Abstract
Stress granules (SG) are membrane-less organelles that are condensates of stalled translation initiation complexes and mRNAs. SG formation is a cytoprotective response to environmental stress and results from protein interactions involving regions of low amino acid complexity and poorly defined post-translational modifications of SG components. Many RNA-binding proteins are methylated, and we previously demonstrated that the potent SG–nucleating protein G3BP1 is methylated by protein arginine methyltransferase 1 and 5 (PRMT1 and PRMT5). G3BP1 methylation represses SG formation and is reversible. Here we functionally link JMJD6 (Jumonji C domain-containing protein 6) to G3BP1 demethylation. Our findings reveal that JMJD6 is a novel SG component that interacts with G3BP1 complexes, and its expression reduces G3BP1 monomethylation and asymmetric dimethylation at three Arg residues. Knockdown of JMJD6 repressed SG formation and G3BP1 demethylation, but SG formation and G3BP1 demethylation were rescued with catalytically active but not mutant JMJD6. These results suggest that JMJD6 functions directly or indirectly as an arginine demethylase of G3BP1 that promotes SG formation.
Keywords: post-translational modification (PTM), protein methylation, RNA-binding protein, stress granule, stress response, demethylase
Introduction
Stress granules (SGs)2 are cytoplasmic mRNP complexes that rapidly formed in response to unfavorable environments in eukaryotic cells. Stalled translation initiation complexes containing translation initiation factors, mRNA, 40S ribosome subunits, and key RNA-binding proteins such as G3BP1, Tia1, HuR, TDP43, FUS, and FRMP concentrate in SGs (1–6). Stress granules are thought to function as short-term repositories for mRNA to prevent mRNA degradation (7), as platforms for innate immune activation of double-stranded RNA-dependent protein kinase PKR (8, 9), and function as mediators of signaling cascades (10–13). SG formation typically follows translation inhibition caused by stress-induced eIF2α phosphorylation (1, 2, 14, 15), disruption of eIF4A and eIF4G function with small molecule inhibitors or virus infection (16) (17), and stress-induced tRNA cleavage (18–21) via an eIF2α-independent pathway.
Some of these RNA-binding proteins have been characterized as SG–nucleating proteins, particularly G3BP1 and Tia1. SG–nucleating proteins can induce SG formation when overexpressed, independent of exogenous stressors (3, 22, 23). Current models of SG structure involve stable cores enriched for G3BP1 surrounded by a dynamic shell that rapidly undergoes assembly and disassembly (24). Consistent with this model, knock-out of G3BP1 and its isoform G3BP2 deplete SG formation (25, 26), thus the classification of G3BP1 as a SG–nucleating protein. G3BP1 is also targeted by many viruses for proteolytic cleavage or sequestration to block SG formation during establishment of successful infections (27, 28).
Two models for SG formation exist. The first model posits that untranslated mRNPs promote liquid–liquid phase separation (LLPS), forming liquid droplets that are held together by weak interactions involving intrinsically disordered regions (IDR) of interacting proteins (29). Core formation then results from higher-affinity interactions that form during a structural transition at the center of the droplet. The second model proposes that stalled 48S preinitiation complexes can serve as a seed for SG condensation (30, 31), which attracts nucleating RNA-binding proteins such as G3BP1 already bound to 40S ribosomes (26), thus increasing their local concentration and favoring LLPS, liquid droplet formation, and nucleation of SGs.
SG assembly is highly dependent on post-translational modification (PTM) of IDRs of key SG–associated RNA-binding proteins (3, 32–37). Many RNA-binding proteins, including G3BP1, are highly enriched with IDR, compared with the rest of the human proteome (38).
Although many PTMs on IDR of SG–nucleating proteins have been proposed to influence SG assembly (3, 32–34, 36, 37), only phosphorylation of G3BP1 at Ser-149 by CKII (3, 39) and arginine methylation of the RGG domain of G3BP1 (25) have been shown to modulate SG assembly. We recently showed that G3BP1 is differentially methylated on Arg-435, Arg-447, and Arg-460 by protein arginine methyltransferase 1 (PRMT1) and PRMT5. Arsenite stress quickly and reversibly decreased asymmetric methylation on G3BP1 promoting SG assembly (25). However, the arginine demethylase responsible for this activity was not identified.
JMJD6 (Jumonji C (JmjC) domain-containing protein 6) was the first arginine demethylase described, capable of demethylating arginine residues on histone H3R2 and histone H4R3 (40). The demethylation activity is driven by the JmjC domain that requires Fe(II) and 2-oxoglutarate to hydroxylate methyl groups followed by deformylation producing formaldehyde (41).
Recently, studies from other groups validated demethylation activity of JMJD6 via biochemical and cell-based assays, confirmed by mass spectrometry (42–46). JMJD6 demethylase activity has also been reported on non-histone proteins, including estrogen receptor ERα (43), RNA helicase A (44), HSP70 (45), PAX3paired box 3) (47), and TNF receptor-associated Factor 6 (46), indicating that JMJD6 also demethylates non-histone substrates.
Our previous work demonstrated rapid demethylation of G3BP1 following arsenite stress. The emerging role of JMJD6 as a demethylase prompted us to investigate JMJD6 activity on G3BP1. Here we show that JMJD6 is a new component of SGs that directly interacts with G3BP1. Expression of JMJD6 specifically reduced the levels of ω-NG,NG-asymmetric dimethylarginine (ADMA) modifications on proteins, including on G3BP1, and promoted SG assembly during arsenite stress. Furthermore, shRNA silencing of JMJD6 or chemical inhibition with a pan-Jmjc domain inhibitor decreased SG formation during stress. Mass spectrometry confirmed that G3BP1 was demethylated at Arg-435, Arg-447, and Arg-460 by overexpression of JMJD6. Lastly, rescue experiments showed that expression of wild-type JMJD6, but not a catalytically inactive mutant, caused demethylation of G3BP1 and promoted SG assembly during arsenite stress. Collectively, these data indicate that catalytic activity of JMJD6 results in G3BP1 demethylation that triggers SG assembly during oxidative stress and strongly suggest methylated G3BP1 is a substrate of JMJD6.
Results
Arginine demethylase JMJD6 associated with stress granule complexes
G3BP1 is a dominant SG–nucleating protein regulated by phosphorylation and arginine methylation. Recently, the kinase responsible for G3BP1 phosphorylation was identified (39), but the demethylase(s) that governs SG condensation has not been resolved. G3BP1 is arginine-methylated, and JMJD6 was reported to demethylate methylarginine substrates, suggesting that JMJD6 might catalyze G3BP1 demethylation (40, 43–46). To investigate whether JMJD6 is involved in the cellular stress granule response, we visualized the distribution of endogenous JMJD6 in U2OS and HeLa cells during arsenite stress. In unstressed cells, immunofluorescence analysis (IFA) indicated that JMJD6 was most strongly localized in the nucleus (Fig. 1a, top panels), which agreed with previous studies from other groups (48, 49). However, we found much of the cytoplasmic fraction of JMJD6 concentrated in cytoplasmic punctate foci after 30 min of arsenite treatment, which colocalized with the SG marker Tia1 in HeLa cells (Fig. 1a, bottom panels). To confirm that these JMJD6 foci are stress granules, we costained cells for JMJD6 and the SG marker G3BP1, during either arsenite, thapsigargin, or heat shock stress (Fig. 1b). We also confirmed that canonical SGs formed in these conditions by staining for translation factors eIF3B, eIF3c, and RNA-HuR (data not shown). Under all conditions, JMJD6 concentrated and colocalized in stress granules as indicated by this marker. To test whether G3BP1 interacts with JMJD6, we immunoprecipitated G3BP1 from unstressed and arsenite-stressed cells followed by Western blotting to detect JMJD6 in G3BP1 complexes. Our results indicated that JMJD6 associated with G3BP1 protein complexes under unstressed conditions, and there was an increase in association of these proteins during arsenite stress (Fig. 1c). The recruitment of JMJD6 into SGs induced by treatment with arsenite, thapsigargin, and heat shock suggested that JMJD6 is a new component of SGs.
Figure 1.
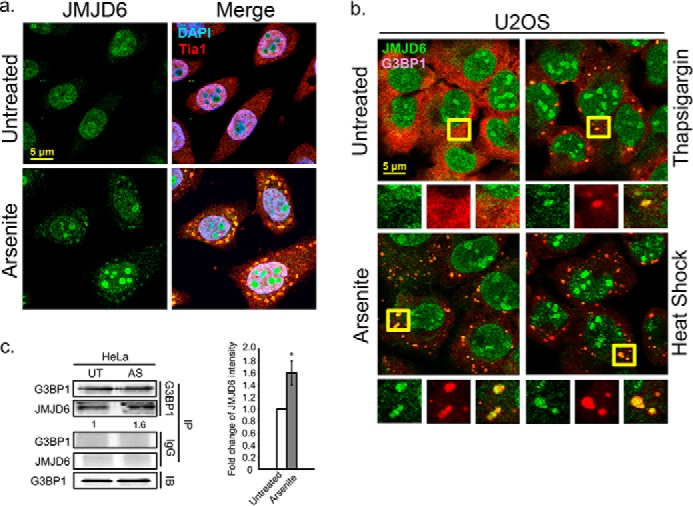
Arginine demethylase JMJD6 plays a role in the cellular stress granule response. a, IFA of endogenous JMJD6 localization in HeLa cells after arsenite stress. The cells were treated with arsenite 30 min to induce SGs, and cells were counterstained against the SG marker Tia1 (red) and DAPI for nuclei (blue). b, SGs formed by thapsigargin-induced (4 h) ER stress or heat shock (60 min) in U2OS cells were visualized by IFA showing endogenous JMJD6 (green) and G3BP1 (red). Yellow squares indicate regions depicted in vignettes. c, pulldown of endogenous G3BP1 in unstressed (UT) and arsenite-treated (AS) HeLa cells. JMJD6 was detected by Western blot analysis. IgG was antibody control for pulldown assay. The quantification of JMJD6 intensity is shown relative to untreated condition and normalized versus G3BP1 levels in the pulldown. The results are representative of three independent experiments that were conducted, with 100 cells counted in each. *, p < 0.05 versus untreated control. Original magnification was 63×. IB, immunoblot; IP, immunoprecipitation.
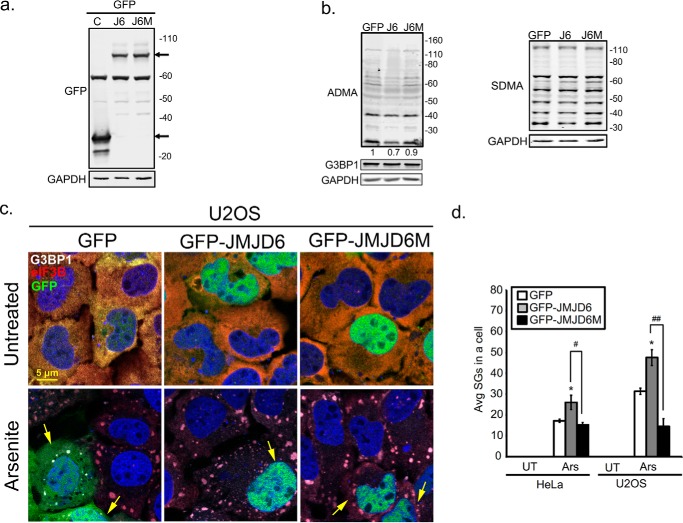
Expression of catalytically active JMJD6 promotes SG formation
To understand the role of JMJD6 in stress responses, we used methods to manipulate JMJD6 activity to examine methylation levels in cells and how it impacts SG assembly. Overexpression of GFP-JMJD6 in U2OS cells resulted in 30% reduction of global ADMA signal in cells compared with a GFP-transfected control (Fig. 2b, left panel). Symmetric dimethyl arginine (SDMA) is another common arginine methylation pattern on proteins, but global SDMA signals are unaffected by JMJD6 overexpression (Fig. 2b, right panel), indicating that JMJD6 bears a substrate preference for ADMA-modified proteins.
Figure 2.
Overexpression of JMJD6 promotes SG formation. U2OS cells were transfected with GFP (C), GFP-JMJD6 (J6), or GFP-catalytic domain mutant JMJD6 (J6M) for 24 h. before analysis. a and b, antibodies specific for GFP (a), asymmetric (ADMA) methyl modification (b, left panel), and symmetric methyl modification (SDMA) (b, right panel) were used in Western blot analysis. Arrows in a indicate GFP or GFP-tagged transgene. c, SG formed in GFP, GFP-JMJD6, and GFP-JMJD6 transfected U2OS cells after treatment with arsenite (60 min). Cells were counterstained with SG markers G3BP1 in gray and eIF3B in red. Yellow arrows point to transfected cells. d, quantification of average SGs/cell in HeLa and U2OS cells expressing either GFP (white bar), GFP-JMJD6 (gray bars), or GFP-JMJD6M (black bars). *, p < 0.05 versus untreated (UT) control. #, p < 0.05; ##, p < 0.01 versus GFP-JMJD6 transfected arsenite (Ars) cells. The results shown in all panels were performed three times, and those in b were performed five times. For IFA, 100 cells were counted in each experimental replicate. Original magnification was 63×.
To further confirm the alteration of ADMA levels in cells correlated with enzymatic activity of JMJD6, we generated dual point mutations H187A/D189A in the catalytic domain of JMJD6, which has been shown to abolish enzymatic activity of JMJD6 (40, 50). U2OS cells expressing JMJD6M showed no alteration of ADMA levels in cells (Fig. 2b, left panel), suggesting that the reduction of ADMA levels of proteins correlated with enzymatic activity of JMJD6. Next, we examined SG abundance in response to expression of JMJD6. In untreated cells, we did not see spontaneous SG formation in cells expressing GFP, GFP-JMJD6, or GFP-JMJD6M (Fig. 2c, top panels). There was an approximately 1.5-fold increase in SG formation in cells that expressed GFP-JMJD6 during arsenite stress compared with GFP transfected cells (Fig. 2, c, bottom panels, and d), whereas cells expressing GFP-JMJD6M were modestly repressed in SG formation (0.9-fold in HeLa, and 0.5-fold in U2OS) (Fig. 2d). Immunoblot analysis indicated that JMJD6 or JMJD6M expression did not alter G3BP1 levels in cells (Fig. 2b). Taken together, these data indicate that expression of JMJD6 modulates ADMA signals on proteins and promotes SG assembly.
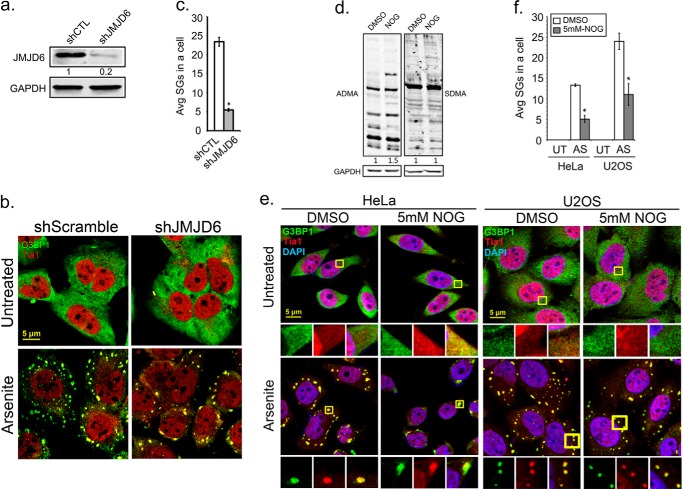
Enzymatic activity of JMJD6 affects SG assembly
To further test the role of JMJD6 in SG assembly, we performed shRNA knockdown of JMJD6 or silenced its enzymatic activity with a pan-spectrum Jmjc family inhibitor. U2OS cells were infected with lentivirus carrying shRNA targeting the 3′-UTR of JMJD6, which effectively depleted JMJD6 (80% reduction) in cells (Fig. 3a). Silencing of JMJD6 did not induce spontaneous SGs in cells (Fig. 3b, top panels) but during stress resulted in a 4-fold reduction of SG formation compared with control cells that were infected with control shScramble (shSC) lentivirus (Fig. 3, b and c). Next, we used an inhibitor of α-ketoglutarate-dependent enzymes, N-oxalylglycine (NOG), that is also effective for enzymes with Jmjc domains (40, 51, 52). We observed a 1.5-fold increase in ADMA levels on total proteins after 24 h of NOG treatment in HeLa cells (Fig. 3d, left panel) but no change in SDMA levels (Fig. 3d, right panel). Treatment with NOG did not induce spontaneous SGs in untreated cells (Fig. 3, e, top panels, and f), whereas NOG treatment resulted in ∼3-fold and 2-fold less arsenite-induced SGs in HeLa and U2OS cells, respectively (Fig. 3, e, bottom panels, and f). Collectively, we observed a defect in SG assembly when enzymatic activity of JMJD6 was impaired either by knockdown or with an inhibitor, suggesting that JMJD6 is an important protein that promotes SG assembly.
Figure 3.
Enzymatic activity of JMJD6 affects SG assembly. U2OS cells were transduced with Lentivirus expressing shRNA against the 3′-UTR region of JMJD6 (shJMJD6), to silence JMJD6, or control shScramble (shSC). a, 36 h later knockdown efficiency of JMJD6 was validated by Western analysis. b, JMJD6 depleted cells were examined by IFA to examine SGs in arsenite stress (AS). G3BP1 (green) and Tia1 (red) were used to mark SGs. c, quantification of average SGs/cell is shown. *, p < 0.05 versus untreated control (UT). d and e, HeLa cells were treated with the JMJD family inhibitor NOG at 5 mm for 24 h prior to further analysis. Antibodies specific for asymmetric (ADMA) methyl modifications (d, left panel) and symmetric methyl modifications (SDMA) (d, right panel) were used in Western blot analysis to examine methylation levels in inhibitor-treated HeLa cells. SGs were induced by arsenite treatment (e). Cells are labeled for G3BP1 (green) and Tia1 (red) for SGs and DAPI (blue) for nuclei. Yellow squares indicate regions depicted in vignettes. f, the quantification of average SGs/cell in U2OS and HeLa cells under untreated and arsenite stress conditions are shown in the bar graph. White bars indicate DMSO treated control cells, and gray bars indicate NOG-treated cells. *, p < 0.05 versus untreated control. At least three independent experiments were conducted for all panels, in which 100 cells were counted in each for IFA. Original magnification was 63×.
To further determine whether JMJD6 modulates SGs through G3PB1 we tested SG formation in JMJD6-silenced G3BP1 KO cells that were rescued with either GFP or GFP-G3BP1 constructs. In agreement with our previous study and those from other groups (25, 26), ablation of G3BP1 (Fig. 4a) strongly inhibited SG formation in untreated and arsenite-treated cells, and further, SGs were not rescued by JMJD6 KD in the G3BP1-null condition (Fig. 4b). Restoration of G3BP1 expression by transfecting GFP-G3BP1 (Fig. 4a) effectively rescued SG assembly in untreated and arsenite-stressed shScramble cells (Fig. 4b). However, this expression of GFP-G3BP1 was ∼2-fold less effective in restoring SGs in G3BP1KO cells where JMJD6 was also knocked down (Fig. 4, c, top panels, and b). In these cells, JMJD6 knockdown also resulted in a ∼2-fold reduction of SG assembly during arsenite stress (Fig. 4, c, bottom panels, and b), suggesting a link between JMJD6 and G3BP1 in promoting SG assembly.
Figure 4.
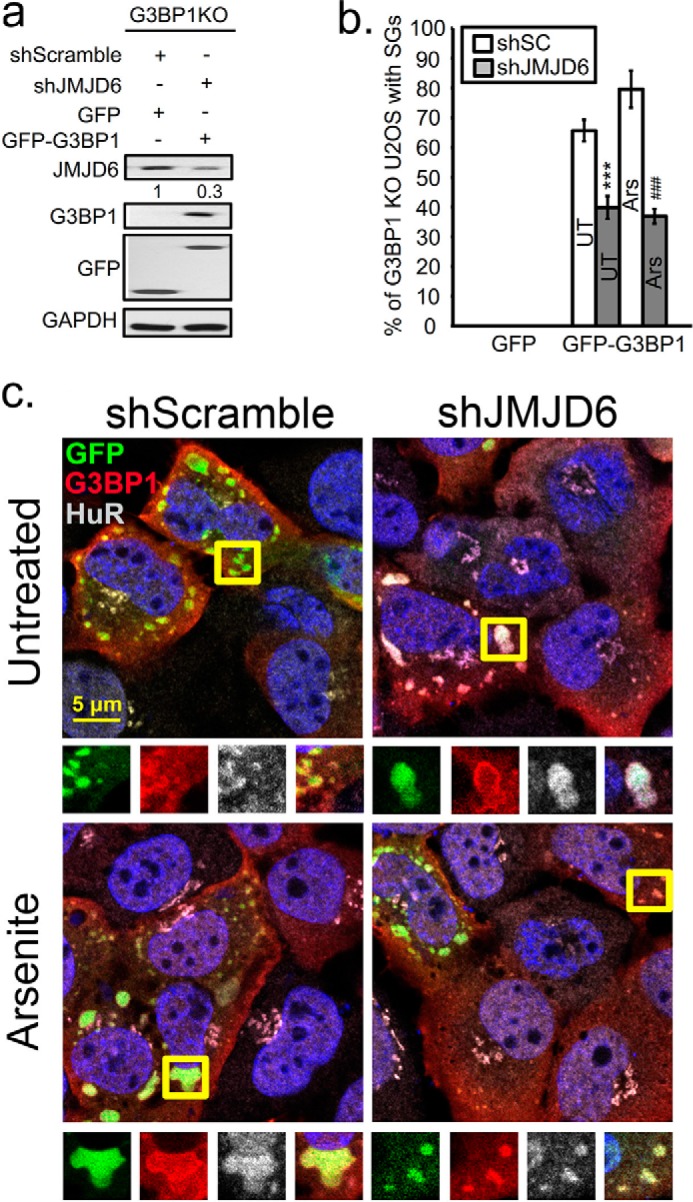
Knockdown of JMJD6 expression represses G3BP1-dependent SG formation. U2OS cells with CRISPR/Cas9 KO of G3BP1 were rescued with transfection of plasmids for GFP-G3BP1 or GFP control 18 h before analysis. G3BP1-KO cells rescued by GFP-G3BP1 expression were treated for JMJD6 KD (shJMJD6) or off-target knockdown (shScramble). a, immunoblot analysis of JMJD6-silenced G3BP1 KO cells transfected with GFP or with GFP-G3BP1 constructs. b, quantification of percentage of cells with SGs under untreated (UT) or arsenite stress conditions (Ars) is shown in the bar graph. No SGs were observed in control cells transfected with GFP expression plasmid in either condition. At least three independent experiments were conducted for each panel, 100 cells counted for IFA in each. ***, p < 0.001 versus untreated control; ###, p < 0.001 versus arsenite-treated cells. c, IFA for SGs induced by GFP-G3BP1 by labeling G3BP1 in red and HuR in gray, and nuclei are labeled with DAPI. Yellow squares indicate regions depicted in vignettes.
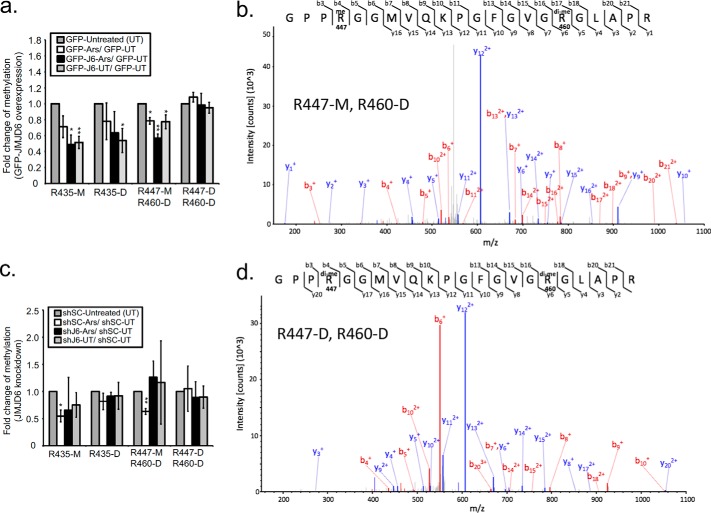
JMJD6 expression results in G3BP1 demethylation
To test whether changes of ADMA signals on total proteins reflect levels of ADMA on G3BP1, we performed LC-MS analysis of changes in G3BP1 methylation after manipulating JMJD6 expression by plasmid overexpression or shRNA knockdown and stressing with oxidative reagents. Consistent with our previous work (25), we saw a decrease in methylation on Arg-435 (∼30% and ∼40%), ArG-447, and Arg-460 (∼25% and 50%) in GFP- or shSC-expressing control cells during arsenite stress (Fig. 5, a, and c, white bars). Both Arg-447 and Arg-460 consistently appear on the same tryptic peptide; however, analysis of fragment ion (MS-MS) spectra can effectively determine, which residue is mono- or dimethylated (Fig. 5, b and d). Expression of JMJD6 reduced mono- and dimethylation on Arg-435 (∼50%), and monomethylation on Arg-477 (∼50%) in arsenite-stressed cells (Fig. 5a, black bars). Demethylation of G3BP1 was reduced (∼50% at Arg-435, ∼20% at Arg-447), even during untreated conditions by expressing JMJD6 (Fig. 5a, light gray bars). Interestingly, G3BP1 containing dimethyl marks on both Arg-447 and Arg-460 were not demethylated under these conditions, suggesting a substrate preference for methyl arginines at Arg-435 and Arg-447, which are both ADMA modified by PRMT1 (25). We assume that the reduction in G3BP1 methylation during arsenite is nearing the sensitivity of detection using LC-MS at these sites on G3BP1.
Figure 5.
Mass spectrometry analysis of G3BP1 demethylation linked to JMJD6 expression. a, summary of mass spectrometry analysis from endogenous G3BP1 immunoprecipitated from U2OS cells expressing either GFP, GFP-JMJD6, or GFP-JMJD6M. Bar graphs indicate the fold change of methylation signal at the indicated arginine residues with monomethylation (M) or dimethylation (D) under the indicated conditions. The fold change for a given condition is normalized to unmodified G3BP1 peptides. c, mass spectrometry analysis of G3BP1 in cells treated as in a together with JMJD6 KD after transduction with shJMJD6 (shJ6) or shScramble (shSC) (control) lentiviruses. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus GFP-untreated or shSC expressed cells. b and d, differential fragment ion (MS-MS) spectra isolated from excised G3BP1 bands from peptides containing Arg-447-M and Arg-460-D or Arg-447-D and Arg-460-D. Mass spectrographic analysis was performed three separate times with both biological and technical replicates.
In contrast to JMJD6 expression, knockdown of JMJD6 resulted in more sustained monomethylation on Arg-447 in response to arsenite stress (Fig. 5c, compare white and black bars on the Arg-447-M, Arg-460-D peptide) and a slight shift on Arg-435. As we expected, in most cases JMJD6 silencing did not significantly change G3BP1 methylation status at the three methylation sites under unstressed conditions (Fig. 5c, light gray bars). Collectively, these data strongly suggest that expression of JMJD6 is linked to G3BP1 demethylation at residues Arg-435 and Arg-447.
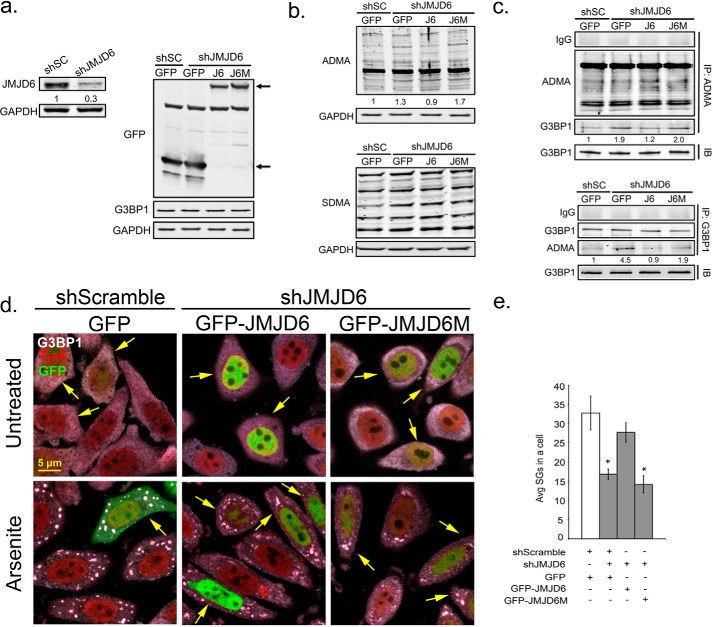
JMJD6 promotes SG formation and demethylation of G3BP1
To confirm that demethylase activity of JMJD6 promotes SG assembly, we performed a JMJD6 rescue experiment in JMJD6 silenced cells to monitor ADMA, SDMA levels on G3PB1, and how SG assembly was affected under these conditions. shJMJD6 expression resulted in a 70% knockdown of JMJD6 compared with shSC control, and transgenes were expressed equivalently (Fig. 6a). We examined the expression levels of JMJD6 transgene, ADMA, and SDMA signals after 24 h of transient transfection of HeLa cells with silencing of endogenous JMJD6 (shJMJD6). Silencing of JMJD6 modestly increased ADMA levels in cells (1.3-fold), but cells with endogenous JMJD6 silenced and rescued with GFP-JMJD6 had almost the same levels of ADMA signals compared with GFP transfected control shRNA. However, cells expressing GFP-J6M failed to rescue the demethylation phenotype and contained 1.7-fold higher ADMA (Fig. 6b, top panel). In agreement with previous results, we did not observe any significant changes of SDMA signals in these experiments (Fig. 6b, bottom panel).
Figure 6.
JMJD6 is a potential demethylase for G3BP1 to promote SG formation. Shown is reconstitution of JMJD6 by wild-type GFP-JMJD6 (J6) or catalytic mutant GFP-JMJD6 (J6M) during knockdown of endogenous JMJD6 in HeLa cells. a, immunoblot analysis for validation of JMJD6 knockdown efficiency (left panel) and GFP, J6, or J6M transgene expression levels (right panel) gene products marked by arrows. b, antibodies specific for asymmetric (ADMA) methyl modifications (top panel), and symmetric methyl modifications (SDMA) (bottom panel) were used in Western blot to examine methylation levels of total endogenous proteins in JMJD6 rescued cells. Relative densitometric analysis of total ADMA signal is shown below immunoblot. c, immunoprecipitation with ADMA antibodies (top panel) or G3BP1 antibodies (bottom panel) to analyze ADMA levels on G3BP1. d, SGs were induced by arsenite and visualized in HeLa cells under the same treatment conditions. SG markers are G3BP1 (gray) and Tia1 (red). Arrows in the panels indicate transfected cells. e, quantification of average SGs/cell in JMJD6 rescued HeLa cells. The white bar indicates shSC control cells, and gray bars indicate JMJD6 knockdown cells. *, p < 0.05 versus shSC control. The results shown are representative of three independent experiments that were conducted in which 100 cells were counted in each for IFA. Original magnification was 63×.
To examine the levels of ADMA signals on G3PB1, we immunoprecipitated total ADMA modified proteins followed by G3BP1 Western blotting in cells depleted of JMJD6 and rescued with GFP, GFP-JMJD6, or GFP-JMJD6M. The ADMA levels on G3BP1 were increased ∼2-fold in shJMJD6 cells. GFP-JMJD6 reconstitution in shJMJD6 silenced cells reduced methylation of G3BP1 (∼35%). In contrast, there was no difference when GFP-JMJD6M was expressed in shJMJD6 cells when compared with the GFP alone rescue in shJMJD6 cells, both of which displayed ∼2-fold higher ADMA than the control (Fig. 6c, top panel). We also performed a reciprocal immunoprecipitation with G3BP1 and found similar results where silencing of JMJD6 increased ADMA signal on G3PB1. There was a 80% reduction of ADMA levels on G3BP1 in shJMJD6 cells rescued with JMJD6 compared with cells rescued with GFP, but not when GFP-JMJD6M was expressed (Fig. 6c, bottom panel).
Finally, we examined whether the changes in ADMA observed on G3BP1 resulting from manipulation of JMJD6 were sufficient to alter SG assembly. In agreement with previous results, expression of JMJD6 and JMJD6M did not induce SGs under unstressed condition (Fig. 6d, top panel). Knockdown of JMJD6 impaired SG formation (∼46%) during arsenite stress (Fig. 6, d, bottom panel, and e). Interestingly, there was a 1.6-fold increase in SG formation when GFP-JMJD6 was expressed in shJMJD6 cells (yellow arrows in Fig. 6, d, bottom panels, and e). However, GFP-JMJD6M expression did not rescue SG assembly in response to arsenite stress (Fig. 6, d, bottom panels, and e). Together, these results indicate that JMJD6 demethylates G3BP1 at Arg-435, Arg-447, and Arg-460 to nucleate SG assembly in response to arsenite stress.
Discussion
SG assembly is a dynamic process regulated by protein–protein and protein–RNA interactions that may involve LLPSs (23, 31, 53–57). SG–nucleating proteins are thought to play critical roles in SG condensation by recruiting other SG constituents, thus acting as platforms for SG assembly (31). SG–nucleating proteins are enriched in IDRs that might facilitate the dynamic interactions between other SG constituents and the cellular milieu and are decorated by PTMs that potentially regulate SG assembly (3, 32–34, 36, 37). Few studies describe how PTMs serve as molecular switches to govern SG–nucleating protein activity in SG assembly (3, 25, 35). In our previous study, we reported that arginine demethylation of G3BP1 promotes SG assembly. Here, we extend our work to identify a candidate G3BP1 demethylase that promotes SG formation. Our data strongly suggests that asymmetric dimethyl argine modification on G3BP1 is reversible, similar to DNA methylation. Given the location of the methylation sites on G3BP1 within an IDR, this work emphasizes the importance of IDRs in SG proteins. Dephosphorylation of Ser-149 is another example of a PTM within an IDR that promotes SG assembly. We propose that phosphate- and methyl-groups are removed during stress to expose IDR, perhaps promoting interaction with other SGs proteins or 40s ribosomes (26). This raises the question of possible synergy between demethylation and dephosphorylation of G3BP1 to promote SG-related functions.
The RGG domain in the C terminus of G3PB1 has critical functions in SG assembly and interacts with 40S ribosomal subunits during translation inhibition (26). The N-terminal NTF2 domain of G3BP1 interacts with other SG components, such as Caprin1, and brings together RGG-bound 40S subunits (26). Therefore, SG assembly requires both the NTF2 and RGG domains of G3BP1. Post-translational modification of G3BP1 could alter SG condensation by changing the affinity of G3BP1 for other SG constituents through the N terminus or the 40S ribosome through the C-terminal RGG domain.
Here, we show that the enzymatic activity of JMJD6 is also positively correlated with SG assembly. Together, these results suggest that JMJD6 enhances SG condensation by demethylating the RGG domain and possibly promoting interaction with the 40S subunit. Deletion of the RGG domain from G3BP1 abolishes association of JMJD6 in G3BP1 complexes (data not shown).
Previous work has suggested a two-staged assembly of SGs, nucleated in the first stage by G3BP1-formation of many small granules that then merge into larger granules in the second stage (24, 58). Formation of small SGs seems to be independent of translational repression in other studies but disrupts SG-PB interactions (15, 59). We noticed the size of SGs is smaller when JMJD6 is depleted in arsenite-stressed cells, despite the increased number per cell (Figs. 3b, 4b, and 5d). SGs in PRMT1 and PRMT5 knock-out cells also showed a small SG phenotype, and those small SGs still correlated with translational repression in response to arsenite stress (25). These data indicate that the link between SGs and the translational apparatus is more complicated than previously thought and is probably a context-dependent relationship.
JMJD6 has been reported to be involved in an array of biological processes, but to our knowledge nobody has shown a link to SG biology. Our data support a role for JMJD6 in SGs because we show cytoplasmic JMJD6 concentrates in SGs and is involved in SG assembly by demethylating G3BP1 at Arg-435, Arg-447, and Arg-460 during arsenite stress. FMDV causes SG formation early in infection but then eliminates SGs for viral replication (60). Intriguingly, Lawrence et al. (44) has observed JMJD6 foci in the cytoplasm during FMDV and BEV-1 infection early in infection, which could be SGs as many enteroviruses initially promote SG formation (61). This idea is supported by our findings that JMJD6 is recruited to SGs during stress. JMJD6 has also been shown to interact with RNA in vitro (62), suggesting that JMJD6 interacts with other RNA-binding proteins or is an RNA-binding protein itself. Indeed, mass spectrometry data from Poulard et al. (43) reveals that JMJD6 interacts with SG components Tia1, PABP1, PABP4, and DDX3. These proteins or G3BP1 itself might be involved in recruiting JMJD6 to SGs. Collectively, these data strongly implicate JMJD6 in SG biology. HSP70 is a substrate of JMJD6, suggesting that JMJD6 may have other roles in the stress response (45). JMJD6 is also involved in regulation of a multitude of biological processes, including embryonic development (63–65), cell proliferation (66), cell cycle (50), cell mobility (67), adipocyte differentiation (68), and the emerging roles in human cancers (50, 67, 69, 70). Despite this, however, further work is required to characterize the function of JMJD6 in the stress response.
Our results demonstrate that the enzymatic activity of JMJD6 promotes G3BP1 demethylation. JMJD6 expression only alters ADMA modified proteins, but not SDMA. Similar results have been shown by other groups on different proteins such as ERα, RNA helicase A, HSP70, and TRAF6 (43–46). Interestingly, these proteins are all substrates of PRMT1, which is the major type I PRMT that generates ADMA. G3BP1 is also a substrate of PRMT1 (25), supporting the notion that JMJD6 is an important regulator of a stress-dependent G3BP1 methylation cycle. The literature has debated whether JMJD6 is a true arginine demethylase or a hydroxylase because it has a JmjC domain similar to lysine hydroxylases and lacks a consensus arginine demethylase domain. However, evidence from some studies also point out that JMJD6 requires other proteins as cofactors (42, 71). This is also reported for PRMT5 and for the lysine demethylase LSD1, which require MEP50 and Co-REST to become fully active (72, 73). Additionally, two methylation readers TDRD3 (74) and Tudor-SN (75) are components of SGs and could direct JMJD6 to methylated residues in SGs. Although we detected demethylation of G3BP1 during JMJD6 overexpression, our data do not distinguish between the effects of JMJD6 itself or as a complex requiring the use of cofactors.
In conclusion, our findings link the arginine demethylase JMJD6 with SG assembly by modulating the SG–nucleating protein G3BP1. Gain- and loss-of-function assays, MS analysis and data from other groups strongly suggest that JMJD6 mediates demethylation of G3BP1 and promotes SG formation. However, arginine demethylation is not sufficient to induce SGs because we did not observe any SG formation in untreated GFP-JMJD6 overexpression cells. Similarly, dephosphorylation of G3BP1 under unstressed conditions cannot drive SG assembly. These data suggest that multiple PTMs on G3BP1 are required to effectively drive SG formation.
Experimental procedures
Cell culture, transfections, and JMJD family inhibitor
The cells were cultured under standard conditions of 10% FBS in DMEM. All expression constructs were transfected into cells by Lipofectamine 3000 (Thermo Fisher Scientific) in accordance with the manufacturer's instructions. The JMJD inhibitor NOG was purchased from SIGMA (o-9390). The cells were pretreated with 5 mm NOG for 24 h before use in experiments.
RNA interference with shJMJD6
HeLa, U2OS, or G3BP1-KO U2OS cells were plated and infected with lentiviruses expressing shJMJD6 (TRCN0000303286, SIGMA) or shScramble in the presence of 8 g/ml protamine sulfate for 24 h followed by puromycin (2 g/ml; 48 h) selection. RT-PCR and/or Western blotting were performed to validate the knockdown efficiency.
Plasmid constructs
JMJD6 cDNA was amplified by PCR from HEK293 cell cDNA and then subcloned into pAC-GFP plasmid. JMJD6 catalytic domain mutant was generated by site-directed mutagenesis at H187A and D189A (forward primer, CTCCGGAACTGGGATTGCGATCGCACCTCTGGGAACCAGTG; reverse primer, CAGGCACTGGTTCCCAGAGGTGCGATCGCAATCCCAGTTCCGGAGCGTG). Site-directed mutagenesis was performed with Herculase II fusion DNA polymerase (Agilent Technologies) as described by the manufacturer.
Mass spectrometry analysis
G3BP1 was immunoprecipitated from untreated and arsenite-treated cells that were overexpressing or silencing JMJD6, subjected to SDS-PAGE, and excised for trypsin digestion. The resulting peptide fragments were analyzed by LC-MS using an LTQ ion-trap mass spectrometer (Orbitrap EliteTM; Thermo Fisher Scientific) equipped with a nano LC electrospray ionization source. Obtained MS/MS spectra were searched in Proteome Discoverer 1.4 interface (Thermo Fisher Scientific) with Mascot algorithm (Mascot 2.4; Matrix Science) to determine arginine methylation of G3BP1 peptides. Approximately 85% of total G3BP1 peptide coverage was achieved in each independent experiment.
Immunofluorescence assay
Microscopy was performed essentially as described previously (25). Primary antibodies were incubated overnight at 4 °C. The primary antibodies used were anti-JMJD6 (Cell Signaling, catalog no. 2449), anti-G3BP1 (28) (Abcam, catalog no. ab56574), anti-Tia1 (Santa Cruz, catalog no. SC11386), anti-HuR (Santa Cruz, catalog no. SC-5261), anti-eIF3b (Santa Cruz, catalog no. SC-28857), and anti-eIF3C (Santa Cruz, catalog no. SC-28858). All secondary antibodies (Molecular Probes) were used at 1:1,000 for 30 min at 25 °C. All images were taken with a Zeiss LSM 710 or 880 with a 63× oil immersion objective equipped with Zen software and processed with ImageJ and Adobe Photoshop CS6.
Immunoblots
Protein lysates (50 μg) were separated and transferred according to standard procedures. Membranes were blocked with Sea Block (Thermo Fisher Scientific) for 30 min at 25 °C and then incubated with primary antibodies overnight at 4 °C or 3 h at 25 °C. Fluorescent conjugated secondary antibodies were anti-mouse IgG (Dylight 680, catalog no. 5470) and anti-rabbit IgG (Dylight 800, catalog no. 5151) (Cell Signaling) and were incubated for 30 min. Primary antibodies used herein not listed under the immunofluorescence heading above were anti-G3BP1 (Abcam, catalog no. ab181149), anti-GFP (Santa Cruz, catalog no. SC-9996), anti-GAPDH (Millipore, catalog no. MAB374), anti-JMJD6 (Abcam, catalog no. ab64575) anti-ADMA (EpiCypher, catalog no. 12-0011), and anti-SDMA (EpiCypher, catalog no. 13-0012). Signals were detected using an Odyssey CLx (LI-COR). The relative levels of protein expression were normalized against GAPDH. Where indicated densitometry of ADMA or SDMA immunostaining of whole blot lanes was determined with NIH Image.
Immunoprecipitation
U2OS or HeLa cells were harvested in RIPA buffer and then quantified by a bicinchoninic acid assay (Thermo Fisher Scientific). 1 mg of cleared protein lysates from each condition were diluted in 60% RIPA, 40% NETN buffer and then incubated with monoclonal primary antibody against G3BP1 (Abcam, 1:100) or polyclonal primary antibody against ADMA proteins for 18 h at 4 °C. Protein A-Sepharose beads (Thermo Fisher Scientific) were equilibrated with NETN buffer, and 40 μl of beads were added into each reaction for 1 h at 4 °C. Samples were washed with 1 ml of NETN buffer five times and eluted with 2× Laemmli sample buffer (Bio-Rad).
Statistical analysis
All primary experiments were performed three to five times. For quantification, data are expressed as the means ± standard deviation and compared between groups using the Student's t test. p values of <0.05 were considered statistically significant (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Author contributions
W.-C. T. and R. E. L. conceived the experiment; W.-C. T., L. C. R., A. J., S. Y. J., and R. E. L. designed and conducted the experiments; L. C. R. provided the reagents, participated in discussions, and edited the paper; W.-C. T, L. C. R., and R. E. L. wrote and edited the paper; and R. E. L. was responsible for funding acquisition.
Acknowledgments
We thank Dr. Mark Bedford for fruitful discussions and support. The Integrated Microscopy Core at Baylor College of Medicine with funded through National Institutes of Health Grants HD007495, DK56338, and CA125123.
This work was supported by National Institutes of Health Public Health Service Grant AI50237 and NCI Cancer Center Support Grant P30CA125123. Additional support was provided by the Dan L. Duncan Cancer Center and the John S. Dunn Gulf Coast Consortium for Chemical Genomics. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- SG
- stress granule
- G3BP1
- RasGAP-SH3-binding protein 1
- PRMT
- protein arginine methyltransferase
- ADMA
- asymmetric dimethylarginine
- SDMA
- symmetric diemethylarginine
- LLPS
- liquid–liquid phase separation
- IDR
- intrinsically disordered region
- PTM
- post-translational modification
- IFA
- immunofluorescence analysis
- NOG
- N-oxalylglycine
- RIPA
- radioimmune precipitation assay.
References
- 1. Kedersha N. L., Gupta M., Li W., Miller I., and Anderson P. (1999) RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 147, 1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M. J., Scheuner D., Kaufman R. J., Golan D. E., and Anderson P. (2005) Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J. M., Bertrand E., and Tazi J. (2003) The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160, 823–831 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4. McDonald K. K., Aulas A., Destroismaisons L., Pickles S., Beleac E., Camu W., Rouleau G. A., and Vande Velde C. (2011) TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum. Mol. Genet. 20, 1400–1410 [DOI] [PubMed] [Google Scholar]
- 5. Kimball S. R., Horetsky R. L., Ron D., Jefferson L. S., and Harding H. P. (2003) Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 284, C273–C284 [DOI] [PubMed] [Google Scholar]
- 6. Anderson P., and Kedersha N. (2006) RNA granules. J. Cell Biol. 172, 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kedersha N., and Anderson P. (2002) Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30, 963–969 [DOI] [PubMed] [Google Scholar]
- 8. Oh S.-W., Onomoto K., Wakimoto M., Onoguchi K., Ishidate F., Fujiwara T., Yoneyama M., Kato H., and Fujita T. (2016) Leader-containing uncapped viral transcript activates RIG-I in antiviral stress granules. PLoS Pathog. 12, e1005444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reineke L. C., Kedersha N., Langereis M. A., van Kuppeveld F. J., and Lloyd R. E. (2015) Stress granules regulate double-stranded RNA-dependent protein kinase activation through a complex containing G3BP1 and Caprin1. MBio 6, e02486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eisinger-Mathason T. S., Andrade J., Groehler A. L., Clark D. E., Muratore-Schroeder T. L., Pasic L., Smith J. A., Shabanowitz J., Hunt D. F., Macara I. G., and Lannigan D. A. (2008) Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol. Cell 31, 722–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takahashi M., Higuchi M., Matsuki H., Yoshita M., Ohsawa T., Oie M., and Fujii M. (2013) Stress granules inhibit apoptosis by reducing reactive oxygen species production. Mol. Cell Biol. 33, 815–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kobayashi T., Winslow S., Sunesson L., Hellman U., and Larsson C. (2012) PKCα binds G3BP2 and regulates stress granule formation following cellular stress. PLoS One 7, e35820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brown J. A., Roberts T. L., Richards R., Woods R., Birrell G., Lim Y. C., Ohno S., Yamashita A., Abraham R. T., Gueven N., and Lavin M. F. (2011) A novel role for hSMG-1 in stress granule formation. Mol. Cell Biol. 31, 4417–4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kedersha N., Chen S., Gilks N., Li W., Miller I. J., Stahl J., and Anderson P. (2002) Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 13, 195–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reineke L. C., Dougherty J. D., Pierre P., and Lloyd R. E. (2012) Large G3BP-induced granules trigger eIF2α phosphorylation. Mol. Biol. Cell. 23, 3499–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mokas S., Mills J. R., Garreau C., Fournier M.-J., Robert F., Arya P., Kaufman R. J., Pelletier J., and Mazroui R. (2009) Uncoupling stress granule assembly and translation initiation inhibition. Mol. Biol. Cell 20, 2673–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Emara M. M., Fujimura K., Sciaranghella D., Ivanova V., Ivanov P., and Anderson P. (2012) Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation. Biochem. Biophys. Res. Commun. 423, 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lyons S. M., Achorn C., Kedersha N. L., Anderson P. J., and Ivanov P. (2016) YB-1 regulates tiRNA-induced stress granule formation but not translational repression. Nucleic Acids Res. 44, 6949–6960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamasaki S., Ivanov P., Hu G.-F., and Anderson P. (2009) Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 185, 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ivanov P., Emara M. M., Villen J., Gygi S. P., and Anderson P. (2011) Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 43, 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Emara M. M., Ivanov P., Hickman T., Dawra N., Tisdale S., Kedersha N., Hu G.-F., and Anderson P. (2010) Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 285, 10959–10968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L. M., and Anderson P. (2004) Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15, 5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kato M., Han T. W., Xie S., Shi K., Du X., Wu L. C., Mirzaei H., Goldsmith E. J., Longgood J., Pei J., Grishin N. V., Frantz D. E., Schneider J. W., Chen S., Li L., et al. (2012) Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jain S., Wheeler J. R., Walters R. W., Agrawal A., Barsic A., and Parker R. (2016) ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tsai W.-C., Gayatri S., Reineke L. C., Sbardella G., Bedford M. T., and Lloyd R. E. (2016) Arginine demethylation of G3BP1 promotes stress granule assembly. J. Biol. Chem. 291, 22671–22685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kedersha N., Panas M. D., Achorn C. A., Lyons S., Tisdale S., Hickman T., Thomas M., Lieberman J., McInerney G. M., Ivanov P., and Anderson P. (2016) G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 212, 845–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reineke L. C., and Lloyd R. E. (2013) Diversion of stress granules and P-bodies during viral infection. Virology 436, 255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. White J. P., Cardenas A. M., Marissen W. E., and Lloyd R. E. (2007) Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2, 295–305 [DOI] [PubMed] [Google Scholar]
- 29. Protter D. S., and Parker R. (2016) Principles and properties of stress granules. Trends Cell Biol. 26, 668–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Panas M. D., Ivanov P., and Anderson P. (2016) Mechanistic insights into mammalian stress granule dynamics. J. Cell Biol. 215, 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kedersha N., Ivanov P., and Anderson P. (2013) Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 38, 494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stoecklin G., Stubbs T., Kedersha N., Wax S., Rigby W. F., Blackwell T. K., and Anderson P. (2004) MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23, 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Courchet J., Buchet-Poyau K., Potemski A., Brès A., Jariel-Encontre I., and Billaud M. (2008) Interaction with 14-3-3 adaptors regulates the sorting of hMex-3B RNA-binding protein to distinct classes of RNA granules. J. Biol. Chem. 283, 32131–32142 [DOI] [PubMed] [Google Scholar]
- 34. Leung A. K., Vyas S., Rood J. E., Bhutkar A., Sharp P. A., and Chang P. (2011) Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell 42, 489–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kwon S., Zhang Y., and Matthias P. (2007) The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 21, 3381–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ohn T., Kedersha N., Hickman T., Tisdale S., and Anderson P. (2008) A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 10, 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. De Leeuw F., Zhang T., Wauquier C., Huez G., Kruys V., and Gueydan C. (2007) The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp. Cell Res. 313, 4130–4144 [DOI] [PubMed] [Google Scholar]
- 38. Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B. M., Strein C., Davey N. E., Humphreys D. T., Preiss T., Steinmetz L. M., Krijgsveld J., and Hentze M. W. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 [DOI] [PubMed] [Google Scholar]
- 39. Reineke L. C., Tsai W.-C., Jain A., Kaelber J. T., Jung S. Y., and Lloyd R. E. (2017) Casein kinase 2 is linked to stress granule dynamics through phosphorylation of the stress granule nucleating protein G3BP1. Mol. Cell Biol. 37, e00596–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chang B., Chen Y., Zhao Y., and Bruick R. K. (2007) JMJD6 is a histone arginine demethylase. Science 318, 444–447 [DOI] [PubMed] [Google Scholar]
- 41. Poulard C., Corbo L., and Le Romancer M. (2016) Protein arginine methylation/demethylation and cancer. Oncotarget 7, 67532–67550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu W., Ma Q., Wong K., Li W., Ohgi K., Zhang J., Aggarwal A., and Rosenfeld M. G. (2013) Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell 155, 1581–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Poulard C., Rambaud J., Hussein N., Corbo L., and Le Romancer M. (2014) JMJD6 regulates ERα methylation on arginine. PLoS One 9, e87982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lawrence P., Conderino J. S., and Rieder E. (2014) Redistribution of demethylated RNA helicase A during foot-and-mouth disease virus infection: role of Jumonji C-domain containing protein 6 in RHA demethylation. Virology 452–453, 1–11 [DOI] [PubMed] [Google Scholar]
- 45. Gao W.-W., Xiao R.-Q., Peng B.-L., Xu H.-T., Shen H.-F., Huang M.-F., Shi T.-T., Yi J., Zhang W.-J., Wu X.-N., Gao X., Lin X.-Z., Dorrestein P. C., Rosenfeld M. G., and Liu W. (2015) Arginine methylation of HSP70 regulates retinoid acid-mediated RARβ2 gene activation. Proc. Natl. Acad. Sci. U.S.A. 112, E3327–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tikhanovich I., Kuravi S., Artigues A., Villar M. T., Dorko K., Nawabi A., Roberts B., and Weinman S. A. (2015) Dynamic arginine methylation of tumor necrosis factor (TNF) receptor-associated factor 6 regulates Toll-like receptor signaling. J. Biol. Chem. 290, 22236–22249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu T.-F., Yao Y.-L., Lai I.-L., Lai C.-C., Lin P.-L., and Yang W.-M. (2015) Loading of PAX3 to mitotic chromosomes is mediated by arginine methylation and associated with Waardenburg syndrome. J. Biol. Chem. 290, 20556–20564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hahn P., Wegener I., Burrells A., Böse J., Wolf A., Erck C., Butler D., Schofield C. J., Böttger A., and Lengeling A. (2010) Analysis of Jmjd6 cellular localization and testing for its involvement in histone demethylation. PLoS One 5, e13769–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Heim A., Grimm C., Müller U., Häussler S., Mackeen M. M., Merl J., Hauck S. M., Kessler B. M., Schofield C. J., Wolf A., and Böttger A. (2014) Jumonji domain containing protein 6 (Jmjd6) modulates splicing and specifically interacts with arginine–serine-rich (RS) domains of SR- and SR-like proteins. Nucleic Acids Res. 42, 7833–7850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang F., He L., Huangyang P., Liang J., Si W., Yan R., Han X., Liu S., Gui B., Li W., Miao D., Jing C., Liu Z., Pei F., Sun L., and Shang Y. (2014) JMJD6 promotes colon carcinogenesis through negative regulation of p53 by hydroxylation. PLoS Biol. 12, e1001819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Suzuki T., and Miyata N. (2011) Lysine demethylases inhibitors. J. Med. Chem. 54, 8236–8250 [DOI] [PubMed] [Google Scholar]
- 52. Han G., Li J., Wang Y., Li X., Mao H., Liu Y., and Chen C. D. (2012) The hydroxylation activity of Jmjd6 is required for its homo-oligomerization. J. Cell Biochem. 113, 1663–1670 [DOI] [PubMed] [Google Scholar]
- 53. Han T. W., Kato M., Xie S., Wu L. C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., and McKnight S. L. (2012) Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779 [DOI] [PubMed] [Google Scholar]
- 54. Elbaum-Garfinkle S., Kim Y., Szczepaniak K., Chen C. C., Eckmann C. R., Myong S., and Brangwynne C. P. (2015) The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U.S.A. 112, 7189–7194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nott T. J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T. D., Bazett-Jones D. P., Pawson T., Forman-Kay J. D., and Baldwin A. J. (2015) Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lin Y., Protter D. S., Rosen M. K., and Parker R. (2015) Formation and maturation of phase-Separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A. P., Kim H. J., Mittag T., and Taylor J. P. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Aulas A., Stabile S., and Vande Velde C. (2012) Endogenous TDP-43, but not FUS, contributes to stress granule assembly via G3BP. Mol. Neurodegener. 7, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Aulas A., Caron G., Gkogkas C. G., Mohamed N.-V., Destroismaisons L., Sonenberg N., Leclerc N., Parker J. A., and Vande Velde C. (2015) G3BP1 promotes stress-induced RNA granule interactions to preserve polyadenylated mRNA. J. Cell Biol. 209, 73–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rai D. K., Lawrence P., Kloc A., Schafer E., and Rieder E. (2015) Analysis of the interaction between host factor Sam68 and viral elements during foot-and-mouth disease virus infections. Virol. J. 12, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lloyd R. E. (2016) Enterovirus control of translation and RNA granule stress responses. Viruses 8, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hong X., Zang J., White J., Wang C., Pan C.-H., Zhao R., Murphy R. C., Dai S., Henson P., Kappler J. W., Hagman J., and Zhang G. (2010) Interaction of JMJD6 with single-stranded RNA. Proc. Natl. Acad. Sci. U.S.A. 107, 14568–14572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Böse J., Gruber A. D., Helming L., Schiebe S., Wegener I., Hafner M., Beales M., Köntgen F., and Lengeling A. (2004) The phosphatidylserine receptor has essential functions during embryogenesis but not in apoptotic cell removal. J. Biol. 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kunisaki Y., Masuko S., Noda M., Inayoshi A., Sanui T., Harada M., Sasazuki T., and Fukui Y. (2004) Defective fetal liver erythropoiesis and T lymphopoiesis in mice lacking the phosphatidylserine receptor. Blood 103, 3362–3364 [DOI] [PubMed] [Google Scholar]
- 65. Li M. O., Sarkisian M. R., Mehal W. Z., Rakic P., and Flavell R. A. (2003) Phosphatidylserine receptor is required for clearance of apoptotic cells. Science 302, 1560–1563 [DOI] [PubMed] [Google Scholar]
- 66. Chen C.-F., Feng X., Liao H.-Y., Jin W.-J., Zhang J., Wang Y., Gong L.-L., Liu J.-J., Yuan X.-H., Zhao B.-B., Zhang D., Chen G.-F., Wan Y., Guo J., Yan H.-P., and He Y.-W. (2014) Regulation of T cell proliferation by JMJD6 and PDGF-BB during chronic hepatitis B infection. Sci. Rep. 4, 6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lee Y. F., Miller L. D., Chan X. B., Black M. A., Pang B., Ong C. W., Salto-Tellez M., Liu E. T., and Desai K. V. (2012) JMJD6 is a driver of cellular proliferation and motility and a marker of poor prognosis in breast cancer. Breast Cancer Res. 14, R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hu Y.-J., Belaghzal H., Hsiao W.-Y., Qi J., Bradner J. E., Guertin D. A., Sif S., and Imbalzano A. N. (2015) Transcriptional and post-transcriptional control of adipocyte differentiation by Jumonji domain-containing protein 6. Nucleic Acids Res. 43, 7790–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Poulard C., Rambaud J., Lavergne E., Jacquemetton J., Renoir J.-M., Trédan O., Chabaud S., Treilleux I., Corbo L., Le Romancer M., and Romancer M. L. (2015) Role of JMJD6 in breast tumourigenesis. PLoS One 10, e0126181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhang J., Ni S.-S., Zhao W.-L., Dong X.-C., and Wang J.-L. (2013) High expression of JMJD6 predicts unfavorable survival in lung adenocarcinoma. Tumor Biol. 34, 2397–2401 [DOI] [PubMed] [Google Scholar]
- 71. Unoki M., Masuda A., Dohmae N., Arita K., Yoshimatsu M., Iwai Y., Fukui Y., Ueda K., Hamamoto R., Shirakawa M., Sasaki H., and Nakamura Y. (2013) Lysyl 5-hydroxylation, a novel histone modification, by Jumonji domain containing 6 (JMJD6). J. Biol. Chem. 288, 6053–6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shi Y.-J., Matson C., Lan F., Iwase S., Baba T., and Shi Y. (2005) Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell 19, 857–864 [DOI] [PubMed] [Google Scholar]
- 73. Friesen W. J., Wyce A., Paushkin S., Abel L., Rappsilber J., Mann M., and Dreyfuss G. (2002) A novel WD repeat protein component of the methylosome binds Sm proteins. J. Biol. Chem. 277, 8243–8247 [DOI] [PubMed] [Google Scholar]
- 74. Goulet I., Boisvenue S., Mokas S., Mazroui R., and Côté J. (2008) TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum. Mol. Genet. 17, 3055–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Su C., Gao X., Yang W., Zhao Y., Fu X., Cui X., Zhang C., Xin L., Ren Y., Li L., Shui W., Yang X., Wei M., and Yang J. (2017) Phosphorylation of Tudor-SN, a novel substrate of JNK, is involved in the efficient recruitment of Tudor-SN into stress granules. Biochim. Biophys. Acta 1864, 562–571 [DOI] [PubMed] [Google Scholar]