Abstract
In green algae, light-harvesting complex stress-related 3 (LHCSR3) is responsible for the pH-dependent dissipation of absorbed light energy, a function vital for survival under high-light conditions. LHCSR3 binds the photosystem II and light-harvesting complex II (PSII–LHCII) supercomplex and transforms it into an energy-dissipative form under acidic conditions, but the molecular mechanism remains unclear. Here we show that in the green alga Chlamydomonas reinhardtii, LHCSR3 modulates the excitation energy flow and dissipates the excitation energy within the light-harvesting complexes of the PSII supercomplex. Using fluorescence decay–associated spectra analysis, we found that, when the PSII supercomplex is associated with LHCSR3 under high-light conditions, excitation energy transfer from light-harvesting complexes to chlorophyll-binding protein CP43 is selectively inhibited compared with that to CP47, preventing excess excitation energy from overloading the reaction center. By analyzing femtosecond up-conversion fluorescence kinetics, we further found that pH- and LHCSR3-dependent quenching of the PSII—LHCII–LHCSR3 supercomplex is accompanied by a fluorescence emission centered at 684 nm, with a decay time constant of 18.6 ps, which is equivalent to the rise time constant of the lutein radical cation generated within a chlorophyll–lutein heterodimer. These results suggest a mechanism in which LHCSR3 transforms the PSII supercomplex into an energy-dissipative state and provide critical insight into the molecular events and characteristics in LHCSR3-dependent energy quenching.
Keywords: algae, bioenergetics, light-harvesting complex (antenna complex), photobiology, photosynthesis, photosystem II, quenching
Introduction
Although light is an essential energy source for photosynthetic organisms, light beyond their photosynthetic capacity tends to generate harmful reactive oxygen species (1). To protect against excessive light, photoprotective mechanisms, such as non-photochemical quenching (NPQ)2 of chlorophyll (Chl) fluorescence (2–5), are critical. Over the last decade, intensive study of NPQ revealed two effector proteins crucial for energy-dependent quenching (qE), which is the most effective and fastest photoprotective mechanism (6–8): PsbS in mosses and vascular plants and light-harvesting complex stress-related protein (LHCSR) in algae and mosses. PsbS, which is composed of four transmembrane helices and does not bind pigments, also functions as a sensor of pH change, which serves as a direct indicator of excess light conditions (9–14). Although the molecular mechanisms of PsbS-dependent quenching have not been fully clarified, several hypotheses have been proposed wherein PsbS is a modulator for the organization of photosynthetic proteins and thylakoid membranes (15–18), the protonation of light-harvesting complex proteins (19), and excitation energy quenching via interaction with Lhcb1 in photosystem II (PSII) supercomplexes (20). In contrast to PsbS in mosses and vascular plants, where its expression is constitutive, the LHCSRs (LHCSR3 and LHCSR1) in algae are pigment-binding proteins containing 6–7 Chls and 2–3 carotenoids whose expression is dependent upon environmental cues (7, 21, 22). LHCSR3 is expressed under high light in the presence of blue light (23), low CO2 (24), calcium signaling mediated by CAS (25, 26), and/or various stress conditions (27–30) and LHCSR1 are expressed under UV-B radiation (31, 32). We therefore expected LHCSR proteins to serve as an energy quencher, unlike PsbS, which does not serve as an energy quencher.
Recent spectroscopic studies on reconstituted LHCSR3 protein (rLHCSR3) revealed that it has shorter fluorescence lifetimes (<100 ps in Ref. 21, ∼300 ps in Ref. 33, and 140–190 ps in Ref. 34) than other light-harvesting complexes (LHCs) under acidic conditions. Also, its transient absorption kinetics at near-IR wavelengths (800–1100 nm) indicate that carotenoid radical cations are generated during the charge transfer process of the Chl-carotenoids heterodimer in rLHCSR3 (21) and rLHCSR1 from moss (35). Those findings demonstrate that rLHCSR3 is not only a pH sensor but also a conductor of charge transfer quenching and/or excitation energy quenching (21). Further studies revealed that a C terminus subdomain in the lumenal loop of rLHCSR3 acts as a sensor of the lumenal pH for the transformational quenching form of rLHCSR3 (33, 34). In addition, aggregation of rLHCSR3 enhances the quenching process of rLHCSR3, indicating that protein–protein interactions inducing conformational changes might facilitate the quenching of LHCSR3 in vivo (34). Indeed, an in vivo study using a fluorescence lifetime snapshot technique revealed that a fluorescence lifetime component of 65 ps was accompanied by qE, whose lifetime was faster than that observed in rLHCSR3 (36). Hence, it is important to study in situ LHCSR3 interacting with other photosynthetic proteins. Although investigating the localization of LHCSR3 found that LHCSR3 binds to the PSII supercomplex and transforms it into its energy dissipative form (37), the mechanism how LHCSR3 dissipates the excitation energy of PSII, however, remains to be clarified. In particular, the fast time domain (<50 ps) has not been fully studied by the fluorescence lifetime analysis.
To investigate how LHCSR3 dissipates excitation energy, thereby protecting the PSII supercomplex, we applied fluorescence decay–associated spectra (FDAS) analysis to a purified PSII–LHCII supercomplex with and without LHCSR3 from Chlamydomonas reinhardtii. FDAS analysis is a versatile method to examine excitation energy dynamics within pigment complexes such as photosystem complexes (38–40).
Here, the FDAS at 77 K revealed that when LHCSR3 binds to the PSII supercomplex, excitation energy transfer from LHCs to CP43 is redirected to CP47. LHCSR3-dependent quenching within the LHCs of the PSII supercomplex is also observed under acidic conditions. We then determined the decay time constant of the quenching by femtosecond up-conversion fluorescence spectroscopy so that we could infer the identity of the quencher. Finally, we proposed a model for the energy dissipation process within the PSII—LHCII–LHCSR3 supercomplex in C. reinhardtii.
Results
Fluorescence decay–associated spectra (FDAS) at 77 K
To investigate energy transfer dynamics in PSII supercomplexes, we isolated PSII supercomplexes from low-light (LL) and high-light (HL) grown wild-type C. reinhardtii cells (Fig. 1). To induce expression of LHCSR3, cells were cultured under 500 μmol of photons m−2 s−1 of white light, where LHCSR3, but not PsbS or LHCSR1 was dominantly accumulated (41, 42). PSII supercomplexes were purified by sucrose density gradient (SDG) ultracentrifugation of the solubilized protein complexes as previously described in Ref. 37. Fig. 1, A and B, respectively, show expression levels of LHCSR3 and separated green bands by SDG ultracentrifugation. In this study, we investigated the PSII supercomplex bands from LL and HL grown cells (PSII-LL and PSII-HL, respectively), which have been identified as dominantly PSII–LHCII and PSII—LHCII–LHCSR3, respectively (37). In absorption spectra at 77 K, the Qx- and Qy-band region from 600 to 700 nm is almost identical for PSII-LL and PSII-HL (Fig. 1C), which indicates that the S1 energy levels of chlorophylls are analogous in both forms. In fluorescence emission spectra at 77 K (Fig. 1D and supplemental Fig. S1), both PSII-LL and PSII-HL show maximum peaks at 685 nm and shoulders at 680 and 695 nm, which are associated with the emissions from CP43, LHCII, and CP47, respectively. In addition, the purity of PSII supercomplexes against PSI was confirmed by absence of the PSI fluorescence emission peak in the 720–740-nm region. The ratio of the PSII—LHCII–LHCSR3 supercomplex to the PSII–LHCII supercomplex in the PSII-HL preparation has been estimated to ∼0.28 by autoradiography as described in a previous study (37), whereas the PSII—LHCII–LHCSR3 supercomplex was not detected in the PSII-LL preparation (Fig. 1A).
Figure 1.
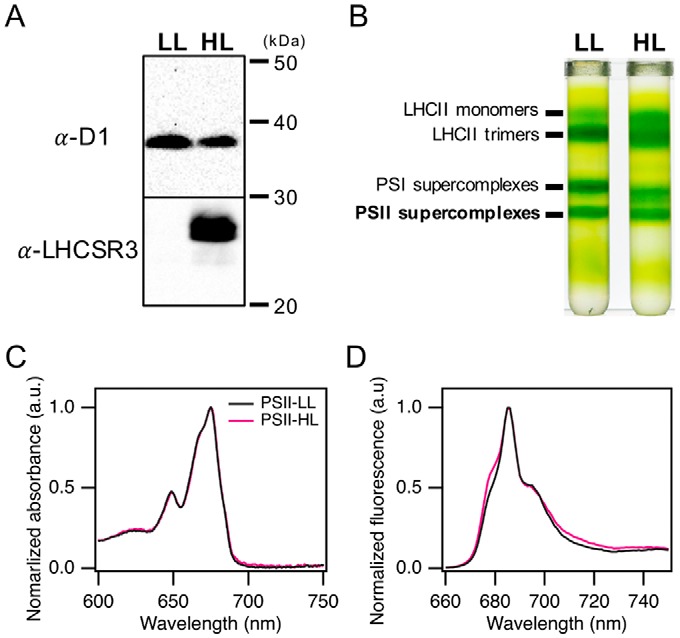
Isolation of PSII supercomplexes and its absorption and fluorescence emission spectra at 77 K. A, thylakoids membranes were isolated from low-light (20 μE m−2 s−1) and high-light (500 μE m−2 s−1) grown C. reinhardtii. For each lane, 2.0 μg of Chl of thylakoids membranes were subjected to immunoblotting analyses with antibodies against LHCSR3 and D1. B, the thylakoid membranes (250 μg of Chl) were solubilized with dodecyl α-maltoside and loaded on SDG. Four bands were identified as LHCII monomers, LHCII trimers, PSI supercomplexes, and PSII supercomplexes, as indicated. C, absorption spectra of PSII supercomplexes at pH 7.5 were obtained from reflectance spectra of frozen samples in NMR glass tubes at 77 K and normalized at the maximum peak. D, fluorescence emission spectra of PSII supercomplexes at pH 7.5 were obtained by excitation at 440 nm and normalized at the maximum peak.
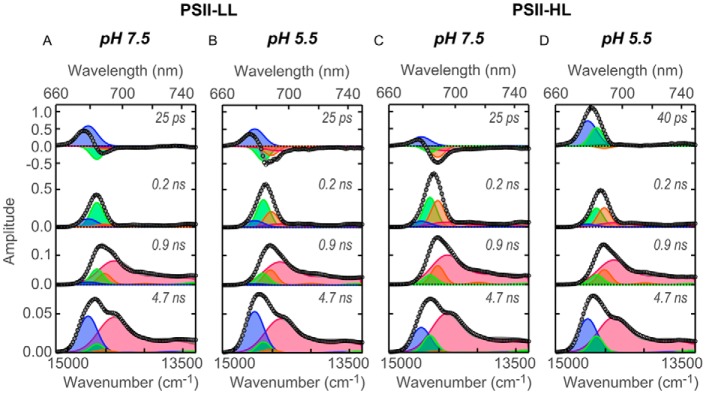
The global fitting analysis of the time-resolved fluorescence curves shows a series of spectra corresponding to individual lifetime components (Fig. 2). Four lifetime components (25–40 ps, 0.2 ns, 0.9 ns, and 4.7 ns) were required for fitting in the 0–10 ns range, as with PSII supercomplexes from other organisms (38, 39). Each of the four FDAS was decomposed by four spectral components peaking at 679, 684, 689, and 694 nm, which were assigned to LHCs, CP43 and/or LHCSR3, CP47, and the lowest-energy Chl(s) in CP47, respectively (43–48). The fluorescence from the primary donor (P680) appears at 682–683 nm, but was negligible in the fluorescence from the PSII supercomplex at 77 K due to its relatively low quantum yield (49).
Figure 2.
Fluorescence decay–associated spectra of PSII supercomplexes at 77 K. FDAS were derived from the time-resolved fluorescence profiles of PSII supercomplexes obtained by excitation at 408 nm. Spectral components of global analysis were shown as blue (679 nm), green (684 nm), orange (689 nm), and red (694 nm). Open circles and gray lines represent FDAS spectra and fitting curve, respectively. A, PSII-LL at pH 7.5; B, PSII-LL at pH 5.5; C, PSII-HL at pH 7.5; and D, PSII-HL at pH 5.5.
In the first FDAS (25–40 ps), a positive peak appeared at 679 nm in all the complexes, reflecting the relaxation of LHCs excitation as its energy was transferred to other component(s) (Fig. 2). Negative peaks appeared at 684 and 689 nm in PSII-LL at pH 7.5, PSII-LL at pH 5.5, and PSII-HL at pH 7.5; the former peaks correspond to the energy transfer to CP43, the latter to CP47 (Fig. 2, A–C). Since the relative contribution of CP43 versus CP47, to which excitation energy is delivered, was higher for PSII-LL at pH 7.5 than it was for PSII-LL at pH 5.5, the excitation energy delivery to CP47 was promoted in the acidic environment (Fig. 2, A and B). Moreover, the shift of excitation energy delivery toward CP47 was more prominent in the presence of LHCSR3 (Fig. 2C).
The most striking feature in this FDAS analysis is seen in PSII-HL at pH 5.5 (Fig. 2D). Under those conditions, when LHCSR3 was bound to the PSII supercomplex as well as activated by the protonation, negative peaks at 684 and 689 nm were significantly reduced, indicating that excitation energy localized on LHCII was barely transferred to the core complex (Fig. 2D). Instead, another positive peak emerged at 684 nm in addition to the one at 679 nm. This is important because such an unpaired positive spectral component like the component at 684 nm (Fig. 2D) indicates energy transfer to a non-fluorescent acceptor, namely, quenched. Thus, the excitation energy on this fluorescence component at 684 nm in PSII-HL was quenched at pH 5.5 rather than transferred to the core complex. The very minor negative peaks probably reflect the energy transfer to CP43 or CP47 in the populations that do not bind LHCSR3 (Fig. 2D). It should be noted that the fluorescence amplitude of a quenching component in this fast lifetime component could be larger than its relative population because the quenching component decays faster so it strongly emits fluorescence with the fast lifetime component.
The quenching component (at 684 nm) had three noteworthy features. First, the excitation energy transfer to this quenching component was not detected in the FDAS analysis indicating that this energy transfer was faster than our system could measure (full width at half-maximum (FWHM) of instrumental response function (IRF) ∼ 75 ps). Second, the quenching occurred specifically within LHCs as revealed by its appearing in the first FDAS only, which reflects the relaxation of LHCs excitation. Third, the peak and decay time scale of the quenching component are analogous to previously reported features of LHCSR3, whose features are a fluorescence peak at 685 nm at 77 K and a pH-dependent quenching capability with a tens of picosecond time constant (21, 22, 34). In addition, LHCSR3 is presumably capable of efficient excitation energy transfer similar to that of the other LHCIIs because of their structural similarities (24). Therefore, the excitation energy transfer between LHCII and LHCSR3 could be as fast as it is between LHCIIs. Moreover, the npq4 mutant lacking LHCSR3 (37) did not show the quenching component even under high-light conditions (Fig. 3). Thus, we concluded that the quenching component is most likely due to LHCSR3 and not caused by other high light-induced acclimations such as pigment changes. This result indicates that the LHCSR3 dissipates the excitation energy of LHCs within a PSII supercomplex.
Figure 3.
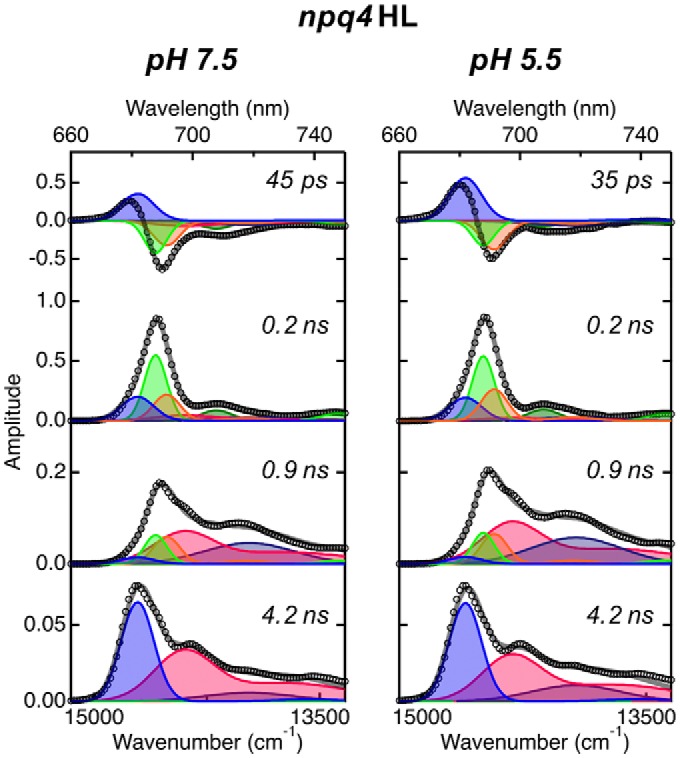
Fluorescence decay–associated spectra of PSII supercomlexes isolated from npq4 mutant grown under high-light conditions at 77 K. Time-resolved fluorescence profiles were obtained at each wavelength by excitation at 408 nm and analyzed to generate FDAS using the same procedure as described in the legend to Fig. 2. Spectral components of global analysis were shown as blue (680 nm), green (685 nm), orange (689 nm), red (694 nm), dark green (705 nm), and navy (715 nm). Open circles and gray lines represent FDAS spectra and fitting curve, respectively.
The shift of energy delivery toward CP47 in an acidic environment or by the LHCSR3 binding as described above was consistently shown in the second FDAS. The ratio of positive amplitudes at 689 to 684 nm in the second FDAS (0.2 ns) was larger in PSII-LL at pH 5.5 and PSII-HL at pH 7.5 than in PSII-LL at pH 7.5 (Fig. 2). Because the second FDAS corresponds to the excitation energy trapped by the special pair in the reaction center or the lowest-energy Chl(s) in CP47, the larger amplitudes at 689 nm in PSII-LL at pH 5.5 and in PSII-HL at pH 7.5 indicate that under these conditions, more excitation energy was populated in CP47 and trapped by the special pair in the reaction center or the lowest-energy Chl(s) in CP47. The long-lived excitation energy within the core complex, which was reflected in the third FDAS (0.9 ns), consistently behaved with the idea described above.
In the fourth FDAS (4.7 ns), we observed two major fluorescence peaks at 679 and 694 nm in all samples (Fig. 2). Because the fourth component reflects long-lived excitation energy within the entire PSII supercomplex after energy transfer, this finding indicates that the untrapped excitation energy was primarily populated on either LHCII or CP47 in all samples. Furthermore, at 684 nm, the PSII-HL spectra show a larger contribution than the PSII-LL spectra, suggesting HL affected the population of untrapped energy on the supercomplexes.
In the second and third lifetime components of PSII isolated from high light-grown npq4 (Fig. 3), new peaks (705 and 715 nm) appeared that were not observed in PSII-LL and PSII-HL (wild-type). Because these are negligible in 4.2-ns lifetime components, therefore, we can exclude the possibility of PSI contamination and assume that peaks are attributable to differences of LHCII-binding patterns, induced by the lack of LHCSR3 under high-light conditions, as observed in the BBY (Berthold, Babcock, and Yocum (73)) particle and LHCII oligomer of Arabidopsis thaliana (50).
Femtosecond up-conversion fluorescence kinetics
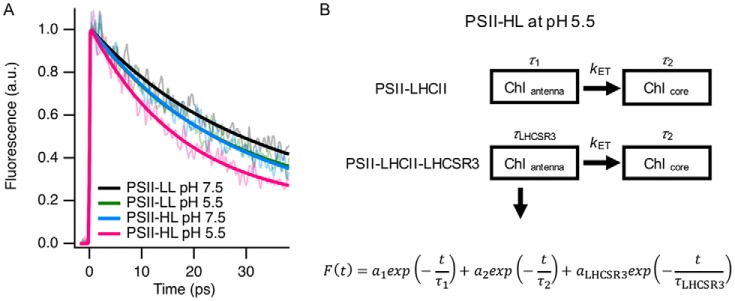
The two fluorescence decay components at 679 and 684 nm in the first FDAS in PSII-HL at pH 5.5 (Fig. 2D) presumably differed in origin and lifetime, but those were not technically resolved by our FDAS measuring system. To study the lifetime of the quenching component in the ultrafast time domain in more detail (<40 ps), we obtained fluorescence decay kinetics of the PSII supercomplexes by a femtosecond fluorescence up-conversion method, which has higher time resolution (∼230-fs IRF (FWHM), 0.20 ps/channel) than the FDAS measuring system (∼75-ps IRF (FWHM), 2.4 ps/channel). As expected, fluorescence decayed faster from PSII-HL at pH 5.5 than from the other supercomplexes (Fig. 4). For exponential fitting, we needed only two lifetime components for PSII-LL, but an additional component was necessary for PSII-HL at pH 5.5. We applied target fitting analysis based on the heterogeneity of the PSII supercomplexes (PSII-LHCII and PSII-LHCII-LHCSR3) in PSII-HL (Fig. 4). The fitting showed the LHCSR3-dependent lifetime components (τLHCSR3) are 18.2 ± 1.9 ps at pH 7.5 and 9.2 ± 2.2 ps at pH 5.5. These lifetime components were faster than the linked fast lifetime components (τ1), such as 24.9 ± 0.3 ps at pH 7.5 and 20.9 ± 0.2 ps at pH 5.5 (Table 1). Upon protonation of LHCSR3, the LHCSR3-dependent lifetime (τLHCSR3) component was shifted from 18.2 ps (at pH 7.5) to 9.2 ps (at pH 5.5) in PSII-HL. Therefore, we estimated the decay time constant for the LHCSR3- and pH-dependent quenching process to be 18.6 ps (1/τ = 1/(9.2 ps) − 1/(18.2 ps)). The relative amplitude ratio of the LHCSR3-dependent component (aLHCSR3) in the fast components here (a1 + aLHCSR3), 0.27 (Table 1), which reflects the relative contribution of the fluorescence emission from the PSII—LHCII–LHCSR3 supercomplex to that from all the PSII–LHCII supercomplex in fast time domain, is indeed comparable with the amplitude ratio of the 684-nm component in the two spectral components in the first FDAS at 679 and 684 nm (Fig. 2D). Although this ratio is larger than the ratio of LHCSR3 to CP26 (0.14 as shown in Ref. 37), it can be accounted for by the fact that the relative amplitude of the LHCSR3-dependent component is larger in the first FDAS of PSII-HL at pH 5.5.
Figure 4.
Up-conversion fluorescence decay curves of PSII supercomplexes at room temperature. A, fluorescence decay curves were obtained with an up-conversion system by excitation at 425 nm. Each fluorescence decay curve is an average of 16∼24 measurements. Fresh samples were used for every four measurements. Fluorescence decay curves were normalized at the peak of fitting line. B, the decay fitting was carried out with a heterogeneous-PSII fitting model considering heterogeneous populations of PSII supercomplexes in PSII-HL. aLHCSR3 represents the amplitude of the LHCSR3-related component in PSII-HL. Details are as described under “Experimental procedures.”
Table 1.
Lifetime components of up-conversion fluorescence decay curves from PSII-LL and PSII-HL at either pH 7.5 and 5.5
| pH | a1 | τ1a | a2 | τ2b | aLHCSR3 | τLHCSR3c | |
|---|---|---|---|---|---|---|---|
| % | ps | % | ns | % | ps | ||
| PSII-LL | 7.5 | 70 | 24.9 ± 0.3 | 30 | 0.2 | ||
| PSII-LL | 5.5 | 73 | 20.9 ± 0.2 | 27 | 0.2 | ||
| PSII-HL | 7.5 | 56 | 24.9 ± 0.3 | 23 | 0.2 | 20.7 ± 3.6 | 18.2 ± 1.9 |
| PSII-HL | 5.5 | 61 | 20.9 ± 0.2 | 17 | 0.2 | 22.3 ± 3.9 | 9.2 ± 2.2 |
a Linked based on pH.
b Fixed to 0.2 ns.
c Linked based on the existence of LHCSR3.
A minor degree of pH-dependent but LHCSR3-independent quenching was observed as a difference between the decay time constants for PSII-LL at pH 7.5 (24.9 ps) and 5.5 (20.9 ps), namely, 130.1 ps. Together, we identified LHCSR3-dependent qE quenching within the PSII—LHCII–LHCSR3 supercomplex, which was a different kinetic component from the pH-dependent quenching within the PSII–LHCII supercomplex.
In addition, it is important to consider the relationship between the antenna size and the fluorescence lifetime. The fluorescence lifetime is normally extended when antenna size increases (51), and the antenna size of PSII—LHCII–LHCSR3 was slightly larger than that of PSII–LHCII due to the addition of LHCSR3. However, the fluorescence lifetime of the PSII–LHCII–LHCSR3 supercomplex was shorter than that of the PSII–LHCII supercomplex, even at pH 7.5 (Fig. 4 and Table 1), suggesting LHCSR3 acting as not just an additional antenna molecule.
However, the antenna sizes of the supercomplexes were comparable based on the similar Chl a/b ratio of the samples (PSII-LL, 2.25, and PSII-HL, 2.27). The slight difference (0.02) in the Chl a/b ratio is most likely due to the high Chl a/b ratios of LHCSR3 (5.1 in Ref. 21, 5.5 in Ref. 22, and 8.86 in Ref. 34) compared with that of LHCII (1.27 ± 0.03). Thus, we conclude that the binding of LHCSR3 to the PSII supercomplex without protonation also decreases the fluorescence lifetime, which is consistent with previous reports, where LHCSR3 shows a shorter fluorescence lifetime than other LHCs at neutral pH (21, 33, 34).
The fate of excitation energy from LHCs
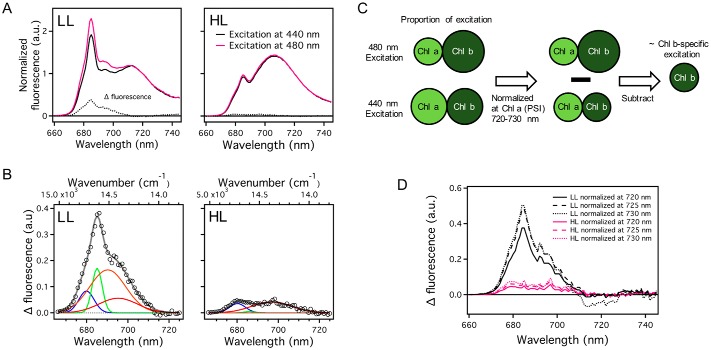
The outer antenna LHCs contain Chl b, whereas the core complexes do not (52). We took advantage of this feature to further investigate the fate of the energy harvested by the outer antenna when the cells show qE quenching under HL conditions. Because Chl b has a distinct absorption peak at 480 nm, the Chl b-specific fluorescence emission spectra can be determined by subtracting fluorescence emission spectra excited at 440 nm, which excites both Chl a and b, from those excited at 480 nm, after normalization in the 720–730 nm range. These wavelengths are considered to be only slightly influenced by PSII fluorescence and PSI fluorescence is dominant (Fig. 5). The obtained difference spectra were markedly different between LL and HL cells, as manifested by Gaussian peak decomposition at 679 (LHCII), 684 (CP43), 689 (CP47), and 689 (lowest-energy Chl(s) in CP47) (Fig. 5B). Interestingly, the peak corresponding to CP43 was markedly reduced in HL cells, compared with the decrease of the peak corresponding to LHCII and CP47. It indicates that the energy transfer from LHCII to CP43 was specifically suppressed in HL cells. This result is consistent in the difference spectra derived by different normalization wavelengths at 720, 725, and 730 nm (Fig. 5D).
Figure 5.
Steady-state fluorescence emission spectra at 77 K of low light- and high light-grown C. reinhardtti. A, the spectra were normalized at 720 nm, which is believed to not contribute to PSII-related fluorescence. The difference spectra were derived by subtraction of the spectrum obtained by 440 nm excitation from the spectrum obtained by 480 nm excitation. B, the spectra were globally fitted with four Gaussian functions. The amplitudes of the Gaussian functions are independent of other spectra but their center and width are linked in the two spectra. Spectral components of global analysis were shown as blue (679 nm), green (685 nm), orange (689 nm), and red (694 nm). Open circles and gray lines represent FDAS spectra and fitting curve, respectively. LL, low light-grown cells; HL, high light-grown cells. C, an illustration of the analysis procedure to investigate fluorescence spectrum by Chl b-specific excitation. 480-nm light preferentially excites Chl b, whereas 440-nm light equally excites Chl a and Chl b. To investigate the fluorescence spectrum by Chl b-specific excitation, we subtract the fluorescence spectrum obtained by 440 nm light from that obtained by the 480-nm light after normalization at a Chl a-specific fluorescence peak such as a PSI fluorescence peak. D, three types of difference-spectra with normalization wavelengths at 720, 725, and 730 nm were derived by subtraction of the spectrum obtained by 440 nm excitation from the spectrum obtained by 480 nm excitation in A.
Discussion
In contrast to PsbS, LHCSR3 binds pigments and has quenching capability. LHCSR3-dependent quenching is induced by protonation and protein–protein interaction (21, 22, 33, 34), and the PSII supercomplex can be transformed into an energy-dissipative state upon binding to LHCSR3 under acidic conditions (37). In this study, using the green alga C. reinhardtii, we studied how LHCSR3 affects the PSII supercomplex when it enters the energy-dissipative state. Our results (Figs. 2 and 3) clearly indicate that LHCSR3 dissipates excitation energy of LHCs within the PSII supercomplex. Although there occur multiple effects under high-light conditions, in this study we focus on the effects due to the presence or absence of LHCSR3.
A LHCSR3-dependent quenching component appeared at an almost identical spectral position (684 nm) as reported in in vitro reconstitution studies (21), indicating that LHCSR3 is likely the quencher itself. On the other hand, the decay time constant (18.6 ps) of the quenching component, determined by the fs up-conversion fluorescence system, was faster than that previously observed in rLHCSR3 (21, 22, 33, 34) and in cells (36). We speculate that these differences were caused by the different conditions between isolated LHCSR3 and associated LHCSR3 with the PSII supercomplex. Previous fluorescence lifetime measurements on isolated PSII–LHCII–LHCSR3 supercomplexes (37) also did not reveal the short lifetime component (<50 ps), but a lower fluorescence amplitude at the initial time (<50 ps) after excitation was observed in the PSII-HL supercomplex at pH 5.5, indicating the presence of fast quenching beyond the temporal resolution. In addition, the fast decay time constant determined in this study is indeed more reasonable because the quenching should compete with the intrinsic energy transfer to the core complex (∼25 ps).
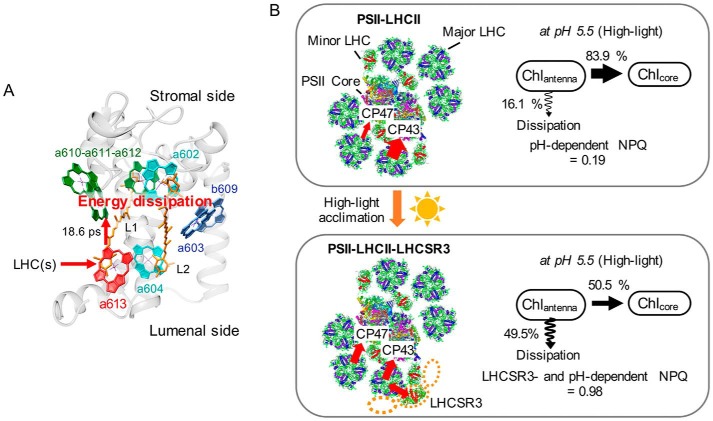
Intriguingly, the decay time constant of the quenching component was analogous to the rise-time constant (∼14 ± 3 ps) of the lutein (Lut) radical cation determined by transient absorption spectroscopy from rLHCSR3 (21). The Lut radical cation may be generated by electron transfer within a Chl–Lut heterodimer, resulting in charge transfer quenching (53–55). The rise-time constant of the Lut radical cation has been determined by simulations that rely on the time constant of excitation energy transfer from bulk Chls to a Chl–Lut heterodimer (56). In the case of LHCSR3, Chl a603, b609, and a Lut in the L2 site may constitute a Chl–Lut heterodimer (21, 34). Interestingly, the decay time constant of the quenching component is also comparable with the time constant (∼21 ps) of energy transfer from Chl a613 to other Chls in rLHCSR3 (22). Moreover Chl a613 reportedly has the lowest energy state in rLHCSR3, which is remarkable because the lowest energy state for most LHC proteins is on the Chl a610-a611-a612 trimer (57). In this regard, we suggest that this Chl a613 is a possible intermediate in the efficient excitation energy transfer from LHCs to LHCSR3 (Fig. 6A), which presumably have an advantage in funneling excitation energy for LHCs.
Figure 6.
A proposed model of LHCSR3-dependent NPQ in PSII—LHCII–LHCSR3 supercomplexes of C. reinhardtii. A, a proposed molecular model of LHCSR3-dependent quenching within the PSII—LHCII–LHCSR3 supercomplex. The structural model of LHCSR3 is a sequence homology-based structural model of LHCSR3 generated by SWISS-MODEL and the orientations of chlorophylls are from a crystal structure of LHCII (70). B, LHCSR3 is expressed and associated with the PSII supercomplex under high-light conditions. The circles of orange dots represent the presumable binding site of LHCSR3. The binding of LHCSR3 to the PSII supercomplex inhibits the excitation energy transfer to CP43. At acidic pH, LHCSR3-dependent NPQ is activated, and it quenches excitation energy of LHCs. NPQ values of PSII–LHCII and PSII—LHCII–LHCSR3 were calculated from the difference between pH 7.5 and 5.5 conditions. The illustration of PSII supercomplex is based on data from Refs. 71 and 72.
Furthermore, our FDAS and in vivo fluorescence study indicated that LHCSR3 interacts with LHCs that are in the vicinity of CP43 (Figs. 2 and 5), which is consistent with one of the averaged single particle images recently reported showing that LHCSR3 binds to CP26 and the LHCII trimer near CP43 (58).
To reveal the significance of the LHCSR3-dependent NPQ process in vivo, we derived NPQ parameters of the isolated PSII supercomplexes by computing changes in the proportion of excitation energy transferred to the core complexes upon protonation (Fig. 6B and supplemental Fig. S2). In the case of the PSII–LHCII supercomplex without LHCSR3, the proportion of excitation energy to the core complexes decreased at pH 5.5 to 84% of the proportion at pH 7.5. We then estimated the NPQ value to be 0.19, which is about 48% of the NPQ value (about 0.4 under 750 μmol of photons m−2 s−1 of actinic light) in the npq4 mutant (37). For the PSII—LHCII–LHCSR3 supercomplex, the NPQ value was 0.98, which is about 82% of the NPQ value (about 1.2 under 750 μmol of photons m−2 s−1 of actinic light) of HL grown cells. In conclusion, we suggest that the binding of LHCSR3 to PSII supercomplexes can contribute up to 82% of the entire NPQ in cells. The remaining 18% of NPQ is pH-dependent and LHCSR3-independent, which may be due to the interactions between LHCSR3 and LHCII and/or LHCSR1 and LHCII, as reported recently (59).
An interesting feature of LHCSR3-dependent photoprotection is that LHCSR3 binding inhibits excitation energy transfer to CP43. CP43 is known to deliver excitation energy to the special pair more efficiently than CP47, which has a potential to form a quenching state (60, 61). Accordingly, under low-light conditions, dominant excitation energy transfer to CP43 may be more suitable for increasing light-harvesting efficiency, which is what we observed by FDAS analysis (Fig. 2). On the other hand, under high-light conditions, inhibition of excitation energy transfer to CP43 by binding of LHCSR3 could be a safety valve to prevent overload of the reaction center by excess excitation energy. Furthermore, such a safety valve for CP43 might also inhibit the generation of harmful reactive oxygen species following from an extended lifetime of the excited Chls within CP43 when CP43 is dissociated from the PSII supercomplex, which inevitably occurs when D1 protein is degraded under high-light conditions and the entire complex is engaged in the repair cycle (62).
Thus, we propose the following model for the LHCSR3-dependent NPQ in C. reinhardtii (Fig. 6). Under high light that includes blue light, LHCSR3 protein is expressed and bound to LHCs near CP43, S-trimer (strongly bound) of LHCII, or CP26 within the PSII supercomplex. That binding alters the excitation energy pathways so that energy transfer to CP43 is inhibited. When LHCSR3 is protonated by lumenal acidification, it quenches the excitation energy harvested by LHCs within the PSII supercomplex. For efficient energy funneling of LHCSR3, the lowest energy state Chl (Chl a613) in LHCSR3 mediates the excitation energy transfer from LHCs to LHCSR3. We assume the transfer is very fast because the fluorescence decay observed in this study (18.6 ps), which could reflect the overall energy transfer from LHCs to Chl–Lut, was not much different from the rise of the Lut radical cation measured by the transient absorption (14 ± 3 ps) (21). The molecular mechanism of the quenching in LHCSR3 has been addressed and it was suggested that multiple quenching mechanisms including charge-transfer quenching occurred (21). To further clarify the molecular mechanism, ultrafast transient absorption and/or fluorescence spectroscopy analysis accompanied by structural studies is required. Also, further structural data on the PSII—LHCII–LHCSR3 supercomplex as determined by crystallography or high-resolution cryoelectron microscopy single particle analysis will clarify the precise binding site of LHCSR3.
Experimental procedures
C. reinhardtii strains and growth conditions
LL grown and HL grown C. reinhardtii cells were prepared from the identical cell 137c line (wild-type) and npq4 (7). The LL and HL cells were precultured in high-salt minimal medium (63) under 50 μmol of photons m−2 s−1 light with 5% CO2 bubbling and stirring and subsequently cultured, respectively, under 20 (LL cells) or 500 (HL cells) μmol of photons m−2 s−1 of white light, with air bubbling for 15 h at 25 °C, as described in Ref. 37.
Isolation of photosystem II supercomplexes and immunoblot analysis
Thylakoid membranes were isolated from C. reinhardtii cells and solubilized with dodecyl α-maltoside (Affymetrix) in 25 mm MES buffer (pH 6.5), as described in Ref. 64. PSII supercomplexes were purified from the solubilized thylakoid membranes by sucrose density gradient centrifugation, as described in Refs. 37 and 64. Betaine (1 m) was added to the buffer for stabilizing the PSII supercomplexes at room temperature (supplemental Fig. S3), especially for the up-conversion fluorescence measurement. Immunoblot analyses were conducted with the antibodies against LHCSR3 (kind gift from Dr. Michael Hippler, University of Münster) or D1 (anti-PsbA, Agrisera, Sweden) polypeptide as described previously (37).
Steady-state absorption and fluorescence measurements
Steady-state absorption spectra were measured by a spectrometer equipped with an integrating sphere (JASCO V-650/ISVC-747) at 77 K, as described in Ref. 65. Steady-state emission spectra were measured by a FluoroMax spectrofluorometer (HORIBA Jobin-Yvon) at 77 K. Samples diluted to 1 μg of Chl/ml were excited at 440 nm (bandwidth = 2 nm) and emission was monitored between 660 and 760 nm with a 2-nm bandwidth.
Time-resolved fluorescence measurements and analysis
Time-resolved fluorescence spectra were obtained from 660 to 750 nm at 1-nm intervals using a 408-nm diode pulse laser and time-correlated single photon counting (TCSPC) system at 77 K (2.4 ps/channel) (40, 65, 66). Fluorescence kinetics were analyzed by a convolution calculation with an instrumental response function of 75 ps (FWHM). FDAS were constructed by global analysis of fluorescence decay curves and global decomposition to four spectral components having vibrational bands, as described in Ref. 38 and 40).
Fluorescence rise and decay curves at the femtosecond domain were measured with a fluorescence up-conversion system at 293 K (0.20 ps/channel) (67, 68). The second harmonic (425 nm) of a Ti:Sapphire laser (Tsunami, Spectra-Physics, Mountain View, CA) was used as the excitation source, and its fundamental (850 nm) served as the gate pulse. The excitation pulse intensity was set to lower than 6.2 × 1010 photons/(pulse/cm2), which was low enough to ignore the annihilation effect (69). All pigments were simultaneously excited, and fluorescence rise and decay curves were obtained at 680 nm (bandwidth = 10 nm). Fluorescence kinetics were analyzed by a convolution calculation with an instrumental response function of 0.23 ps (FWHM).
The fluorescence decay curves were globally fitted with a heterogeneous-PSII fitting model because the PSII-HL fraction contains both PSII—LHCII–LHCSR3 and PSII–LHCII at a ratio of ∼3:7 (37), which probably results in a heterogeneous lifetime on the fast scale. Deconvolution of the exponentials and the Gaussian function (FWHM ∼ 230 fs) were carried out with Equation 1,
| (Eq. 1) |
where, F(t) is the fluorescence decay curve, α1 is the amplitude and τ1 is the lifetime of a decay component mainly associated with the first lifetime component of the FDAS spectra in Fig. 1. aLHCSR3 and τLHCSR3 are the amplitude and the lifetime of the LHCSR3-related component, respectively. a2 and τ2 are, respectively, the amplitude and the lifetime of long decay components related with the second lifetime components of FDAS spectra in Fig. 2. τ1 of PSII-LL at pH 7.5 and PSII-LL at pH 5.5 were, respectively, linked to τ1 of PSII-HL at pH 7.5 and PSII-HL at pH 5.5, because some proportion of PSII in PSII-HL is presumably the same as PSII-LL based on the ratio of LHCSR3 to CP26 (LHCSR3/CP26 = 0.14) and it is also supported by the identical first lifetime components between PSII-LL at pH 7.5 and PSII-HL at pH 7.5 in FDAS in Fig. 2. The ratio of aLHCSR3 to a1 (aLHCSR3/a1) was linked between PSII-HL at pH 7.5 and PSII-HL at pH 5.5 because the ratio of LHCSR3 is identical, whereas τLHCSR3 was not linked. τ2 was fixed to 200 ps based on the FDAS results in Fig. 2. It also has been confirmed that τ2 has negligible influence on short lifetime components (supplemental Table S1).
Author contributions
J. M., R. T., and E. K. conceived the work and designed the experiments. E. K., R. T., and S. A. performed the experiments. E. K., S. A., and M. Y. analyzed the spectroscopic data. All authors contributed to writing and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Profs. Shigeichi Kumazaki (Kyoto University), Yutaka Shibata (Tohoku University), and Shunichi Takahashi (National Institute for Basic Biology) for helpful discussions.
This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant-in-Aids JP16H06553 (to J. M., R. T., S. A., and E. K.), JP26251033 (to J. M. and R. T.), and JP16F16087 (to E. K.), and a International Research Fellowship of the JSPS (to E. K.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Table S1 and Figs. S1–S3.
- NPQ
- non-photochemical quenching
- Chl
- chlorophyll
- qE
- energy-dependent quenching
- LHCSR
- light-harvesting complex stress-related
- PSII
- photosystem II
- LHC
- light-harvesting complex
- rLHCSR3
- reconstituted LHCSR3
- FDAS
- fluorescence decay–associated spectra
- SDG
- sucrose density gradient
- HL
- high light
- LL
- low light
- FWHM
- full width at half-maximum
- IRF
- instrumental response function
- Lut
- lutein.
References
- 1. Li Z., Wakao S., Fischer B. B., and Niyogi K. K. (2009) Sensing and responding to excess light. Annu. Rev. Plant Biol. 60, 239–260 [DOI] [PubMed] [Google Scholar]
- 2. Müller P., Li X. P., and Niyogi K. K. (2001) Non-photochemical quenching: a response to excess light energy. Plant Physiol. 125, 1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Horton P. (2012) Optimization of light harvesting and photoprotection: molecular mechanisms and physiological consequences. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 3455–3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Erickson E., Wakao S., and Niyogi K. K. (2015) Light stress and photoprotection in Chlamydomonas reinhardtii. Plant J. 82, 449–465 [DOI] [PubMed] [Google Scholar]
- 5. Minagawa J., and Tokutsu R. (2015) Dynamic regulation of photosynthesis in Chlamydomonas reinhardtii. Plant J. 82, 413–428 [DOI] [PubMed] [Google Scholar]
- 6. Li X. P., Björkman O., Shih C., Grossman A. R., Rosenquist M., Jansson S., and Niyogi K. K. (2000) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403, 391–395 [DOI] [PubMed] [Google Scholar]
- 7. Peers G., Truong T. B., Ostendorf E., Busch A., Elrad D., Grossman A. R., Hippler M., and Niyogi K. K. (2009) An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462, 518–521 [DOI] [PubMed] [Google Scholar]
- 8. Niyogi K. K., and Truong T. B. (2013) Evolution of flexible non-photochemical quenching mechanisms that regulate light harvesting in oxygenic photosynthesis. Curr. Opin. Plant Biol. 16, 307–314 [DOI] [PubMed] [Google Scholar]
- 9. Li X. P., Gilmore A. M., and Niyogi K. K. (2002) Molecular and global time-resolved analysis of a psbS gene dosage effect on pH- and xanthophyll cycle-dependent nonphotochemical quenching in photosystem II. J. Biol. Chem. 277, 33590–33597 [DOI] [PubMed] [Google Scholar]
- 10. Li X. P., Müller-Moule P., Gilmore A. M., and Niyogi K. K. (2002) PsbS-dependent enhancement of feedback de-excitation protects photosystem II from photoinhibition. Proc. Natl. Acad. Sci. U.S.A. 99, 15222–15227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li X. P., Gilmore A. M., Caffarri S., Bassi R., Golan T., Kramer D., and Niyogi K. K. (2004) Regulation of photosynthetic light harvesting involves intrathylakoid lumen pH sensing by the PsbS protein. J. Biol. Chem. 279, 22866–22874 [DOI] [PubMed] [Google Scholar]
- 12. Niyogi K. K., Li X. P., Rosenberg V., and Jung H. S. (2005) Is PsbS the site of non-photochemical quenching in photosynthesis? J. Exp. Bot. 56, 375–382 [DOI] [PubMed] [Google Scholar]
- 13. Fan M., Li M., Liu Z., Cao P., Pan X., Zhang H., Zhao X., Zhang J., and Chang W. (2015) Crystal structures of the PsbS protein essential for photoprotection in plants. Nat. Struct. Mol. Biol. 22, 729–735 [DOI] [PubMed] [Google Scholar]
- 14. Gerotto C., Franchin C., Arrigoni G., and Morosinotto T. (2015) In vivo identification of photosystem II light harvesting complexes interacting with photosystem II subunit S. Plant Physiol. 168, 1747–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Betterle N., Ballottari M., Zorzan S., de Bianchi S., Cazzaniga S., Dall'osto L., Morosinotto T., and Bassi R. (2009) Light-induced dissociation of an antenna hetero-oligomer is needed for non-photochemical quenching induction. J. Biol. Chem. 284, 15255–15266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goral T. K., Johnson M. P., Duffy C. D., Brain A. P., Ruban A. V., and Mullineaux C. W. (2012) Light-harvesting antenna composition controls the macrostructure and dynamics of thylakoid membranes in Arabidopsis. Plant J. 69, 289–301 [DOI] [PubMed] [Google Scholar]
- 17. Kiss A. Z., Ruban A. V., and Horton P. (2008) The PsbS protein controls the organization of the photosystem II antenna in higher plant thylakoid membranes. J. Biol. Chem. 283, 3972–3978 [DOI] [PubMed] [Google Scholar]
- 18. Ruban A. V. (2016) Nonphotochemical chlorophyll fluorescence quenching: mechanism and effectiveness in protecting plants from photodamage. Plant Physiol. 170, 1903–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnson M. P., and Ruban A. V. (2011) Restoration of rapidly reversible photoprotective energy dissipation in the absence of PsbS protein by enhanced ΔpH. J. Biol. Chem. 286, 19973–19981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Correa-Galvis V., Poschmann G., Melzer M., Stühler K., and Jahns P. (2016) PsbS interactions involved in the activation of energy dissipation in Arabidopsis. Nat. Plants 2, 15225. [DOI] [PubMed] [Google Scholar]
- 21. Bonente G., Ballottari M., Truong T. B., Morosinotto T., Ahn T. K., Fleming G. R., Niyogi K. K., and Bassi R. (2011) Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLos Biol. 9, e1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liguori N., Novoderezhkin V., Roy L. M., van Grondelle R., and Croce R. (2016) Excitation dynamics and structural implication of the stress-related complex LHCSR3 from the green alga Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1857, 1514–1523 [DOI] [PubMed] [Google Scholar]
- 23. Petroutsos D., Tokutsu R., Maruyama S., Flori S., Greiner A., Magneschi L., Cusant L., Kottke T., Mittag M., Hegemann P., Finazzi G., and Minagawa J. (2016) A blue-light photoreceptor mediates the feedback regulation of photosynthesis. Nature 537, 563–566 [DOI] [PubMed] [Google Scholar]
- 24. Miura K., Yamano T., Yoshioka S., Kohinata T., Inoue Y., Taniguchi F., Asamizu E., Nakamura Y., Tabata S., Yamato K. T., Ohyama K., and Fukuzawa H. (2004) Expression profiling-based identification of CO2-responsive genes regulated by CCM1 controlling a carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol. 135, 1595–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Petroutsos D., Busch A., Janssen I., Trompelt K., Bergner S. V., Weinl S., Holtkamp M., Karst U., Kudla J., and Hippler M. (2011) The chloroplast calcium sensor CAS is required for photoacclimation in Chlamydomonas reinhardtii. The Plant Cell 23, 2950–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang L., Yamano T., Takane S., Niikawa Y., Toyokawa C., Ozawa S. I., Tokutsu R., Takahashi Y., Minagawa J., Kanesaki Y., Yoshikawa H., and Fukuzawa H. (2016) Chloroplast-mediated regulation of CO2-concentrating mechanism by Ca2+-binding protein CAS in the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 113, 12586–12591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gagné G., and Guertin M. (1992) The early genetic response to light in the green unicellular alga Chlamydomonas eugametos grown under light/dark cycles involves genes that represent direct responses to light and photosynthesis. Plant Mol. Biol. 18, 429–445 [DOI] [PubMed] [Google Scholar]
- 28. Richard C., Ouellet H., and Guertin M. (2000) Characterization of the LI818 polypeptide from the green unicellular alga Chlamydomonas reinhardtii. Plant Mol. Biol. 42, 303–316 [DOI] [PubMed] [Google Scholar]
- 29. Naumann B., Busch A., Allmer J., Ostendorf E., Zeller M., Kirchhoff H., and Hippler M. (2007) Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 7, 3964–3979 [DOI] [PubMed] [Google Scholar]
- 30. Maruyama S., Tokutsu R., and Minagawa J. (2014) Transcriptional regulation of the stress-responsive light harvesting complex genes in Chlamydomonas reinhardtii. Plant Cell Physiol. 55, 1304–1310 [DOI] [PubMed] [Google Scholar]
- 31. Allorent G., Lefebvre-Legendre L., Chappuis R., Kuntz M., Truong T. B., Niyogi K. K., Ulm R., and Goldschmidt-Clermont M. (2016) UV-B photoreceptor-mediated protection of the photosynthetic machinery in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 113, 14864–14869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tilbrook K., Dubois M., Crocco C. D., Yin R., Chappuis R., Allorent G., Schmid-Siegert E., Goldschmidt-Clermont M., and Ulm R. (2016) UV-B perception and acclimation in Chlamydomonas reinhardtii. The Plant Cell 28, 966–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liguori N., Roy L. M., Opacic M., Durand G., and Croce R. (2013) Regulation of light harvesting in the green alga Chlamydomonas reinhardtii: the C-terminus of LHCSR is the knob of a dimmer switch. J. Am. Chem. Soc. 135, 18339–18342 [DOI] [PubMed] [Google Scholar]
- 34. Ballottari M., Truong T. B., De Re E., Erickson E., Stella G. R., Fleming G. R., Bassi R., and Niyogi K. K. (2016) Identification of pH-sensing sites in the light harvesting complex stress-related 3 protein essential for triggering non-photochemical quenching in Chlamydomonas reinhardtii. J. Biol. Chem. 291, 7334–7346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pinnola A., Staleva-Musto H., Capaldi S., Ballottari M., Bassi R., and Polívka T. (2016) Electron transfer between carotenoid and chlorophyll contributes to quenching in the LHCSRI. protein from Physcomitrella patens. Biochim. Biophys. Acta 1857, 1870–1878 [DOI] [PubMed] [Google Scholar]
- 36. Amarnath K., Zaks J., Park S. D., Niyogi K. K., and Fleming G. R. (2012) Fluorescence lifetime snapshots reveal two rapidly reversible mechanisms of photoprotection in live cells of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 109, 8405–8410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tokutsu R., and Minagawa J. (2013) Energy-dissipative supercomplex of photosystem II associated with LHCSR3 in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 110, 10016–10021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yokono M., Nagao R., Tomo T., and Akimoto S. (2015) Regulation of excitation energy transfer in diatom PSII dimer: how does it change the destination of excitation energy? Biochim. Biophys. Acta 1847, 1274–1282 [DOI] [PubMed] [Google Scholar]
- 39. Nagao R., Yokono M., Tomo T., and Akimoto S. (2014) Control mechanism of excitation energy transfer in a complex consisting of photosystem II and fucoxanthin chlorophyll a/c-binding protein. J. Phys. Chem. Lett. 5, 2983–2987 [DOI] [PubMed] [Google Scholar]
- 40. Yokono M., Tomo T., Nagao R., Ito H., Tanaka A., and Akimoto S. (2012) Alterations in photosynthetic pigments and amino acid composition of D1 protein change energy distribution in photosystem II. Biochim. Biophys. Acta 1817, 754–759 [DOI] [PubMed] [Google Scholar]
- 41. Tibiletti T., Auroy P., Peltier G., and Caffarri S. (2016) Chlamydomonas reinhardtii PsbS protein is functional and accumulates rapidly and transiently under high light. Plant Physiol. 171, 2717–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Correa-Galvis V., Redekop P., Guan K., Griess A., Truong T. B., Wakao S., Niyogi K. K., and Jahns P. (2016) Photosystem II Subunit PsbS Is Involved in the Induction of LHCSR protein-dependent energy dissipation in Chlamydomonas reinhardtii. J. Biol. Chem. 291, 17478–17487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van Dorssen R. J., Plijter J. J., Dekker J. P., Denouden A., Amesz J., and van Gorkom H. J. (1987) Spectroscopic properties of chloroplast grana membranes and of the core of Photosystem II. Biochim. Biophys. Acta 890, 134–143 [Google Scholar]
- 44. van Dorssen R. J., Breton J., Plijter J. J., Satoh K., van Gorkom H. J., and Amesz J. (1987) Spectroscopic properties of the reaction center and of the 47-kDa chlorophyll protein of Photosystem II. Biochim. Biophys. Acta 893, 267–274 [Google Scholar]
- 45. De Weerd F. L., Palacios M. A., Andrizhiyevskaya E. G., Dekker J. P., and Van Grondelle R. (2002) Identifying the lowest electronic states of the chlorophylls in the CP47 core antenna protein of photosystem II. Biochemistry 41, 15224–15233 [DOI] [PubMed] [Google Scholar]
- 46. Groot M. L., Frese R. N., de Weerd F. L., Bromek K., Pettersson A., Peterman E. J., van Stokkum I. H., van Grondelle R., and Dekker J. P. (1999) Spectroscopic properties of the CP43 core antenna protein of photosystem II. Biophys. J. 77, 3328–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Andrizhiyevskaya E. G., Chojnicka A., Bautista J. A., Diner B. A., van Grondelle R., and Dekker J. P. (2005) Origin of the F685 and F695 fluorescence in Photosystem II. Photosynth. Res. 84, 173–180 [DOI] [PubMed] [Google Scholar]
- 48. Komura M., Shibata Y., and Itoh S. (2006) A new fluorescence band F689 in photosystem II revealed by picosecond analysis at 4–77 K: function of two terminal energy sinks F689 and F695 in PSII. Biochim. Biophys. Acta 1757, 1657–1668 [DOI] [PubMed] [Google Scholar]
- 49. Mimuro M., Akimoto S., Tomo T., Yokono M., Miyashita H., and Tsuchiya T. (2007) Delayed fluorescence observed in the nanosecond time region at 77 K originates directly from the photosystem II reaction center. Biochim. Biophys. Acta 1767, 327–334 [DOI] [PubMed] [Google Scholar]
- 50. Kirchhoff H., Hinz H.-J., and Rösgen J. (2003) Aggregation and fluorescence quenching of chlorophyll a of the light-harvesting complex II from spinach in vitro. Biochim. Biophys. Acta 1606, 105–116 [DOI] [PubMed] [Google Scholar]
- 51. Caffarri S., Broess K., Croce R., and van Amerongen H. (2011) Excitation energy transfer and trapping in higher plant photosystem II complexes with different antenna sizes. Biophys. J. 100, 2094–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Caffarri S., Kouril R., Kereïche S., Boekema E. J., and Croce R. (2009) Functional architecture of higher plant photosystem II supercomplexes. EMBO J. 28, 3052–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Holt N. E., Zigmantas D., Valkunas L., Li X. P., Niyogi K. K., and Fleming G. R. (2005) Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science 307, 433–436 [DOI] [PubMed] [Google Scholar]
- 54. Ahn T. K., Avenson T. J., Ballottari M., Cheng Y. C., Niyogi K. K., Bassi R., and Fleming G. R. (2008) Architecture of a charge-transfer state regulating light harvesting in a plant antenna protein. Science 320, 794–797 [DOI] [PubMed] [Google Scholar]
- 55. Avenson T. J., Ahn T. K., Niyogi K. K., Ballottari M., Bassi R., and Fleming G. R. (2009) Lutein can act as a switchable charge transfer quencher in the CP26 light-harvesting complex. J. Biol. Chem. 284, 2830–2835 [DOI] [PubMed] [Google Scholar]
- 56. Cheng Y. C., Ahn T. K., Avenson T. J., Zigmantas D., Niyogi K. K., Ballottari M., Bassi R., and Fleming G. R. (2008) Kinetic modeling of charge-transfer quenching in the CP29 minor complex. J. Phys. Chem. B 112, 13418–13423 [DOI] [PubMed] [Google Scholar]
- 57. Novoderezhkin V., Marin A., and van Grondelle R. (2011) Intra- and inter-monomeric transfers in the light harvesting LHCII complex: the Redfield-Förster picture. Phys. Chem. Chem. Phys. 13, 17093–17103 [DOI] [PubMed] [Google Scholar]
- 58. Semchonok D. A., Sathish Yadav K. N., Xu P., Drop B., Croce R., and Boekema E. J. (2017) Interaction between the photoprotective protein LHCSR3 and C2S2 photosystem II supercomplex in Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1858, 379–385 [DOI] [PubMed] [Google Scholar]
- 59. Dinc E., Tian L., Roy L. M., Roth R., Goodenough U., and Croce R. (2016) LHCSR1 induces a fast and reversible pH-dependent fluorescence quenching in LHCII in Chlamydomonas reinhardtii cells. Proc. Natl. Acad. Sci. U.S.A. 113, 7673–7678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shibata Y., Nishi S., Kawakami K., Shen J. R., and Renger T. (2013) Photosystem II does not possess a simple excitation energy funnel: time-resolved fluorescence spectroscopy meets theory. J. Am. Chem. Soc. 135, 6903–6914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Raszewski G., and Renger T. (2008) Light harvesting in photosystem II core complexes is limited by the transfer to the trap: can the core complex turn into a photoprotective mode? J. Am. Chem. Soc. 130, 4431–4446 [DOI] [PubMed] [Google Scholar]
- 62. Järvi S., Suorsa M., and Aro E. M. (2015) Photosystem II repair in plant chloroplasts-regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim. Biophys. Acta 1847, 900–909 [DOI] [PubMed] [Google Scholar]
- 63. Sueoka N. (1960) Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 46, 83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tokutsu R., Kato N., Bui K. H., Ishikawa T., and Minagawa J. (2012) Revisiting the supramolecular organization of photosystem II in Chlamydomonas reinhardtii. J. Biol. Chem. 287, 31574–31581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hamada F., Yokono M., Hirose E., Murakami A., and Akimoto S. (2012) Excitation energy relaxation in a symbiotic cyanobacterium, Prochloron didemni, occurring in coral-reef ascidians, and in a free-living cyanobacterium, Prochlorothrix hollandica. Biochim. Biophys. Acta 1817, 1992–1997 [DOI] [PubMed] [Google Scholar]
- 66. Akimoto S., Yokono M., Hamada F., Teshigahara A., Aikawa S., and Kondo A. (2012) Adaptation of light-harvesting systems of Arthrospira platensis to light conditions, probed by time-resolved fluorescence spectroscopy. Biochim. Biophys. Acta 1817, 1483–1489 [DOI] [PubMed] [Google Scholar]
- 67. Akimoto S., Shinoda T., Chen M., Allakhverdiev S. I., and Tomo T. (2015) Energy transfer in the chlorophyll f-containing cyanobacterium, Halomicronema hongdechloris, analyzed by time-resolved fluorescence spectroscopies. Photosynth. Res. 125, 115–122 [DOI] [PubMed] [Google Scholar]
- 68. Hamada F., Murakami A., and Akimoto S. (2015) Comparative analysis of ultrafast excitation energy-transfer pathways in three strains of divinyl chlorophyll a/b-containing cyanobacterium, Prochlorococcus marinus. J. Phys. Chem. B 119, 15593–15600 [DOI] [PubMed] [Google Scholar]
- 69. Connelly J. P., Müller M. G., Hucke M., Gatzen G., Mullineaux C. W., Ruban A. V., Horton P., and Holzwarth A. R. (1997) Ultrafast spectroscopy of trimeric light-harvesting complex II from higher plants. J. Phys. Chem. B 101, 1902–1909 [Google Scholar]
- 70. Standfuss J., Terwisscha van Scheltinga A. C., Lamborghini M., and Kühlbrandt W. (2005) Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5-Å resolution. EMBO J. 24, 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wei X., Su X., Cao P., Liu X., Chang W., Li M., Zhang X., and Liu Z. (2016) Structure of spinach photosystem II-LHCII supercomplex at 3.2-Å resolution. Nature 534, 69–74 [DOI] [PubMed] [Google Scholar]
- 72. van Bezouwen L. S., Caffarri S., Kale R. S., Kouřil R., Thunnissen A. W. H., Oostergetel G. T., and Boekema E. J. (2017) Subunit and chlorophyll organization of the plant photosystem II supercomplex. Nat. Plants 3, 17080. [DOI] [PubMed] [Google Scholar]
- 73. Berthold D. A., Babcock G. T., and Yocum C. F. (1981) A highly resolved, oxygen-evolving photosystem II preparation from spinach thylakoid membranes: EPR and electron-transport properties. Febs. Lett. 134, 231–234 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.