Abstract
We report here an approach to redirecting somatic cell fate under chemically defined conditions without transcription factors. We start by converting mouse embryonic fibroblasts to epithelial-like cells with chemicals and growth factors. Subsequent cell fate mapping reveals a robust induction of SOX17 in the resulting epithelial-like cells that can be further reprogrammed to endodermal progenitor cells. Interestingly, these cells can self-renew in vitro and further differentiate into albumin-producing hepatocytes that can rescue mice from acute liver injury. Our results demonstrate a rational approach to convert mouse embryonic fibroblasts to hepatocytes and suggest that this mechanism-driven approach may be generalized for other cells.
Keywords: cell signaling, chemical biology, hepatocyte, reprogramming, small molecule, MET, endoderm progenitor cells
Introduction
Cell fate is determined during development to ensure proper formation of tissues and organs in an individual. Amid intrinsic and extrinsic signals, transcription factors play a primary role in controlling cell fate by specifying the expression of genes unique to a particular cell type in vivo and in vitro. This remarkable insight led to the development of iPSC3 reprogramming almost 10 years ago (i.e. four transcription factors, Oct4, Sox2, Myc, and Klf4, capable of converting embryonic fibroblasts back to the pluripotent state found only in cells derived from blastocysts) (1). In addition, various transcription factors have been discovered to convert fibroblasts into neurons (2–5), hepatocytes (6–8), cardiomyocytes (9), and hematopoietic cells (10).
Despite the ease and remarkable advances achieved with transcription factor-based cell fate reprogramming, safety concerns associated with the insertion of retroviral vectors and the potential reactivation of exogenous transcription factors may hamper any attempt to deploy this technology therapeutically (11–15). Therefore, alternative approaches have been developed to ameliorate this problem. For example, an episome-based delivery system has been used to reprogram cells without the insertion of exogenous factors (16–18). Recently, small chemicals have been used to replace transcription factors and convert somatic cells into ciPSCs (19–21) and chemically induced neurons (22, 23), suggesting a way to reprogram cell fate.
Unlike transcription factors, chemicals are mostly synthetic and designed originally to regulate biological activities through cellular targets, mostly receptors and enzymes. Although designed as potential therapeutics, only a limited number of them eventually became registered drugs. The remaining pool of chemicals has now become a vast resource for biomedical research. Recent success in utilizing chemicals to reprogram cell fate demonstrates the utility of chemicals in stem cell and cell fate research (21). Therefore, it is plausible that further research could be able to develop ways to reprogram cell fate as reliably and rationally as transcription factors, without the safety concerns.
The main challenges ahead can be categorized into two main areas. First, there is a knowledge gap between the biological activities regulated by chemicals and their relevance to cell fate decision. Second, chemicals are less specific than transcription factors and have off-target effects well-known in the pharmaceutical research as side effects. For example, vitamin C is a small molecule found in nature. It is best known as an antioxidant important for human health. Recently, we have shown that vitamin C can enhance somatic cell reprogramming by promoting histone and DNA demethylation through histone and DNA demethylases (24–27). As such, the relationship between chemicals and cell fates can become a fertile ground for further investigations. New tools may be developed such that cell fate can be reprogrammed with chemicals with relative ease. In this report, we attempted to develop a rational approach to convert one cell type to another in a chemically defined and mechanistically understood manner.
Results
Chemical induction of epithelial-like cells (ELCs) from mouse embryonic fibroblast (MEFs)
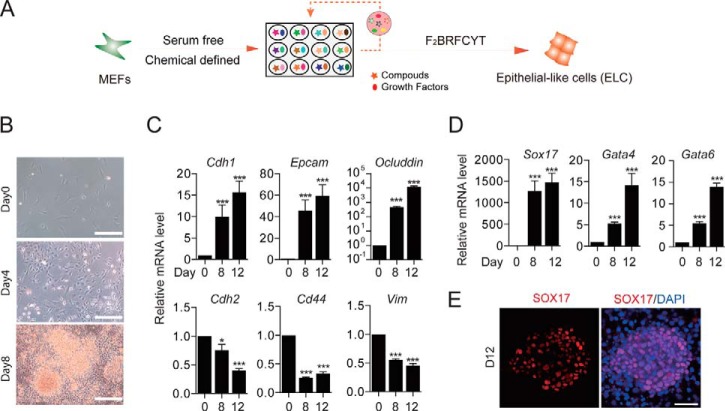
One of the earlier insights we gained in analyzing the reprogramming of MEFs into iPSCs by the Yamanaka factors is the realization that the starting fibroblasts undergo a mesenchymal–epithelial transition (MET) process to become epithelial cells. This is accomplished by the suppression of the fibroblastic characteristics reinforced mostly through the TGFβ signaling pathway with Oct4, Sox2, and Myc and then the activation of E-cad and other epithelial features by Klf4 (28). Inspired by this mechanistic insight, we wished to formulate a chemical recipe that could reliably convert fibroblasts into epithelial cells. To this end, we have developed a mixture called F2BRFCYT that can convert MEFs into ELCs in a chemically defined medium (Fig. 1A). The starting rationale is to first inhibit TGFβ signaling with RepSox (R) and then activate epithelial characteristics with BMP4 (B) based on our studies (28, 29). In addition to RB, we also included Fgf2, CHIR, and Y27632 used previously in our reprogramming medium (30). Furthermore, through compound screening, we also determined that Forskolin (F) and TTNPB (T), two ingredients important for ciPSC generation (21), are very helpful in promoting the MET process. Specifically, we observed cell morphological changes typical of MET when MEFs were treated with F2BRFCYT for 4 and 8 days (Fig. 1B). Accordingly, mesenchymal genes, such as Cdh2, Cd44, and Vim, were inhibited (Fig. 1C, bottom), whereas epithelial genes, such as Cdh1, Epcam, and Ocluddin, were up-regulated (Fig. 1C, top), confirming at the molecular level a MET process. We then wished to determine the cellular state for the emerging ELCs and therefore performed RNA-seq to map their molecular signatures. Interestingly, the ELCs appear to have acquired endoderm characteristics as demonstrated by robust induction of Sox17, Gata4, and Gata6 at the mRNA level (Fig. 1D), apparently without appreciable induction of pluripotency genes (supplemental Fig. S1). These ELCs showed positive staining for SOX17 in the nuclei (Fig. 1E), further confirming that they have transitioned into the endoderm lineage.
Figure 1.
Chemical induction of MET from MEFs. A, schematics of chemically induced MET (ciMET). F2, FGF2; B, BMP4; R, RepSox; F, Forskolin; C, CHIR99021; Y, Y27632; T, TTNPB. B, cell morphological changes during ciMET. Scale bars, 250 μm. C, qRT-PCR analysis of epithelial and mesenchymal genes during ciMET. Data are means ± S.D. (error bars); n = 3 independent experiments. D, qRT-PCR analysis of endoderm marker Sox17 and lineage-specific genes Gata4 and Gata6 during ciMET. Data are means ± S.D.; n = 3 independent experiments. E, immunostaining of SOX17 in cells at day 12 during ciMET. Scale bars, 100 μm. *, p < 0.05; ***, p < 0.001
Chemically induced endoderm progenitor cells (ciEPCs) from MEF-derived ELCs
Given the fact that chemicals are not designed to mediate cell fate changes as specifically as transcription factors have been evolved to do, we were surprised by the acquisition of endodermal markers, such as Sox17 and Gata4/6, in the ELCs (Fig. 1E). To understand and improve this induction process further, we systematically optimized the induction process (supplemental Fig. S2A). We first performed drop-out experiments to assess the role of each component in mediating the induction of individual markers, such as Sox17, Foxa2, Hnf4a, and Cdh1. It became apparent that RepSox plays a critical role for the induction of Sox17, Foxa2, Hnf4a, and Cdh1 (supplemental Fig. S2B). As for Sox17, TTNPB and BMP4 are also critical (supplemental Fig. S2B). On the other hand, all except for RepSox appear to be dispensable for Foxa2, Hnf4a, and Cdh1 (supplemental Fig. S2B). Furthermore, small chemicals, such as TTNPB, appear to be detrimental for the induction of Cdh1, Foxa2, and Hnf4a (supplemental Fig. S2B). In addition to these markers, we also analyzed their impact on cell proliferation and determined that Fgf2 and, to a lesser degree, TTNPB affect cell proliferation as expected (supplemental Fig. S2C) (30).
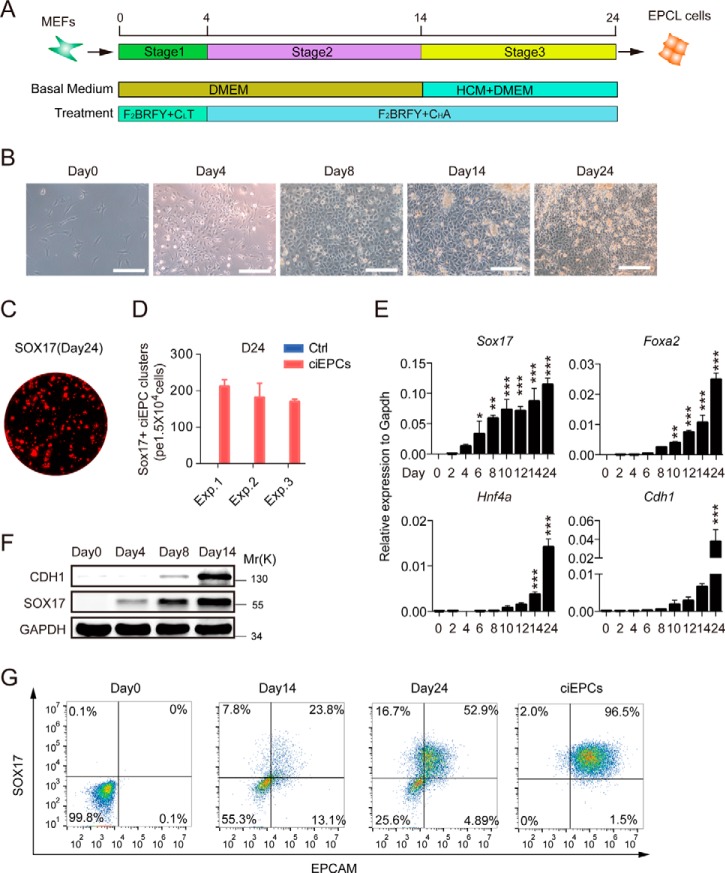
Based on the role of each component in mediating marker induction and also cell proliferation, we decided to further optimize the induction process toward the endodermal lineage. We first performed dose-response experiments for BMP4 (0–50 ng/ml), CHIR99021 (0–20 μm), RepSox (0–20 μm), TTNPB (0–5 μm), FGF2 (0–50 ng/ml), and FSK (0–50 μm) and analyzed the induction of Cdh1, Sox17, Foxa2, and Hnf4a. As shown in supplemental Fig. S2D, it is apparent that each component must be titrated carefully. For example, CHIR appears to work optimally at 10 μm. We then performed time-course experiments as shown in supplemental Fig. S2E and demonstrated that TTNPB is time-sensitive for Sox17 and Hnf4a induction and detrimental to Cdh1 and Foxa2, yet CHIR is time-sensitive for Sox17 and Foxa2 and detrimental to Cdh1 and Hnf4a. Based on these results and insights, we developed an optimized schedule as shown in Fig. 2A. We reduced the concentration of CHIR based on its inhibitory effect on Cdh1 for the first 4 days and removed TTNPB after day 4 to allow the induction of Hnf4a, Cdh1, and Foxa2 (supplemental Fig. S2B). To facilitate the induction of endoderm markers, we also included activin A based on its known inductive activity. Based on this improved protocol, we can consistently induce the formation of ELCs and the further reprogramming of these cells morphologically (Fig. 2B). To quantify the reprogramming efficiency, we stained the cells with SOX17 antibody in site (Fig. 2C and supplemental Fig. S3A) and counted the Sox17+ cell clusters. More than 150 Sox17+ cell clusters were formed per 1.5 × 104 cells at day 24, thus with an induction efficiency of >1% (Fig. 2D). Furthermore, to analysis the optimized protocol in dynamics, we show by qRT-PCR that Sox17 is induced rapidly, followed by Foxa2, Hnf4a, and Cdh1 (Fig. 2E), and also a robust induction of Gata6 early and Epcam and Gata4 relatively late (supplemental Fig. S3B). Western blot analysis confirmed the early induction of SOX17 protein ahead of CDH1 (Fig. 2F). We further showed by flow cytometry that more than 50% of the chemically induced cells in the dishes were Sox17+/Epcam+ at day 24, indicating a robust endoderm cell induction (Fig. 2G).
Figure 2.
Chemically induced endoderm progenitor-like cells (ciEPCLs). A, schedule of an optimized ciEPCL protocol. F2, FGF2; B, BMP4; R, RepSox; F, Foskolin; CL, low dose of CHIR99021 (3 μm); CH, high dose of CHIR99021 (10 μm); Y, Y27632; T, TTNPB. A, activin A. B, representative images of the ciEPCL protocol at different time points. Scale bars, 250 μm. C, image of SOX17-positive endoderm progenitor-like cell clusters by immunostaining. Scale bars, 2 mm. D, the efficiency of ciEPCL induction was determined by counting the SOX17+ cell clusters at day 24, with medium-retreated chemicals as control. Data are means ± S.D. (error bars) (n = 3 wells). E, qRT-PCR analysis of endoderm and epithelial markers during optimized ciEPCLs between days 0 and 24. Data are means ± S.D. (n = 3 independent experiments). F, immunoblotting of SOX17 and CDH1 during ciEPCL induction. G, flow cytometry analysis of the expression of EPCAM and SOX17 during the ciEPCL induction process. ciEPCL were cell lines established by passage the SOX17+ cells in the induction medium. See also “Experimental procedures.” *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Characterization of ciEPCs
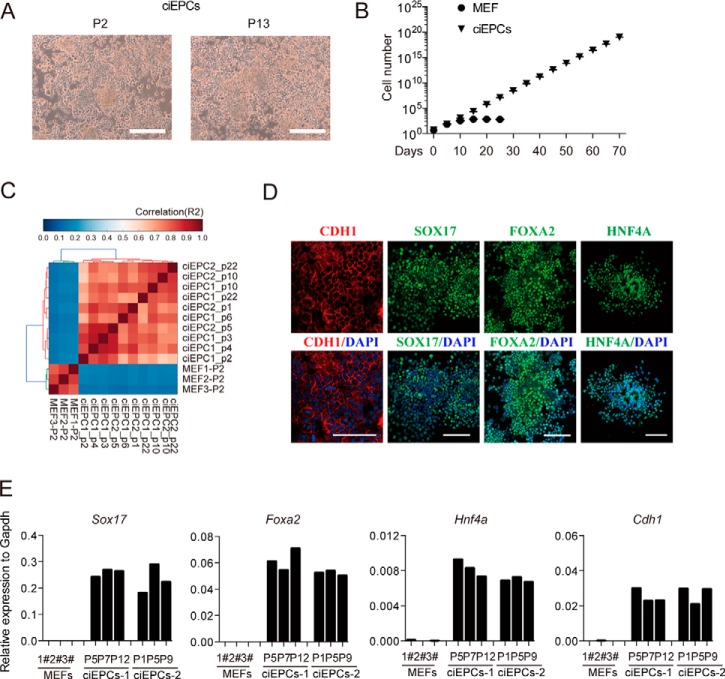
To see whether ciEPCs could self-renew, we derived stable lines and characterized them in depth. As shown in Fig. 3A, these ciEPCs can be passaged without any appreciable differentiation based on cell morphology, as passages 2 and 13 are essentially the same. We then cultured and passaged them continuously for 70 days and can demonstrate that they proliferated by ∼1014 times to ∼1020 cells from a starting cell population of 500,000 cells, whereas the MEFs can grow for 5–10 days and cannot be maintained for more than 30 days (Fig. 3B). We then performed RNA-seq for cells collected between passages 1 and 22 and demonstrated that they maintain very stable transcriptomes (Fig. 3C). Consistently, we could detect SOX17, FOXA2, HNF4A, and CDH1 with immunostaining for two independently derived ciEPC lines (Fig. 3D and supplemental Fig. S4A). We also performed qRT-PCR for MEFs and ciEPCs at different passages to show that they were quite stable in expressing the key endoderm markers, such as Sox17, Foxa2, Hnf4a, and the epithelial marker Cdh1 (Fig. 3E).
Figure 3.
Characterization of ciEPCs. A, represented images of ciEPCs at different passages. Scale bars, 250 μm. B, growth curve of MEF and ciEPCLs. Cells were seeded at 5 × 105 cells/well (6-well plate) and passaged every 5 days. Data are means ± S.D. (error bars) (n = 3 wells). C, R2 correlation coefficient matrix of all versus all samples. D, immunostaining of CDH1, SOX17, FOXA2, and HNF4A in ciEPCLs. Scale bars, 100 μm. E, qRT-PCR analysis of endoderm marker genes in MEFs and ciEPCs.
To evaluate the potential risk of tumorigenesis, we injected the ciEPCs into an SCID mouse and found no teratoma formed in the ciEPC groups during a 6–10-week period, whereas all of the ESC/iPSC controls could form teratoma (supplemental Fig. S4, B and C). We then tested the karyotypes of ciEPCs at different passages and found that the ciEPCs have normal karyotypes when cultured for more then 30 passages (supplemental Fig. S4D). Taken together, these data indicated that the ciEPCs could maintain in vitro stability.
The ciEPCs have differentiation potential restricted to endoderm
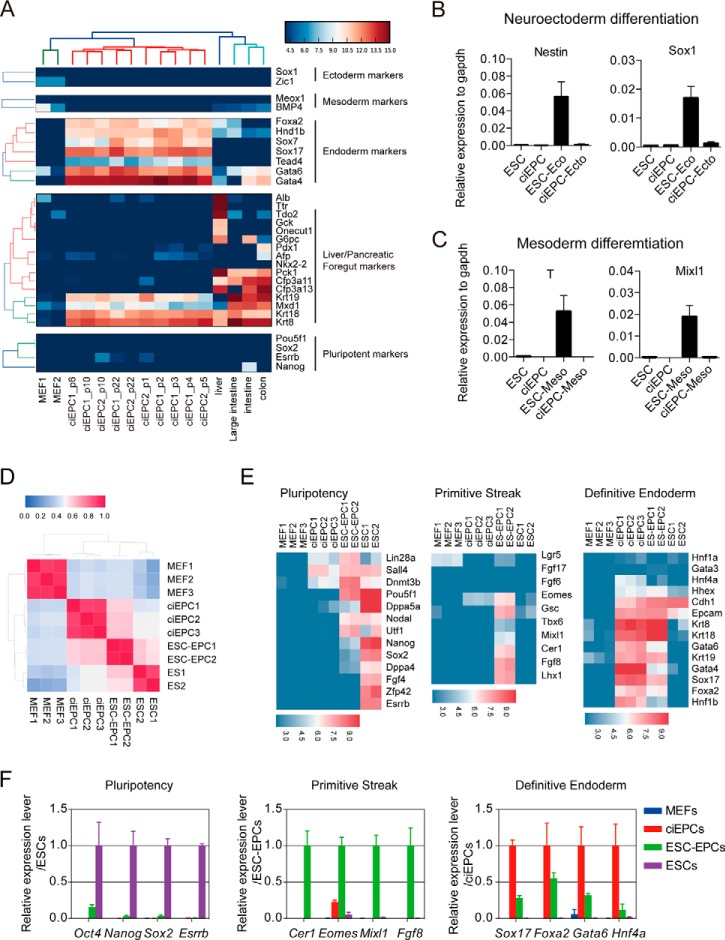
To evaluate the differentiation potential of the ciEPCs, we first compared the RNA-seq data sets from ciEPCs generated in different experiments with MEFs, liver, intestines, and colon. The data show that the ciEPCs are positive for additional endoderm markers Hind1b and Tead4, in addition to Sox17, Foxa2, Gata4/6, and Sox7, while lacking ectoderm or mesoderm markers (Fig. 4A). Interestingly, the ciEPCs also express Krt8, Krt18, and Krt19 strongly, markers known for liver/foregut. We then induced ciEPCs and mouse ESCs with conditions established for promoting mESCs differentiation toward neuroectoderm (31) or mesoderm. We showed that the neuroectoderm markers Nestin and Sox1 (Fig. 4B) or mesoderm markers T and Mixl1 (Fig. 4C) were up-regulated significantly in the ES group, but not in the ciEPC group.
Figure 4.
ciEPCs have the potential for endoderm differentiation. A, RNA-seq analysis of ciEPCs from different experiments to indicate their consistency and variations in comparison with MEFs, liver, intestines, and colon. B and C, ciEPCs and mouse ESCs were induced with conditions established for promoting mESC differentiation toward neuroectoderm (B) or mesoderm (C), and gene expression was analyzed by qRT-PCR (day 8 for neuroectoderm differentiation; day 4 for mesoderm differentiation). Data are means ± S.D. (error bars) (n = 3 independent experiments). D, heat maps of RNA-seq data from MEFs, ciEPCs, ESC-EPCs, and ESCs. E, heat maps for expression levels of genes related to pluripotency, primitive streak, and definitive endoderm markers. F, qRT-PCR analysis of the expression levels of the indicated genes related to pluripotency, primitive streak, and definitive endoderm markers. Data are means ± S.D. (n = 3 independent experiments).
EPCs could be derived from ES cells by a stepwise differentiation protocol (32–34). We then compared the gene expression profile of the ESC-derived EPCs (ESC-EPCs) with the protocol published previously by Sakano et al. (35), with our ciEPCs. We showed that ESC-EPCs were quite similar to ciEPCs in gene expression (Fig. 4D). However, the ESC-EPCs have residual pluripotent property, as indicated by Pou5f1, and also primitive stick or mesoderm markers, such as Cer1, Fgf8, and Mixl1, which is consistent with the recent report by Cheng et al. (36) (Fig. 4E). Compared with ESC-EPCs, the ciEPCs have a higher level of expression for definitive endoderm markers, such as Sox17, Foxa2, and Hnf4a (Fig. 4E). We then further confirmed the RNA-seq data by qPCR and observed the same result (Fig. 4F). These data suggest that the ciEPCs are restricted to the endoderm lineage and closely resemble endoderm progenitor cells.
Differentiation of ciEPCs toward hepatocytes (ciHeps)
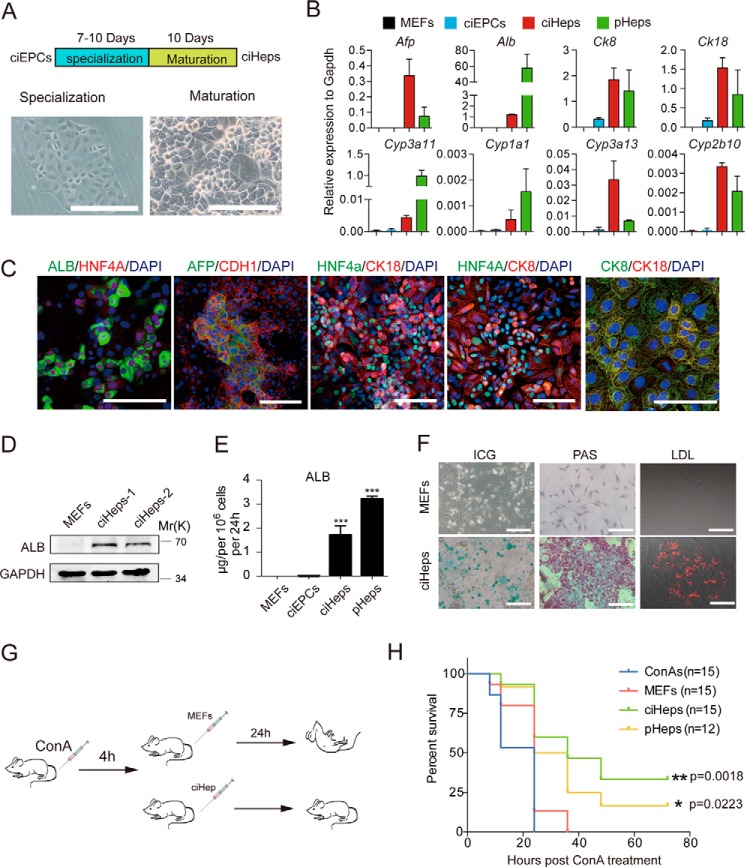
We then attempted to direct the ciEPCs toward hepatic lineage. To this end, we devised a two-step process consisting of a specification phase between 7 and 10 days and a maturation phase of 10 days (Fig. 5A). At the end of the specification phase, we obtained a quite homogeneous cell population with flat cell morphology (Fig. 5A, left). After a maturation period of 10 days, these cells give rise to cells of typical hepatocytic morphology (Fig. 5A, right). We then performed RNA-seq to show that the resulting ciHeps have acquired expression profiles similar to primary hepatocytes (supplemental Fig. S5A). We then checked the expression of hepatocyte markers by qPCR. Specifically, ciHeps express higher levels of Afp and Cyp3a13 than primary hepatocytes and comparable levels of Ck8, Ck18, and Cyp2b10 but lower levels of Alb, Cyp3a11, and Cyp1a1 (Fig. 5B). Compared with starting MEFs, ciHeps express ALB, AFP, HNF4A, CK18, CK8, CDH1, and ZO-1, as demonstrated by immune staining (Fig. 5C and supplemental Fig. S5B). Given the fact that albumin is a hallmark for hepatocytes, we further characterized ciHeps for their expression of albumin and demonstrated that albumin can be detected by both Western blotting (Fig. 5D) and ELISA (Fig. 5E). We further detected the expression of ALB by FACS and found that 66.7% of the ciHep cells were ALB-positive (supplemental Fig. S5C). We further analyzed the liver-specific function of ciHeps and showed that they are capable of storing glycogen as well as taking up LDL and indocyanine green (ICG) (Fig. 5F).
Figure 5.
Maturation of ciEPCs toward hepatocytes. A, schedule and representative images for the conversion of ciEPCs to ciHeps by specialization and maturation lineage. Scale bars, 250 μm. B, qRT-PCR analysis of hepatocytic markers in MEFs, ciEPCs, ciHeps, and primary hepatocytes (pHeps). Primary hepatocytes were freshly isolated from mouse liver without culture in dishes. Data are means ± S.D. (error bars) (n = 3 independent experiments). C, immunostaining of hepatocyte markers in ciHeps. Scale bars, 100 μm. D, immunoblotting of ALB in MEFs and ciHeps. E, ELISA analysis of ALB in culture medium secreted by MEFs, ciEPCLs, ciHeps, and pHeps. Data are means ± S.D. (n = 3 independent experiments). F, glycogen storage (PAS), DiI-ac-LDL, and ICG uptake in MEFs and MEF-derived ciHeps. Scale bars, 250 μm. G, experimental design for treatment of acute liver failure with ciHeps in mice. C57Bl6/J mice injected with ConA develop fulminant hepatitis and acute liver failure that cause death within 12–24 h in mice. Four hours after ConA treatment, 5 × 106 ciHep, primary hepatocytes, MEFs, or saline solution were injected into mice through the tail vein with acute liver failure. H, survival curve of G. ***, p < 0.001.
To test their function in vivo, we injected ciHeps into concanavalin A (ConA)-treated mice and showed that they can rescue mice from liver failure-related death (Fig. 5, G and H). As shown in supplemental Fig. S5 (D and E), ciHeps are as effective as primary hepatocytes in reducing the serum levels of alanine aminotransferase and aspartate aminotransferase in mice.
Chemical reprogramming of fibroblasts into EPCs and hepatocytes
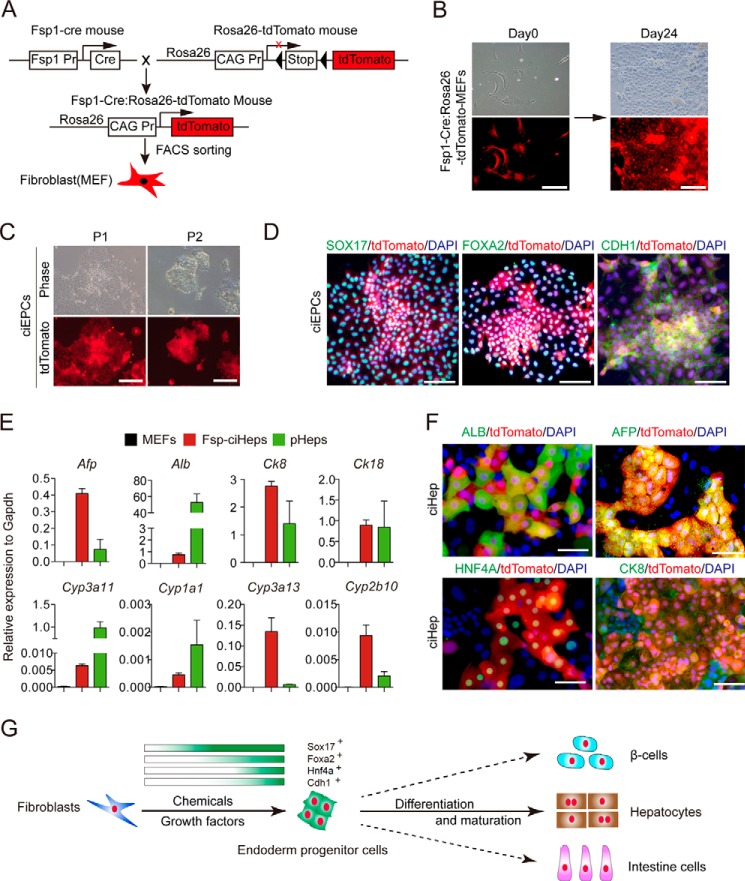
To ultimately confirm that the ciEPCs were induced from fibroblast, we used a lineage tracing system to track the origin of the ciEPCs. As shown in Fig. 6A, transgene mice that express Cre recombinase under the control of fibroblast-specific protein 1 (Fsp1 or S100A4) promoter were crossed with the R26RtdTomato mice. Because Fsp1 is specifically expressed in the fibroblast, the offspring of the mice (Fsp1-Cre:R26RtdTomato) would express tdTomato in the fibroblast. The Fsp1-Cre:R26RtdTomato MEFs, which were isolated from Fsp1-Cre:R26RtdTomato mice and purified by FACS sorting, were used for EPC induction (Fig. 6B). The tdTomato-MEFs underwent a similar morphology change and could form EPC-like colonies as wild-type MEFs when induced toward EPCs by chemicals at day 24 (Fig. 6B). The tdTomato-ciEPC could passage continuously (Fig. 6C) and express key endoderm markers, such as SOX17, FOXA2, and epithelial marker CDH1 (Fig. 6D). We then differentiated the tdTomato-ciEPCs into hepatic lineage following the protocol described in MEF-derived ciEPCs. Consistent with the MEF-ciHep, the tdTomato-ciHeps widely express the hepatocytes markers, such as Afp, Alb, CK8, CK18, Cyp3a11, Cyp1a1, Cyp3a13, and Cyp2b10 (Fig. 6E). The expression of ALB, AFP, HNF4A, and CK8 were further confirmed by immune staining (Fig. 6F). These data definitely demonstrate that fibroblasts could be induced into EPCs by chemicals.
Figure 6.
Chemical reprogramming of FSP-fibroblast toward endoderm lineage. A, schematic diagram of the obtaining of Fsp1-Cre:R26RtdTomato MEFs. B, induction of ciEPCs from tdTomato-MEFs. Scale bars, 250 μm. C, morphology of tdTomato-ciEPCs at different passages. Scale bars, 250 μm. D, co-localization of tdTomato with endoderm markers SOX17 and FOXA2 and epithelium marker CDH1 by immune staining. Scale bars, 100 μm. E, qPCR analysis of the expression of hepatocyte makers in FSP-ciHeps with MEFs as a negative control and pHeps as a positive control. Primary hepatocytes were freshly isolated from mouse liver without culture in dishes. The samples of MEFs and primary hepatocytes are the same as in Fig. 5B. Data are means ± S.D. (error bars) (n = 3 independent experiments). F, co-localization of tdTomato with hepatocyte markers ALB, AFP, HNF4A, and CK8 by immunostaining. Scale bars, 100 μm. G, a model for reprogramming fibroblasts into functional endoderm cell types with chemicals and growth factors.
We also showed that mouse neonatal dermal fibroblasts (MNFs), defined adult fibroblast cell, could be induced to EPCs and hepatocytes in the same chemical mixture. As shown in supplemental Fig. S6A, MNF cells had a morphologic change similar to that of MEFs when induced by chemicals. The induction of SOX17 expression was confirmed by immune staining at day 24 (supplemental Fig. S6B). The derived MNF-ciEPCs cell lines could be passaged stably (supplemental Fig. S6C) and express a high level of endoderm markers Sox17, Foxa2, Hnf4a, Gata4, and Gata6 and the epithelial markers Cdh1 and Epcam but no appreciable level of mesenchyme cell marker Chd2 or liver makers Alb and Afp (supplemental Fig. S6D). The expression of SOX17 and FOXA2 was further confirmed by immune staining (supplemental Fig. S6E). We then differentiated the MNF-ciEPCs into hepatic lineage following the protocol described in MEF-derived ciEPCs. RNA-seq data showed that the MNF-ciHeps were very similar to the MEF-ciHeps in whole-gene expression profiles (supplemental Fig. S6F). The expression of hepatocyte markers ALB, AFP, HNF4A, CDH1, CK8, and CK18 in MNF-ciEPCs were confirmed by immune straining (supplemental Fig. S6G). Functionally, the MNF-ciEPCs were proven to be capable of storing glycogen, taking up LDL and ICG (supplemental Fig. S6H). We further detected the expression of albumin by FACS and found that 64.3% of the MNF-ciHep cells were ALB-positive (supplemental Fig. S6I).
Discussion
In the last 10 years, the iPSC field has generated key insights into cell fate decisions. One such insight is the realization that cell fate can be reprogrammed if the tissue-specific or lineage-specific transcription factors have been identified or characterized through developmental biology or genetics. However, these factors must be delivered through efficient methods, such as retroviral transduction, and integrated into the genomes of recipient cells, thus raising serious safety issues that may prevent their ultimate use in regenerative medicine (37). With high-throughput screening and chemical biology, the role of transcription factors may be gradually replaced by chemicals (small chemicals and growth factors). To this end, several groups have succeeded in converting somatic cells to iPSCs and neural stem cells/neurons by small molecules (19, 21–23, 38). In this report, we show that a similar approach can be used to convert fibroblasts into self-renewing ciEPCs and then eventually functional ciHeps. Furthermore, we were also able to differentiate the ciEPCs toward cells of the pancreatic lineage that express markers such as Pdx1, Nkx2.2, Neurod1, and Hnf6 by qRT-PCR (supplemental Fig. S7, A and B) and PDX1 and HNF6 by immune staining (supplemental Fig. S7C). On the whole, we established a feasible method to generate the main cell types of endoderm by chemicals and growth factors (Fig. 6G). This approach may have several implications.
First, our study suggest that cell fate can be manipulated rationally based on the molecular signatures of the starting cells and the induction conditions defined by chemicals and growth factors. Whereas the initial morphological transformation of fibroblasts into epithelial-like cells (i.e. MET) provided the first informative guide, detailed transcriptomic signatures obtained by RNA-seq provided the most critical parameters to design and optimize the induction process. By recognizing that the cells start to assume endodermal characteristics, we then initiated subsequent work to direct them toward endoderm progenitors and hepatocytes. Similar approaches should be considered for the development of progenitors of the mesoderm or ectoderm lineages.
Second, we were surprised to find that TTNPB, an agonist of the retinoic acid receptor, could activate endoderm marker Sox17 robustly but impair the activation of the other two endoderm markers, Foxa2 and Hnf4a, indicating a complex regulating pattern in chemical-triggered cell fate transition. Given this, the mechanism by which one compound exhibits a different, even contrasting, effect on genes belonging to the same lineage requires further investigation. To obtain a designed cell type with special expression of a group of biomarkers together, we rationally optimized the time window for treatment at the beginning to guarantee the expression of Sox17, and then we substituted TTNPB with activin A in the mixture to allow the successful activation of two other endoderm marker genes. This method could be a general way to guide the cell fate transition by chemicals.
Third, in this study, to rule out the possible contamination of progenitor cells and non-fibroblast cells in the embryo, we used MEFs, MNFs, and fibroblast-specific protein (FSP) MEFs for chemical induction, respectively. All of the three cell types showed a similar response to the chemicals, indicating the universality of our protocol for cell fate transition. In a future study, we will extend this protocol for the transition of other special fibroblasts, such as those in monkeys and humans. Collectively, this discovery has promised application in future drug screening and regeneration medicine.
Last, in our study, we have identified a critical factor in the induction process, SOX17. SOX17 has been implicated in many cell fate decisions, including in endoderm progenitors and other cells (39–43). The activation of Sox17 by chemicals offered a new platform to understand the mechanism of endoderm development and will offer new insights into cell lineage transition. In future studies, we will continue to analyze the induction process for SOX17 and use this as a marker to further improve the process in terms of timing as well as simplicity.
Experimental procedures
MEF cell isolation and culture
MEFs were derived from 13.5-day postcoitum mouse embryos from a cross of male Oct4-GFP transgenic allele-carrying mice (CBA/CaJ X C57BL/6J) to 129Sv/Jae female mice and maintained in DMEM supplemented with 10% FBS, GlutaMAX, and NEAA.
Primary hepatocyte isolation and culture
Primary hepatocytes were isolated with the standard two-step collagenase perfusion method. Briefly, the mouse liver was preperfused through the portal vein with calcium-free buffer (0.5 mm EGTA, Hanks' balanced salt solution without Ca2+ and Mg2+) and then perfused with collagenase (0.1 mg/ml collagenase type IV (Sigma), Hanks' balanced salt solution with Ca2+ and Mg2+). After perfusion, the livers were excised and pelleted into small pieces in DMEM (Hyclone, high glucose) with an additional 9 mg/ml glucose. Then the pellets were washed and centrifuged at low speed (1,500 rpm, 10 min) 2–3 times. The purified primary hepatocytes were strained with 0.04% trypan blue to evaluate the activity, which should exceed 80%, and cultured in HCM medium (Lonza, CC-3198) in Matrigel-coated dishes for 4 days before harvesting for RNA extraction or other analysis.
Induction of ELCs
MEFs were seeded in a 12-well plate (1 × 104 cells/well). For the first 4 days, cells were cultured in N2B27 medium supplemented with bFGF (10 ng/ml; Peprotech), BMP4 (10 ng/ml; R&D Systems), RepSox (5 μm; Sigma), CHIR (3 μm; Sigma), FSK (10 μm; Selleck), TTNPB (1 μm; Selleck), Y27632 (10 μm; Selleck). For the following 10 days (days 4–14), cells were cultured with a modified medium, for which the concentration of CHIR was adjusted to 10 μm and TTNPB was deleted from and activin A (100 ng/ml) was added to the mixtures.
Induction and expansion of endoderm progenitor cells (ciEPCs)
Induced ELCs were continually cultured in a modified medium, which mixed the HCM medium (Lonza, CC-3198) with the induction medium described above (days 4–14), for another 10 days. Then cells were harvested with 0.25% trypsin and passaged in Matrigel-coated dishes at 5,000–6,000 cell/cm2 in the same medium.
The differentiation of ciEPCs into hepatocytes (ciHeps)
ciEPCs were subjected to known hepatocyte specialization and maturation protocols as described. Briefly, ciEPCs were cultured in specialization medium (DMEM + N2 + B27 + bFGF (20 ng/ml) + BMP4 (20 ng/ml)) for 7–10 days and then changed to maturation medium (HCM supplemented with oncostatin M (OSM) (20 ng/ml), DAPT (10 μm; Selleck), dexamethasone (0.1 μm; Selleck), A83-01 (0.5 μm; StemRD), hepatocyte growth factor (HGF) (25 ng/ml)) for another 10 days.
Differentiation of mouse ESCs into EPCs
The differentiation of mouse ESCs into EPCs was performed as described previously (35) with slight modifications. Briefly, mouse ESCs were trypsinized and seeded onto a gelatin-coated plate at 50,000 cells/ml in mouse ESC-2i medium (DMEM, 15% FBS, NEAA, GlutaMAX, PD0325901, Chir99021, LIF). After overnight incubation, the medium was switched to 1:1 mixture of neurobasal medium and DMEM/F-12 medium supplemented with 0.5× N2 and 0.5× B27 supplements, 1% NEAA, 1% GlutMax, 0.1 mm β-mercaptoethanol, 20 ng/ml activin A, 10 ng/ml bFGF, 1 μm XAV939, and 3 μm Chir99021 for 3 days. Then the medium was changed to DMEM supplemented with 1% NEAA, 1% GlutaMAX, 1% sodium pyruvate, 1% ITS, 0.25% BSA, 10 ng/ml activin A, and 5 ng/ml bFGF for another 4 days.
The differentiation of ciEPCs into pancreatic progenitor cells
ciEPCs were subject to known pancreatic progenitor differentiation protocols as described previously (44, 45) with slight modifications. Briefly, ciEPCs were cultured in pancreatic differentiation medium (DMEM, 1% NEAA, 1% GlutaMAX, 1% sodium pyruvate, 0.5× B27, 0.1 mm β-mercaptoethanol) supplemented with 50 ng/ml vitamin C, 50 ng/ml FGF10, 2 μm LDE225, and 2 μm retinoic acid for 4 days, and then the medium supplement was exchanged with 50 ng/ml vitamin C, 20 ng/ml EGF, and 10 ng/ml bFGF for another 5 days. The medium was changed every 2 days.
The differentiation of mouse ESCs and ciEPCs into neuroectoderm and mesoderm
For neuroectoderm differentiation, ciEPCs or mESCs were cultured in N2B27 medium for 8 days (31). N2B27 medium is a 1:1 mixture of DMEM/F-12 supplemented with modified N2 (25 μg/ml insulin, 100 μg/ml apotransferrin, 6 ng/ml progesterone, 16 μg/ml putrescine, 30 nm sodium selenite, and 50 μg/ml bovine serum albumin fraction V) and neurobasal medium supplemented with B27.
For mesoderm differentiation, trypsinized ciEPCs or mESCs were aggregated and cultured as a suspension in embryoid body medium (DMEM + 15% FBS + 1% GlutMAX + 1% NEAA + 1% sodium pyruvate + 0.1 mm β-mercaptoethanol) for 4 days. Then EBs were collected by brief centrifugation.
Immunofluorescence
Cells growing on a confocal dish (NEST catalog no. 801002) or coverslips were washed three times with PBS and then fixed with 4% PFA for 30 min and subsequently penetrated and blocked with 0.1% Triton X-100 and 3% BSA for 30 min at room temperature. Then the cells were incubated with primary antibody for 2 h. After three washes in PBS and 1 h of incubation in second antibodies, cells were then incubated in DAPI for 2 min. Then the coverslips were mounted on slides for observation on the confocal microscope (Zeiss, 710 NLO). The following antibodies were used in this project: rat anti-E-cad (1:100; Abcam, ab11512), rabbit anti-β-catenin (1:200; Santa Cruz Biotechnology, Inc., sc7199), goat anti-Sox17 (1:200; R&D Systems, AF1924), goat anti-Foxa2 (1:200; Santa Cruz Biotechnology, sc6554), rabbit anti-HNF4a (1:100; Sigma-Aldrich, SAB2104352), rabbit anti-Ck8 (1:200; Abcam, ab53280), mouse anti-Ck18 (1:200; Abcam, ab31844), goat anti-albumin (1:200; Bethyl, A90-134A), and mouse anti-Afp (1:100; R&D Systems, MAB1368).
Immunoblotting
Cells were collected and lysed in lysis buffer supplemented with protease inhibitor mixture (Roche Applied Science) on ice for 15 min, and then cells were boiled at 100 °C for 10 min. After centrifugation, the cell supernatants were subjected to SDS-PAGE and detected with corresponding primary antibody and second antibodies. The following antibodies were used in the project: anti-E-cad (1:1,000; CST3195), anti-c-Sox17 (1:200; R&D Systems, AF1924), and anti-GAPDH (1:5,000; Bioworld, AP2063).
qRT-PCR and RNA-seq
Total RNAs were prepared with TRIzol. For quantitative PCR, cDNAs were synthesized with ReverTra Ace (Toyobo) and oligo(dT) (Takara) and then analyzed by qPCR with Premix Ex Taq (Takara). The TruSeq RNA Sample Prep Kit (RS-122-2001, Illumina) was used for library constructions, and sequencing was done with Miseq Reagent Kit V2 (MS-102-2001, Illumina) for RNA-seq. The qPCR primers used in this research can be found in supplemental Table 1.
FACS analyses
For intracellular staining of albumin, 106 cells were harvested and fixed with 4% PFA for 15 min and then permeabilized in Perm/Wash buffer (BD Biosciences, 557885) for 10 min. Cells were then incubated with primary antibody (anti-albumin-488, R&D Systems) for 30 min in staining buffer (BD Biosciences, 554656). After two washes in Perm/Wash buffer (BD Biosciences, 557885), cells were analyzed by the Calibur flow cytometer (BD Biosciences).
Periodic acid–Schiff (PAS) stain, DiI-ac-LDL, and ICG uptake assays
Cells were stained by PAS (Sigma) and DiI-ac-LDL (Invitrogen) following the manufacturer's instructions. For the ICG (Sigma) uptake assay, ICG was dissolved in double-distilled H2O and added to culture medium to a final concentration of 1 mg/ml. After cells were cultured at 37 °C for 1 h, they were washed with PBS three times, and then uptake of ICG was examined by microscopy.
ALB ELISA
To determine ALB secretion, MEFs, ciEPCs, ciHeps, and primary hepatocytes were cultured in HCM medium (Lonza, CC-3198). Culture supernatant was collected 24 h after medium change. The amount of ALB in the supernatant was determined by the mouse albumin ELISA kit (Bethyl Laboratory) according to the manufacturer's instructions.
Transplantation of ciHep to ConA-induced acute liver failure mice
C57B16/J mice were injected with ConA through the tail vein at a dose of 35 mg/kg body weight. 4 h later, MEF cells, ciHep cells, or primary hepatocytes were injected into the acute liver failure mice through the tail vein. Mice in the control group were dead at 24 h after the ConA injection. Blood samples were collected from the orbital vein of the surviving mice. Liver samples were collected from the surviving animals after transplantation. All of the animal experiments were performed with the approval of and according to the guidelines of the Animal Care and Use Committee of the Guangzhou Institutes of Biomedicine and Health.
Statistics and reproducibility
Data are presented as mean ± S.D., as indicated in the figure legends. For unpaired two-tailed Student's t tests, the p values were calculated with Prism version 6 software. A p value < 0.05 was considered statistically significant (*, p < 0.05; **, p < 0.01; ***, p < 0.001). No statistical method was used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during the experiment and outcome assessment.
Author contributions
J. L. designed and performed the experiments and analyzed the data. S. C. performed the cell experiments with S. Y. S. C. and Y. C. performed primary hepatocyte isolation experiments. X. W., Y. Q., and H. L. performed RNA-seq experiment and analyzed the data. C. Z., Y. Liu, L. W., L. G., and J. K. identified the ciEPC and ciHep cell lines. D. L. and C. S. performed the cell transplantation experiment. Y. Li supervised the hepatocyte transplantation experiment, X. S. supervised the hepatocyte differentiation experiments, and J. C. and D. P. supervised the whole study. D. P. conceived the whole study, wrote the manuscript, and approved the final version.
Supplementary Material
This work was supported by Strategic Priority Research Program of the Chinese Academy of Sciences Grants XDA01020307 and XDA01020401; National Key R&D Program of China Grants 2017YFJC040188 and 2016YFA0101800; National Natural Science Foundation of China Grants 31421004, 31530038, 31522033, and 91419310; National Basic Research Program of China Grant 2014CB965200; and Science and Technology Planning Project of Guangdong Province Grants 2015B020228003, 2014B020225002, 2014B030301058, and 2014B05052012. The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as one of our Editors' Picks.
This article contains supplemental Table 1 and Figs. S1–S7.
- iPSC
- induced pluripotent stem cell
- ciPSC
- chemically induced pluripotent stem cell
- ciEPCL
- chemically induced endoderm progenitor-like cell
- ELCs
- epithelial-like cells
- MEF
- mouse embryo fibroblast
- MET
- mesenchymal–epithelial transition
- E-cad
- E-cadherin
- ciEPC
- chemically induced endoderm progenitor cell
- qRT-PCR
- quantitative RT-PCR
- qPCR
- quantitative PCR
- ESC
- embryonic stem cell
- mESC
- mouse ESC
- EPC
- endoderm progenitor cell
- ciHep
- chemically induced hepatocyte
- pHep
- primary hepatocyte
- MNF
- mouse neonatal dermal fibroblast
- ICG
- indocyanine green
- FSP
- fibroblast-specific protein
- NEAA
- nonessential amino acids
- bFGF
- basic fibroblast growth factor
- PAS
- periodic acid–Schiff
- ciMET
- chemically induced MET
- ConA
- concanavalin A.
References
- 1. Takahashi K., and Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 2. Vierbuchen T., Ostermeier A., Pang Z. P., Kokubu Y., Südhof T. C., and Wernig M. (2010) Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ambasudhan R., Talantova M., Coleman R., Yuan X., Zhu S., Lipton S. A., and Ding S. (2011) Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell 9, 113–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caiazzo M., Dell'Anno M. T., Dvoretskova E., Lazarevic D., Taverna S., Leo D., Sotnikova T. D., Menegon A., Roncaglia P., Colciago G., Russo G., Carninci P., Pezzoli G., Gainetdinov R. R., Gustincich S., Dityatev A., and Broccoli V. (2011) Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 476, 224–227 [DOI] [PubMed] [Google Scholar]
- 5. Pang Z. P., Yang N., Vierbuchen T., Ostermeier A., Fuentes D. R., Yang T. Q., Citri A., Sebastiano V., Marro S., Südhof T. C., and Wernig M. (2011) Induction of human neuronal cells by defined transcription factors. Nature 476, 220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang P., He Z., Ji S., Sun H., Xiang D., Liu C., Hu Y., Wang X., and Hui L. (2011) Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 475, 386–389 [DOI] [PubMed] [Google Scholar]
- 7. Sekiya S., and Suzuki A. (2011) Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475, 390–393 [DOI] [PubMed] [Google Scholar]
- 8. Du Y., Wang J., Jia J., Song N., Xiang C., Xu J., Hou Z., Su X., Liu B., Jiang T., Zhao D., Sun Y., Shu J., Guo Q., Yin M., et al. (2014) Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell Stem Cell 14, 394–403 [DOI] [PubMed] [Google Scholar]
- 9. Ieda M., Fu J. D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B. G., and Srivastava D. (2010) Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Szabo E., Rampalli S., Risueño R. M., Schnerch A., Mitchell R., Fiebig-Comyn A., Levadoux-Martin M., and Bhatia M. (2010) Direct conversion of human fibroblasts to multilineage blood progenitors. Nature 468, 521–526 [DOI] [PubMed] [Google Scholar]
- 11. Mikkelsen T. S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B. E., Jaenisch R., Lander E. S., and Meissner A. (2008) Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Okita K., Ichisaka T., and Yamanaka S. (2007) Generation of germline-competent induced pluripotent stem cells. Nature 448, 313–317 [DOI] [PubMed] [Google Scholar]
- 13. Ohi Y., Qin H., Hong C., Blouin L., Polo J. M., Guo T., Qi Z., Downey S. L., Manos P. D., Rossi D. J., Yu J., Hebrok M., Hochedlinger K., Costello J. F., Song J. S., and Ramalho-Santos M. (2011) Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat. Cell Biol. 13, 541–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lister R., Pelizzola M., Kida Y. S., Hawkins R. D., Nery J. R., Hon G., Antosiewicz-Bourget J., O'Malley R., Castanon R., Klugman S., Downes M., Yu R., Stewart R., Ren B., Thomson J. A., et al. (2011) Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hawley R. G. (2008) Does retroviral insertional mutagenesis play a role in the generation of induced pluripotent stem cells? Mol. Ther. 16, 1354–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I. I., and Thomson J. A. (2009) Human induced pluripotent stem cells free of vector and transgene sequences. Science 324, 797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheng L., Hansen N. F., Zhao L., Du Y., Zou C., Donovan F. X., Chou B. K., Zhou G., Li S., Dowey S. N., Ye Z., NISC Comparative Sequencing Program, Chandrasekharappa S. C., Yang H., Mullikin J. C., and Liu P. P. (2012) Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell Stem Cell 10, 337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K., Shibata T., Kunisada T., Takahashi M., Takahashi J., Saji H., and Yamanaka S. (2011) A more efficient method to generate integration-free human iPS cells. Nat. Methods 8, 409–412 [DOI] [PubMed] [Google Scholar]
- 19. Zhao Y., Zhao T., Guan J., Zhang X., Fu Y., Ye J., Zhu J., Meng G., Ge J., Yang S., Cheng L., Du Y., Zhao C., Wang T., Su L., et al. (2015) A XEN-like state bridges somatic cells to pluripotency during chemical reprogramming. Cell 163, 1678–1691 [DOI] [PubMed] [Google Scholar]
- 20. Long Y., Wang M., Gu H., and Xie X. (2015) Bromodeoxyuridine promotes full-chemical induction of mouse pluripotent stem cells. Cell Res. 25, 1171–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hou P., Li Y., Zhang X., Liu C., Guan J., Li H., Zhao T., Ye J., Yang W., Liu K., Ge J., Xu J., Zhang Q., Zhao Y., and Deng H. (2013) Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 341, 651–654 [DOI] [PubMed] [Google Scholar]
- 22. Zhang L., Yin J. C., Yeh H., Ma N. X., Lee G., Chen X. A., Wang Y., Lin L., Chen L., Jin P., Wu G. Y., and Chen G. (2015) Small molecules efficiently reprogram human astroglial cells into functional neurons. Cell Stem Cell 17, 735–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu W., Qiu B., Guan W., Wang Q., Wang M., Li W., Gao L., Shen L., Huang Y., Xie G., Zhao H., Jin Y., Tang B., Yu Y., Zhao J., and Pei G. (2015) Direct conversion of normal and Alzheimer's disease human fibroblasts into neuronal cells by small molecules. Cell Stem Cell 17, 204–212 [DOI] [PubMed] [Google Scholar]
- 24. Chen J., Liu H., Liu J., Qi J., Wei B., Yang J., Liang H., Chen Y., Chen J., Wu Y., Guo L., Zhu J., Zhao X., Peng T., Zhang Y., et al. (2013) H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 45, 34–42 [DOI] [PubMed] [Google Scholar]
- 25. Wang T., Chen K., Zeng X., Yang J., Wu Y., Shi X., Qin B., Zeng L., Esteban M. A., Pan G., and Pei D. (2011) The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin C-dependent manner. Cell Stem Cell 9, 575–587 [DOI] [PubMed] [Google Scholar]
- 26. Chen J., Guo L., Zhang L., Wu H., Yang J., Liu H., Wang X., Hu X., Gu T., Zhou Z., Liu J., Liu J., Wu H., Mao S. Q., Mo K., et al. (2013) Vitamin C modulates TET1 function during somatic cell reprogramming. Nat. Genet. 45, 1504–1509 [DOI] [PubMed] [Google Scholar]
- 27. Esteban M. A., Wang T., Qin B., Yang J., Qin D., Cai J., Li W., Weng Z., Chen J., Ni S., Chen K., Li Y., Liu X., Xu J., Zhang S., et al. (2010) Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell 6, 71–79 [DOI] [PubMed] [Google Scholar]
- 28. Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q., Qin B., Xu J., Li W., Yang J., Gan Y., et al. (2010) A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7, 51–63 [DOI] [PubMed] [Google Scholar]
- 29. Chen J., Liu J., Yang J., Chen Y., Chen J., Ni S., Song H., Zeng L., Ding K., and Pei D. (2011) BMPs functionally replace Klf4 and support efficient reprogramming of mouse fibroblasts by Oct4 alone. Cell Res. 21, 205–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen J., Liu J., Chen Y., Yang J., Chen J., Liu H., Zhao X., Mo K., Song H., Guo L., Chu S., Wang D., Ding K., and Pei D. (2011) Rational optimization of reprogramming culture conditions for the generation of induced pluripotent stem cells with ultra-high efficiency and fast kinetics. Cell Res. 21, 884–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ying Q. L., Stavridis M., Griffiths D., Li M., and Smith A. (2003) Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 21, 183–186 [DOI] [PubMed] [Google Scholar]
- 32. Gouon-Evans V., Boussemart L., Gadue P., Nierhoff D., Koehler C. I., Kubo A., Shafritz D. A., and Keller G. (2006) BMP-4 is required for hepatic specification of mouse embryonic stem cell-derived definitive endoderm. Nat. Biotechnol. 24, 1402–1411 [DOI] [PubMed] [Google Scholar]
- 33. Li F., He Z., Li Y., Liu P., Chen F., Wang M., Zhu H., Ding X., Wangensteen K. J., Hu Y., and Wang X. (2011) Combined activin A/LiCl/Noggin treatment improves production of mouse embryonic stem cell-derived definitive endoderm cells. J. Cell. Biochem. 112, 1022–1034 [DOI] [PubMed] [Google Scholar]
- 34. Gadue P., Huber T. L., Paddison P. J., and Keller G. M. (2006) Wnt and TGF-β signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 103, 16806–16811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sakano D., Shiraki N., Kikawa K., Yamazoe T., Kataoka M., Umeda K., Araki K., Mao D., Matsumoto S., Nakagata N., Andersson O., Stainier D., Endo F., Kume K., Uesugi M., and Kume S. (2014) VMAT2 identified as a regulator of late-stage beta-cell differentiation. Nat. Chem. Biol. 10, 141–148 [DOI] [PubMed] [Google Scholar]
- 36. Cheng X., Ying L., Lu L., Galvão A. M., Mills J. A., Lin H. C., Kotton D. N., Shen S. S., Nostro M. C., Choi J. K., Weiss M. J., French D. L., and Gadue P. (2012) Self-renewing endodermal progenitor lines generated from human pluripotent stem cells. Cell Stem Cell 10, 371–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robinton D. A., and Daley G. Q. (2012) The promise of induced pluripotent stem cells in research and therapy. Nature 481, 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li X., Zuo X., Jing J., Ma Y., Wang J., Liu D., Zhu J., Du X., Xiong L., Du Y., Xu J., Xiao X., Wang J., Chai Z., Zhao Y., and Deng H. (2015) Small-molecule-driven direct reprogramming of mouse fibroblasts into functional neurons. Cell Stem Cell 17, 195–203 [DOI] [PubMed] [Google Scholar]
- 39. Séguin C. A., Draper J. S., Nagy A., and Rossant J. (2008) Establishment of endoderm progenitors by SOX transcription factor expression in human embryonic stem cells. Cell Stem Cell 3, 182–195 [DOI] [PubMed] [Google Scholar]
- 40. Engert S., Burtscher I., Liao W. P., Dulev S., Schotta G., and Lickert H. (2013) Wnt/β-catenin signalling regulates Sox17 expression and is essential for organizer and endoderm formation in the mouse. Development 140, 3128–3138 [DOI] [PubMed] [Google Scholar]
- 41. Irie N., Weinberger L., Tang W. W., Kobayashi T., Viukov S., Manor Y. S., Dietmann S., Hanna J. H., and Surani M. A. (2015) SOX17 is a critical specifier of human primordial germ cell fate. Cell 160, 253–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McDonald A. C., Biechele S., Rossant J., and Stanford W. L. (2014) Sox17-mediated XEN cell conversion identifies dynamic networks controlling cell-fate decisions in embryo-derived stem cells. Cell Rep. 9, 780–793 [DOI] [PubMed] [Google Scholar]
- 43. Nobuhisa I., Osawa M., Uemura M., Kishikawa Y., Anani M., Harada K., Takagi H., Saito K., Kanai-Azuma M., Kanai Y., Iwama A., and Taga T. (2014) Sox17-mediated maintenance of fetal intra-aortic hematopoietic cell clusters. Mol. Cell. Biol. 34, 1976–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. D'Amour K. A., Bang A. G., Eliazer S., Kelly O. G., Agulnick A. D., Smart N. G., Moorman M. A., Kroon E., Carpenter M. K., and Baetge E. E. (2006) Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat. Biotechnol. 24, 1392–1401 [DOI] [PubMed] [Google Scholar]
- 45. Zhang D., Jiang W., Liu M., Sui X., Yin X., Chen S., Shi Y., and Deng H. (2009) Highly efficient differentiation of human ES cells and iPS cells into mature pancreatic insulin-producing cells. Cell Res. 19, 429–438 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.