutionary implications. J Proteome. 2017;164:19–32. doi: 10.1016/j.jprot.2017.05.024. [DOI] [PubMed] [Google Scholar]
- 29.dos Santos LD, da Silva Menegasso AR, dos Santos Pinto JR, Santos KS, Castro FM, Kalil JE, et al. Proteomic characterization of the multiple forms of the PLAs from the venom of the social wasp Polybia paulista. Proteomics. 2011;11(8):1403–1412. doi: 10.1002/pmic.201000414. [DOI] [PubMed] [Google Scholar]
- 30.Lebbe EKM, Peigneur S, Brullot W, Verbiest T, Titgat J. Ala-7, His-10 and Arg-12 are crucial amino acids for activity of a synthetically engineered μ-conotoxin. Peptides. 2014;53:300–306. doi: 10.1016/j.peptides.2013.07.005. [DOI] [PubMed] [Google Scholar]


