Abstract
Anoxygenic photosynthetic prokaryotes arose in ancient oceans ~3.5 billion years ago. Evolution of oxygenic photosynthesis by cyanobacteria followed soon after, enabling eukaryogenesis and the evolution of complex life. The Archaeplastida lineage dates back ~1.5 billion years to the domestication of a cyanobacterium. Eukaryotic algae have subsequently radiated throughout oceanic/fresh-water/terrestrial environments, adopting distinctive morphological and developmental strategies for adaptation to diverse light environments. Descendants of the ancestral photosynthetic alga remain challenged by a typical diurnally fluctuating light supply ranging from ~0 to ~2000 μE m−2 s−1. Such extreme changes in light intensity and variations in light quality have driven evolution of novel photoreceptors, light harvesting complexes and photoprotective mechanisms in photosynthetic eukaryotes. This minireview focuses on algal light sensors, highlighting the unexpected roles for linear tetrapyrroles (bilins) in the maintenance of functional chloroplasts in chlorophytes, sister species to streptophyte algae and land plants.
Keywords: biliverdin, Chlamydomonas reinhardtii, Micromonas pusilla, photoacclimation, phycobilin, phytochrome, prasinophyte
INTRODUCTION
Sunlight is the most abundant energy resource on our planet. The evolution of many life forms that harvest solar energy and fix carbon dioxide through the photosynthetic process has been, and continues to be, essential for the maintenance of the biosphere. Extant photosynthetic organisms are found in a wide variety of light environments, typically serving as keystone species in diverse ecosystems. The origins of photosynthesis trace back to anaerobic bacterial phototrophs that used bacteriochlorophyll (Bchl)-based reaction center complexes to oxidize inorganic materials such as sulfide or metals for generation of reducing power for carbon dioxide fixation (Gupta 2012; Fischer et al. 2016). Descendants of these phototrophs still inhabit low light, (semi)-anaerobic niches where DNA-damaging UV light fluences are minimal (Bryant & Frigaard 2006; Gupta 2010; Nowicka & Kruk 2016). Evolution of oxygenic photosynthesis by cyanobacteria facilitated life at or near the earth’s surface and also supported development of complex life. These newly habitable ecological niches required new regulatory systems to cope with the transient, diurnal and seasonal fluctuating light environments. Such environments also supported evolution of diverse light harvesting systems to fully utilize the visible light spectrum of sunlight suitable for photosynthesis (Buchel 2015; Dall’Osto et al. 2015). Functional diversification of light sensors along with development of novel photoprotective mechanisms also remains ongoing as competition for light from new neighbors has increased.
Primary endosymbiosis, a process by which a colorless eukaryotic host converted a free-living cyanobacterium into a plastid organelle, occurred ~1.5 billion years ago and led to nearly all species of photosynthetic eukaryotes on earth. Organisms with primary plastids comprise the kingdom Archaeplastida, which includes glaucophytes, rhodophytes (red algae) and the green lineage Viridiplantae (comprising prasinophytes, chlorophytes, streptophyte algae and land plants). Photosynthesis further spread laterally to other algal species by secondary, tertiary or perhaps even higher order endosymbioses, arising via eukaryote-eukaryote phagotrophic endosymbiotic events. Engulfment of ancestral green algal species led to chlorarachniophytes and euglenophytes (Archibald & Keeling 2002), whereas other algae are likely derived from secondary endosymbiosis of red algae (Burki et al. 2016). With the notable exception of Paulinella spp., all plastids are derived from this single endosymbiotic event. Paulinella’s discovery shows that evolution of photosynthetic organelles from cyanobacteria is ongoing (Nowack et al. 2008).
After evolution of blue-shifted chlorophyll (Chl)-based photosynthetic reaction centers capable of water oxidation, cyanobacteria evolved phycobiliprotein antennae complexes with reduced linear tetrapyrrole (phycobilin) chromophores (Mullineaux 2008; Watanabe & Ikeuchi 2013). Derived from the oxygenation of heme, phycobilins are responsible for the expanded light harvesting and light sensing range of most extant cyanobacteria. This expanded spectral range has enabled cyanobacteria to acclimate to a wide diversity of both aquatic and terrestrial environments. Due to cyanobacterial endosymbiosis and horizontal gene transfer (HGT) events, phycobilin-based antennae and light sensing systems subsequently radiated amongst oxygenic photosynthetic eukaryotes, which in some lineages were later lost and/or replaced by new innovations.
Light harvesting diversity has been shaped by diverse light environments found in aquatic systems that are now inhabited by many photosynthetic species. Compared to terrestrial environments, oceanic and freshwater environments provide unique challenges for aquatic phototrophs. Wavelengths of solar radiation available for light sensing and harvesting are dramatically different from those of terrestrial environments, because the water column effectively attenuates longer wavelengths of visible and near infrared light. Indeed, red light poorly penetrates below ~15 meters in water. Hence, the near surface of the open ocean (above 30 m) becomes progressively enriched in blue/green light with depth. Selective absorption of light by water and by other competing photosynthetic organisms has been argued to be strong selective pressures for evolution of new light harvesting and sensing systems (Grossman et al. 1995; Hohmann-Marriott & Blankenship 2011; Rockwell et al. 2014a). In this perspective, we focus on eukaryotic algal light sensing, particularly highlighting our emerging knowledge of the important roles that linear tetrapyrroles (bilins) play in chlorophyte photoacclimation under diurnal growth conditions.
LIGHT HARVESTING AND PHOTOPROTECTIVE SYSTEMS DIFFER AMONGST EUKARYOTIC ALGAE
High-resolution crystal structures of the two photosystems from cyanobacteria (Jordan et al. 2001; Umena et al. 2011; Suga et al. 2015) and plants (Rhee et al. 1998; Mazor et al. 2015; Qin et al. 2015) have provided many details on pigments and protein arrangement, mechanisms of oxygen evolution and energy transfer pathways in these multi-subunit pigment-protein supercomplexes. Despite independent evolution of eukaryote photosystems for ~1.5 billion years, plant PSII and PSI reaction centers (RCs) remain quite similar to those of their cyanobacterial counterparts, including structures that support binding and orientation of Chls (Nelson & Yocum 2006). The core antennae of PSII and PSI of eukaryotic phototrophs are also associated with accessory light-harvesting complexes (LHCs) to increase their absorption cross section. Compared with the highly conserved PSII or PSI RCs and core antennae, LHCs exhibit a high degree of variability, reflecting gene duplication/divergence, gene losses and horizontal gene transfer in different algal lineages (Hoffman et al. 2011).
Light harvesting complexes
A broad window of the sunlight spectrum can be efficiently harvested by algae, i.e., from near UV (~350 nm) to the IR (700~1000 nm). This versatility reflects diverse antenna systems that have been adapted to their unique and changing ecological niches for many millions of years. The phycobiliproteins possess the broadest spectral coverage, as the bilin chromophores can be fine-tuned to harvest light efficiently at both the planet surface and under water. Phycobiliproteins are retained in chloroplasts of glaucophyte, rhodophyte (red) algae, and red-derived cryptophyte algae but have been replaced with Chl-based antennae in most other eukaryotic algal lineages.
Chl a-based LHCs are the most prevalent antennae proteins present in green algae, i.e., prasinophyte, chlorophyte and streptophyte algal lineages, and in land plants. Such LHCs consist of three thylakoid membrane-spanning α-helices. This protein family also includes one helix-, two helix- as well as four helix-proteins (Engelken et al. 2012). Other structurally related LHC proteins also bind Chl b and lutein in green algae. LHCs in Chromalveolates usually bind Chl c and other carotenoids instead (Büchel 2015). Interestingly, red algal LHCaR1 can bind all major Chls and carotenoids found in plants and in other algae in vitro, suggesting a certain level of plasticity of pigment binding inherent in LHCs (Grabowski et al. 2001). Besides the most intensively studied higher plant LHCs (Ballottari et al. 2012; Pagliano et al. 2013), biochemical and functional data are available for only a few algal lineages, i.e., green algae and diatoms (Koziol et al. 2007; Neilson & Durnford 2010).
LHCs from the green alga Chlamydomonas reinhardtii differ significantly from those of plants with respect to the number of genes and biochemical properties of the complexes (Neilson & Durnford 2010). Land plants and mosses possess only five Lhca genes (Lhca1-5). By comparison with Arabidopsis, C. reinhardtii contains nine Lhca genes, resulting in a significantly larger PSI-LHCI complex and a ~41% greater photon harvesting efficiency. The enlarged antenna complex of Chlamydomonas PSI-LHCI exhibits similar energy trapping efficiency as that of plants, largely due to the presence of more blue-shifted pigments (compared to plants) in Lhca2 and Lhca9 and more Chl b (Le Quiniou et al. 2015). Larger PSI-LHCI complexes were also observed in Ostreococcus tauri and high-light grown O. tauri only contains a smaller PSI-LHCI that lacks Lhca6 and Lhcp2 subunits present in the larger complex (Swingley et al. 2010). However, this highly-derived species is likely not to be typical for prasinophyte lineage as a whole (Duanmu et al. 2014).
Photoprotective LHCs
Although light is essential for photosynthesis, absorption of too much light can lead to production of highly reactive molecules or byproducts that cause photodamage to the photosynthetic machinery lowering its efficiency (photoinhibition). Plants and algae have evolved a plethora of photoprotective mechanisms to prevent damage by excess light. One of the most important short-term acclimation responses is non-photochemical quenching (NPQ) of Chl a fluorescence, consisting of high-energy-state quenching (qE), photo-inhibitory quenching (qI) and state transitions (qT) (Ruban 2016). Excess excitation energy is dissipated rapidly as heat by qE, the main component of NPQ (Goss & Lepetit 2015). Mechanistic analyses of NPQ in photosynthetic eukaryotes have revealed special types of photoprotective LHCs as essential components in the qE process. In the green lineage, these include the well characterized PSBS (a four-helix protein in the LHC superfamily) and LHCSR (stress-induced LHC protein), which exhibit uneven distribution among green algae, mosses and vascular plants (Niyogi & Truong 2013).
C. reinhardtii possesses two LHCSR proteins, LHCSR1 and LHCSR3, which exhibit ~82% sequence identity. Chlamydomonas LHCSR3, formally called LI818, is an early diverged member of the LHC superfamily with 3 helices. The mutant lacking LHCSR3 protein shows a strong reduction of NPQ when transferred from a low-light acclimated state to high light (Peers et al. 2009). LHCSR3 protein can bind Chl a/b and xanthophylls in vitro and also appears to be able to sense low pH and perform energy-quenching simultaneously (Bonente et al. 2011). In the LHCSR3-less mutant, LHCSR1 is sufficient for pH sensing and could induce a fast and pH-dependent quenching of LHCII (Dinc et al. 2016). In contrast, plants use PSBS and monomeric/trimeric Lhcb proteins for pH sensing and energy-quenching, respectively (Li et al. 2004; Holt et al. 2004; Pérez-Bueno et al. 2008).
The diatom Phaeodactylum tricornutum also possesses an LHCSR gene family member, LHCX1, which functions as a molecular gauge to regulate NPQ quantitatively. LHCX1 regulation of NPQ differs from that of green algal LHCSR proteins, i.e. LHCX1 is already highly expressed in low light-acclimated cultures and is not further upregulated by high light stress, and LHCX1 protein does not contain the amino acids thought to be essential for low pH sensing (Bailleul et al. 2010). Homologs of LHCSR/LHCX proteins are present in green algae, mosses and diatoms, but apparently have been lost in red algae and vascular plants (Depauw et al. 2012). It is therefore probable that the LHCSR/LHCX protein family evolved prior to the split of Viridiplantae and rhodophytes.
DIVERSITY AND DISTRIBUTION OF LIGHT SENSORS AMONGST PHOTOSYNTHETIC EUKARYOTES
Owing to their dependence on light as an energy source, photosynthetic species use light sensors to optimize photosynthetic light capture and light energy conversion in response to changing light conditions in the environment (Ruban 2015; Galvão & Fankhauser 2015; Parihar et al. 2016). Such light quality and quantity perception is independent of active photosynthesis or other photosynthesis-derived signals, i.e., reactive oxygen species (ROS), plastoquinone reduction state, chloroplast energy status, etc. All photoreceptors require a protein scaffold that has evolved to optimize the signal-activating photochemical event to effectively compete with non-productive energy-dissipating pathways (Möglich et al. 2010). Photoreceptor activation relies upon the light-dependent photochemistry of their prosthetic chromophores, such as isomerization around a double bond (phytochromes and rhodopsins), electron transfer [cryptochromes and BLUF (blue-light sensors using flavin adenine dinucleotide) proteins], or formation or cleavage of a covalent bond [light-oxygen-voltage (LOV) domains] (Möglich et al. 2010). The recently identified UV-B specific photoreceptor in plants and green algae (UVR8) utilizes a cluster of conserved Trp amino acids as chromophore (Rizzini et al. 2011; Jenkins 2014; Tilbrook et al. 2016). Transmission of the UV light signal involves subunit dissociation, although the molecular mechanism of light activation by UVR8 remains enigmatic. The number and diversity of putative light sensors can be astonishingly large in algae and our understanding of their unique and overlapping regulatory roles remains a sparsely explored frontier (see Table 1 for list of representative photoreceptors in eukaryotic algae).
Table 1.
Representative photoreceptors in eukaryotic algae
| Photoreceptor classes | Chromophore | Domain composition | Representative algae | Reference |
|---|---|---|---|---|
| Phytochrome | Bilins | PAS-GAF-PHY-(PAS)2-HKD-REC | Micromonas pusilla | Duanmu et al. 2014 |
| PAS-GAF-PHY-PAS-HKD-REC | Cyanophora paradoxa | Duanmu et al. 2014 | ||
| PAS-GAF-PHY-PAS-PKC-RING | Guillardia theta | Duanmu et al. 2014 | ||
| PAS-GAF-PHY-HKD-REC | Thalassiosira pseudonana | Fortunato et al. 2016 | ||
|
| ||||
| Cryptochrome | FAD+MTHF(?) | Cry-DASH: PHR | Chlamydomonas reinhardtii | Beel et al. 2012 |
| FAD+MTHF (?) | Plant Cry: PHR-DAS | Coccomyxa subellipsoidea | Fortunato et al. 2015 | |
| FAD+MTHF | Plant Cry-like: dAS-PHR | Coccomyxa subellipsoidea | Fortunato et al. 2015 | |
|
| ||||
| Phototropin | FMN | LOV-LOV-STKD | Chlamydomonas reinhardtii | Huang & Beck 2003 |
|
| ||||
| BLUF-protein | FMN | Aureochrome: bZIP-LOV | Vaucheria frigida | Takahashi et al. 2007 |
| Bilin+FMN(?) | Neochrome: PHY-LOV-LOV-STKD | Mougeotia scalaris | Suetsugu et al. 2005 | |
| FAD | PAC: BLUF-AC-BLUF-AC | Euglena gracilis | Iseki et al. 2002 | |
|
| ||||
| Rhodopsin | Retinal | Channelrhodopsin: Rh/ChR-Reg.domain | Chlamydomonas reinhardtii | Nagel et al. 2002 |
| Enzymerhodopsin: Rh-HisK-REC-AC/GC | Chlamydomonas reinhardtii | Kateriya et al. 2004 | ||
| Light driven ion pump: Rh/AR | Acetabularia acetabulum | Tsunoda et al. 2006 | ||
|
| ||||
| UVR8 | Trp | (RCC1)7 | Chlamydomonas reinhardtii | Tilbrook et al. 2016 |
Representative photoreceptor classes and the domain composition according to the Pfam database (due to space limit, not all characterized algal photoreceptors are included in this table). Domain abbreviations are: AC/GC, adenylyl or guanylyl cyclase; AR, Acetabularia rhodopsins; BLUF, blue-light sensors using flavin adenine dinucleotide; bZIP, basic region/leucine zipper; CCE, cryptochrome C-terminal extension; ChR, channelrhodopsin; Cry-DASH, cryptochrome of Drosophila, Arabidopsis, Synechococcus, Homo; DAS: DQXVP Acidic STAESS motif; dAS, refers to a DAS motif missing the DQXVP; FAD, flavin adenine dinucleotide; FMN, flavin mononucleotide; GAF, cGMP-specific phosphodiesterases, adenylyl cyclases and FhlA; HisK, histidine kinase; HKD, Histidine Kinase A domain and H-ATPase-c domain; LOV, light-oxygen-voltage; MTHF: 5-methenyltetrahydrofolate; PAC, photoactivated adenylyl cyclase; PAS, Period-Arnt-Sim domain; PHY, phytochrome photosensory domain; PHR, photolyase homology region; RCC1, regulator of chromosome condensation repeat 1; REC, response regulator receiver; Reg.domain, regulatory domain; Rh, rhodopsin; RING, really interesting new gene; STKD, serine/threonine kinase domain; Trp, tryptophan; UVR8, Ultraviolet Resistance Locus 8.
Flavin-based photoreceptors constitute the most diversified classes of light sensors in algae
Shorter wavelength blue and green light penetrate more deeply into the water column and are more accessible to aquatic algae. Blue-light sensing photoreceptors, such as the extensively studied phototropin (PHOT) and cryptochrome (CRY), utilize flavin mononucleotide (FMN) or flavin adenine dinucleotide (FAD) as chromophores. Photoexcitation of flavin chromophores induces redox reactions (CRYs), covalent bond formation (LOV domains of PHOTs), or hydrogen-bond rearrangements (BLUF) (Losi & Gärtner 2012). The rich chemistry of flavins may have facilitated expansion of flavin-based light sensors in UVA/blue/green enriched environments and their integration with the redox status of the cell (Conrad et al. 2014).
Flavin-binding LOV sensor domains most likely originated from bacteria before mitochondrial and plastid endosymbioses (Krauss et al. 2009). LOV domains are tethered to other functional modules and thus give rise to unique photoreceptors in eukaryotes. Aureochromes with N-terminal DNA-binding domains use blue light to regulate cell branching and differentiation in photosynthetic stramenopile algae (Takahashi et al. 2007; Takahashi 2016). Class I aureochromes of the diatom P. tricornutum also bind canonical aureo-box sequences but form enhancer-like loop-structured DNA complexes in a light-independent manner (Banerjee et al. 2016). In Euglena gracilis, a photoactivated adenylyl cyclase domain in the blue-light sensor PAC catalyzes cAMP synthesis to mediate photophobic responses (Iseki et al. 2002). Such novel LOV-based photoreceptors are restricted to select species of aquatic algae and are absent from land plants, suggesting that the light sensor LOV domains have been repeatedly incorporated into pre-existing cellular signaling molecules for environmental acclimation.
PHOTs are tandem LOV domain proteins that undergo autophosphorylation at several serine and threonine residues when excited by blue light (Christie 2007). Plant phototropins are used to regulate chloroplast movement and other responses, i.e., photomorphogenesis, stomatal opening and leaf flattening to optimize light capture for photosynthesis (Li et al. 2015b; Okajima 2016). By contrast, the green alga C. reinhardtii contains only one phototropin that controls primarily developmental processes, i.e., gametogenesis, maintenance of mating competence and zygote germination (Huang & Beck 2003). Several recent studies have identified other biological functions of CrPHOT, such as regulation of Chl and carotenoid biosynthesis related genes, regulation of eyespot size and phototaxis, and induction of LHCSR3 expression for energy-dependent NPQ (qE) in high light stress (Im et al. 2006; Trippens et al. 2012; Petroutsos et al. 2016). It is therefore surprising that Chlamydomonas phototropin rescues the Arabidopsis phot-deficient phenotypes, suggesting conserved downstream signaling mechanisms between plant and green algal phototropins (Onodera et al. 2005). Phototropin genes are present in prasinophyte algae, but we do not yet know the processes they regulate. Phototropin homologs are absent in glaucophyte, rhodophyte, cryptophyte and haptophyte algae, implicating the origin of phototropins in a common ancestor of Viridiplantae (Li et al. 2015b).
Cryptochromes, another class of flavin-based blue light sensors related to DNA-photolyase 1 family, were first discovered in Arabidopsis thaliana but appear to be widely distributed in Archaea, Eubacteria and Eukaryota (Ahmad & Cashmore 1993; Chaves et al. 2011). Biochemical and phenotypic analyses of CRYs in algae have revealed novel functions distinct from those in plants, suggesting that these sensors arose independently in different photosynthetic lineages (Beel et al. 2013). It is also possible that CRYs arose via fusion of a sensor domain to different signaling output domains as proposed for LOV domain-based photosensors (see above). C. reinhardtii contains a plant-type CRY (CPH1, Chlamydomonas photolyase homolog 1), an animal-type CRY (aCRY), and two CRY-DASH (Drosophila, Arabidopsis, Synechococcus, Homo) proteins (Merchant et al. 2007). In addition to a plastid-localized CRY-DASH, Arabidopsis and other dicots possess two CRYs – one of which participates in photoperiod sensing (Chaves et al. 2011; Liu et al. 2011). The C. reinhardtii CrCPH1 protein tracks diurnal dark/light cycle and is rapidly degraded in the presence of red or blue light via a proteasome-dependent pathway (Reisdorph & Small 2004). By comparison, Chlamydomonas aCRY is relatively light stable, but can also be activated by either red or blue light to impact transcription of genes involved in photosynthesis, pigment biosynthesis, cell cycle control and circadian clock (Beel et al. 2012). The diatom cryptochrome PtCPF1 (Cry/photolyase family 1) exhibits both DNA repair activity and blue light-dependent transcriptional regulation of genes in vivo (Coesel et al. 2009). Dual enzymatic activities of CPF were also found in the prasinophyte Ostreococcus tauri, highlighting the important role of this bifunctional photoreceptor in coordinating circadian rhythms with cell-cycle control and DNA-damage repair (Heijde et al. 2010; Fortunato et al. 2015).
Retinal-based rhodopsins are not just photoreceptors for vision in algae
Distinctive from other soluble light sensors, rhodopsins are membrane proteins with covalently attached retinal as the chromophore. Like phytochromes, rhodopsins transduce light signals via photoisomerization of their bound chromophores. C. reinhardtii harbors rich subsets of rhodopsin photoreceptors, including two animal rhodopsin-related proteins (chlamyrhodopsin-1 and chlamyrhodopsin-2), two microbial-related channelrhodopsins (ChR1 and ChR2), and the chlamyopsins, COP3 and COP4 (Hegemann 2008). ChR1 is a light-activated proton channel involved in phototaxis of Chlamydomonas and its homologs may play similar roles in other green algae (Nagel et al. 2002). New subfamilies of rhodopsins were also identified in certain algal groups with no clear biological functions, such as two-component rhodopsin systems with a histidine kinase, a response regulator and an adenylyl/guanylyl cyclase domain (Kateriya et al. 2004), a proton-pumping rhodopsin in the marine chlorophyte alga Acetabularia acetabulum (Tsunoda et al. 2006), and green light-absorbing chloride-conducting channelrhodopsins from cryptophyte algae (Wietek et al. 2016).
UV-B photoreceptors are functionally conserved in the green lineage
First characterized in plants, UVR8 (UV Resistance Locus 8) mediates light sensing in the ultraviolet-B region (280~315 nm) via an intrinsic tryptophan-based mechanism (Rizzini et al. 2011; Christie et al. 2012; Wu et al. 2012). The evolutionary history of UV-B photoreceptors can be traced back to ancestral Viridiplantae, because both streptophyte land plants and chlorophyte algae such as C. reinhardtii and Volvox carteri possess apparent UVR8 orthologs. CrUVR8 contains conserved Trp residues essential for UV-B perception; photoexcitation has been shown to induce conformational changes and subsequent monomerization of the protein (Tilbrook et al. 2016). Like plant UVR8, light-activated CrUVR8 interacts with CrCOP1 (Constitutively Photomorphogenic 1) and induces expression of photosynthesis and photoprotection-related genes. Successful complementation of the Arabidopsis uvr8 mutant by ectopic expression of CrUVR8 further suggests the functional conservation of UV-B signaling and acclimation in the green lineage even after ~100 million years of independent evolution (Tilbrook et al. 2016). UV-B-dependent behavioral changes, i.e., photoavoidance or photosystem restructuring responses, were also observed in several cyanobacteria (Bebout & Garcia-Pichel 1995; Campbell et al. 1998). Although not yet identified, the putative UV-B photoreceptor of cyanobacteria is hypothesized to use a reduced pterin as chromophore to regulate biosynthesis of UV-absorbing, mycosporine-like amino acids (Portwich & Garcia-Pichel 2000). The wide distribution of UVR8 homologs within Viridiplantae (green algae, lower and higher plants) suggests the necessity of UV-B sensing during water to land colonization (Parihar et al. 2016).
Bilin-based phytochromes in algae
Phytochrome was the first bilin-based photoreceptor to be discovered. As photoswitchable red/far-red photosensors, plant phytochromes are master regulators of plant growth and development that use phytobilins as chromophores (Rockwell et al. 2006; Chen & Chory 2011). Canonical phytochromes have a conserved N-terminal photosensory core module (PCM), consisting of PAS (Per-ARNT-Sim domain), GAF (cGMP-specific phosphodiesterases, adenylyl cyclases and formate hydrogen lyase) and PHY (phytochrome specific) domains. Cyanobacteria possess the largest diversity of phytochromes, which include the phytochrome-derived cyanobacteriochrome (CBCR) family that have proliferated in many cyanobacterial species (Ikeuchi & Ishizuka 2008; Rockwell & Lagarias 2010; Anders & Essen 2015). CBCRs exhibit a broad range of photocycles, encompassing the near-UV to the far red region of the visible light spectrum (Ikeuchi & Ishizuka 2008; Rockwell et al. 2011; Rockwell et al. 2016). However, CBCRs are not found in eukaryotic algae, indicating that they either evolved after primary endosymbiosis or were lost during endosymbiosis. By contrast, phytochromes are widely distributed in the Archaeplastida indicating their presence in the founding member of this lineage. Nevertheless, many chlorophyte (green) algae lack phytochrome genes altogether (see below). The wide distribution of phytochromes within the Viridiplantae indicates that phytochromes were selectively lost from the chlorophyte lineage (Duanmu et al. 2014; Li et al. 2015a).
The enigmatic evolutionary origin of algal and plant phytochromes
Structural, evolutionary and functional studies of phytochromes have been greatly aided by characterization of cyanobacterial and eukaryotic algal phytochromes. The cyanobacterial phytochrome Cph1 was the first prokaryotic algal phytochrome identified. Cph1 uses phycocyanobilin (PCB) as chromophore and exhibits a red/far-red photocycle similar to higher plant phytochromes that use phytochromobilin (PΦB) as chromophore (Hughes et al. 1997; Yeh et al. 1997). Cph1 arose within the widely distributed bacteriophytochrome (BphP) family of biliverdin (BV)-binding photoreceptors with identical domain architecture (Karniol & Vierstra 2006; Auldridge & Forest 2011). Since plant and cyanobacterial phytochromes share nearly identical PCMs and a conserved bilin-binding cysteine residue, it was proposed that eukaryote phytochrome PCMs arose by gene transfer from the cyanobacterial endosymbiont (Buchberger & Lamparter 2015).
However, recently released algal genomic and transcriptomic data support the alternative hypothesis that Archaeplastida phytochromes arose independently from those found in extant cyanobacteria (Duanmu et al. 2014; Li et al. 2015a; Fortunato et al. 2016). It has thus been proposed that the Archaeplastida phytochrome PCM originated from the pre-photosynthetic eukaryotic host via acquisition of a phytochrome gene from a non-cyanobacterial source (Duanmu et al. 2014). The evidence that phytochromes found in other eukaryotes, e.g., heterokonts (diatom and brown algae), algal viruses and fungi, possess PCMs more similar to those of phytochromes in the Archaeplastida lineage, is consistent with this hypothesis (Montsant et al. 2007; Duanmu et al. 2014; Fortunato et al. 2016).
In addition to the evolutionary implications of PCM comparisons, phylogenetic analyses of the C-terminal histidine kinase-related output modules (HKMs; i.e. H-accepting and ATPase domains) of phytochromes are consistent with the repeated and independent acquisition hypothesis (Duanmu et al. 2014). While the origin of Archaeplastida phytochrome HKMs from a proteobacterial ancestor protein with multiple PAS domain and a C-terminal HK has been proposed (Buchberger & Lamparter 2015), the appearance of this module clearly coincides with emergence of the Viridiplantae (Li et al. 2015a). These studies also reveal that the plant phytochrome lineage arose within the streptophyte algae at the same time they were beginning to colonize land environments.
Diverse photocycles of algal phytochromes
Several recent studies reveal that algal phytochromes sense a wider wavelength range than that of classic red/far-red phytochromes from plants. Expression of representative prasinophyte, glaucophyte, and heterokont phytochromes in engineered Escherichia coli cells that coproduce phytobilin chromophores has revealed diverse photocycles spanning blue to far-red light in different algal lineages, presumably reflecting depth, ocean mixing, or other environmental parameters associated with different algal species (Duanmu et al. 2014; Rockwell et al. 2014a). The blue-shifted dark state of algal phytochromes could be associated with detecting the green and blue light present in seawater at depth. EsilPHL1, a phytochrome from the heterokont alga Ectocarpus siliculosus, yielded a novel far-red/green photocycle distinct from that of plant phytochromes (Rockwell et al. 2014a). However, the related phytochromes from two diatoms (Phaeodactylum tricornutum and Thalassiosira pseudonana) exhibit red/far-red photocycles slightly red-shifted from those of plant phytochromes due to their more oxidized BV chromophores (Fortunato et al. 2016). Due to the production of both phycobilins and BV in these heterokont species, the native chromophores of these interesting proteins remain to be determined. Based on these studies, the broad spectral diversity of eukaryotic algal phytochromes appears comparable to those of the CBCRs. Thus, these distantly related protein families appear to have independently evolved new light sensing ranges better suited to aquatic environments than are the classical red/far-red responses plant phytochromes use to assess competition with neighboring plants in terrestrial environments (Casal 2013).
The diversity of algal phytochrome signaling mechanisms
The N-terminal light sensory module of phytochromes is most commonly associated with a C-terminal histidine kinase-related output module (HKM) (Schneider-Poetsch 1992), implicating phosphotransfer to be the mechanism of signal transduction. Biochemical analyses confirmed that Cph1 functions as a light-regulated two-component sensor (TCS). The red light absorbing Pr form of Cph1 autophosphorylates the conserved histidine residue in the HKM, supporting phosphotransfer to an aspartate residue of the cognate Rcp1 response regulator (Yeh et al. 1997). This C-terminal regulatory module has undergone frequent domain replacements and/or losses after primary endosymbiosis. For example, some cyanobacterial phytochromes and glaucophyte phytochrome sensors (GPS) possess distinct C-terminal regulatory domains with extra PAS domains between the PHY domain and HKM, whereas cryptophyte algae have phytochromes in which the HKM has been replaced with canonical Ser/Thr kinase domains (Duanmu et al. 2014; Li et al. 2015a). Similar changes of kinase domains have occurred in phytochromes in streptophyte algae, hornworts, mosses and ferns (Thümmler et al. 1992; Nozue et al. 1998; Suetsugu et al. 2005; Li et al. 2015a). These results implicate organism-specific functions for these phytochromes that are absent in other lineages.
Surprisingly, the domain organizations of phytochromes from prasinophytes are similar to plant phytochromes except for the presence of one or more C-terminal TCS receiver (REC) domains (Duanmu et al. 2014; Rockwell et al. 2014a). Light-regulated histidine kinase activity was confirmed biochemically in one prasinophyte alga, Dolichomastix tenuilepis (Duanmu et al. 2014), suggesting that all prasinophyte phytochromes function as light regulated two component sensors. During the emergence of streptophyte phytochromes, both histidine phosphotransfer activity and C-terminal REC domain(s) have been lost. Plant phytochromes have also acquired new regulatory functions, such as PIF (phytochrome interacting factor) binding and/or serine/threonine phosphotransfer activities (Yeh & Lagarias 1998; Li et al. 2015a).
Functional analyses of several algal phytochromes has shed further insights into their signaling mechanisms. The prasinophyte alga Micromonas pusilla possesses one phytochrome gene that encodes a protein with a functional histidine kinase coupled with a REC domain (Worden et al. 2009). The abundance of MpPHY protein was quite stable throughout the light-dark cycle, similar to the light-stable phytochromes of plants. Another hallmark property of plant phytochromes, relocalization to the nucleus in the light period of the diurnal cycle, is conserved in MpPHY (Duanmu et al. 2014). RNA-seq measurements revealed that transcripts of bilin biosynthetic genes and of MpPHY itself were light-regulated and preceded those of most photosynthesis related genes, suggesting a shared signaling mechanism for prasinophyte and plant phytochromes (Duanmu et al. 2014).
By contrast with prasinophyte phytochromes, the abundance of diatom phytochromes gradually decreased in the light and reaccumulated after transfer to darkness. Transcriptomic measurements implicated a regulatory role for diatom phytochromes in a far-red light response that targets ~80 mostly diatom-specific genes of unknown function (Fortunato et al. 2016). These studies suggest that diatom phytochromes target a unique pathway distinct from that of green algal and plant phytochromes, consistent with the evolutionary divergence of heterokont and Viridiplantae species. The authors propose that the diatom phytochrome might function to detect changes in the red/far red ratio with depth and/or the presence of other Chl-containing photosynthetic organisms (Fortunato et al. 2016).
Although phytochromes are present in many primary and secondary algal species (Figure 1), rhodophytes such as Cyanidioschyzon merolae and Porphyridium purpureum and chlorophytes such as Chlamydomonas reinhardtii, Volvox carteri and Chlorella variabilis NC64A lack phytochrome genes (Duanmu et al. 2014; Li et al. 2015a). Prasinophytes typically possess one phytochrome gene or none at all. All streptophyte species have at least one phytochrome gene, and many species have two or more phytochromes. This includes streptophyte algae, in which three distinct families have been identified, and land plants, which can contain five or more phytochrome genes (Li et al. 2015a).
Figure 1.
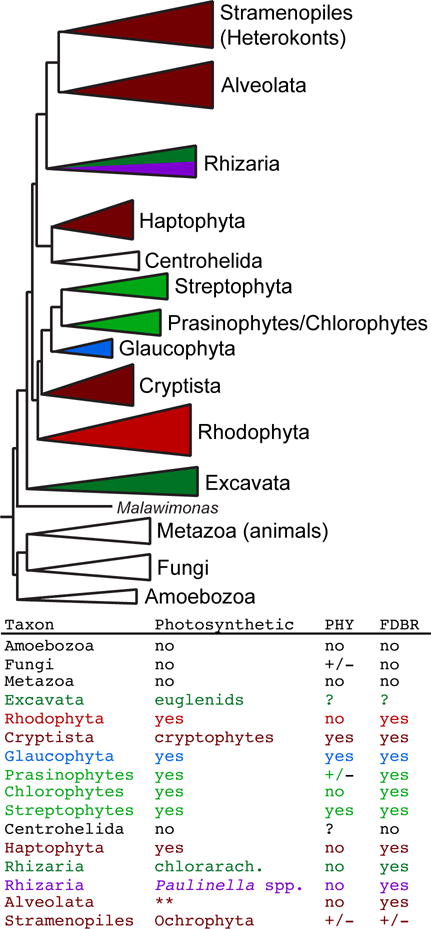
Distribution of phytochromes (PHY) and FDBRs (ferredoxin-dependent bilin reductases) in eukaryotes. (top) A simplified cartoon of eukaryotic evolution is shown, following the scheme of Keeling and colleagues (Burki et al. 2016). (bottom) The presence of photosynthesis, PHY and FDBRs in various taxa is indicated. Photosynthetic taxa are color-coded by plastid lineage as follows: rhodophyte, red; red-derived, dark red; glaucophyte, blue; Viridiplantae, green; green-derived, dark green. The independent primary endosymbiosis in Paulinella chromatophora and related species (Yoon et al. 2009; Kim & Park 2016) is color-coded in purple. Non-photosynthetic taxa (Amoebozoa, Fungi, Metazoa, Centrohelida) are in white and lack FDBRs. For photosynthetic taxa, the presence or absence of phytochromes and FDBRs was assessed previously (Rockwell et al. 2014b). Photosynthetic stramenopiles (Ochrophyta) have been proposed to be monophyletic (e.g., Derelle et al. 2016). The photosynthetic stramenopile Aureococcus anophagefferens lacks FDBRs. Photosynthetic alveolates (double asterisk: dinoflagellates and chromerids) contain FDBR genes, whereas nonphotosynthetic alveolates do not. Prasinophytes + streptophytes comprise the Viridiplantae. Stramenopiles, Alveolata, and Rhizaria comprise the SAR clade. +/−, variable presence within photosynthetic organisms.
Gene duplication followed by functional specialization probably results in expansion of phytochromes with diverse functions in seed plants (Rensing et al. 2016). Interestingly, multiple copies of phytochromes are also present in many multicellular algae, such as the heterokont alga Ectocarpus siliculosus and the related phaeophyte alga Saccharina japonica (Wang et al. 2013). These may reflect sub- or neo-functionalization events in multicellular algae. Further investigation is needed to address the biological functions of these interesting photoreceptors.
EMERGING SIGNALING ROLES FOR BILINS
As breakdown products from heme, linear tetrapyrroles (bilins) are well known cofactors of either light harvesting phycobiliproteins (in cyanobacteria, glaucophytes, rhodophytes, and cryptophytes) and of light-sensing phytochromes of cyanobacteria, land plants and many, but not all eukaryotic algae. Although BV is sometimes used, the more reduced phytobilins, PCB, phycoerythrobilin (PEB) and PΦB, are more commonly used as precursors of the covalently bound chromophores of both classes of biliprotein. Due to feedback inhibition of heme oxygenase, BV is only produced in small amounts in algae and plants. Ferredoxin-dependent bilin reductases (FDBRs) are responsible for the reduction of BV that regenerates active heme oxygenase for a new round of heme turnover while also yielding phytobilin products. FDBRs to date have only been found in oxygenic phototrophs, and the phytobilins they produce are the major bilin metabolites found in algal and plant cells (Dammeyer & Frankenberg-Dinkel 2008; Rockwell et al. 2014b).
Apart from their role as chromophore precursors, bilins have been implicated as signaling molecules in some systems. In Xenopus laevis, BV acts as a cytoplasmic determinant that is involved in the regulation of dorsal axis development in the embryo (Falchuk et al. 2002). Novel light sensing roles for bilins have also been implicated by studies on several phycobiliproteins. The allophycocyanin-like (AplA) protein from the cyanobacterium Fremyella diplosiphon contains a covalently attached bilin chromophore yet lacks conserved amino acids critical for known phycobiliprotein structure. The absence of AplA from purified phycobilisomes (PBS) suggests that biological functions of these proteins are not related to light harvesting. It has been proposed that AplA might instead play a role in light sensing for regulation of PBS transcription and protein abundance (Montgomery et al. 2004). Another cyanobacterium, Prochlorococcus sp. MED4, depends on phycoerythrin as a green light-absorbing photoreceptor for high-light acclimation (Steglich et al. 2005). Moreover, although phycobiliprotein-deficient cyanobacteria remain fully viable, FDBR knockout experiments suggest that bilin biosynthesis is essential for phototrophic growth of Synechococcus sp. PCC 7002 (Alvey et al. 2011). However, it remains possible that FDBRs perform other functions that are essential to viability, because exogenous phytobilin rescue experiments are difficult to interpret when the results are negative. Through inactivation of bilin biosynthesis in C. reinhardtii, our recent studies on the hmox1 mutant have revealed a potential signaling role for bilins (Duanmu et al. 2013). As we will discuss in the succeeding text, this signaling role impacts the processes needed to cope with dark-to-light transitions that are one of the most dangerous environmental challenges faced by photosynthetic organisms.
Diversity of bilin biosynthesis in eukaryotic algae
All oxygenic phototrophs possess plastid-localized heme oxygenases (HOs) that catalyze the oxygen-dependent conversion of heme to BV (Tanaka & Tanaka 2007; Terry & Smith 2013). To support high throughput HO turnover, two class of BV reductases evolved in nature, i.e., the NAD(P)H-dependent BVR/BvdRs (Kutty & Maines 1981; Schluchter & Glazer 1997) and the FDBR family of phytobilin synthases (Dammeyer & Frankenberg-Dinkel 2008). In contrast with the limited distribution of BvdRs, FDBRs are found in all extant cyanobacteria, including prochlorophytes which lack phycobiliprotein antennae. Eukaryotic FDBRs are restricted to photosynthetic eukaryotes and are exclusively localized to plastids. It is therefore likely that eukaryotic HOs and FDBRs arose via gene transfer from an ancestral cyanobacterial endosymbiont to the host nucleus, resulting in a phytobilin biosynthetic pathway indispensable to extant photosynthetic eukaryotes (Rockwell et al. 2014b). Interestingly, several heterotrophic dinoflagellates apparently lack FDBRs (Jeong et al. 2010), consistent with an essential function for bilin biosynthesis in oxygenic photosynthetic organisms.
Genomic and transcriptomic data has confirmed the presence of FDBRs in all phototrophic eukaryotic taxa, including species that are derived from secondary endosymbiosis, with the possible exception of euglenids (Rockwell et al. 2014a, b; Figure 1). Different FDBRs or combinations of FDBRs are found in different algal lineages. Consistent with the presence of three FDBR lineages, PcyA (ferredoxin:phycocyanobilin oxidoreductase), PebA (ferredoxin: 15, 16-dihydrobiliverdin oxidoreductase) and PebB (ferredoxin: phycoerythrobilin oxidoreductase) in extant cyanobacteria (Frankenberg et al. 2001), all Archaeplastida lineages have retained at least one descendant derived from one or more of these lineages (Figure 1). These include PCYA, PUBS and HY2 enzymes that are responsible for the respective synthesis of phycocyanobilin (PCB), phycourobilin (PUB) and phytochromobilin (PΦB) in photosynthetic eukaryotes. Representatives of the PebA-related PUBS family can be found in the Viridiplantae in both chlorophyte and streptophyte lineages (Chen et al. 2012), whereas HY2 is only found in streptophytes (Rockwell et al. 2017). It is notable that no single FDBR is indispensable across the highly diverged algal lineages (Rockwell et al. 2014b). Bilin products of FDBRs generally serve as chromophore cofactors for phytochromes or phycobiliproteins. However, many eukaryotic algae (i.e., chlorophytes and some prasinophytes) lack both classes of biliproteins (Duanmu et al. 2014; Rockwell et al. 2014a). Such observations raise a key question: what are the biological functions of bilins in these organisms?
Retrograde bilin signals are important for the dark-to-light transition in Chlamydomonas
Recent studies have addressed this question in the green alga Chlamydomonas, a chlorophyte alga lacking phycobiliproteins and phytochromes. The bilin biosynthetic pathway of C. reinhardtii consists of a heme oxygenase (HMOX1) producing BV from heme and an FDBR (PCYA1) producing PCB from BV. Both proteins are encoded in the nucleus and targeted to the chloroplast (Merchant et al. 2007; Duanmu et al. 2013). Chlamydomonas also contains an animal type heme oxygenase (HMOX2), with a putative C-terminal transmembrane helix that appears to anchor the protein to the endoplasmic reticulum (Duanmu et al. 2013). Homologs of the animal type HMOX2 are present in other chlorophytes (V. carteri and Chlorella sp NC64A) but are absent in cyanobacteria and plants, e.g., all four Arabidopsis heme oxygenases are targeted to the chloroplast (Gisk et al. 2010). Different from mammalian heme oxygenases that convert heme to biliverdin IXα, Chlamydomonas HMOX2 cleaves heme nonspecifically and makes several biliverdin isomers. While the origin and biological function of HMOX2 remain unclear, putative roles for the cytosolic membrane-associated HMOX2 include heme detoxification and iron recycling (Duanmu et al. 2013).
Using a cyanobacterial bilin-binding GAF domain as reporter, these studies also showed that PCB biosynthesis in vivo required a functional HMOX1 gene (Duanmu et al. 2013). Disruption of PCB biosynthesis in the knock-out hmox1 mutant compromised light-dependent Chl accumulation and photoautotrophic growth, phenotypes which could be rescued by in vitro feeding of BV. Transcriptomic analysis uncovered bilin-regulated gene networks essential for the dark to light transition in Chlamydomonas. Significantly different from phytochrome regulation of photosynthesis-associated nuclear genes (PhANGs) in plants, the majority of the bilin-responsive genes in Chlamydomonas have unknown functions. As such, this discovery holds considerable potential to help understand the molecular mechanisms governing dark/light transition in green algae. Since the hmox1 mutant is unable to grow in minimal medium containing no organic carbon source, this genetic program appears to be essential for maintaining photoautotrophic growth (Duanmu et al. 2013).
As a semi-autonomous organelle, the chloroplast is able to relay its developmental and functional states to nucleus for real-time coordination of nuclear genome expression for optimal photosynthetic light energy capture. The informational flow from chloroplast to nucleus is called retrograde signaling (Singh et al. 2015). Among the most well documented plastid-derived retrograde signals are intermediates of the tetrapyrrole biosynthetic pathway; however, signals derived from reactive oxygen species (ROS), chloroplast redox state and chloroplast gene expression have also received experimental support (Nagahatenna et al. 2015). Bilins are promising candidates as retrograde signaling molecules because they are synthesized in the chloroplast, they are exported to the cytoplasm and the genes for key enzymes in their synthesis are regulated by the clock. These attributes make them desirable candidates as signals for reprogramming nuclear transcription, especially during the dark/light transition. The presence of bilin biosynthetic enzymes in all eukaryotic algae might thus be explained by an essential role for bilins in anticipating oxidative stress during the transition from dark/heterotrophic to light/phototrophic growth (Figure 2). Bilins could be also involved in sustaining functional chloroplasts under continuous light illumination (Duanmu et al. 2013; Rochaix 2013). However, more work is needed to elucidate the molecular basis of bilin-dependent photosynthesis regulation in green algae.
Figure 2.
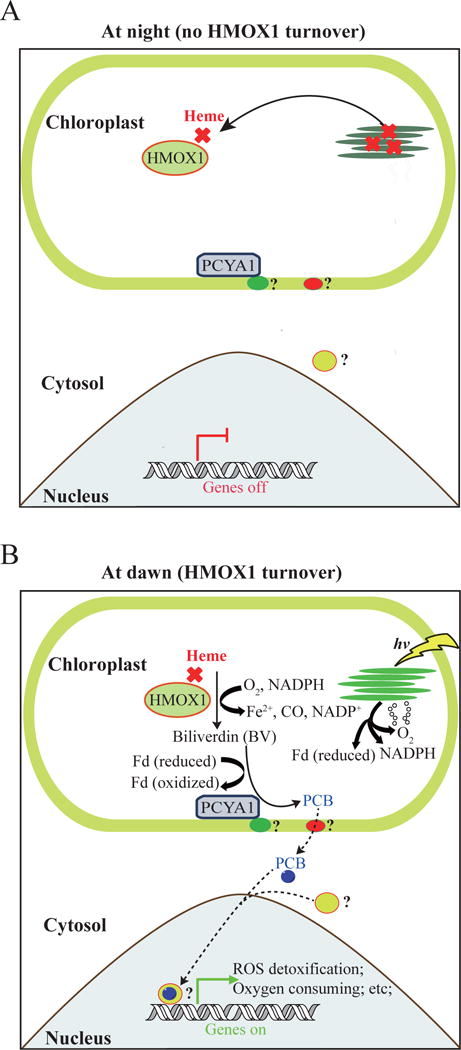
Schematic illustration of hypothetical retrograde bilin signaling in Chlamydomonas reinhardtii. (A) At night, repair of damaged photosynthetic apparatus afects heme release and HMOX1 capture. (B) At dawn, oxygen-dependent heme turnover and PCB (phycocyanobilin) biosynthesis occurs. PCB might be exported to the cytosol and finally to the nucleus to regulate gene expression and mitigate oxidative stress during dark to light transition. Fd, ferredoxin; HMOX1, heme oxygenase; PCYA1, phycocyanobilin ferredoxin oxidoreductase. Association of PCYA1 with the chloroplast envelope membrane could be due to its interaction with an unknown membrane protein (green oval). PCB is expected to be exported out of the chloroplast by putative bilin transporter (red oval). In the cytosol, a putative bilin sensor (yellow oval) binds PCB and the holoprotein moves to the nucleus to regulate gene expression.
Perspectives on bilin signaling in phototrophic eukaryotes
The diurnal light/dark cycle imposes serious challenges to survival of oxygenic photosynthetic organisms. Oxidative stress in chloroplasts at dawn and throughout the day requires both rapid acclimation and slower adaptive responses that include activation of nucleus-encoded stress responsive proteins and other photoprotective components to facilitate the daily transition from dark/heterotrophic to light/phototrophic growth. Bilins are well suited to integrate such environmental signals, because their biosynthesis by heme oxygenase and FDBRs is tied to production of oxygen and reductant via photosynthesis (Figure 2). Moreover, oxygenic eukaryotes arose via capture of an ancestral cyanobacterial cell that was filled with bilins. It is therefore plausible that bilins have gained important signaling functions during the ~1.5 billion years of evolution since endosymbiosis (Montgomery & Lagarias 2002).
Besides bilins, other tetrapyrrole intermediates have been hypothesized to function as putative chloroplast signals in plants. Magnesium protoporphyrin IX (MgPPIX) appears to represent a negative “operational control” signal mediating photo-oxidative damage response when plastid integrity is disrupted by herbicides or plastid translation inhibitors. In contrast, a specific non-photosynthetic heme pool generated by ferrochelatase-1 in plants appears to function as a positive “biogenic control” signal to up-regulate PhANG expression (Woodson et al. 2011; Terry & Smith 2013). Heme is a bilin precursor, and phytochromes promote PhANG expression when activated by light; therefore, bilins clearly function as positive retrograde signals in plant and algal species that retain phytochromes. Finally, phyllobilins, a class of bilin-type Chl breakdown products structurally related to bilins derived from heme, have also been proposed to play important biological functions besides their well-known roles as signs of leaf senescence and fruit ripening (Kräutler 2014). Although the metabolic pathways are poorly understood, phyllobilins were found in some green algae and marine dinoflagellates (Engel et al. 1991). In addition to phyllobilins, it is tempting to speculate that other bilins derived from Chl breakdown, such as those observed in phagotrophic protists (Kashiyama et al. 2012), are also good candidate signaling molecules in both photosynthetic and non-photosynthetic species.
In plants, bilin-based phytochromes are master regulators of PhANG expression and many other light-dependent processes. The putative retrograde signaling activity of bilins might thus be masked by phytochrome signaling (Duanmu et al. 2013). Interestingly, targeted expression of mammalian biliverdin reductase in Arabidopsis suggested a regulatory function of bilins within the chloroplast which is distinct from cytosolic phytochrome signaling (Lagarias et al. 1997; Montgomery et al. 1999; Warnasooriya & Montgomery 2009). It thus seems plausible that bilin-dependent retrograde signaling will prove important to all photosynthetic eukaryotes.
Many questions, however, still remain to be answered. How are bilins trafficked out of the chloroplast? What are the identities of putative bilin sensors or receptors and where are they localized in the cell? How are bilin signals relayed to the nucleus and resulting in expression of specific genes? Mechanistic investigations of bilin signaling in model algae such as C. reinhardtii is expected to provide novel insights into strategies phototrophic eukaryotes developed to optimize their photosynthetic activity in natural settings in the absence of phytochrome.
CONCLUDING REMARKS
In the last two decades, we have gained a deeper understanding of photosynthesis and light signaling mechanisms in aquatic eukaryotic algae, which have evolved for over ~1 billion years and are now present in almost every ecological niche on earth. The number of extant algal species has been estimated between 30,000 to more than 1 million (Guiry 2012). So far, more than 146,000 species have been described in AlgaeBase (www.algaebase.org). With high throughput sequencing technologies and sophisticated “-omics” tools now available, we are confident that significant progress will be made in the next decade towards characterization of light harvesting, light sensing and other photoacclimation processes in a number of model algal species.
SUMMARY STATEMENT.
Aquatic environments pose daunting challenges for efficient light harvesting and photosynthesis, requiring novel photoacclimation strategies in eukaryotic algae. Recent studies provide new insight into the loss and repurposing of bilin-based photoreceptors in algae that facilitate responses to inevitable changes in light fluences inherent to their complex light environments.
Acknowledgments
We thank Professor Jean-David Rochaix (University of Geneva, Switzerland) for critical evaluation of this manuscript. Research in the Duanmu laboratory was supported by the National Natural Science Foundation of China (grant no. 31570233), Huazhong Agricultural University Scientific and Technological Self-innovation Foundation (Program No.2014RC018) and the Fundamental Research Funds for the Central Universities (Program No. 2662015PY171). We also acknowledge financial support from NIH GM068552 and NSF-MCB-0843625 (both to J.C.L.).
Footnotes
The authors declare no conflict of interest.
References
- Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- Alvey RM, Biswas A, Schluchter WM, Bryant DA. Effects of modified phycobilin biosynthesis in the cyanobacterium Synechococcus sp. Strain PCC 7002. Journal of Bacteriology. 2011;193:1663–1671. doi: 10.1128/JB.01392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders K, Essen LO. The family of phytochrome-like photoreceptors: diverse, complex and multi-colored, but very useful. Current Opinion in Structural Biology. 2015;35:7–16. doi: 10.1016/j.sbi.2015.07.005. [DOI] [PubMed] [Google Scholar]
- Archibald JM, Keeling PJ. Recycled plastids: a ‘green movement’ in eukaryotic evolution. Trends in Genetics. 2002;18:577–584. doi: 10.1016/s0168-9525(02)02777-4. [DOI] [PubMed] [Google Scholar]
- Auldridge ME, Forest KT. Bacterial phytochromes: More than meets the light. Critical Reviews in Biochemistry and Molecular Biology. 2011;46:67–88. doi: 10.3109/10409238.2010.546389. [DOI] [PubMed] [Google Scholar]
- Bailleul B, Rogato A, de Martino A, Coesel S, Cardol P, Bowler C, Finazzi G. An atypical member of the light-harvesting complex stress-related protein family modulates diatom responses to light. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18214–18219. doi: 10.1073/pnas.1007703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballottari M, Girardon J, Dall’osto L, Bassi R. Evolution and functional properties of photosystem II light harvesting complexes in eukaryotes. Biochimica et Biophysica Acta. 2012;1817:143–157. doi: 10.1016/j.bbabio.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Herman E, Serif M, Maestre-Reyna M, Hepp S, Pokorny R, Kottke T. Allosteric communication between DNA-binding and light-responsive domains of diatom class I aureochromes. Nucleic Acids Research. 2016;44:5957–5970. doi: 10.1093/nar/gkw420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebout BM, Garcia-Pichel F. UV B-Induced vertical migrations of cyanobacteria in a microbial mat. Applied and Environmental Microbiology. 1995;61:4215–4222. doi: 10.1128/aem.61.12.4215-4222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beel B, Müller N, Kottke T, Mittag M. News about cryptochrome photoreceptors in algae. Plant Signaling & Behavior. 2013;8:e22870. doi: 10.4161/psb.22870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beel B, Prager K, Spexard M, Sasso S, Weiss D, Müller N, Mittag M. A flavin binding cryptochrome photoreceptor responds to both blue and red light in Chlamydomonas reinhardtii. Plant Cell. 2012;24:2992–3008. doi: 10.1105/tpc.112.098947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonente G, Ballottari M, Truong TB, Morosinotto T, Ahn TK, Fleming GR, Bassi R. Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLoS Biology. 2011;9:e1000577. doi: 10.1371/journal.pbio.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant DA, Frigaard NU. Prokaryotic photosynthesis and phototrophy illuminated. Trends in Microbiology. 2006;14:488–496. doi: 10.1016/j.tim.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Buchberger T, Lamparter T. Streptophyte phytochromes exhibit an N-terminus of cyanobacterial origin and a C-terminus of proteobacterial origin. BMC Research Notes. 2015;8:144. doi: 10.1186/s13104-015-1082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büchel C. Evolution and function of light harvesting proteins. Journal of Plant Physiology. 2015;172:62–75. doi: 10.1016/j.jplph.2014.04.018. [DOI] [PubMed] [Google Scholar]
- Burki F, Kaplan M, Tikhonenkov DV, Zlatogursky V, Minh BQ, Radaykina LV, Keeling PJ. Untangling the early diversification of eukaryotes: a phylogenomic study of the evolutionary origins of Centrohelida, Haptophyta and Cryptista. Proceedings of the Royal Society B Biological Sciences. 2016;283 doi: 10.1098/rspb.2015.2802. pii: 20152802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D, Eriksson MJ, Oquist G, Gustafsson P, Clarke AK. The cyanobacterium Synechococcus resists UV-B by exchanging photosystem II reaction-center D1 proteins. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:364–369. doi: 10.1073/pnas.95.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. Photoreceptor signaling networks in plant responses to shade. Annual Review of Plant Biology. 2013;64:403–427. doi: 10.1146/annurev-arplant-050312-120221. [DOI] [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Ahmad M. The cryptochromes: blue light photoreceptors in plants and animals. Annual Review of Plant Biology. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J. Phytochrome signaling mechanisms and the control of plant development. Trends in Cell Biology. 2011;21:664–671. doi: 10.1016/j.tcb.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YR, Su YS, Tu SL. Distinct phytochrome actions in nonvascular plants revealed by targeted inactivation of phytobilin biosynthesis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8310–8315. doi: 10.1073/pnas.1201744109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM. Phototropin blue-light receptors. Annual Review of Plant Biology. 2007;58:21–45. doi: 10.1146/annurev.arplant.58.032806.103951. [DOI] [PubMed] [Google Scholar]
- Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O’Hara A, Getzoff ED. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science. 2012;335:1492–1496. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coesel S, Mangogna M, Ishikawa T, Heijde M, Rogato A, Finazzi G, Falciatore A. Diatom PtCPF1 is a new cryptochrome/photolyase family member with DNA repair and transcription regulation activity. EMBO Reports. 2009;10:655–661. doi: 10.1038/embor.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad KS, Manahan CC, Crane BR. Photochemistry of flavoprotein light sensors. Nature Chemical Biology. 2014;10:801–809. doi: 10.1038/nchembio.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Osto L, Bressan M, Bassi R. Biogenesis of light harvesting proteins. Biochimica et Biophysica Acta. 2015;1847:861–871. doi: 10.1016/j.bbabio.2015.02.009. [DOI] [PubMed] [Google Scholar]
- Dammeyer T, Frankenberg-Dinkel N. Function and distribution of bilin biosynthesis enzymes in photosynthetic organisms. Photochemical & Photobiological Sciences. 2008;7:1121–1130. doi: 10.1039/b807209b. [DOI] [PubMed] [Google Scholar]
- Derelle R, López-García P, Timpano H, Moreira D. A phylogenomic framework to study the diversity and evolution of stramenopiles (=heterokonts) Molecular Biology and Evolution. 2016;33:2890–2898. doi: 10.1093/molbev/msw168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depauw FA, Rogato A, Ribera d’Alcalá M, Falciatore A. Exploring the molecular basis of responses to light in marine diatoms. Journal of Experimental Botany. 2012;63:1575–1591. doi: 10.1093/jxb/ers005. [DOI] [PubMed] [Google Scholar]
- Dinc E, Tian L, Roy LM, Roth R, Goodenough U, Croce R. LHCSR1 induces a fast and reversible pH-dependent fluorescence quenching in LHCII in Chlamydomonas reinhardtii cells. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:7673–7678. doi: 10.1073/pnas.1605380113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu D, Bachy C, Sudek S, Wong CH, Jiménez V, Rockwell NC, Worden AZ. Marine algae and land plants share conserved phytochrome signaling systems. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:15827–15832. doi: 10.1073/pnas.1416751111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu D, Casero D, Dent RM, Gallaher S, Yang W, Rockwell NC, Lagarias JC. Retrograde bilin signaling enables Chlamydomonas greening and phototrophic survival. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3621–3626. doi: 10.1073/pnas.1222375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel N, Jenny TA, Mooser V, Gossauer A. Chlorophyll catabolism in Chlorella protothecoides. Isolation and structure elucidation of a red bilin derivative. FEBS Letters. 1991;293:131–133. doi: 10.1016/0014-5793(91)81168-8. [DOI] [PubMed] [Google Scholar]
- Engelken J, Funk C, Adamska I. The Extended Light-Harvesting Complex (LHC) Protein Superfamily: Classification and Evolutionary Dynamics. In: Burnap RI, Vermaas WJF, editors. Functional Genomics and Evolution of Photosynthetic Systems. Dordrecht, The Netherlands: 2012. pp. 265–284. [Google Scholar]
- Falchuk KH, Contin JM, Dziedzic TS, Feng Z, French TC, Heffron GJ, Montorzi M. A role for biliverdin IXα in dorsal axis development of Xenopus laevis embryos. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:251–256. doi: 10.1073/pnas.012616099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer WW, Hemp J, Johnson JE. Evolution of Oxygenic Photosynthesis. Annual Review of Earth and Planetary Sciences. 2016;44:647–683. [Google Scholar]
- Fortunato AE, Annunziata R, Jaubert M, Bouly JP, Falciatore A. Dealing with light: the widespread and multitasking cryptochrome/photolyase family in photosynthetic organisms. Journal of Plant Physiology. 2015;172:42–54. doi: 10.1016/j.jplph.2014.06.011. [DOI] [PubMed] [Google Scholar]
- Fortunato AE, Jaubert M, Enomoto G, Bouly JP, Raniello R, Thaler M, Falciatore A. Diatom phytochromes reveal the existence of far-red-light-based sensing in the ocean. Plant Cell. 2016;28:616–628. doi: 10.1105/tpc.15.00928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenberg N, Mukougawa K, Kohchi T, Lagarias JC. Functional genomic analysis of the HY2 family of ferredoxin-dependent bilin reductases from oxygenic photosynthetic organisms. Plant Cell. 2001;13:965–978. doi: 10.1105/tpc.13.4.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão VC, Fankhauser C. Sensing the light environment in plants: photoreceptors and early signaling steps. Current Opinion in Neurobiology. 2015;34:46–53. doi: 10.1016/j.conb.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Gisk B, Yasui Y, Kohchi T, Frankenberg-Dinkel N. Characterization of the haem oxygenase protein family in Arabidopsis thaliana reveals a diversity of functions. Biochemical Journal. 2010;425:425–434. doi: 10.1042/BJ20090775. [DOI] [PubMed] [Google Scholar]
- Goss R, Lepetit B. Biodiversity of NPQ. Journal of Plant Physiology. 2015;172:13–32. doi: 10.1016/j.jplph.2014.03.004. [DOI] [PubMed] [Google Scholar]
- Grabowski B, Cunningham FX, Jr, Gantt E. Chlorophyll and carotenoid binding in a simple red algal light-harvesting complex crosses phylogenetic lines. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:2911–2916. doi: 10.1073/pnas.031587198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AR, Bhaya D, Apt KE, Kehoe DM. Light-harvesting complexes in oxygenic photosynthesis: diversity, control, and evolution. Annual Review of Genetics. 1995;29:231–288. doi: 10.1146/annurev.ge.29.120195.001311. [DOI] [PubMed] [Google Scholar]
- Guiry MD. How many species of algae are there? Journal of Phycology. 2012;48:1057–1063. doi: 10.1111/j.1529-8817.2012.01222.x. [DOI] [PubMed] [Google Scholar]
- Gupta RS. Molecular signatures for the main phyla of photosynthetic bacteria and their subgroups. Photosynthesis Research. 2010;104:357–372. doi: 10.1007/s11120-010-9553-9. [DOI] [PubMed] [Google Scholar]
- Gupta RS. Origin and spread of photosynthesis based upon conserved sequence features in key bacteriochlorophyll biosynthesis proteins. Molecular Biology and Evolution. 2012;29:3397–3412. doi: 10.1093/molbev/mss145. [DOI] [PubMed] [Google Scholar]
- Hegemann P. Algal sensory photoreceptors. Annual Review of Plant Biology. 2008;59:167–189. doi: 10.1146/annurev.arplant.59.032607.092847. [DOI] [PubMed] [Google Scholar]
- Heijde M, Zabulon G, Corellou F, Ishikawa T, Brazard J, Usman A, Bowler C. Characterization of two members of the cryptochrome/photolyase family from Ostreococcus tauri provides insights into the origin and evolution of cryptochromes. Plant, Cell and Environment. 2010;33:1614–1626. doi: 10.1111/j.1365-3040.2010.02168.x. [DOI] [PubMed] [Google Scholar]
- Hoffman GE, Sanchez Puerta MV, Delwiche CF. Evolution of light-harvesting complex proteins from Chl c-containing algae. BMC Evolutionary Biology. 2011;11:101. doi: 10.1186/1471-2148-11-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann-Marriott MF, Blankenship RE. Evolution of photosynthesis. Annual Review of Plant Biology. 2011;62:515–548. doi: 10.1146/annurev-arplant-042110-103811. [DOI] [PubMed] [Google Scholar]
- Holt NE, Fleming GR, Niyogi KK. Toward an understanding of the mechanism of nonphotochemical quenching in green plants. Biochemistry. 2004;43:8281–8289. doi: 10.1021/bi0494020. [DOI] [PubMed] [Google Scholar]
- Huang K, Beck CF. Phototropin is the blue-light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:6269–6274. doi: 10.1073/pnas.0931459100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J, Lamparter T, Mittmann F, Hartmann E, Gärtner W, Wilde A, Börner T. A prokaryotic phytochrome. Nature. 1997;386:663. doi: 10.1038/386663a0. [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Ishizuka T. Cyanobacteriochromes: a new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochemical & Photobiological Sciences. 2008;7:1159–1167. doi: 10.1039/b802660m. [DOI] [PubMed] [Google Scholar]
- Im CS, Eberhard S, Huang K, Beck CF, Grossman AR. Phototropin involvement in the expression of genes encoding chlorophyll and carotenoid biosynthesis enzymes and LHC apoproteins in Chlamydomonas reinhardtii. Plant Journal. 2006;48:1–16. doi: 10.1111/j.1365-313X.2006.02852.x. [DOI] [PubMed] [Google Scholar]
- Iseki M, Matsunaga S, Murakami A, Ohno K, Shiga K, Yoshida K, Watanabe M. A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature. 2002;415:1047–1051. doi: 10.1038/4151047a. [DOI] [PubMed] [Google Scholar]
- Jenkins GI. The UV-B photoreceptor UVR8: from structure to physiology. Plant Cell. 2014;26:21–37. doi: 10.1105/tpc.113.119446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong HJ, Yoo YD, Kim JS, Seong KA, Kang NS, Kim TH. Growth, feeding and ecological roles of the mixotrophic and heterotrophic dinoflagellates in marine planktonic food webs. Ocean Science Journal. 2010;45:65–91. [Google Scholar]
- Jordan P, Fromme P, Witt HT, Klukas O, Saenger W, Krauss N. Three-dimensional structure of cyanobacterial photosystem I at 2.5 Aresolution. Nature. 2001;411:909–917. doi: 10.1038/35082000. [DOI] [PubMed] [Google Scholar]
- Karniol B, Vierstra RD. Structure, function, and evolution of microbial phytochromes. In: Schäfer E, Nagy F, editors. Photomorphogenesis in Plants and Bacteria: Function and Signal Transduction Mechanisms. Springer; The Netherlands: 2006. pp. 65–98. [Google Scholar]
- Kashiyama Y, Yokoyama A, Kinoshita Y, Shoji S, Miyashiya H, Shiratori T, Tamiaki H. Ubiquity and quantitative significance of detoxification catabolism of chlorophyll associated with protistan herbivory. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17328–17335. doi: 10.1073/pnas.1207347109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kateriya S, Nagel G, Barnberg E, Hegemann P. “Vision” in single-celled algae. News in Physiological Sciences. 2004;19:133–137. doi: 10.1152/nips.01517.2004. [DOI] [PubMed] [Google Scholar]
- Kim S, Park MG. Paulinella longichromatophora sp. nov., a new marine photosynthetic testate amoeba containing a chromatophore. Protist. 2016;167:1–12. doi: 10.1016/j.protis.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Koziol AG, Borza T, Ishida K, Keeling P, Lee RW, Durnford DG. Tracing the evolution of the light-harvesting antennae in chlorophyll a/b-containing organisms. Plant Physiology. 2007;143:1802–1816. doi: 10.1104/pp.106.092536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss U, Minh BQ, Losi A, Gartner W, Eggert T, von Haeseler A, Jaeger KE. Distribution and phylogeny of light-oxygen-voltage-blue-light-signaling proteins in the three kingdoms of life. Journal of Bacteriology. 2009;191:7234–7242. doi: 10.1128/JB.00923-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kräutler B. Phyllobilins–the abundant bilin-type tetrapyrrolic catabolites of the green plant pigment chlorophyll. Chemical Society Reviews. 2014;43:6227–6238. doi: 10.1039/c4cs00079j. [DOI] [PubMed] [Google Scholar]
- Kutty RK, Maines MD. Purification and characterization of biliverdin reductase from rat liver. Journal of Biological Chemistry. 1981;256:3956–3962. [PubMed] [Google Scholar]
- Lagarias DM, Crepeau MW, Maines MD, Lagarias JC. Regulation of photomorphogenesis by expression of mammalian biliverdin reductase in transgenic Arabidopsis plants. Plant Cell. 1997;9:675–688. doi: 10.1105/tpc.9.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Quiniou C, Tian L, Drop B, Wientjes E, van Stokkum IH, van Oort B, Croce R. PSI-LHCI of Chlamydomonas reinhardtii: Increasing the absorption cross section without losing efficiency. Biochimica et Biophysica Acta. 2015;1847:458–467. doi: 10.1016/j.bbabio.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FW, Melkonian M, Rothfels CJ, Villarreal JC, Stevenson DW, Graham SW, Mathews S. Phytochrome diversity in green plants and the origin of canonical plant phytochromes. Nature Communications. 2015a;6:7852. doi: 10.1038/ncomms8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FW, Rothfels CJ, Melkonian M, Villarreal JC, Stevenson DW, Graham SW, Pryer KM. The origin and evolution of phototropins. Frontiers in Plant Science. 2015b;6:637. doi: 10.3389/fpls.2015.00637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XP, Gilmore AM, Caffarri S, Bassi R, Golan T, Kramer D, Niyogi KK. Regulation of photosynthetic light harvesting involves intrathylakoid lumen pH sensing by the PsbS protein. Journal of Biological Chemistry. 2004;279:22866–22874. doi: 10.1074/jbc.M402461200. [DOI] [PubMed] [Google Scholar]
- Liu H, Liu B, Zhao C, Pepper M, Lin C. The action mechanisms of plant cryptochromes. Trends in Plant Science. 2011;16:684–691. doi: 10.1016/j.tplants.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losi A, Gärtner W. The evolution of flavin-binding photoreceptors: an ancient chromophore serving trendy blue-light sensors. Annual Review of Plant Biology. 2012;63:49–72. doi: 10.1146/annurev-arplant-042811-105538. [DOI] [PubMed] [Google Scholar]
- Mazor Y, Borovikova A, Nelson N. The structure of plant photosystem I super-complex at 2.8 A resolution. eLife. 2015;4:e07433. doi: 10.7554/eLife.07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Grossman AR. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A, Yang X, Ayers RA, Moffat K. Structure and function of plant photoreceptors. Annual Review of Plant Biology. 2010;61:21–47. doi: 10.1146/annurev-arplant-042809-112259. [DOI] [PubMed] [Google Scholar]
- Montgomery BL, Casey ES, Grossman AR, Kehoe DM. AplA, a member of a new class of phycobiliproteins lacking a traditional role in photosynthetic light harvesting. Journal of Bacteriology. 2004;186:7420–7428. doi: 10.1128/JB.186.21.7420-7428.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery BL, Lagarias JC. Phytochrome ancestry: sensors of bilins and light. Trends in Plant Science. 2002;7:357–366. doi: 10.1016/s1360-1385(02)02304-x. [DOI] [PubMed] [Google Scholar]
- Montgomery BL, Yeh KC, Crepeau MW, Lagarias JC. Modification of distinct aspects of photomorphogenesis via targeted expression of mammalian biliverdin reductase in transgenic Arabidopsis plants. Plant Physiology. 1999;121:629–639. doi: 10.1104/pp.121.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montsant A, Allen AE, Coesel S, De Martino A, Falciatore A, Mangogna M, Bowler C. Identification and comparative genomic analysis of signaling and regulatory components in the diatom Thalassiosira pseudonana. Journal of Phycology. 2007;43:585–604. [Google Scholar]
- Mullineaux CW. Phycobilisome-reaction centre interaction in cyanobacteria. Photosynthesis Research. 2008;95:175–182. doi: 10.1007/s11120-007-9249-y. [DOI] [PubMed] [Google Scholar]
- Nagahatenna DS, Langridge P, Whitford R. Tetrapyrrole-based drought stress signalling. Plant Biotechnology Journal. 2015;13:447–459. doi: 10.1111/pbi.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, Bamberg E, Hegemann P. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- Neilson JA, Durnford DG. Structural and functional diversification of the light-harvesting complexes in photosynthetic eukaryotes. Photosynthesis Research. 2010;106:57–71. doi: 10.1007/s11120-010-9576-2. [DOI] [PubMed] [Google Scholar]
- Nelson N, Yocum CF. Structure and function of photosystems I and II. Annual Review of Plant Biology. 2006;57:521–565. doi: 10.1146/annurev.arplant.57.032905.105350. [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Truong TB. Evolution of flexible non-photochemical quenching mechanisms that regulate light harvesting in oxygenic photosynthesis. Current Opinion in Plant Biology. 2013;16:307–314. doi: 10.1016/j.pbi.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Nowack EC, Melkonian M, Glockner G. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Current Biology. 2008;18:410–418. doi: 10.1016/j.cub.2008.02.051. [DOI] [PubMed] [Google Scholar]
- Nowicka B, Kruk J. Powered by light: Phototrophy and photosynthesis in prokaryotes and its evolution. Microbiological Research. 2016;186–187:99–118. doi: 10.1016/j.micres.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Nozue K, Kanegae T, Imaizumi T, Fukuda S, Okamoto H, Yeh KC, Wada M. A phytochrome from the fern Adiantum with features of the putative photoreceptor NPH1. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:15826–15830. doi: 10.1073/pnas.95.26.15826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okajima K. Molecular mechanism of phototropin light signaling. Journal of Plant Research. 2016;129:149–157. doi: 10.1007/s10265-016-0783-6. [DOI] [PubMed] [Google Scholar]
- Onodera A, Kong SG, Doi M, Shimazaki K, Christie J, Mochizuki N, Nagatani A. Phototropin from Chlamydomonas reinhardtii is functional in Arabidopsis thaliana. Plant & Cell Physiology. 2005;46:367–374. doi: 10.1093/pcp/pci037. [DOI] [PubMed] [Google Scholar]
- Pagliano C, Saracco G, Barber J. Structural, functional and auxiliary proteins of photosystem II. Photosynthesis Research. 2013;116:167–188. doi: 10.1007/s11120-013-9803-8. [DOI] [PubMed] [Google Scholar]
- Parihar P, Singh R, Singh S, Tripathi DK, Chauhan DK, Singh VP, Prasad SM. Photoreceptors mapping from past history till date. Journal of Photochemistry and Photobiology B. 2016;162:223–231. doi: 10.1016/j.jphotobiol.2016.06.020. [DOI] [PubMed] [Google Scholar]
- Peers G, Truong TB, Ostendorf E, Busch A, Elrad D, Grossman AR, Niyogi KK. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature. 2009;462:518–521. doi: 10.1038/nature08587. [DOI] [PubMed] [Google Scholar]
- Pérez-Bueno ML, Johnson MP, Zia A, Ruban AV, Horton P. The Lhcb protein and xanthophyll composition of the light harvesting antenna controls the DeltapH-dependency of non-photochemical quenching in Arabidopsis thaliana. FEBS Letters. 2008;582:1477–1482. doi: 10.1016/j.febslet.2008.03.040. [DOI] [PubMed] [Google Scholar]
- Petroutsos D, Tokutsu R, Maruyama S, Flori S, Greiner A, Magneschi L, Minagawa J. A blue-light photoreceptor mediates the feedback regulation of photosynthesis. Nature. 2016;537:563–566. doi: 10.1038/nature19358. [DOI] [PubMed] [Google Scholar]
- Portwich A, Garcia-Pichel F. A novel prokaryotic UVB photoreceptor in the cyanobacterium Chlorogloeopsis PCC 6912. Photochemistry and Photobiology. 2000;71:493–498. doi: 10.1562/0031-8655(2000)071<0493:anpupi>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Qin X, Suga M, Kuang T, Shen JR. Photosynthesis. Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science. 2015;348:989–995. doi: 10.1126/science.aab0214. [DOI] [PubMed] [Google Scholar]
- Reisdorph NA, Small GD. The CPH1 gene of Chlamydomonas reinhardtii encodes two forms of cryptochrome whose levels are controlled by light-induced proteolysis. Plant Physiology. 2004;134:1546–1554. doi: 10.1104/pp.103.031930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Sheerin DJ, Hiltbrunner A. Phytochromes: More Than Meets the Eye. Trends in Plant Science. 2016;21:543–546. doi: 10.1016/j.tplants.2016.05.009. [DOI] [PubMed] [Google Scholar]
- Rhee KH, Morris EP, Barber J, Kühlbrandt W. Three-dimensional structure of the plant photosystem II reaction centre at 8 A resolution. Nature. 1998;396:283–286. doi: 10.1038/24421. [DOI] [PubMed] [Google Scholar]
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Ulm R. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332:103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- Rochaix JD. Surprising roles for bilins in a green alga. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3218–3219. doi: 10.1073/pnas.1300399110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Duanmu D, Martin SS, Bachy C, Price DC, Bhattacharya D, Lagarias JC. Eukaryotic algal phytochromes span the visible spectrum. Proceedings of the National Academy of Sciences of the United States of America. 2014a;111:3871–3876. doi: 10.1073/pnas.1401871111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Lagarias JC. A brief history of phytochromes. Chemphyschem. 2010;11:1172–1180. doi: 10.1002/cphc.200900894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Lagarias JC, Bhattacharya D. Primary endosymbiosis and the evolution of light and oxygen sensing in photosynthetic eukaryotes. Frontiers in Ecology and Evolution. 2014b;2 doi: 10.3389/fevo.2014.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Feoktistova K, Lagarias JC. Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11854–11859. doi: 10.1073/pnas.1107844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Lagarias JC. Identification of cyanobacteriochromes detecting far-red light. Biochemistry. 2016;55:3907–3919. doi: 10.1021/acs.biochem.6b00299. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Martin SS, Li FW, Mathews S, Lagarias JC. The phycocyanobilin chromophore of streptophyte algal phytochromes is synthesized by HY2. New Phytologist. 2017 doi: 10.1111/nph.14422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annual Review of Plant Biology. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruban AV. Evolution under the sun: optimizing light harvesting in photosynthesis. Journal of Experimental Botany. 2015;66:7–23. doi: 10.1093/jxb/eru400. [DOI] [PubMed] [Google Scholar]
- Ruban AV. Nonphotochemical chlorophyll fluorescence quenching: mechanism and effectiveness in protecting plants from photodamage. Plant Physiology. 2016;170:1903–1916. doi: 10.1104/pp.15.01935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluchter WM, Glazer AN. Characterization of cyanobacterial biliverdin reductase. Conversion of biliverdin to bilirubin is important for normal phycobiliprotein biosynthesis. Journal of Biological Chemistry. 1997;272:13562–13569. doi: 10.1074/jbc.272.21.13562. [DOI] [PubMed] [Google Scholar]
- Schneider-Poetsch HAW. Signal Transduction by Phytochrome – Phytochromes have a module related to the transmitter modules of bacterial sensor proteins. Photochemistry and Photobiology. 1992;56:839–846. doi: 10.1111/j.1751-1097.1992.tb02241.x. [DOI] [PubMed] [Google Scholar]
- Singh R, Singh S, Parihar P, Singh VP, Prasad SM. Retrograde signaling between plastid and nucleus: A review. Journal of Plant Physiology. 2015;181:55–66. doi: 10.1016/j.jplph.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Steglich C, Frankenberg-Dinkel N, Penno S, Hess WR. A green light-absorbing phycoerythrin is present in the high-light-adapted marine cyanobacterium Prochlorococcus sp MED4. Environmental Microbiology. 2005;7:1611–1618. doi: 10.1111/j.1462-2920.2005.00855.x. [DOI] [PubMed] [Google Scholar]
- Suetsugu N, Mittmann F, Wagner G, Hughes J, Wada M. A chimeric photoreceptor gene, NEOCHROME, has arisen twice during plant evolution. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13705–13709. doi: 10.1073/pnas.0504734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga M, Akita F, Hirata K, Ueno G, Murakami H, Nakajima Y, Shen JR. Native structure of photosystem II at 1.95 Aresolution viewed by femtosecond X-ray pulses. Nature. 2015;517:99–103. doi: 10.1038/nature13991. [DOI] [PubMed] [Google Scholar]
- Swingley WD, Iwai M, Chen Y, Ozawa S, Takizawa K, Takahashi Y, Minagawa J. Characterization of photosystem I antenna proteins in the prasinophyte Ostreococcus tauri. Biochimica et Biophysica Acta. 2010;1797:1458–1464. doi: 10.1016/j.bbabio.2010.04.017. [DOI] [PubMed] [Google Scholar]
- Takahashi F. Blue-light-regulated transcription factor, Aureochrome, in photosynthetic stramenopiles. Journal of Plant Research. 2016;129:189–197. doi: 10.1007/s10265-016-0784-5. [DOI] [PubMed] [Google Scholar]
- Takahashi F, Yamagata D, Ishikawa M, Fukamatsu Y, Ogura Y, Kasahara M, Kataoka H. Aureochrome, a photoreceptor required for photomorphogenesis in stramenopiles. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19625–19630. doi: 10.1073/pnas.0707692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Tanaka A. Tetrapyrrole biosynthesis in higher plants. Annual Review of Plant Biology. 2007;58:321–346. doi: 10.1146/annurev.arplant.57.032905.105448. [DOI] [PubMed] [Google Scholar]
- Terry MJ, Smith AG. A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Frontiers in Plant Science. 2013;4:14. doi: 10.3389/fpls.2013.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thümmler F, Dufner M, Kreisl P, Dittrich P. Molecular cloning of a novel phytochrome gene of the moss Ceratodon purpureus which encodes a putative light-regulated protein kinase. Plant Molecular Biology. 1992;20:1003–1017. doi: 10.1007/BF00028888. [DOI] [PubMed] [Google Scholar]
- Tilbrook K, Dubois M, Crocco CD, Yin R, Chappuis R, Allorent G, Ulm R. UV-B perception and acclimation in Chlamydomonas reinhardtii. Plant Cell. 2016;28:966–983. doi: 10.1105/tpc.15.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippens J, Greiner A, Schellwat J, Neukam M, Rottmann T, Lu Y, Kreimer G. Phototropin influence on eyespot development and regulation of phototactic behavior in Chlamydomonas reinhardtii. Plant Cell. 2012;24:4687–4702. doi: 10.1105/tpc.112.103523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda SP, Ewers D, Gazzarrini S, Moroni A, Gradmann D, Hegemann P. H+-pumping rhodopsin from the marine alga Acetabularia. Biophysical Journal. 2006;91:1471–1479. doi: 10.1529/biophysj.106.086421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umena Y, Kawakami K, Shen JR, Kamiya N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature. 2011;473:55–60. doi: 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- Wang WJ, Wang FJ, Sun XT, Liu FL, Liang ZR. Comparison of transcriptome under red and blue light culture of Saccharina japonica (Phaeophyceae) Planta. 2013;237:1123–1133. doi: 10.1007/s00425-012-1831-7. [DOI] [PubMed] [Google Scholar]
- Warnasooriya SN, Montgomery BL. Detection of spatial-specific phytochrome responses using targeted expression of biliverdin reductase in Arabidopsis. Plant Physiology. 2009;9:424–433. doi: 10.1104/pp.108.127050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Ikeuchi M. Phycobilisome: architecture of a light-harvesting supercomplex. Photosynthesis Research. 2013;116:265–276. doi: 10.1007/s11120-013-9905-3. [DOI] [PubMed] [Google Scholar]
- Wietek J, Broser M, Krause BS, Hegemann P. Identification of a natural green light absorbing chloride conducting channelrhodopsin from Proteomonas sulcata. Journal of Biological Chemistry. 2016;291:4121–4127. doi: 10.1074/jbc.M115.699637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Chory J. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Current Biology. 2011;21:897–903. doi: 10.1016/j.cub.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden AZ, Lee JH, Mock T, Rouzé P, Simmons MP, Aerts AL, Grigoriev IV. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- Wu D, Hu Q, Yan Z, Chen W, Yan C, Huang X, Shi Y. Structural basis of ultraviolet-B perception by UVR8. Nature. 2012;484:214–219. doi: 10.1038/nature10931. [DOI] [PubMed] [Google Scholar]
- Yeh KC, Lagarias JC. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh KC, Wu SH, Murphy JT, Lagarias JC. A cyanobacterial phytochrome two-component light sensory system. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Nakayama T, Reyes-Prieto A, Andersen RA, Boo SM, Ishida K, Bhattacharya D. A single origin of the photosynthetic organelle in different Paulinella lineages. BMC Evolutionary Biology. 2009;9:98. doi: 10.1186/1471-2148-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]


