Abstract
9-O-Acetylation is a common natural modification on sialic acids (Sias) that terminate many vertebrate glycan chains. This ester group has striking effects on many biological phenomena, including microbe-host interactions, complement action, regulation of immune responses, sialidase action, cellular apoptosis, and tumor immunology. Despite such findings, 9-O-acetyl sialoglycoconjugates have remained largely understudied, primarily because of marked lability of the 9-O-acetyl group to even small pH variations, and/or the action of mammalian or microbial esterases. Our current studies involving 9-O-acetylated sialoglycans on glycan microarrays revealed that even the most careful precautions cannot assure complete stability of the 9-O-acetyl group. We now demonstrate a simple chemical biology solution to many of these problems by substituting the oxygen atom in the ester with a nitrogen atom, resulting in sialic acids with chemically and biologically stable 9-N-acetyl group. We present an efficient one-pot multienzyme method to synthesize a sialoglycan containing 9-acetamido-9-deoxy-N-acetylneuraminic acid (Neu5Ac9NAc) and compare it to the one with naturally occurring 9-O-acetyl-N-acetylneuraminic acid (Neu5,9Ac2). Conformational resemblance of the two molecules was confirmed by computational molecular dynamics simulations. Microarray studies showed that the Neu5Ac9NAc-sialoglycan is a ligand for viruses naturally recognizing Neu5,9Ac2, with a similar affinity but with much improved stability in handling and study. Feeding of Neu5Ac9NAc or Neu5,9Ac2 to mammalian cells resulted in comparable incorporation and surface expression as well as binding to 9-O-acetyl-Sia-specific viruses. However, cells fed with Neu5Ac9NAc remained resistant to viral esterases and showed a slower turnover. This simple approach opens numerous research opportunities that have heretofore proved intractable.
Keywords: carbohydrate, chemical glycobiology, chemoenzymatic synthesis, one-pot multienzyme (OPME), sialic acid
Graphical abstract
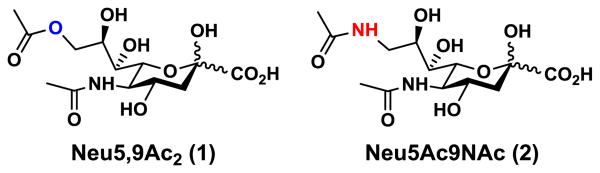
The first sialic acid discovered (crystallized by Gunner Blix from a hot mild acid extract of bovine submaxillary mucin in 1936) contained two acetyl groups, only one possibly attached to nitrogen.1 In retrospect, Blix likely isolated a 9-O-acetyl variant of the common sialic acid N-acetylneuraminic acid (Neu5Ac), 9-O-acetyl-N-acetylneuraminic acid (Neu5,9Ac2) (1, Figure 1). Neu5,9Ac2 along with Neu5Ac and N-glycolylneuraminic acid (Neu5Gc) are the three most frequently occurring sialic acid forms in mammals.2 It has long been known that 9-O-acetylation of sialic acids masks the recognition by influenza A virus hemagglutinin3 and by other lectins such as Factor H,4,5 CD22/Siglec-2,5-7 and Sialoadhesin/Siglec-1,7 while being required for the binding of influenza C virus hemagglutinin.3,8-11 Despite such early recognition and understanding of their importance, studies of the biological significance of sialoglycans presenting this common O-acetylated form of sialic acid have lagged far behind those of the parent molecules. The reasons are many, and have been detailed elsewhere, including their lability to both acidic and basic conditions (often used in standard purification methods for glycans and glycoconjugates), their propensity to migrate from one position to another, and their relative or absolute effects in blocking sialidase action.2,3,12 However, when it has been further studied, such O-acetyl group modification turns out to be a key determinant modulating recognition by viruses,3,13,14 antibodies,15-18 and mammalian lectins,6,7,17,19 as well as modulating sialidase action2,3,12 and cellular apoptosis.20-24
Figure 1.
Structures of 9-O-acetyl-N-acetylneuraminic acid (Neu5,9Ac2) (1) and its N-acetyl analog Neu5Ac9NAc (2).
Overall, exploration of the functions of Neu5,9Ac2 and its biological and pathological interactions has been greatly hampered by the chemical instability of the O-acetyl group and/or the esterase cleavage of such a group.8 Taken together with the fact that it is eliminated by basic conditions during conventional approaches to glycomic analysis such as beta-elimination and permethylation, it has come to the point where 9-O-acetylation tends to be simply ignored in many studies. Synthetic analogs of Neu5,9Ac2 with chemical modifications of the 9-position of Neu5Ac generated included a 9-N-acetyl analog, 9-acetamido-9-deoxy-N-acetylneuraminic acid (Neu5Ac9NAc) (2, Figure 1), which was shown to mimic Neu5,9Ac2 in binding to influenza C virus without being destroyed by esterase activity of the hemagglutinin-esterase.25 This observation was not however further explored for chemical and biological studies of Neu5,9Ac2. Herein, we report a more efficient chemoenzymatic method for synthesizing Neu5Ac9NAc and a sialoside containing this Neu5,9Ac2 analog. Applications in glycan microarray and cell feeding studies are presented, laying the foundation for a new approach to elucidate the important roles of 9-O-acetylation of sialic acids.
Results
Problems with Instability of O-Acetyl Sialic Acids in Preparing and Using Glycan Microarrays
The early described glycan microarrays did not have much representation of O-acetyl sialic acids.26 In an attempt to address this deficiency, our groups have been collaborating to synthesize, print, and study matched pairs of sialoglycans that either have or do not have O-acetyl esters. Due to their relative ease of synthesis and handling compared to 7-O- or 8-O-acetyl analogs, sialosides with 9-O-acetylated sialic acids were the ones studied first. Over the last few years we have been collaborating with several other groups to study the binding of various proteins to such glycans using glycan microarray technology.16-19,27-32 In revisiting some of these studies, we noticed that some of the 9-OAc-glycans used for microarray printing were partially de-O-acetylated.
Investigating and Minimizing 9-O-Acetyl Group Loss at Various Steps in Sample Preparation, Preparing and Using of Glycan Microarrays
Based on the above observations, we investigated the extent of de-O-acetylation at the final steps of the synthesis and entire glycan arraying process for the model 9-OAc-glycan Neu5,9Ac2α3Galβ4GlcβProN3 and its reduced form Neu5,9Ac2α3Galβ4GlcβProNH2. The azide-containing glycan was analyzed using nuclear magnetic resonance (NMR) spectrometry and high resolution mass spectrometry (HRMS). It was found to be pure, without any loss of OAc group. The compound was then subjected to hydrogenolysis using H2 and Pd-C in water with a drop of glacial acetic acid. The reaction was completed in one hour as detected using MS. To minimize de-O-acetylation during the celite filtration step which uses Na2CO3 as a mild base,33 filtration after catalytic hydrogenation was replaced by passing the reaction solution through a nylon syringe filter (0.2 μm). The product was then purified using a short C-18 cartridge (water as an eluant) instead of the commonly and previously used Bio-Gel purification, as it is faster. The fractions containing the pure product were combined and lyophilized. No noticeable loss of the O-acetyl group was observed using the improved procedures described above.
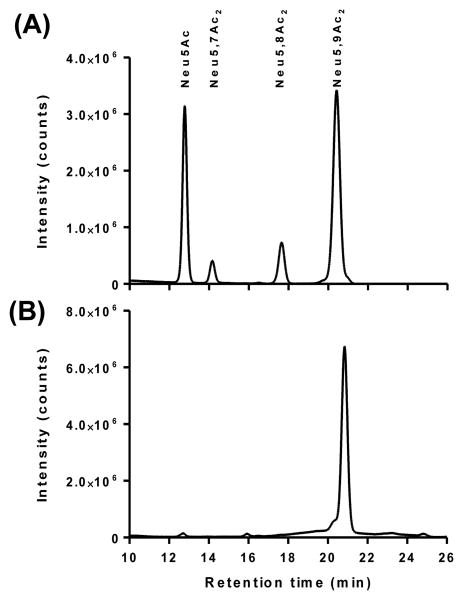
Then, the reduced glycan was quantified for the OAc loss at each step of glycan microarray process. Our results revealed that ∼45% of the OAc group was either lost from the 9-position of Neu5Ac or migrated to 7- and 8-positions of the sialic acid upon conventional analysis procedures including release of sialic acids with acetic acid, derivatization with 1,2-diamino-4,5-methylenedioxybenzene (DMB), followed by high performance liquid chromatography (HPLC) analysis of fluorescent adducts (Figure 2A). In comparison, Arthrobacter ureafaciens sialidase (AUS)34 treatment followed by DMB derivatization at low temperature35 showed only 1.5% loss of the OAc group (Figure 2B). This means there is no significant de-O-acetylation during initial preparation of stock solution in water (it should be noted that the final synthetic product is a neutral sodium salt - earlier studies in which the acid form was prepared was associated with some de-O-acetylation, data not shown) and AUS treatment with low temperature DMB derivatization. However, about 3% loss of OAc was observed during storage in phosphate buffer (pH 8.4) at room temperature, a condition popular for current microarray printing. Furthermore, about 5% of the sialic acid was de-O-acetylated under the standard blocking condition used for glycan arrays (ethanolamine in Tris-HCl buffer, pH 9.0). The glycan stock solution was also analyzed again after three months of storage at -20 °C and a few freeze-thaw cycles. It was noticed that another 4% of the OAc group was released over three months. Therefore, a small amount of de-O-acetylation occurred under the storage conditions and under conditions used for microarray printing and slide blocking. Any further loss of O-acetylation after printing during the binding and analysis, however, could not be directly monitored.
Figure 2.
DMB derivatization and HPLC analysis of sialic acids released from Neu5,9Ac2α3Galβ4GlcβProNH2 using acetic acid (A) and AUS (B).
Overall, definitive conclusions about binding of probes can therefore only be made where binding is exclusively to the O-acetylated sialoglycan spots and not to the corresponding non-O-acetylated ones, as discussed later. A more general conclusion is that, even under the most carefully managed handling by skilled experimentalists, some loss of 9-O-acetylation appears inevitable. Taken together with the knowledge that esterases of bacterial36-40 and vertebrate41-43 origin are very common in biological systems, there is an intractable problem facing the systematic study of the chemistry and biology of sialic acid 9-O-acetylation.
Chemical Synthesis of Neu5Ac9NAc Precursor 6-Acetamido-6-deoxy-N-acetylmannosamine (ManNAc6NAc) and Enzymatic Synthesis of Neu5Ac9NAc
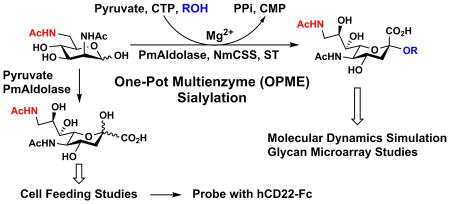
A new efficient chemoenzymatic synthetic strategy was developed for the synthesis of Neu5Ac9NAc. The design was to chemically synthesize ManNAc6NAc, a precursor of Neu5Ac9NAc, followed by a sialic acid aldolase-catalyzed reaction44,45 for the synthesis of desired Neu5Ac9NAc. ManNAc6NAc was synthesized in a 82% yield from previously obtained 6-azido-6-deoxy-N-acetylmannosamine (ManNAc6N3)46 using a simple one step reduction and simultaneous acetylation process achieved by adding thioacetic acid to a pyridine solution containing ManNAc6N3.47 Neu5Ac9NAc was readily obtained from ManNAc6NAc and pyruvate with a 78% yield using a reaction catalyzed by a sialic acid aldolase from Pasteurella multocida (PmAldolase).45
One-pot Multienzyme (OPME) Synthetic Approach for Facile Production of Neu5Ac9NAcα3Galβ4GlcβProNH2 for Glycan Microarray Studies
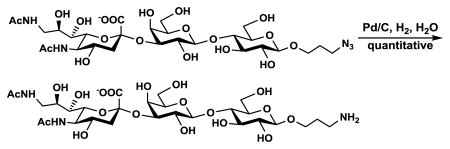
ManNAc6NAc was also used directly for synthesizing a sialoside analog Neu5Ac9NAcα3Galβ4GlcβProN3 via an efficient one-pot multienzyme (OPME) sialylation system46,48,49 containing PmAldolase,45 a CMP-sialic acid synthetase from Neisseria meningitidis (NmCSS),44 and Pasteurella multocida sialyltransferase 1 M144D mutant (PmST1 M144D).50 Neu5Ac9NAcα3Galβ4GlcβProN3 was obtained in 84% yield and was readily converted in a quantitative yield to Neu5Ac9NAcα3Galβ4GlcβProNH2 by catalytic hydrogenation using H2 in the presence of Pd/C.51
Glycan Microarray Studies of Viral Proteins and Human Siglec-9 Binding to Sialosides Containing Neu5,9Ac2 and Neu5Ac9NAc
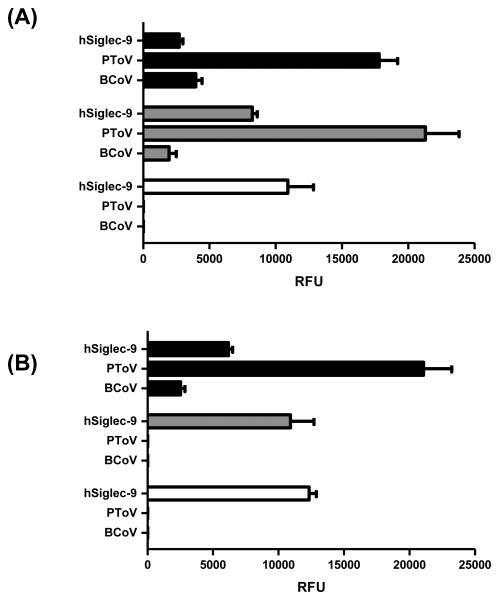
The synthesized glycan Neu5Ac9NAcα3Galβ4GlcβProNH2 was added to our previously described glycan library for glycan microarray studies.28 Proteins known to recognize Neu5,9Ac2 were tested including immunoglobulin Fc-fused human Siglec-9-Fc (hSiglec-9),52 as well as viral proteins (hemagglutinins in an esterase inactivated form) of porcine torovirus (PToV-P4-Fc, PToV) and bovine coronavirus (BCoV-Mebus-Fc, BCoV).13 The results were compared to those obtained using Neu5,9Ac2α3Galβ4GlcβProNH2, and Neu5Acα3Galβ4GlcβProNH2 lacking 9-modification at sialic acid as probes on the same slides (Figure 3). Results from glycan microarray studies (Figure 3A) showed that hSiglec-9 prefers binding to Neu5Ac-glycan without 9-modification at sialic acid (white column), but with observable binding to both Neu5Ac9NAc (black column) and Neu5,9Ac2 (grey column)-containing glycans. In comparison, both PToV and BCoV prefer binding to both sialoglycans with 9-modifications such as Neu5Ac9NAc and Neu5,9Ac2. BCoV is a known viral protein probe for 9OAc sialic acids (both Neu5,9Ac2 and Neu5Gc9Ac) and especially 7,9-di-OAc sialic acids.13 The more specific 9OAc-sialic acid viral probe PToV (prefers Neu5,9Ac2 over Neu5Gc9Ac)13 binds to both 9OAc and 9NAc glycans although it shows slightly stronger binding to the 9OAc derivative. The specific recognition of 9-modified sialic acid by esterase-inactive PToV and BCoV, the stability of 9NAc, and the esterase lability of 9OAc were evident by comparing the binding study results of esterase active PToV-treated samples (Figure 3B) with those non-treated ones (Figure 3A).
Figure 3.
Sialoglycan microarray binding specificity studies of human Siglec-9 (hSiglec-9-Fc, hSiglec-9), porcine torovirus hemagglutinin-esterase (PToV), and bovine coronavirus hemagglutinin-esterase (BCoV) (both PToV and BCoV were mutated to ablate their esterase activity) towards Neu5Ac9NAcα3Galβ4GlcβProNH2 (black columns), Neu5,9Ac2α3Galβ4GlcβProNH2 (gray columns), and Neu5Acα3Galβ4GlcβProNH2 (white columns) without esterase treatment (A) or treated with esterase active PToV (B).
Feeding of Free Neu5Ac9NAc and Neu5,9Ac2 to Human Cells Followed by Measuring Cell Surface Incorporation with PToV Probes
Human Burkitt lymphoma B (BJA-B K20) cells are hypo-sialylated due to the lack of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase,53 therefore maximizing exogenous sialic acid incorporation. After three days of feeding with either 1 mM of free Neu5Ac9NAc or Neu5,9Ac2, cells were probed with PToV-P4-Fc (PToV) with or without active esterase to measure cell surface incorporation of the sugars. Both Neu5Ac9NAc and Neu5,9Ac2 were detected on the cell surface, but only Neu5Ac9NAc-fed cells were resistant to virus-hemagglutinin-esterase activity (Figure 4).
Figure 4.
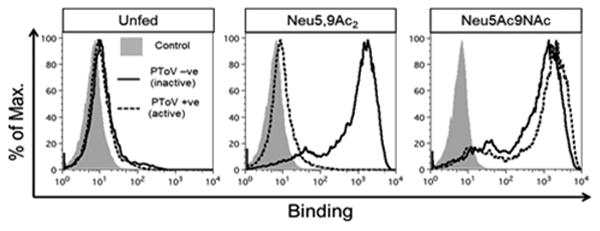
Detection of incorporation of Neu5,9Ac2 or Neu5Ac9NAc into hypo-sialylated human lymphoma BJA-B K20 cells. Cells were fed (or not) for 3 days with free Sias (1 mM) and then stained with PToV in inactive (mutated, solid black line) or active (dashed black line) O-acetylesterase forms. The binding was analyzed by flow cytometry.
Feeding of Free Neu5Ac, Neu5,9Ac2, Neu5Ac9NAc to Human BJA-B K20 Cells Followed by Measuring Cell Surface Incorporation of Sialic Acids with DMB Derivatization and HPLC Analysis
BJA-B K20 cells were fed with 1 mM of Neu5Ac (Figure 5A), Neu5,9Ac2 (Figure 5B), or Neu5Ac9NAc (Figure 5C) for 3 days. The cell membranes were recovered and sialic acids were released with Vibrio cholerae neuraminidase.34,54 The released sialic acids were derivatized with DMB and subjected to HPLC analysis which showed incorporation of each sugar on the cell membrane. Although BJA-B K20 cells are hypo-sialylated, they still express some Neu5Ac as can be observed on the cell membrane of Neu5,9Ac2 or Neu5Ac9NAc-fed cells (Figure 5B and 5C).
Figure 5.
DMB derivatization and HPLC analysis of sialic acids released from cell membranes of BJA-B K20 fed for 3 days with 1 mM of Neu5Ac (A), Neu5,9Ac2 (B) or Neu5Ac9NAc (C).
Monitoring Turnover of Neu5Ac9NAc and Neu5,9Ac2 on the Cell Surface
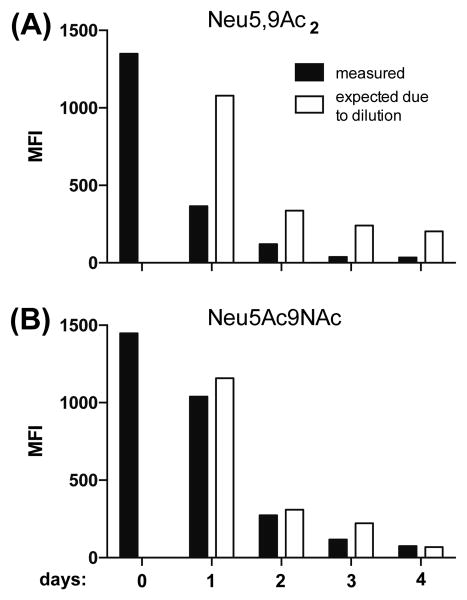
After feeding BJA-B K20 cells for two days with either 1 mM of free Neu5Ac9NAc or Neu5,9Ac2, the sugars were removed from the cell culture medium (day 0) and the presence of the sialic acids on the cell surface was monitored for 4 days with the PToV (esterase inactive) probe (Figure 6). The turnover of Neu5,9Ac2 on cell surface was shown to be faster than that of Neu5Ac9NAc.
Figure 6.
Monitoring turnover of incorporated Neu5,9Ac2 (A) and Neu5Ac9NAc (B) in BJA-B K20 cells. Cells were fed for two days with 1 mM of Neu5,9Ac2 or Neu5Ac9NAc. After two days, the feeding with Neu5,9Ac2 or Neu5Ac9NAc was stopped and the turnover of these sugars was measured with PToV (esterase inactive) probe by flow cytometry in the course of 4 days. The expected amount of sugar was calculated based on the initial MFI (Mean Fluorescence Intensity) at day 0 and the following cell doubling.
Feeding of Neu5Ac, Neu5,9Ac2, and Neu5Ac9NAc to Human BJA-B K20 Cells Followed by Measuring Cell Surface Incorporation with Human CD22-Fc
BJA-B K20 cells were fed with 3 mM of Neu5Ac, Neu5Ac9NAc, or Neu5,9Ac2. After 3 days of feeding, BJA-B K20 cells were probed with human CD22-Fc/Siglec-2 to detect ligands (Figure 7). It was found that there was a high expression of CD22 ligand when the cells were fed with Neu5Ac (Figure 7A). However, when the cells were grown in the presence of Neu5Ac9NAc, the expression of the CD22 ligand was minimal (Figure 7C). The level of CD22 ligand expressed on the cells fed with Neu5,9Ac2 (Figure 7B) was between the two which could be explained by possible partial de-O-acetylation of Neu5,9Ac2. This supports previous observations that human CD22-Fc recognizes α2–6-linked sialosides with Neu5Ac or N-glycolylneuraminic acid (Neu5Gc) as the preferred naturally existing sialic acid form and 9-O-acetyl modification on sialic acid blocks the binding.6,7 In addition, it demonstrates further that Neu5Ac9NAc can be a suitable substituent for natural Neu5,9Ac2 while has improved stability.
Figure 7.
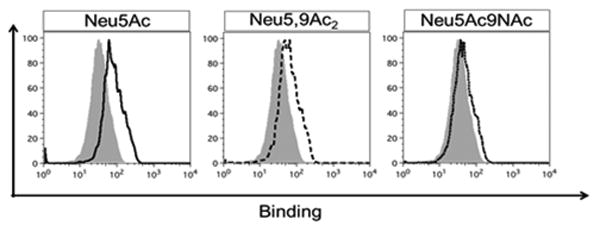
Probing Neu5Ac, Neu5,9Ac2 or Neu5Ac9NAc-fed human lymphoma BJA-B K20 cells with CD22-Fc to detect ligands. Cells were fed for 3 days with 3 mM of Neu5Ac (solid black line), Neu5,9Ac2 (dashed black line) or Neu5Ac9NAc (dotted line) and then stained with human CD22-Fc to detect ligands (and compared with non-fed cells, gray). The binding was analyzed by flow cytometry.
Comparison of Sialosides Containing Neu5Ac9NAc and Neu5,9Ac2 Using Molecular Dynamics
Classical molecular dynamics (MD) simulations of Neu5,9Ac2α3Galβ4GlcβProN3 and Neu5Ac9NAcα3Galβ4GlcβProN3 were carried out to investigate any differences that the chemical modification would introduce into the conformational ensemble in aqueous solution. The simulations were based on the GLYCAM carbohydrate force field55 and TIP3P water model,56 with added electrostatic parameters for describing the N-acetylation. The total length of the simulation exceeded 2 μs for each of the two sialosides studied, which has previously been noted to provide good sampling of oligosaccharide conformational degrees of freedom.57
Figure 8 shows the results of our simulations projected onto the (ϕ, ψ) dihedral angles of the glycosidic linkage between Sia and Gal. The main observation is that the conformational ensembles of the N-acetylated and O-acetylated sialosides are highly equivalent. There are three free energy basins on the positive ψ half of the plot with the same shape and the relative free energies between the basins are similar to within 1 kcal/mol.
Figure 8.
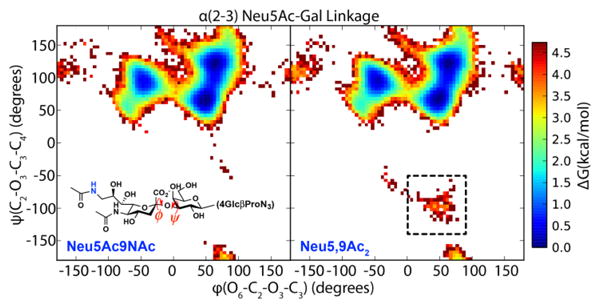
Free energy diagram of glycosidic linkage in solution for Neu5Ac9NAcα3Galβ4GlcβProN3 and the corresponding Neu5,9Ac2α3Galβ4GlcβProN3. The 9NAc and 9OAc analogs are compared in the left and right panels. The predominant states are almost identical for the two molecules, indicating that the 9NAc and 9OAc sialosides have similar structures in solution. The 9OAc sialoside has a sparsely populated state not seen in 9NAc (dotted box).
The classical MD simulation approximates the effect of the chemical modification as changing the force field parameters at the acetylation site. In particular, only the partial charges on the N-acetyl functional group are modified. To test this assumption, we used density functional theory to calculate the difference in electrostatic potential as a result of changing O-acetylation to N-acetylation.58 The results are shown in Figure 9 which confirm that the change in electrostatic potential is entirely localized to the acetylation site, which lends credence to the results of the classical simulations.
Figure 9.
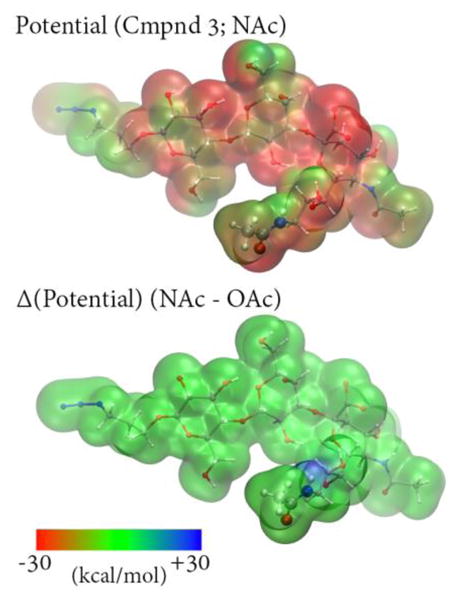
Top: Calculated electrostatic potential for a representative sialoside structure extracted from simulations of Neu5Ac9NAcα3Galβ4GlcβProN3 using the B3LYP/6-31G* level of theory60,61 and plotted on the ρ=0.002 density isosurface. Bottom: Electrostatic potential difference computed between Neu5Ac9NAcα3Galβ4GlcβProN3 and Neu5,9Ac2α3Galβ4GlcβProN3 plotted on the same isosurface; the O-acetylated potential was calculated by replacing NH with O in the N-acetylated structure. The difference between the potentials is very small, although NH group has a slightly positive potential relative to O.
The O-acetylated sialoside appears to display a slightly higher amount of flexibility, as there exists a cluster of conformations at 3–4 kcal/mol higher in energy than the minimum (dotted box in Figure 8) where ψ adopts negative values. These conformations represent far less than 1% of the whole ensemble, but are nonetheless interesting to investigate for understanding the effects of chemical modification. The overall conformational ensemble is shown in the top panel of Figure 10, and the negative- ψ conformations are shown in the bottom panel, indicating that the position and orientation of Sia differ significantly between the full set vs. subset of conformations. The sialoside spends most of its time in an extended conformation, but occasionally adopts a hairpin-like structure where the C7–C9 part of Sia bends back towards Glc. These hairpin conformations were not observed in the N-acetylated ensemble. The sialosides formed few intramolecular hydrogen bonds, which suggests that the chemical modification alters the conformational ensemble via the local hydrogen bonding network of the solvent;59 a more detailed analysis of solvent degrees of freedom is needed to test this hypothesis.
Figure 10.
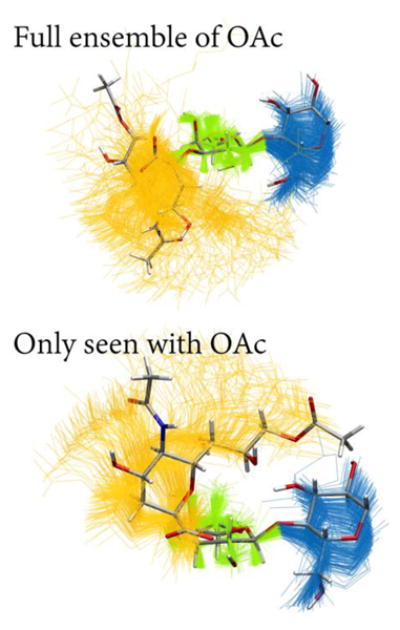
Superimposed conformations of the 9OAc sialoside simulations, showing only carbohydrate residues and aligned using galactose heavy atoms. One representative conformation is shown in licorice representation (C, gray; O, red; N, blue; H, white. Top: The full conformational ensemble of Neu5,9Ac2α3Galβ4GlcβProN3 with snapshots taken 10 ns apart (Neu5,9Ac2, yellow; Gal, green; Glc, blue). Bottom: The conformations corresponding to the dotted box in Figure 8 with snapshots taken 10 ps apart. These rarely seen conformations were only visited in the O-acetylated simulations but are only a small part of the ensemble, being higher in free energy.
Discussion
This study began with the realization that even with the best of precautions, some de-O-acetylation occurs during preparation of sialoglycan arrays. In retrospect, definite conclusions to date by us and by others about 9-O-acetylation on such arrays can only be made in instances where binding is exclusively to the O-acetylated sialoglycan spot, and not to the corresponding non-O-acetylated one.13 And even in this instance the ideal conclusive proof is to show loss of binding upon pretreatment with a Nidovirus 9-O-acetylesterase (such as PToV and BCoV).13 To be clear this does not overtly contradict our previous studies16-19,27-32 as we rarely commented about the role of O-acetylation except when selective binding occurred.13 But it is important to prevent inadvertent misinterpretation of some of our published raw data such as binding patterns and heat maps, by readers who may not know the subtleties and to alert others who may be studying these fascinating acetyl esters that modify important members of the vertebrate sialome.
Here we have presented a very practical solution to long-standing technical problems that have markedly inhibited progress in studies of the biology of 9-O-acetylated sialic acids. The strategy is replacing the labile OAc group with a more stable NAc group, which otherwise behaved functionally in a generally similar manner on microarrays, incorporation into cells after feeding, and subsequent expression on cell surfaces. The single obvious difference in both the array and cell feeding studies with or without binding studies using human CD22-Fc as a probe was the general resistance of the NAc group to both viral esterases and to endogenous cellular esterases. Of course in both instances we cannot rule out a very low level of esterase cleavage, and further studies are required.
The lack of cell lines that consistently express 9OAc sialic acids has also been a great limitation with cellular studies. The incorporation of the 9NAc molecules by feeding now allows one to ask if the cells become targets for infection by viruses that normally recognize the 9OAc group. Many other previously well-recognized but poorly studied biological or pathological roles of sialic acid O-acetylation may now become accessible. For example, while the disialoganglioside GD3 appears to protect cells from apoptosis, the unstable 9OAcGD3 is said to enhance apoptosis.20-24 If feeding of cells expressing GD3 results in expression of 9NAc-GD3, this phenomenon can now be better studied. Likewise the potential role of O-acetylation in regulating Siglec functions could also now be better explored.
The rapid turnover of the O-acetyl groups in fed cells is very likely due to the action of a cytosolic sialic acid esterase we previously described.42,62-64 We predict the NAc-form to be resistant to this enzyme. The slower but continued turnover of the N-acetyl form is likely due to continued dilution by cell doubling. Assuming that the Neu5Ac9NAc is first degraded into ManNAc6NAc, the question also arises as to whether this molecule can be further metabolized. Biochemical analysis can be used to track possible outcomes.
In principle, the same approach could be tried to address difficulties in the studies of 4-O-acetyl sialic acid and even more labile esters known to occur naturally at the 7- or 8- positions of sialic acids.2 A paper recently appeared showing an example of how this approach can work for 4-O-acetyl group.65 The approach could also potentially address the strong propensity of O-acetyl groups to migrate from one position to another.2,54,66,67 Even at near neutral/physiological pH values under which no significant de-O-acetylation occurred, the 7-O-acetyl group of Neu5,7Ac2 can easily migrate to the 9-position, producing Neu5,9Ac2 (possibly via an 8-position intermediate). Even more remarkably, the di-O-acetyl form Neu5,7,9Ac3 quickly yields a dynamic equilibrium mixture of Neu5,7,9Ac3 and Neu5,8,9Ac3, in a molar ratio of ∼1:1. Both the di-O-acetyl forms and the transient mono-8-O-acetyl form have been virtually impossible to study. While even more challenging to synthesize, N-acetyl versions of the 7,9-di-O-acetyl, 8,9-di-O-acetyl, 4,7-di-O-acetyl and 4,9-di-O-acetyl molecules that are known to occur in nature, as well as the 7,8,9-tri-O-acetyl entity, could be considered. Preparation of the NAc versions of these molecules could give the first hope of potentially understanding functions of the evanescent 8-O-acetyl group, and of differentiating between the recognition functions of 7,9 and 8,9 di-O-acetyl sialic acids, which are normally in a dynamic equilibrium state. Nevertheless, it remains to be seen if NAc analogs of such molecules recapitulate the conformation and biology of their natural counterparts, and also if they can all be efficiently incorporated by feeding the cells.
Conclusions
We present an efficient one-pot multienzyme method to synthesize a sialoglycan containing Neu5Ac9NAc and demonstrate that substituting the C9-oxygen atom in Neu5,9Ac2 with a nitrogen atom is an effective approach to study sialosides containing unstable 9-O-acetyl sialic acid. Computational molecular dynamics simulations, mammalian cell feeding with or without binding studies using human CD22-Fc as a probe, and glycan microarray studies showed that Neu5Ac9NAc-sialoglycan structurally and conformationally resembles Neu5,9Ac2-sialoglycan but with much improved stability in handling and study. This simple approach therefore opens up numerous research opportunities to gain a new understanding of the biology that depends on the modification of sialic acids with O-acetyl group at various positions that have heretofore proved intractable.
Methods
Materials
Cy3 and PE affinipure goat anti-human IgG (H+L) antibodies were obtained from Jackson ImmunoResearch Laboratories. Arthrobacter ureafaciens sialidase (AUS) and Vibrio cholerae neuraminidase were purchased from EY Labs and Roche, respectfully. Chemical and biological material were purchased from commercial sources and used as received. Nuclear magnetic resonance (NMR) spectra were recorded in the NMR facility of the University of California, Davis on a Bruker Avance-400 NMR spectrometer (400 MHz for 1H, 100 MHz for 13C). Chemical shifts are reported in parts per million (ppm) on the δ scale. High resolution electrospray ionization (ESI) mass spectra were obtained using a Thermo Electron LTQ-Orbitrap Hybrid MS at the Mass Spectrometry Facility at the University of California, Davis. Column chromatography was performed using RediSep Rf silica columns or an ODS-SM column (51 g, 50 μm, 120 Å, Yamazen) on the CombiFlash® Rf 200i system. Thin layer chromatography (TLC) was performed on silica gel plates (Sorbent Technologies) using anisaldehyde sugar stain for detection. Gel filtration chromatography was performed with a column (100 cm × 2.5 cm) packed with Bio-Gel P-2 Fine resins (Bio-Rad). Pasteurella multocida sialic acid aldolase (PmNanA), Neisseria meningitidis CMP-sialic acid synthetase (NmCSS), and Pasteurella multocida sialyltransferase 1 M144D mutant (PmST1 M144D) were expressed and purified as described previously.
Preparation of Hemagglutinin-Esterase Probes for 9-OAc Sia
The sequences for expressing the ectodomains of the hemagglutinin-esterase (HE) of two nidoviruses (PToV-P4 and BCoV-Mebus) were synthesized by GenScript (Piscataway, NJ), with varied degrees of insect codon optimization. To generate the probe molecules, the esterase domains were inactivated by changing the active site of the Ser residue to Ala by site-directed mutagenesis using Q5 mutagenesis (New England Biolabs). The HE proteins were linked to the baculovirus gp64 signal sequence peptide at their N-termini, and the C-terminus fused to a linker containing a thrombin cleavage sequence, the Fc domain of Human IgG1, and a 6-His sequence. Constructs were cloned into pFastBac-1 (Life Technologies) to generate recombinant bacmids following the manufacturer's protocol. Recombinant baculoviruses were recovered by transfection of the bacmids into Sf9 insect cells using Cellfectin II (Life Technologies). Viruses were used to infect suspension High Five cells and the supernatant harvested 2–3 days post-infection. The proteins were purified by binding to a HiTrap ProteinG HP 5ml column (GE Healthcare Life Sciences, Piscataway, NJ) and eluted with 0.1M citrate, pH 3.0 (pH neutralization to 7.8 with 1 M of Tris-HCl, pH 9.0) using ÄKTA FPLC system (GE Healthcare Life Sciences). The HE-Fc containing fractions were dialyzed in PBS and concentrated using 30 kD Amicon Ultra-15 filters (EMD Millipore). Purified proteins were stored at -80 °C in aliquots.
Cell Culture and Sugar Supplementation
BJA-B K20 cells were propagated as suspension with RPMI 1640 medium, supplemented with 10% heat-inactivated fetal calf serum, 2 mM glutamine, 100 U of penicillin per mL, 100 μg of streptomycin per mL in a humidified 5% CO2, 37 °C atmosphere as described previously.53 For medium supplementation, Neu5Ac9NAc and Neu5,9Ac2 were dissolved in PBS, titrated to a neutral pH and filter sterilized. The sugars were added at the indicated concentrations. For the sugar turnover experiments, the cells were fed as mentioned above and on day 0, the cell culture media was switched to RPMI 1640 medium, supplemented with 1% Nutridoma (Roche), 100 U of penicillin per mL and 100 μg of streptomycin without Neu5Ac9NAc and Neu5,9Ac2.
Flow Cytometry
Cells were washed with PBS and incubated with 10 μg/mL of PToV probes in PBS with 1% BSA and 10 mM EDTA for 30 minutes on ice. Subsequently, cells were washed with PBS and incubated with PE goat anti-human IgG antibody for 30 minutes on ice. After an additional washing step, the cells were analyzed by FACSCalibur (BD).
Synthesis of 2,6-Diacetamido-2,6-dideoxy-D-mannopyranose (ManNAc6NAc) from 2-Acetamido-6-azido-2,6-dideoxy-D-mannopyranose (ManNAc6N3)
To a solution of ManNAc6N346 (300 mg, 1.14 mmol) in pyridine (8 mL), thioacetic acid (2 mL) was added and the mixture was stirred at room temperature for 24 h. The solvent was concentrated in vacuo. The crude product was purified by column chromatography (ethyl acetate/methanol = 3:1) to produce ManNAc6NAc (262 mg, 82%) as a colorless amorphous solid. 1H NMR (400 MHz, D2O) α-isomer: δ 5.10 (d, J = 1.5 Hz, 1H), 4.31 (dd, J = 4.7, 1.5 Hz, 1H), 4.04 (dd, J = 9.8, 4.7 Hz, 1H), 3.96–3.86 (m, 1H), 3.68–3.33 (m, 3H), 2.07 (s, 3H), 2.03 (s, 3H); β-isomer: δ 5.00 (d, J = 1.7 Hz, 1H), 4.46 (dd, J = 4.5, 1.7 Hz, 1H), 3.82 (dd, J = 9.0, 4.5 Hz, 1H), 3.68–3.33 (m, 4H), 2.11 (s, 3H), 2.04 (s, 3H); 13C NMR (100 MHz, D2O) δ 175.70, 174.77, 174.54, 174.53, 93.01, 74.43, 71.73, 70.24, 68.58, 68.49, 68.20, 53.98, 53.21, 40.27, 40.16, 22.04, 21.90, 21.77; HRMS (ESI) Anal. Calcd for C10H19N2O2 [M+H]+: 263.1243, Found: 263.1241.
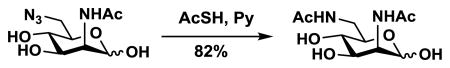
Synthesis of 5,9-Diacetamido-3,5,9-Trideoxy-D-glycero-D-galacto-2-nonulopyranosylonic Acid (Neu5Ac9NAc)
To a solution (10 mL) containing ManNAc6NAc (50 mg, 0.19 mmol), sodium pyruvate (210 mg, 1.9 mmol), and Tris-HCl buffer (100 mM, pH 7.5), PmAldolase (3.0 mg) was added and the reaction was incubated in an isotherm incubator for 48 h at 37 °C with agitation at 100 rpm. The reaction was quenched by adding the same volume of ice-cold ethanol and incubating at 4 °C for 1 h. The solvent was removed and the crude product was purified by column chromatography (ethyl acetate/methanol/water = 4:2:1) and followed by a short BioGel P-2 gel filtration column to give Neu5Ac9NAc (55.4 mg, 78%) as a colorless amorphous solid. 1H NMR (400 MHz, D2O) δ 4.06–3.92 (m, 3H, H-4, H-6, H-5), 3.80 (ddd, J = 9.0, 7.7, 3.1 Hz, 1H, H-8), 3.59 (dd, J = 14.1, 3.1 Hz, 1H, H-9), 3.44 (dd, J = 9.0, 1.1 Hz, 1H, H-7), 3.24 (dd, J = 14.1, 7.6 Hz, 1H, H-9), 2.22 (dd, J = 12.9, 4.8 Hz, 1H, H-3e), 2.06 (s, 3H, COCH3), 2.02 (s, 3H, COCH3), 1.83 (dd, J = 12.9, 11.4 Hz, 1H, H-3a); 13C NMR (100 MHz, D2O) δ 176.64, 174.68, 174.56, 96.36, 70.09, 69.77, 68.77, 67.25, 52.26, 42.76, 39.35, 22.08, 21.82; HRMS (ESI) Anal. Calcd for C13H21N2O9 [M-H]-: 349.1247, Found: 349.1255.
Synthesis of 3-Azidopropyl O-(5,9-Diacetamido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-D-galactopyranosyl-(1→4)-β-D-glucopyranoside (Neu5Ac9NAcα3Galβ4GlcβProN3)
Neu5Ac9NAcα3Galβ4GlcβProN3 was synthesized following the general procedure for enzymatic synthesis of sialosides. Galβ4GlcβProN346 (46 mg, 0.11 mmol), ManNAc6NAc (43 mg, 0.16 mmol), sodium pyruvate (91 mg, 0.83 mmol) and CTP (87 mg, 0.16 mmol) were dissolved in Tris-HCl buffer (100 mM, pH 8.5, 10 mL) containing 20 mM of MgCl2. The pH of the solution was further adjusted to 8.5 with 4 M NaOH. PmAldolase (1.5 mg), NmCSS (2.5 mg) and PmST1 M144D (2.5 mg) were added. The reaction was incubated in an isotherm incubator for 24 h at 37 °C with agitation at 100 rpm. The reaction was quenched by adding the same volume of ice-cold ethanol and incubating at 4 °C for 1 h. The formed precipitates were removed by centrifugation and the supernatant was concentrated. The residue was purified by passing through a BioGel P-2 gel filtration column followed by a C18 column (H2O/CH3CN = 10:1) (70.9 mg, Yield 84%). 1H NMR (400 MHz, D2O) δ 4.52 (dd, J = 12.8, 7.9 Hz, 2H), 4.10 (dd, J = 9.9, 3.2 Hz, 1H), 4.06–3.86 (m, 4H), 3.88–3.53 (m, 13H), 3.54–3.43 (m, 3H), 3.37–3.23 (m, 2H), 2.76 (dd, J = 12.4, 4.6 Hz, 1H), 2.04 (s, 3H), 2.04 (s, 3H), 1.92 (p, J = 6.6 Hz, 2H), 1.81 (t, J = 12.1 Hz, 1H); 13C NMR (100 MHz, D2O) δ 174.92, 174.39, 173.81, 102.60, 102.11, 99.87, 78.17, 75.53, 75.14, 74.78, 74.33, 72.79, 72.73, 69.96, 69.58, 69.36, 68.30, 67.46, 67.34, 61.01, 60.01, 51.66, 47.86, 42.11, 39.61, 28.22, 22.02, 21.81; HRMS (ESI) Anal. Calcd for C28H46N5O19 [M-H]-: 756.2787, Found: 756.2792.
Synthesis of 3-Aminopropyl O-(5,9-Diacetamido-3,5,9-trideoxy-D-glycero-α-D-galacto-2-nonulopyranosylonic acid)-(2→3)-O-β-D-galactopyranosyl-(1→4)-β-D-glucopyranoside (Neu5Ac9NAcα3Galβ4GlcβProNH2)
A catalytic amount of 10% palladium on charcoal (Pd/C) was added to the solution of Neu5Ac9NAcα3Galβ4GlcβProN3 (5.6 mg) in H2O (1 mL). The mixture was stirred under hydrogen atmosphere for 3 h. The solution was diluted with MeOH (2 mL) and passed through a filter to remove the catalyst. The solvent was concentrated in vacuo to give Neu5Ac9NAcα3Galβ4GlcβProNH2 (5.4 mg, quant.) as a colorless amorphous solid. HRMS (ESI) Anal. Calcd for C28H48N3O19 [M-H]-: 730.2882, Found: 730.2914.
Reduction of Neu5,9Ac2α3Galβ4GlcβProN3 to Neu5,9Ac2α3Galβ4GlcβProNH2
Neu5,9Ac2α3Galβ4GlcβProN3 (20 mg) was dissolved in H2O/MeOH (3 mL, 2:1 by volume) and a drop of glacial acetic acid was added. The mixture was stirred under hydrogen atmosphere in the presence of 10% palladium on charcoal for 1 h. The reaction mixture was then passed through a HyperSep C18 cartridge (1 g, 40–60 μm, Thermo) and eluted with water. The collection fraction containing the desired product was collected and lyophilized. The obtained white power was stored at -20 °C for long-term storage.
HPLC Analysis of Neu5,9Ac2α3Galβ4GlcβProNH2
Neu5,9Ac2α3Galβ4GlcβProNH2 was first hydrolyzed in glacial acetic acid (2 M) for 3 h at 80 °C. AUS hydrolysis was performed based on the company protocol. DMB derivatization was performed as it is reported previously.68 The DMB-derivatized samples were analyzed on a Dionex Ultra3000 HPLC System using a Phenomenex Gemini 5 μm C18 250 × 4.6 mm HPLC column at room temperature. The fluorescence was detected at 448 nm using excitation at 373 nm.
Glycan Microarray Screening
Glycan microarrays were fabricated using epoxide-derivatized slides (Corning by Thermo Fisher Scientific) and Arrayit SpotBot® Extreme Microarray Spotter as previously described.17,28,53 The arrays were printed with Stealth SMP3 microarray spotting pins from ArrayIt (Sunnyvale, CA, USA) generating 100 μm diameter spots. 4 pins were used; with each pin printing 4 replicate spots/well. The glycoconjugates were prepared at 100 μM concentration in an optimized printing buffer (300 mM phosphate buffer, pH 8.4). They were then distributed into a 384-well source plate in 20 μL per well. To monitor printing quality, 4 replicate-wells of human IgG (Jackson ImmunoResearch) was used at 100 μg/ml (in PBS) for each printing-pin. One complete array was printed on each slide (within approx 12 hour/∼28 slides). The humidity level in the arraying chamber was maintained at about 60–65% during printing. Printed slides were left on spotter deck overnight, allowing humidity to drop to ambient levels. Printed glycan microarray slides were blocked by 50 °C pre-warmed blocking solution (0.1 M Tris-HCl, 0.05 M ethanolamine, pH 9.0, 1 h). They were then washed twice with 50 °C pre-warmed water and dried. Slides were packed, vacuum-sealed and stored at RT until used.
To assure the quality of the printed slides, we used one or two slides for quality control using antibodies, lectins and serum samples with known glycan binding specificity. For glycan array binding, slides were fitted in a multi-well microarray hybridization cassette (AHC4X8S, ArrayIt, Sunnyvale, CA, USA) to divide into 8 subarrays. The subarrays were blocked with Ovalbumin (1% w/v) in PBS (pH 7.4) for 1 h at RT, with gentle shaking. For the esterase activity studies on the array, after removing the blocking solution, slides were first treated with 20 μg/mL PToV-P4-Fc (esterase active) in blocking buffer for 2 h at RT and then washed with PBS in 0.1% Tween 20 and then PBS (10 min/wash with shaking). Subsequently, diluted protein samples in blocking solution with various concentrations were added to each subarray (both esterase treated and non-treated slides). After incubating the protein samples for 2 h at RT with gentle shaking, the slides were washed with PBS in 0.1% Tween 20 and then PBS (10 min/wash with shaking). Cy3 affinipure goat anti-human IgG (H+L) antibody (Jackson ImmunoResearch) was used for the detection of BCoV-Mebus-Fc, PToV-P4-Fc and human Siglec-9-Fc. Diluted antibody in PBS was added to the subarrays and incubated for 1 h at RT. They were washed with PBS in 0.1% Tween 20, PBS and water (10 min/wash with shaking) and dried. The microarray slides were scanned using a Genepix 4000B microarray scanner (Molecular Devices Corp., Union City, CA, USA) at 100% laser power, PMT Gain 450 and 10 μM pixels. Data analysis was performed using Genepix Pro 7.0 analysis software (Molecular Devices Corp., Union City, CA) and the outputs were saved as gpr and jpg files. The gpr files then were saved as xls. The data were further analyzed with Excel. Local background subtraction was performed and data were plotted separately for each subarray. The binding specificity to glycoconjugates for each protein was plotted based on the RFU (Relative Fluorescence Units), average fluorescence value for 4 replicates, versus glycan IDs. The standard deviations were calculated and the error bars were found for each glycan binding. The final graphs (Figure 3) were plotted based on the tested proteins versus RFU for each glycan in GraphPad Prism 5.
Comparison of N-Acetylated and O-Acetylated Sialosides Using Molecular Dynamics
The molecular dynamics simulations were carried out using the AMBER15 software suite, principally using tleap for setup and pmemd.cuda for dynamics.69 The simulations employed the GLYCAM force field and TIP3P water model; the sialoside was solvated in a rectangular water box with roughly 1000 water molecules. The simulations used a 2.0 fs time step and a Langevin thermostat set to 298.15 K and a collision frequency of 1.0 ps-1. The equilibration simulations used a Berendsen barostat set to 1.0 atm, a compressibility of 44.6 × 10-6 bar-1 and a time constant of 1.0 ps-1; the production simulations were carried out at constant volume. The particle mesh Ewald method was used to treat long range electrostatics with a real-space cutoff of 8.0 Å. All input files and parameters are provided as part of the Supporting Information.
The parameters for the N-acetyl functional group were developed following the procedure outlined in Reference 70. The initial guess force field for Neu5Ac9NAc was constructed by replacing the 9-O-acetyl group with a copy of the 5-N-acetyl group and the corresponding force field parameters. After 100 ps of equilibration in the NPT ensemble, we generated 50 ns of NVT dynamics, and saved 50 snapshots at 1 ns intervals. These snapshots were energy-minimized in the TeraChem software package with the dihedral angles constrained, employing the restricted Hartree-Fock method and 6-31G* basis set.71-74 The energy-minimized geometries were used as input to a restrained electrostatic potential calculation carried out using the R.E.D. server using the RESP-C2 charge model as appropriate for the GLYCAM force field.55 In the charge optimization, the charges for the terminal hydroxyl group were kept at the initial values and all hydrogen charges set to zero. The resulting final set of charges on the N-acetyl group has a slightly different total charge from the original O-acetyl group; the charge on the carbon atom that the N-acetyl group is bonded to is increased by +0.0014, instead of the +0.008 value used for O-acetyl. An analogous calculation was carried out for the ProN3 functional group to obtain a complete set of charges for Neu5,9Ac2α3Galβ4GlcβProN3. The valence force field parameters for ProN3 were copied over from the general AMBER force field (GAFF)75 by comparing GLYCAM atom types to GAFF atom types.
After the parameterization was finished, the production simulations of Neu5,9Ac2α3Galβ4GlcβProN3 and Neu5Ac9NAcα3Galβ4GlcβProN3 were carried out to collect data for the free energy plots. The simulations were unbiased and each simulation ran for 2.5 μs, and configurations were saved every 10 ps. The free energy plot was constructed by projecting the ensemble of configurations onto the selected dihedral degrees of freedom.
Supplementary Material
Acknowledgments
This work was supported by United States National Institutes of Health grants R01HD065122 (to X.C.) and R01GM32373 (to A.V.).
Abbreviations
- AUS
Arthrobacter ureafaciens sialidase
- DMB
1,2-diamino-4,5-methylenedioxybenzene
- ESI
electrospray ionization
- HE
hemagglutinin-esterase
- HPLC
high performance liquid chromatography
- HRMS
hig resolution mass spectrometry
- ManNAc6NAc
6-acetamido-6-deoxy-N-acetylmannosamine
- MD
molecular dynamics
- MS
mass spectrometry
- Neu5Ac
N-acetylneuraminic acid
- Neu5,9Ac2
9-O-acetyl-N-acetylneuraminic acid
- Neu5Ac9NAc
9-acetamido-9-deoxy-N-acetylneuraminic acid
- NMR
nuclear magnetic resonance
- OPME
one-pot multienzyme
- Sia
sialic acid
- TLC
thin layer chromatography
Footnotes
Supporting Information. NMR spectra for products as well and computational residue files, parameter file, and input files for molecular dynamics simulations. This material is available free of charge via the Internet at http://pubs.acs.org.
H. Y. and X. C. are co-founders of Glycohub, Inc., a company focused on the development of carbohydrate based reagents, diagnostics, and therapeutics. Glycohub, Inc. played no role in the design, execution, interpretation, or publication of this study.
References
- 1.Lundblad A. Gunnar Blix and his discovery of sialic acids. Fascinating molecules in glycobiology. Upsala J Med Sci. 2015;120:104–112. doi: 10.3109/03009734.2015.1027429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schauer R. Chemistry, metabolism, and biological functions of sialic acids. Adv Carbohydr Chem Biochem. 1982;40:131–234. doi: 10.1016/s0065-2318(08)60109-2. [DOI] [PubMed] [Google Scholar]
- 3.Rogers GN, Herrler G, Paulson JC, Klenk HD. Influenza C virus uses 9-O-acetyl-N-acetylneuraminic acid as a high affinity receptor determinant for attachment to cells. J Biol Chem. 1986;261:5947–5951. [PubMed] [Google Scholar]
- 4.Varki A, Kornfeld S. An autosomal dominant gene regulates the extent of 9-O-acetylation of murine erythrocyte sialic acids. A probable explanation for the variation in capacity to activate the human alternate complement pathway. J Exp Med. 1980;152:532–544. doi: 10.1084/jem.152.3.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi WX, Chammas R, Varki NM, Powell L, Varki A. Sialic acid 9-O-acetylation on murine erythroleukemia cells affects complement activation, binding to I-type lectins, and tissue homing. J Biol Chem. 1996;271:31526–31532. doi: 10.1074/jbc.271.49.31526. [DOI] [PubMed] [Google Scholar]
- 6.Sjoberg ER, Powell LD, Klein A, Varki A. Natural ligands of the B cell adhesion molecule CD22 beta can be masked by 9-O-acetylation of sialic acids. J Cell Biol. 1994;126:549–562. doi: 10.1083/jcb.126.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelm S, Schauer R, Manuguerra JC, Gross HJ, Crocker PR. Modifications of cell surface sialic acids modulate cell adhesion mediated by sialoadhesin and CD22. Glycoconj J. 1994;11:576–585. doi: 10.1007/BF00731309. [DOI] [PubMed] [Google Scholar]
- 8.Herrler G, Rott R, Klenk HD, Muller HP, Shukla AK, Schauer R. The receptor-destroying enzyme of influenza C virus is neuraminate-O-acetylesterase. EMBO J. 1985;4:1503–1506. doi: 10.1002/j.1460-2075.1985.tb03809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrler G, Reuter G, Rott R, Klenk HD, Schauer R. N-acetyl-9-O-acetylneuraminic acid, the receptor determinant for influenza C virus, is a differentiation marker on chicken erythrocytes. Biol Chem Hoppe-Seyler. 1987;368:451–454. doi: 10.1515/bchm3.1987.368.1.451. [DOI] [PubMed] [Google Scholar]
- 10.Muchmore EA, Varki A. Selective inactivation of influenza C esterase: a probe for detecting 9-O-acetylated sialic acids. Science. 1987;236:1293–1295. doi: 10.1126/science.3589663. [DOI] [PubMed] [Google Scholar]
- 11.Enard W, Khaitovich P, Klose J, Zollner S, Heissig F, Giavalisco P, Nieselt-Struwe K, Muchmore E, Varki A, Ravid R, Doxiadis GM, Bontrop RE, Paabo S. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- 12.Schauer R. Characterization of sialic acids. Meth Enzymol. 1978;50:64–89. doi: 10.1016/0076-6879(78)50008-6. [DOI] [PubMed] [Google Scholar]
- 13.Langereis MA, Bakkers MJ, Deng L, Padler-Karavani V, Vervoort SJ, Hulswit RJ, van Vliet AL, Gerwig GJ, de Poot SA, Boot W, van Ederen AM, Heesters BA, van der Loos CM, van Kuppeveld FJ, Yu H, Huizinga EG, Chen X, Varki A, Kamerling JP, de Groot RJ. Complexity and diversity of the mammalian sialome revealed by nidovirus virolectins. Cell Rep. 2015;11:1966–1978. doi: 10.1016/j.celrep.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song H, Qi J, Khedri Z, Diaz S, Yu H, Chen X, Varki A, Shi Y, Gao GF. An Open Receptor-Binding Cavity of Hemagglutinin-Esterase-Fusion Glycoprotein from Newly-Identified Influenza D Virus: Basis for Its Broad Cell Tropism. PLoS Pathog. 2016;12:e1005411. doi: 10.1371/journal.ppat.1005411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheresh DA, Reisfeld RA, Varki AP. O-Acetylation of disialoganglioside GD3 by human melanoma cells creates a unique antigenic determinant. Science. 1984;225:844–846. doi: 10.1126/science.6206564. [DOI] [PubMed] [Google Scholar]
- 16.Padler-Karavani V, Tremoulet AH, Yu H, Chen X, Burns JC, Varki A. A simple method for assessment of human anti-Neu5Gc antibodies applied to Kawasaki disease. PloS One. 2013;8:e58443. doi: 10.1371/journal.pone.0058443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Padler-Karavani V, Song X, Yu H, Hurtado-Ziola N, Huang S, Muthana S, Chokhawala HA, Cheng J, Verhagen A, Langereis MA, Kleene R, Schachner M, de Groot RJ, Lasanajak Y, Matsuda H, Schwab R, Chen X, Smith DF, Cummings RD, Varki A. Cross-comparison of protein recognition of sialic acid diversity on two novel sialoglycan microarrays. J Biol Chem. 2012;287:22593–22608. doi: 10.1074/jbc.M112.359323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Q, Padler-Karavani V, Yu H, Chen X, Wu SL, Varki A, Hancock WS. LC-MS analysis of polyclonal human anti-Neu5Gc xeno-autoantibodies immunoglobulin G Subclass and partial sequence using multistep intravenous immunoglobulin affinity purification and multienzymatic digestion. Ana Chem. 2012;84:2761–2768. doi: 10.1021/ac2030893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Padler-Karavani V, Hurtado-Ziola N, Chang YC, Sonnenburg JL, Ronaghy A, Yu H, Verhagen A, Nizet V, Chen X, Varki N, Varki A, Angata T. Rapid evolution of binding specificities and expression patterns of inhibitory CD33-related Siglecs in primates. FASEB J. 2014;28:1280–1293. doi: 10.1096/fj.13-241497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birks SM, Danquah JO, King L, Vlasak R, Gorecki DC, Pilkington GJ. Targeting the GD3 acetylation pathway selectively induces apoptosis in glioblastoma. Neuro Oncol. 2011;13:950–960. doi: 10.1093/neuonc/nor108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malisan F, Franchi L, Tomassini B, Ventura N, Condo I, Rippo MR, Rufini A, Liberati L, Nachtigall C, Kniep B, Testi R. Acetylation suppresses the proapoptotic activity of GD3 ganglioside. J Exp Med. 2002;196:1535–1541. doi: 10.1084/jem.20020960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malisan F, Testi R. GD3 in cellular ageing and apoptosis. Exp Gerontol. 2002;37:1273–1282. doi: 10.1016/s0531-5565(02)00138-9. [DOI] [PubMed] [Google Scholar]
- 23.Bhunia AK, Schwarzmann G, Chatterjee S. GD3 recruits reactive oxygen species to induce cell proliferation and apoptosis in human aortic smooth muscle cells. J Biol Chem. 2002;277:16396–16402. doi: 10.1074/jbc.M200877200. [DOI] [PubMed] [Google Scholar]
- 24.Chen HY, Varki A. O-Acetylation of GD3: an enigmatic modification regulating apoptosis? J Exp Med. 2002;196:1529–1533. doi: 10.1084/jem.20021915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrler G, Gross HJ, Imhof A, Brossmer R, Milks G, Paulson JC. A synthetic sialic acid analogue is recognized by influenza C virus as a receptor determinant but is resistant to the receptor-destroying enzyme. J Biol Chem. 1992;267:12501–12505. [PubMed] [Google Scholar]
- 26.Feizi T, Fazio F, Chai W, Wong CH. Carbohydrate microarrays - a new set of technologies at the frontiers of glycomics. Curr Opin Struct Biol. 2003;13:637–645. doi: 10.1016/j.sbi.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Feng KH, Gonzalez G, Deng L, Yu H, Tse VL, Huang L, Huang K, Wasik BR, Zhou B, Wentworth DE, Holmes EC, Chen X, Varki A, Murcia PR, Parrish CR. Equine and canine influenza H3N8 viruses show minimal biological differences despite phylogenetic divergence. J Virol. 2015;89:6860–6873. doi: 10.1128/JVI.00521-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng L, Bensing BA, Thamadilok S, Yu H, Lau K, Chen X, Ruhl S, Sullam PM, Varki A. Oral streptococci utilize a Siglec-like domain of serine-rich repeat adhesins to preferentially target platelet sialoglycans in human blood. PLoS Pathog. 2014;10:e1004540. doi: 10.1371/journal.ppat.1004540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laubli H, Pearce OM, Schwarz F, Siddiqui SS, Deng L, Stanczak MA, Deng L, Verhagen A, Secrest P, Lusk C, Schwartz AG, Varki NM, Bui JD, Varki A. Engagement of myelomonocytic Siglecs by tumor-associated ligands modulates the innate immune response to cancer. Proc Natl Acad Sci U S A. 2014;111:14211–14216. doi: 10.1073/pnas.1409580111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng L, Song J, Gao X, Wang J, Yu H, Chen X, Varki N, Naito-Matsui Y, Galan JE, Varki A. Host adaptation of a bacterial toxin from the human pathogen Salmonella Typhi. Cell. 2014;159:1290–1299. doi: 10.1016/j.cell.2014.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scobie L, Padler-Karavani V, Le Bas-Bernardet S, Crossan C, Blaha J, Matouskova M, Hector RD, Cozzi E, Vanhove B, Charreau B, Blancho G, Bourdais L, Tallacchini M, Ribes JM, Yu H, Chen X, Kracikova J, Broz L, Hejnar J, Vesely P, Takeuchi Y, Varki A, Soulillou JP. Long-term IgG response to porcine Neu5Gc antigens without transmission of PERV in burn patients treated with porcine skin xenografts. J Immunol. 2013;191:2907–2915. doi: 10.4049/jimmunol.1301195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Padler-Karavani V, Hurtado-Ziola N, Pu M, Yu H, Huang S, Muthana S, Chokhawala HA, Cao H, Secrest P, Friedmann-Morvinski D, Singer O, Ghaderi D, Verma IM, Liu YT, Messer K, Chen X, Varki A, Schwab R. Human xeno-autoantibodies against a non-human sialic acid serve as novel serum biomarkers and immunotherapeutics in cancer. Cancer Res. 2011;71:3352–3363. doi: 10.1158/0008-5472.CAN-10-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chokhawala HA, Huang S, Lau K, Yu H, Cheng J, Thon V, Hurtado-Ziola N, Guerrero JA, Varki A, Chen X. Combinatorial chemoenzymatic synthesis and high-throughput screening of sialosides. ACS Chem Biol. 2008;3:567–576. doi: 10.1021/cb800127n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chokhawala HA, Yu H, Chen X. High-throughput substrate specificity studies of sialidases by using chemoenzymatically synthesized sialoside libraries. Chembiochem. 2007;8:194–201. doi: 10.1002/cbic.200600410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samraj AN, Pearce OM, Laubli H, Crittenden AN, Bergfeld AK, Banda K, Gregg CJ, Bingman AE, Secrest P, Diaz SL, Varki NM, Varki A. A red meat-derived glycan promotes inflammation and cancer progression. Proc Natl Acad Sci U S A. 2015;112:542–547. doi: 10.1073/pnas.1417508112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corfield AP, Williams AJ, Clamp JR, Wagner SA, Mountford RA. Degradation by bacterial enzymes of colonic mucus from normal subjects and patients with inflammatory bowel disease: the role of sialic acid metabolism and the detection of a novel O-acetylsialic acid esterase. Clin Sci. 1988;74:71–78. doi: 10.1042/cs0740071. [DOI] [PubMed] [Google Scholar]
- 37.Corfield AP, Wagner SA, O'Donnell LJ, Durdey P, Mountford RA, Clamp JR. The roles of enteric bacterial sialidase, sialate O-acetyl esterase and glycosulfatase in the degradation of human colonic mucin. Glycoconj J. 1993;10:72–81. doi: 10.1007/BF00731190. [DOI] [PubMed] [Google Scholar]
- 38.Steenbergen SM, Jirik JL, Vimr ER. YjhS (NanS) is required for Escherichia coli to grow on 9-O-acetylated N-acetylneuraminic acid. J Bacteriol. 2009;191:7134–7139. doi: 10.1128/JB.01000-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rangarajan ES, Ruane KM, Proteau A, Schrag JD, Valladares R, Gonzalez CF, Gilbert M, Yakunin AF, Cygler M. Structural and enzymatic characterization of NanS (YjhS), a 9-O-acetyl N-acetylneuraminic acid esterase from Escherichia coli O157:H7. Protein Sci. 2011;20:1208–1219. doi: 10.1002/pro.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phansopa C, Kozak RP, Liew LP, Frey AM, Farmilo T, Parker JL, Kelly DJ, Emery RJ, Thomson RI, Royle L, Gardner RA, Spencer DI, Stafford GP. Characterization of a sialate-O-acetylesterase (NanS) from the oral pathogen Tannerella forsythia that enhances sialic acid release by NanH, its cognate sialidase. Biochem J. 2015;472:157–167. doi: 10.1042/BJ20150388. [DOI] [PubMed] [Google Scholar]
- 41.Hayes BK, Varki A. O-Acetylation and de-O-acetylation of sialic acids. Sialic acid esterases of diverse evolutionary origins have serine active sites and essential arginine residues. J Biol Chem. 1989;264:19443–19448. [PubMed] [Google Scholar]
- 42.Takematsu H, Diaz S, Stoddart A, Zhang Y, Varki A. Lysosomal and cytosolic sialic acid 9-O-acetylesterase activities can Be encoded by one gene via differential usage of a signal peptide-encoding exon at the N terminus. J Biol Chem. 1999;274:25623–25631. doi: 10.1074/jbc.274.36.25623. [DOI] [PubMed] [Google Scholar]
- 43.Schauer R, Shukla AK. Isolation and properties of two sialate-O-acetylesterases from horse liver with 4- and 9-O-acetyl specificities. Glycoconj J. 2008;25:625–632. doi: 10.1007/s10719-008-9109-9. [DOI] [PubMed] [Google Scholar]
- 44.Yu H, Yu H, Karpel R, Chen X. Chemoenzymatic synthesis of CMP-sialic acid derivatives by a one-pot two-enzyme system: comparison of substrate flexibility of three microbial CMP-sialic acid synthetases. Bioorg Med Chem. 2004;12:6427–6435. doi: 10.1016/j.bmc.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Yu H, Cao H, Lau K, Muthana S, Tiwari VK, Son B, Chen X. Pasteurella multocida sialic acid aldolase: a promising biocatalyst. Appl Microbiol Biotechnol. 2008;79:963–970. doi: 10.1007/s00253-008-1506-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu H, Chokhawala H, Karpel R, Yu H, Wu B, Zhang J, Zhang Y, Jia Q, Chen X. A multifunctional Pasteurella multocida sialyltransferase: a powerful tool for the synthesis of sialoside libraries. J Am Chem Soc. 2005;127:17618–17619. doi: 10.1021/ja0561690. [DOI] [PubMed] [Google Scholar]
- 47.Shangguan N, Katukojvala S, Greenberg R, Williams LJ. The reaction of thio acids with azides: a new mechanism and new synthetic applications. J Am Chem Soc. 2003;125:7754–7755. doi: 10.1021/ja0294919. [DOI] [PubMed] [Google Scholar]
- 48.Yu H, Chokhawala HA, Huang S, Chen X. One-pot three-enzyme chemoenzymatic approach to the synthesis of sialosides containing natural and non-natural functionalities. Nat Protoc. 2006;1:2485–2492. doi: 10.1038/nprot.2006.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu H, Huang S, Chokhawala H, Sun M, Zheng H, Chen X. Highly efficient chemoenzymatic synthesis of naturally occurring and non-natural alpha-2,6-linked sialosides: a P. damsela alpha-2,6-sialyltransferase with extremely flexible donor-substrate specificity. Angew Chem Int Ed. 2006;45:3938–3944. doi: 10.1002/anie.200600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugiarto G, Lau K, Qu J, Li Y, Lim S, Mu S, Ames JB, Fisher AJ, Chen X. A sialyltransferase mutant with decreased donor hydrolysis and reduced sialidase activities for directly sialylating LewisX. ACS Chem Biol. 2012;7:1232–1240. doi: 10.1021/cb300125k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu H, Chokhawala HA, Varki A, Chen X. Efficient chemoenzymatic synthesis of biotinylated human serum albumin-sialoglycoside conjugates containing O-acetylated sialic acids. Org Biomol Chem. 2007;5:2458–2463. doi: 10.1039/b706507h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang JQ, Nicoll G, Jones C, Crocker PR. Siglec-9, a novel sialic acid binding member of the immunoglobulin superfamily expressed broadly on human blood leukocytes. J Biol Chem. 2000;275:22121–22126. doi: 10.1074/jbc.M002788200. [DOI] [PubMed] [Google Scholar]
- 53.Hinderlich S, Berger M, Keppler OT, Pawlita M, Reutter W. Biosynthesis of N-acetylneuraminic acid in cells lacking UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. Biol Chem. 2001;382:291–297. doi: 10.1515/BC.2001.036. [DOI] [PubMed] [Google Scholar]
- 54.Varki A, Diaz S. The release and purification of sialic acids from glycoconjugates: methods to minimize the loss and migration of O-acetyl groups. Ana Biochem. 1984;137:236–247. doi: 10.1016/0003-2697(84)90377-4. [DOI] [PubMed] [Google Scholar]
- 55.Vanquelef E, Simon S, Marquant G, Garcia E, Klimerak G, Delepine JC, Cieplak P, Dupradeau FY. RED Server: a web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 2011;39:W511–W517. doi: 10.1093/nar/gkr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 57.Sattelle BM, Almond A. Shaping up for structural glycomics: a predictive protocol for oligosaccharide conformational analysis applied to N-linked glycans. Carbohydr Res. 2014;383:34–42. doi: 10.1016/j.carres.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haranczyk M, Gutowski M. Differences in electrostatic potential around DNA fragments containing guanine and 8-oxo-guanine. Theor Chem Acc. 2007;117:291–296. doi: 10.1016/j.jmgm.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 59.Ren PY, Ponder JW. Polarizable atomic multipole water model for molecular mechanics simulation. J Phys Chem B. 2003;107:5933–5947. [Google Scholar]
- 60.Becke AD. Density-functional thermochemistry. 3. The role of exact exchange. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 61.Binkley JS, Pople JA, Hehre WJ. Self-consistent molecular-orbital methods. 21. Small split-valence basis-sets for 1st-row elements. J Am Chem Soc. 1980;102:939–947. [Google Scholar]
- 62.Varki A, Muchmore E, Diaz S. A sialic acid-specific O-acetylesterase in human erythrocytes: possible identity with esterase D, the genetic marker of retinoblastomas and Wilson disease. Proc Natl Acad Sci U S A. 1986;83:882–886. doi: 10.1073/pnas.83.4.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Higa HH, Diaz S, Varki A. Biochemical and genetic evidence for distinct membrane-bound and cytosolic sialic acid O-acetyl-esterases: serine-active-site enzymes. Biochem Biophys Res Commun. 1987;144:1099–1108. doi: 10.1016/0006-291x(87)91425-2. [DOI] [PubMed] [Google Scholar]
- 64.Butor C, Higa HH, Varki A. Structural, immunological, and biosynthetic studies of a sialic acid-specific O-acetylesterase from rat liver. J Biol Chem. 1993;268:10207–10213. [PubMed] [Google Scholar]
- 65.Bakkers MJ, Zeng Q, Feitsma LJ, Hulswit RJ, Li Z, Westerbeke A, van Kuppeveld FJ, Boons GJ, Langereis MA, Huizinga EG, de Groot RJ. Coronavirus receptor switch explained from the stereochemistry of protein-carbohydrate interactions and a single mutation. Proc Natl Acad Sci U S A. 2016;113:E3111–3119. doi: 10.1073/pnas.1519881113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kamerling JP, Schauer R, Shukla AK, Stoll S, Van Halbeek H, Vliegenthart JF. Migration of O-acetyl groups in N,O-acetylneuraminic acids. Eur J Biochem. 1987;162:601–607. doi: 10.1111/j.1432-1033.1987.tb10681.x. [DOI] [PubMed] [Google Scholar]
- 67.Diaz S, Higa HH, Hayes BK, Varki A. O-Acetylation and de-O-acetylation of sialic acids. 7- and 9-O-acetylation of alpha 2,6-linked sialic acids on endogenous N-linked glycans in rat liver Golgi vesicles. J Biol Chem. 1989;264:19416–19426. [PubMed] [Google Scholar]
- 68.Hara S, Yamaguchi M, Takemori Y, Furuhata K, Ogura H, Nakamura M. Determination of mono-O-acetylated N-acetylneuraminic acids in human and rat sera by fluorometric high-performance liquid chromatography. Ana Biochem. 1989;179:162–166. doi: 10.1016/0003-2697(89)90218-2. [DOI] [PubMed] [Google Scholar]
- 69.Salomon-Ferrer R, Case DA, Walker RC. An overview of the Amber biomolecular simulation package. Wiley Interdiscipl Rev Comput Mol Sci. 2013;3:198–210. [Google Scholar]
- 70.Kirschner KN, Yongye AB, Tschampel SM, Gonzalez-Outeirino J, Daniels CR, Foley BL, Woods RJ. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J Comput Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang LP, Song C. Geometry optimization made simple with translation and rotation coordinates. J Chem Phys. 2016;144:214108. doi: 10.1063/1.4952956. [DOI] [PubMed] [Google Scholar]
- 72.Ufimtsev IS, Martinez TJ. Quantum chemistry on graphical processing units. 1. Strategies for two-electron integral evaluation. J Chem Theory Comput. 2008;4:222–231. doi: 10.1021/ct700268q. [DOI] [PubMed] [Google Scholar]
- 73.Ufimtsev IS, Martinez TJ. Quantum chemistry on graphical processing units. 3. Analytical energy gradients, geometry optimization, and first principles molecular dynamics. J Chem Theory Comput. 2009;5:2619–2628. doi: 10.1021/ct9003004. [DOI] [PubMed] [Google Scholar]
- 74.Ufimtsev IS, Martinez TJ. Quantum chemistry on graphical processing units. 2. Direct self-consistent-field implementation. J Chem Theory Comput. 2009;5:1004–1015. doi: 10.1021/ct800526s. [DOI] [PubMed] [Google Scholar]
- 75.Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.