Abstract
Technological advances in liquid chromatography and tandem mass spectrometry (LC-MS/MS) have enabled comprehensive analyses of proteins and their post translational modifications from cell culture and tissue samples. However, sample complexity necessitates offline prefractionation via a chromatographic method that is orthogonal to online reversed phase high performance liquid chromatography (RP-HPLC). This additional fractionation step improves target identification rates by reducing the complexity of the sample as it is introduced to the instrument. A commonly employed offline prefractionation method is high pH reversed phase (Hi-pH RP) chromatography. Though highly orthogonal to online RP-HPLC, Hi-pH RP relies on buffers that interfere with electrospray ionization (ESI). Thus, samples that are prefractionated using Hi-pH RP are typically desalted prior to LC-MS/MS. In the present work, we evaluate an alternative offline prefractionation method, pentafluorophenyl (PFP)-based reversed phase chromatography. Importantly, PFP prefractionation results in samples that are dried prior to analysis by LC-MS/MS. This reduction in sample handling relative to Hi-pH RP results in time savings, and could facilitate higher target identification rates. Here, we have compared the performances of PFP and Hi-pH RP in offline prefractionation of peptides and phosphopeptides that have been isolated from human cervical carcinoma (HeLa) cells. Given the prevalence of isobaric mass tags for peptide quantification, we evaluated PFP chromatography of peptides labeled with tandem mass tags (TMT). Our results suggest that PFP is a viable alternative to Hi-pH RP for both peptide and phosphopeptide offline prefractionation.
Keywords: LC-MS/MS, PFP, chromatography, prefractionation, TMT, phosphopeptides
Introduction
Biological research has benefited from advances made in the fields of proteomics and phosphoproteomics. “Shotgun/bottom-up” liquid chromatography and tandem mass spectrometry (LC-MS/MS) has become the principal means through which proteins and their post translational modifications are analyzed in complex cell culture and tissue samples [1]. In this platform, a protein or protein mixture is digested into peptides with a protease, typically trypsin, and the resulting peptides are separated by microcapillary low pH (formic acid as an ion-pairing agent) reversed phase high performance liquid chromatography (RP-HPLC) coupled to mass spectrometry. Peptides eluting from the online RP-HPLC column are ionized by electrospray ionization (ESI), scanned, fragmented and detected to generate parent and fragment ion mass spectra. These spectra are then compared to theoretical spectra produced in silico from a translated organism-specific database, which enables identification of the protein or proteins from which the peptides were derived [2].
Shotgun proteomics has become prominent because peptides are readily amenable to chromatographic separation and efficiently ionized by ESI [3]. Effective chromatography is essential for in-depth proteome coverage as even the most advanced mass spectrometers are typically limited by duty cycle (MS1 and MS2 scanning cycles) and dynamic range of peptide ion detection for generating informative mass spectra, and thus undersample unfractionated whole proteome samples [4–5]. Furthermore, advances in chemical labeling strategies for isobaric multiplex quantification, including tandem mass tag (TMT) chemistry, which enables multiplex analyses of multiple samples in a single LC-MS/MS run, have increased the complexity of a typical proteomics experiment [6]. Thus, offline prefractionation using a chromatographic technique that is orthogonal to C18-based online RP-HPLC improves the depth and coverage of the proteome under investigation [7].
A variety of prefractionation approaches have been exploited for proteomics and phosphoproteomics analyses, including strong cation exchange (SCX), hydrophilic interaction chromatography (HILIC) and high pH C18-based reversed phase (Hi-pH RP) [8]. The high degree of orthogonality and peak capacity of efficient Hi-pH RP systems have led to their integration into a number of proteomics and phosphoproteomics workflows [8–11]. One disadvantage of Hi-pH RP prefractionation is that even nominally volatile buffers used to maintain basic pH conditions are usually removed by solid-phase extraction prior to LC-MS/MS analysis. This additional step results in increased sample handling as well as processing time, which can result in reduced identifications in proteomics analyses [12].
In the present work, we have explored an alternative method of proteomic and phosphoproteomic sample complexity reduction through the use of offline pentafluorophenyl (PFP)-reversed phase chromatographic prefractionation. Similar to SCX, HILIC, and Hi-pH RP, these methods are orthogonal to online C18-based reversed-phase separation [8]. A benefit of PFP prefractionation is that samples are separated using trifluoroacetic acid (TFA) as an ion-pairing agent and thus do not require solid-phase extraction prior to LC-MS/MS. The utility of TMT in quantitative analyses has led to their widespread adoption in a number of proteomics laboratories, leading us to compare the chromatographic efficiency of PFP and Hi-pH RP of TMT labeled peptides derived from human cervical carcinoma (HeLa) cells [13–15]. Additionally, we explored the use of PFP prefractionation for separating phosphopeptides. We assert that offline PFP fractionation of complex peptide and phosphopeptide samples reduces sample handling and processing time while maintaining or exceeding current expectations for sample complexity reduction by other off-line chromatographic methods.
Materials and Methods
Reagents
Modified trypsin was from Promega (Madison, WI). Urea, Tris-HCL, ammonium bicarbonate (NH4HCO3), sodium fluoride (NaF), dibasic potassium phosphate (K2HPO4), sodium ortho-vanadate, sodium molybdate, beta-glycerophosphate, DL-dithiothreitol, thymidine, nocodazole and iodoacetamide were from Sigma-Aldrich (St. Louis, MO). HPLC-MS grade acetonitrile (ACN), acetone, trifluoroacetic acid (TFA) and water were from Honeywell Burdick and Jackson (Morristown, NJ). Methanol and SepPak C18 solid phase extraction cartridges were from Fisher (Pittsburgh, PA). High purity formic acid was from EMD (Gibbstown, NJ). Lactic acid was from Lee Biosolutions, Inc (St. Louis, MO). TiO2 beads were from GL Sciences (Tokyo, Japan). Dulbecco’s modified Eagle’s medium (DMEM), PBS, and penicillin-streptomycin were from Invitrogen (Carlsbad, CA). Hyclone fetal bovine serum (FBS), BCA protein assays, SOLAμ HRP desalting plates, and TMTzero Reagent (TMT0; hereafter “TMT”) were purchased from ThermoFisher Scientific (Pittsburgh, PA). HEPES free acid was from Amresco (Dallas, TX). Protease inhibitor tablets were from Roche (Basel, Switzerland). Calyculin A was from TOCRIS (Bristol, UK). Lysyl endopeptidase (Lys-C) was from Wako (Osaka, Japan).
Cell culture
HeLa cells were maintained in DMEM supplemented with 10% FBS and penicillin-streptomycin (100 U/ml and 100 μg/ml) at 37°C under humidification with 5% CO2.
Peptide preparation and TMT labeling
Asynchronously growing HeLa cells were washed in PBS and lysed in 8.5 M urea, 50 mM Tris pH 8.1 and protease inhibitors. Lysate was sonicated using a Branson sonicator equipped with a micro-tip three times at 50% power for 15 seconds each on ice. Protein concentration of the lysate was determined by BCA protein assay. Proteins were reduced with 5 mM DTT at 55°C for 30 minutes, cooled to room temperature and alkylated with 15 mM iodoacetamide at room temperature for 1 hour in the dark. The alkylation reaction was quenched by the addition of another 5 mM DTT. After 15 minutes of incubation at room temperature, the lysate was diluted 6-fold in 25 mM Tris pH 8.1, 1 mg sequencing-grade trypsin per 200 mg total protein was added, and incubated overnight at 37° C. The digest was acidified to pH 3 by addition of TFA to 0.2% and allowed to stand at room temperature for 15 minutes; the resulting precipitates were removed by centrifugation at 7100 RCF for 15 minutes. The acidified lysate was then desalted using a C18 solid-phase extraction (SPE) cartridge and the eluate was separated into 100 μg aliquots and dried by vacuum centrifugation.
Peptide aliquots were dissolved in a buffer containing 20% ACN/HEPES pH 8.5 and 250 μg TMT reagent. After 1 hour of incubation at room temperature, the labeling reaction was quenched with 50 mM DTT and kept at room temperature for 10 minutes. TFA was added to 0.5% and the samples were vacuum centrifuged for 15 minutes. The labeled peptides were then desalted on a SOLAμ HRP desalting plate. The liquid eluate from the desalting procedure was dried in a 1.5 ml Eppendorf tube by vacuum centrifugation and stored at −80°C.
Phosphopeptide preparation and enrichment
HeLa cells were arrested in mitosis using 100 ng/ml nocodazole 3 hours after release from a 20 hour 2 mM thymidine block. Mitotically arrested cells were treated with 25 nM calyculin A for 1 hour and collected by mitotic shake-off. Cells were washed in PBS, frozen in liquid nitrogen and stored at −80°C.
Cell pellets were lysed in 8.5 M urea, 50 mM Tris pH 8.1, protease inhibitors and the phosphatase inhibitors: sodium ortho-vanadate, sodium molybdate, beta-glycerophosphate and sodium fluoride. The lysates were reduced and alkylated as described above. Lysates were incubated with 1 mg Lys-C per 100 mg total protein for 4 hours at 37°C. The digests were then diluted 6-fold in 25 mM Tris pH 8.1, 1 mg sequencing-grade trypsin per 200 mg total protein was added, and the digests were incubated overnight at 37° C. In order to ensure complete digestion, the following morning 1 mg additional trypsin was added per 800 mg total protein for an additional 3 hours. Digests were then acidified to pH 3 by addition of TFA to 0.2%; the resulting precipitates were removed by centrifugation at 7100 RCF for 15 minutes. The acidified lysates were then desalted using a C18 solid-phase extraction (SPE) cartridge and the eluate (~1.2 mL) was vacuum centrifuged for 30 minutes. Desalted peptides were frozen in liquid nitrogen and lyophilized overnight.
Phosphopeptide enrichment was performed using titanium dioxide microspheres as described [16]. Briefly, lyophilized peptides were dissolved in 50% ACN / 2M lactic acid, incubated with 1.25 mg TiO2 microspheres per 1 mg peptide digest and vortexed at 75% power for 1 hour. Microspheres were washed twice with 50% ACN / 2M lactic acid and once with 50% ACN / 0.1% TFA. Phosphopeptides were eluted with 50 mM K2HPO4 pH 10 (adjusted with ammonium hydroxide). Formic acid was added to the eluates to 1.7%. The acidified phosphopeptides were desalted using a C18 solid-phase extraction (SPE) cartridge and the eluate was vacuum centrifuged to dryness.
Hi-pH RP HPLC fractionation and desalting (pH 8)
Approximately 40 μg (PFP vs. Hi-pH (pH 8) RP comparison) or 25 μg (Hi-pH (pH 8) vs. Hi-pH (pH 10) comparison) of TMT labeled peptides were fractionated per replicate using an Agilent 300 Extend-C18 3.5 μm particle, 300Å pore size 2.1 x 150 mm column attached to an Agilent 1200 series HPLC and fraction collector. Buffer A was 3% ACN in 10 mM NH4HCO3, buffer B was 95% ACN in 10mM NH4HCO3. Buffers were confirmed to be at pH 8, which is consistent with Hi-pH RP buffers used in recent publications [17–18]. The flow rate was 0.15 ml/minute and the column was maintained at 20°C throughout the run. The LC gradient is described in the Electronic Supplementary Material (ESM) in ESM_1. The fraction collection window was established empirically; 48 fractions were collected evenly between 11 and 80 minutes. Fractions were combined to 12 (Fig. 1). Fractions were dried by vacuum centrifugation.
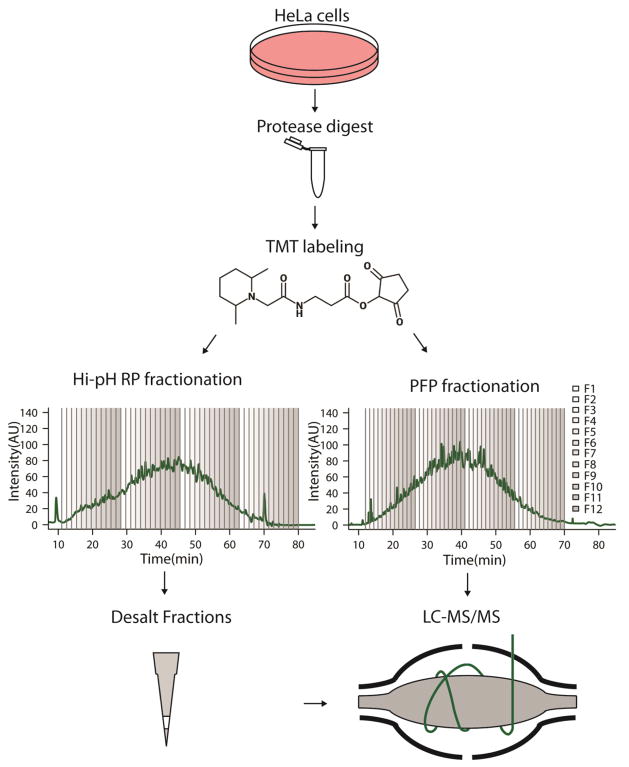
Fig. 1.
Scheme of comparative sample processing workflows Proteins from HeLa cells were digested with trypsin and labeled with TMT reagent. Labeled peptides were then fractionated offline with a PFP column and acidic mobile phase (PFP), or a RP column and basic mobile phase (Hi-pH RP). Chromatograms are of TMT labeled peptides with fractional combinations overlaid; every 12th fraction was combined. Samples derived from RP fractionation were desalted prior to LC-MS/MS, whereas PFP fractions were not.
Approximately 65 μg phosphopeptides were fractionated using the same RP column and buffers as indicated for TMT-labeled peptides, except that the flow rate was 0.135 ml/minute. The phosphopeptide LC gradient is described in ESM_1. 48 fractions were collected between 10 and 80 minutes, and combined to 12. Fractions were dried by vacuum centrifugation.
TMT and phosphopeptide fractions were re-suspended in 0.1% TFA and desalted using a C18 solid-phase extraction (SPE) cartridge and the eluates were vacuum centrifuged to dryness.
Hi-pH RP HPLC fractionation and desalting (pH 10)
Approximately 25 μg of TMT labeled peptides were fractionated using an Agilent 300 Extend-C18 3.5 μm particle, 300Å pore size 2.1 x 150 mm column attached to an Agilent 1200 series HPLC and fraction collector. Buffer A was 3% ACN in 10 mM NH4HCO2, buffer B was 90% ACN in 10mM NH4HCO2. Buffers were confirmed to be at pH ≥ 9.8. The flow rate was 0.15 ml/minute and the column was maintained at 20°C throughout the run. The LC gradient is described in ESM_1. The fraction collection window was established empirically; 48 fractions were collected evenly between 11 and 80 minutes. Fractions were combined to 12 (ESM_2, Fig. S1a). Fractions were dried by vacuum centrifugation.
TMT fractions were re-suspended in 0.1% TFA and desalted using a C18 solid-phase extraction (SPE) cartridge and the eluates were vacuum centrifuged to dryness.
PFP HPLC fractionation
Approximately 40 μg of TMT labeled peptides were fractionated per replicate using a Waters XSelect HSS PFP 2.5 μm 2.1 x 150 mm column and the same HPLC and fraction collector indicated above. Buffer A was 3% ACN in 0.1% TFA, buffer B was 95% ACN in 0.1% TFA. Buffers had a pH less than 3. The flow rate was 0.135 ml/minute and the column was maintained at 20°C throughout the run. The LC gradient is described in ESM_1. The fraction collection window was established empirically; 48 fractions were collected between 12 and 70 minutes. Fractions were combined to 12 (Fig. 1). Fractions were dried by vacuum centrifugation.
Approximately 65 μg phosphopeptides were fractionated using the same PFP column, using methanol as an organic solvent: buffer A contained 3% methanol and 0.1% TFA, and buffer B contained 95% methanol and 0.1% TFA. The flow rate for the column was 0.135 ml/minute and the column was maintained at 40°C throughout the run. The LC gradient is described in ESM_1. After fractionation, the system was washed as indicated above. 48 fractions were collected between 11 and 70 minutes and were combined to 12. Fractions were dried by vacuum centrifugation.
LC-MS/MS analysis
LC-MS/MS analysis was performed on an Orbitrap Fusion Tribrid mass spectrometer (ThermoFisher Scientific, San Jose, CA) equipped with EASY-nLC 1000 ultra-high pressure liquid chromatograph (ThermoFisher Scientific, Waltham, MA). Peptides and phosphopeptide fractions were dissolved in 5% methanol / 1.5 % formic acid and loaded directly onto an in-house pulled and packed polymer coated fritless fused silica analytical resolving column (40 cm length, 100μm inner diameter, ReproSil, C18 AQ 1.9 μm 120 Å pore (Dr. Maisch GMBH, Ammerbuch, Germany))[19]. Peptides (in 1 – 5μl loading buffer) were loaded at 650 bar pressure by chasing on to the column with 10μl loading buffer (5% methanol / 1.5% formic acid).
Phosphopeptide samples were separated with a 90 minute gradient of 2 to 33% LC-MS buffer B (LC-MS buffer A: 0.125% formic acid, 3% ACN; LC-MS buffer B: 0.125% formic acid, 95% ACN) at a flow rate of 330 nl/minutes. For phosphopeptides, the Orbitrap Fusion was operated with an Orbitrap MS1 scan at 120K resolution and an AGC target value of 500K. The maximum injection time was 100 milliseconds, the scan range was 350 to 1500 m/z and the dynamic exclusion window was 15 seconds (+/− 15 ppm from precursor ion m/z). Precursor ions were selected for MS2 using quadrupole isolation (0.7 m/z isolation width) in a “top speed” (2 second duty cycle), data-dependent manner. MS2 scans were generated through higher energy collision-induced dissociation (HCD) fragmentation (29% HCD energy) and Orbitrap analysis at 15 K resolution. Ion charge states of +2 through +4 were selected for HCD MS2. The MS2 scan maximum injection time was 60 milliseconds and AGC target value was 60K.
TMT labeled peptides were separated with a 120 minute gradient of 3 to 33% LC-MS buffer B at a flow rate of 330 nl/minute. Buffers were the same as indicated above. For TMT peptides, the Orbitrap Fusion was operated with an Orbitrap MS1 scan at 120K resolution and an AGC target value of 500K. The maximum injection time was 100 milliseconds, the m/z range was 350 to 1300 and the dynamic exclusion window was 30 seconds. Precursor ions were selected for MS2 using quadrupole isolation (0.6 m/z isolation width) in a “top speed” (3 second duty cycle), data-dependent manner. Ion charge states of +2 through +5 were selected for MS2 by collision induced dissociation (CID) fragmentation (32% CID energy) and ion trap analysis. The MS2 scan maximum injection time was 60 milliseconds and AGC target value was 8K. MS2 fragment ions were selected for synchronous precursor selection (SPS)-MS3 analysis in a top 10 data-dependent manner. MS3 scans were generated through HCD fragmentation (55% HCD energy) and Orbitrap analysis at 30K resolution, with a scan range of 110 to 750 m/z. The MS3 scan maximum injection time was 60 milliseconds and AGC target value was 50K. Two injections were performed for fractions 3 and 9 in each replicate of TMT-labeled peptide analyses. The first injection was used for fractional overlap comparisons; results from both injections were included in all other peptide and protein comparisons.
Peptide spectral matching and bioinformatics
Raw data were searched using COMET [20] against a target-decoy version of the human (Homo sapiens) proteome sequence database (UniProt; downloaded 2013; 20,241 total proteins) with a precursor mass tolerance of +/− 1.00 Da and requiring fully tryptic peptides with up to 3 missed cleavages, carbamidomethyl cysteine as a fixed modification and oxidized methionine as a variable modification. In phosphopeptide analyses phosphorylation of serine, threonine and tyrosine were searched with up to 3 variable modifications per peptide. In TMT-labeled peptide analyses, the TMT reagent mass was searched as a static modification on lysine residues and peptide N-termini. The resulting peptide spectral matches were filtered to <1% false discovery rate (FDR) by defining thresholds of decoy hit frequencies at particular mass measurement accuracy (measured in parts-per-million from theroretical), XCorr and delta-XCorr (dCn) values. Comparative analyses were performed using the R statistical programming language (http://www.R-project.org). All assertions of statistical significance that are not explicitly described were determined by Wilcoxon rank-sum tests wherein the calculated p-values were less than 0.05. Isoelectric point calculations were derived from (http://isoelectric.ovh.org/).
Results
Analytical comparison of Hi-pH RP methodologies
Recent reports in the proteomics literature suggest that higher pH reverse-phase separations have been performed at pH 8 (ammonium bicarbonate buffer) and pH 10 (ammonium formate buffer) [9,11]. In order to arrive at a final Hi-pH RP method for comparison with PFP, we compared the two higher pH methods against each other. To do this, TMT-labeled HeLa peptides (25 μg) were fractionated using either pH 8 or pH 10 buffers. The 48 fractions collected in each method were then reduced to 12 by combining every 12th fraction (ESM_2, Fig. S1a); these fractions were dried by vacuum centrifugation and desalted by C18 solid-phase extraction prior to Orbitrap LC-MS/MS. The number of peptides and proteins identified through fractionation at pH 8 were 38505, and 5870, respectively; at pH 10, the number of peptides identified was 38397, and the number of proteins identified was 5845. The percentages of total peptides and proteins that were identified in both methods were 63% (29609/47293), and 80% (5209/6506), respectively (ESM_2, Fig. S1b; ESM_3 & ESM_4).
The method of fraction combination described above allowed for the preservation of adjacency between fractions and the relevance of fractional peptide overlap between adjacent fractions. Fractional peptide overlap was expressed as the percentage of peptides that were shared between two adjacent fractions. The mean fractional peptide overlap for pH 8 and pH 10 Hi-pH RP HPLC was 8.93% and 8.02%, respectively (ESM_3 & ESM_5, Fig. S2a). Although this difference was considered statistically significant (Wilcoxon rank -sum test p-value < 0.05), the practical impact of this difference is considered relatively minor.
In order to compare the orthogonality of online RP-HPLC to Hi-pH RP at pH 8 versus pH 10, peptides uniquely identified in each fraction were assigned to bins corresponding to their average retention time. Sequentially aligned fractions from both Hi-pH RP prefractionation methods were plotted with peptide retention time distributions (ESM_5, Fig. S2b). Methods with low orthogonality would be predicted to result in narrowly distributed bands of peptides increasing in retention time across sequentially collected fractions, with the number of bands matching the number of combined fractions. Peptide distributions from highly orthogonal methods should be more evenly distributed within and between fractions [11]. This comparison of pH 8 and pH 10 prefractionation revealed a similarly high orthogonality between both pH methods and online RP-HPLC The equivocal performance and orthogonality between pH 8 and pH 10 Hi-pH RP enabled the use of pH 8 buffers as the method we chose for comparison of PFP with Hi-pH RP prefractionation.
Comparison of identification rates for PFP and Hi-pH RP fractionated, TMT-labeled peptides
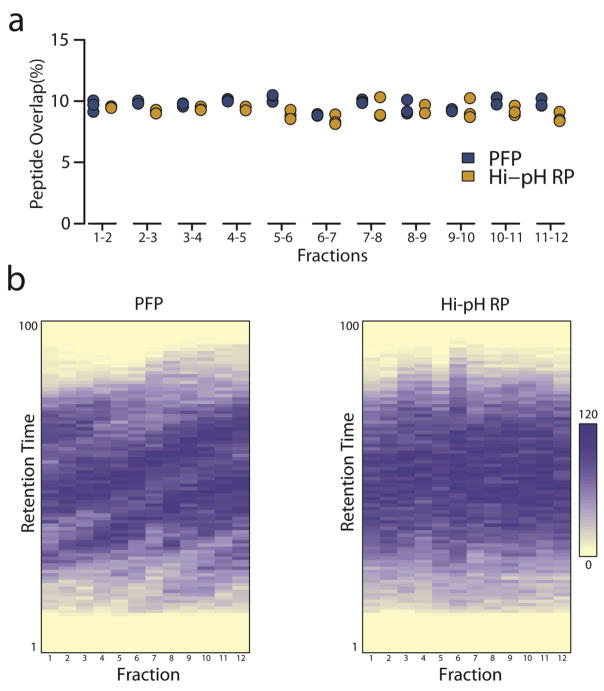
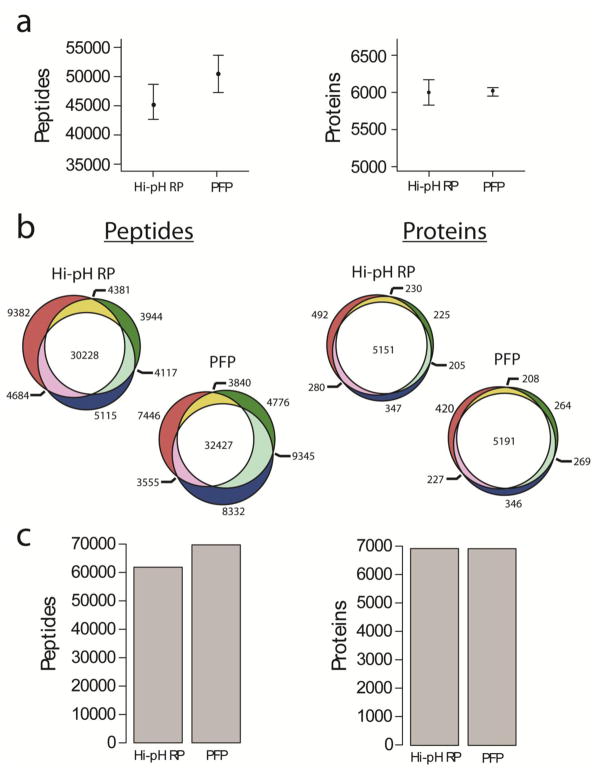
TMT-labeled HeLa peptides were used to optimize PFP and Hi-pH RP separation conditions (gradient elution) to produce chromatograms with similar intensity distributions and maxima (Fig. 1). Equal quantities of TMT-labeled peptides (45 μg) were then fractionated by either PFP or Hi-pH RP chromatography. The 48 fractions collected in each method were then reduced to 12 by combining every 12th fraction (Fig. 1); these fractions were then dried by vacuum centrifugation and either directly analyzed by Orbitrap Fusion mass spectrometry (PFP; see ESM_6 & ESM_7), or first desalted by C18 solid-phase extraction prior to Orbitrap LC-MS/MS (Hi-pH RP; see ESM_8 & ESM_9). The mean fractional peptide overlap for PFP and Hi-pH RP HPLC was 9.71% and 9.13%, respectively. Though modest, this difference was considered statistically significant, possibly owing to the higher degree of orthogonality between Hi-pH RP and online LC-MS/MS compared to the orthogonality between PFP and online LC-MS/MS (Wilcoxon rank -sum test p-value < 0.05) (Fig. 2a)[8]. A plot of peptide retention time distributions by fraction illustrates a modest reduction in orthogonality of PFP relative to Hi-pH RP (Fig. 2b). Triplicate analyses produced higher median replicate peptide and protein counts in PFP versus Hi-pH RP, although these differences were not considered statistically significant (Fig. 3a). The percentage of peptides that were identified in all three replicates was 46.5% for PFP (32427/69721) and 48.9% for Hi-pH RP (30228/61851). The percentage of proteins that were identified in all three replicates was 75.0% for PFP (5191/6925) and 74.3% for Hi-pH RP (5151/6930) (Fig. 3b; ESM_6, ESM_7, ESM_8 & ESM_9). The total unique peptide identifications across all three replicates were 69721 and 61851 for PFP and Hi-pH RP, respectively. Unique protein identification totals for all replicates was 6925 for PFP and 6930 for Hi-pH RP (Fig. 3c). These results suggested that PFP and Hi-pH RP are comparable in their ability to facilitate protein and peptide identifications, as well as their capacity to effectively and reproducibly reduce fractional peptide overlap, which we consider a proxy for chromatographic efficiency.
Fig. 2.
Fractional peptide overlap (A) Overlap between each adjacent fraction is expressed as a percentage of the total peptides identified between the pair of fractions. The fractional peptide overlap for each of the three replicate experiments is displayed. Wilcoxon rank-sum tests revealed that there was significantly (p < 0.05) less overlap between Hi-pH RP fractions than PFP fractions. (B) Heat maps of unique peptide retention time distributions plotted for each combined fraction in PFP and Hi-pH RP analyses. Each peptide was allocated to 1 of 100 bins based on its average retention time between 1 and 138 minutes. The white-blue scale represents 0–120 peptides in each bin. Replicate 1 is depicted. Dark blue diagonal bands across fractions are indicative of reduced orthogonality.
Fig. 3.
Comparison of method performance and reproducibility (A) Box and whisker plots of maximum, minimum and median peptide and protein identifications in triplicate experiments. Wilcoxon rank-sum tests were performed and differences in peptide (p = 0.2) and protein (p = 1) identifications between methods were not considered statistically significant. (B) Proportional Venn diagrams illustrating the reproducibility of peptide and protein identifications from each of the three replicate experiments for both Hi-pH RP and PFP. (C) Total peptide and protein identifications from the union of all three replicate experiments.
Physicochemical comparison of TMT labeled peptides derived from PFP or Hi-pH RP
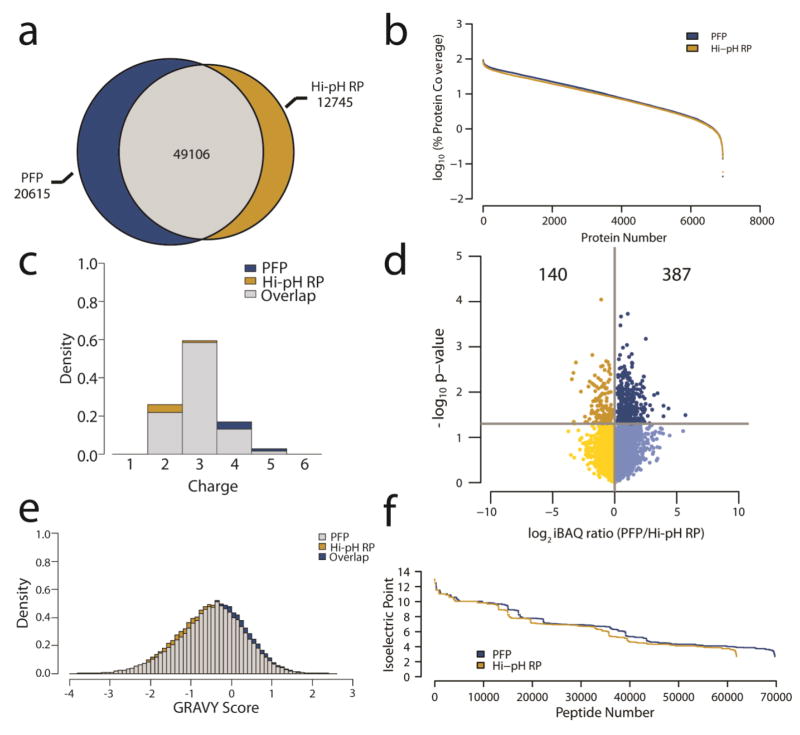
Direct comparison of TMT labeled peptide identities revealed that of the 82466 unique peptides identified in all three replicate PFP and Hi-pH RP experiments, 49106 (60%) were identified in both methods. Notably, 85% (69721) of all the peptides identified were identified via PFP fractionation, whereas this was 75% (61851) for Hi-pH RP (Fig. 4a). As noted above, the higher number of unique peptides identified through PFP fractionation did not result in a higher number of total proteins identified. This suggested that total protein coverage could be higher in PFP versus Hi-pH RP fractionation. Although the average protein coverage via PFP fractionation was 16.0% versus 14.0% by Hi-pH RP fractionation, this difference was not considered statistically significant (Fig. 4b).
Fig. 4.
Physicochemical properties of identified peptides (A) Proportional Venn diagram of the total unique peptide identifications from both fractionation methods. (B) Waterfall plot of protein coverage expressed as a log10 ratio of the percent of each protein sequence for which there were peptides identified. (C) Overlapping density histograms of peptide gas phase ion charge states identified in the analyses. (D) Volcano plot of the average log2 ratio of protein abundance (iBAQ) of three replicates in PFP versus Hi-pH RP. The y-axis is the -log10 of the p-value for each ratio derived from a Student’s t-test. Of the iBAQ ratios with p-values < 0.05, 387 are more abundant by PFP and 140 are more abundant by Hi-pH RP (E) Overlapping density histograms of the hydropathy (GRAVY score) of the peptides identified in each method. (F) Waterfall plot of isoelectric points for the peptides identified in PFP and Hi-pH RP fractionation. Differences between gas phase ion charge state (p = 0.1), isoelectric point (p = 1), GRAVY score (p = 0.1) and protein coverage (p = 0.2) were not considered statistically significant by Wilcoxon rank-sum tests.
While these data suggest that total peptide and protein identifications are similar, they do not provide insight into the physical and chemical characteristics of the peptides identified. These characteristics had the potential to be method specific, which could limit the utility of PFP as an alternative to Hi-pH RP fractionation. To evaluate this possibility, peptide ion gas phase charge states were compared between all TMT labeled peptides identified through each fractionation method. Ion charge states ranged from +2 to +5 for both fractionation methods, and there were no significant differences in charge state distribution from PFP to Hi-pH RP (Fig. 4c).
Intensity based absolute quantification (iBAQ) is an effective method for the estimation of protein abundances in proteomics experiments [1, 21]. In order to compare protein abundance estimates between PFP and Hi-pH RP fractionation, iBAQ ratios were compared for all 4671 proteins commonly identified in at least two of the three replicates in both methods. The requirement of multiple replicate identifications enabled the calculation of a p-value via a Student’s t-test to establish the reproducibility of each comparison. This assessment revealed that offline fractionation via PFP resulted in higher abundance estimates than Hi-pH RP fractionation for 73% (387/527) of the protein comparisons that were deemed reproducible (p-value < 0.05) (Fig. 4d).
In order to further assess whether any physical or chemical features are distinct to peptides fractionated via PFP or Hi-pH RP, the Kyte-Doolittle hydrophobicity index was used to compare total peptide hydrophobicity between methods [22]. The hydrophobicity score (GRAVY) distributions were almost entirely overlapping between PFP and Hi-pH RP fractionated peptides (Fig. 4e). Differences in isoelectric point distributions were also not significant between peptides identified using either method (Fig. 4f). These analyses suggest that the physicochemical characteristics of TMT labeled peptides fractionated with a PFP column were effectively indistinguishable from those fractionated via Hi-pH RP chromatography. Furthermore, PFP and Hi-pH RP offline fractionation methods result in similar protein identifications and are similarly reproducible. While the increase in total peptide identifications observed upon fractionation with PFP as opposed to Hi-pH RP did not result in a corresponding increase in protein identifications, there was a resultant increase in protein coverage, as well as total abundance, as approximated by iBAQ.
Comparisons of phosphopeptides fractionated by PFP and Hi-pH RP
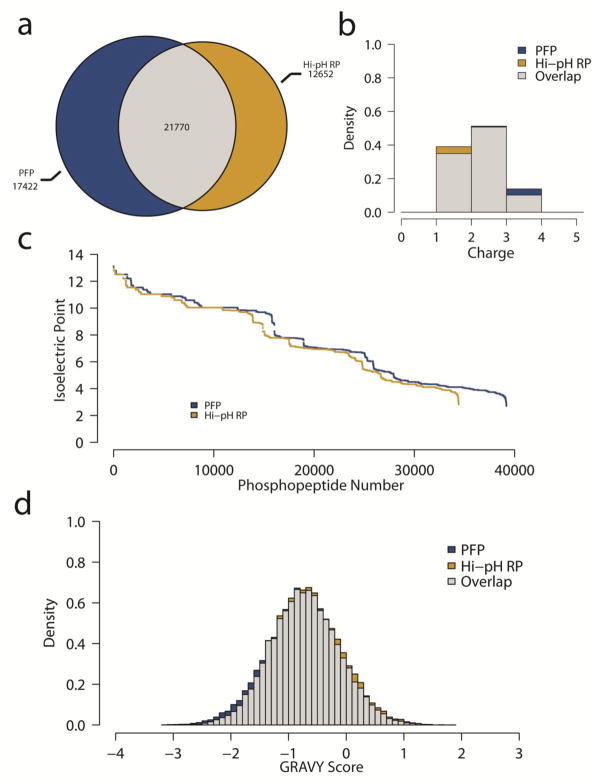
Peptides were isolated from phosphatase inhibitor treated HeLa cells that had been arrested in mitosis and further treated with phosphoprotein phosphatase inhibitors to increase global phosphorylation levels. Samples were enriched for phosphopeptides using TiO2 beads [16] and fractionated with either PFP or Hi-pH RP chromatography in a similar manner as indicated previously for TMT-labeled peptides to generate 12 combined fractions of phosphopeptides for each method. Also as before, Hi-pH RP fractions were desalted prior to LC-MS/MS, whereas PFP-separated phosphopeptides were dried to remove organic solvent and excess TFA, then resuspended and analyzed directly by Orbitrap Fusion mass spectrometry (ESM_10). Of the 51844 total phosphopeptides identified using either fractionation method, 75.6% (39192) were identified through PFP, whereas 66.4% (34422) were identified via Hi-pH RP (Fig. 5a). To assess any potential differences in the physicochemical characteristics of the phosphopeptides, gas phase ion charge state, isoelectric point and hydrophobicity were calculated and were virtually identical between separation methods (Fig. 5b, 5c, and 5d). These analyses revealed that fractionation via PFP is at least equally effective in facilitating the identification of phosphopeptides when compared to Hi-pH RP chromatography. Furthermore, complementarity of the phosphopeptide physicochemical characteristics between methods indicate that there is a general increase in phosphopeptide identifications using PFP versus Hi-pH RP, as opposed to a bias towards a particular chemical subset.
Fig. 5.
Phosphopeptide analysis (A) Proportional Venn diagram of the total phosphopeptide identifications from both fractionation methods. (B) Overlapping density histograms of phosphopeptide gas phase ion charge states (C) Waterfall plot of isoelectric points for the phosphopeptides identified in PFP and Hi-pH RP fractionation. (D) Overlapping density histograms of the hydropathy (GRAVY score) of the phosphopeptides identified in each method.
Discussion
Offline chromatographic fractionation prior to analysis by LC-MS/MS has proven to be an effective method for biological sample complexity reduction in proteome-wide shotgun sequencing experiments. However, the most frequently employed offline fractionation approaches (SCX and Hi-pH RP) require samples to be desalted before being analyzed by LC-MS/MS [16–17, 23]. This additional processing step is time consuming and can result in sample loss [12]. Here, we have evaluated PFP as an alternative to Hi-pH RP for offline fractionation of large scale proteomic and phosphoproteomic analyses. The compatibility of TFA as an ion-pairing agent in PFP chromatography allows for LC-MS/MS analysis without a post-fractionation desalting step.
We have observed PFP prefractionation to be at least as effective as Hi-pH RP in comprehensive shotgun proteomics analyses. Specifically, we determined that while total protein identifications via TMT labeled HeLa peptide spectral matching were equivalent using either PFP or Hi-pH RP, average protein coverage and abundance was higher for PFP fractionated samples (Fig. 4b, 4d). In addition, we observed higher numbers of phosphopeptide identifications when using PFP prefractionation versus Hi-pH RP.
Previous research has revealed a higher degree of orthogonality between Hi-pH RP and formic acid-based low-pH reverse-phase (RP-HPLC) than between PFP and RP-HPLC [8]. Our observations are consistent with these findings. We note that while PFP is orthogonal to RP-HPLC, Hi-pH RP does exhibit greater orthogonality (Fig. 2b). Since higher orthogonality should result in peak capacity increases in two-dimensional LC-MS/MS experiments, a reasonable expectation would be that Hi-pH RP HPLC would outperform PFP as an offline prefractionation method. However, the peptide and protein identification performances of the compared prefractionation methods were highly comparable. A possible explanation for the similar performance of Hi-pH RP and PFP in this report is that combining fractions from 48 to 12 in a manner intended to sample the full range of peptide hydrophobicity could normalize the effective peak capacity of the two methods, as has been suggested previously [8, 11]. Additionally, as mentioned previously, each sample handling step in a workflow can result in sample losses [12]. The potential losses from additional sample manipulations during desalting after Hi-pH RP prefractionation could offset gains in performance due to orthogonality. Ultimately, the experiments and data presented here allow us to assert that PFP is a viable alternative to Hi-pH RP for both proteomics and phosphoproteomics workflows, with equivalent or better performance and fewer processing steps.
Supplementary Material
Acknowledgments
The authors would like to thank Hildreth R. Frost for help with statistical considerations, Richard Avonti, Jason Gilmore and Sameh Magdeldin for help in generating Venn diagrams. The authors would like to acknowledge funding from the National Institutes of Health (R01-CA155260) to S.A.G.
Footnotes
Compliance with Ethical Standards
The authors declare no conflicts of interest.
References
- 1.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 2.Yates JR. Mass Spectrometry and the Age of the Proteome. Journal of Mass Spectrometry. 1998;33:1–19. doi: 10.1002/(SICI)1096-9888(199801)33:1<1::AID-JMS624>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Fonslow BR, Shan B, Moon-Chang B, Yates JR. Protein Analysis by Shotgun/Bottom-up Proteomics. Chem Rev. 2013;113(4):2343–2394. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malmstrom J, Lee H, Aebersold R. Advances in proteomic workflows for systems biology. Curr Opin Biotechnol. 2007;18(4):378–384. doi: 10.1016/j.copbio.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michalski A, Cox J, Mann M. More than 100,000 Detectable Peptide Species Elute in Single Shotgun Proteomics Runs but the Majority is Inaccessible to Data-Dependent LC-MS/MS. J Proteome Res. 2011;10:1785–1793. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- 6.McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, Rogers JC, Kuhn K, Pike I, Grothe RA, Blethrow JD, Gygi SP. Increasing the Multiplexing Capacity of TMTs Using Reporter Ion Isotopologues with Isobaric Masses. Anal Chem. 2012;84:7469–7478. doi: 10.1021/ac301572t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anagnostopoulos AK, Stravopodis DJ. TSANGARIS GT. Yield of 6,000 proteins by 1D nLC-MS/MS without pre-fractionation. Journal of Chromatography B. 2016:1–5. doi: 10.1016/j.jchromb.2016.08.031. [DOI] [PubMed] [Google Scholar]
- 8.Gilar M, Olivova P, Daly AE, Gebler JC. Orthogonality of Separation in Two-Dimensional Liquid Chromatography. Anal Chem. 2005;77:6426–6434. doi: 10.1021/ac050923i. [DOI] [PubMed] [Google Scholar]
- 9.Paulo JA, Gaun A, Gygi SP. Global Analysis of Protein Expression and Phosphorylation Levels in Nicotine-Treated Pancreatic Stellate Cells. J Proteome Res. 2015;14:4246–4256. doi: 10.1021/acs.jproteome.5b00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baths TS, Olsen JV. Offline Hi pH Reversed-Phase Peptide Fractionation for Deep Phosphoproteome Coverage. Methods Mol Biol. 2016;1355:179–192. doi: 10.1007/978-1-4939-3049-4_12. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Yang F, Gritsenko MA, Wang Y, Clauss T, Liu T, Shen Y, Monroe ME, Lopez-Ferrer D, Reno T, Moore RJ, Klemke RL, Camp DG, Smith RD. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics. 2011;11:2019–2026. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magdeldin S, Moresco JJ, Yamamoto T, Yates JR. Off-Line Multidimensional Liquid Chromatography and Auto Sampling Result in Sample Loss in LC/LC-MS/MS. J Proteome Res. 2014;13(8):3826–3836. doi: 10.1021/pr500530e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rauniyar N, Gao B, McClatchy DB, Yates JR. Comparison of protein expression ratios observed by sixplex and duplex TMT labeling method. J Proteome Res. 2013;12:1031–1039. doi: 10.1021/pr3008896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards A, Haas W. Multiplexed Quantitative Proteomics for High-Throughput Comprehensive Proteome Comparisons of Human Cell Lines. Methods Mol Biol. 2016;1394:1–13. doi: 10.1007/978-1-4939-3341-9_1. [DOI] [PubMed] [Google Scholar]
- 15.Cao JY, Xu YP, Cai XZ. TMT-based quantitative proteomics analyses reveal novel defense mechanisms of Brassica napus against the devastating necrotrophic pathogen Sclerotinia scleriotorum. J Proteomics. 2016;143:265–277. doi: 10.1016/j.jprot.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Kettenbach AN, Gerber SA. Rapid and reproducible single stage phosphopeptide enrichment of complex peptide mixtures: application to general and phosphotyrosine-specific phosphoproteomics experiments. Anal Chem. 2011;83:7635–7644. doi: 10.1021/ac201894j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paulo JA, O’Connell JD, Everley RA, O’Brien J, Gygi MA, Gygi SP. Quantitative mass spectrometry-based multiplexing compares the abundance of 5000 S. cerevisiae proteins across 10 carbon sources. Journal of Proteomics. 2016;148:85–93. doi: 10.1016/j.jprot.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chick JM, Munger SC, Simecek P, Huttlin EL, Choi K, Gatti DM, Raghupathy N, Svenson KL, Churchull GA, Gygi SP. Defining the consequences of genetic variation on a proteome-wide scale. Nature. 2016;534:500–505. doi: 10.1038/nature18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie R, Oleschuk R. Photoinduced polymerization for entrapping of octadecylsilane microsphere columns for capillary electrochromatography. Anal Chem. 2007;79(4):1529–1535. doi: 10.1021/ac061349t. [DOI] [PubMed] [Google Scholar]
- 20.Eng JK, Jahan TA, Hoopmann MR. Comet: An open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 21.Arike L, Valgepea K, Peil L, Nahku R, Adamberg K, Vilu R. Comparison and applications of label-free absolute proteome quantification methods on Escherichia coli. Journal of Proteomics. 2012;75:5437–5448. doi: 10.1016/j.jprot.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 22.Kyte J, Doolittle R. A simple method for displaying the hydropathic character of a protein. Journal of Molecular Biology. 1982;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 23.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR. Direct analysis of protein complexes using mass spectrometry. Nature Biotechnology. 1999;17(7):676–692. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 24.Senko MW, Remes PM, Canterbury JD, Mathur R, Song Q, Eliuk SM, Mullen C, Earley L, Hardman M, Blethrow JD, Bui H, Specht A, Lange O, Denisov E, Makarov A, Horning S, Zabrouskov V. Novel Parallelized Quadrupole/Linear Ion Trap/ Orbitrap Tribrid Mass Spectrometer Improving Proteome Coverage and Peptide Identification Rates. Analytical Chemistry. 2013;85(24):11710–1171. doi: 10.1021/ac403115c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.