Abstract
Observed variations in drug responses among patients may result from differences in heritable genetic traits or from alterations in the epigenetic regulation of drug metabolizing enzymes and transporters (DMETs). MicroRNAs (miRNAs), a group of small non-coding RNAs, provide an epigenetic mechanism for fine-tuning the expression of targeted DMET genes by regulating the efficiency of protein translation and by decreasing mRNA stability via enhanced degradation. In the current study we systematically screened 374 important genes encoding DMETs for potential response elements to hsa-miR-29a-3p, a highly abundant miRNA in human liver. RNA electrophoresis mobility shift assays displayed direct interactions between hsa-miR-29a-3p and its cognate targets within the mRNA transcripts for the ABCC6, SLC22A7 and ALDH5A1 genes. The expression of luciferase reporter genes containing the 3′-UTRs of SLC22A7 or ALDH5A1 and the expression of endogenous SLC22A7 and ALDH5A1 were each suppressed by transfection with hsa-miR-29a-3p mimics. Importantly, chemically-induced up-regulation of hsa-miR-29a-3p correlated inversely with the expression of SLC22A7 and ALDH5A1. However, our studies failed to detect suppressive effects of hsa-miR-29a-3p on ABCC6 expression, which might be explained by the notion that the interaction of hsa-miR-29a-3p and ABCC6 mRNA was unable to recruit ribonucleoproteins to form a RNA-induced silencing complex.
Keywords: microRNA, hsa-miR-29a-3p, ALDH5A1, SLC22A7, Drug metabolizing enzymes and transporters, Pharmacogenomics
1. Introduction
Individual differences in drug efficacy and safety are a serious concern for public health. In the United States, over 770,000 serious adverse drug reactions (ADRs), including more than 111,000 deaths, were reported during 2013 in the FDA Adverse Events Reporting System (http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects/ucm070461.htm). Results from numerous studies published during last half century show that variable drug responses in the human population are associated with functional genetic variants (i.e., single nucleotide polymorphisms, SNPs) that affect the expression of drug metabolizing enzymes and transporters (DMETs), key proteins for the uptake, metabolism, and efflux of drugs [1,2]. In addition, individual variations in epigenetic regulatory machinery also contribute to variable DMET expression via altered DNA methylation, histone modifications, and other post-transcriptional effects [3,4]. Furthermore, endogenous factors [5,6], environmental factors, and drug-drug interactions are also involved in ADRs [7,8].
It is well known that inter-individual variability in DMET expression has a major impact on drug efficacy and side effects [1,9–11]; and the functional genomics of DMETs has been applied in rational therapeutics [12]. However, the prediction of drug response based on pharmacogenomics is not yet available or is far from reliable for most clinical drugs, indicating the complexity of individual differences in drug efficacy and safety.
Epigenetic regulation of DMET gene expression provides an additional genotype-independent molecular mechanism that may contribute to individual differences in drug response [13]. MicroRNAs (miRNAs), a group of small non-coding RNAs with a length of approximately 22 nucleotides, are a newly recognized type of epigenetic post-transcriptional regulator. Typically, miR-NAs suppress the expression of targeted genes either by inhibiting protein translation or by stimulating mRNA degradation via sequence–specific interactions with response elements present in the 3′-UTRs or coding regions of the targeted mRNA transcripts [4,14]. Mechanistic studies have demonstrated the involvement of miRNAs in regulating the expression of a number of DMET genes, such as CYP1B1 [15], CYP2E1 [16], CYP2C9 [17], CYP3A4 [18], and SULT1A1 [19]. However, additional studies are needed to elucidate specific mechanisms underlying the regulation of DMET gene expression by miRNAs to provide a more complete pharmacogenomic perspective on individual drug responses.
The miR-29 family, containing miR-29a, miR-29b and miR-29c members, shares a common seed region sequence that is predicted to target a large number of genes associated with diverse biological functions, including the regulation of cell proliferation, secretion and maintenance of extracellular matrix, cell differentiation and apoptosis [20], adaptive immune system functions [21], and bone remodeling [22]. miR-29 has also been reported to play critical roles in the pathogenesis of a variety of human diseases, including cancer [23], liver fibrosis [24], heart injury [20], and pulmonary fibrosis [25]. Liver fibrosis, a common feature of liver dysfunction caused by liver diseases (e.g., hepatitis C viral infection) or drug-induced liver injury (DILI), is usually associated with down-regulated DMET gene expression, resulting in decreased hepatic capacity for drug metabolism and disposition [26]. Therefore, miR-29 could serve as a therapeutic target for these diseases [27]. For example, recent evidence proved that the miR-29 family is activated by celecoxib [28] and by other chemotherapeutics that are involved in the suppression of metastasis in gastric cancer [29].
We recently showed that hsa-miR-29a-3p suppresses CYP2C19 expression in liver cells by targeting its coding region [30], suggesting that the miR-29 family might also modulate the expression of additional DMET genes. In this study, putative hsa-miR-29a-3p response elements in 374 commonly expressed DMET genes [31], were screened systematically using in silico analyses; and then a series of biochemical assays were applied to characterize the interactions between hsa-miR-29a-3p and the cognate sequences detected within DMET genes.
2. Materials and methods
2.1. Culture of cell lines
HepaRG cells, which express many key DMETs highly at levels similar to those found in primary hepatocytes, were selected to detect the regulatory roles of hsa-miR-29a-3p on endogenous DMETs production, while HepG2 and 293T cells were used to test the roles of hsa-miR-29a-3p on exogenous reporter genes expression. All media and supplements for cell culture were obtained from Life Technologies (Carlsbad, CA).
HepaRG cells (terminally differentiated hepatic cells) were obtained from Life Technologies. Terminally differentiated HepaRG cells were cultured in Williams’ E medium supplemented with thaw, plate, & general purpose medium supplement for one day, and then maintained in Williams’ E medium supplemented with the maintenance/metabolism medium supplement for additional seven days to prepare them for further experiments.
HepG2 cells were purchased from the American Type Culture Collection (ATCC; Manassas, VA), and grown in Rosewell Park Memorial Institute 1640 (RPMI 1640) medium supplemented with 10% fetal bovine serum (FBS). The 293T cells were obtained from Biosettia (San Diego, CA) and maintained in Dulbecco’s Modified Eagle medium (DMEM) supplemented with 10% FBS, 1 mM sodium pyruvate, and non-essential amino acids.
2.2. In silico analyses
A total of 374 DMET genes [31] were screened using the CellMiner™ database (version 1.5.1, http://discover.nci.nih.gov/cellminer) to select candidate genes whose expression levels correlated negatively (r <−0.250) with hsa-miR-29a-3p levels among the panel of NCI-60 cell lines [32]. Potential hsa-miR-29a-3p response elements were detected in the candidate DMET genes using the databases miRTar.human (http://mirtar.mbc.nctu.edu.tw/human/) and microRNA.org (http://www.microrna.org/). RNA-hybrid, a RNA hybridization algorithm (http://bibiserv2.cebitec.uni-bielefeld.de/rnahybrid), was used to predict the free energy of potential miRNA:mRNA duplexes. The GSE22058 public dataset was used to extract the RNA expression data for selected DMET genes and the expression level for hsa-miR-29a-3p in a set of 96 non-tumor liver tissues.
2.3. RNA electrophoretic mobility shift assays
All synthetic oligonucleotides or primers, conventional or covalently modified used in this study were obtained from Integrated DNA Technologies (Coraville, IA). The miRNA oligonucleotide hsa-miR-29a-3p: 5′-UAG CAC CAU CUG AAA UCG GUU A-3′ was labeled with cy5.5™ dye on its 5′ end, while the 2′-O-methyl-modified mRNA oligonucleotides, miR-29a-ABCC6-target: 5′-CAA AGC CAA GAU GGU GCU U-3′, miR-29a-ALDH5A1-target: 5′-UUU CUG UCC UGG UGU GGU GCU G-3′, and miR-29a-SLC22A7-target: 5′-GCU UCU UCU AGA GAU GGU GCU A-3′, corresponding to hsa-miR-29a-3p response elements in ABCC6, ALDH5A1, and SLC22A7, were labeled with IRDye®800 dye on their 5′ ends. Unlabeled miRNA negative control Cold-NC: 5′-UCA CAA CCU CCU AGA AAG AGU AGA-3′ or unlabeled hsa-miR-29a-3p oligonucleotides were utilized in the competition assays. Cytoplasmic extracts were prepared from terminally differentiated HepaRG cells using NE-PER Nuclear and Cytoplasmic extraction reagents (Thermo Scientific, Tewksbury, MA), following the manufacturer’s instructions.
As described in our previous study [17], RNA electrophoretic mobility shift assays (RNA EMSAs) were carried out according to the manufacturer’s protocol for the LightShift Chemiluminescent RNA EMSA Kit (Thermo Scientific). Briefly, the basic reaction mixtures contained 1× REMSA Binding Buffer, 5% glycerol, 200 mM KCl, 100 mM MgCl2, and 2 mM miRNA or/and cognate mRNA oligonucleotides. HepaRG cytoplasmic extracts (2 μg) and non-specific tRNA (1 μg) were added to the 20 μL basic reaction mixtures to form the RNA:protein complexes. Antibodies against Ago1 and Ago2 were obtained from Abcam (Cambridge, MA) and utilized in the supershift assays. Unlabeled oligonucleotides were used at 50-fold molar excesses in competition reactions. The reaction mixtures were incubated at room temperature for 20 min, separated by 12% native polyacrylamide gel electrophoresis (PAGE) at 4 °C, and labeled complexes were detected according to the manufacturer’s protocol for the Odyssey CLx Infrared Imaging System (LI-COR Biosciences, Lincoln, NE).
2.4. Luciferase reporter gene assays
The pGL3-CU plasmid containing the firefly luciferase reporter gene [17] was used to create reporter gene vectors. Briefly, the core 3′-UTRs of ALDH5A1 or SLC22A7, which harbor the response elements for hsa-miR-29a-3p, were created by PCR with the cloning primers ALDH5A1-F: 5′-GGG AAA GU TCC CAT CCC ATG GAT ACA GT-3′ and ALDH5A1-R: 5′-GGA GAC AU AGG TGC TTC GTC TCC ACC TA-3′, or SLC22A7-F: 5′-GGG AAA GU TGA CTT CCC AGA ATG CAG TG -3′ and SLC22A7-R: 5′-GGA GAC AU ATG AGA CCA GTG GGT TGG AG -3′, with extension oligonucleotides 5′-GGA GAC AU-3′ or 5′-GGG AAA GU-3′ at their 5′ end, and then subcloned into the linearized nicked pGL3-CU vector to prepare ALDH5A1-CU or SLC22A7-CU constructs, following the Universal USER Cassette protocol (New England Biolabs, Beverly, MA). In addition, the mutant constructs ALDH5A1-Mut and SLC22A7-Mut, with site-specific mutations in the hsa-miR-29a-3p response elements of the ALDH5A or SLC22A7 genes were created by site-directed mutagenesis using ALDH5A1-Mut-F: 5′-TGT CCT GGT GTA GTG CTG CTC CCT CAC CTC C-3′ and ALDH5A1-Mut-R: 5′-AGC AGC ACT ACA CCA GGA CAG AAA GAA CAG-3′ primers, or SLC22A7-Mut-F: 5′-TCT TCT AGA GAT GGT ACT AAA GAA AGG ACT AGC ATA TG-3′ and SLC22A7-Mut-R: 5′-CTT TAG TAC CAT CTC TAG AAG AAG CAG AAG CAC CTT A-3′ primers, respectively. The nucleotide sequences of all plasmid constructs used in this study were determined to confirm their authenticity.
HepG2 or 293Tcells were seeded into 96-multiwell plates. After reaching approximately 80% confluence, cells were transfected with constructed reporter gene plasmids (100 ng/well), together with hsa-miR-29a-3p mimic (final concentration: 50 nmol/L) or miRNA negative control (final concentration: 50 nmol/L) (Thermo Scientific), using the Lipofectamine 2000 reagent (Life Technologies). Transfection efficiencies were normalized to Renilla luciferase activity measurements expressed by co-transfection with 1 ng/well pRL-SV40 plasmid (Promega, Madison, WI). Firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter 1000 Assay System (Promega). Three independent transfection experiments were performed in triplicate for each combination of nucleic acids.
2.5. Exogenous ABCC6 expression assays
The pCMV6-ABCC6 plasmid was purchased from OriGene Technologies (Rockville, MD) and used to express the ABCC6 protein in transfected HepG2 and 293T cells. Cells were seeded in 24-multiwell plates and propagated until the coverage reached 70% and then transiently transfected with pCMV6-ABCC6 plasmid (300 ng) along with 50 nmol/L (final concentration) hsa-miR-29a-3p mimic or miRNA negative control using Lipofect-amine reagent. Each assay was performed at least three times.
2.6. Transfection assays and chemical treatments in HepaRG cells
HepaRG cells were seeded in 24-multiwell plates (6 × 105 cells per well), cultured for eight days as described above, and then transiently transfected with the hsa-miR-29a-3p mimic (final concentration: 50 nmol/L) or miRNA negative control (final concentration: 50 nmol/L) using Lipofectamine 2000 reagent (Life Technologies), and cells were harvested 48 h after transfection. N-(4-(9-acridinylamino)-2-methoxyphenyl)methanesulfonamide methanesulfonate (NSC-156306) and ((3aR,4S,9bS)-8-chloro-2,3,3a,4,5,9b-hexahydro-1H-cyclopenta[c]quinolin-4-yl)-phenyl-methanone (NSC-642957) were obtained from the Developmental Therapeutics Program (DTP) of the National Cancer Institute (NCI). Chemical compounds were dissolved in dimethyl sulfoxide (DMSO) to form 20 μmol/L NSC-156306 or NSC-6642957 stock solutions that were added to the culture medium of HepaRG cells to produce final concentrations of 0, 10, or 100 nmol/L. Cells were harvested to extract total RNA or proteins for further experiments 48 h after transfection or chemical treatments. Each assay was performed at least three times.
2.7. Real-time PCR assays of DMET RNA
The miRNeasy Mini Kit (Qiagen, Valencia, CA) was used to isolate total RNA from HepG2, 293T or HepaRG cells. QuantiTect Reverse Transcription Kit (Qiagen) and NCode™ microRNA First-Strand cDNA Synthesis Kit (Life Technologies) were used to create the first-strand cDNA from isolated mRNA and miRNA samples. Real-time quantitative reverse transcription-PCR (qRT-PCR) was carried out using the QuantiFast SYBR®Green RT-PCR Kit (Qiagen) with an ABI Prism7900 Sequence Detection System (Applied Biosystems) according to the manufacturer’s recommendations, with the primers ABCC6-RT-F: 5′-GGC TTG AAA AGG AGT GGA TG-3′ and ABCC6-RT-R: 5′-CTC GGT CTC TGG AGC CTT C-3′ for the ABCC6 gene, primers ALDH5A1-RT-F:5′-GCT GCC TAC GAG GCT TTC T-3′ and ALDH5A1-RT-R: 5′-GGC AAG GTC ATC CTT ATT TTG-3′ for the ALDH5A1 gene, primers SLC22A7-RT-F: 5′-TTG TAC CCT ACG GTG CTC AG-3′ and SLC22A7-RT-R: 5′-CAC ACT CCA TCC AGC AAG G-3′ for the SLC22A7 gene, or primers GAPDH-RT-F: 5′-GAA ATC CCA TCA CCA TCT TCC AGG-3′ and GAPDH-RT-R: 5′-GAG CCC CAG CCT TCT CCA TG-3′ for the GAPDH gene. Besides, the miR-29a-RT-F: 5′-TAG CAC CAT CTG AAA TCG GTT A-3′ and U6-F: 5′-CTC GCT TCG GCA GCA CA-3′ primers were used to detect the hsa-miR-29a-3p and U6 levels, respectively, according to the NCode™ miRNA First-Strand cDNA Synthesis Kit protocol. The relative RNA levels for DMET genes (ABCC6, ALDH5A1, and SLC22A7) or hsa-miR-29a-3p were calculated compared to GAPDH or U6 RNA levels.
2.8. Western blotting
Detergent lysis buffer (RIPA, Thermo Scientific) was used to isolate total proteins from HepG2, 293T and HepaRG cells. Antibodies against the DDK tag or GAPDH were purchased from Abcam (Cambridge, MA), while antibodies against ABCC6, ALDH5A1, and SLC22A7 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Quantitative analyses for western blotting were carried out using the Odyssey CLx Infrared Imaging System.
2.9. Statistical analyses
Pearson correlation analysis was used to calculate the correlation between DMET genes and hsa-miR-29a-3p RNA levels in the GSE22058 public dataset. Differences between subgroups in luciferase activity and protein or RNA levels for DMET genes were detected by t test, and P <0.05 was considered statistically significant.
3. Results
3.1. Selection of candidate DMET genes modulated by hsa-miR-29a-3p
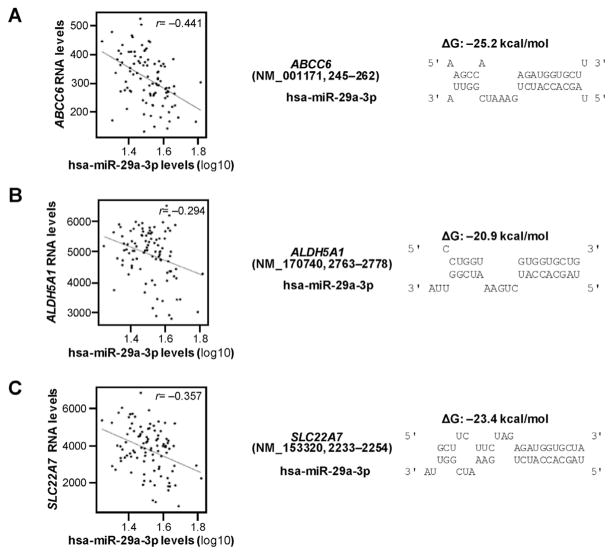
The correlations between mRNA expression levels for the 374 DMET genes and the expression of hsa-miR-29a-3p in NCI-60 cell lines using the CellMiner™ database were tested in the first step in our selection process. This procedure indicated that the expression levels for 56 DMET genes were negatively correlated with the expression of hsa-miR-29a-3p (detailed list available on request). Next, the microRNA.org database and the miRTar Human Program were applied to test whether or not these 56 DMET genes contained potential hsa-miR-29a-3p response elements in their 3′-UTRs or elsewhere in their full length mRNA transcripts. As shown in Table 1, our approach predicted that 11 genes were likely to be regulated by hsa-miR-29a-3p based on matches from at least one database. We then calculated the free energy of binding between hsa-miR-29a-3p and its potential response elements in the mRNA transcripts for each of these genes using RNAhybrid software. Next, we evaluated the correlation between the expression of the 11 selected DMET genes and the expression of hsa-miR-29a-3p in liver tissues (non-tumor). In addition to CYP2C19 that was reported in our previous manuscript, we identified three additional DMET genes, ABCC6, ALDH5A1, and SLC22A7, as potential hsa-miR-29a-3p targeted genes based on two criteria: (1) free energy of hybridization less than −20 kcal/mol (suggesting a high binding affinity, Fig. 1) and (2) mRNA expression for that gene is negatively correlated (r <−0.2) with the expression of hsa-miR-29a-3p level in liver tissues (Fig. 1 and Table 1). Thus, our multi-step selection process predicted that hsa-miR-29a-3p might regulate the expression of ABCC6, ALDH5A1 and SLC22A7 by targeting cognate miRNA response elements located in these genes.
Table 1.
DMET genes potentially targeted by hsa-miR-29a-3p.
| Gene symbol | Location | hsa-miR-29a-3p targeting site
|
Free energy (kcal/mol)a | Correlation (r)
|
||
|---|---|---|---|---|---|---|
|
|
|
|||||
| miRTar.Human | microRNA.org | CellMiner™ | GSE22058 | |||
| ABCC6 | 16p13.11 | Exon | N/A | −25.2 | −0.330 | −0.441 |
| SLC22A7 | 6p21.1 | 3′-UTR | 3′-UTR | −23.4 | −0.318 | −0.357 |
| ALDH5A1 | 6p22.3 | 3′-UTR | 3′-UTR | −20.9 | −0.423 | −0.294 |
| CYP2C19 | 10q23.33 | Exon | N/A | −23.3 | −0.292 | −0.214 |
| SLC22A2 | 6q25.3 | 3′-UTR | N/A | −20.6 | −0.309 | −0.185 |
| SLC16A14 | 2q36.3 | 3′-UTR | N/A | −21.6 | −0.308 | −0.142 |
| ALDH3B2 | 11q13.2 | Exon | N/A | −22.6 | −0.500 | 0.068 |
| SLC22A14 | 3p22.2 | Exon | N/A | −22.3 | −0.315 | 0.090 |
| SLC29A2 | 11q13.2 | N/A | 3′-UTR | −17.0 | −0.273 | 0.119 |
| NAT1 | 8p22 | N/A | 3′-UTR | −19.6 | −0.288 | 0.167 |
| CYP11B2 | 8q24.3 | Exon | N/A | −25.0 | −0.435 | 0.250 |
N/A: not available.
Calculated by RNAhybrid software.
Fig. 1. Correlation and free energy analyses between hsa-miR-29a-3p and candidate DMET genes.
(A) ABCC6, (B) ALDH5A1, and (C) SLC22A7 mRNA levels were negatively correlated to hsa-miR-29a-3p levels in 96 non-tumor tissues in GSE22058 public dataset (left). In silico analysis showed that free energies of binding between hsa-29a-3p and cognate mRNAs are −25.2 kcal/mol, −20.9 kcal/mol and −23.4 kcal/mol (right), respectively. ΔG, Gibbs energy.
3.2. hsa-miR-29a-3p interacts in vitro with putative response elements in ABCC6, ALDH5A1, and SLC22A7 transcripts
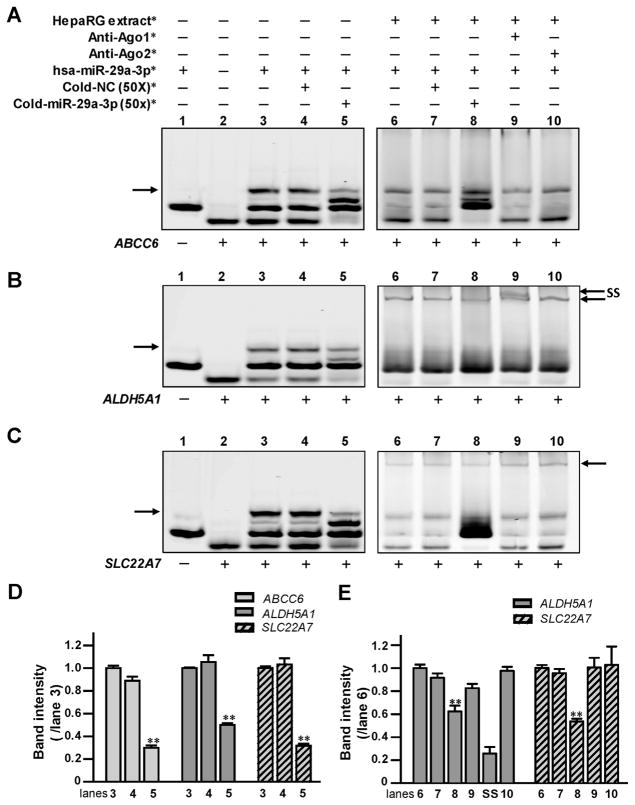
RNA EMSAs were performed to show a direct interaction between hsa-miR-29a-3p and its cognate mRNA targets in vitro. Under our experimental conditions, hsa-miR-29a-3p formed electrophoretically stable complexes with the targeted sequences from the ABCC6, ALDH5A1, and SLC22A7 genes (Fig. 2A, lane 3; B, lane 3; and C, lane 3). Further, mRNA oligonucleotides derived from ALDH5A1 or SLC22A7 could interact with hsa-miR-29a-3p oligo-nucleotides and HepaRG cytoplasmic extracts to form protein-RNA complexes (Fig. 2B, lane 6; C, lane 6). However, a protein–RNA complex was not detected for reactions containing the hsa-miR29a-3p mimic, mRNA oligonucleotides derived from the predicted response element of the ABCC6 gene, and HepaRG protein extracts (Fig. 2A, lanes 6–10). Competition assays showed a sequence-specific interaction between hsa-miR-29a-3p and its cognate mRNA targets in the ABCC6, ALDH5A1, and SLC22A7 genes, since the decreases of miRNA–mRNA complexes (70% in Fig. 2A, lane 5; 50% in Fig. 2B, lane 5; and 68% in Fig. 2C, lane 5; All data histogramed in Fig. 2D, all P <0.001) occurred when excess unlabeled hsa-miR-29a-3p oligonucleotides were added to the reactions. Moreover, the elimination of the miRNA–mRNA complex was not observed when excess unlabeled nonspecific oligonucleotides were added (Fig. 2A, lane 4; B, lane 4; and C, lane 4). Likewise, the sequence–specific interaction between miRNA and mRNA was further confirmed by competition assays using excess unlabeled miR-29a-3p in the presence of protein extracts (38% reduction of protein-RNA complex in Fig. 2B, lane 8; and 47% reduction of those complex in Fig. 2C, lane 8; all data histogramed in Fig. 2E, all P <0.001). Furthermore, we observed that anti-Ago1 antibody could form a supershift band together with protein-RNA complexes consisting of HepaRG cytoplasmic extracts, hsa-miR-29a-3p oligonucleotides, and ALDH5A1 mRNA oligonucleotides (Fig. 2B, lane 9). For unknown reasons, a supershift band was not observed using either antibody (anti-Ago1 or anti-Ago2 antibody) with reactions containing SLC22A7 mRNA oligonucleotides, HepaRG cytoplasmic extracts, and hsa-miR-29a-3p oligonucleotides (Fig. 2C, lane 9), suggesting that the microRNA ribonucleoprotein (miRNP) complex formed using SLC22A7 and hsa-miR-29a-3p oligonucleotides might contain other Ago family members. Our results indicate that hsa-miR-29a-3p RNA mimics are capable of forming miRNP complexes containing ALDH5A1 and SLC22A7 mRNA target sequences and that Argo1 is also detectable within hsa-miR-29a-3p/ALDH5A1 mRNA miRNP complexes.
Fig. 2. The hsa-miR-29a-3p oligonucleotides directly interacted with (A) ABCC6, (B) ALDH5A1, and (C) SLC22A7 mRNA oligonucleotides in vitro.
*The reagents were used in RNA EMSA containing ABCC6,ALDH5A1, and SLC22A7 mRNA oligonucleotides, under the same experimental conditions. NC, nonspecific competitor. Lanes 1 and 2 indicated the mobility of each type of oligonucleotide; lane 3 indicated the mobility status of the miRNA:mRNA complex formed by the interaction of hsa-miR-29a-3p oligonucleotides with ABCC6, ALDH5A1, or SLC22A7 mRNA oligonucleotides; lanes 4 and 5 revealed the mobility shift status of miRNA:mRNA complex in the presence of excess unlabeled nonspecific competitors and specific competitors (hsa-miR-29a-3p). Lane 6 showed complexes formed using the cytoplasmic extracts from HepaRG cells incubated with hsa-miR-29a-3p oligonucleotides and ABCC6, ALDH5A1, and SLC22A7 mRNA oligonucleotides. Lanes 7 and 8 showed the mobility shift status of protein: miRNA:mRNA complexes in the presence of excess unlabeled nonspecific competitors and specific hsa-miR-29a-3p competitors. Lanes 9 and 10 indicated the mobility status of miRNA/mRNA/protein complexes with Ago1 or Ago2 antibodies. Arrows (left) indicate the oligonucleotide complexes in lane 3. Arrows (right) indicate the miRNA/mRNA/protein complexes formed by oligonucleotides and cytoplasmic proteins in lanes 6–10 in (B) and (C). SS indicates the supershift complex formed by the miRNA/mRNA/protein and antibody against Ago1 in lane 9 in (B). (D and E) Histogram corresponds to the relative densitometry quantification of the key miRNA/mRNA complexes or miRNA/mRNA/protein complexes observed in (A, B or C), compared to those in lane 3 or lane 6, respectively. **P <0.001.
3.3. hsa-miR-29a-3p suppresses reporter gene expression for constructs containing ALDH5A1 and SLC22A7 response elements
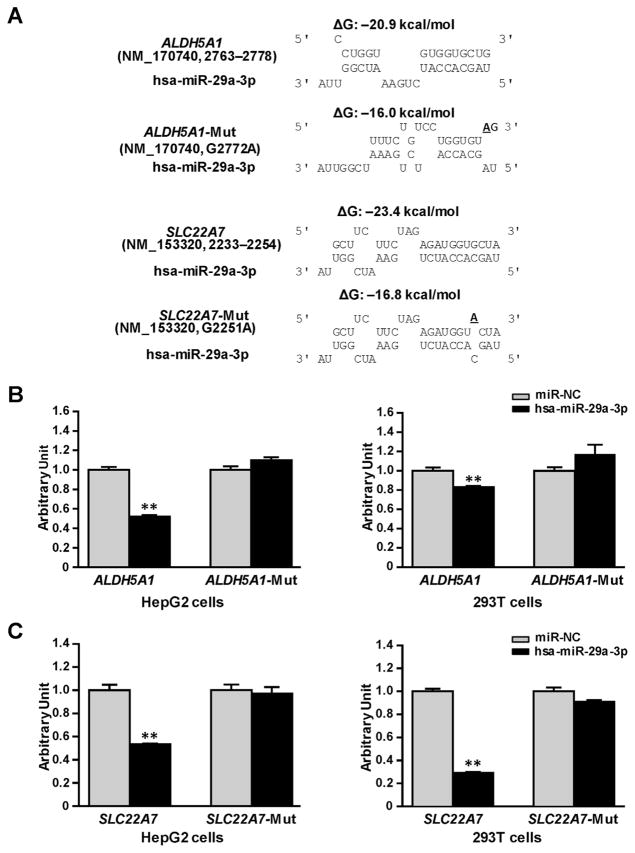
We constructed reporter gene vectors containing the 3′-UTRs of ALDH5A1 or SLC22A7, and mutant vectors containing base-mutated response elements for hsa-miR-29a-3p, which elevates the free energy for miRNA–mRNA interaction (Fig. 3A), and then transiently co-transfected HepG2 and 293T cell lines with these vectors combined with the miRNA negative control or the hsa-miR-29a-3p mimic. As shown in Fig. 3B, in both HepG2 and 293T cells, hsa-miR-29a-3p mimics reduced reporter gene activity for constructs containing the ALDH5A1 3′-UTR (48% and 18%, respectively; all P <0.001) compared with the miRNA negative control. Shown in Fig. 3C, a similar suppressive effect of the hsa-miR-29a-3p mimic was also observed in luciferase assays using the SLC22A7 3′-UTR (47% in HepG2 cells and 70% in 293Tcells, all P <0.001). We created mutants containing base-mutated response elements for hsa-miR-29a-3p in the 3′-UTRs of ALDH5A1 and SLC22A7 and then applied luciferase reporter gene assays to evaluate the specificity of suppression effect. The hsa-miR-29a-3p mimic did not suppress luciferase activities for the mutated-ALDH5A1 or mutated-SLC22A7 constructs, indicating that hsa-miR-29a-3p targets the 3′-UTRs of ALDH5A1 and SLC22A7 mRNA in a sequence–specific manner.
Fig. 3. hsa-miR-29a-3p inhibited luciferase reporter gene expression controlled by the 3′-UTRs of ALDH5A1 or SLC22A7.
(A) Free energy analyses for the interactions between hsa-miR-29a-3p and the targeting sequences or site-mutants present the in ALDH5A1 3′-UTR, or SLC22A7 3′-UTR. ΔG, free energy; the underlined letter, the mutated base. (B) ALDH5A1-CU and ALDH5A1-Mut plasmids or (C) SLC22A7-CU and SLC22A7-Mut plasmids were transiently transfected into 293T and HepG2 cells, together with 50 nmol/L hsa-miR-29a-3p mimic or miRNA negative control. Cells were harvested 48 h after transfection. Three independent experiments, each in triplicate, were performed, and fold changes of luciferase activity were calculated by defining the activity of ALDH5A1-CU plasmid, or SLC22A7-CU, together with miRNA negative control as unity. **P <0.001; NC, miRNA negative control.
Considering that miRNA complexes containing the ABCC6 response element were not detected for hsa-miR-29a-3p using RNA EMSA and that the potential miRNA target sequence is located within the coding region, we speculated that the luciferase reporter gene strategy used with the 3′-UTRs of ALDH5A1 and SLC22A7 was less appropriate to determine if the putative response element in the ABCC6 coding region is truly not a bona fide target for regulation by hsa-miR-29a-3p. Instead, we tested the ability of hsa-miR-29a-3p to interfere with ABCC6 expression using the cytomegovirus promoter. Unfortunately, we found that exogenous ABCC6 was not expressed efficiently in HepG2 or 293T cells transfected with the pCMV6-ABCC6 plasmid under our experimental conditions (data not shown). However, endogenous ABCC6 was found to be expressed in HepaRG cells, although the transfection of HepaRG cells with the hsa-miR-29a-3p mimic did not show any suppression effect on the expression of ABCC6 mRNA or the production of ABCC6 protein (data not shown).
3.4. hsa-miR-29a-3p inhibits endogenous ALDH5A1 and SLC22A7 expression
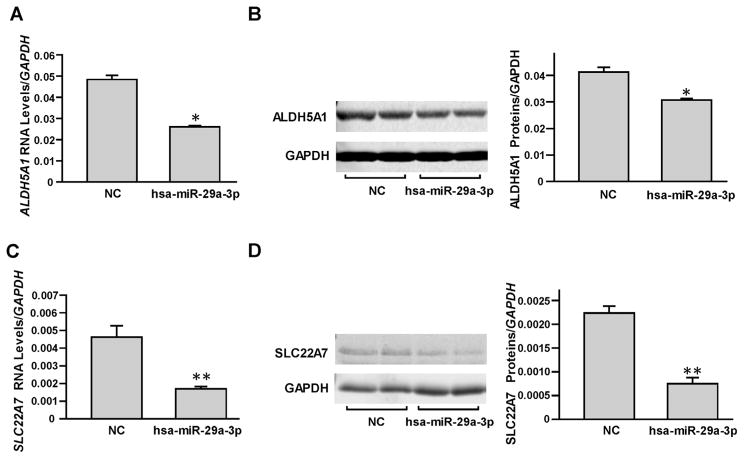
As described in our previous report [17], we selected HepaRG cells to study the suppression effects of hsa-miR-29a-3p on endogenous DMET expression. First, the miRNA negative control or hsa-miR-29a-3p mimic was transiently transfected into HepaRG cells and the RNA levels and protein levels of ALDH5A1 and SLC22A7 were examined. As shown in Fig. 4, hsa-miR-29a-3p was able to suppress endogenous ALDH5A1 and SLC22A7 production (46% and 63% for RNA levels and 27% and 66% for protein levels, respectively; all P <0.05).
Fig. 4. miRNA hsa-miR-29a-3p inhibited endogenous ALDH5A1 and SLC22A7 expression in HepaRG cells.
Differentiated HepaRG cells were transiently transfected using 50 nmol/L miRNA negative control or hsa-miR-29a-3p mimic. Each assay was carried out in triplicate. *P <0.05; **P <0.001; NC, miRNA negative control. Down-regulated mRNA levels (A) and protein levels (B) of ALDH5A1 were observed by hsa-miR-29a-3p mimic transfection. Down-regulated mRNA levels (C) and protein levels (D) of SLC22A7 were observed by hsa-miR-29a-3p mimic transfection. Data are shown as relative ALDH5A1 or SLC22A7 levels normalized by GAPDH levels.
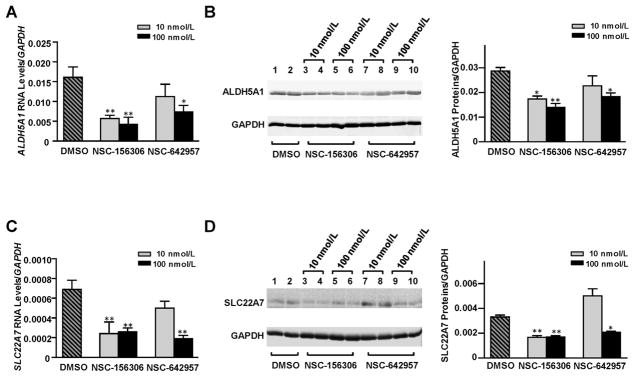
NSC-156306 and NSC-642957, which were shown to increase hsa-miR-29a-3p expression in HepaRG cells in our previous study [30], were also applied in the current study to test if chemically-induced increases of hsa-miR-29a-3p levels in HepaRG cells would also result in decreased expression of the endogenous ALDH5A1 and SLC22A7 genes. We found that both chemical inducers of hsa-miR-29a-3p caused signicant reductions in ALDH5A1 and SLC22A7 expression, although NSC-156306 appeared to be more potent than NSC-642957 in our studies.
As shown in Fig. 5A, a significant dose-dependent decrease in ALDH5A1 mRNA levels was detected in cells treated with NSC-156306 (64% and 73% reductions with 10 nM and 100 nM NSC-156306 treatments, respectively). Treatments with 100 nM NSC-642957 also caused a 54% decrease in the level of ALDH5A1 mRNA. Fig. 5B indicates similar decreases in ALDH5A1 protein levels occurred in response to treatment with NSC-156306 or NSC-642957. Western blotting showed that ALDH5A1 protein levels were diminished by 40, 51, and 36% in cells treated with 10 nM or 100 nM NSC-156306 or with 100 nM NSC-642957, respectively.
Fig. 5. Chemical compounds NSC-156306 and NSC-642957 down-regulated ALDH5A1 and SLC22A7 expression in HepaRG cells.
Differentiated HepaRG cells were treated with 0, 10, or 100 nmol/L NSC-156306 or NSC-642957. Cells were harvested 48 h after treatments. Each assay was done at least 3 times. *P <0.05; **P <0.001; DMSO, no chemical compound in DMSO reagent. Down-regulated mRNA levels (A) and protein levels (B) for ALDH5A1 were observed after NSC-156306 or NSC-642957 treatments. Down-regulated mRNA levels (C) and protein levels (D) for SLC22A7 were observed after NSC-156306 or NSC-642957 treatments. Data were shown as relative ALDH5A1 or SLC22A7 levels normalized by GAPDH expression.
The expression of SLC22A7 also decreased in cells treated with NSC-156306 and NSC-642957. Fig. 5C and D show that SLC22A7 mRNA levels were reduced by 65, 62, or 72% and that SLC22A7 protein levels decreased by 46, 50, or 60% in cells treated with 10 nM or 100 nM NSC-156306 or with 100 nM NSC-642957, respectively.
4. Discussion
Gaining a better understanding of inter-individual variability in the expression of drug metabolizing enzymes and transporters (DMETs) is a fundamentally important objective for improving drug efficacy and avoiding adverse reactions. The results of many studies have revealed relationships between genetic polymorphisms and DMET expression [12,33,34]. However, known genetic variations alone appear to be inadequate to account for the extent of inter-individual variability in DMET expression observed among patients. The influence of epigenetic factors on gene expression offers a new avenue for research to elucidate mechanisms underlying the inter-individual variability in the expression of DMETs. As epigenetic modulators, miRNAs can provide a mechanism to fine-tune gene expression precisely, thus playing an important role in gene regulatory networks that are relevant to drug absorption, metabolism, and disposition [35–38].
Elucidating the mechanistic roles of miRNAs in DMET expression is challenging due to the complexity of regulatory networks involving miRNAs and their targeted genes. Each miRNA may regulate a number of targets owing to its relaxed target sequence complementarity, with some estimates reaching over 200 mRNA transcripts regulated by a single miRNA. Furthermore, the expression of a single gene might be modulated by multiple miRNAs [39]. Moreover, miRNAs may regulate the expression of a DMET gene, such as a transcription factor, so that the action of these miRNAs is indirect with respect to DMET gene expression.
In the present study, we selected hsa-miR-29a-3p as a candidate miRNA, evaluated the interactions between hsa-miR-29a-3p and 374 DMET genes by in silico analysis, found negative correlations in vivo between this miRNA and the expression of 11 DMET genes in human liver samples, and showed that hsa-miR-29a-3p suppressed ALDH5A1 and SLC22A7 production in liver cells using in vitro assays.
The human miR-29 family includes hsa-miR-29a, hsa-miR-29b, and hsa-miR-29c, which are encoded in gene loci on chromosomes 1 and 7 [20]. In liver tissues, hsa-miR-29a-3p levels account for more than 70% of the expression of the miR-29 family (data from TCGA database, not shown), suggesting that hsa-miR-29a-3p might be the most important of these miRNAs for regulating hepatic functions. Accumulating evidence has proved that the miR-29 family is associated with multiple functions relevant to a large group of cellular events. For example, miR-29s have anti-fibrotic effects on heart, kidney and liver; miR-29s exhibited regulatory roles in cell differentiation; as a tumor suppressor, miR-29s can promote cell apoptosis to constrain tumor growth; and as a tumor promoter, miR-29s can facilitate epithelial-mesenchymal transition and mediate tumor metastasis [20,23]. We believe that the overall effects of miR-29s on cells are a result of molecular interactions in specific tissues under specific environmental conditions.
miR-29s have been considered as potential epigenetic therapeutics for the treatment of several diseases, including hematological malignancies [40] and fibrosis [41]. In addition, miR-29b was reported to facilitate cancer therapy by increasing the sensitivity of cancer cells toward the chemotherapeutic drugs decitabine and bortezomib [42]. Further study revealed that bortezomib upregulated the expression of miR-29b and increased the sensitivity of tumors to decitabine through down-regulation of FLT3 and SP1 and disruption of the SP1/Nf-kB complex [43].
To interrogate the potential effects of miR-29a on the regulation of DMETs, we systematically investigated potential miR-29a targets among 374 commonly expressed DMET genes. In silico analyses showed that, besides CYP2C19 that was identified in our previous study [30], three additional DMET genes, ABCC6, SLC22A7 and ALDH5A1, might be targeted by hsa-miR-29a-3p. The predicted hsa-miR-29a-3p target sequences for the SLC22A7 and ALDH5A1 genes were found within their 3′-UTRs, while the target sequence of hsa-miR-29a-3p for ABCC6 transcript was predicted inside its coding region. In selecting of miRNA targeted genes, free energy for the binding between miRNA/mRNA duplexes was considered as one of the pivotal parameters [17]. The calculated Gibbs energies for the formation of the predicted miRNA/mRNA duplexes were all favorable (ΔG <−20 kcal/mol), indicating that stable hybrids should form. Further, using archived data from the CellMiner™ database and the GSE22058 dataset, significant negative correlations between the expression of hsa-miR-29a-3p and the mRNA levels for ABCC6, SLC22A7, and ALDH5A1 were observed in cells and liver tissues, which encouraged us to investigate the suppression effects of hsa-miR-29a-3p on these genes. The highlight of our strategy was to integrate the free energy prediction and the correlation analyses between miRNA levels and the expression of targeted gene in cells and liver tissues that are archived from publically available databases, which could efficiently and precisely identify the targets of miRNAs.
In vitro assays proved that hsa-miR-29a-3p indeed suppressed the expression of SLC22A7 and ALDH5A1 in liver cells, but failed to inhibit ABCC6 expression. Intriguingly, ABCC6 mRNA oligonucleotides were observed to form an mRNA/miRNA complex with hsa-miR-29a-3p in vitro, as predicted, but the mRNA/miRNA complex failed to recruit any cellular proteins, which might be a plausible reason for the observed failure of hsa-miR-29a-3p to regulate ABCC6 expression. Furthermore, we observed that hsa-miR-29a-3p showed a selective advantage to bind Ago1 protein in the modulation of ALDH5A1 expression, but not in the suppression of SLC22A7 expression. Although miRNAs have been reported to have less sorting ability (distinct binding preferences to different Ago proteins) to distinguish Ago-RISC (RNA-induced silencing complex) in mammals compared with those in lower organisms [44], other studies [45,46] and our results supported the notion that miRNAs still might exhibit Ago sorting ability in mammals, at least under some conditions.
As a phase I drug metabolizing enzyme, ALDH5A1 encodes a mitochondrial NAD(+)-dependent succinic semialdehyde dehydrogenase that catalyzes the degradation of the endogenous neurotransmitter 4-aminobutyricacid (GABA). ALDH5A1 deficiency causes a rare inborn disorder in the metabolism of GABA, resulting in accumulation of GABA and gamma-hydroxybutyric acid (GHB) which have many neuro-modulatory properties [47]. With its hypnotic, anxiolytic, anti-depressive and sedative effects, exogenous GHB has been widely used as a therapeutic for anesthesia, narcolepsy, sleep apnea, schizophrenia and alcoholism and opiate withdrawal [48]; however, the pharmaceutical activities of GHB also led to its abuse as an illicit drug [48]. ALDH5A1 enzymatic activity is a key factor determining the pharmaceutical effectiveness of GHB and its biological effects when abused. In addition, ALDH5A1 is involved in the detoxification of some cancer therapeutics, such as busulfan and cyclophosphamide, and increased ALDH5A1 activity contributes to cancer cell survival and disease progression [49].
SLC22A7 encodes organic anion transporter 2, a transport protein responsible for the uptake of many xenobiotics and endogenous substrates and for the excretion of organic anions [50,51]. Specifically, SLC22A7 is involved in the transport of many drugs. For example, SLC22A7 is important for the transport of the angiotensin-converting enzyme inhibitors captopril and quinapril; the β-lactam-antibiotics benzylpenicillin and cephalosporins; otherantibiotics, such as doxycycline, minocycline and tetracycline; the antiviral agents valacyclovir and zidovudine, the antineoplastic agent methotrexate, and H2 receptor antagonists [51]. Therefore, genetic or epigenetic factors that influence SLC22A7 expression could have an impact on the pharmacokinetics and pharmacodynamics of a wide variety of drugs, including those listed above.
Our strategy for this study has several limitations. First, owing to the heterogeneity of tumor cells and relatively high inter-individual variability in the expression of DMETs among the population, archived gene/miRNA expression data from public databases might not reflect precisely the actual correlation between levels of miRNAs and mRNAs; therefore, it is possible that the selection criteria for targeting prediction of hsa-miR-29a-3p might not be reliable. Second, it has been reported that miRNAs can decrease, increase or not change the expression level of its targeted genes [52]. Our strategy selected candidate genes based on the reverse correlation between miRNA and mRNA and therefore might overlook other potential candidates that showed positive correlation or no correlation. Third, although we observed the sorting ability of hsa-miR-29a-3p to recruit Ago1 into RISC in the modulation of ALDH5A1 production, the precise mechanisms for its role in gene expression have not been elucidated.
In conclusion, we have shown that hsa-miR-29a-3p modulates the production of SLC22A7 and ALDH5A1 in liver cells. These results provide new insights regarding the regulatory roles and molecular mechanisms of miRNAs in the modulation of DMET activity, which may help to explain better the diversity of drug responses among patients.
Acknowledgments
This study was supported and funded by the National Center for Toxicological Research (Project E0752601), U.S. Food and Drug Administration.
Abbreviations
- miRNA
microRNA
- DMETs
drug metabolizing enzymes and transporters
- 3′-UTR
3′-untranslated region
- miRNP
microRNA ribonucleoprotein
- GABA
neurotransmitter 4-aminobutyric acid
- GHB
gamma-hydroxybutyric acid
- Ago
argonaute RISC catalytic component
- RISC
RNA-induced silence complex
- FBS
fetal bovine serum
- EMSA
electrophoretic mobility shift assay
Footnotes
Conflict of interest
The authors have no conflict of interest.
Disclaimer
The views presented in this paper are those of the authors and do not necessarily represent those of the U.S. Food and Drug Administration.
Author contributions
Participated in study design: Yu, Tolleson, and Ning.
Conducted experiments: Yu, Knox, and Jin.
Performed data analysis: Yu, Tolleson, L. Guo and Ning.
Wrote or contributed to the writing of the manuscript: Yu, Tolleson, Y. Guo, Kadlubar, and Ning.
References
- 1.Evrard A, Mbatchi L. Genetic polymorphisms of drug metabolizing enzymes and transporters: the long way from bench to bedside. Curr Top Med Chem. 2012;12:1720–1729. doi: 10.2174/156802612803531388. [DOI] [PubMed] [Google Scholar]
- 2.Lima LO, Bruxel EM, Hutz MH, Van der Sand CR, Van der Sand LC, Ferreira ME, et al. Influence of PPARA, RXRA, NR1I2 and NR1I3 gene polymorphisms on the lipid-lowering efficacy and safety of statin therapy. Arq Bras Endocrinol Metabol. 2013;57:513–519. doi: 10.1590/s0004-27302013000700003. [DOI] [PubMed] [Google Scholar]
- 3.Hirota T, Takane H, Higuchi S, Ieiri I. Epigenetic regulation of genes encoding drug-metabolizing enzymes and transporters; DNA methylation and other mechanisms. Curr Drug Metab. 2008;9:34–38. doi: 10.2174/138920008783331130. [DOI] [PubMed] [Google Scholar]
- 4.Yokoi T, Nakajima M. microRNAs as mediators of drug toxicity. Annu Rev Pharmacol Toxicol. 2013;53:377–400. doi: 10.1146/annurev-pharmtox-011112-140250. [DOI] [PubMed] [Google Scholar]
- 5.Franconi F, Brunelleschi S, Steardo L, Cuomo V. Gender differences in drug responses. Pharmacol Res. 2007;55:81–95. doi: 10.1016/j.phrs.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Marinelli M, Piazza PV. Interaction between glucocorticoid hormones, stress and psychostimulant drugs. Eur J Neurosci. 2002;16:387–394. doi: 10.1046/j.1460-9568.2002.02089.x. [DOI] [PubMed] [Google Scholar]
- 7.Juurlink DN, Gomes T, Ko DT, Szmitko PE, Austin PC, Tu JV, et al. A population-based study of the drug interaction between proton pump inhibitors and clopidogrel. CMAJ. 2009;180:713–718. doi: 10.1503/cmaj.082001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silvestri L, Sonzogni L, De Silvestri A, Gritti C, Foti L, Zavaglia C, et al. CYP enzyme polymorphisms and susceptibility to HCV-related chronic liver disease and liver cancer. Int J Cancer. 2003;104:310–317. doi: 10.1002/ijc.10937. [DOI] [PubMed] [Google Scholar]
- 9.Scott SA. Personalizing medicine with clinical pharmacogenetics. Genet Med. 2011;13:987–995. doi: 10.1097/GIM.0b013e318238b38c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosch TM. Pharmacogenomics of drug-metabolizing enzymes and drug transporters in chemotherapy. Methods Mol Biol. 2008;448:63–76. doi: 10.1007/978-1-59745-205-2_5. [DOI] [PubMed] [Google Scholar]
- 11.Motulsky AG. Drug reactions enzymes, and biochemical genetics. J Am Med Assoc. 1957;165:835–837. doi: 10.1001/jama.1957.72980250010016. [DOI] [PubMed] [Google Scholar]
- 12.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 13.Kim IW, Han N, Burckart GJ, Oh JM. Epigenetic changes in gene expression for drug-metabolizing enzymes and transporters. Pharmacotherapy. 2014;34:140–150. doi: 10.1002/phar.1362. [DOI] [PubMed] [Google Scholar]
- 14.Ikemura K, Iwamoto T, Okuda M. MicroRNAs as regulators of drug transporters, drug-metabolizing enzymes, and tight junctions: implication for intestinal barrier function. Pharmacol Ther. 2014;143:217–224. doi: 10.1016/j.pharmthera.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66:9090–9098. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- 16.Mohri T, Nakajima M, Fukami T, Takamiya M, Aoki Y, Yokoi T. Human CYP2E1 is regulated by miR-378. Biochem Pharmacol. 2010;79:1045–1052. doi: 10.1016/j.bcp.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 17.Yu D, Green B, Marrone A, Guo Y, Kadlubar S, Lin D, et al. Suppression of CYP2C9 by microRNA hsa-miR-128-3p in human liver cells and association with hepatocellular carcinoma. Sci Rep. 2015;5:8534. doi: 10.1038/srep08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan YZ, Gao W, Yu AM. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab Dispos. 2009;37:2112–2117. doi: 10.1124/dmd.109.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu X, Dhakal IB, Beggs M, Edavana VK, Williams S, Zhang X, et al. Functional genetic variants in the 3′-untranslated region of sulfotransferase isoform 1A1 (SULT1A1) and their effect on enzymatic activity. Toxicol Sci. 2010;118:391–403. doi: 10.1093/toxsci/kfq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kriegel AJ, Liu Y, Fang Y, Ding X, Liang M. The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol Genom. 2012;44:237–244. doi: 10.1152/physiolgenomics.00141.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liston A, Papadopoulou AS, Danso-Abeam D, Dooley J. MicroRNA-29 in the adaptive immune system: setting the threshold. Cell Mol Life Sci. 2012;69:3533–3541. doi: 10.1007/s00018-012-1124-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapinas K, Delany AM. MicroRNA biogenesis and regulation of bone remodeling. Arthritis Res Ther. 2011;13:220. doi: 10.1186/ar3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang H, Zhang G, Wu JH, Jiang CP. Diverse roles of miR-29 in cancer (review) Oncol Rep. 2014;31:1509–1516. doi: 10.3892/or.2014.3036. [DOI] [PubMed] [Google Scholar]
- 24.Bian EB, Li J, Zhao B. miR-29, a potential therapeutic target for liver fibrosis. Gene. 2014;544:259–260. doi: 10.1016/j.gene.2014.04.076. [DOI] [PubMed] [Google Scholar]
- 25.Cushing L, Kuang P, Lu J. The role of miR-29 in pulmonary fibrosis. Biochem Cell Biol. 2015;93:109–118. doi: 10.1139/bcb-2014-0095. [DOI] [PubMed] [Google Scholar]
- 26.Nakai K, Tanaka H, Hanada K, Ogata H, Suzuki F, Kumada H, et al. Decreased expression of cytochromes P450 1A2, 2E1, and 3A4 and drug transporters Na+-taurocholate-cotransporting polypeptide, organic cation transporter 1, and organic anion-transporting peptide-C correlates with the progression of liver fibrosis in chronic hepatitis C patients. Drug Metab Dispos. 2008;36:1786–1793. doi: 10.1124/dmd.107.020073. [DOI] [PubMed] [Google Scholar]
- 27.He Y, Huang C, Lin X, Li J. MicroRNA-29 family, a crucial therapeutic target for fibrosis diseases. Biochimie. 2013;95:1355–1359. doi: 10.1016/j.biochi.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Saito Y, Suzuki H, Imaeda H, Matsuzaki J, Hirata K, Tsugawa H, et al. The tumor suppressor microRNA-29c is downregulated and restored by celecoxib in human gastric cancer cells. Int J Cancer. 2013;132:1751–1760. doi: 10.1002/ijc.27862. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Liu C, Luo M, Zhang Z, Gong J, Li J, et al. Chemotherapy-induced miRNA-29c/catenin-delta signaling suppresses metastasis in gastric cancer. Cancer Res. 2015;75:1332–1344. doi: 10.1158/0008-5472.CAN-14-0787. [DOI] [PubMed] [Google Scholar]
- 30.Yu D, Green B, Tolleson W, Jin Y, Mei N, Guo Y, et al. MicroRNA hsa-miR-29a-3p modulates CYP 2C19 production in human liver cells. Biochem Pharmacol. 2015;98:215–223. doi: 10.1016/j.bcp.2015.08.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L, Price ET, Chang CW, Li Y, Huang Y, Guo LW, et al. Gene expression variability in human hepatic drug metabolizing enzymes and transporters. PLoS One. 2013;8:e60368. doi: 10.1371/journal.pone.0060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reinhold WC, Sunshine M, Liu H, Varma S, Kohn KW, Morris J, et al. CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 2012;72:3499–3511. doi: 10.1158/0008-5472.CAN-12-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma MK, Woo MH, McLeod HL. Genetic basis of drug metabolism. Am J Health Syst Pharm. 2002;59:2061–2069. doi: 10.1093/ajhp/59.21.2061. [DOI] [PubMed] [Google Scholar]
- 34.Zhou SF, Liu JP, Chowbay B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab Rev. 2009;41:89–295. doi: 10.1080/03602530902843483. [DOI] [PubMed] [Google Scholar]
- 35.Ingelman-Sundberg M, Zhong XB, Hankinson O, Beedanagari S, Yu AM, Peng L, et al. Potential role of epigenetic mechanisms in the regulation of drug metabolism and transport. Drug Metab Dispos. 2013;41:1725–1731. doi: 10.1124/dmd.113.053157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu AM. Role of microRNAs in the regulation of drug metabolism and disposition. Expert Opin Drug Metab Toxicol. 2009;5:1513–1528. doi: 10.1517/17425250903307448. [DOI] [PubMed] [Google Scholar]
- 37.Nakajima M, Yokoi T. MicroRNAs from biology to future pharmacotherapy: regulation of cytochrome P450s and nuclear receptors. Pharmacol Ther. 2011;131:330–337. doi: 10.1016/j.pharmthera.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 38.He Y, Chevillet JR, Liu G, Kim TK, Wang K. The effects of microRNA on the absorption, distribution, metabolism and excretion of drugs. Br J Pharmacol. 2014;172:2733–2747. doi: 10.1111/bph.12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 40.Amodio N, Rossi M, Raimondi L, Pitari MR, Botta C, Tagliaferri P. miR-29s: a family of epi-miRNAs with therapeutic implications in hematologic malignancies. Oncotarget. 2015;6:12837–12861. doi: 10.18632/oncotarget.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knabel MK, Ramachandran K, Karhadkar S, Hwang HW, Creamer TJ, Chivukula RR. Systemic delivery of scAAV8-encoded miR-29a ameliorates hepatic fibrosis in carbon tetrachloride-treated mice. PLoS One. 2015;10:e0124411. doi: 10.1371/journal.pone.0124411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blum W, Garzon R, Klisovic RB, Schwind S, Walker A, Geyer S, et al. Clinical response and miR-29b predictive significance in older AML patients treated with a 10-day schedule of decitabine. Proc Natl Acad Sci U S A. 2010;107:7473–7478. doi: 10.1073/pnas.1002650107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blum W, Schwind S, Tarighat SS, Geyer S, Eisfeld AK, Whitman S, et al. Clinical and pharmacodynamic activity of bortezomib and decitabine in acute myeloid leukemia. Blood. 2012;119:6025–6031. doi: 10.1182/blood-2012-03-413898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamakawa N, Okuyama K, Ogata J, Kanai A, Helwak A, Takamatsu M. Novel functional small RNAs are selectively loaded onto mammalian Ago1. Nucleic Acids Res. 2014;42:5289–5301. doi: 10.1093/nar/gku137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turchinovich A, Burwinkel B. Distinct AGO1 and AGO2 associated miRNA profiles in human cells and blood plasma. RNA Biol. 2012;9:1066–1075. doi: 10.4161/rna.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tay CG, Ariffin H, Yap S, Rahmat K, Sthaneshwar P, Ong LC. Succinic semialdehyde dehydrogenase deficiency in a Chinese boy: a novel ALDH5A1 mutation with severe phenotype. J Child Neurol. 2015;30:927–931. doi: 10.1177/0883073814540523. [DOI] [PubMed] [Google Scholar]
- 48.Galloway GP, Frederick-Osborne SL, Seymour R, Contini SE, Smith DE. Abuse and therapeutic potential of gamma-hydroxybutyric acid. Alcohol. 2000;20:263–269. doi: 10.1016/s0741-8329(99)00090-7. [DOI] [PubMed] [Google Scholar]
- 49.Hassan M, Andersson BS. Role of pharmacogenetics in busulfan/cyclophosphamide conditioning therapy prior to hematopoietic stem cell transplantation. Pharmacogenomics. 2013;14:75–87. doi: 10.2217/pgs.12.185. [DOI] [PubMed] [Google Scholar]
- 50.Kobayashi Y, Ohshiro N, Sakai R, Ohbayashi M, Kohyama N, Yamamoto T. Transport mechanism and substrate specificity of human organic anion transporter 2 (hOat2 [SLC22A7]) J Pharm Pharmacol. 2005;57:573–578. doi: 10.1211/0022357055966. [DOI] [PubMed] [Google Scholar]
- 51.Burckhardt G. Drug transport by organic anion transporters (OATs) Pharmacol Ther. 2012;136:106–130. doi: 10.1016/j.pharmthera.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 52.Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genom. 2014;2014:970607. doi: 10.1155/2014/970607. [DOI] [PMC free article] [PubMed] [Google Scholar]