Abstract
GAC repeat expansion from five to seven in the exonic region of the gene for cartilage oligomeric matrix protein (COMP) leads to pseudoachondroplasia, a skeletal abnormality. However, the molecular mechanism by which GAC expansions in the COMP gene lead to skeletal dysplasias is poorly understood. Here we used molecular dynamics simulations, which indicate that an A … A mismatch in a d(GAC)6·d(GAC)6 duplex induces negative supercoiling, leading to a local B-to-Z DNA transition. This transition facilitates the binding of d(GAC)7·d(GAC)7 with the Zα-binding domain of human adenosine deaminase acting on RNA 1 (ADAR1, hZαADAR1), as confirmed by CD, NMR, and microscale thermophoresis studies. The CD results indicated that hZαADAR1 recognizes the zigzag backbone of d(GAC)7·d(GAC)7 at the B–Z junction and subsequently converts it into Z-DNA via the so-called passive mechanism. Molecular dynamics simulations carried out for the modeled hZαADAR1–d(GAC)6.d(GAC)6 complex confirmed the retention of previously reported important interactions between the two molecules. These findings suggest that hZαADAR1 binding with the GAC hairpin stem in COMP can lead to a non-genetic, RNA editing–mediated substitution in COMP that may then play a crucial role in the development of pseudoachondroplasia.
Keywords: CD, molecular dynamics, NMR, RNA editing, trinucleotide repeat disease, cartilage oligomeric matrix protein, human adar1 protein, pseudoachdroplasia
Introduction
Expansion of GAC repeat sequences can be observed in exonic regions of the genome, which can lead to a poly-Asp track in the cartilage oligomeric matrix protein (COMP)3 (1). COMP is a noncollagenous pentameric extracellular matrix protein that is localized in chromosome 19p13.1 (2). It is expressed predominantly in cartilage as well as transiently in tendons, ligaments, smooth muscles, etc. (3, 4) and is important for growth plate organization and its function (5). The COMP gene contains five tandem GAC repeats, and expansion of even one or two repeats causes multiple epiphyseal dysplasia or pseudoachondroplasia, respectively. Such expansion in pseudoachondroplasia results in short stature, early-onset osteoarthritis, and limb dwarfism (3). Mutations in COMP also cause disruption of calcium/ligand binding, intramolecular interactions, and disulfide bond formation (5).
Although an in vitro study has shown that GAC repeats exhibit orientation-dependent instability that subsequently leads to repeat deletion and expansion during replication and transcription, respectively (6), the exact mechanism of how GAC expansion in the COMP gene leads to skeletal dysplasias is poorly understood. Interestingly, a very recent molecular dynamics (MD) study indicates that nonisostericity of an A … A mismatch with respect to the flanking canonical base pairs provokes a left-handed Z-DNA conformation in CAG repeat expansion (7). As d(GAC) repeats can also have similar periodic A … A mismatches, we investigated its effect on DNA conformation using MD simulation and CD studies. Indeed, earlier investigations report that d(GAC)15 (8) and r(CGA)17 (9) form stable hairpin structures that may have periodic A … A mismatches. Previous CD studies also indicate that d(GAC) repeats exhibit multiple conformations and even form parallel duplexes in acidic environments (10, 11). Nonetheless, the stereochemical rationale behind such a non-B-DNA secondary structural preference by GAC repeats and the consequent biological significance are unknown. The MD simulations performed here to explore this clearly underpin that the nonisosteric character of the A … A mismatch leads to a B–Z junction by inducing negative supercoiling akin to CAG repeats (7), as also confirmed by CD. Further, CD and NMR titration experiments reveal for the first time that GAC interacts with the Zα binding domain of human adenosine deaminase acting on RNA (ADAR1) (hereafter hZαADAR1, human Zα-binding domain of ADAR1) through specific recognition of B–Z junctions. A microscale thermophoresis (MST) experiment further reveals that the d(GAC)7·d(GAC)7 duplex interacts with hZαADAR1 with nanomolar binding affinity.
ADAR family proteins mainly catalyze the adenosine-to-inosine editing process in pre-mRNA substrates (12). In fact, the role of hyper/altered A-to-I editing mediated by ADAR in several neurological disorders is well-established (13–16). The N terminus of ADAR1 contains a Z-DNA–binding winged helix–turn–helix domain. Using the MD-derived structure of d(GAC)6·d(GAC)6, the published crystal structure of hZαADAR1 (PDB code 2ACJ), NMR chemical shift mapping of hZαADAR1, and d(GAC)7·d(GAC)7 titration, a complex model is proposed here. Subsequent MD simulation of the complex model confirms the importance of certain amino acids in hZαADAR1 recognizing the B-Z/Z-DNA conformation. Based on hZαADAR1 and d(GAC)·d(GAC) binding studies, a model of how hZαADAR1 can anchor to the Z-philic GAC repeat, facilitate A-to-I editing of the corresponding mRNA transcript of COMP, and lead to pseudoachondroplasia is also proposed.
Results
An A … A mismatch amid G … C and C … G base pairs imposes B–Z junction formation
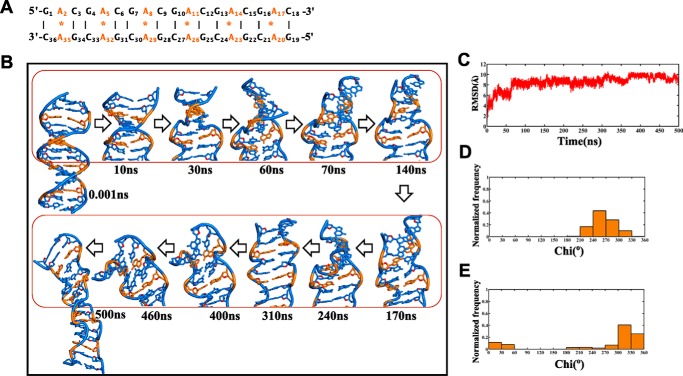
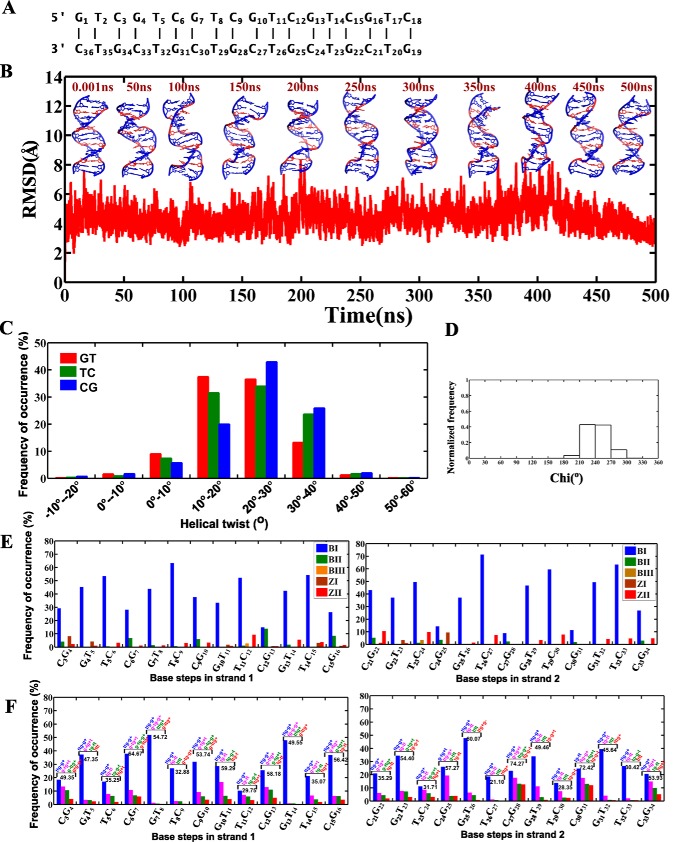
For MD simulation, the d(GAC)6·d(GAC)6 repeat sequence has been considered to complete one helical turn of a DNA duplex (i.e. 10 bases per turn in a normal B-DNA). Flanking sequences (one GAC repeat) on either sides are added to avoid the end-fraying effect during MD simulation. The effect of the A … A mismatch in the d(GAC)6·d(GAC)6 duplex (Fig. 1A) has been investigated at the atomistic level using MD simulations by considering two different starting glycosyl conformations for the mismatch, following earlier studies (7, 17). In the first model, both As are chosen to have an anti conformation (anti … anti). On the other hand, one of the two As in the second model is chosen to be in an anti glycosyl conformation, and the other is chosen to be in a +syn glycosyl conformation (+syn … anti).
Figure 1.
B-DNA to B–Z junction transition in the d(GAC)6.d(GAC)6 duplex with the anti … anti starting glycosyl conformation for the A … A mismatch. A, sequence of the 18-mer DNA duplex that is subjected to MD simulation in this investigation. B, schematic of the duplex at various time intervals during the 500-ns simulation, which indicates formation of a left-handed conformation (shown in boxes). C, time versus RMSD profile showing significant conformational changes in the duplex, as indicated by a high RMSD value with respect to the starting model. D and E, glycosyl torsion (Chi) showing high-anti and ±syn conformational preference for As and Gs, respectively. The noncanonical A … A mismatches are indicated in orange in B.
Starting model with anti … anti glycosyl conformation
Analysis of the 500-ns simulation of the d(GAC)6·d(GAC)6 duplex that comprises six A … A mismatches with anti … anti glycosyl conformation shows that the nonisostericity of the A … A mismatch with respect to canonical G … C and C … G base pairs induces distortions in the helix. Distortions are seen within ∼2 ns of the simulation, become prominent ∼63 ns through the unwinding of the helix, and stay in the same conformation until the end of the simulation (Fig. 1B and supplemental Movie S1). This eventually reflects in the root mean square deviation (RMSD), whose average value stays at ∼4.5 Å between 0.5 to 10.5 ns and ∼6.5 Å between 10.5–60.3 ns before finally reaching the highest value of ∼9 Å (Fig. 1C).
The conformational features that are associated with such helix unwinding are as follows. First, As take up a high anti glycosyl conformation (61%) and exhibit a preference for -syn (39%) transiently during the simulation (Fig. 1D), and Gs that are engaged in canonical hydrogen bonding with Cs profoundly favor a ±syn glycosyl conformation (92%) after 50 ns (Fig. 1E).
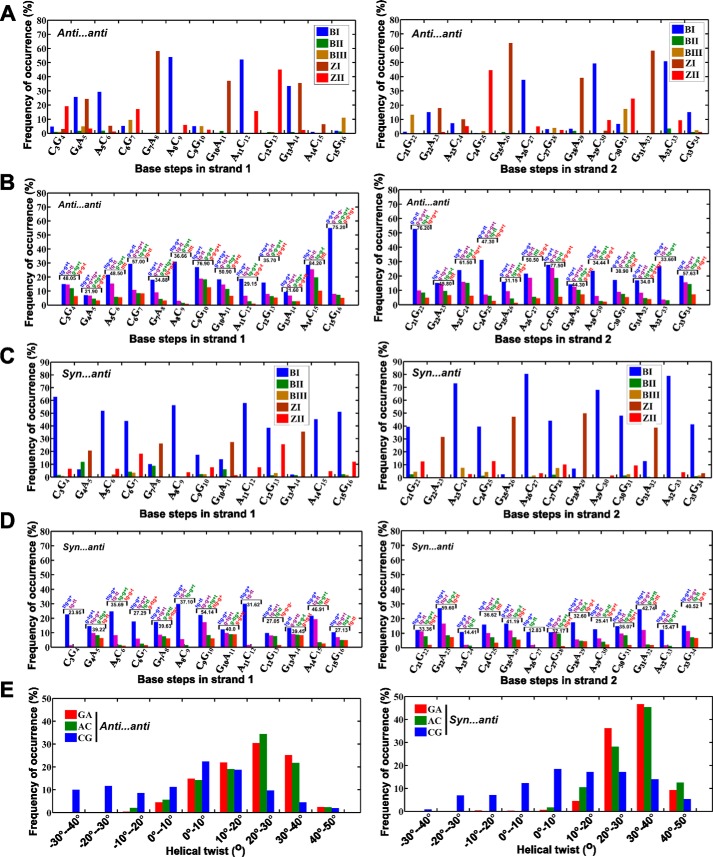
Second, backbone torsion angles (ϵ, ζ, α, γ) that are calculated for the last 400 ns (after reaching the equilibration state) show the possibility of BIII(g−,g−,g−,g+) (18) and Z(ZI = (g−,g+,g+,t)/ZII = (t,g−,t,g+)) (19) conformations at base steps such as GA (44%) and CG (23%), leading to a zigzag backbone (Fig. 2A). Interestingly other unusual conformations, mainly (t,t,g−,g+), (g−,g−,t,t), (t,g−,g+,t), (t,g−,t,t), and (g−,t,g+,t), are also seen predominantly (58%) (Fig. 2B). Such unusual conformations can be attributed to the interaction of cations with the duplex. However, the mechanistic effect arising from the nonisomorphic nature of the A … A mismatch dominates over the effect of counterion interaction with the duplex (20) (supplemental Fig. S1A). AC steps have shown 36% preference for BI(t,g−,g−,g+)/BII(g−,t,g−,g+) conformations, which is higher than GA (11%) and CG (5%) steps (Fig. 2A). Such a mixed occurrence of a variety of conformations at different steps leads to a B–Z junction in the duplex. As a result, the final conformation of the duplex deviates significantly from the starting B-form conformation (Fig. 1B), as also reflected in the RMSD.
Figure 2.
Parameters associated with B–Z junction formation during 101–500-ns simulation time in the duplex containing A … A mismatches with anti … anti and +syn … anti starting glycosyl conformations. A and C, frequency of occurrence of BI, BII, BIII, ZI, and ZII conformations defined in terms of (ϵ,ζ,α,γ) at different steps of the A … A mismatch–containing duplex. Preponderance for BIII/Z conformations compared with BI/BII at GA/CG steps can be seen irrespective of the starting glycosyl conformation. B and D, frequency of occurrence of (ϵ,ζ,α,γ) conformations other than BI, BII, BIII, and Z during two different MD simulations of d(GAC)6·d(GAC)6. Populations corresponding to top four conformations are alone shown. (ϵ,ζ,α,γ) conformational preference and the corresponding frequency of occurrence of the top four populations (%) are given for each base step. E, histogram showing the distribution of helical twists at different steps of the DNA duplex. Strikingly, CG steps exhibit a tendency for lower twists (between −40° to 10°) compared with GA and AC steps. The percentage of such low twists is significantly higher in A … A mismatches compared with the canonical duplex (Fig. 4C).
Concomitantly, helical twist angles at GA, AC, and CG steps also exhibit variations. Among the three steps, CG steps exhibit a higher population of low helical twists (64% of helical twists lower than 10°) compared with AC (22%) and GA (20%) steps (Fig. 2E, left panel). Yet another property that can support the formation of Z-DNA in the midst of B-DNA in the d(GAC)6·d(GAC)6 duplex is the angle formed by three adjacent phosphates. For the average structure calculated over the last 10 ns, the angles at the central phosphate in the following steps are below 110°, which further supports the formation of local Z-DNA (∼110°) (21): G4pA5 (119°), C6pG7 (96°), G7pA8 (90°), C9pG10 (81°), G10pA11 (99°), C12pG13 (92°), G13pA14 (108°), A14pC15 (111°), C15pG16 (85°), A20pC21 (104°), C21pG22 (88°), C24pG25 (93°), G25pA26 (102°), A26pC27 (117°), C27pG28 (87°), G28pA29 (99°), C30pG31 (90°), and G31pA32 (104°). The remaining steps are confined to a B-DNA conformation with an angle at the central phosphate close to ∼150° (supplemental Fig. S2A).
Starting model with +syn … anti glycosyl conformation
To further explore the global conformational preference for the A … A mismatch, another starting conformation, the +syn … anti glycosyl conformation, is considered for the mismatch. This conformation is specifically chosen based on the glycosyl angle preference for the A … A mismatch in the CAG-containing RNA duplex (17, 22).
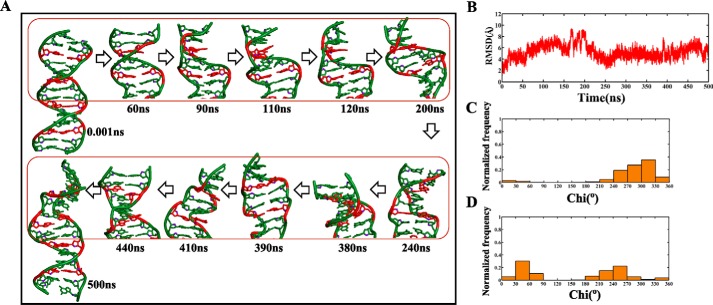
As seen above, the nonisosteric character of the A … A mismatch with respect to the flanking canonical G … C/C … G base pairs triggers unwinding of the helix after ∼90 ns of simulation (Fig. 3A and supplemental Movie S2). Time versus RMSD profile, calculated with respect to the initial model (Fig. 3B), was also indicative of significant deviation from initial model (∼5 Å). The associated conformational changes are as follows: Gs predominantly taking the ±syn glycosyl conformation (76%) (Fig. 3C) along with As in anti/+syn conformation (Fig. 3D). Exceptionally, some of the Gs and As briefly take a high-anti and -syn conformation, respectively. Backbone torsion angles such as ϵ, ζ, α, and γ in GA steps exhibit the characteristics of Z-DNA (35%), whereas AC (64%) and CG (45%) steps favor forming BI and BII conformations (Fig. 2C). CG (37%), GA (52%), and AC (28%) steps are also populated by other conformations such as (t,t,g−,g+), (g−,g−,t,t), (t,g−,g+,t), (t,g−,t,t), and (g−,t,g+,t) (Fig. 2D). As discussed earlier, these unusual conformations may be due to the interaction of counterions with the duplex (supplemental Fig. S1B). As before, CG steps (46% of helical twists less than 10°) exhibit a lower twist angle compared with GA (2%) and AC (2%) steps (Fig. 2E, right panel). The angles at the central phosphate in the steps C6pG7 (90°), G7pA8 (90°), G22pA23 (89°), C24pG25 (93°), G25pA26 (102°), A26pC27 (117°), C27pG28 (87°), A29pC30 (105°), and G31pA32 (85°) exhibit a very low value during the last 10 ns, yet more supportive evidence for the presence of Z-DNA (supplemental Fig. S2B).
Figure 3.
Evolution of the B–Z junction in the d(GAC)6.d(GAC)6 duplex with +syn … anti glycosyl conformation for the mismatch. A, snapshots showing the unwinding of the helix because of Z-DNA evolution, resulting in negative supercoiling. The A … A mismatch is colored red. B, time versus RMSD profile illustrating (RMSD > 4Å) significant conformational changes taking place with respect to the starting model. C and D, glycosyl torsions showing (C) Gs favoring ±syn and (D) As favoring high-anti/+syn conformations, respectively.
Together, these properties confirm the presence of the B–Z junction in the GAC repeat–containing duplex with a +syn … anti starting glycosyl conformation for the A … A mismatch. Nonetheless, the preference for the Z conformation is less prominent compared with the anti … anti starting glycosyl conformation.
Canonical base pairs containing the d(GAC)6·d(GTC)6duplex and a T … T mismatch containing the d(GTC)6·d(GTC)6 duplex retain B-form geometry
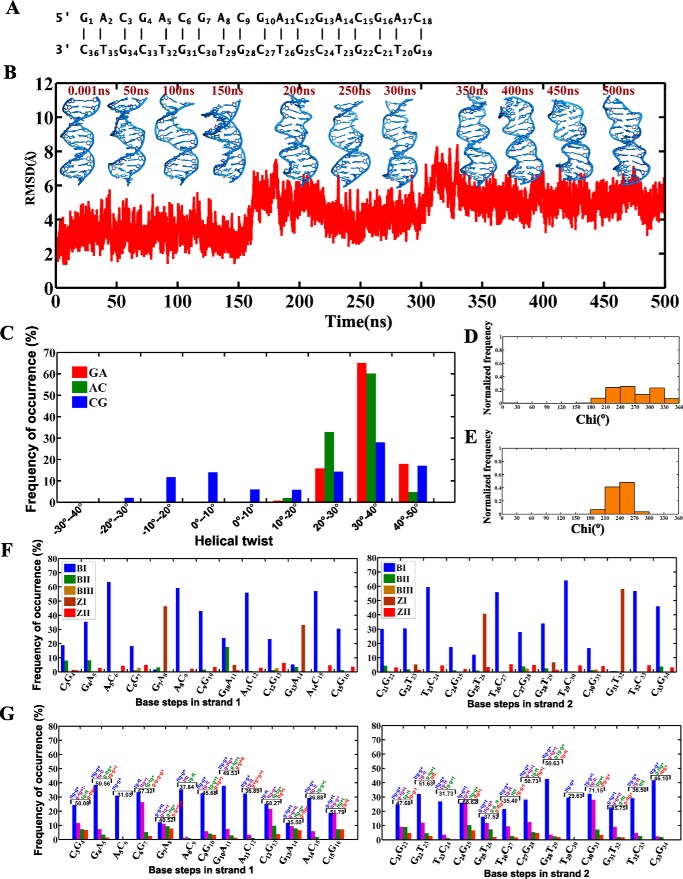
Control simulations carried out for the d(GAC)6·d(GTC)6 duplex with G … C and A … T canonical base pairs (Fig. 4A) to pinpoint that the B–Z junction formation observed in d(GAC)6·d(GAC)6 is purely due to nonisomorphism of the A … A mismatch indicate the dominance of B-form geometry (Fig. 4B). Although Z-DNA characteristics are observed during the simulation by 34% CG steps having helical twists lower than 10° (Fig. 4C) along with 42% of Gs preferring the ±syn glycosyl conformation (Fig. 4D), this is comparatively lower than in the mismatch situations (Figs. 1E and 3C). It is noteworthy that As (96%) prefer predominantly anti/high-anti glycosyl conformations (Fig. 4E). Few GA/GT steps also show Z-DNA backbone conformation (Fig. 4, F and G). In fact, such a minor population can be attributed to the presence of cations in the minor groove, as pointed out in an earlier study (supplemental Fig. S1C) (20).
Figure 4.
The d(GAC)6·d(GTC)6 duplex containing canonical base pairs retains B-form geometry. A, sequence of the 18-mer DNA duplex containing canonical base pairs used for MD simulations. B, time versus RMSD profile along with a schematic of the d(GAC)6·d(GTC)6 duplex at various time intervals. C, histograms corresponding to helical twist values of GA, AC, and CG steps over the last 400 ns. D and E, glycosyl chi angle values for guanine (favoring the ±syn conformation, D) and adenine (favoring the anti conformation, E) residues. F and G, backbone conformational angles for canonical base pairs over the last 400 ns of simulations. (ϵ,ζ,α,γ) conformational preference and the corresponding frequency of occurrence of the top four populations (percent) other than BI, BII, BIII, ZI, and ZII are given for each base step (G).
Similarly, MD simulation of the d(GTC)6·d(GTC)6 duplex, which contains six T … T mismatches in the place of six A … A mismatches in d(GAC)6·d(GAC)6 duplex (Fig. 5A), clearly indicates the preponderance for B-form geometry (Fig. 5B). The overall percentages of helical twists below 10° are 5%, 9%, and 11% at the CG, TC, and GT steps, respectively (Fig. 5C). Unlike the A … A mismatch, Gs (90%) favor anti/high-anti glycosyl conformations (Fig. 5D). Thus, the Z-DNA backbone conformation is less observed here (Fig. 5, E and F).
Figure 5.
The d(GTC)6·d(GTC)6 duplex containing the T … T mismatch retains B-form geometry. A, sequence of the 18-mer DNA duplex containing the T … T mismatch used for MD simulation. B, time versus RMSD profile along with a schematic of the d(GTC)6·d(GTC)6 duplex at various time intervals. C, histograms corresponding to helical twists at GT, TC, and CG steps over the last 400 ns. D, glycosyl chi angle values for guanine residues (favoring the anti/high-anti conformation). E, backbone conformational (ϵ,ζ,α,γ) angles for canonical base pairs over the last 400 ns of simulations. F, frequency of occurrence of the top four populations other than BI, BII, BIII, ZI, and ZII are given for each base step. The d(GTC)6·d(GTC)6 sequences were carried out using pmemd.cuda of the AMBER 16 suite.
In summary, only a minor population of Z-DNA is observed in d(GAC)6·d(GTC)6 and d(GTC)6·d(GTC)6 duplexes compared with d(GAC)6·d(GAC)6 duplexes. To further validate that B–Z junction formation is mainly induced by an A … A mismatch, CD studies were carried out (see below).
CD confirms B–Z junction formation in the d(GAC)7·d(GAC)7 duplex
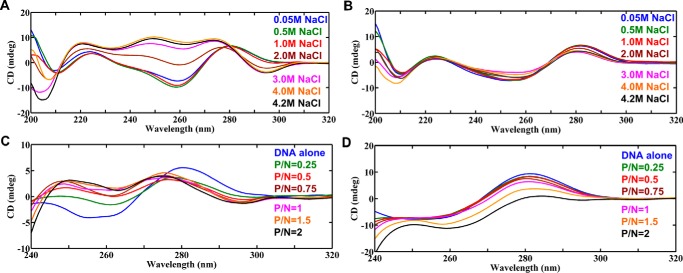
At 50 mm NaCl salt concentration, the CD spectrum of d(GAC)7·d(GAC)7 shows a positive peak between 270–280 nm and a negative peak around 260 nm, a typical characteristic of B-form DNA (Fig. 6A). However, with an increase in NaCl concentration in the range of 0.05 m to 4.2 m, the negative ellipticity around 260 nm moves toward positive ellipticity. Additionally, the spectra start developing two negative peaks (∼290 nm and ∼205 nm) with respect to the increase in salt concentration, which are all Z-DNA signature peaks (Fig. 6A). Nevertheless, canonical base pairs containing the d(GAC)7·d(GTC)7 duplex do not exhibit any B-to-Z transition with respect to the increase in NaCl concentration and stay in B-form, with positive and negative peaks around 285 nm and 260 nm, respectively (Fig. 6B). The d(GTC)7·d(GTC)7 duplex that has seven T … T mismatches that also exhibit same tendency as the d(GAC)7·d(GTC)7 duplex (supplemental Fig. S3). Thus, salt-dependent CD spectra clearly indicate that the A … A mismatch dictates B–Z junction formation, which subsequently converts the duplex to complete Z-form at a higher salt concentration.
Figure 6.
CD spectra showing the role of the A … A mismatch in promoting Z-phillicity in the d(GAC)7.d(GAC)7 duplex. A and B, salt-dependent (A) B-to-Z transition in d(GAC)7·d(GAC)7 (contains seven A … A mismatches) and (B) absence of the same in d(GAC)7·d(GTC)7 (contains only canonical base pairs). C and D, titration of (C) d(GAC)7·d(GAC)7 and (D) d(GAC)7·d(GTC)7 with hZαADAR1, indicating complete B-Z–to–Z transition in the former and absence of the same in the latter.
The Zα domain of human ADAR1 binds with the d(GAC)7·d(GAC)7 duplex
The CD spectra of the d(GAC)7·d(GAC)7 duplex (N) and hZαADAR1 protein (P) titration clearly show that increasing the concentration of P (viz. increasing the P/N ratio by keeping N as a constant) completely changes the duplex to the left-handed Z-form. As the concentration of P increases, the negative peak at ∼255 nm gradually diminishes, accompanied by the appearance of a new negative peak at ∼295 nm and a shift in the positive peak from 280 nm to 275 nm, characteristic features of the Z-DNA conformation (Fig. 6C). In contrast, the d(GAC)7·d(GTC)7 duplex, which contains only canonical base pairs, does not exhibit such a tendency for B-to-Z transition (Fig. 6D). Such a scenario is seen irrespective of the number of repeats in the duplex. For instance, the d(GAC)6·d(GAC)6 duplex, which has 6 A … A mismatches, also takes up the Z-form upon increasing the P/N ratio (supplemental Fig. S4A), whereas the corresponding canonical duplex does not exhibit such characteristics (supplemental Fig. S4B). In fact, d((GAC)3T4(GAC)3) (wherein one GAC in the d(GAC)7 is replaced by T4 to facilitate the hairpin formation), which is expected to form a hairpin with three A … A mismatches, also exhibits B-to-Z transition upon titration with hZαADAR1 (supplemental Fig. S5). This situation mimics hairpin formation in d(GAC)7 by having one GAC repeat in the hairpin loop and six GAC repeats in the stem with three A … A mismatches. Although there is a possibility that d(GAC)7 can take up either an intramolecular hairpin conformation (with three A … A mismatches) or an intermolecular duplex conformation (with seven A … A mismatches) in solution (supplemental Fig. S6), it is difficult to identify the preferred conformation from CD data. Indeed, both conformations may equally be populated in vitro, unlike in vivo, wherein it can take up only the hairpin conformation. Thus, d((GAC)3T4(GAC)3) titration with hZαADAR1 confirms that d(GAC)7 can adopt a stable hairpin conformation with three A … A mismatches, which subsequently facilitates binding with the protein.
hZαADAR1 binds d(GAC)7·d(GAC)7 with nanomolar affinity
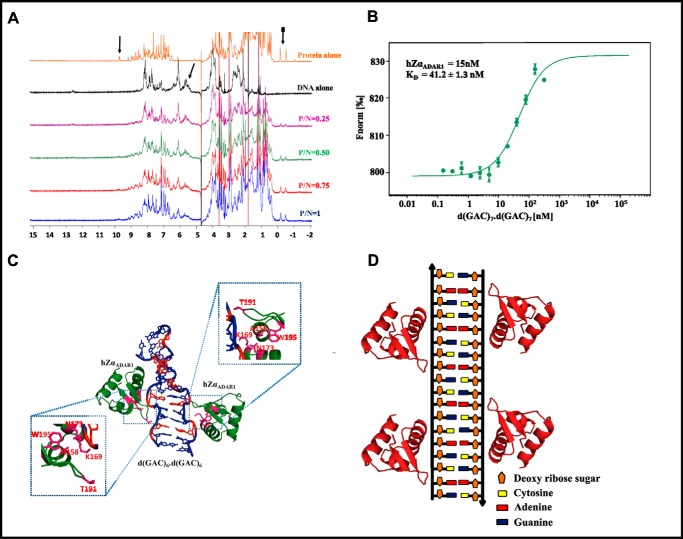
In accordance with the CD results, 1D proton NMR spectra of hZαADAR1 and d(GAC)7·d(GAC)7 duplex titration also confirm the interaction between these two. Overall, the spectra show a gradual reduction in peak intensity as the concentration of hZαADAR1 increases. Although it may be difficult to identify the amino acids/nucleotides that are associated with the proton chemical shifts simply from the 1D spectra, the signature chemical shifts around 0 to −1 ppm, 9.8 ppm, and 9.6 ppm can be assigned to protons corresponding to Thr-191 (23), the Hϵ1 proton of Trp-195 and the amide proton of Ala-158 (24), respectively, which are located in and around the binding site of hZαADAR1 (PDB code 2ACJ). Likewise, the chemical shifts between 5 to 6 ppm belong to backbone protons of the DNA duplex (25). Notably, the protein and DNA chemical shifts in these regions do not overlap with each other (Fig. 7A). MST exhibits a nanomolar binding affinity between hZαADAR1 and the d(GAC)7·d(GAC)7 duplex, with a dissociation constant (KD) of 41 nm (Fig. 7B).
Figure 7.
D(GAC)7·d(GAC)7–hZαADAR1 complex model. A, 1H NMR spectra corresponding to d(GAC)7·d(GAC)7 duplex titration with hZαADAR1. Arrows indicate the reduction in peak intensities (peak broadening) as the concentration of protein increases, suggestive of an intermediate chemical exchange between the two. B, DNA concentration–dependent (the protein concentration is kept constant, whereas the DNA concentration is varied, as described under “Experimental Procedures”) binding isotherms obtained from the microscale thermophoresis assay indicate that d(GAC)7·d(GAC)7 and hZαADAR1 exhibit nanomolar binding affinity with a KD of 41 nm. C, 1H NMR–based docked model of hZαADAR1 (PDB code 2ACJ)–d(GAC)6·d(GAC)6 (MD-derived) complex (red represents the A … A mismatch). The important interactions are enlarged and boxed. D, schematic of hZαADAR1 binding at multiple mismatch sites of the d(GAC)6·d(GAC)6 duplex.
The d(GAC)7·d(GAC)7–hZαADAR1 complex model
The aforementioned information about hZαADAR1 amino acids (Thr-191, Trp-195, and Ala-158) that may be involved in interaction with the d(GAC)7·d(GAC)7 duplex (Fig. 7A), along with the readily available complex structure of hZαADAR1 and a B–Z junction (PDB code 2ACJ), have been used to model the hZαADAR1–d(GAC)6·d(GAC)6 complex. Fig. 7C shows the modeled complex derived from X-ray (former) and MD (latter) structures. As more than one hZαADAR1 can bind to a single duplex, depending on the availability of Z-philic centers (PDB code 2ACJ) (21), d(GAC)6·d(GAC)6 can also accommodate more than one hZαADAR1 molecule (Fig. 7D).
MD simulation retains the conserved interactions between hZαADAR1 and the DNA duplex
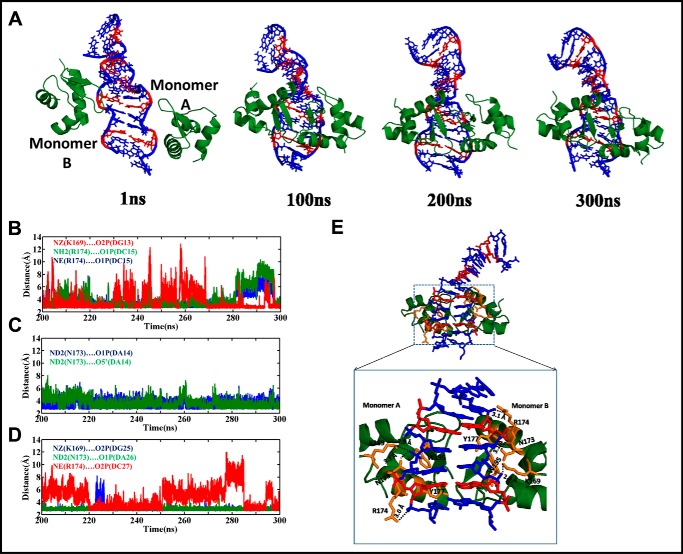
The modeled hZαADAR1 … d(GAC)6·d(GAC)6 complex has been subjected to 300-ns MD simulations to optimize the interaction between the two. It is noteworthy that the complex has been modeled so that two monomers of hZαADAR1 interact with two different strands of the duplex (Fig. 7C), as reported earlier (PDB code 2ACJ). Analysis of the MD trajectories reveals that hZαADAR1 interacts with the duplex through its minor groove (Fig. 8A). The Lys-169, Asn-173, and Arg-174 residues of hZαADAR1 monomers participate in a hydrogen-bonding interaction with the duplex backbone atoms (like O5′, O1P, and O2P) either transiently or persistently (Fig. 8, B–D). This is consistent with previous mutagenesis and NMR studies (26, 27) that show the importance of the above mentioned residues in facilitating the interaction between the two. However, a minor difference in the nature of interaction is also seen. For instance, Tyr-177, which is involved in a stacking interaction in the crystal structure (PDB code 2ACJ), is engaged in a transient hydrogen-bonding interaction with the sugar-phosphate backbone atoms (Fig. 8E). Similarly, Trp-195 does not participate in any direct hydrogen-bonding interaction with the duplex, although it lies in the proximity of the duplex (Fig. 8E). Thus, the unwinding of the d(GAC)6·d(GAC)6 duplex because of the presence of the A … A mismatch (Figs. 1B and 3A) facilitates the interaction of the hZαADAR1 protein at the minor groove.
Figure 8.
B-Z junction formation in the d(GAC)6·d(GAC)6 duplex facilitates the accommodation of hZαADAR1 in the minor groove. A, snapshots of d(GAC)6·d(GAC)6–hZαADAR1 complex MD simulation reveal that the β hairpins of the hZαADAR1 dimer interact with the duplex through its minor groove (A … A mismatches are colored red). B–D, time versus hydrogen bond distance profile corresponding to hZαADAR1 monomers A (B and C) and B (D). See text for details. E, snapshot illustrating all hydrogen bonding interactions between d(GAC)6·d(GAC)6 and hZαADAR1 during the simulation.
Discussion
Left-handed Z-DNA has a higher-energy conformation compared with the canonical B-DNA conformation (28), and in vitro, d(GC)n sequences are shown to choose the Z-form under extreme conditions, like high salt concentrations (29). There is increasing evidence regarding the participation of Z-DNA in gene regulation, the formation of which is believed to relieve stress on the DNA structure through negative supercoiling (30). Proteins that specifically recognize and bind to Z-DNA are also identified: hZαADAR1 (27), E3L (31), DLM1 (32), and PKZ (33). Interconversion between B- and Z-DNA is believed to take place either through a “stretch–collapse mechanism” or a “zipper mechanism” (34, 35), facilitated by base extrusion and base and/or backbone flipping. Intriguingly, the A … A mismatch in the hairpin stem of the CAG repeat readily exhibits a preponderance for the Z-DNA conformation through the zipper mechanism (7). As GAC repeats that are responsible for pseudoachondroplasia also contain periodic A … A mismatches, we investigate here the ability of the same to adopt a Z-form structure by employing MD simulation, CD, MST, and NMR techniques. Subsequently, its ability to bind with the hZαADAR1 protein is also explored.
The A … A mismatch induces local B-to-Z transition through backbone flipping
MD simulations carried out by considering two different models to explore all possible conformational preference for the d(GAC)6·d(GAC)6 duplex reveal that the A … A mismatch leads to local Z-DNA formation irrespective of anti … anti and +syn … anti starting glycosyl conformations. Such a conformational change is facilitated by backbone flipping through the base steps taking local Z-DNA and other non-B-DNA backbone conformations. This is further concomitant with Gs and a few As favoring the ±syn glycosyl conformation (Figs. 1D and 3C). Not surprisingly, pyrimidines favor the anti glycosyl conformation. Indeed, such a mixed occurrence of syn and anti glycosyl conformations, together with the aforementioned non-B-DNA backbone conformations, leads to Z-DNA features alongside the B-DNA conformation. Normalized frequencies of occurrence of Gs falling in ±syn conformation in the d(GAC)6·d(GAC)6 (+syn … anti), d(GAC)6·d(GTC)6, and d(GTC)6·d(GTC)6 duplexes with respect to the d(GAC)6·d(GAC)6 (anti … anti) duplex are 0.8, 0.4, and 0, respectively. This clearly indicates the influence of the A … A mismatch in inducing B-to-Z transition in d(GAC)6·d(GAC)6 in contrast to d(GAC)6·d(GTC)6 and d(GTC)6·d(GTC)6 duplexes. A minor population of B–Z junctions observed in d(GAC)6·d(GTC)6 may also be attributed to the interaction with counterions (supplemental Fig. S1C). This eventually reflects in the CG step taking a low twist in the midst of high twists at the AC and GA steps in d(GAC)6·d(GAC)6 duplexes (64 and 46% in anti … anti and +syn … anti glycosyl conformations, respectively) (Fig. 2E), leading to a B–Z junction in the vicinity of the mismatch. Such an occurrence of high and low twists in the duplex leads to unwinding of the helix, a typical characteristic of B–Z junctions, as observed in the crystal structures (PDB codes 1FV7 and 2ACJ) (supplemental Fig. S7). It is noteworthy that the B–Z junction does not show alternating glycosyl (±syn and anti) and backbone conformations as in the Z-form, wherein alternating glycosyl conformations lead to a zigzag backbone (36, 37). Instead, the B–Z junction possesses the characteristics of both B- and Z-forms.
In sharp contrast to the d(GAC)6·d(GAC)6 duplex, the d(GAC)6·d(GTC)6 duplex with canonical base pairs and the d(GTC)6·d(GTC)6 duplex with a T … T mismatch have a preference for B-form geometry. This finding is further confirmed by CD spectroscopy by titrating NaCl with the d(GAC)7·d(GAC)7, d(GTC)7·d(GTC)7, and d(GAC)7·d(GTC)7 duplexes; although d(GAC)7·d(GAC)7 clearly displays B-Z–to–Z transition with respect to the increase in salt concentration, the other two do not exhibit such a transition (Figs. 6, A and B, and supplemental Fig. S3). Such an inclination of the A … A mismatch toward the Z-form is due to its nonisostericity that is exemplified by a high residual twist and radial difference with the flanking C … G/G … C base pairs (18). This provides discomfort for the A … A mismatch to get accommodated in a B-DNA. Thus, it unwinds the helix to relieve the mechanistic effect arising from the nonisostericity of the A … A mismatch with respect to the flanking canonical base pairs as well as to retain the backbone connectivity (18, 38–40). As seen in the d(CAG)6·d(CAG)6 duplex (7), B-to-Z transition takes place through a zipper mechanism rather than a stretch–collapse mechanism. One can envisage a similar situation in the case of the (GA)n homoduplex, where the nonisostericity between G … G and A … A may provoke parallel duplex formation (41).
Inclination of the A … A mismatch toward Z-DNA leads to passive binding with hZαADAR1
The mechanism of recognition and binding of hZαADAR1 protein with the B–Z junction/Z-DNA is still a matter of debate. According to the active mechanism, hZαADAR1 binds to B-DNA and subsequently converts it into Z-DNA (24). Nevertheless, the passive mechanism suggests that hZαADAR1 traps the transient B–Z junction/Z-DNA and subsequently converts it into Z-DNA (42). The MD simulation (Figs. 1 and 3), CD (Fig. 6C), NMR (Fig. 7A), and microscale thermophoresis data presented here (Fig. 7B) conjointly identify that hZαADAR1 binds to d(GAC)7·d(GAC)7 in a “passive mechanism” because of the formation of a B–Z junction induced by the A … A mismatch.
As discussed above, MD simulation clearly shows the preference for B–Z junction formation in the d(GAC)6·d(GAC)6 duplex (Figs. 1B and 3A), in accordance with the CD spectra of NaCl titration with the d(GAC)7·d(GAC)7 duplex (Fig. 6, A and B). Although the mismatch-containing duplex (former) has proclivity toward Z-DNA transition (Fig. 6A), the canonical base pair–containing duplex (latter) does not possess such a property (Fig. 6B). In line with this, titration of hZαADAR1 with d(GAC)7·d(GAC)7 converts the duplex completely to the Z-form irrespective of duplex length (Fig. 6C and supplemental Figs. S4 and S5). Although some studies have shown that the GAC sequence is prone to form Z-DNA (10, 11), the rationale behind such a conformational preference is unknown. For the first time, it has been shown here that the nonisosteric A … A mismatch provokes B–Z transition in GAC repeats, which subsequently facilitates binding with the hZαADAR1 protein through a passive mechanism. This is further confirmed by 1D proton NMR spectroscopy, which indicates tighter affinity between the two (Fig. 7A). Additionally, KD measured by MST also indicates that the d(GAC)7·d(GAC)7 duplex binds with hZαADAR1 with nanomolar affinity (Fig. 7B). MD simulation carried out on the modeled hZαADAR1–d(GAC)6·d(GAC)6 complex (Figs. 7, C and D, and 8A) subsequently confirmed that the protein residues interact with the duplex through the minor groove, in accordance with earlier studies (PDB codes 2ACJ and 3IRQ).
Model for pathogenicity in d(GAC)n expansion disorders through RNA editing mediated by hADAR1
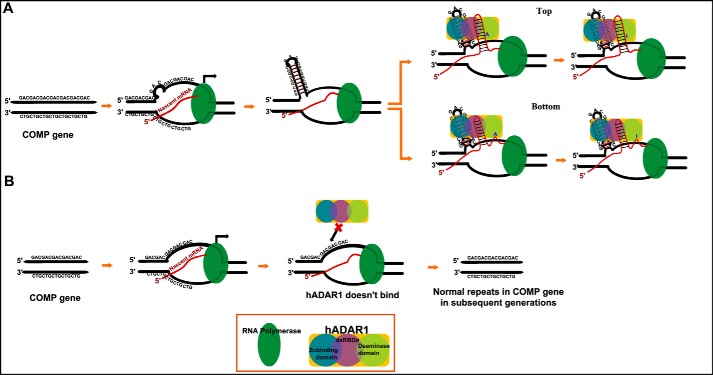
This study clearly shows that the d(GAC)7·d(GAC)7 duplex is not only prone to form Z-DNA but also binds to the Z-DNA binding domain of the human ADAR1 protein. Intriguingly, expansion in the d(GAC) trinucleotide repeat is shown to cause skeletal dysplasias, such as multiple epiphyseal dysplasia and pseudoachondroplasia (3, 6). Hence, based on the results of this study, we propose a model that explains how d(GAC) trinucleotide expansion in the COMP gene may lead to skeletal dysplasia, such as pseudoachondroplasia. According to our model (Fig. 9), d(GAC)7, which can form a hairpin structure with the stem possessing a B–Z junction (induced by A … A mismatches), facilitates anchorage of the Z-DNA binding domain of hADAR1 onto the DNA during transcription. Succeeding this event, the double-stranded RNA–specific deaminase domain of hADAR1 performs A-to-I editing in GAC either in the corresponding nascent RNA duplex (Fig. 9A, top panel) or downstream (Fig. 9A, bottom panel). This eventually codes for Gly instead of Asp in COMP. In fact, samples isolated from pseudoachondroplasia patients show that genomic point mutations in the d(GAC) track of the COMP gene that can code for Gly instead of Asp-473/Asp-482 are among the 70 possible mutations in the COMP gene (2, 43). According to the current model, A-to-I editing can lead to such Asp-to-Gly mutation at the protein level during transcription and, thus, can reflect the effect of genomic point mutations, as mentioned above. Such A-to-I editing downstream of the GAC repeat expansion (Asp-482) can also take place (Fig. 9A, bottom panel), resulting in Asp-to-Gly in COMP, which has already been shown to be deleterious (44, 45). On the other hand, when the d(GAC) repeat does not undergo expansion, hairpin formation may not take place. Thus, hADAR1 may not be able to bind to the DNA duplex, and A-to-I editing may not occur (Fig. 9B). Its noteworthy that, although direct evidence for the role of hADAR1 in pseudoachondroplasia is not well-established, its hyper/altered editing in several neurodegenerative disorders has been well-documented (13, 14, 16). In line with this, the hypothesis presented here offers new insight into the role of non-genetic A-to-I mutation in pseudoachondroplasia.
Figure 9.
Proposed model for RNA editing by neuronal hADAR1 in COMP in the perspective of pseudoachondroplasia disease. A, during transcription, formation of the d(GAC) hairpin containing the Z-conformation facilitates hZαADAR1 to anchor to the hairpin stem and aids in A-to-I editing either in the corresponding nascent RNA (top) or downstream (bottom). B, under normal conditions, hZαADAR1 does not bind to the duplex because of the absence of B-Z/Z conformations, resulting in wild-type protein expression.
We have shown here that the nonisomorphic nature of the A … A mismatch with respect to the flanking base pairs is the underlying factor for the Z-philic nature observed in the d(GAC)7·d(GAC)7 repeat expansion that is found in pseudoachondroplasia. We have shown here, for the first time, that such a structural trait of the A … A mismatch facilitates binding of hZαADAR1 to the d(GAC))n = 6,7·d(GAC)n = 6,7 duplex irrespective of the repeat length. A model for the complex of hZαADAR1–d(GAC)7·d(GAC)7 duplex and the consequent A-to-I editing during transcription by the double-stranded RNA–specific deaminase domain of hADAR1 under the disease condition are also presented.
Experimental procedures
Molecular dynamics simulations
Starting models of d(GAC)6·d(GAC)6 DNA duplexes were manually modeled using the PyMOL suite (57). Subsequently, the models were refined using constrained–restrained molecular geometry optimization using XPLOR-NIH (46). MD simulations of the modeled duplexes (Fig. 1A) were carried out in an explicit solvent environment following the protocol described earlier (7) using the AMBER 12 suite (47). FF99SB forcefield was used during the simulation. The systems were initially equilibrated for 50 ps, and then production runs were extended to 0.5 μs for each system using isobaric and isothermal conditions (NPT), 2-fs integration time, and 9-Å cutoff distance for the Lennard–Jones interaction. Following the above procedure, MD simulations of the d(GTC)6·d(GTC)6 and d(GAC)6·d(GTC)6 duplexes were carried out for 0.5 μs each. The 3D-NuS web server was used to build these models (48).
Analysis of the trajectories
The Ptraj module of Amber 12 was used to post-process the trajectories corresponding to d(GAC)6·d(GAC)6, d(GTC)6·d(GTC)6, and d(GAC)6·d(GTC)6 simulations. RMSD was calculated to acquire quantitative information about the deviation or proximity of the trajectories from the initial structure. Backbone conformational angles and helical parameters were extracted from 3DNA (49) output using in-house programs. PyMOL (57) and VMD (50) were used for visualization, and MATLAB software (The MathWorks Inc., Natick, MA) was used for plotting the graphs. Note that for the analysis, the central 14mer alone was considered.
Docking of the d(GAC)6·d(GAC)6 DNA duplex with the hZαADAR1 protein
The complex structure of the d(GAC)6·d(GAC)6 DNA duplex and hZαADAR1 protein was manually modeled by replacing the duplex present in the crystal structure (PDB code 2ACJ) with our MD-derived d(GAC)6·d(GAC)6 duplex. Subsequently, the complex model was subjected to 0.3-μs MD simulations using the pmemd.cuda module of the AMBER 16 suite. Analysis was carried out by using the cpptraj module of AMBER 16.
Duplex preparation
HPLC-grade d(GAC)n = 6,7 and d(GTC))n = 6,7 oligonucleotides were purchased from Sigma-Aldrich. The oligonucleotides were dissolved in 50 mm Tris-HCl and 50 mm NaCl (pH 7.4). The DNA duplex with canonical base pairs was formed by annealing (GAC)7 and the complementary (GTC)7 oligonucleotides at 95 °C and cooling them down to room temperature for 3 h. On the other hand, only the former was considered for formation of the duplex with the A … A mismatch, and the latter was used for formation of the T … T mismatch. Subsequently, duplex formation was verified by acquiring the CD spectrum (see below). Likewise, hairpin formation of d((GAC)3T4(GAC)3) was carried out. It is noteworthy that, to investigate the salt-dependent behavior of the duplex, the above process was repeated in the presence of appropriate salt concentrations (0.05 m, 0.5 m, 1 m, 2 m, 3 m, 4 m, and 4.2 m). Baseline correction was done using 50 mm Tris-HCl along with the corresponding salt concentration (pH 7.4).
Subcloning of the hZαADAR1 gene into the pET21a expression vector
The hZαADAR1 gene cloned in the pMAT cloning vector was acquired from Invitrogen with Ndel and Sal1 restriction sites at the 5′ and 3′ ends, respectively. Subsequently, the PCR-amplified, double-digested hZαADAR1 gene was subcloned into the ampicillin-resistant pDZ1 expression vector, a modified form of the pET-21a vector with a T7 promoter (51–53). The construct was organized in the following order: an N-terminal His6 tag, GB1 solubility tag, and tobacco etch virus protease cleavage site that were followed by the hZαADAR1 gene (225 bp).
Protein expression and purification
The pDZ1 expression vector was transformed into Escherichia coli BL21 (DE3) (Bioline) cells for overexpression of the hZαADAR1 protein. Preinoculum cells grown overnight were transferred into LB medium containing 100 μg/ml ampicillin and incubated at 37 °C until the optical density reached 0.6 at A600. Protein expression was induced by 1 mm isopropyl 1-thio-β-d-galactopyranoside, followed by overnight incubation at 15 °C to attain the optical density at A600 in the range of 1.4 to 1.6. Cells were then harvested and sonicated in binding buffer containing 20 mm Tris-HCl, 500 mm NaCl, 5 mm imidazole (pH 8.0), and 0.1 mm PMSF. The hZαADAR1 protein was eluted in a buffer containing 20 mm Tris-HCl, 500 mm NaCl, and 200 mm imidazole (pH 8.0) using nickel-nitrilotriacetic acid affinity column chromatography that was treated with 50 mm NiSO4 solution.
Purification involved two steps. First, the hZαADAR1 protein tagged with GB1 protein was purified as described above (supplemental Fig. S8A), followed by removal of the GB1 tag through overnight digestion with tobacco etch virus protease. During the second round of purification, the hZαADAR1 protein was isolated from the cleaved GB1 tag, and the fractions were collected (supplemental Fig. S8B) in binding buffer. Finally, the protein was dialyzed in NMR buffer (10 mm phosphate buffer and 10 mm NaCl (pH 7.4)). Protein concentration was measured by UV absorption at 280 nm using an extinction coefficient value of 8480 m−1 cm−1.
d((GAC)n·(GAC)n)n = 6,7–hZαADAR1 complex formation
The d((GAC)n·(GAC)n)n = 6,7–hZαADAR1 complex was prepared by changing the concentration of hZαADAR1 while retaining the concentration of DNA. For NMR experiments, the following P/N ratios were used by keeping the DNA concentration at 120 μm: 0.25, 0.5, 0.75, and 1. For CD experiments, P/N ratios of 0.25, 0.5, 0.75, 1, 1.5, and 2 were used by keeping the DNA concentration at 40 μm. The samples were prepared in buffer containing 10 mm sodium phosphate and 10 mm NaCl (pH 7.4), and 10% of D2O was added to the NMR sample. The complex was prepared by adding the protein to the DNA sample in fractions of 10 μl at 2-min intervals and incubated for 1 h at 25 °C.
CD spectroscopy
All CD spectra reported here were acquired in JASCO-1500 and processed by Spectra Manager software. The data were collected in triplicate in the wavelength region of 320 nm to 200 nm, and baseline correction was done with respect to the appropriate buffer. The average of triplicate spectra is reported here.
NMR spectroscopy
1D proton NMR experiments were performed on a Bruker 700-MHz instrument equipped with a room temperature probe. The Zggpw5 pulse sequence (54) was used to acquire data at 25 °C. All acquisition parameters were kept identical for all experiments: 768 scans and 32768 1H complex points. Bruker Top Spin was used for data processing and analysis.
Dissociation constant measurement using microscale thermophoresis
The dissociation constants of hZαADAR1 binding with the d(GAC)7·d(GAC)7 duplex were estimated using a microscale thermophoresis assay (55, 56). The assay was carried out using His6-GB1-hZαADAR1–tagged protein. The MST assay required one fluorescent binding partner (protein) and one non-fluorescent binding partner (DNA). Prior to titration, NT-647 fluorescent dye was non-covalently attached to the histidine residues of hZαADAR1. DNA was titrated to hZαADAR1 in serial dilutions, with concentrations ranging from 0.313 μm to 0.000153 μm. Subsequently, the assay was carried out in 10 mm phosphate buffer by keeping the concentration of labeled hZαADAR1 protein as a constant (15 nm). These samples were loaded into Monolith NT.115 MST Premium–coated capillaries, and the MST analysis was performed using 100% light-emitting diode (LED) power and 60% MST power in NanoTemper Monolith NT.115 at 24 °C. Using NanoTemper software, KD was calculated using the mass action equation from triplicate experiments.
Author contributions
N. K. carried out molecular dynamics simulations, analyzed the data, and performed salt-dependent CD experiments. Y. A. carried out subcloning, expression, and purification of hZαADAR1 along with CD, NMR titration, and KD experiments of the hZαADAR1-DNA complex, followed by docking and MD simulations of the hZαADAR1-DNA complex. T. R. designed and supervised the entire project. N. K., Y. A., and T. R. wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Prof. Roberto De Guzman (University of Kansas) for the generously gift of the pDZ1 construct. The High Performance Computing Facility of the Indian Institute of Technology Hyderabad, Center for Development of Advance Computing (Government of India) and Inter University Accelerator Center (Government of India) are acknowledged. We thank Tata Institute of Fundamental Research (TIFR) Mumbai and TIFR Centre for Interdisciplinary Sciences (TCIS)-Hyderabad for help with preliminary NMR data collection and Drs. Saji Menon and Sivaramaiah Nallapeta for KD measurements. We also thank Venkata Subbaiah, Komal Ghule, and Kripi Tomar for technical support.
This work was supported by the Department of Biotechnology, Government of India IYBA-2012 (D.O.No.BT/06/IYBA/2012), BIO-CaRE (SAN.No.102/IFD/SAN/1811/2013–2014), and R&D (SAN.No.102/IFD/SAN/3426/2013–2014) and the Indian Institute of Technology Hyderabad (to T. R.). The Ministry of Human Resources Development, Government of India provided fellowships to N. K. and Y. A. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S8, Movies S1 and S2, and References.
- COMP
- cartilage oligomeric matrix protein
- MD
- molecular dynamics
- MST
- microscale thermophoresis
- ADAR
- adenosine deaminase acting on RNA
- RMSD
- root mean square deviation
- P
- protein
- N
- nucleic acid.
References
- 1. Pearson C. E., Nichol Edamura K., and Cleary J. D. (2005) Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 6, 729–742 [DOI] [PubMed] [Google Scholar]
- 2. Briggs M. D., Hoffman S. M., King L. M., Olsen A. S., Mohrenweiser H., Leroy J. G., Mortier G. R., Rimoin D. L., Lachman R. S., and Gaines E. S. (1995) Pseudoachondroplasia and multiple epiphyseal dysplasia due to mutations in the cartilage oligomeric matrix protein gene. Nat. Genet. 10, 330–336 [DOI] [PubMed] [Google Scholar]
- 3. Délot E., King L. M., Briggs M. D., Wilcox W. R., and Cohn D. H. (1999) Trinucleotide expansion mutations in the cartilage oligomeric matrix protein (COMP) gene. Hum. Mol. Genet. 8, 123–128 [DOI] [PubMed] [Google Scholar]
- 4. Tufan A. C., Satiroglu-Tufan N. L., Jackson G. C., Semerci C. N., Solak S., and Yagci B. (2007) Serum or plasma cartilage oligomeric matrix protein concentration as a diagnostic marker in pseudoachondroplasia: differential diagnosis of a family. Eur. J. Hum. Genet. 15, 1023–1028 [DOI] [PubMed] [Google Scholar]
- 5. Tan K., Duquette M., Joachimiak A., and Lawler J. (2009) The crystal structure of the signature domain of cartilage oligomeric matrix protein: implications for collagen, glycosaminoglycan and integrin binding. FASEB J. 23, 2490–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mochmann L. H., and Wells R. D. (2004) Transcription influences the types of deletion and expansion products in an orientation-dependent manner from GAC*GTC repeats. Nucleic Acids Res. 32, 4469–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khan N., Kolimi N., and Rathinavelan T. (2015) Twisting right to left: A.A mismatch in a CAG trinucleotide repeat overexpansion provokes left-handed Z-DNA conformation. PLoS Comput. Biol. 11, e1004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mitas M. (1997) Trinucleotide repeats associated with human disease. Nucleic Acids Res. 25, 2245–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sobczak K., Michlewski G., de Mezer M., Kierzek E., Krol J., Olejniczak M., Kierzek R., and Krzyzosiak W. J. (2010) Structural diversity of triplet repeat RNAs. J. Biol. Chem. 285, 12755–12764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kejnovská I., Tmová M., and Vorlícková M. (2001) (CGA)(4): parallel, anti-parallel, right-handed and left-handed homoduplexes of a trinucleotide repeat DNA. Biochim. Biophys. Acta 1527, 73–80 [DOI] [PubMed] [Google Scholar]
- 11. Vorlícková M., Kejnovská I., Tumová M., and Kypr J. (2001) Conformational properties of DNA fragments containing GAC trinucleotide repeats associated with skeletal dysplasias. Eur. Biophys. J. 30, 179–185 [DOI] [PubMed] [Google Scholar]
- 12. Barraud P., and Allain F. H. (2012) ADAR proteins: double-stranded RNA and Z-DNA binding domains. Curr. Top. Microbiol. Immunol. 353, 35–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meier J. C., Kankowski S., Krestel H., and Hetsch F. (2016) RNA editing: systemic relevance and clue to disease mechanisms? Front. Mol. Neurosci. 9, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Singh M. (2012) Dysregulated A to I RNA editing and non-coding RNAs in neurodegeneration. Front. Genet. 3, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nishikura K. (2016) A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 17, 83–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farajollahi S., and Maas S. (2010) Molecular diversity through RNA editing: a balancing act. Trends Genet. 26, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yildirim I., Park H., Disney M. D., and Schatz G. C. (2013) A dynamic structural model of expanded RNA CAG repeats: a refined X-ray structure and computational investigations using molecular dynamics and umbrella sampling simulations. J. Am. Chem. Soc. 135, 3528–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thenmalarchelvi R., and Yathindra N. (2005) New insights into DNA triplexes: residual twist and radial difference as measures of base triplet non-isomorphism and their implication to sequence-dependent non-uniform DNA triplex. Nucleic Acids Res. 33, 43–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krepl M., Zgarbová M., Stadlbauer P., Otyepka M., Banáš P., Koča J., Cheatham T. E. 3rd, Jurečka P., and Sponer J. (2012) Reference simulations of noncanonical nucleic acids with different chi variants of the AMBER force field: quadruplex DNA, quadruplex RNA and Z-DNA. J. Chem. Theory Comput. 8, 2506–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dans P. D., Faustino I., Battistini F., Zakrzewska K., Lavery R., and Orozco M. (2014) Unraveling the sequence-dependent polymorphic behavior of d(CpG) steps in B-DNA. Nucleic Acids Res. 42, 11304–11320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ha S. C., Lowenhaupt K., Rich A., Kim Y. G., and Kim K. K. (2005) Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature 437, 1183–1186 [DOI] [PubMed] [Google Scholar]
- 22. Kiliszek A., Kierzek R., Krzyzosiak W. J., and Rypniewski W. (2010) Atomic resolution structure of CAG RNA repeats: structural insights and implications for the trinucleotide repeat expansion diseases. Nucleic Acids Res. 38, 8370–8376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ulrich E. L., Akutsu H., Doreleijers J. F., Harano Y., Ioannidis Y. E., Lin J., Livny M., Mading S., Maziuk D., Miller Z., Nakatani E., Schulte C. F., Tolmie D. E., Kent Wenger R., Yao H., and Markley J. L. (2008) BioMagResBank. Nucleic Acids Res. 36, D402–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kang Y. M., Bang J., Lee E. H., Ahn H. C., Seo Y. J., Kim K. K., Kim Y. G., Choi B. S., and Lee J. H. (2009) NMR spectroscopic elucidation of the B-Z transition of a DNA double helix induced by the Z α domain of human ADAR1. J. Am. Chem. Soc. 131, 11485–11491 [DOI] [PubMed] [Google Scholar]
- 25. Wijmenga S. S., and van Buuren B. N. M. (1998) The use of NMR methods for conformational studies of nucleic acids. Prog. Nucl. Magn. Reson. Spectrosc. 32, 287–387 [Google Scholar]
- 26. Jeong M., Lee A. R., Kim H. E., Choi Y. G., Choi B. S., and Lee J. H. (2014) NMR study of the Z-DNA binding mode and B-Z transition activity of the Zα domain of human ADAR1 when perturbed by mutation on the α3 helix and β-hairpin. Arch. Biochem. Biophys. 558, 95–103 [DOI] [PubMed] [Google Scholar]
- 27. Schade M., Turner C. J., Kühne R., Schmieder P., Lowenhaupt K., Herbert A., Rich A., and Oschkinat H. (1999) The solution structure of the Zα domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA. Proc. Natl. Acad. Sci. U.S.A. 96, 12465–12470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., and Rich A. (1979) Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 282, 680–686 [DOI] [PubMed] [Google Scholar]
- 29. Pohl F. M., and Jovin T. M. (1972) Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J. Mol. Biol. 67, 375–396 [DOI] [PubMed] [Google Scholar]
- 30. Nordheim A., Lafer E. M., Peck L. J., Wang J. C., Stollar B. D., and Rich A. (1982) Negatively supercoiled plasmids contain left-handed Z-DNA segments as detected by specific antibody binding. Cell 31, 309–318 [DOI] [PubMed] [Google Scholar]
- 31. Ha S. C., Lokanath N. K., Van Quyen D., Wu C. A., Lowenhaupt K., Rich A., Kim Y. G., and Kim K. K. (2004) A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a Yatapoxvirus Zα bound to DNA. Proc. Natl. Acad. Sci. U.S.A. 101, 14367–14372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schwartz T., Behlke J., Lowenhaupt K., Heinemann U., and Rich A. (2001) Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 8, 761–765 [DOI] [PubMed] [Google Scholar]
- 33. Kim D., Hur J., Park K., Bae S., Shin D., Ha S. C., Hwang H. Y., Hohng S., Lee J. H., Lee S., Kim Y. G., and Kim K. K. (2014) Distinct Z-DNA binding mode of a PKR-like protein kinase containing a Z-DNA binding domain (PKZ). Nucleic Acids Res. 42, 5937–5948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moradi M., Babin V., Roland C., and Sagui C. (2013) Reaction path ensemble of the B-Z-DNA transition: a comprehensive atomistic study. Nucleic Acids Res. 41, 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim D., Reddy S., Kim D. Y., Rich A., Lee S., Kim K. K., and Kim Y. G. (2009) Base extrusion is found at helical junctions between right- and left-handed forms of DNA and RNA. Nucleic Acids Res. 37, 4353–4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berger I., Winston W., Manoharan R., Schwartz T., Alfken J., Kim Y. G., Lowenhaupt K., Herbert A., and Rich A. (1998) Spectroscopic characterization of a DNA-binding domain, Z α, from the editing enzyme, dsRNA adenosine deaminase: evidence for left-handed Z-DNA in the Z α-DNA complex. Biochemistry 37, 13313–13321 [DOI] [PubMed] [Google Scholar]
- 37. Kimura T., Kawai K., Tojo S., and Majima T. (2004) One-electron attachment reaction of B- and Z-DNA modified by 8-bromo-2′-deoxyguanosine. J. Org. Chem. 69, 1169–1173 [DOI] [PubMed] [Google Scholar]
- 38. Rathinavelan T., and Yathindra N. (2006) Base triplet nonisomorphism strongly influences DNA triplex conformation: effect of nonisomorphic G* GC and A* AT triplets and bending of DNA triplexes. Biopolymers 82, 443–461 [DOI] [PubMed] [Google Scholar]
- 39. Goldsmith G., Rathinavelan T., and Yathindra N. (2016) Correction: selective preference of parallel DNA triplexes is due to the disruption of Hoogsteen hydrogen bonds caused by the severe nonisostericity between the G*GC and T*AT triplets. PLoS ONE 11, e0155090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ananth P., Goldsmith G., and Yathindra N. (2013) An innate twist between Crick's wobble and Watson-Crick base pairs. RNA 19, 1038–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vorlícková M., Kejnovská I., Kovanda J., and Kypr J. (1999) Dimerization of the guanine-adenine repeat strands of DNA. Nucleic Acids Res. 27, 581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bae S., Kim D., Kim K. K., Kim Y. G., and Hohng S. (2011) Intrinsic Z-DNA is stabilized by the conformational selection mechanism of Z-DNA-binding proteins. J. Am. Chem. Soc. 133, 668–671 [DOI] [PubMed] [Google Scholar]
- 43. Hecht J. T., Nelson L. D., Crowder E., Wang Y., Elder F. F., Harrison W. R., Francomano C. A., Prange C. K., Lennon G. G., and Deere M. (1995) Mutations in exon 17B of cartilage oligomeric matrix protein (COMP) cause pseudoachondroplasia. Nat. Genet. 10, 325–329 [DOI] [PubMed] [Google Scholar]
- 44. Jackson G. C., Mittaz-Crettol L., Taylor J. A., Mortier G. R., Spranger J., Zabel B., Le Merrer M., Cormier-Daire V., Hall C. M., Offiah A., Wright M. J., Savarirayan R., Nishimura G., Ramsden S. C., Elles R., et al. (2012) Pseudoachondroplasia and multiple epiphyseal dysplasia: a 7-year comprehensive analysis of the known disease genes identify novel and recurrent mutations and provides an accurate assessment of their relative contribution. Hum. Mutat. 33, 144–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Susic S., Ahier J., and Cole W. G. (1998) Pseudoachondroplasia due to the substitution of the highly conserved Asp482 by Gly in the seventh calmodulin-like repeat of cartilage oligomeric matrix protein. Hum. Mutat. 1, S125–127 [DOI] [PubMed] [Google Scholar]
- 46. Schwieters C. D., Kuszewski J. J., Tjandra N., and Clore G. M. (2003) The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 47. Case D. A., Darden T. A., Cheatham T. E., Simmerling C. L., Wang J., Duke R. E., Luo R., Walker R. C., Zhang W., Merz K. M., Roberts B., Hayik S., Roitberg A., Seabra G., Swails J., et al. (2012) AMBER 12. University of California, San Francisco [Google Scholar]
- 48. Patro L. P. P., Kumar A., Kolimi N., and Rathinavelan T. (2017) 3D-NuS: a web server for automated modeling and visualization of non-canonical 3-dimensional nucleic acid structures. J. Mol. Biol. 429, 2438–2448 [DOI] [PubMed] [Google Scholar]
- 49. Lu X. J., and Olson W. K. (2003) 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 31, 5108–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Humphrey W., Dalke A., and Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- 51. Wang Y., Ouellette A. N., Egan C. W., Rathinavelan T., Im W., and De Guzman R. N. (2007) Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH. J. Mol. Biol. 371, 1304–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rathinavelan T., Tang C., and De Guzman R. N. (2011) Characterization of the interaction between the Salmonella type III secretion system tip protein SipD and the needle protein PrgI by paramagnetic relaxation enhancement. J. Biol. Chem. 286, 4922–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rathinavelan T., Lara-Tejero M., Lefebre M., Chatterjee S., McShan A. C., Guo D. C., Tang C., Galan J. E., and De Guzman R. N. (2014) NMR model of PrgI-SipD interaction and its implications in the needle-tip assembly of the Salmonella type III secretion system. J. Mol. Biol. 426, 2958–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu M., Mao X.-a., Ye C., Huang H., Nicholson J. K., and Lindon J. C. (1998) Improved WATERGATE pulse sequences for solvent suppression in NMR spectroscopy. J. Magn. Reson. 132, 125–129 [Google Scholar]
- 55. Parker J. L., and Newstead S. (2014) Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature 507, 68–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jerabek-Willemsen M., Wienken C. J., Braun D., Baaske P., and Duhr S. (2011) Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 9, 342–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 1.5.0.1, Schroedinger, LLC, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.