Abstract
A better understanding of neuromodulation in a behavioral system requires identification of active modulatory transmitters. Here, we used identifiable neurons in a neurobiological model system, the mollusc Aplysia, to study neuropeptides, a diverse class of neuromodulators. We took advantage of two types of feeding neurons, B48 and B1/B2, in the Aplysia buccal ganglion that might contain different neuropeptides. We performed a representational difference analysis (RDA) by subtraction of mRNAs in B48 versus mRNAs in B1/B2. The RDA identified an unusually long (2025 amino acids) peptide precursor encoding Aplysia leucokinin-like peptides (ALKs; e.g. ALK-1 and ALK-2). Northern blot analysis revealed that, compared with other ganglia (e.g. the pedal-pleural ganglion), ALK mRNA is predominantly present in the buccal ganglion, which controls feeding behavior. We then used in situ hybridization and immunohistochemistry to localize ALKs to specific neurons, including B48. MALDI-TOF MS on single buccal neurons revealed expression of 40 ALK precursor–derived peptides. Among these, ALK-1 and ALK-2 are active in the feeding network; they shortened the radula protraction phase of feeding motor programs triggered by a command-like neuron. We also found that this effect may be mediated by the ALK-stimulated enhancement of activity of an interneuron, which has previously been shown to terminate protraction. We conclude that our multipronged approach is effective for determining the structure and defining the diverse functions of leucokinin-like peptides. Notably, the ALK precursor is the first verified nonarthropod precursor for leucokinin-like peptides with a novel, marked modulatory effect on a specific parameter (protraction duration) of feeding motor programs.
Keywords: mass spectrometry (MS), neurobiology, neuropeptide, neuroscience, neurotransmitter, Aplysia, Leucokinin, arthropods, feeding, neuromodulation
Introduction
In both invertebrates and vertebrates, neuropeptides are the most diverse class of neuromodulators (1–4). This makes it challenging to identify the full complement of peptides that modulate a particular network. Much progress along these lines has, however, been made in experiments that have investigated peptidergic neurotransmission in experimentally advantageous model organisms. The present study was conducted in one such preparation, the mollusc Aplysia californica.
A number of bioactive neuropeptides have been identified in Aplysia using biochemical techniques to purify substances and molecular biological tools to characterize neuropeptides and their precursors. In particular, many peptides that have been characterized are present in connectives and identified neurons, including motoneurons, of the feeding network (5–13). This network has been extensively characterized (e.g. sensory neurons, motoneurons, and interneurons have been identified) (14–25). The Aplysia feeding network is, therefore, ideally suited for functional studies of peptidergic modulation. In this report, we demonstrate that it contains leucokinin-like peptides.
Previous studies of the leucokinin family of peptides have been primarily conducted in arthropods. Members of this family were initially purified from the cockroach Leucophaera maderae using biochemical methods (26–30). Subsequently, leucokinin-like peptides were identified in a number of other insect species (31). Insect leucokinins have been studied in physiological experiments, with most studies focusing on their myoactivity in the hindgut and increasing diuretic activity via the insect Malpighian tubules (31–33). Further, more recent work has demonstrated that leucokinin signaling pathways play a role in regulating feeding behavior in Drosophila (34). Despite this progress, there are still major gaps in knowledge. For example, bioinformatic studies have predicted the amino acid (aa)4 sequences for leucokinin precursors in non-arthropods (35, 36). However, these gene products have not been experimentally verified. It is therefore not clear how extensive the leucokinin family actually is in metazoa. Also, the descriptions of actual expression patterns of leucokinins in the nervous systems and their functional roles are unavailable in non-arthropods and are poorly understood even in some arthropod species (37). Importantly, although neuropeptides can alter feeding responses by modulating the feeding central pattern generator (CPG) that controls the feeding movements (2–4, 38), no prior studies have examined the effects of leucokinins on any feeding CPG, and this is one of our main objectives in this work.
Although there have been relatively few leucokinin-like peptides predicted in non-arthropod species, Cox et al. (39) did biochemically identify a single leucokinin-like peptide in Lymnaea. This suggested that leucokinin-like peptides could be present in Aplysia, which, like Lymnaea, is a gastropod mollusc. To determine whether there are novel peptides, such as leucokinins, in Aplysia, we performed representational difference analysis (RDA) (11, 40, 41) by subtraction of mRNAs in two types of identified neurons in the feeding circuit. This is a nontargeted approach that can potentially identify any mRNA that is unique to one neuron. RDA in various forms has been successfully utilized in other model systems, including vertebrates (42, 43).
Using RDA, we identified an Aplysia leucokinin-like peptide (ALK) precursor. The precursor that we identify is one of the longest neuropeptide precursors characterized to date. Additionally, we mapped the distribution of ALK-containing neurons in the Aplysia CNS using immunohistochemistry and in situ hybridization and demonstrated that ALKs are indeed expressed in single neurons using single-cell MALDI-TOF mass spectrometry. Finally, we showed that ALKs are bioactive in the Aplysia feeding network, where they modulate a specific parameter of feeding motor programs partly through a direct action on an identified element of the feeding CPG, interneuron B64. Thus, the ALK precursor is the first verified non-arthropod leucokinin precursor. Together with previous work in Drosophila (34), these results suggest that the leucokinins may play an important role in regulating feeding behavior.
Results
Identification of the ALK precursor by RDA, cDNA library screening and RACE, and bioinformatics
RDA takes advantage of the fact that two types of neurons (i.e. a tester and a driver) may express different complements of mRNA, which may code proteins, including neuropeptide precursors. To identify neuron-specific proteins, cDNA is amplified, and the cDNA from one neuron is subtracted from the cDNA of the second neuron. This technique is non-targeted, meaning that it can potentially identify any proteins/peptides that are present in the tester, but not in the driver, and has been used successfully to identify several novel proteins in Aplysia (11, 40, 41).
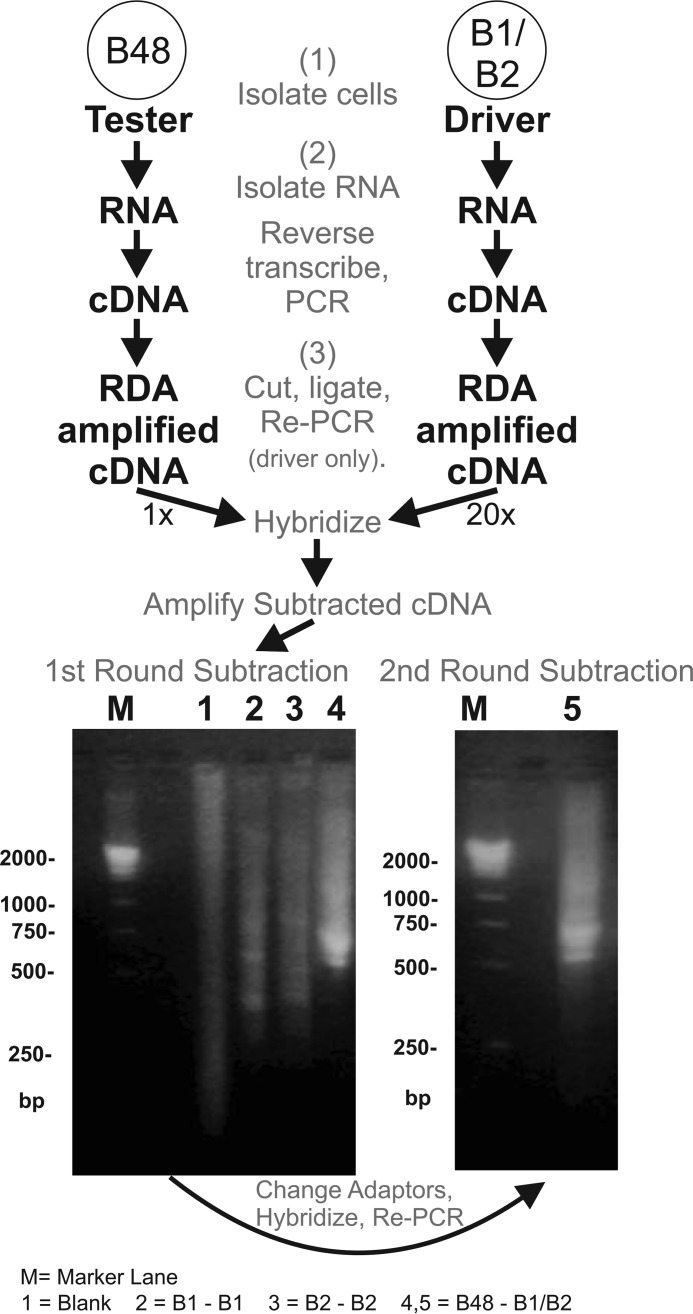
In the current work, we used buccal motoneuron B48 (14, 44) as a tester and two large neurons, B1/B2, as a driver. The first round of subtraction between B48 and B1/B2 allowed abundant RNAs that are unique to B48 to appear as prominent difference bands between 500 and 750 bp (Fig. 1). In some cases, one round of subtraction permits identification of clones that contain unique products (11, 41). In other cases (40) and in the present study, this was not the case. Therefore, a second round of subtraction was performed by hybridizing the subtracted cDNA from the first round with the driver's DNA. In addition, cDNAs corresponding to peptide precursors known to be expressed by the tester neuron B48 (i.e. myomodulin cDNA) were added to exclude these cDNAs. Subcloning the products from the second subtraction produced many colonies that contained unique products (as determined by differential screening). The difference clones obtained from B48 were sequenced to reveal an open reading frame that contained partial sequences of several hundred bp. Sequences were considered partial because at the 5′-end, there was no start codon, and at the 3′ end, there was no poly(A) addition site.
Figure 1.
RDA procedure used to identify the ALK precursor. Illustrated is the RDA between a buccal motoneuron, B48, and B1/B2 neurons. The results of the subtractions performed are shown in the gels at the bottom. Bottom left, results of the first subtraction performed. Lane 1, a blank subtraction. Lanes 2 and 3, additional control subtractions using the B1 or B2 neurons subtracted against themselves, respectively. These lanes only show a weak smear with some faint bands. Lane 4, a B48 that was subtracted against neurons B1 and B2. This lane shows some intense bands that do not appear in the control subtractions. M, marker lane showing cDNA masses in base pairs. Bottom right, lane 5 shows the results of a second round of subtraction. One difference clone obtained from B48 was sequenced to reveal an open reading frame that coded for an incomplete ALK transcript. The complete transcript was subsequently obtained with RACE (see “Results”).
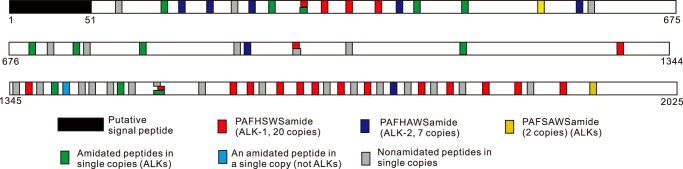
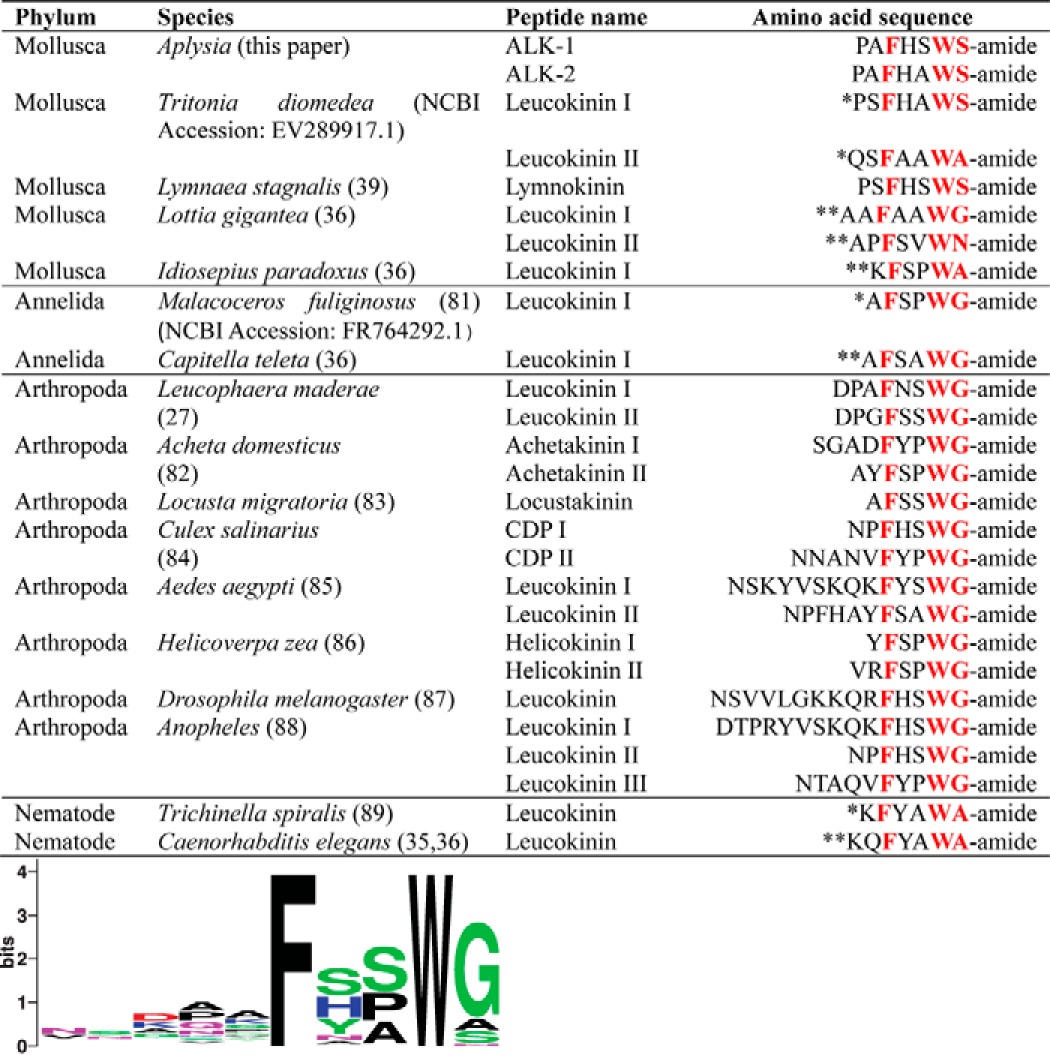
To obtain a complete sequence, we used the RDA fragments to screen a poly(T) pBluescript library and then used RACE to extend the sequence. We successfully obtained a clone with 6591 bp, which contained a complete protein of 2025 aa in length (Fig. 2). The gene sequence has been deposited into the NCBI database (GenBankTM accession number MF664476). The precursor is predicted by NeuroPred (http://stagbeetle.animal.uiuc.edu/cgi-bin/neuropred.py)5 (45, 46) to code a number of putative neuropeptides, some of which appear in multiple copies. Some of these sequences are homologous to the insect leucokinins and have an amidated C-terminal pentapeptide motif: FXXWX-amide. Consequently, we putatively named the peptides Aplysia leucokinin-like peptides (ALKs). We named the most abundant ALK on the precursor ALK-1. Its sequence is PAFHSWS-amide, and it is present in 20 copies. There are seven copies of ALK-2 (PAFHAWS-amide), and two copies of PAFSAWS-amide (Fig. 2). There is only one copy of the 13 other amidated peptides (see supplemental Table S2). A comparison of ALK-1 and ALK-2 with leucokinin-like peptides in various species is shown in Table 1. Leucokinin-like peptides in insects typically have a Phe amino acid residue near the N terminus and have a C-terminal WG-amide. Gastropod molluscs differ in that the C terminus is WS-amide. In addition, we compared some selected leucokinin-like peptide precursors, which are listed in supplemental Table S1. For these precursors, we generated a sequence alignment (supplemental Fig. S1) and a hypothetical phylogenetic tree (supplemental Fig. S2).
Figure 2.
Schematic diagram of the ALK precursor, which is composed of 2025 aa. Illustrated are the approximate locations of a putative signal peptide, 16 amidated peptides (shown in colors), and 24 nonamidated peptides (shown in gray) (see supplemental Table S2). Among the 16 amidated peptides, all but one (DSPRMFAFNSLS-amide, shown in light blue) have a C-terminal pentapeptide motif (FXXWX-amide) with the last aa being Ser, Gly, Ala, or Asn and are classified as ALKs. Note that several peptides at three locations have overlapping sequences, indicating alternative cleavage sites.
Table 1.
Comparison of leucokinin-like neuropeptides in representative species
*, Predicted; **, Predicted and the precursor is incomplete. Additional peptides included for the frequency plot under the table but not in the table for clarity are as follows: in L. maderae, Leucokinin III (DQGFNSWG-amide), Leucokinin IV (DASFHSWG-amide) (28); Leucokinin V (GSGFSSWG-amide), Leucokinin VI (pESSFHSWG-amide) (29); Leucokinin VII (DPAFSSWG-amide), Leucokinin VIII (GADFYSWG-amide) (30); in A. domesticus Achetakinin III (ALPFSPWG-amide), Achetakinin IV (NFKFNPWG-amide), Achetakinin V (AFHSWG-amide) (82); in A. domesticus, Achetakinin III (ALPFSPWG-amide), Achetakinin IV (NFKFNPWG-amide), and Achetakinin V (AFHSWG-amide) (82); in Culex salinarius CDP III (TKYVSKQFFSWG-amide) (84); in A. aegypti, Leucokinin III (NNPNVFYPWG-amide) (85); and in Helicoverpa zea Helicokinin III (KVKFSAWG-amide) (86). The sequence letters shared between different species are highlighted in red.
Whereas the ALK preproprotein has a structure characteristic of the neuropeptide prohormone, encodes numerous putative peptides, and has a priming methionine, its signal peptide cannot be identified reliably. Using PrediSi (http://www.predisi.de/)5 prediction (47), there is a 57% chance of a signal peptide cleavage site between Tyr50 and Ala51 in the preproprotein (supplemental Table S1). Our MS analysis later indicated that the peptide detected closest to the N terminus was Ser75–Val98. Therefore, the exact length of the ALK precursor signal peptide remains unknown.
We searched the NCBI database (including the EST database) and found several short matching nucleotide sequences ranging from 218 to 781 bp that matched 20% of the ALK precursor sequence in the beginning and the end portions of the precursor. These short sequences are EST deposits from an earlier Aplysia genome project (48). We then searched a recently completed Aplysia EST database (http://www.aplysiagenetools.org/),5 which has become a more popular source for Aplysia sequences. Indeed, we found several long matching sequences that showed >99% similarity with the ALK mRNA sequence in the following ranges: 37–990 (comp80863_c0_seq1); 1119–5215 (comp127554_c0_seq3), and 5216–6578 (comp88691_c3_seq1) (supplemental Fig. S3). Overall, these three sequences matched 81.52% of the ALK precursor, providing independent support for the ALK precursor sequence. The search results are consistent with the idea that there is a single Aplysia gene that encodes leucokinin-like peptides. However, we cannot rule out the possibility that an additional gene exists.
Distribution of ALK mRNA and immunoreactivity in the Aplysia CNS
We performed a Northern blot analysis to determine the distributions of ALK mRNA in specific ganglia of Aplysia. Fig. 3 shows that ALK mRNA is predominantly expressed in buccal ganglia. ALK mRNA is ∼8 kb in length (of which ∼6 kb is the coding sequence), which is in a range similar to the long mRNA (6591 bp) that we identified. ALK mRNA was not detected in other ganglia. It is likely that this is due to the fact that the overall mRNA level in these ganglia was too low to be detected.
Figure 3.
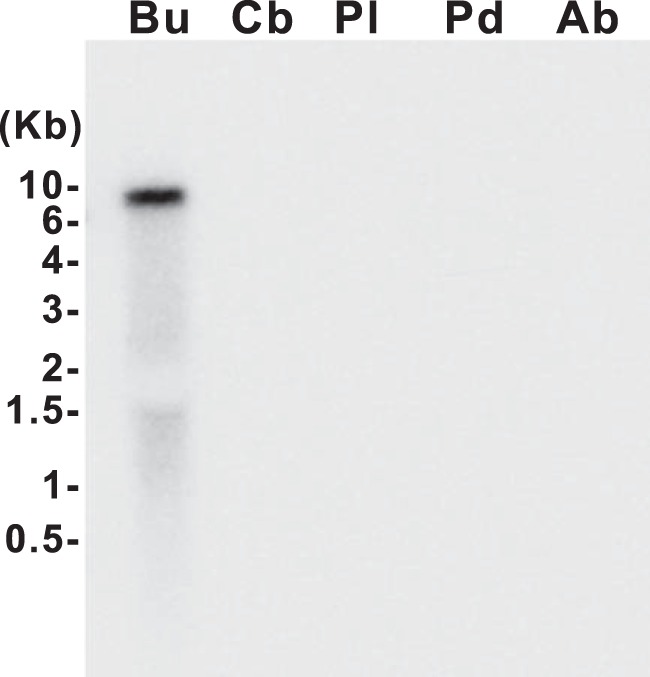
Northern blot gel of ALK mRNA in the Aplysia CNS. In situ hybridization of total RNA isolated from the different Aplysia ganglia with an ALK cDNA probe. Northern analysis only detected ALK mRNA in the buccal ganglion. ALK mRNA in other ganglia was below the level of detection. ALK mRNA is about 8 kb in length. Bu, buccal ganglion; Cb, cerebral ganglion; Pl, pleural ganglion; Pd, pedal ganglion; Ab, abdominal ganglion.
To map ALK-positive neurons and their processes in the CNS of Aplysia, we used two methods: in situ hybridization performed on whole mounts obtained from small size animals (∼10 g) (n = 3 for each ganglion) and immunohistochemistry performed on whole mounts obtained from larger animals (>90 g) (n = 3 for each ganglion). Smaller animals are preferred for in situ hybridization experiments, as they tend to generate less background. For immunohistochemistry experiments, we raised a polyclonal antibody against ALK-1. The specificity of the antibody was demonstrated by preabsorption experiments that abolished staining (supplemental Fig. S4B). We detected ALK-positive neurons in all central ganglia (i.e. in the buccal ganglion (Fig. 4, A and B), the cerebral ganglion (Fig. 4, C and D), the pleural-pedal ganglion (Fig. 5, A–D), and the abdominal ganglion (Fig. 5, E and F)). Notably, the patterns of cell body staining by both in situ hybridization and immunostaining are similar, providing further support for the specificity of the antibody that we generated.
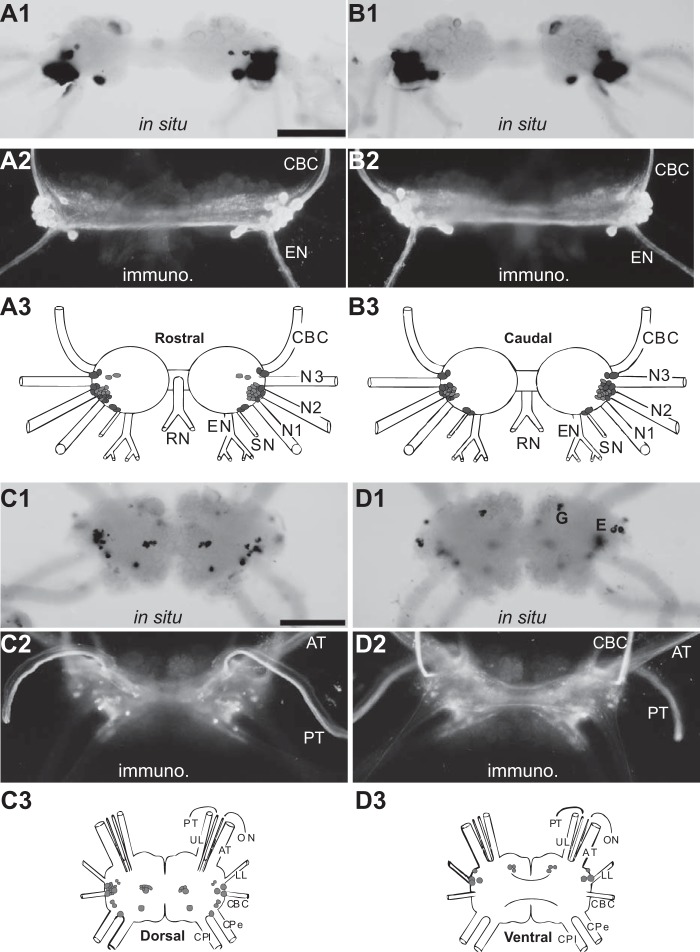
Figure 4.
Distribution of ALK-positive neurons and fibers in the buccal and cerebral ganglia (whole mounts). A and B, rostral (A) and caudal (B) buccal ganglia. Note the immunoreactive axons in the CBC. C and D, dorsal (C) and ventral (D) cerebral ganglia. 1, in situ hybridization (in situ); 2, immunohistochemistry (immuno.); 3, composite drawings of ALK neurons. In situ hybridization and immunohistochemistry reveal that the majority of ALK neuropeptides are located in the buccal ganglion. Note the staining of G cluster and E cluster neurons on the cerebral ventral surface (D1). Darker shades of gray indicate more intense staining. Scale bar, 500 μm (in A1 and C1). Buccal abbreviations are as follows. N1, nerve 1; N2, nerve 2; N3, nerve 3; SN, salivary nerve; EN, esophageal nerve; RN, radula nerve. Cerebral abbreviations are as follows. UL, upper labial nerve; PT, posterior tentacular nerve; ON, optic nerve; AT, anterior tentacular nerve; LL, lower labial nerve; CPe, cerebropedal connective; CPl, cerebropleural connective.
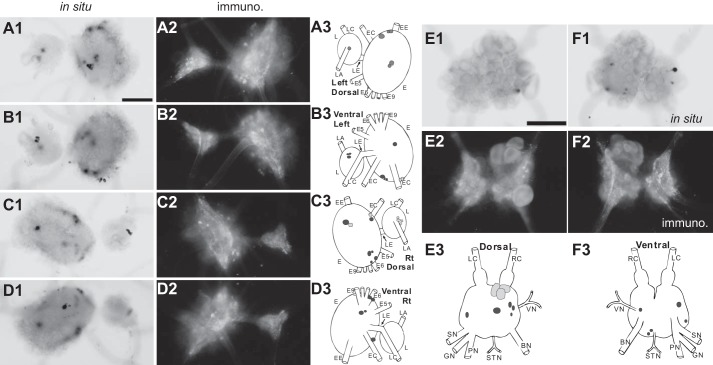
Figure 5.
Distribution of ALK-positive neurons and fibers in the pleural and pedal ganglia and the abdominal ganglion (whole mounts). A–D, left dorsal (A), left ventral (B), right dorsal (C), and right ventral (D) pedal ganglia. E and F, dorsal (E) and ventral (F) abdominal ganglia. 1, in situ hybridization (in situ); 2, immunohistochemistry (immuno.); 3, composite drawings of ALK neurons. Darker shades of gray indicate more intense staining. Scale bar (in A1 and E1), 500 μm. Pleural-pedal abbreviations are as follows. Rt, right; L, pleural ganglion; E, pedal ganglion; LE, pleuropedal connective; EE, pedal commissure; EC, cerebropedal connective; LC, cerebropleural connective; LA, pleuroabdominal connective; E5, posterior tegumentary nerve (P5); E6, anterior parapodial nerve (P6); E9, posterior pedal nerve (P9). Abdominal abbreviations are as follows. LC, left pleuroabdominal connective; RC, right pleuroabdominal connective; VN, vulvar nerve; BN, branchial nerve; STN, spermathecal nerve; PN, pericardial nerve; GN, genital nerve; SN, siphon nerve. For simplicity, not all nerves in the pleural and pedal ganglia were drawn.
Consistent with Northern blot data, the heaviest labeling was observed in the buccal ganglion. Particularly well labeled was a cluster of dorsal-lateral cells that can be seen in both the rostral and caudal surfaces near the roots of the esophageal nerve (EN) and the buccal nerve 1 (Fig. 4, A2 and B2). Some neurons in this location project their axons in the cerebro-buccal connective (CBC). To determine whether this is the case for the ALK-positive neurons, we backfilled the buccal end of the CBC (Fig. 6B1) and processed preparations for immunohistochemistry (Fig. 6B2). Within the limits of our resolution, 12 of the 13 ALK-immunoreactive neurons were double-labeled, and therefore their axons are present in the CBCs. Note that the giant, largest B1/B2 neurons on the caudal surface (Fig. 4B), used as the driver for RDA, were negative in both in situ hybridization and immunohistochemistry experiments. This also indicates that the antibody we used is specific.
Figure 6.
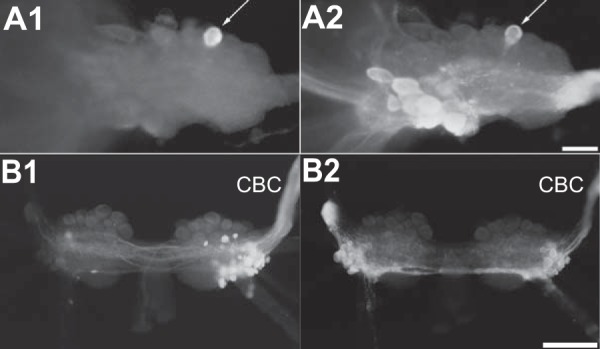
ALK is present in B48 and several buccal neurons that project to the CBC. A1, B48 injected with carboxyfluorescein. A2, immunostaining with an antibody to ALK-1. The figure shows that B48 (arrows) contains the ALKs. The buccal ganglion (one hemi-ganglion) was photographed from the caudal surface. Scale bar, 100 μm. B1, backfill of CBC using biocytin (processed with avidin-fluorescein). B2, immunostaining with an antibody to ALK-1. Some cells are double-labeled. The buccal ganglion was photographed from the rostral surface. Scale bar, 500 μm.
In the cerebral ganglion, we observed ALK-positive neurons scattered on the dorsal surface and within the G and E clusters of the ventral surface. Some neurons in the E cluster may be cerebral-buccal interneurons (CBIs) (21, 49–51). In immunostaining, we observed axons of at least some of these neurons that appeared to continue out of the anterior and posterior tentacular nerves (AT and PT) (Fig. 4, C2 and D2). Additionally, numerous immunoreactive axons were present in the CBC (Fig. 4, A2, B2, and D2). These results are consistent with the idea that ALKs play a role in feeding, as feeding motoneurons and pattern-generating interneurons are located in the buccal ganglion, and feeding higher-order interneurons (i.e. CBIs) are located in the cerebral ganglion (52–54).
Other ganglia, including the pleural-pedal and abdominal ganglion (Fig. 5), did show staining of some scattered cells, but the overall staining was relatively weak. Moreover, immunostaining showed little staining of axons.
Because the buccal motoneuron B48 was the original source of the tester material used for RDA, we performed double-labeling experiments to determine whether B48 is ALK-immunoreactive (n = 3). We physiologically identified B48 neurons, injected carboxyfluorescein (Fig. 6A1), and then performed immunohistochemistry (Fig. 6A2). B48 was indeed ALK-immunopositive.
Mass spectrometric analysis of prohormone processing and posttranslational modifications (PTMs)
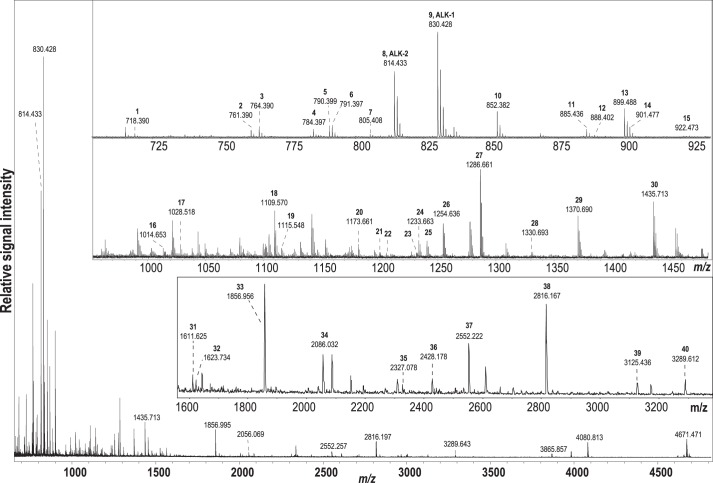
MALDI-TOF MS is well suited for profiling neuropeptides from small samples, such as individual Aplysia neurons (55, 56). Here, we isolated eight individual buccal neurons from left and right dorsal–lateral clusters expressing ALK mRNA and showing immunoreactivity and profiled them using MALDI-TOF MS to determine the processing of the ALK precursor and the structure of the final peptides expressed. We detected a total of 40 peptides that had masses that matched in silico-predicted (45, 46) ALK precursor-derived peptides. The most intense signal corresponded to the amidated form of ALK-1, which is present in 20 copies on the prohormone, at m/z 830.402 ± 10 ppm (Fig. 7 and supplemental Table S2). Another amidated peptide, ALK-2, was also detected at high intensity. In total, 16 predicted amidated peptides and 24 linker peptides have been detected (supplemental Table S2). Among the 16 amidated peptides, all but one (DSPRMFAFNSLS-amide) have a C-terminal pentapeptide motif (FXXWX-amide) (see “Discussion”) with the last aa being Ser, Gly, Ala, or Asn, and thus are classified as ALKs (Fig. 2). Isotopic pattern fit has been calculated to verify the assignment of select peptides with close molecular weights and to resolve their overlapping isotopic clusters in the mass spectrum (PAFSPWS-NH2, m/z 790; PAFSAWN-NH2, m/z 791; PAFHSWS-NH2, ALK-1, m/z 830; PAAFHAWS-NH2, m/z 885; PAFHSWSG, m/z 888; PRFHAWS-NH2, m/z 899; LAAFHAWS-NH2, m/z 901). Interestingly, among detected peptides, four are presumably processing intermediates (supplemental Table S2).
Figure 7.
Processing of the ALK prohormone as detected by MALDI-TOF MS in individual buccal neurons. The main panel shows a representative spectrum over a wide mass range from one of the eight neurons; insets show enlarged views into mass range segments from spectra obtained on different neurons because not all reported peptides were detected in a single spectrum (see supplemental Table S2). Detected peptides are labeled according to their numerical order in supplemental Table S2.
We also performed MALDI-TOF MS analysis of the CBC to determine whether it contains ALK precursor-derived peptides (i.e. whether ALK precursor-derived peptides are present in cerebral–buccal and/or buccal–cerebral interneurons). These two types of interneurons play an important role in the generation of feeding motor programs (16–18, 22, 49). We did indeed detect ALK precursor-derived peptides, including ALK-1 and ALK-2, in the CBC. These data are consistent with the idea that the ALKs may influence feeding.
Modulatory actions of ALKs in the feeding circuit
Feeding in Aplysia has both appetitive and consummatory phases (57). Because food is ingested during the consummatory phase, it has been the subject of much research (49, 58, 59). Consummatory behaviors are mediated by the radula, which protracts and retracts and opens and closes. Behaviors are ingestive if the radula closes during retraction (to pull food into the buccal cavity) and are egestive if the radula closes during protraction (which pushes food out) (58, 59).
Previous studies have shown that fictive feeding or feeding motor programs, which are an in vitro representation of consummatory feeding, can be elicited in the isolated CNS by stimulation of a command-like interneuron, CBI-2 (14, 16–18, 20, 22, 49, 60). CBI-2 evokes motor programs through its excitatory actions on pattern-generating interneurons in the buccal ganglion, such as B34, which in turn provide inputs to other pattern-generating neurons. Pattern-generating interneurons drive motoneurons in the buccal ganglion, which ultimately produce the behavior.
In the isolated CNS, radula protraction is monitored by recording extracellularly from the I2 nerve, which contains axons of protraction motoneurons (15). Radula retraction can be indirectly monitored by recording intracellularly from protraction interneurons (i.e. they are hyperpolarized during retraction). Radula closing is monitored by recording intracellularly from the B8 motoneurons. Motor programs are ingestive when B8 is predominantly active during retraction, egestive when B8 is active during protraction, and intermediate when B8 is active during both protraction and retraction.
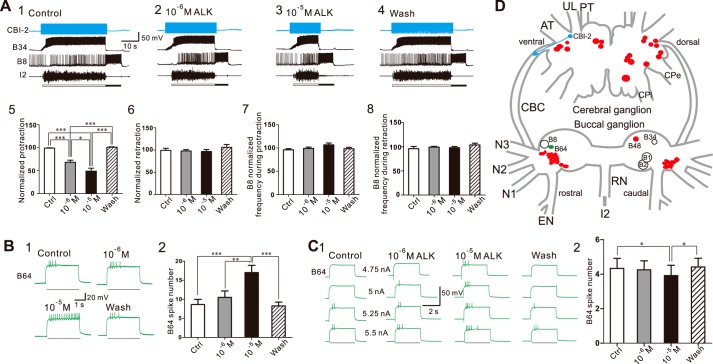
We sought to determine whether the ALKs are bioactive in the feeding circuit. We tested both ALK-1 and ALK-2 and found that their effects were similar, so we pooled the results from ALK-1 and ALK-2 together. In the first set of experiments, we examined whether the ALKs can modulate parameters of motor programs elicited by CBI-2. We elicited a single cycle of a motor program by stimulating CBI-2 at 8–10 Hz. The interstimulation interval was 1.5 min. One such experiment is shown in Fig. 8 (A1–4). Under control conditions (Fig. 8A1), CBI-2 stimulation elicited an ingestive motor program (i.e. B8 was predominantly active during retraction). When the preparation was superfused with 10−6 m ALK (Fig. 8A2) and 10−5 m ALK (Fig. 8A3), there was a concentration-dependent shortening of protraction (Fig. 8A5, F(3, 21) = 40.93, p < 0.001, n = 8). Other phases of the motor program were not significantly altered (i.e. there was no effect on retraction duration (Fig. 8A6, F(3, 21) = 0.63, p > 0.05, n = 8), or on B8 activity during protraction (Fig. 8A7, F(3, 21) = 1.68, p > 0.05, n = 8) or retraction (Fig. 8A8, F(3, 21) = 0.79, p > 0.05, n = 8)).
Figure 8.
ALKs modulate feeding motor programs elicited by CBI-2 partly through their actions on the interneuron B64. A, ALKs shorten protraction duration in a concentration-dependent manner. A1–4, a representative example; a single cycle of a motor program was elicited by simulating CBI-2 at 10 Hz until the end of the protraction phase, detected by the onset of the sharp synaptic inhibition of interneuron B34 and the abrupt ending of I2 nerve activity. Protraction phase (open bar) is defined by the activity in the I2 nerve. Retraction phase (filled bar) is defined by a period of hyperpolarization of B34 after the protraction phase is terminated and also by a period of high-frequency activity of the radula closing motoneuron B8. Upon wash, protraction duration returned to its control value. A5, group data on protraction duration. In contrast, the ALKs had no significant effect on the duration of retraction (A6) or B8 activity during either protraction (A7) or retraction (A8). B and C, ALKs increase B64 excitability (B1) and decrease B64 threshold to spike (C1). B2 and C2, group data. Bars in B1 and C1 denote current injections in B64. Recordings in A were made in ASW, whereas recordings in B and C were made in high divalent saline. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (Bonferroni post hoc tests). Error bars, S.E. D, schematic drawing of the Aplysia cerebral and buccal ganglia, illustrating the locations of the neurons recorded from and the source (ALK neurons, red) and target (B64, green) of ALKs. B1/B2, used as a driver for the RDA, are also illustrated. For the cerebral ganglion, neurons on the ventral surface are shown on the left, dorsal on the right. For the buccal ganglion, neurons on the rostral surface are shown on the left, caudal on the right. Nerve abbreviations are as in Fig. 4.
Similar to most rhythmic behaviors, Aplysia feeding behavior is controlled by a CPG. In a subsequent set of experiments, therefore, we sought to determine whether the ALKs shorten the duration of the protraction phase via modulation of the CPG. Specifically, we examined the effects of ALKs on the excitability of the CPG element, interneuron B64. Previous studies have shown that B64 acts as a terminator of protraction and thus as a shortener of protraction (15, 61). We found that the ALKs increased B64 excitability in a concentration-dependent manner (Fig. 8B, F(3, 21) = 12.97, p < 0.001, n = 8). In addition, the ALKs decreased B64 threshold to spike (Fig. 8C, F(3, 6) = 10.37, p < 0.01, n = 3).
Discussion
Here, we used RDA and RACE to discover a novel, unusually long neuropeptide precursor that encodes ALKs. Below, we discuss our approach and evolutionary and functional aspects of leucokinin-like peptides.
Utility of our approach and properties of the ALK precursor
It is most notable that the ALK precursor is the first verified and functionally characterized leucokinin-like precursor outside of arthropods (see supplemental Figs. S1 and S2). Importantly, RDA proved to be critical for the identification of this protein. Querying the NCBI public depositories with ALK precursor, we did not find any leucokinin precursors from other animals. This is not surprising, given the fact that shared motifs (Table 1) that interact with their cognate receptor are short (i.e. ∼5 amino acids). Large sections of leucokinin-like prohormones are often species-specific (see supplemental Figs. S1 and S2). Thus, the homology of the full-length prohormone to well-studied species is not long enough to offer accurate neuropeptide gene annotation by similarity across species. In addition, searches of NCBI EST (48) and of a recently completed, more complete transcriptome database (Aplysia gene tools) (supplemental Fig. S3) did come up with some matching sequences, but they were all shorter pieces, none of them encoding the complete ALK precursor. The absence of the ALK precursor in any currently available database reinforces the utility of our RDA-based approach in identifying long sequences like the ALK precursor.
RDA is a non-targeted approach to identify potentially novel peptide precursors that are present in motoneuron B48 but not in neurons B1/B2. RDA generated incomplete transcripts. Our subsequent usage of RACE enabled us to generate the complete precursor (i.e. the unusually long ALK precursor). By contrast, all currently identified/predicted leucokinin-like peptide precursors, including several unverified molluscan leucokinin-like peptide precursors (supplemental Fig. S1 and Table S1) range from 97 to 347 aa. At present, it is not clear why the ALK precursor is so long. This may, however, become clearer as additional leucokinin-like peptide precursors are identified in other molluscs, including other Aplysia species. Regardless, it is likely that the ALKs play critical regulatory roles in Aplysia, given that a number of copies of ALK-1 and ALK-2 can be expressed and secreted through a single translation.
We had difficulty in determining the signal peptide for the ALK precursor. Perhaps the occurrence of numerous basic and acidic amino acid residues in the predicted amino terminus of the prohormone precursor made it less likely to form hydrophobic α-helix (H-region) in the center of a signal peptide. In fact, the exact length and location of the cleavage site of the signal peptide cannot be confidently predicted by prediction tools (47, 62) (supplemental Table S1). We found a similar challenge with the leucokinin-like peptide precursor of Lucilia cuprina (supplemental Table S1), where signal peptide cleavage is predicted at a low 71% probability. This suggests that signal peptide predictions may be challenging for some neuropeptide precursors. Regardless, the presence of ALKs in the CNS is indicated by our immunostaining and direct mass spectrometric measurement. The latter experiments detected multiple processed peptides in individual neurons and in the CBC. Thus, our data indicate that ALK transcript is expressed, translated, and posttranslationally processed like a typical neuropeptide prohormone. Moreover, numerous processed peptides undergo further PTM (i.e. amidation of C terminus), which often renders biological activity to mature peptides.
Evolutionary relationship of leucokinin-like peptides
To compare peptides, we selected a number of identified and predicted leucokinins from other species (Table 1). We included peptides from molluscs, annelids, and nematodes. Some of the included peptides are predicted in silico from putative precursors, and some of the predicted precursors are incomplete. Leucokinin-like peptides in arthropods (mostly insects) and annelids share an amidated C-terminal pentapeptide motif called FXXWG-amide. In nematodes, the motif becomes FXXWA-amide. In molluscs, leucokinin-like peptides showed some variability in the last amino acid. In most cases (including Lymnaea (39) and Aplysia), this motif is FXXWS-amide. In fact, the Lymnaea leucokinin and ALK-1 only differ by a single amino acid. Other motifs, such as FXXWG-amide and FXXWA-amide, are observed. Indeed, Aplysia also express an FXXWG-amide peptide (PAFHAWG-amide) and an FXXWA-amide peptide (AGFAPWA-amide), and both are present in single copies (Fig. 2 and supplemental Table S2). Thus, the ALK precursor encodes peptides matching most motifs currently identified for leucokinin-like peptides, which is perhaps not surprising, given its unusually long sequence. This also suggests that the ALK precursor may reflect the structure of an ancient leucokinin-like peptide prohormone.
Initially, leucokinins were thought to be orthologues of the vertebrate tachykinins (which include substance P) (e.g. see Ref. 34). Tachykinins have a somewhat similar C-terminal pentapeptide motif, FXGLM-amide. The similarity between the Drosophila leucokinin receptor (Lkr) and vertebrate tachykinin receptors appears to support this notion (63). However, the Drosophila genome encodes several other peptides with a C-terminal motif (FXGXR-amide) that are more similar to tachykinins than leucokinins (31, 64). More recently, two extensive bioinformatic studies of a large number of verified and predicted neuropeptide precursors and G-protein–coupled receptors (GPCRs) (35, 36) have demonstrated that tachykinin orthologues are present in both protostomes and deuterostomes (see Data Set S1 of Ref. 36). On the other hand, leucokinin orthologues are only present in protostomes (including arthropods and molluscs) (35, 36, 65). Thus, it is likely that leucokinin orthologues are not orthologues of the tachykinins and are not even present in vertebrates.
In protostomes, most of the known leucokinins and their precursors are from arthropods. The ALK precursor is the first complete, structurally and functionally characterized precursor that is not present in an arthropod. The two bioinformatic studies mentioned above (35, 36) did, however, identify in silico leucokinin-like precursors that are non-arthropod. Some of these are incomplete transcripts. Others are complete and include precursors from the annelid Malacoceros fuliginosus, the mollusc Tritonia diomedea, and the nematode Trichinella spiralis (supplemental Fig. S1 and Table S1; some of the predicted leucokinin-like peptides are listed in Table 1).
Diverse functions of leucokinin-like peptides: Roles in feeding
Before our study, functional studies of leucokinin-like peptides were almost entirely conducted in insects. An exception, however, is research that has been conducted in Lymnaea. The GPCR for the Lymnaea leucokinin has been identified, and one biochemically isolated Lymnaea leucokinin (i.e. lymnokinin) exerts effects via this receptor (39). However, the lymnokinin distribution in the CNS has not been determined using either in situ hybridization or immunohistochemistry.
In insects, although earlier work focused on the roles of leucokinin-like peptides in the digestive system and fluid secretion (31, 34), a more recent study conducted in Drosophila demonstrated that leucokinin signaling pathways regulate meal size, indicating a major role in feeding (34). This is the first and only functional study of the role of the leucokinins in feeding.
In the present work, we provide multiple lines of evidence that clearly show that ALKs play a marked role in Aplysia feeding (Fig. 8D). First, Northern blots, in situ hybridization, immunohistochemistry, and MALDI-TOF MS all demonstrate that the ALKs are predominantly located in the buccal ganglion, which directly controls feeding. Immunostaining also shows the ALKs are present in the CBC, which links the buccal ganglion to the cerebral ganglion (which also contains feeding circuitry). Thus, the ALKs are present in feeding neurons and processes.
Second, we directly demonstrate that ALKs modulate one specific parameter of the protraction-retraction sequence of consummatory feeding (i.e. they shorten protraction in motor programs evoked by a command-like neuron in the isolated CNS). Previous studies have demonstrated that identical patterns of motor activity can be recorded from both behaving animals (59, 66) and in vitro ganglia (17, 60). Thus, the in vitro effects of ALKs probably translate to effects on consummatory feeding behavior. In addition, we identified a potential mechanism by which ALKs shorten protraction because ALKs enhance the excitability and decrease the spiking threshold of an identified interneuron (B64). This neuron is normally active during retraction and serves to terminate protraction (15, 61). An increase in its excitability and a decrease of threshold would therefore be expected to shorten protraction.
Future studies may identify the ALK-positive neurons that release ALKs and shorten protraction. For example, it would be of interest to identify the ALK-positive neurons that send their axons to the CBCs. Regardless of which neurons are the sources of ALKs, the current study and the work on Drosophila (34) both support the idea that leucokinin-like peptides play a critical role in various aspects of consummatory feeding. Importantly, in both Drosophila and Aplysia, leucokinin-like peptides do not prevent the execution of feeding behavior or the generation of motor programs. Rather, they exert modulatory actions. Specifically, our study suggests that ALKs may promote faster consumption of food because a shorter protraction is likely to result in a shorter cycle period. In contrast, in Drosophila, leucokinins function to reduce meal size and increase meal frequency, possibly by enhancing a gut distension signal. The Drosophila work used a gross measure of feeding (i.e. the amount of food intake) but did not monitor the duration of individual feeding movements. Consequently, it is presently unclear how the feeding effects in the two species may be related. Importantly, to the best of our knowledge, our findings represent the first demonstration of modulation of the feeding CPG by leucokinin-like peptides, as this has not been previously demonstrated in any animals, including arthropods.
In addition, given the presence of a number of ALK-positive neurons in the buccal and cerebral ganglia, it is likely that ALKs may also play a role in other aspects of feeding, including regulation of meal size and frequency, as is the case in Drosophila, which would be another interesting subject for future studies. In this context, it is notable that Aplysia Neuropeptide Y plays a role during the transition from hunger to satiation and reduces meal size (67). Effects of Aplysia Neuropeptide Y are exerted by reconfiguring the feeding CPG and converting ingestive programs to egestive activity. In contrast, if ALKs also reduce meal size, it is unlikely that they do so by reconfiguring the feeding circuit. This is suggested by the fact that we demonstrate that patterns of B8 activity during protraction and retraction did not change (Fig. 8, A7–8). This indicates that program type was not altered.
Finally, wide distributions of ALKs in the Aplysia CNS also suggest that ALKs may have broader functions in the modulation of other behavioral networks. Although not as prominent, ALK-positive neurons are present in the pleural-pedal and abdominal ganglia, which mediate other behaviors, such as locomotion and visceral functions. Indeed, some other roles of leucokinins in insects have been reviewed recently (68).
In summary, we used RDA to identify ALK mRNA. We mapped the distribution of ALKs and characterized proteolytic processing and PTM of the ALK precursor using single-cell mass spectrometry. Finally, we demonstrated a novel role of ALKs in modulating a movement parameter of feeding motor programs. Importantly, we identified a CPG element as a target of leucokinin modulation that may carry out leucokinin action on the duration of the protraction phase of feeding responses. The ALK precursor represents a first verified precursor for leucokinin-like peptides outside of arthropods. Our functional studies of ALKs in the feeding system taken together with previous work in Drosophila establish that leucokinin-like peptides play a regulatory role in different aspects of feeding, supporting their diverse functions in protostomes. Moreover, our multidisciplinary approach based on RDA is also likely to be useful in other model systems in which specific neurons are identifiable.
Experimental procedures
Animals
Experiments were performed on A. californica (10–300 g), purchased from Marinus Scientific (Long Beach, CA) and the Aplysia Research Facility (Miami, FL). Aplysia are hermaphroditic (i.e. each animal has reproductive organs normally associated with both male and female sexes). Animals were kept in an aquarium containing aerated and filtered artificial seawater (Instant Ocean, Aquarium Systems Inc., Mentor, OH) at 14–16 °C until use. The animal room was equipped with a 24-h light/dark cycle with light period from 6:00 a.m. to 6:00 p.m. Before dissection, animals were anesthetized by injection of isotonic 333 mm MgCl2 (about 50% of body weight) into the body cavity.
Reagents and peptides
All reagents were purchased from Sigma-Aldrich unless otherwise indicated. We synthesized two Aplysia leucokinin-like peptides: ALK-1 (PAFHSWS-amide) by ChinaPeptides Co., Ltd., and ALK-2 (PAFHAWS-amide) by PeptidoGenic Research Co., Inc. The peptide standards for MS calibration were supplied by Bruker Daltonics (Bremen, Germany).
RDA
Previously, we modified the RDA approach (69, 70) to identify novel peptides from single identified neurons in the Aplysia CNS, as described in detail elsewhere (11).
Briefly, the entire procedure can be divided into three steps (Fig. 1). 1) Two types of cells are isolated, where the first is the cell of interest, or “tester,” whose proteins are to be identified. The second cell, or “driver,” is used to subtract sequences that are shared with the tester. The rationale behind this RDA approach is that the tester contains one or more proteins not present in the driver. In this study, the buccal motoneuron B48 of the feeding network was the tester, whereas buccal neurons B1/B2 were the drivers. Because any peptides unique to B48 can potentially be identified, this method can be considered a non-targeted approach for peptide identification. Multiple cells from each cell type were collected in a solution of ice-cold 50% propylene glycol and 1.2 m NaCl in diethylpyrocarbonate-treated H2O and stored at −80 °C. 2) Amplification of the cDNA from the RNA of the tester and the driver was done as described previously (71). 3) RDA (69, 70) with the amplified cDNA of the tester and the driver was performed. The cDNAs of the driver and tester were digested with DpnII, and the driver cDNA was ligated to R-Bam adaptors, whereas the tester cDNA was ligated to N-Bam adaptors. DpnII was used to cut cDNAs with an average length of 256 bp that contained overhangs to facilitate ligation of adapters for PCR and to clone amplified fragments, because the overhangs are complementary to BamHI-digested plasmids. The driver DNA was amplified with biotinylated R-Bam primer and then hybridized with unamplified tester cDNA. Here, we modified the RDA method by including a physical subtraction based on biotinylated driver primers, streptavidin incubation, and phenol/chloroform extraction (72). This procedure removed the driver and with it any tester cDNA that is complementary and bound to it. We then performed the final amplification with tester N-Bam primer to further enrich the protein sequences that were unique to the tester.
Cloning
Standard molecular techniques were used except where noted. We used random primers to generate 32P-labeled fragments from the incomplete mRNA transcripts derived from the above RDA procedure. These fragments were then used to screen the poly(T) pBluescript library. The isolated clones were subcloned and sequenced. Then RACE was performed to obtain the complete sequence of the ALK precursor. RACE includes the following steps. First, total RNA isolated from the buccal ganglion was reverse transcribed to cDNA. Next, 5′-RACE and 3′-RACE were implemented using gene-specific primers.
Sequence comparison
We compared leucokinin-like peptides (Table 1) and the ALK precursor and 16 leucokinin-like peptide precursors (supplemental Figs. S1 and S2). For the peptide comparison in Table 1, we generated a frequency plot of each aa (aligned from the C terminus) using Weblogo software (http://weblogo.berkeley.edu/logo.cgi)5 with default parameters except that bitmap resolution was changed to 300. The larger the character for each aa is, the higher was the frequency of the aa appearing in the group of the peptides.
For precursor comparison in supplemental Figs. S1 and S2, the 16 precursor sequences were downloaded from the NCBI protein database (https://www.ncbi.nlm.nih.gov/protein) (supplemental Table S1). We generated the sequence alignment (supplemental Fig. S1) using BioEdit Software. We used MEGA7 software to generate a phylogenetic tree. First, the 17 sequences were aligned by ClustalW, using default parameters. Second, the alignment result was used to perform phylogenetic analysis (supplemental Fig. S2) with a construct/test maximum likelihood tree, of which a phylogeny was chosen using the Bootstrap method. The other parameters were set as default.
Antibodies
Antibodies were generated to ALK-1 (PAFHSWS-amide). The peptide antigens were prepared as described in detail previously (9, 73, 74). Briefly, the antigen was made by coupling ALK-1 to BSA (Sigma, catalogue no. A0281), using 1-ethyl-3-(dimethylaminopropyl)carbodiimide (Sigma, catalogue no. E7750), and then purified. For each antigen, two male Sprague-Dawley rats (250–300 g; Taconic Farms) were immunized by intraperitoneal injections. At days 21 and 42, the rats were boosted by intraperitoneal injections. Animals were killed by decapitation at 49 d, and the blood was harvested and processed for serum. Sera were aliquoted, frozen, and lyophilized or stored at 4 °C with EDTA (25 mm final concentration), and thimerosal (0.1% final concentration) was added as a stabilizer. For antibodies that produced immunostaining, specificity was confirmed by preincubation overnight of the primary antibodies with the corresponding synthetic peptide (i.e. ALK-1 (100 μm)), which abolished the staining (supplemental Fig. S4).
Northern blot analysis
Northern blot analysis was performed as described previously (11, 73, 74). Briefly, RNA was isolated from homogenized pooled ganglia using the acid-phenol method (75). RNA from each ganglion type (buccal, cerebral, pleural, pedal, and abdominal) was fractionated separately using denaturing agarose gels (1.5%) and transferred to nylon membranes (Biodyne B, Invitrogen). The RNA was immobilized with Stratalinker UV (Stratagene) and visualized by staining with 0.02% methylene blue in 0.3 m sodium acetate, pH 5.5. After washing out the excess stain with DEPC-treated water, the blot was scanned to document the amount of RNA transferred from each lane. After destaining with 1% SDS, 1 mm EDTA, and 50 mm Na3PO4, pH 7.2, the blot was prehybridized for 1 h at 50 °C using 50% formamide, 10% dextran sulfate, 7% SDS, 10 mm EDTA, 50 μg/ml salmon sperm DNA, and 250 mm Na3PO4, pH 7.2. Heat-denatured, random-primed (New England Biolabs), [32P]dCTP-labeled cDNA probe (ALK precursor (4848–5235 bp), 388 bp in length) was added, and hybridization was continued overnight at 50 °C. Blots were washed twice for 15 min at room temperature with 2× standard saline phosphate EDTA (SSPE) and 0.1% SDS, washed for 60 min at 50 °C with 0.1× SSPE and 0.1% SDS, and exposed to film. Autoradiographs and methylene blue–stained blots were scanned into Photoshop (Adobe Systems) and compiled into figures.
In situ hybridization
In situ hybridization was performed as described previously (40, 71). Ganglia were digested with 1% protease type IX (Sigma-Aldrich) in 10 ml of artificial seawater (ASW) for 3 h at room temperature (with rocking) to facilitate the removal of the sheath. After digestion, ganglia were washed with ASW and fixed overnight at 4 °C with 4% paraformaldehyde (Electron Microscopy Sciences) in PBS. Ganglia were then washed, desheathed, and dehydrated in an ascending ethanol series. After rehydration in a descending ethanol series, the ganglia were prehybridized for 6 h and then hybridized overnight at 50 °C in hybridization buffer (50% formamide, 5 mm EDTA, 5× SSC, 1× Denhardt's solution, 0.1% Tween 20, and 0.5 mg/ml yeast tRNA) containing 2 μg/ml digoxigenin-labeled cRNA probes made from a partial clone from RDA, which corresponded to ALK precursor (4848–5235 bp) (388 bp in length). After washout of the probes, ganglia were then incubated overnight at 4 °C with a 1:200 dilution of alkaline phosphatase–conjugated anti-digoxigenin antibody (Roche Applied Science) in PBS containing 0.1% Tween 20 (PBT), 0.2% bovine serum albumin (BSA) (catalogue no. A0281, Sigma-Aldrich), and 1% normal goat serum. After washes with PBT to remove unbound antibody, ganglia were washed with detection buffer (100 mm NaCl, 50 mm MgCl2, 0.1% Tween 20, 1 mm levamisol, and 100 mm Tris-HCl, pH 9.5) and developed with 4.5 μl of nitro blue tetrazolium and 3.5 μl of 5-bromo-4-chloro-3-indolyl phosphate (Roche Applied Science) in 1 ml of detection buffer. The staining reaction was monitored visually and stopped by washing with PBT when the level of staining was adequate. The stained ganglia were observed and photographed using a fluorescence microscope (Nikon) with epiillumination against a white background. Photographs were taken with a Nikon CoolPix 990 digital camera, imported into Photoshop, and compiled into figures.
Immunohistochemistry
Immunohistochemistry was performed as described previously (67, 74). The tissue was fixed in a buffer (4% paraformaldehyde, 0.2% picric acid, 25% sucrose, and 0.1 m NaH2PO4, pH 7.6) for either 3 h at room temperature or overnight at 4 °C. All subsequent incubations were done at room temperature. The tissue was washed with PBS and was permeabilized and blocked by overnight incubation in blocking buffer (10% normal goat serum, 2% Triton X-100, 1% BSA, 154 mm NaCl, 50 mm EDTA, 0.01% thimerosal, and 10 mm Na2HPO4, pH 7.4). The primary antibody was diluted 1:250 in blocking buffer and incubated with the tissue for 4–7 days. The tissue was then washed twice per day for 2–3 days with washing buffer (2% Triton X-100, 1% BSA, 154 mm NaCl, 50 mm EDTA, 0.01% thimerosal, and 10 mm Na2HPO4, pH 7.4). After washing, the tissue was incubated with a 1:500 dilution of secondary antibody (lissamine rhodamine goat anti-rat; Jackson ImmunoResearch) for 2–3 days and then washed again two times with washing buffer for 1 day and four times with storage buffer (1% BSA, 154 mm NaCl, 50 mm EDTA, 0.01% thimerosal, and 10 mm Na2HPO4, pH 7.4) for 1 day. We observed and photographed the tissue under a fluorescence microscope.
For double labeling of physiologically identified B48 cells with ALK immunohistochemistry, neurons were identified based on morphology and electrophysiological characters and injected with carboxyfluorescein (76). For double labeling with backfills, CBCs of live buccal ganglia were cut, and the buccal ends of the CBCs were incubated overnight in a Vaseline well containing biocytin. This allowed biocytin transport to the somata of neurons with axons in the CBC. Ganglia were then fixed and processed with avidin-fluorescein so that somata were green. Tissues were also processed for immunocytochemistry as described above.
Mass spectrometric analysis of peptides
To characterize ALKs in the Aplysia CNS, we used MALDI-TOF MS on individual ALK-expressing neurons from the buccal ganglion and the CBCs. Buccal hemi-ganglia were treated with 1% protease IX in ASW with antibiotics (100 units/ml penicillin G, 100 μg/ml streptomycin, and 100 μg/ml gentamicin) for 45 min at 34 °C and then desheathed. Individual buccal neurons were manually isolated according to the ALK in situ hybridization staining map using electrolytically sharpened tungsten needles. Isolated neurons were transferred one by one onto a stainless steel MALDI sample plate (Bruker Daltonics) using homemade plastic micropipettes filled with deionized water. Excess liquid was aspirated from the sample plate, and 0.5 μl of saturated DHB matrix (DHB: 2,5-dihydroxybenzoic acid, 20 mg/ml deionized water) was applied onto the sample spots. Peptide profiles were measured using an ultrafleXtreme MALDI-TOF/TOF mass spectrometer (Bruker Daltonics) equipped with a Smartbeam II frequency tripled Nd:YAG solid-state laser. Mass spectra were manually acquired in positive reflectron mode and calibrated externally using Bruker Peptide Mix II standards. Signals from 600–1800 laser shots fired at 2000-Hz frequency at multiple locations within each sample spot were summed into a representative cell spectrum. Obtained mass spectra were processed using flexAnalysis version 3.4 software (Bruker Daltonics).
To aid interpretation of the MS results, we generated a library of putative peptides encoded by the ALK prohormone using the NeuroPred prediction tool (http://stagbeetle.animal.uiuc.edu/cgi-bin/neuropred.py)5 (45, 46). When predicted masses of select peptides were very similar and signals in the mass spectrum were expected to overlap, the theoretical isotopic ratios of the accurate mass formula for each of the conflicting peptides were compared with the experimentally determined isotopic patterns using the Simulated Isotopic Pattern function in flexAnalysis version 3.4, and assignment was based on the best fit. Location of the putative signal peptide cleavage site was inferred from the PrediSi (http://www.predisi.de/)5 model (47).
In the single-cell MS experiments (Fig. 7), endogenous processed peptides were identified using a peptide mass fingerprint (PMF) approach (77). The PMF approach is effective when numerous peptides are encoded by a specific prohormone and are detected experimentally. We verified that the masses of experimentally detected peptides were within the acceptable margin of error from theoretical masses of predicted peptides derived from the ALK precursor. In other words, we fingerprinted ALK precursor-derived peptides by detecting the predicted processed peptides. The fact that we detected 40 peptides from the ALK prohormone (supplemental Table S2) increased the confidence of our assignments. The PMF method is accepted in protein characterization studies, and its accuracy is determined by both the mass assignment errors and the number of predicted peptides that are detected (both parameters are included in supplemental Table S2).
Electrophysiology
Intracellular and extracellular recording techniques were utilized as described previously (13, 25, 40, 78, 79). Briefly, ganglia were desheathed, transferred to a recording chamber containing 1.5 ml of ASW (460 mm NaCl, 10 mm KCl, 11 mm CaCl2, 55 mm MgCl2, and 10 mm HEPES, pH 7.6), continuously perfused at 0.3 ml/min, and maintained at 14–17 °C. Peptides, ALK-1 or ALK-2, were dissolved in ASW immediately before each physiological test, and the peptide/ASW solution was perfused into the recording chamber. Some experiments were also performed in a high divalent saline (368 mm NaCl, 8 mm KCl, 13.8 mm CaCl2, 115 mm MgCl2, and 10 mm HEPES, pH 7.6), which increases the spiking threshold of neurons and therefore curtails polysynaptic influences. Intracellular recordings were obtained using 5–10-megaohm sharp microelectrodes filled with an electrolyte (0.6 m K2SO4 plus 60 mm KCl). Extracellular recordings were acquired from polyethylene suction electrodes. Grass S88 and WPI Pulsemaster A300 stimulators were used to provide timing signals for intracellular stimulation.
To test the effects of ALK-1 or ALK-2 on the feeding circuit, we performed experiments in the cerebral and buccal ganglia. The buccal ganglion innervates the feeding organ (radula). Feeding motor programs were elicited by stimulation of command-like neuron CBI-2 at 8–10 Hz and were monitored by cyclic bursts in the I2 nerve of the buccal ganglion (17, 80). Electrophysiological recordings were digitized online using AxoScope software version 10.7 (Molecular Devices, LLC, Sunnyvale, CA) and were plotted by CorelDraw version X7 (Corel Corp., Ottawa, Canada). Bar graphs are plotted with Prism version 5 (GraphPad Software, La Jolla, CA). Data are expressed as mean ± S.E. All statistical tests (e.g. repeated measures one-way analysis of variance) were performed using Prism. When the data showed significant effects in analysis of variance, further individual comparisons were performed with Bonferroni's correction.
Author contributions
G. Z., F. S. V., E. V. R., K. R. W., J. V. S., and J. J. conceived and designed the experiments. G. Z., F. S. V., D.-D. L., E. V. R., K. Y., W.-D. Y., H. X., A. B. H., T.-T. C., V. A., S.-Y. Y., S.-A. C., E. C. C., K. R. W., J. V. S., and J. J. performed the experiments and analyzed the data. G. Z., F. S. V., E. V. R., E. C. C., J. V. S., K. R. W., and J. J. wrote the paper.
Supplementary Material
This work was supported by National Natural Science Foundation of China Grants 31671097, 31371104, J1103512, and J1210026; NINDS, National Institutes of Health, Grants RO1 NS066587, RO1 NS070583, and RO1 NS031609; NIDA, National Institutes of Health, Grant P30 DA018310; and National Science Foundation Grant CHE 16-067915. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health and other funding agencies.
This article contains supplemental Tables S1 and S2 and Figs. S1–S4.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) MF664476.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- aa
- amino acid(s)
- CPG
- central pattern generator
- RDA
- representational difference analysis
- ALK
- Aplysia leucokinin-like peptide
- EST
- expressed sequence tag
- CBC
- cerebro-buccal connective
- CBI
- cerebral-buccal interneuron
- PTM
- posttranslational modification
- GPCR
- G-protein–coupled receptor
- SSPE
- standard saline phosphate EDTA
- ASW
- artificial seawater
- PMF
- peptide mass fingerprint.
References
- 1. Strand F. L. (1999) Neuropeptides: Regulators of Physiological Processes, MIT Press, Cambridge, MA [Google Scholar]
- 2. Nusbaum M. P., and Blitz D. M. (2012) Neuropeptide modulation of microcircuits. Curr. Opin. Neurobiol. 22, 592–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Taghert P. H., and Nitabach M. N. (2012) Peptide neuromodulation in invertebrate model systems. Neuron 76, 82–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marder E., O'Leary T., and Shruti S. (2014) Neuromodulation of circuits with variable parameters: single neurons and small circuits reveal principles of state-dependent and robust neuromodulation. Annu. Rev. Neurosci. 37, 329–346 [DOI] [PubMed] [Google Scholar]
- 5. Cropper E. C., Lloyd P. E., Reed W., Tenenbaum R., Kupfermann I., and Weiss K. R. (1987) Multiple neuropeptides in cholinergic motor neurons of Aplysia: evidence for modulation intrinsic to the motor circuit. Proc. Natl. Acad. Sci. U.S.A. 84, 3486–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cropper E. C., Price D., Tenenbaum R., Kupfermann I., and Weiss K. R. (1990) Release of peptide cotransmitters from a cholinergic motor neuron under physiological conditions. Proc. Natl. Acad. Sci. U.S.A. 87, 933–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Church P. J., and Lloyd P. E. (1991) Expression of diverse neuropeptide cotransmitters by identified motor neurons in Aplysia. J. Neurosci. 11, 618–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li L., Moroz T. P., Garden R. W., Floyd P. D., Weiss K. R., and Sweedler J. V. (1998) Mass spectrometric survey of interganglionically transported peptides in Aplysia. Peptides 19, 1425–1433 [DOI] [PubMed] [Google Scholar]
- 9. Sweedler J. V., Li L., Rubakhin S. S., Alexeeva V., Dembrow N. C., Dowling O., Jing J., Weiss K. R., and Vilim F. S. (2002) Identification and characterization of the feeding circuit-activating peptides, a novel neuropeptide family of Aplysia. J. Neurosci. 22, 7797–7808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Proekt A., Vilim F. S., Alexeeva V., Brezina V., Friedman A., Jing J., Li L., Zhurov Y., Sweedler J. V., and Weiss K. R. (2005) Identification of a new neuropeptide precursor reveals a novel source of extrinsic modulation in the feeding system of Aplysia. J. Neurosci. 25, 9637–9648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jing J., Sweedler J. V., Cropper E. C., Alexeeva V., Park J. H., Romanova E. V., Xie F., Dembrow N. C., Ludwar B. C., Weiss K. R., and Vilim F. S. (2010) Feedforward compensation mediated by the central and peripheral actions of a single neuropeptide discovered using representational difference analysis. J. Neurosci. 30, 16545–16558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vilim F. S., Sasaki K., Rybak J., Alexeeva V., Cropper E. C., Jing J., Orekhova I. V., Brezina V., Price D., Romanova E. V., Rubakhin S. S., Hatcher N., Sweedler J. V., and Weiss K. R. (2010) Distinct mechanisms produce functionally complementary actions of neuropeptides that are structurally related but derived from different precursors. J. Neurosci. 30, 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bai L., Livnat I., Romanova E. V., Alexeeva V., Yau P. M., Vilim F. S., Weiss K. R., Jing J., and Sweedler J. V. (2013) Characterization of GdFFD, a D-amino acid-containing neuropeptide that functions as an extrinsic modulator of the Aplysia feeding circuit. J. Biol. Chem. 288, 32837–32851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Church P. J., and Lloyd P. E. (1994) Activity of multiple identified motor neurons recorded intracellularly during evoked feedinglike motor programs in Aplysia. J. Neurophysiol. 72, 1794–1809 [DOI] [PubMed] [Google Scholar]
- 15. Hurwitz I., and Susswein A. J. (1996) B64, a newly identified central pattern generator element producing a phase switch from protraction to retraction in buccal motor programs of Aplysia californica. J. Neurophysiol. 75, 1327–1344 [DOI] [PubMed] [Google Scholar]
- 16. Hurwitz I., Kupfermann I., and Susswein A. J. (1997) Different roles of neurons B63 and B34 that are active during the protraction phase of buccal motor programs in Aplysia californica. J. Neurophysiol. 78, 1305–1319 [DOI] [PubMed] [Google Scholar]
- 17. Jing J., and Weiss K. R. (2001) Neural mechanisms of motor program switching in Aplysia. J. Neurosci. 21, 7349–7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jing J., and Weiss K. R. (2002) Interneuronal basis of the generation of related but distinct motor programs in Aplysia: implications for current neuronal models of vertebrate intralimb coordination. J. Neurosci. 22, 6228–6238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Evans C. G., Jing J., Rosen S. C., and Cropper E. C. (2003) Regulation of spike initiation and propagation in an Aplysia sensory neuron: gating-in via central depolarization. J. Neurosci. 23, 2920–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hurwitz I., Kupfermann I., and Weiss K. R. (2003) Fast synaptic connections from CBIs to pattern-generating neurons in Aplysia: initiation and modification of motor programs. J. Neurophysiol. 89, 2120–2136 [DOI] [PubMed] [Google Scholar]
- 21. Jing J., Cropper E. C., Hurwitz I., and Weiss K. R. (2004) The construction of movement with behavior-specific and behavior-independent modules. J. Neurosci. 24, 6315–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jing J., and Weiss K. R. (2005) Generation of variants of a motor act in a modular and hierarchical motor network. Curr. Biol. 15, 1712–1721 [DOI] [PubMed] [Google Scholar]
- 23. Sasaki K., Brezina V., Weiss K. R., and Jing J. (2009) Distinct inhibitory neurons exert temporally specific control over activity of a motoneuron receiving concurrent excitation and inhibition. J. Neurosci. 29, 11732–11744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sasaki K., Cropper E. C., Weiss K. R., and Jing J. (2013) Functional differentiation of a population of electrically coupled heterogeneous elements in a microcircuit. J. Neurosci. 33, 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu J. S., Wang N., Siniscalchi M. J., Perkins M. H., Zheng Y. T., Yu W., Chen S. A., Jia R. N., Gu J. W., Qian Y. Q., Ye Y., Vilim F. S., Cropper E. C., Weiss K. R., and Jing J. (2014) Complementary interactions between command-like interneurons that function to activate and specify motor programs. J. Neurosci. 34, 6510–6521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Holman G. M., and Cook B. J. (1983) Isolation and partial characterization of a second myotropic peptide from the hindgut of the cockroach, Leucophaea maderae. Comp. Biochem. Physiol. C 76, 39–43 [DOI] [PubMed] [Google Scholar]
- 27. Holman G. M., Cook B. J., and Nachman R. J. (1986) Primary structure and synthesis of a blocked myotropic neuropeptide isolated from the cockroach, Leucophaea maderae. Comp. Biochem. Physiol. C 85, 219–224 [DOI] [PubMed] [Google Scholar]
- 28. Holman G. M., Cook B. J., and Nachman R. J. (1986) Primary structure and synthesis of 2 additional neuropeptides from Leucophaea-Maderae: members of a new family of Cephalomyotropins. Comp. Biochem. Physiol. C 84, 271–276 [Google Scholar]
- 29. Holman G. M., Cook B. J., and Nachman R. J. (1987) Isolation, primary structure, and synthesis of Leukokinin-V and Leukokinin-Vi: myotropic peptides of Leucophaea-Maderae. Comp. Biochem. Physiol. C 88, 27–30 [Google Scholar]
- 30. Holman G. M., Cook B. J., and Nachman R. J. (1987) Isolation, primary structure and synthesis of Leukokinin-Vii and Leukokinin-Viii: the final members of this new family of cephalomyotropic peptides isolated from head extracts of Leucophaea-Maderae. Comp. Biochem. Physiol. C 88, 31–34 [Google Scholar]
- 31. Nässel D. R. (2002) Neuropeptides in the nervous system of Drosophila and other insects: multiple roles as neuromodulators and neurohormones. Prog. Neurobiol. 68, 1–84 [DOI] [PubMed] [Google Scholar]
- 32. Hayes T. K., Pannabecker T. L., Hinckley D. J., Holman G. M., Nachman R. J., Petzel D. H., and Beyenbach K. W. (1989) Leucokinins, a new family of ion transport stimulators and inhibitors in insect Malpighian tubules. Life Sci. 44, 1259–1266 [DOI] [PubMed] [Google Scholar]
- 33. Coast G. M., Holman G. M., and Nachman R. J. (1990) The diuretic activity of a series of cephalomyotropic neuropeptides, the achetakinins, on isolated malpighian tubules of the house cricket, Acheta-Domesticus. J. Insect Physiol. 36, 481–488 [Google Scholar]
- 34. Al-Anzi B., Armand E., Nagamei P., Olszewski M., Sapin V., Waters C., Zinn K., Wyman R. J., and Benzer S. (2010) The leucokinin pathway and its neurons regulate meal size in Drosophila. Curr. Biol. 20, 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jékely G. (2013) Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc. Natl. Acad. Sci. U.S.A. 110, 8702–8707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mirabeau O., and Joly J. S. (2013) Molecular evolution of peptidergic signaling systems in bilaterians. Proc. Natl. Acad. Sci. U.S.A. 110, E2028–E2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nässel D. R., and Homberg U. (2006) Neuropeptides in interneurons of the insect brain. Cell Tissue Res. 326, 1–24 [DOI] [PubMed] [Google Scholar]
- 38. Cropper E. C., Friedman A. K., Jing J., Perkins M. H., and Weiss K. R. (2014) Neuromodulation as a mechanism for the induction of repetition priming. Curr. Opin. Neurobiol. 29, 33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cox K. J., Tensen C. P., Van der Schors R. C., Li K. W., van Heerikhuizen H., Vreugdenhil E., Geraerts W. P., and Burke J. F. (1997) Cloning, characterization, and expression of a G-protein-coupled receptor from Lymnaea stagnalis and identification of a leucokinin-like peptide, PSFHSWSamide, as its endogenous ligand. J. Neurosci. 17, 1197–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jing J., Alexeeva V., Chen S. A., Yu K., Due M. R., Tan L. N., Chen T. T., Liu D. D., Cropper E. C., Vilim F. S., and Weiss K. R. (2015) Functional characterization of a vesicular glutamate transporter in an interneuron that makes excitatory and inhibitory synaptic connections in a molluscan neural circuit. J. Neurosci. 35, 9137–9149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Romanova E. V., Sasaki K., Alexeeva V., Vilim F. S., Jing J., Richmond T. A., Weiss K. R., and Sweedler J. V. (2012) Urotensin II in invertebrates: from structure to function in Aplysia californica. PLoS One 7, e48764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dong X., Han S., Zylka M. J., Simon M. I., and Anderson D. J. (2001) A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell 106, 619–632 [DOI] [PubMed] [Google Scholar]
- 43. Pfisterer P., Ehlermann J., Hegen M., and Schorle H. (2002) A subtractive gene expression screen suggests a role of transcription factor AP-2 α in control of proliferation and differentiation. J. Biol. Chem. 277, 6637–6644 [DOI] [PubMed] [Google Scholar]
- 44. Evans C. G., Rosen S., Kupfermann I., Weiss K. R., and Cropper E. C. (1996) Characterization of a radula opener neuromuscular system in Aplysia. J. Neurophysiol. 76, 1267–1281 [DOI] [PubMed] [Google Scholar]
- 45. Southey B. R., Amare A., Zimmerman T. A., Rodriguez-Zas S. L., and Sweedler J. V. (2006) NeuroPred: a tool to predict cleavage sites in neuropeptide precursors and provide the masses of the resulting peptides. Nucleic Acids Res. 34, W267–W272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Southey B. R., Sweedler J. V., and Rodriguez-Zas S. L. (2008) A python analytical pipeline to identify prohormone precursors and predict prohormone cleavage sites. Front. Neuroinform. 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nielsen H., Engelbrecht J., von Heijne G., and Brunak S. (1996) Defining a similarity threshold for a functional protein sequence pattern: the signal peptide cleavage site. Proteins 24, 165–177 [DOI] [PubMed] [Google Scholar]
- 48. Moroz L. L., Edwards J. R., Puthanveettil S. V., Kohn A. B., Ha T., Heyland A., Knudsen B., Sahni A., Yu F., Liu L., Jezzini S., Lovell P., Iannucculli W., Chen M., Nguyen T., et al. (2006) Neuronal transcriptome of Aplysia: neuronal compartments and circuitry. Cell 127, 1453–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rosen S. C., Teyke T., Miller M. W., Weiss K. R., and Kupfermann I. (1991) Identification and characterization of cerebral-to-buccal interneurons implicated in the control of motor programs associated with feeding in Aplysia. J. Neurosci. 11, 3630–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Perrins R., and Weiss K. R. (1998) Compartmentalization of information processing in an Aplysia feeding circuit interneuron through membrane properties and synaptic interactions. J. Neurosci. 18, 3977–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sasaki K., Due M. R., Jing J., and Weiss K. R. (2007) Feeding CPG in Aplysia directly controls two distinct outputs of a compartmentalized interneuron that functions as a CPG element. J. Neurophysiol. 98, 3796–3801 [DOI] [PubMed] [Google Scholar]
- 52. Cropper E. C., Evans C. G., Hurwitz I., Jing J., Proekt A., Romero A., and Rosen S. C. (2004) Feeding neural networks in the mollusc Aplysia. Neurosignals 13, 70–86 [DOI] [PubMed] [Google Scholar]
- 53. Jing J. (2009) Command systems. in Encyclopedia of Neuroscience, Vol. 2 (Squire L. R., ed) pp. 1149–1158, Academic Press, Inc., Oxford, UK [Google Scholar]
- 54. Jing J., Cropper E. C., and Weiss K. R. (2017) Network functions of electrical coupling present in multiple and specific sites in behavior-generating circuits. in Network Functions and Plasticity: Perspectives from Studying Neuronal Electrical Coupling in Microcircuits (Jing J., ed) pp. 79–107, Academic Press, Inc., London, UK [Google Scholar]
- 55. Romanova E. V., Aerts J. T., Croushore C. A., and Sweedler J. V. (2014) Small-volume analysis of cell-cell signaling molecules in the brain. Neuropsychopharmacology 39, 50–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Comi T. J., Do T. D., Rubakhin S. S., and Sweedler J. V. (2017) Categorizing cells on the basis of their chemical profiles: progress in single-cell mass spectrometry. J. Am. Chem. Soc. 139, 3920–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kupfermann I. (1974) Dissociation of the appetitive and consummatory phases of feeding behavior in Aplysia: a lesion study. Behav. Biol. 10, 89–97 [DOI] [PubMed] [Google Scholar]
- 58. Morton D. W., and Chiel H. J. (1993) The timing of activity in motor neurons that produce radula movements distinguishes ingestion from rejection in Aplysia. J. Comp. Physiol. A 173, 519–536 [DOI] [PubMed] [Google Scholar]
- 59. Morton D. W., and Chiel H. J. (1993) In vivo buccal nerve activity that distinguishes ingestion from rejection can be used to predict behavioral transitions in Aplysia. J. Comp. Physiol. A 172, 17–32 [DOI] [PubMed] [Google Scholar]
- 60. Morgan P. T., Jing J., Vilim F. S., and Weiss K. R. (2002) Interneuronal and peptidergic control of motor pattern switching in Aplysia. J. Neurophysiol. 87, 49–61 [DOI] [PubMed] [Google Scholar]
- 61. Wu J. S., Due M. R., Sasaki K., Proekt A., Jing J., and Weiss K. R. (2007) State dependence of spike timing and neuronal function in a motor pattern generating network. J. Neurosci. 27, 10818–10831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 63. Radford J. C., Davies S. A., and Dow J. A. (2002) Systematic G-protein-coupled receptor analysis in Drosophila melanogaster identifies a leucokinin receptor with novel roles. J. Biol. Chem. 277, 38810–38817 [DOI] [PubMed] [Google Scholar]
- 64. Siviter R. J., Coast G. M., Winther A. M., Nachman R. J., Taylor C. A., Shirras A. D., Coates D., Isaac R. E., and Nässel D. R. (2000) Expression and functional characterization of a Drosophila neuropeptide precursor with homology to mammalian preprotachykinin A. J. Biol. Chem. 275, 23273–23280 [DOI] [PubMed] [Google Scholar]
- 65. Bauknecht P., and Jékely G. (2015) Large-scale combinatorial deorphanization of Platynereis neuropeptide GPCRs. Cell Rep. 12, 684–693 [DOI] [PubMed] [Google Scholar]
- 66. Lum C. S., Zhurov Y., Cropper E. C., Weiss K. R., and Brezina V. (2005) Variability of swallowing performance in intact, freely feeding Aplysia. J. Neurophysiol. 94, 2427–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jing J., Vilim F. S., Horn C. C., Alexeeva V., Hatcher N. G., Sasaki K., Yashina I., Zhurov Y., Kupfermann I., Sweedler J. V., and Weiss K. R. (2007) From hunger to satiety: reconfiguration of a feeding network by Aplysia neuropeptide Y. J. Neurosci. 27, 3490–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schoofs L., De Loof A., and Van Hiel M. B. (2017) Neuropeptides as regulators of behavior in insects. Annu. Rev. Entomol. 62, 35–52 [DOI] [PubMed] [Google Scholar]
- 69. Lisitsyn N., Lisitsyn N., and Wigler M. (1993) Cloning the differences between two complex genomes. Science 259, 946–951 [DOI] [PubMed] [Google Scholar]
- 70. Hubank M., and Schatz D. G. (1994) Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 22, 5640–5648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vilim F. S., Alexeeva V., Moroz L. L., Li L., Moroz T. P., Sweedler J. V., and Weiss K. R. (2001) Cloning, expression and processing of the CP2 neuropeptide precursor of Aplysia. Peptides 22, 2027–2038 [DOI] [PubMed] [Google Scholar]
- 72. Sive H. L., and St John T. (1988) A simple subtractive hybridization technique employing photoactivatable biotin and phenol extraction. Nucleic Acids Res. 16, 10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fujisawa Y., Furukawa Y., Ohta S., Ellis T. A., Dembrow N. C., Li L., Floyd P. D., Sweedler J. V., Minakata H., Nakamaru K., Morishita F., Matsushima O., Weiss K. R., and Vilim F. S. (1999) The Aplysia mytilus inhibitory peptide-related peptides: identification, cloning, processing, distribution, and action. J. Neurosci. 19, 9618–9634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Furukawa Y., Nakamaru K., Wakayama H., Fujisawa Y., Minakata H., Ohta S., Morishita F., Matsushima O., Li L., Romanova E., Sweedler J. V., Park J. H., Romero A., Cropper E. C., Dembrow N. C., Jing J., Weiss K. R., and Vilim F. S. (2001) The enterins: a novel family of neuropeptides isolated from the enteric nervous system and CNS of Aplysia. J. Neurosci. 21, 8247–8261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chomczynski P., and Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159 [DOI] [PubMed] [Google Scholar]
- 76. Jing J., Vilim F. S., Wu J. S., Park J. H., and Weiss K. R. (2003) Concerted GABAergic actions of Aplysia feeding interneurons in motor program specification. J. Neurosci. 23, 5283–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yates J. R. 3rd, Speicher S., Griffin P. R., and Hunkapiller T. (1993) Peptide mass maps: a highly informative approach to protein identification. Anal. Biochem. 214, 397–408 [DOI] [PubMed] [Google Scholar]
- 78. Livnat I., Tai H. C., Jansson E. T., Bai L., Romanova E. V., Chen T. T., Yu K., Chen S. A., Zhang Y., Wang Z. Y., Liu D. D., Weiss K. R., Jing J., and Sweedler J. V. (2016) A d-amino acid-containing neuropeptide discovery funnel. Anal. Chem. 88, 11868–11876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yang C. Y., Yu K., Wang Y., Chen S. A., Liu D. D., Wang Z. Y., Su Y. N., Yang S. Z., Chen T. T., Livnat I., Vilim F. S., Cropper E. C., Weiss K. R., Sweedler J. V., and Jing J. (2016) Aplysia locomotion: network and behavioral actions of GdFFD, a d-amino acid-containing neuropeptide. PLoS One 11, e0147335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hurwitz I., Neustadter D., Morton D. W., Chiel H. J., and Susswein A. J. (1996) Activity patterns of the B31/B32 pattern initiators innervating the I2 muscle of the buccal mass during normal feeding movements in Aplysia californica. J. Neurophysiol. 75, 1309–1326 [DOI] [PubMed] [Google Scholar]
- 81. Struck T. H., Paul C., Hill N., Hartmann S., Hösel C., Kube M., Lieb B., Meyer A., Tiedemann R., Purschke G., and Bleidorn C. (2011) Phylogenomic analyses unravel annelid evolution. Nature 471, 95–98 [DOI] [PubMed] [Google Scholar]
- 82. Holman G. M., Nachman R. J., and Wright M. S. (1990) A strategy for the isolation and structural characterization of certain insect myotropic peptides that modify the spontaneous contractions of the isolated cockroach hindgut. in Chromatography and isolation of insect hormones and pheromones (McCaffery A. R., and Wilson I. D., eds) pp. 195–204, Plenum, New York [Google Scholar]
- 83. Schoofs L., Holman G. M., Proost P., Van Damme J., Hayes T. K., and De Loof A. (1992) Locustakinin, a novel myotropic peptide from Locusta-Migratoria, isolation, primary structure and synthesis. Regul. Pept. 37, 49–57 [DOI] [PubMed] [Google Scholar]
- 84. Clottens F. L., Meola S. M., Coast G. M., Hayes T. K., Wright M. S., Nachman R. J., and Holman G. M. (1993) Characterization of an antiserum against an achetakinin-I analog and its use for the localization of culekinin depolarizing peptide-Ii in the mosquito, Culex-Salinarius. Regul. Pept. 49, 145–157 [DOI] [PubMed] [Google Scholar]
- 85. Veenstra J. A. (1994) Isolation and identification of three leucokinins from the mosquito Aedes aegypti. Biochem. Biophys. Res. Commun. 202, 715–719 [DOI] [PubMed] [Google Scholar]
- 86. Blackburn M. B., Wagner R. M., Shabanowitz J., Kochansky J. P., Hunt D. F., and Raina A. K. (1995) The isolation and identification of three diuretic kinins from the abdominal ventral nerve cord of adult Helicoverpa-Zea. J. Insect Physiol. 41, 723–730 [Google Scholar]
- 87. Terhzaz S., O'Connell F. C., Pollock V. P., Kean L., Davies S. A., Veenstra J. A., and Dow J. A. T. (1999) Isolation and characterization of a leucokinin-like peptide of Drosophila melanogaster. J. Exp. Biol. 202, 3667–3676 [DOI] [PubMed] [Google Scholar]
- 88. Radford J. C., Terhzaz S., Cabrero P., Davies S. A., and Dow J. A. (2004) Functional characterisation of the Anopheles leucokinins and their cognate G-protein coupled receptor. J. Exp. Biol. 207, 4573–4586 [DOI] [PubMed] [Google Scholar]
- 89. Mitreva M., Jasmer D. P., Zarlenga D. S., Wang Z., Abubucker S., Martin J., Taylor C. M., Yin Y., Fulton L., Minx P., Yang S. P., Warren W. C., Fulton R. S., Bhonagiri V., Zhang X., et al. (2011) The draft genome of the parasitic nematode Trichinella spiralis. Nat. Genet. 43, 228–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.