Abstract
During tissue development, transcription factors bind regulatory DNA regions called enhancers, often located at great distances from the genes they regulate, to control gene expression. The enhancer landscape during embryonic stem cell differentiation has been well characterized. By contrast, little is known about the shared and unique enhancer regulatory mechanisms in different ectodermally derived epithelial cells. Here we use ChIP sequencing (ChIP-seq) to identify domains enriched for the histone marks histone H3 lysine 4 trimethylation, histone H3 lysine 4 monomethylation, and histone H3 lysine 27 acetylation (H3K4me3, H3K4me1, and H3K27ac) and define, for the first time, the super enhancers and typical enhancers active in primary human corneal epithelial cells. We show that regulatory regions are often shared between cell types of the ectodermal lineage and that corneal epithelial super enhancers are already marked as potential regulatory domains in embryonic stem cells. Kruppel-like factor (KLF) motifs were enriched in corneal epithelial enhancers, consistent with the important roles of KLF4 and KLF5 in promoting corneal epithelial differentiation. We now show that the Kruppel family member KLF7 promotes the corneal progenitor cell state; on many genes, KLF7 antagonized the corneal differentiation–promoting KLF4. Furthermore, we found that two SNPs linked previously to corneal diseases, astigmatism, and Stevens-Johnson syndrome fall within corneal epithelial enhancers and alter their activity by disrupting transcription factor motifs that overlap these SNPs. Taken together, our work defines regulatory enhancers in corneal epithelial cells, highlights global gene-regulatory relationships shared among different epithelial cells, identifies a role for KLF7 as a KLF4 antagonist in corneal epithelial cell differentiation, and explains how two SNPs may contribute to corneal diseases.
Keywords: cell proliferation, chromatin, epithelial cell, eye, gene regulation, Kruppel-like factor 4 (KLF4), KLF7, corneal epithelium, superenhancer
Introduction
During tissue development, transcription factors bind distal regulatory regions called enhancers to control and coordinate gene expression in a temporally and spatially specific manner. Unlike gene regulation by transcription factors binding at proximal promoters, enhancer-binding transcription factors regulate genes at long distances through chromatin looping that brings together distal enhancers and target promoters (1, 2).
Enhancers and promoters can be identified through posttranslational histone modification signatures created by the histone-modifying enzymes that control enhancer and promoter function. Enhancers have high levels of H3K4me1 and lower levels of H3K4me3 (3); when active, they are additionally marked by high levels of H3K27ac, and when repressed, they are marked by H3K27me3 (4). By contrast, promoters are marked by H3K4me3, H3K9ac, and H3K27ac.
Contained within the library of enhancers of each cell type is a smaller group of enhancers that are unusually long (more than 12.5 kb), with an unusually high density of transcription factor binding and with unusually active enhancer histone signature marks (5–8). These enhancers, termed super enhancers (SEs)2 to distinguish them from typical enhancers (TEs), are frequently tissue-specific; they are thought to preferentially regulate the unique gene expression programs in each cell type.
The chromatin regulatory landscape in embryonic stem cells, including during their commitment to distinct embryonic cell lineages, has been well defined (9–11). By contrast, the locations and roles of TEs and SEs in corneal epithelial cells remain to be defined. Furthermore, the extent of shared TEs and SEs in different cell types of the ectodermal embryonic lineage remains poorly understood.
The corneal epithelium is an excellent tissue to study how enhancers regulate genes during development and homeostasis. This stratified squamous epithelium undergoes constant cell turnover: cells that slough off from the corneal surface are replenished by proliferating, limbally located progenitor cells (12, 13). As the progenitor cells divide and move toward the center and surface of the corneal epithelium, they repress progenitor genes and activate differentiation genes. Hence, precise and dynamic gene expression that balances proliferation, migration, and differentiation is required to maintain a functioning corneal epithelium. The underlying gene-regulatory program is not well understood (14).
To gain better understanding of gene regulation in corneal epithelial cells and to provide insights into how their gene-regulatory mechanisms relate to other epithelial cell types, we identified enhancers in primary human corneal epithelial cells through ChIP-seq, using antibodies detecting characteristic histone modifications. These data reveal a complex enhancer relationship between distinct epithelial cell types. We identified shared regulatory regions among different epithelial cell types, but we also found cell type–specific regulatory regions that control unique corneal epithelial functions. In addition, our data provide insight into corneal diseases as we find that two GWAS-discovered SNPs lie within and alter the function of corneal enhancers. Motif analysis for enriched transcription factor binding sites within these enhancers led to the discovery of a previously undescribed role for KLF7 in corneal epithelial progenitor cells. KLF7 acts to prevent corneal epithelial differentiation, functioning in part antagonistically to the related pro-differentiation factor KLF4. We propose that the competing functions of KLF7 and KLF4 balance proliferation and differentiation during corneal epithelial development.
Results
Both typical enhancers and super enhancers associate with corneal epithelial identity genes
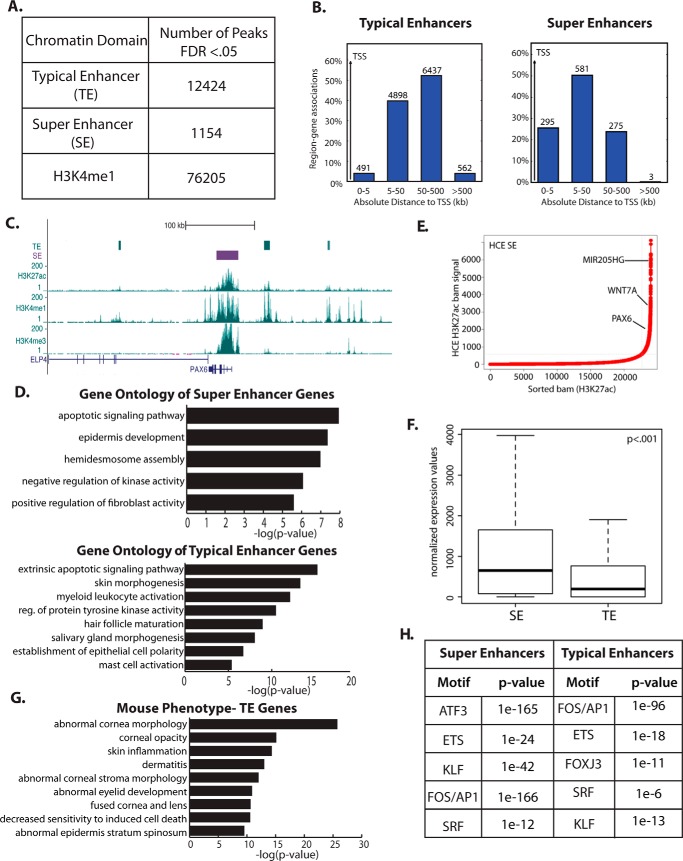
To gain insights into the gene-regulatory landscape in corneal epithelial cells, we performed ChIP-seq with antibodies to the histone marks H3K4me3, H3K4me1, and H3K27ac in primary human corneal epithelial (HCE) cells. We identified active enhancers as having high levels of H3K27ac and H3K4me1 and relatively low levels of H3K4me3 (4). Because of their importance in controlling cell type–specific gene expression, we also defined SEs, using the strength of the H3K27ac mark as described previously (5, 6). We identified 1154 SEs and 12,424 TEs in corneal epithelial cells as well as 76,205 distal regulatory regions, marked by H3K4me1 alone (Fig. 1A).
Figure 1.
TEs and SEs in primary HCE cells are linked to epithelial identity genes. A, quantification of chromatin regulatory domains in HCE cells. FDR, false discovery rate. B, distribution of TEs and SEs around transcription start sites. C, TEs and an SE near the corneal identity regulator PAX6. D, GO for genes linked to SEs (top panel) and TEs (bottom panel). E, plot of the H3K27ac signal, highlighting selective genes linked to top-ranked SEs. F, comparison of the expression levels of the nearest gene to TEs and SEs in HCE cells (t test). G, mouse phenotype analysis for the nearest genes to HCE TEs. H, top enriched motifs in HCE TEs and SEs.
As expected, the majority of enhancers was found at distal sites in the genome rather than in proximal promoters (Fig. 1B). Although TEs showed a trend toward being localized near genes, SEs were even more strongly enriched near genes; most SEs were found within 50 kb of a transcriptional start site.
Both TEs and SEs were located near key corneal epithelial identity genes, including the gene encoding the transcription factor PAX6, a master regulator of eye development (15, 16). An SE is located directly over the PAX6 gene body, and three TEs are found upstream and downstream of the gene, in regions that have been linked to the regulation of PAX6 mRNA expression (17), suggesting that both TEs and an SE regulate this gene (Fig. 1C).
Gene Ontology (GO) analysis showed that corneal epithelial SE-linked genes were involved in the regulation of apoptosis, epidermal development (encompassing many epithelial factors), hemidesmosome assembly, and regulation of fibroblast proliferation (Fig. 1D). TE-linked genes were enriched for similar categories (Fig. 1D). Highly ranked SEs were linked to genes with crucial roles in corneal epithelial identity, including PAX6, WNT7A, and MIR205HG (Fig. 1E). Although both TEs and SEs fell near highly expressed corneal epithelial genes, SE-associated genes are expressed at a significantly higher level than TE-associated genes (Fig. 1F). Although TEs may be general regulators of genes expressed in numerous tissues, the genes linked to TEs in HCE cells are still important to corneal and epithelial function, as evidenced by mouse phenotypes linked to these genes: abnormal corneal morphology, corneal opacity, fused cornea and lens, and numerous epidermal abnormalities (Fig. 1G).
The two types of enhancers were enriched for similar transcription factor motifs, with only subtle differences between the two. Motifs enriched in both SEs and TEs included those for AP1, KLF, and ETS factor family members (Fig. 1H), all known to have important roles in corneal epithelial differentiation (18–21). Interestingly, TEs showed a low but significant enrichment for forkhead box (FOX) family motifs that was not found in SEs (Fig. 1H). Together, these data suggest that both TEs and SEs are important in driving corneal epithelial cell identity.
Ectodermally derived epithelial cells use a common set of regulatory regions marked by H3K4me1; cell type specificity is conferred by enhancer activation and SE formation
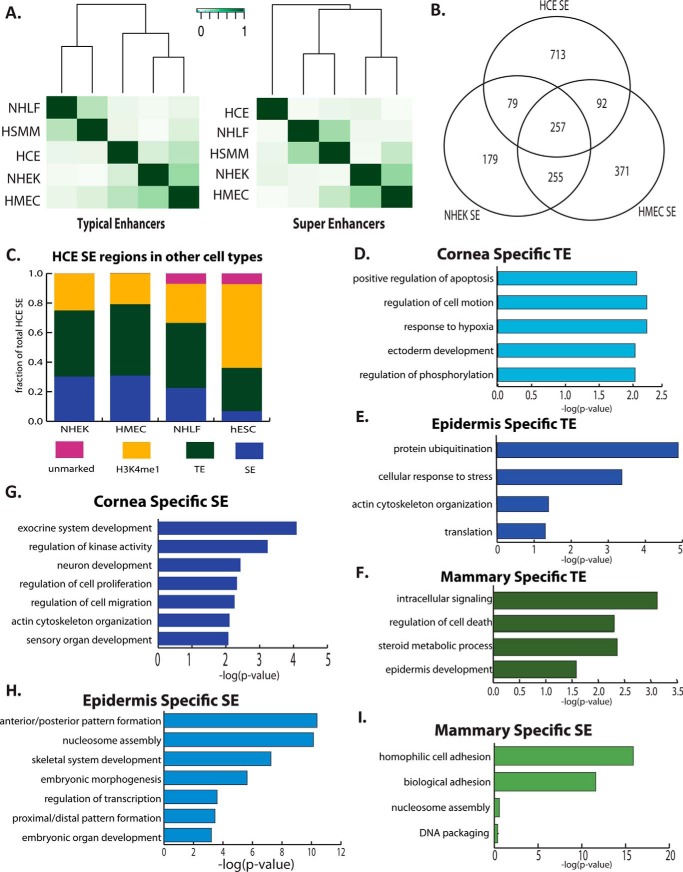
We next compared our enhancer data for corneal epithelial cells with the publicly available Encyclopedia of DNA Elements (ENCODE) data for histone modifications in four additional primary human cell types: human mammary epithelial cells (HMECs), epidermal keratinocytes (NHEK), lung fibroblasts (NHLF), and human skeletal muscle myoblasts (HSMM) (22). Clustering these datasets based on overlap of the TEs, we found that the epithelial cell types clustered together, indicative of their related functions and gene expression. Among the epithelial cells, NHEK cells and HMECs were more similar to each other than each of them was to HCE cells (Fig. 2A).
Figure 2.
Gene-regulatory regions in corneal epithelial cells are related to those in other epithelial cell types. A, clustering of primary cell types based on similarities in TEs (left panel) and SEs (right panel). B, overlap between SEs in primary epithelial cell types. C, assessment of the regulatory cell state of HCE SE regions in other epithelial cells (HMEC and NHEK), mesodermally derived cells (NHLF), and hESCs. D, GO for genes linked to cornea-specific TEs. E, enriched GO for epidermis-specific TEs. F, enriched GO for mammary epithelium–specific TEs. G, GO for genes linked to cornea-specific SEs. H, enriched GO for epidermis-specific SEs. I, enriched GO for mammary epithelium-specific SEs.
Analysis of SEs revealed similar relationships among the cell types, with NHEK cells and HMECs having the most overlap in the SE landscape. There was less similarity overall between cell types based on SEs than TEs, indicative of the unique locations and roles of SEs associated with tissue-specific functions in each cell type (Fig. 2A). Unexpectedly, HCE cells no longer clustered with HMEC and NHEK cells based on SE overlap, suggesting that, in the three epithelial cell types, SEs in corneal epithelial cells are most distinct (Fig. 2A). Considering the number of overlapping SEs between the different epithelial cell types, HMEC and NHEK cells also share more overlapping SEs than either one does with HCE cells (Fig. 2B).
We next examined the overlap between SEs and TEs among the different epithelial cell types (Fig. 2C), finding that, although only about a third of SEs were shared between two or more epithelial cell types, SEs in one epithelial cell type were frequently TEs in another, and all SEs in one epithelial cell type were marked by H3K4me1 in the other epithelial cell types. In contrast, there was less overlap between HCE SEs and regions marked as TEs or SEs in the mesenchymal cell type NHLF; some HCE SEs did not even bear any active marks in NHLF cells.
We saw more dramatic differences when we compared HCE SEs with chromatin regions in hESCs: there was very little overlap between HCE SEs and hESC SEs. The majority of HCE SEs, however, were marked with H3K4me1 alone in hESCs, indicative of the potential of these regions to become active enhancers in the future corneal epithelial cell lineage. Together, these data indicate that most epithelial distal regulatory regions are common to multiple epithelial cell types, differing only in the type of enhancer and/or the level of enhancer activity, as indicated by the activation mark H3K27Ac (Fig. 2C).
Enhancers unique to epithelial cell types are close to genes that confer cell type–specific functions
To explore the active epithelial enhancers further, we defined unique TEs for each epithelial cell type. GO analysis for the genes near the TEs unique to each epithelial cell type revealed potential functional differences.
Although GO categories related to epithelial or ectodermal development appeared in many cases, genes involved in the cellular response to hypoxia were only enriched around unique corneal epithelial TEs, perhaps reflecting the importance of coping with hypoxia in this avascular tissue (Fig. 2D). GO analysis for genes found near NHEK-specific TEs revealed enrichment in categories related to the actin cytoskeleton, perhaps reflecting the importance of cell shape changes and migration in the function of the epidermis (Fig. 2E). The genes near HMEC-specific TEs were enriched for the GO category steroid metabolic process, perhaps reflecting mammary gland hormone signaling (Fig. 2F).
Because SEs are thought to be more important for cell identity than TEs, we next studied the function of genes linked to the unique SEs for each cell type: SEs that did not overlap SEs or TEs in the other epithelial cell types. We found that SEs unique to HCE cells were located near genes involved in sensory organ development and cell signaling (Fig. 2G). SEs unique to NHEK cells were found near clusters of histone genes as well as near homeobox cluster A (HOXA) and homeobox cluster B (HOXB) (Fig. 2H), suggesting that these SEs act as sites for coordinated regulation of genes within clusters. SEs unique to HMECs were found near cell adhesion genes and histone cluster genes (Fig. 2I).
De novo motif analysis of the unique corneal epithelial SEs and TEs revealed that the motif for the ETS family member EHF was enriched in both unique SEs and TEs (Fig. 3, A and B). TEs unique to HCE cells were also enriched for the retinoic acid receptor α (RXRA) motif, whereas SEs unique to HCE cells were enriched in AP1 and KLF motifs. These results indicate that ETS and KLF family members, important regulators of corneal epithelial differentiation, carry out their functions in part by binding to and activating cell type–specific enhancers.
Figure 3.
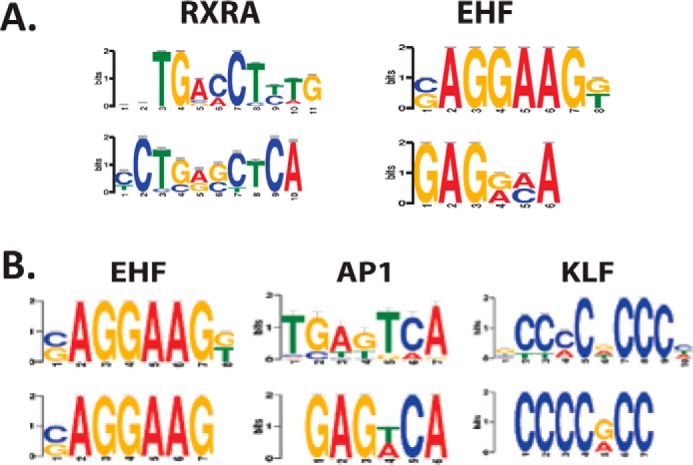
A, enriched motifs in cornea-specific TEs. B, enriched motifs in cornea-specific SEs.
KLF7 and KLF4 exhibit reciprocal expression patterns during corneal epithelial development
The enrichment of KLF motifs in both TEs and SEs in corneal epithelial cells is unsurprising given the well-defined cornea roles of KLF4 and KLF5 (20, 21, 23, 25). KLFs, however, are a large family of transcription factors, many of which are highly expressed in both the mouse and the human corneal epithelium. Hence, we wanted to explore the functions of other KLF family members and determine whether multiple KLFs collaborate to regulate gene expression within corneal epithelial cells.
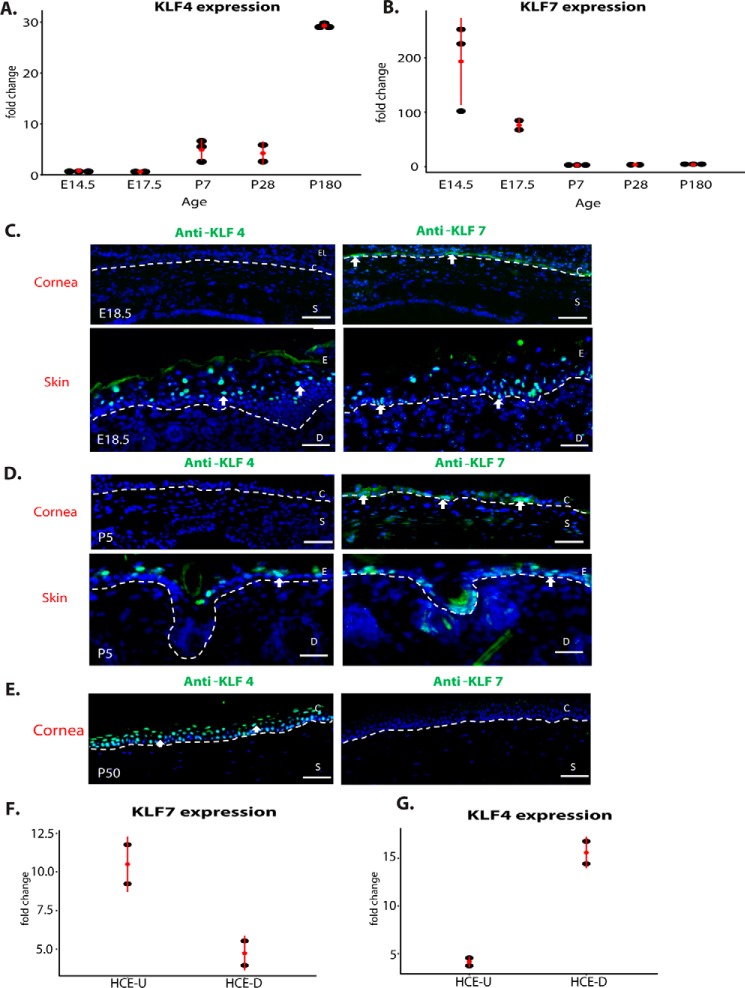
Using our previously published cornea transcriptome data showing global gene expression changes over the lifetime of the mouse (18), we identified two distinct time course patterns of KLF family member expression in the cornea. Although the majority, including KLF4 (Fig. 4A), KLF5, and KLF6, increased in expression as the cornea developed, correlating positively with differentiation, KLF7 (Fig. 4B) and KLF12 were highly expressed initially but decreased in expression as the cornea developed, correlating negatively with differentiation.
Figure 4.
KLF7 and KLF4 have opposing expression patterns in corneal epithelial development. A, qPCR analysis of KLF4 expression in the cornea across the mouse lifespan: E14.5 to 2 years. B, qPCR analysis of KLF7 expression in the cornea across the mouse lifespan. C, immunofluorescence staining of KLF4 (green) on E18.5 embryonic cornea (top left panel) compared with expression in the adjacent eyelid skin (bottom left panel) and immunofluorescence staining of KLF7 (green) on E18.5 embryonic cornea (top right panel) compared with expression in the adjacent skin (bottom right panel). D, immunofluorescence staining of KLF4 in a P5 mouse eye section compared with the adjacent skin (left panels) and immunofluorescence staining of KLF7 (green) in a P5 mouse eye section compared with the adjacent skin (right panels). E, immunofluorescence staining of KLF4 (green) on an adult mouse eye section compared with adjacent skin (left panel) and immunofluorescence staining of KLF7 (green) on adult mouse eye section (right panel). F, expression of KLF7 in undifferentiated (U) or differentiated (D) primary human corneal epithelial cells. G, expression of KLF4 in undifferentiated or differentiated primary human corneal epithelial cells. C, corneal epithelia; S, stroma; E, epidermis; D, dermis.
Based on these expression patterns, we hypothesized that, during corneal development, KLF7 and KLF12 maintain the progenitor state, acting antagonistically to the pro-differentiation factor KLF4. We chose to focus on KLF7, as KLF12 is not detectably expressed in primary human corneal epithelial cells.
By immunofluorescent staining, we observed KLF7 staining in the cornea epithelium at both embryonic (E18.5) and early postnatal (P5) time points (Fig. 4, C and D) but not at P50, when the corneal epithelium is fully differentiated (Fig. 4E). In the same sections, we also observed KLF7 staining primarily in the basal layer of the epidermis, a related epithelial tissue. In contrast, KLF4 staining was not detected in the cornea epithelium at E18.5 or P5 but was easily detected at P50 (Fig. 4, C–E). Consistent with its role in epidermal differentiation, KLF4 was expressed in the suprabasal layers of the epidermis.
For KLF7 and KLF4, we then established that they are expressed similarly vis à vis differentiation in cultured primary human corneal epithelial cells. KLF7 was more highly expressed in proliferating than in differentiating cells; conversely, KLF4 was more highly expressed in differentiating than in proliferating cells (Fig. 4, F and G).
KLF7 antagonizes KLF4 in the control of corneal epithelial cell differentiation
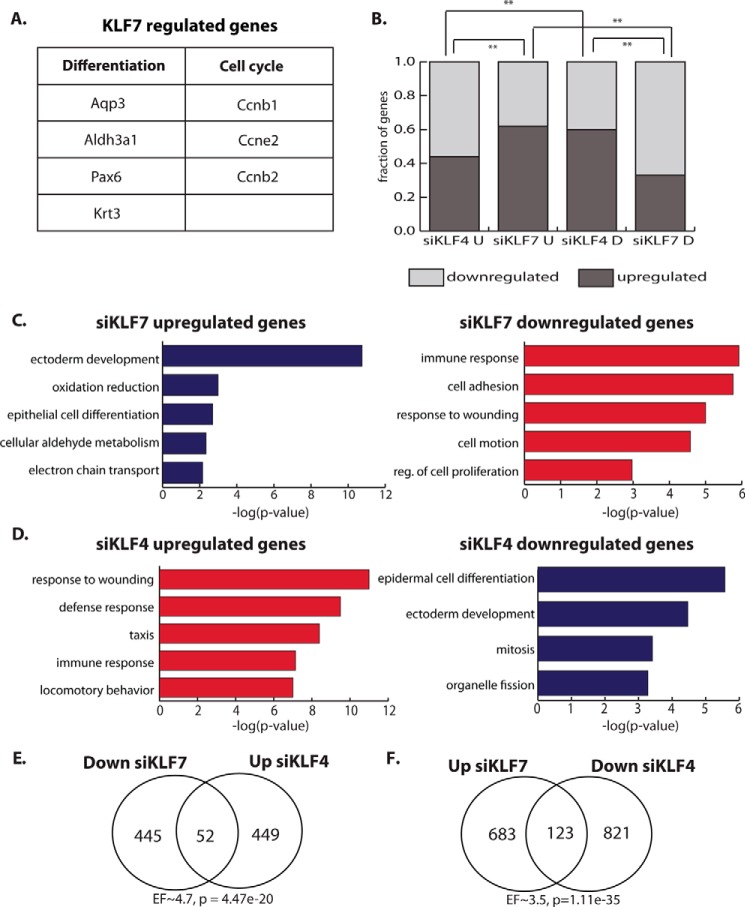
To test the role of KLF7 and KLF4 in corneal epithelia cells, we performed loss-of-function experiments in proliferating HCE cells with siKLF7 or siKLF4, assessing global gene expression changes by microarray analysis. Genes up-regulated by knockdown of KLF7 include the key corneal differentiation genes Pax6 and Aldh3a1 (Fig. 5A), supporting the role of KLF7 as a progenitor factor that represses the differentiation program in corneal epithelial cells. We also identified many cell cycle regulators that are aberrantly expressed in siKLF7 cells (Fig. 5A), indicating that KLF7 may also modulate the proliferation of corneal epithelial progenitor cells.
Figure 5.
KLF7 and KLF4 regulate corneal epithelial genes antagonistically. A, examples of KLF7-regulated genes involved in corneal epithelial differentiation or the cell cycle. B, proportion of up-regulated and down-regulated genes by siKLF4 or siKLF7 in undifferentiated (U) or differentiated (D) HCE cells. C, enriched GO categories for genes up-regulated (blue bars, left panel) or down-regulated (red bars, right panel) by knockdown of KLF7 (siKLF7). D, enriched GO categories for genes up-regulated (red bars, left panel) or down-regulated (blue bars, right panel) by knockdown of KLF4 (siKLF4). E, overlap between genes down-regulated by siKLF7 and genes up-regulated by siKLF4 (hypergeometric test). F, overlap between genes up-regulated by siKLF7 and genes down-regulated by siKLF4 (hypergeometric test). EF, enrichment factor. *, p < 0.05; **, p < 0.01.
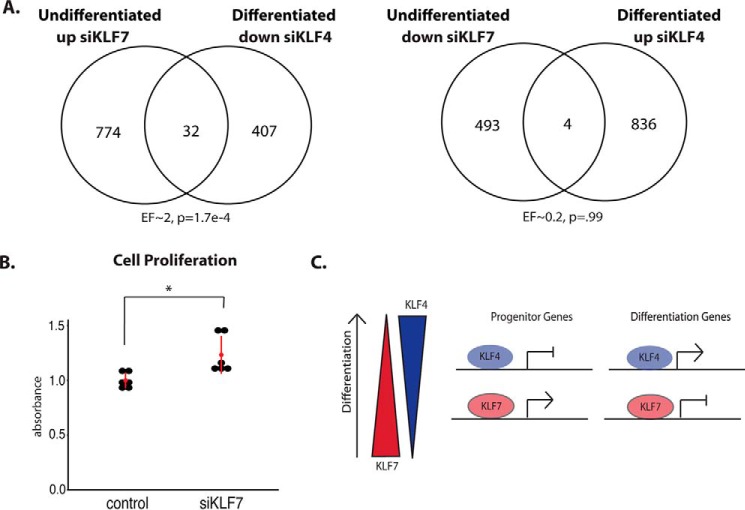
We also found a difference in the proportion of up-regulated and down-regulated genes between siKLF7 and siKLF4. A significantly larger proportion of genes is up-regulated by siKLF7, indicating that KLF7 represses a large number of genes in proliferating HCE cells; in contrast, a significantly larger proportion of genes is down-regulated by siKLF4, indicating that KLF4 activates a large number of genes in proliferating HCE cells (Fig. 5B).
When comparing the two knockdown experiments, we found that KLF7 and KLF4 regulate similar functional categories of genes but in the opposite direction. Genes that are down-regulated by siKLF7 fall into categories that contain genes up-regulated by siKLF4 and vice versa (Fig. 5, C and D). Genes up-regulated by siKLF7 and down-regulated by siKLF4 include those encoding many epithelial differentiation factors, further supporting a progenitor-promoting function for KLF7. Genes down-regulated by siKLF7 and up-regulated by siKLF4 include those conferring immune function and wounding response. Consistent with the overlap in functional categories, we also found a significant overlap in specific genes regulated in opposite directions by KLF7 and KLF4 (Fig. 5, E and F).
We next performed similar loss-of-function experiments in corneal epithelial cells that had been induced to differentiate. In contrast to the results from undifferentiated cells, KLF7 and KLF4 did not regulate a high number of genes in an opposing manner. Such a relationship, however, existed between proliferating and differentiating corneal epithelial cells. Thus, there was a significant overlap in genes down-regulated by siKLF4 in differentiation and those that were up-regulated by siKLF7 in proliferating cells (Fig. 6A, left panel). There was, however, not a significant overlap in genes up-regulated by siKLF4 in differentiation and those down-regulated by siKLF7 in proliferating cells (Fig. 6A, right panel).
Figure 6.
KLF7 and KLF4 regulate corneal epithelial differentiation antagonistically, and KLF7 suppresses proliferation. A, overlap between genes up-regulated by siKLF7 in undifferentiated HCE cells and genes down-regulated by siKLF4 in differentiated HCE cells (left panel) and overlap between genes down-regulated by siKLF7 in undifferentiated HCE cells and up-regulated by siKLF4 in differentiated HCE cells (right panel) (hypergeometric test). B, MTT cell proliferation assays on HCE cells, comparing siRNA control and siKLF7 treated cells (n = 6, t test). C, model for KLF7- and KLF4-mediated regulation of corneal epithelial differentiation. *, p < 0.05; **, p < 0.01.
In differentiating corneal epithelial cells, we also observed a difference in the proportion of up-regulated and down-regulated genes in each siRNA experiment. In contrast to undifferentiated cells, knockdown of KLF7 repressed a large number of genes, and knockdown of KLF4 up-regulated many genes in differentiated cells, suggesting a differentiation-dependent switch in activation–repression regulatory mechanisms for the two factors (Fig. 5B).
Because of the strong enrichment for cell cycle genes upon KLF7 knockdown in proliferating cells, we next wanted to test whether KLF7 regulates corneal epithelial cell proliferation. MTT assays revealed a significant increase in proliferation in undifferentiated HCE cells when KLF7 was knocked down (Fig. 6B), suggesting that, in addition to suppressing differentiation gene expression, KLF7 tempers proliferation, perhaps maintaining progenitor cell quiescence (Fig. 6C). In this instance, then, KLF7 has a similar effect on corneal epithelial cell proliferation as KLF4 (21, 25). Together, these results indicate that, like KLF4, KLF7 tempers cornea epithelial cell proliferation, but unlike KLF4, KFL7 suppresses cornea epithelial cell differentiation.
Single nucleotide polymorphisms linked to corneal diseases map to corneal epithelial enhancers
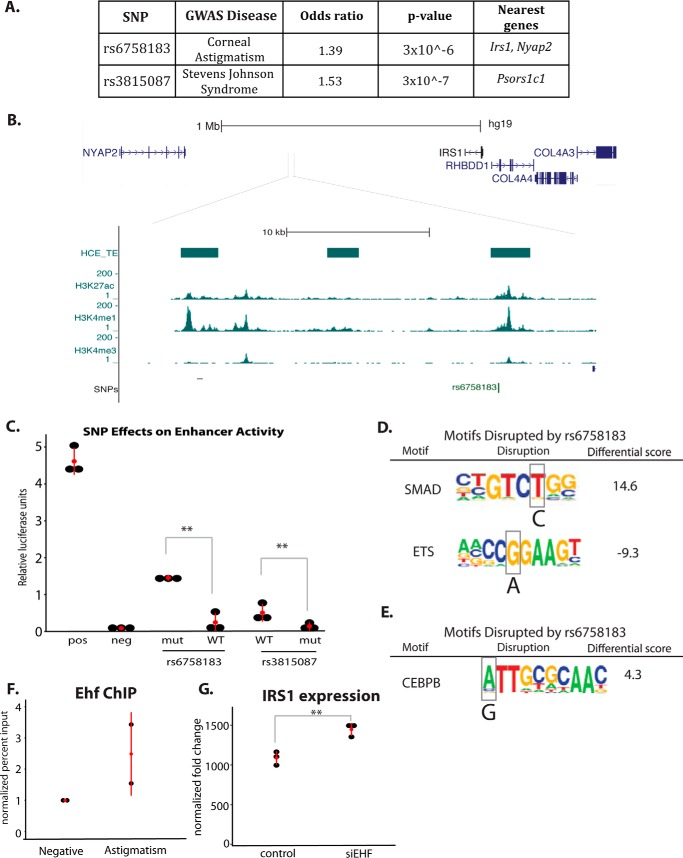
To investigate the functional role of enhancers in corneal diseases, we overlapped our corneal epithelial cell enhancer coordinates with SNPs linked to corneal diseases in GWAS. Two cornea-specific TEs overlapped SNPs associated with disease phenotypes in the cornea: rs3815087, a SNP on chromosome 6 associated with Stevens-Johnson syndrome (26), an immune-mediated condition that affects both the skin and cornea, and rs6758183, a SNP on chromosome 2 associated with corneal astigmatism (27) (Fig. 7A).
Figure 7.
HCE cell enhancers overlap SNPs linked to corneal diseases. A, table of GWAS SNP information. B, genomic view of rs6758183 with HCE enhancer data. C, luciferase reporter assays for WT and mutant (disease-associated allele) SNPs (n = 6, t test). D, analysis of motif disruption by SNP rs6758183. Scores were calculated by TRAP (24): differential score = mutant score − WT score. E, motif disruption analysis for rs3815087. Scores were calculated by TRAP. F, EHF ChIP in HCE cells at the rs6758183 (astigmatism) region compared with a negative control region. G, IRS1 expression in siRNA control and siEHF-treated HCE cells, detected by microarray as in Ref.18 (n = 3; *, p < 0.05; **, p < 0.01.
SNP rs3815087 is located in an exon of PSORS1C (Fig. 7A), a gene identified as a psoriasis susceptibility locus, although evidence points to this SNP being non-coding, as the exon in which it is located is not translated in any of the known isoforms of PSORS1C. Additionally, the enhancer histone modification pattern supports that this SNP may affect disease risk through non-coding mechanisms; even though enhancers are often thought of as distal regulatory regions, many have been found to directly overlap gene exons. SNP rs6758183 is located in a gene desert, ∼500 kb from the nearest coding gene (Fig. 7, A and B).
Single nucleotide polymorphisms linked to corneal diseases affect the function of corneal epithelial enhancers
We cloned the genomic regions containing SNPs rs3815087 and rs6758183 upstream of a neutral promoter in luciferase reporter vectors. When transfected into HCE cells, the genomic region of rs3815087 showed enhancer activity, and rs6758183 showed a trend toward increased activity compared with the negative control (Fig. 7C). We next mutated the SNP nucleotide to the disease-linked variant in each enhancer construct and transfected it into HCE cells. The region containing the disease-linked allele of rs3815087 showed significantly reduced enhancer activity compared with the WT allele. In contrast, the region containing the disease-linked variant of rs6758183 showed significantly increased enhancer activity compared with the WT allele and significantly increased enhancer activity above the negative control (Fig. 7C). These results show that alterations in cornea enhancer function caused by the SNP variants could contribute to the risk of disease by affecting the regulatory ability of these loci.
SNP rs6758183 decreases predicted Ets factor EHF enhancer binding affinity
We hypothesized that these SNP variants could disrupt transcription factor binding, altering enhancer activity for each allele. To study this possibility, we identified transcription factor binding motifs that overlap the SNPs and show significant differences in motif scores between WT and the disease-linked alleles. At rs6758183, the disease-associated A allele increased the strength of a SMAD motif and decreased the strength of an ETS motif (Fig. 7D). The disease-associated allele of rs3815087 strengthened a CCAAT/enhancer-binding protein β (CEBPB) motif, among others (Fig. 7E).
The Ets factor EHF is highly expressed in the corneal epithelium and has been shown to be an important regulator of corneal epithelial cell identity (18), making it a candidate for binding to the disrupted Ets motif. Using ChIP, we found that EHF bound to the WT allele of rs6758183, suggesting that disruption of EHF binding by the SNP could result in the observed differences in enhancer activity (Fig. 7F). Intriguingly, the WT version of the SNP showed less enhancer activity than the mutant, suggesting that EHF binding acts to repress the enhancer region. When this binding is lost, the enhancer becomes overactive, a change that could result in growth imbalances that cause alterations in the curvature and refractive power of the cornea.
Because rs6758183 is located in a gene desert, we looked for studies of the surrounding area of this SNP. We found that a number of SNPs in the vicinity of rs6758183 have been linked to type II diabetes (28, 29). Additionally, ChIA-PET studies in MCF7 cells found that this region (and the diabetes-linked SNPs) looped to make contact with the IRS1 promoter, 500 kb away (30). As insulin signaling is important for the growth and development of both the corneal epithelium and the stroma, IRS1 is a candidate target for the enhancer containing rs6758183. Consistent with this possibility, in previously published siRNA data for EHF in corneal epithelial cells, we found that knockdown of EHF caused a small but significant increase in IRS1 gene expression (Fig. 7G) (18). Together, these data suggest that EHF regulates IRS1 through binding to the rs6758183 enhancer and that disruption of the EHF motif causes reduced affinity of EHF for this site, resulting in aberrant IRS1 expression during corneal development.
Discussion
The identification of enhancers in corneal epithelial cells provides insights into the mechanisms and factors regulating corneal development and homeostasis. We have identified potential roles for SEs and TEs in corneal epithelial cell differentiation and characterized the differences in enhancer landscapes in different epithelial cell types, thus defining the regulatory regions important for cell identity. In combination with publicly available data, we have characterized disease-associated SNPs that lie within corneal enhancers, identifying regulatory regions with a potential role in disease risk. We have also defined a novel role for KLF7 in corneal epithelial cells, where it acts antagonistically to the pro-differentiation factor KLF4. Our work highlights the potential importance of KLF4 and KLF7 in regulating the balance between proliferation and differentiation required for proper corneal epithelial development and homeostasis.
Identification and characterization of SEs and TEs in human corneal epithelial cells
Consistent with results for SEs in other cell types, we found that SE-associated genes are more highly expressed than TE-associated genes. Our data, however, also provide evidence for the importance of TEs for gene regulation in the corneal epithelium; genes near TEs are strongly linked to corneal phenotypes in mouse mutants. We also found both an SE and several TEs in the vicinity of the PAX6 gene; these enhancers are presumed to be important for the proper expression of this key corneal epithelial regulator.
Although SEs confer cell type–specific regulation, the vast majority of SEs in HCE cells are either TEs or marked by H3K4me1, a histone mark of regulatory potential, in other epithelial cell types. As epithelial tissues express a number of common genes, it is unsurprising that different epithelial cells share similar regulatory landscapes. The main differences between different epithelial cell types lie in the type of enhancers and the strength of the H3K27Ac mark, suggesting that, rather than a binary on–off determination, these regulatory regions can be TEs or SEs, depending on the epithelial cell type, perhaps to provide the right level of gene activation for a given epithelial tissue.
SNPs linked to corneal diseases affect the activity of corneal epithelial enhancers
A challenge to understanding the disease-promoting mechanism for noncoding SNPs that map to enhancers, as rs3815087 and rs6758183 do, is that enhancers can be located great distances from the genes they regulate, making the link between SNP variants and gene expression difficult to decipher. This challenge was particularly apparent with SNP rs6758183, which was linked to corneal astigmatism, although extensive studies on the mechanism behind this SNP and the cellular alterations within the tissue that can lead to disease are lacking. In addition, this SNP is more than 500 kb away from the nearest gene, making the link to astigmatism difficult to study. Using a combination of data, from studies on corneal development and disease, published ChIA-PET studies, motif analysis, and siRNA experiments, we have provided insights into the potential molecular mechanisms for the contribution of rs6758183 of SNP to corneal astigmatism.
Based on our motif analysis, SNP rs6758183 is predicted to disrupt an ETS family binding motif. This finding is consistent with the known importance of ETS transcription factors in epithelial development and, specifically, of the ETS family member EHF in the corneal epithelium. Indeed, we were able to show with ChIP assays in corneal epithelial cells that EHF binds to the region of the enhancer containing SNP rs6758183.
Publicly available ChIA-PET data from MCF7 epithelial cells provided additional information, demonstrating that the enhancer region containing SNP rs6758183 contacts the promoter of IRS1, an important insulin and IGF1 signaling component, located 500 kb away from the SNP. This is also consistent with the finding that many SNPs in the vicinity of rs6758183 are associated with increased risk of metabolic syndrome and type 2 diabetes, suggesting that the entire region has regulatory potential toward IRS1.
We also find that SNP rs6758183 increases enhancer activity and that knockdown of EHF increases IRS1 gene expression, consistent with EHF binding to the SNP-containing enhancer to repress IRS1 in corneal epithelial cells. In agreement with this mechanism, a number of Ets family members, including EHF, have been shown previously to be able to repress gene expression (18, 31, 32).
To our knowledge, there are no studies directly linking ETS family members to astigmatism. However, Ets family members are known to regulate insulin and other growth factor signaling pathways (33–35), and there is a strong and well documented link between growth factor signaling and astigmatism (27). Additionally, insulin signaling has been implicated in two other ocular diseases related to the curvature of the cornea: myopia (36) and keratoconus (37), suggesting a general theme that the curvature of the eye is affected by growth factor signaling pathways.
Previous work has shown that both corneal epithelium and stroma contribute to the refractive power of the eye (21, 38), with the stroma being primarily implicated in the refractive errors in corneal astigmatism. Although our studies were carried out in epithelial cells, the enhancer containing SNP rs6758183 could also be active in stromal keratocytes, leading to alterations in insulin signaling in both cell types (27). Additionally, as extensive signaling occurs between the epithelium and the stroma, alterations in IRS1 expression in one cell type could lead to downstream growth effects in both. In summary, we have established a role for the ETS factor EHF in binding to the enhancer containing SNP rs6758183 and in regulating the nearby gene IRS1 in corneal epithelial cells, a mechanism that could underlie corneal astigmatism risk.
KLF7 antagonizes KLF4 in corneal epithelial cells
Motif analysis of corneal epithelial cell enhancers highlighted the important role of KLF transcriptional regulators in corneal epithelium and led to the discovery of a previously undescribed role for KLF7 in corneal epithelial progenitor cells. KLF7 regulates neuronal and olfactory development (39) and has been characterized as an inhibitor of adipocyte differentiation (39–41). Its potential role in corneal epithelial cells, however, has not been investigated before.
We found that KLF7 represses the differentiation gene expression program and reduces proliferation in undifferentiated corneal epithelial cells, potentially acting to maintain progenitor cells in a quiescent and undifferentiated state. We also found that KLF7 maintains the corneal epithelial progenitor state in part through antagonistic regulation of KLF4 gene targets. Thus, the relative expression level of each KLF may determine the transcriptional state of shared target genes; as KLF7 levels drop and KLF4 expression increases during differentiation, KLF4 may be able to outcompete KLF7 and change the response of shared targets (Fig. 6C). The complementary expression and antagonistic functions of KLF7 and KLF4 may be crucial for the balancing of proliferation and differentiation during corneal epithelial development and homeostasis. It is also possible that other mechanisms are at play in the dynamic chromatin landscape of corneal epithelial cell differentiation. A recently published study on chromatin regulatory domains in epidermal keratinocytes demonstrated that the SE and TE landscape changes substantially between undifferentiated keratinocytes and differentiated keratinocytes (42). Based on this, it is likely that the SE and TE landscape in corneal epithelial cells also changes with differentiation, and this may create new binding opportunities for transcription factors. Another possible mechanism for the regulation of KLF binding under the changing conditions of differentiation is the multiprotein complexes in which it participates, which may change based on changing expression patterns of different members during differentiation, potentially influencing binding site selection.
In conclusion, we have defined the chromatin and regulatory landscape in human corneal epithelial cells, advanced mechanistic insights into disease by showing that two SNPs associated with corneal diseases disrupt the activity of corneal epithelial enhancers, and identified the transcription factor KLF7 as an antagonist of its pro-differentiating family member KLF4 and a promoter of the progenitor state of corneal epithelial cells.
Experimental procedures
Cell culture
Normal HCE cells were purchased from LifeLine Technologies and grown according to the instructions of the manufacturer in OcuLife medium (LifeLine Technologies) supplemented with OcuLife growth factors (LifeLine Technologies). To induce differentiation, cells were transferred into CNT-Prime 2D Diff medium (Cell-n-Tech, CNT-PR-D). hTERT immortalized human corneal epithelial (hTCEpi) (43) were grown in KBM medium (Lonza) supplemented with SingleQuots (Lonza).
Chromatin immunoprecipitation assays
ChIP assays were performed as described previously (44, 45) with the following changes: 24 μg of sonicated chromatin was used for each immunoprecipitation, magnetic Dynabeads (Invitrogen) were used for immunoprecipitation, and, for ChIP-qPCR analysis, enrichment was calculated over IgG and normalized to an intergenic negative control region. The following antibodies were used: IgG (Sigma), Ehf (Santa Cruz Biotechnology, Inc.), H3K4me1 (Abcam), H3K27ac (Millipore), and H3K4me3 (Millipore). Primer sequences are listed in supplemental Table S1.
ChIP-Seq
Sequencing libraries were generated for the H3K4me1, H3K4me3, H3K27ac, and input samples using NEB Next reagents and Illumina adaptors and oligos according to the Illumina protocol for ChIP-seq library preparations, with some modification of the protocol by Schmidt et al. (46): after adaptor ligation, PCR amplification was performed prior to size selection of the library. Clustering and 50-cycle single end sequencing were performed on the Illumina Hi-Seq 2000 genome analyzer.
ChIP-Seq analysis
The resulting ChIP-Seq reads were aligned using Bowtie (47), and only uniquely aligning reads were retained. Peaks were called using spatial clustering for identification of ChIP-enriched regions (SICER) (48), and Galaxy (49–51) software was used for further analysis. Super enhancers were identified using ROSE (5, 52).
Quantitative real-time PCR
For mRNA expression analysis, cDNA was prepared using the iScript cDNA kit. RT-PCR was performed using SsoFast for Probes and SsoFast EvaGreen (Bio-Rad) master mixes in the CFX384 real-time PCR detection system (Bio-Rad). GAPDH or ribosomal protein lateral stalk subunit P0 (RPLPO) were used as endogenous controls. Primer sequences are listed in supplemental Table S1.
RNA extraction
Cells and tissues were collected and lysed in TRIzol, followed by chloroform extraction. RNA was extracted from the aqueous phase as described previously (53). RNA concentration and quality were quantified on a NanoDrop instrument.
Luciferase assays
Approximately 1-kb regions surrounding each SNP were cloned into PGL3 reporter vectors. Site-directed mutagenesis was performed following the specifications of the manufacturer (Agilent QuikChange II site-directed mutagenesis kit). Successful mutagenesis was verified by DNA sequencing. Cells were transfected with each plasmid and a Renilla control plasmid and collected after 72 h. Luciferase was quantified using the Promega luciferase assay system.
MTT assays
Cells were plated to a density of 10,000 cells/well in 96-well plates and transfected with siRNA to KLF7 or scramble control the following day. Proliferation was assayed after 72 h using the Cell Titer proliferation assay (Promega).
Immunofluorescent staining of corneal cryosections
Cryosections (6–8 μm thick) from optimal cutting temperature compound–embedded embryo heads and adult mouse eyes were fixed in acetone for 13 min, followed by two PBS washes. Slides were then fixed with 4% paraformaldehyde for 10 min and washed twice with PBS. For permeabilization, slides were incubated for 15 min in permeabilization buffer (0.3% Triton X-100 in 1× PBS). Slides were then blocked for 1 h at room temperature with 0.1%Triton X-100 and 2% BSA in 1× PBS. Primary antibodies were incubated overnight at 4 °C with 1:200 dilution of rabbit anti-mouse KLF4 primary antibody (ab129473, Abcam) and 1:50 dilution of rabbit anti-mouse KLF7 antibody (ab197690, Abcam), washed three times with PBS for 5 min each, incubated with secondary antibody (Alexa Fluor 488-coupled goat anti-rabbit IgG, Life Technologies) at a 1:1000 dilution for 1 h at room temperature in the dark, rinsed with PBS, and mounted with mounting medium with DAPI (Vector Laboratories). Images were collected with a Keyence BZ-X710 All-In-One fluorescence microscope. All images presented within each figure were acquired under identical settings and processed in a similar manner using ImageJ.
Microarray analysis
Gene expression analysis for undifferentiated HCE cells was performed with biological duplicates as described previously (54), except Affymetrix Human Gene 1.0 ST arrays (26,869 probe sets) were used and washed according to the recommendations of the manufacturer (Affymetrix, Santa Clara, CA). Gene expression analysis for differentiated HCE cells used Human Gene 2.0 ST arrays. Plier analysis was performed, and the data were then filtered for expression levels. Probes with raw expression values below 200 were considered not expressed for subsequent analysis. CyberT was used to define statistically significant differentially expressed genes (53). Gene ontology analysis was performed using DAVID (55).
Author contributions
R. H. K. and B. A. conceived and coordinated the study and wrote the paper. R. H. K., W. H., G. K., Z. L., T. N., and M. D. designed, performed, and analyzed the experiments. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This work was supported by National Institutes of Health Grants R01EY019413 and R01AR44882 (to B. A.), National Library of Medicine Grant T15LM00744 (to R. H. K.), and NCI, National Institutes of Health Grant 5T32CA09054–38 (to Z. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Table S1.
- SE
- super enhancer
- TE
- typical enhancer
- HCE
- human corneal epithelial
- GO
- gene ontology
- KLF
- Kruppel-like factor
- HMEC
- human mammary epithelial cell
- E
- embryonic day
- P
- postnatal day
- hESC
- human embryonic stem cell
- H3K4me3
- histone H3 lysine 4 trimethylation
- H3K4me1
- histone H3 lysine 4 monomethylation
- H3K27ac
- histone H3 lysine 27 acetylation
- H3K27me3
- histone H3 lysine 27 trimethylation
- H3K9ac
- histone H3 lysine 9 acetylation
- GWAS
- genome-wide association
- ETS
- E-twenty-six
- NHEK
- neonatal human epidermal keratinocytes
- NHLF
- normal human lung fibroblasts
- EHF
- ETS homologous factor
- ChIA-PET
- Chromatin Interaction Analysis by Paired-End Tag Sequencing
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
References
- 1. Deng W., Lee J., Wang H., Miller J., Reik A., Gregory P. D., Dean A., and Blobel G. A. (2012) Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149, 1233–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Erokhin M., Vassetzky Y., Georgiev P., and Chetverina D. (2015) Eukaryotic enhancers: common features, regulation, and participation in diseases. Cell. Mol. Life Sci. 72, 2361–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., and Ren B. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 [DOI] [PubMed] [Google Scholar]
- 4. Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S. A., Flynn R. A., and Wysocka J. (2011) A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Whyte W. A., Orlando D. A., Hnisz D., Abraham B. J., Lin C. Y., Kagey M. H., Rahl P. B., Lee T. I., and Young R. A. (2013) Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Adam R. C., Yang H., Rockowitz S., Larsen S. B., Nikolova M., Oristian D. S., Polak L., Kadaja M., Asare A., Zheng D., and Fuchs E. (2015) Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521, 366–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heinz S., Romanoski C. E., Benner C., and Glass C. K. (2015) The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 16, 144–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hnisz D., Abraham B. J., Lee T. I., Lau A., Saint-André V., Sigova A. A., Hoke H. A., and Young R. A. (2013) Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown J. D., Lin C. Y., Duan Q., Griffin G., Federation A., Paranal R. M., Bair S., Newton G., Lichtman A., Kung A., Yang T., Wang H., Luscinskas F. W., Croce K., Bradner J. E., and Plutzky J. (2014) NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol. Cell 56, 219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cavazza A., Miccio A., Romano O., Petiti L., Malagoli Tagliazucchi G., Peano C., Severgnini M., Rizzi E., De Bellis G., Bicciato S., and Mavilio F. (2016) Dynamic transcriptional and epigenetic regulation of human epidermal keratinocyte differentiation. Stem Cell Rep. 6, 618–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Loft A., Forss I., Siersbæk M. S., Schmidt S. F., Larsen A. S., Madsen J. G., Pisani D. F., Nielsen R., Aagaard M. M., Mathison A., Neville M. J., Urrutia R., Karpe F., Amri E. Z., and Mandrup S. (2015) Browning of human adipocytes requires KLF11 and reprogramming of PPARγ superenhancers. Genes Dev. 29, 7–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zieske J. D. (2004) Corneal development associated with eyelid opening. Int. J. Dev. Biol. 48, 903–911 [DOI] [PubMed] [Google Scholar]
- 13. Yoon J. J., Ismail S., and Sherwin T. (2014) Limbal stem cells: central concepts of corneal epithelial homeostasis. World J. Stem Cells 6, 391–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eghrari A. O., Riazuddin S. A., and Gottsch J. D. (2015) Overview of the cornea: structure, function, and development. Prog. Mol. Biol. Transl. Sci. 134, 7–23 [DOI] [PubMed] [Google Scholar]
- 15. Ramaesh T., Collinson J. M., Ramaesh K., Kaufman M. H., West J. D., and Dhillon B. (2003) Corneal abnormalities in Pax6+/− small eye mice mimic human aniridia-related keratopathy. Invest. Ophthalmol. Vis. Sci. 44, 1871–1878 [DOI] [PubMed] [Google Scholar]
- 16. Ouyang H., Xue Y., Lin Y., Zhang X., Xi L., Patel S., Cai H., Luo J., Zhang M., Yang Y., Li G., Li H., Jiang W., Yeh E., Lin J., et al. (2014) WNT7A and PAX6 define corneal epithelium homeostasis and pathogenesis. Nature 511, 358–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kleinjan D. A., Seawright A., Schedl A., Quinlan R. A., Danes S., and van Heyningen V. (2001) Aniridia-associated translocations, DNase hypersensitivity, sequence comparison and transgenic analysis redefine the functional domain of PAX6. Hum. Mol. Genet. 10, 2049–2059 [DOI] [PubMed] [Google Scholar]
- 18. Stephens D. N., Klein R. H., Salmans M. L., Gordon W., Ho H., and Andersen B. (2013) The Ets transcription factor EHF as a regulator of cornea epithelial cell identity. J. Biol. Chem. 288, 34304–34324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McConnell B. B., Ghaleb A. M., Nandan M. O., and Yang V. W. (2007) The diverse functions of Kruppel-like factors 4 and 5 in epithelial biology and pathobiology. BioEssays 29, 549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Swamynathan S., Buela K. A., Kinchington P., Lathrop K. L., Misawa H., Hendricks R. L., and Swamynathan S. K. (2012) Klf4 regulates the expression of Slurp1, which functions as an immunomodulatory peptide in the mouse cornea. Invest. Ophthalmol. Vis. Sci. 53, 8433–8446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Swamynathan S. K., Katz J. P., Kaestner K. H., Ashery-Padan R., Crawford M. A., and Piatigorsky J. (2007) Conditional deletion of the mouse Klf4 gene results in corneal epithelial fragility, stromal edema, and loss of conjunctival goblet cells. Mol. Cell. Biol. 27, 182–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. The ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kenchegowda D., Harvey S. A., Swamynathan S., Lathrop K. L., and Swamynathan S. K. (2012) Critical role of Klf5 in regulating gene expression during post-eyelid opening maturation of mouse corneas. PLoS ONE 7, e44771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thomas-Chollier M., Hufton A., Heinig M., O'Keeffe S., Masri N. E., Roider H. G., Manke T., and Vingron M. (2011) Transcription factor binding predictions using TRAP for the analysis of ChIP-seq data and regulatory SNPs. Nat. Protoc. 6, 1860–1869 [DOI] [PubMed] [Google Scholar]
- 25. Swamynathan S. K., Davis J., and Piatigorsky J. (2008) Identification of candidate Klf4 target genes reveals the molecular basis of the diverse regulatory roles of Klf4 in the mouse cornea. Invest. Ophthalmol. Vis. Sci. 49, 3360–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Génin E., Schumacher M., Roujeau J. C., Naldi L., Liss Y., Kazma R., Sekula P., Hovnanian A., and Mockenhaupt M. (2011) Genome-wide association study of Stevens-Johnson syndrome and toxic epidermal necrolysis in Europe. Orphanet J. Rare Dis. 6, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fan Q., Zhou X., Khor C. C., Cheng C. Y., Goh L. K., Sim X., Tay W. T., Li Y. J., Ong R. T., Suo C., Cornes B., Ikram M. K., Chia K. S., Seielstad M., Liu J., et al. (2011) Genome-wide meta-analysis of five Asian cohorts identifies PDGFRA as a susceptibility locus for corneal astigmatism. PLoS Genet. 7, e1002402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Walford G. A., Gustafsson S., Rybin D., Stančáková A., Chen H., Liu C. T., Hong J., Jensen R. A., Rice K., Morris A. P., Mägi R., Tönjes A., Prokopenko I., Kleber M. E., Delgado G., et al. (2016) Genome-wide association study of the modified Stumvoll insulin sensitivity index identifies BCL2 and FAM19A2 as novel insulin sensitivity loci. Diabetes 65, 3200–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Manning A. K., Hivert M. F., Scott R. A., Grimsby J. L., Bouatia-Naji N., Chen H., Rybin D., Liu C. T., Bielak L. F., Prokopenko I., Amin N., Barnes D., Cadby G., Hottenga J. J., Ingelsson E., et al. (2012) A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 44, 659–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li G., Ruan X., Auerbach R. K., Sandhu K. S., Zheng M., Wang P., Poh H. M., Goh Y., Lim J., Zhang J., Sim H. S., Peh S. Q., Mulawadi F. H., Ong C. T., Orlov Y. L., et al. (2012) Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fossum S. L., Mutolo M. J., Tugores A., Ghosh S., Randell S. H., Jones L. C., Leir S. H., and Harris A. (2017) Ets homologous factor (EHF) has critical roles in epithelial dysfunction in airway disease. J. Biol. Chem. 292, 10938–10949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mavrothalassitis G., and Ghysdael J. (2000) Proteins of the ETS family with transcriptional repressor activity. Oncogene 19, 6524–6532 [DOI] [PubMed] [Google Scholar]
- 33. Tugores A., Le J., Sorokina I., Snijders A. J., Duyao M., Reddy P. S., Carlee L., Ronshaugen M., Mushegian A., Watanaskul T., Chu S., Buckler A., Emtage S., and McCormick M. K. (2001) The epithelium-specific ETS protein EHF/ESE-3 is a context-dependent transcriptional repressor downstream of MAPK signaling cascades. J. Biol. Chem. 276, 20397–20406 [DOI] [PubMed] [Google Scholar]
- 34. Wasylyk B., Hagman J., and Gutierrez-Hartmann A. (1998) Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem. Sci. 23, 213–216 [DOI] [PubMed] [Google Scholar]
- 35. Yordy J. S., and Muise-Helmericks R. C. (2000) Signal transduction and the Ets family of transcription factors. Oncogene 19, 6503–6513 [DOI] [PubMed] [Google Scholar]
- 36. Ritchey E. R., Zelinka C. P., Tang J., Liu J., and Fischer A. J. (2012) The combination of IGF1 and FGF2 and the induction of excessive ocular growth and extreme myopia. Exp. Eye Res. 99, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Foster J., Wu W. H., Scott S. G., Bassi M., Mohan D., Daoud Y., Stark W. J., Jun A. S., and Chakravarti S. (2014) Transforming growth factor β and insulin signal changes in stromal fibroblasts of individual keratoconus patients. PLoS ONE 9, e106556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Courville C. B., Smolek M. K., and Klyce S. D. (2004) Contribution of the ocular surface to visual optics. Exp. Eye Res. 78, 417–425 [DOI] [PubMed] [Google Scholar]
- 39. Laub F., Lei L., Sumiyoshi H., Kajimura D., Dragomir C., Smaldone S., Puche A. C., Petros T. J., Mason C., Parada L. F., and Ramirez F. (2005) Transcription factor KLF7 is important for neuronal morphogenesis in selected regions of the nervous system. Mol. Cell. Biol. 25, 5699–5711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Z., Wang H., Sun Y., Li H., and Wang N. (2013) Klf7 modulates the differentiation and proliferation of chicken preadipocyte. Acta Biochim. Biophys. Sin. 45, 280–288 [DOI] [PubMed] [Google Scholar]
- 41. Wu Z., and Wang S. (2013) Role of Kruppel-like transcription factors in adipogenesis. Dev. Biol. 373, 235–243 [DOI] [PubMed] [Google Scholar]
- 42. Klein R. H., Lin Z., Hopkin A. S., Gordon W., Tsoi L. C., Liang Y., Gudjonsson J. E., and Andersen B. (2017) GRHL3 binding and enhancers rearrange as epidermal keratinocytes transition between functional states. PLoS Genet. 13, e1006745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Robertson D. M., Kalangara J. P., Baucom R. B., Petroll W. M., and Cavanagh H. D. (2011) A reconstituted telomerase-immortalized human corneal epithelium in vivo: a pilot study. Curr. Eye Res. 36, 706–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu Z., Mannik J., Soto A., Lin K. K., and Andersen B. (2009) The epidermal differentiation-associated Grainyhead gene Get1/Grhl3 also regulates urothelial differentiation. EMBO J. 28, 1890–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Klein R. H., Stephens D. N., Ho H., Chen J. K., Salmans M. L., Wang W., Yu Z., and Andersen B. (2016) Cofactors of LIM domains associate with estrogen receptor α to regulate the expression of noncoding RNA H19 and corneal epithelial progenitor cell function. J. Biol. Chem. 291, 13271–13285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schmidt D., Wilson M. D., Spyrou C., Brown G. D., Hadfield J., and Odom D. T. (2009) ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods 48, 240–248, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Langmead B., Trapnell C., Pop M., and Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xu S., Grullon S., Ge K., and Peng W. (2014) Spatial clustering for identification of ChIP-enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods Mol. Biol. 1150, 97–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Blankenberg D., Von Kuster G., Coraor N., Ananda G., Lazarus R., Mangan M., Nekrutenko A., and Taylor J. (2010) Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. Chapter 19, Unit 19.10.11–21, 10.1002/0471142727.mb1910s89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Giardine B., Riemer C., Hardison R. C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Miller W., Kent W. J., and Nekrutenko A. (2005) Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15, 1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Goecks J., Nekrutenko A., Taylor J., and Galaxy Team (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lovén J., Hoke H. A., Lin C. Y., Lau A., Orlando D. A., Vakoc C. R., Bradner J. E., Lee T. I., and Young R. A. (2013) Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hopkin A. S., Gordon W., Klein R. H., Espitia F., Daily K., Zeller M., Baldi P., and Andersen B. (2012) GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS Genet. 8, e1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu Z., Lin K. K., Bhandari A., Spencer J. A., Xu X., Wang N., Lu Z., Gill G. N., Roop D. R., Wertz P., and Andersen B. (2006) The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev. Biol. 299, 122–136 [DOI] [PubMed] [Google Scholar]
- 55. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.