Summary
Ten years ago, we discovered that microbiota composition controls intestinal T cell homeostasis and alters T cell responses of mice in different animal facilities. Here I discuss how these discoveries, reported in Cell Host & Microbe in 2008, came to be and contributed to our understanding of microbiota immune effects.
The microbiota is now recognized as an important modifier of host immunity in both steady state and disease. In the last decade, our understanding of this regulation has significantly evolved with multiple examples of microbiota effects on immune cell subset representation and function, and the identification of microbial taxons with immunomodulatory activities. The microbiota is also now de facto considered the default environmental factor to explain variability in immunological experiments.
However, in early 2006, when we initiated our studies, whether microbiota composition could control steady state T cell immune responses was not clear and commensal bacteria were not high on immunologists’ radars. Nevertheless, the microbiota was long known to affect multiple aspects of host physiology and metabolism, and to be required for maturation of the immune system. Pioneering studies with germ-free animals had already shown that immune-system development and functions are compromised at multiple levels in the absence of microbiota. It was also known that commensals contribute to disease phenotypes. For example, all inflammatory bowel disease (IBD) animal models require the presence of microbiota. However, most of these functions of the microbiota were generally ascribed to the presence of a diverse population of microbes that provided immune stimulation mostly through innate immune microbial pattern recognition receptors (PRRs), rather than the presence of unique species or types of microbes. It followed that a heterogeneous microbial community can recover the immune defects in germ-free mice and little was known of how the composition of this community might affect the nature of immune responses. In an extension to the hygiene hypothesis, it was postulated by a number of researchers that the composition of commensal microbiota, and even individual species in this community, might play important beneficial roles in host immunity, such as protection from allergy and asthma. Specific examples of such effects, however, were difficult to ascertain. The first examples came from groundbreaking studies by Sarkis Mazmanian and Dennis Kasper demonstrating that the human commensal Bacteroides fragilis can affect systemic Th1/Th2 balance and mucosal T cell IL-10 production in germ-free mice, thus showing that innocuous intestinal microbes can modulate systemic T cell responses and influence susceptibility to IBD (Mazmanian et al., 2005; Mazmanian et al., 2008). This suggested that immunomodulatory microbes might also be present in the endogenous commensal microbiota and serve as environmental modifiers of immune responses.
Enter Th17 cells
My entry into the microbiota field stemmed from our studies on the newly discovered Th17 cell subset of CD4 T cells. We discovered that RORγt is the master transcriptional regulator of Th17 cells (Ivanov et al., 2006). Th17 cells were, and still are, particularly popular, because they represent a novel helper T cell subset, in addition to Th1 and Th2 cells. However, although Th17 cells had been shown to produce unique effector cytokines and require unique cytokines for differentiation, a transcriptional regulator to establish them as a definitive T-cell subset was missing. RORγt had been a long-term interest in the Littman lab, and over the years the lab had generated a number of knockout and reporter lines for this molecule. These studies demonstrated crucial roles for RORγt in thymocyte and intestinal group 3 innate lymphoid cell (ILC3) development, but did not identify RORγt expression in mature T cells in the periphery (Eberl et al., 2004; Sun et al., 2000). I was interested in studying intestinal immunity and used an optimized protocol for isolation of intestinal lamina propria (LP) lymphocytes to study RORγt expression in RORγt-GFP mice. This revealed unexpected expression of RORγt in a large fraction of small intestinal LP CD4 T cells and γδ T cells. At the same time across the country, Dan Cua’s group at then Schering-Plough had identified RORγt in gene expression arrays of Th17 cells, and together we went ahead to show that the RORγt-expressing T cells in the gut are Th17 cells and IL-17-expressing γδ T cells, and that RORγt controls their differentiation (Ivanov et al., 2006). These experiments also suggested that the gut provides a unique environment for Th17 cell differentiation. To test this hypothesis, I examined RORγt and IL-17 expression in other peripheral tissues, including lung and liver, and showed that they do not contain a significant number of Th17 cells (Ivanov et al., 2008). We then asked what specific intestinal signals could control and restrict this process to the gut mucosa. Being T-cell immunologists, we hypothesized that this is due to the unique cytokine environment of the gut. But what creates this environment? The possibility that microbiota could be involved first occurred to me while looking at the development of Th17 cells during ontogeny. These experiments showed that Th17 cells were not present in neonates and appeared in the LP at around 3–4 weeks of age, shortly after weaning. There are two major environmental changes that occur at this developmental stage—diet and microbiota. Familiar with the work of Ruslan Medzhitov’s group on MyD88 effects on microbiota (Rakoff-Nahoum et al., 2004), I decided to treat my mice with their antibiotic cocktail. The results were encouraging, because there was a significant reduction of Th17 cells in treated mice, albeit only by 50%. Combining the two approaches, I decided to treat mice with the antibiotic cocktail from birth and this completely ablated Th17 cell development. Moreover, when antibiotic-treated mice were co-housed with wild-type mice they re-acquired Th17 cells. At this point we were hot on the microbiota trail. The definitive experiment required confirmation of the phenotype in germ-free animals. In late 2006 germ-free facilities were only available in a handful of places in the country. We approached Balfour Sartor at UNC, who headed one of the largest gnotobiotic facilities, supported by the NIH. Balfour was excited to help and hosted me in his lab to test his germ-free and gnotobiotic animals. Analyzing data at 3 AM is common for intestinal experiments, but this was a truly sleepless week. I will always remember running samples in the middle of the night in a ghost-like Chapel Hill campus the weekend before Christmas. It paid off. Germ-free animals were completely devoid of LP Th17 cells, and Th17 cells were induced by fecal transplants into these animals (Ivanov et al., 2008). Combined, these experiments unequivocally proved that microbiota drive Th17 cell differentiation (Ivanov et al., 2008). Similar results were independently reported, almost at the same time, by Kenya Honda’s group (Atarashi et al., 2008).
The main question remained: how does the microbiota induce Th17 cells? Most reported microbiota-immune effects are mediated by recognition of microbes by PRRs, such as toll-like receptors (Rakoff-Nahoum et al., 2004). However, none of the known PRRs seemed to be required for the Th17 cell response (this remains a mystery even today). So, we decided to first examine whether the phenotype is mediated only by certain bacteria, and therefore depends on microbiota composition rather than the presence of microbiota per se. The problem was how to study this, especially at a time when we knew very little about the commensal community and had no genetic tools or culture methods for isolating individual microbes. Since then, our knowledge about microbiota composition has increased immensely. However, the challenges of culturing and genetically manipulating individual microbes in order to study their immune effects still remain. For us, the actual breakthrough came from experiments aimed at investigating the role of cytokines in intestinal Th17 cell differentiation, in particular IL-23. However, this happened in a way no one could imagine.
IL-23 is a cytokine discovered by Fernando Bazan and Robert Kastelein at the then DNAX Research Institute. Further work on this cytokine, notably at DNAX and Genentech, jump-started the Th17 field when it was shown that IL-23 can induce IL-17 expression by T cells in vitro and that IL-23 knockout animals have defects in Th17-cell differentiation in models of autoimmune inflammation. IL-23 knockout mice were generated by our collaborator Dan Cua at DNAX/Schering-Plough who sent us animals to establish a colony. Notably, as it became important later, we received the breeding pairs from a Schering-Plough contract colony at the Jackson laboratory. As is still a common practice in many laboratories, homozygous knockout male and female mice were set to breed to each other in order to quickly generate large numbers of animals. The first few experiments had a conclusive result. The knockout animals almost completely lacked LP Th17 cells and therefore intestinal Th17 cell differentiation requires IL-23. Despite the fact that I had performed the experiment several times with identical results (and even prepared a manuscript figure to this effect), and despite the fact that this result made sense considering everything we knew about IL-23, I felt uneasy that the control animals I used were C57BL/6 (B6) mice from our colony. I knew that these animals came from Taconic farms, hence not from the same source as the IL-23 knockout animals. I was vaguely thinking that Taconic and Jackson B6 mice might have some genetic differences. Also, we already knew that microbiota was involved in Th17 cell induction, however there was no reason to think that there would be differences between SPF animals based on housing conditions (all animals I had ever examined in multiple laboratories across the country had Th17 cells present). Rather my decision to order Jackson B6 animals as controls for one last experiment was driven by an internal desire, hard-wired by my graduate training, to always use the best possible experimental control. Surprisingly, the first experiment comparing Jackson B6 to IL-23 knockout mice showed very little Th17-cell differentiation in both control and knockout animals. I quickly attributed this to a faulty intracellular IL-17 staining, labeled the experiment as failed, and ordered new Jackson B6 mice to repeat it. However, to control for reagent and/or staining problems I also included a B6 mouse from our colony. The results were both shocking and exciting. Control and knockout mice from Jackson still lacked significant Th17 cell population, in contrast to the Taconic/NYU B6 control, which, as expected, had high Th17 levels. This was the Eureka moment. Because we knew that microbiota is required, the results suggested that Taconic mice may contain — and Jackson mice lack — a Th17 cell-inducing component of the microbiota. In support of this hypothesis, co-housing Jackson and Taconic mice led to Th17 cell induction in the former and fecal transplants from Jackson and Taconic mice into germ-free animals had differential ability to recover mucosal Th17 cells (Ivanov et al., 2008). These results represented one of the first direct demonstrations that microbiota composition can modify steady-state mucosal adaptive immunity. They also gave us a handle for identifying the microbiota components involved.
Immunomodulatory commensal microbes
Of course, the immediate next question (also asked by the reviewers of the Cell Host & Microbe submission) was: what are the Th17 cell-inducing bacteria? 16S rRNA microbiota-sequencing methods were not sufficiently advanced or readily available at the time. With the help of Balfour Sartor, we examined candidate microbes in monocolonized mice, and heroic efforts in Brett Finlay’s laboratory at UBC using FISH identified certain phylum differences in the microbiota of Th17 cell-positive (Taconic) and negative (Jackson) mice. Despite these efforts, we could not answer the question definitively in this study; however, the manuscript drew attention to the role of microbiota composition in immune-cell function. Mazmanian and Kasper had previously demonstrated that a commensal microbe can skew systemic Th responses (Mazmanian et al., 2005). Our work, and the work of Kenya Honda, showed that microbiota composition regulates the level of steady-state mucosal Th17 cell responses in different mouse colonies. Together, these studies ignited an exciting search for various effects of the microbiota on immune cell development and function and for identification of microbiota members with immunomodulatory roles. A year after the Cell Host & Microbe manuscript was published, we reported segmented filamentous bacteria (SFB) as the Th17 cell-inducing component of Taconic microbiota (Ivanov et al., 2009). (This SFB study, which came out as a continuation of the Cell Host & Microbe study, is sometimes incorrectly referenced as showing the Taconic vs. Jackson differences. However, it should be noted that the Cell Host & Microbe manuscript contained the original demonstration). SFB were also independently shown to induce Th17 cells by Nadine Cerf-Bensussan’s group in France (Gaboriau-Routhiau et al., 2009). Shortly after, the Mazmanian group showed that B. fragilis promotes mucosal Treg function (Round and Mazmanian, 2010), and Kenya Honda’s group showed that certain commensal Clostridia induce colonic Tregs (Atarashi et al., 2011). The search for immunomodulatory bacteria is also being applied to human microbiota, and Treg and Th17 cell–inducing human microbes have been reported by several groups (Atarashi et al., 2015; Atarashi et al., 2011; Tan et al., 2016).
The Taconic versus Jackson problem
One of the most consequential observations of our Cell Host & Microbe study was that differences in microbiota between animals of the same genotype can instruct differences in immune responses, even at steady state. As discussed above, microbiota had long been associated with disease in experimental models, e.g. colitis, diabetes, or allergy. Composition of the microbiota had been proposed to affect these models. Studies, in particular in Balfour Sartor’s lab, showed that different commensal bacteria induce phenotypically distinct colitis in germ-free susceptible animals (Kim et al., 2005). However, whether individual bacteria can exert dominant effects in the context of complete microbiota was unclear and specific examples of microbiota composition–directed differences in immune phenotypes were lacking. The drastic difference in Th17 cell levels in B6 mice from two commercial vendors that we reported in the Cell Host & Microbe manuscript was arguably the first example of such effects (Ivanov et al., 2008). This has had long-term implications and has changed immunologists’ experimental practices and interpretations. Non-littermate controls, including commercially purchased animals, are often used for comparison to genetic models and this may lead to erroneous interpretations. In our case, we had initially concluded that all Th17 cells in the LP require IL-23 signaling. However, when IL-23 knockout animals were co-housed with SFB-positive controls (or later examined with littermates), only a slight reduction in Th17 cell numbers was observed — de facto reversing our initial interpretation (Figure 1) (Ivanov et al., 2008). After the Cell Host & Microbe manuscript, we and others showed that the presence of SFB has functional consequences and modifies susceptibility in multiple disease models, including models of mucosal and systemic bacterial, fungal, and protist infections, experimental autoimmune encephalomyelitis, diabetes, and rheumatoid arthritis. Similarly a number of groups have since reported microbiota-dependent differences in immune phenotypes at steady state and in disease models between Taconic and Jackson mice. It is to be expected that microbiota differences will also affect experimental outcomes in different research facilities. In fact, fluctuations in disease and immune phenotypes following changes of animal housing facilities have been ’dirty little secrets’ in the field for a long time. Fluctuations can occur even within a facility and therefore the importance of using littermate controls cannot be overstated. For example, simply assuming that Taconic mice have SFB and Jackson mice do not would be wrong. We have examined multiple rooms that house B6 animals in both Taconic and Jackson for the presence of SFB and found that some rooms in Taconic lack SFB, while some rooms in Jackson contain SFB (our unpublished data). Mostly due to the results of this study and the follow-up Cell study (Ivanov, et al, 2009), Taconic Farms currently includes SFB as part of their health screening. Virtually the only way to deal with this type of variability is to use the fundamental scientific principle of littermate controls. Therefore, co-housed littermate controls are mandatory for all experiments in our laboratory.
Figure 1. Variations in microbiota composition can skew experimental interpretation.
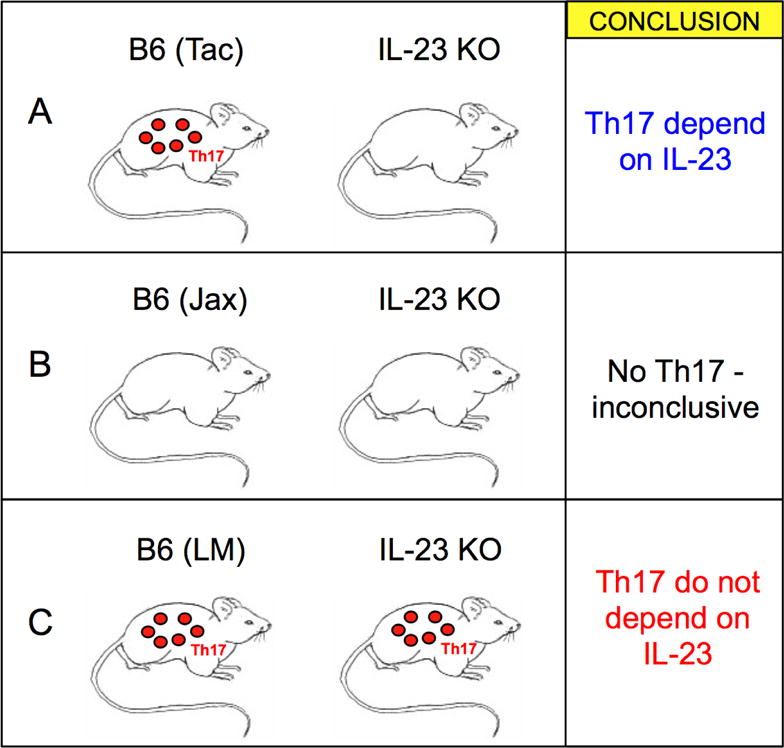
Investigating the role of IL-23 in LP Th17 cell differentiation is used as an example. A) WT B6 animals from Taconic Farms (Tac) compared to IL-23 knockout animals from Jackson Laboratory. B) Jackson (Jax) B6 animals compared to Jackson IL-23 knockout animals. C) Co-housing or crossbreeding of knockout animals to Taconic B6 mice shows little difference in Th17 cells when compared to Littermate controls (LM). Note that diametrically opposite conclusions are reached in A and C as a result of microbiota differences and differences in the experimental setup
Final thoughts
Understanding tissue immune networks and immune responses to resident microbes are fast developing fields of immunology. The knowledge of these interactions has considerably advanced since our Cell Host & Microbe paper in 2008. A multitude of studies explored the role of microbiota in immune defenses around the time of our publication, and we were fortunate to participate in the beginning of this scientific revolution, which, together with major advances in high throughput sequencing and computational biology, has led to a revival and refocusing of microbiota research. In only a few years the field has gone from a few known microbiota effects on host immunity to a profusion of examples of immune and physiological processes regulated by commensal bacteria. The next steps, to characterize the underlying mechanisms and develop strategies to harness them, have already begun.
My infatuation with science started with Paul de Kruif’s Microbe Hunters. The late 19th and early 20th century microbe hunters described in that book raised battles with the most dangerous microbes that wreaked havoc in our world. Today, a new generation of microbe hunters is going after the most domesticated microbes that have arguably even stronger effects on human health. It is a pleasure to be in their ranks.
Acknowledgments
I would like to thank all members of the Ivanov lab for their hard work and dedication through the years. I would particularly like to thank Dan Littman for giving me the opportunity to work in his laboratory and for his scientific input and support, without which these discoveries would not be possible. I would like to thank Kenya Honda who independently discovered the role of microbiota in Th17 cell differentiation and has been a gracious collaborator ever since. I would like to thank Balfour Sartor and Brett Finlay who were instrumental for the success of this study as well as all collaborators throughout the years. I would also like to acknowledge all of the scientists that have made significant contributions to the understanding of microbiota-immune effects and whose work I have not been able to discuss or reference here due to space and reference constraints. Research in my group is supported by NIH grants R01 DK098378, R21 AI126305 and P50 AR070588, The Pew Charitable Trust, Crohn’s and Colitis Foundation of America, and The Schaefer Research Program.
References
- Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, Littman DR. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol. 2004;5:64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J, Huycke MM, Sartor RB. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology. 2005;128:891–906. doi: 10.1053/j.gastro.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, Mebius RE, Littman DR. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- Tan TG, Sefik E, Geva-Zatorsky N, Kua L, Naskar D, Teng F, Pasman L, Ortiz-Lopez A, Jupp R, Wu HJ, et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E8141–E8150. doi: 10.1073/pnas.1617460113. [DOI] [PMC free article] [PubMed] [Google Scholar]


