Abstract
Over the last twenty years, many strategies utilizing sol-gel chemistry to integrate biological cells into silica-based materials have been reported. One such strategy, Sol-Generating Chemical Vapor into Liquid (SG-CViL) deposition, shows promise as an efficient encapsulation technique due to the ability to vary the silica encapsulation morphology obtained by this process through variation of SG-CViL reaction conditions. In this report, we develop SG-CViL as a tunable, multi-purpose silica encapsulation strategy by investigating the mechanisms governing both silica particle generation and subsequent interaction with phospholipid assemblies (liposomes and living cells). Using Dynamic Light Scattering (DLS) measurements, linear and exponential silica particle growth dynamics were observed which were dependent on deposition buffer ion constituents and ion concentration. Silica particle growth followed a cluster-cluster growth mechanism at acidic pH, and a monomer-cluster growth mechanism at neutral to basic pH. Increasing silica sol aging temperature resulted in higher rates of particle growth and larger particles. DLS measurements employing PEG coated liposomes and cationic liposomes, serving as model phospholipid assemblies, revealed electrostatic interactions promote more stable liposome-silica interactions than hydrogen bonding and facilitate silica coating on suspension cells. However, continued silica reactivity leads to aggregation of silica coated suspensions cells, revealing the need for cell isolation to tune deposited silica thickness. Utilizing these mechanistic study insights, silica was deposited onto adherent HeLa cells under biocompatible conditions with micron scale control over silica thickness, minimal cell manipulation steps, and retained cell viability over several days.
Keywords: Sol-gel chemistry, chemical vapor deposition, cationic liposomes, hydrogen bonding, electrostatic interaction, living hybrid biomaterials, cellular encapsulation
Graphical Abstract
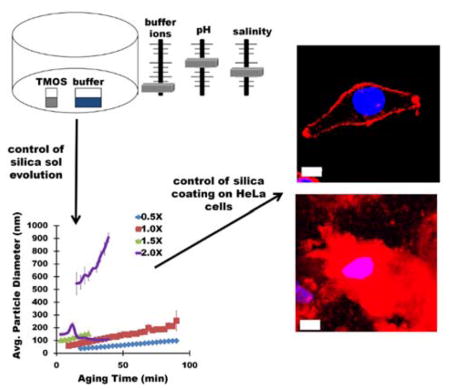
Silica sols are generated via vapor deposition of tetramethylorthosilicate into buffer. By varying the buffer ionic consituents, concentration, pH, and sol aging temperature, silica particle size in silica sols can be controlled, facilitating deposition of silica layers with tunable thickness on mammalian HeLa cells (represented by red fluorescense).
1. INTRODUCTION
The generation of functional hybrid biomaterials for development of whole cell based biosensor1–2 continues to be a challenging research topic. One strategy that has shown promise for generating functional cell based biosensors is encapsulation of biomolecules and whole cells in silica via the sol gel process.3–8 In this strategy, hydrolysis and condensation of silica precursors is utilized to form cell-silica nano-bio interfaces that afford cells protection from harsh ex-vivo conditions with robust control of material structure and properties.9–10 Carturan et al. pioneered silica encapsulation of cells by using the sol–gel process to incorporate genetically engineered Saccharomyces cerevisiae (S. cerevisiae) cells in tetraethyl orthosilicate (TEOS)-based gels. Subsequent studies showing encapsulation of cells in low density silica gels,11 and highly ordered silica thin films12–13, as well as generation of cell template silica-cell composites, showcased that varying sol-gel parameters can facilitate cell encapsulation in diverse silica geometries with wide ranging organic/inorganic interphases for biosensing, bioanalysis, and artificial organ applications.7–8, 14–18
While initial research focused on cell encapsulation in gels and thin film matrices, single cell encapsulation in nanometer scale silica shells has also been realized. Carturan et al. established the concept of single cell encapsulation by exposing cells to an alkoxysilane containing gas stream, generating thin silica shells on the cell surface, the thickness of which could be controlled by varying the exposure time of cells to the gas stream. However, this approach can be technically challenging, requires cells to be attached to a scaffold material, and does not ameliorate compressive stresses associated with silica condensation. Recently, Choi et. al. developed a chemical nano-encapsulation approach where the cell surface is functionalized with polyelectrolyte polymer layers which promote condensation of silica from pre-hydrolyzed sols, coating cells in nanometer scale silica shells while shielding the cell surface from condensation induced compressive stresses. While this method allows encapsulation of a variety of prokaryotic and eukaryotic cell types in both silica and non-silica based shells with control of shell thickness,19–21 the cytotoxic synthetic polycationic polymers and multiple deposition and wash steps employed in these studies limit their application for encapsulating more fragile eukaryotic cell lines (such as CANARY22), with robust sensing capabilities. To encapsulate more sensitive eukaryotic sensing cell lines, a facile silica encapsulation strategy is needed which offers tunable cell encapsulation, while minimizing cell manipulation steps and exposure to cytotoxic polycationic polymers.
One encapsulation process with the potential to meet this need is the Sol-Generating Chemical Vapor into Liquid (SG-CViL) deposition approach.23–24 In SG-CViL, silica sols are generated via deposition of an alkoxysilane vapor into buffer, and then aged. Aging allows evaporation of harmful reaction constituents (i.e. methanol) and polymerization of cytotoxic tetramethylorthosilicate (TMOS) monomers, greatly increasing sol biocompatibility. Subsequent mixing of aged silica sols with cell suspensions results in cell encapsulation in either large silica species, or deposition of thin silica films on cell surfaces, with maintained cell viability and functionality.23 The encapsulation of cells in distinct silica morphologies through variation of SG-CViL parameters indicates the ability; to control the thickness of the cell encapsulating matrix; however, leveraging this ability requires understanding the mechanisms governing SG-CViL silica particle generation and subsequent interaction with phospholipid assemblies. To develop this mechanistic understanding, silica particle growth in SG-CViL silica sols was observed via dynamic light scattering (DLS) using a variety of reaction conditions. These studies show that silica particle size can be controlled by varying reaction buffer composition, ionic concentration, pH, and sol aging temperature. We further show that liposome-silica colloids are more stable when silica interacts with liposomes via electrostatic bonding compared to hydrogen bonding. Ultimately, we utilize insights from these mechanistic studies to define SG-CViL parameters that allow silica deposition on HeLa cell surfaces with micron scale control of layer thickness and maintained cell viability.
2. Methods
2.1. Materials
Aqueous solutions were prepared using deionized water (DI). Calcofluor white stain, 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI), Dulbecco’s Modified Eagle’s Medium (DMEM), Fetal Bovine Serum (FBS), sodium acetate, sodium chloride, sodium phosphate (dibasic, heptahydrate and monobasic, monohydrate), potassium chloride, potassium phosphate, penicillin-streptomycin, Rhodamine B, and tetramethylorthosilicate (TMOS), were purchased from Sigma-Aldrich (St. Louis, MO). 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-(methoxy(polyethylene glycol) 2000) ammonium salt (DSPE-PEG2000), and 1,2-dipalmitoyl-3-trimethylammonium-propane (TAP) lipids were purchased from Avanti Polar Lipids (Alabaster, AL). Carboxyfluorescein Diacetate (CFDA) and propidium iodide (PI) were from Invitrogen and Yeast Peptone Dextrose (YPD) was from Sunrise Scientific products (San Diego, CA). Phosphate buffered saline (PBS 10×) was from Fischer Scientific (Hampton NH).
2.2. Particle Growth Under Various SG-CViL Conditions
Silica particle growth was characterized under various reaction conditions using dynamic light scattering (DLS) measurements conducted with a Malvern Zetasizer DLS instrument. For all experiments, tetramethylorthosilicate (TMOS) was deposited into buffer for 30 min, as shown schematically in Figure 1, step 1. For buffer composition studies (step 1, branch 1, Figure 1), TMOS was deposited into 1× phosphate buffer saline (PBS, 1.05 mM KH2PO4; 155 mM NaCl; 2.97 mM Na2HPO4*7H2O), sodium phosphate buffer (NaPB, 24 mM NaH2PO4; 76 mM Na2HPO4), sodium chloride (NaCl, 100 mM) solution, sodium acetate buffer (NaAc, 1.05 mM KH2PO4; 155 mM sodium acetate; 2.97 mM Na2HPO4), or potassium buffer, (K-buffer, 155 mM KCl; 4.02 mM KH2PO4).
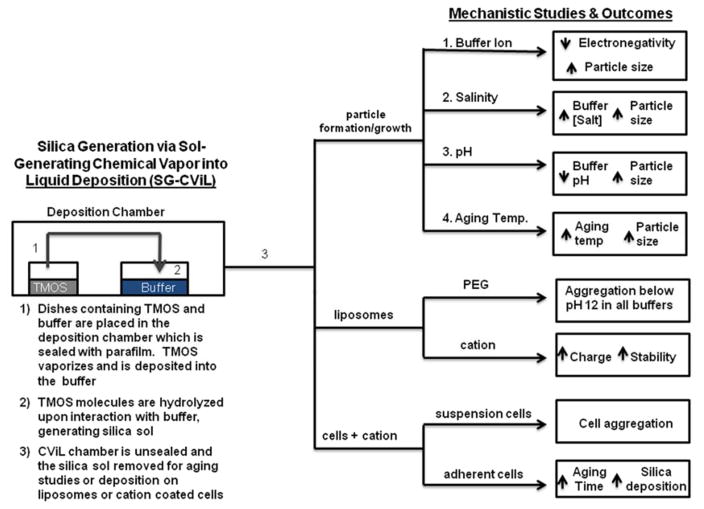
Figure 1.
Schematic of mechanistic studies investigating Sol-Generating Chemical Vapor into Liquid (SG-CViL) reaction conditions impact on silica particle growth and silica-phospholipid assembly interactions.
In salinity experiments (step 1, branch 2, Figure 1), TMOS was deposited into 0.5×, 1×, 1.5×, or 2× concentration PBS. To examine pH effects on particle growth (step 1, branch 3, Figure 1), TMOS was deposited into 1× PBS at pH 6.4, 7.4, 8.4 and 9.8. Post deposition, the resulting sols were diluted with 1 mL deposition buffer (3 mL final volume) and placed in the DLS instrument for particle size analysis. The average particle diameter of silica in the diluted sols was measured for 90 minutes (aging time) at 40°C (aging temperature) using a 3 min measurement interval. For aging temperature studies (step 1 branch 4, Figure 1) TMOS was deposited in 1× PBS, pH 7.4, and particle size was measured as described above, but with aging temperatures of 25°C, 30°C, 35°C and 40°C.
2.3. Liposome Encapsulation with Various SG-CViL Conditions
2.3.1. Liposome Preparation
PEG liposome lipid thin films were prepared by mixing 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-(methoxy(polyethylene glycol) 2000) ammonium salt (DSPE-PEG2000) in a 95:5 molar ratio of DPPC to DSPE. For cationic lipid films 1,2-dipalmitoyl-3-trimethylammonium-propane (TAP) was substituted for DSPE-PEG 2000 at 90:10 and 80:20 molar ratio of DPPC to TAP. Lipids were then dried under nitrogen flow and placed in a vacuum oven at 50°C for 2 hours to completely remove the chloroform solvent. Buffer (1 mL, 1× PBS) was added to lipid films, followed by sonication in a sonicating bath for 30 min at 50°C to generate a liposome suspension (final lipid concentration of 5 mg/mL). Liposome suspensions were subsequently extruded through 200 and 100 nm pore size membranes (11 passages per membrane) to generate monodisperse liposome suspensions for deposition experiments.
2.3.2. Varying Deposition Time
For mechanism studies involving PEG liposomes, SG-CViL silica sols were generated by depositing TMOS in 1.98 mL 1× PBS for 5, 10, 12, or 15 minutes at 23°C. Following unsealing of the CViL chamber, 20 μL of 5 mg/ml liposome solution was added to silica sols and particle size was analyzed using DLS.
2.3.3. Varying pH
SG-CViL silica deposition was performed for 15 min at 30°C as described in section 2.3.2 using 1× PBS at pH 12. Liposome solution (20 μL, 5 mg/mL) was added to the sols and the particle size measured pre and post addition of acetic acid (50 μL, 50% v/v) to the silica-liposome solution.
2.3.4. Varying Surface Charge
For cationic liposome studies, SG-CViL was performed for 10 min at 30°C as described in section 2.3.2, substituting cationic liposomes for PEG liposomes. Additionally, experiments were conducted with liposomes present in the buffer during TMOS deposition (referred to as in-situ CViL) for comparison to SG-CViL. For in-situ CViL deposition, a cationic liposome solution was prepared by adding 20 μL of 5 mg/mL liposome stock solution to 1.98 mL of 1× PBS. Particle size results are averages from 3 independent experiments analyzed using students T-test.
2.4. Whole Cell Encapsulation of Cation Coated-Suspension Cells
2.4.1. Suspension Cell Culture
Saccharamyces cerevisiae (S. cerevisiae,) were inoculated into 5 mL YPD rich media (10 g/L yeast extract, 20 g/L peptone, and 20 g/L dextrose) and incubated under rotary shaking at 30°C 18 to 24 hours. Jurkat cells were cultured in RPMI supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin in a CO2 incubator at 37°C, 5% CO2 in 25 mL culture flasks. Cultures were maintained by diluting cell suspensions 1:10 (v/v) every 3 days using fresh RPMI culture media
2.4.2. Characterization of Silica Interaction in Suspension Cells using Fluorescence Microscopy
S. cerevisiae and Jurkat cells (1×106 cell/mL) were pelleted, washed twice with 1 mL 1× PBS, pH 7.4 and stained with 2% calcofluor white (S. cerevisiae) or 10 μM 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI, Jurkat) for 30 min. Post staining, cells were washed twice with 1× PBS and incubated with 1× PBS, pH 7.4, containing 200 μM spermidine for 5 min at 30°C in a shaking incubator. Silica sols were generated by performing SG-CViL for 20 min at 23°C in 1× PBS pH 7.4, adding 1 μM Rhodamine B to the sample chamber prior to SG-CViL initiation to fluorescently label silica.23 Spermidine coated S. cerevisiae and Jurkat cells were resuspended in 1 mL fluorescently labeled silica sol for 10 min at 30°C in a shaking incubator. Cells were washed twice via centrifugation and resuspension in 1 mL 1× PBS and imaged using Olympus FE10i laser scanning confocal microscope system using a 60× water objective.
2.5. Whole Cell Encapsulation of Adherent Cells Using Tuned SG-CViL Parameters
2.5.1. Cell Culture
HeLa cells from approximately 80% confluent cultures were trypsinized and diluted in cell culture media (10% fetal bovine serum; 1% penicillin/streptomycin in DMEM) to a final concentration of 100,00 cell/mL. Cell suspension (200 μL) was pipetted in the center of tissue culture treated confocal microscopy slides that had been previously attached to cell culture dishes (Matek), and cells were allowed to adhere for 30 min under culture conditions (37°C; 5% CO2). Post adherence, 3 mL of media was added to cell culture dishes and cells were incubated an additional 18 to 24 hours before silica encapsulation (section 2.4.3.
2.5.2. Fluorescence Microscopy Characterization of Silica-HeLa Interaction Under Varying SG-CViL Encapsulation Parameters
HeLa cells were stained with the DNA binding dye, 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI, 10 μM) for 30 min, washed twice with 1× PBS, pH 7.4, and incubated with 1× PBS, pH 7.4, containing 200 μM spermidine for 5 min at 30°C in a shaking incubator. All silica sols used for HeLa encapsulation were generated by performing SG-CViL for 30 min at 40°C. To image silica deposition on HeLa cells silica was fluorescently labeled by adding 1 μM Rhodamine B to the sample chamber prior to initiation of the SG-CViL reaction.23 Spermidine coated HeLa cells were treated with 3 mL of fluorescently labeled SG-CViL silica sols generated using 1× PBS or 1× K-buffer using two aging regimes (unaged, or aged 30 min at 40°C) for 20 min at 30°C in a shaking incubator. Post silica deposition, cells were washed twice with 1× PBS, pH 7.4, and imaged with an Olympus FE10i laser scanning confocal microscope system using a 60× water objective using Fluoview software to measure silica thickness.
2.5.3. Morphology and Viability Analysis of Silica Coated HeLa Cells
To characterize cell morphology and viability post deposition, phase contrast microscopy and vital dye staining were used. HeLa cells were encapsulated in SG-CViL generated silica, as described in section 2.4.3, without fluorescent labeling of cells or silica. Post silica deposition, cells were washed twice with 1× PBS, followed by addition of 3 mL cell culture media, and cells were returned to the incubator. Cells were imaged at 30 min, 48 hours, and 96 hours post encapsulation using a phase contrast 40× objective. To assess cell viability 96 hours post encapsulation, cells were incubated with 1.5 mL of carboxyfluorescein diacetate (CFDA) and propidium iodide (PI) solution (10 μM CFDA, 4 μM PI in 1× PBS, pH 7.4) for 30 min at 30°C. Post staining, cells were imaged using confocal fluorescence microscopy, counting cells that fluoresced green as viable, and cells that fluoresced red or yellow as non-viable. Error bars represent standard deviations of 10 independent regions of interest in a single culture dish.
3. RESULTS AND DISCUSSION
3.1. Influence of SG-CViL Reaction Parameters on Particle Generation
Mechanistic investigations initially focused on examination of silica particle growth in silica sols generated by depositing TMOS into an aqueous buffer system in the absence of biomolecules as detailed schematically in Figure 1. To examine the silica particles size distribution in silica sols, DLS measurements were performed on sols generated using a variety of reaction parameters (Figure 1). For these experiments, sols were generated by depositing TMOS for 30 min at 40°C (Figure 1, steps 1 and 2). Following sol generation, sols were placed in the DLS instrument and aged for 90 minutes in order to examine particle size evolution over time. For all experiments, data are plotted only if there are 10 million or greater photons collected per time point (minimum number required for accurate size determination using cumulant analysis of the scattering data). The effect of different buffer systems, salt concentration, pH and sol-aging temperature are summarized in Figure 1 (step 3). Careful control over these reaction parameters can result in the facile, biocompatible, and tailorable generation of homogenous silica sols with known silica particle size distributions, ideal for encapsulation of phospholipid assemblies, including whole cells, while maintaining desired biological functions.
3.2. Particle Dynamics
3.2.1. Buffer Solution Affects Particle Dynamics
The impact of SG-CViL reaction buffer chemistry (Figure 1, path I) on particle growth is shown in Figure 2. Figure 2A displays the average particle size from silica sols generated using 5 different buffer systems: phosphate buffered saline (PBS), sodium phosphate buffer (NaPB, minus sodium chloride), sodium chloride solution (NaCl, minus phosphate), sodium acetate buffer (NaAc, PBS replacing sodium chloride with NaAc), and potassium buffer (K-buffer, replacing sodium as cation). Data representing both initial and final particle size, particle growth rate, and particle size increase are summarized in Table 1.
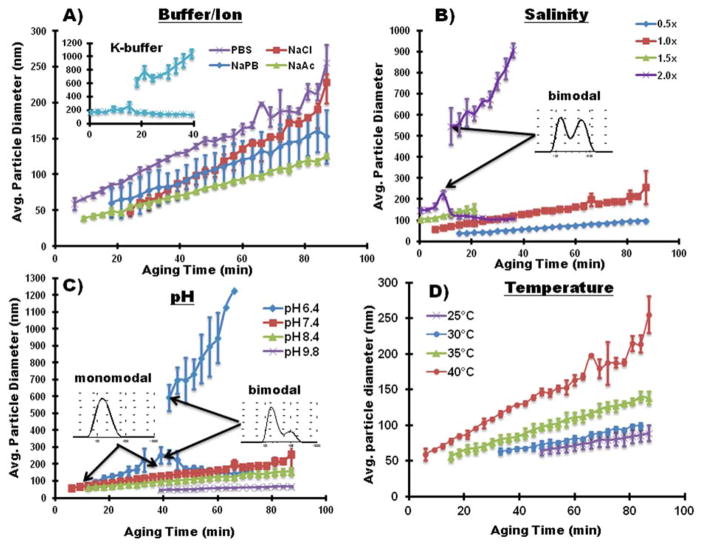
Figure 2.
Particle Growth of SG-CViL generated silica sols using differing ionic solutions (A), varying solution salinity (B), pH (C), and aging temperature (D). Particle size was monitored for 90 minutes using DLS at 40°C aging temperature, except for experiments in (D) where aging temperature was varied. Inset in (A) shows particle size over time of silica generated using K-buffer; insets in (B) and (C) show bimodal size distributions at the referenced data points. In (D), experiments were conducted using 1× PBS, pH 7.4, with the sols aged at 25, 30, 35, or 40°C. Error bars represent the standard deviation of N=3 experiments.
Table 1.
Particle Size Dynamics In SG-CViL Silica Sols Generated Using Different Ionic Solutions
| Buffer | Salt concentration (mM) | Particle size distribution | Initial particle size (nm)a | Final particle size (nm) | Particle growth rate (nm/min)b | Particle Size Increase (%) |
|---|---|---|---|---|---|---|
| Phosphate buffered saline (PBS, 1×) | 2.97 NaPO4 1.05 KH2PO4 155 NaCl |
monomodal | 59.3 ± 3.44 (6 min) | 255.7 ± 78.2 (90 min) | 2.03 | 431 |
| Sodium phosphate buffer (NaPB) | 24 Na2PO4 76 NaH2PO4 |
monomodal | 60.6 ± 20.4 (18 min) | 153.2 ± 37.1 (90 min) | 1.30 | 252 |
| Sodium Acetate (NaAc) | 2.97 Na2PO4 1.05 KH2PO4 155 NaCH3COO |
monomodal | 38.8 ± 5.21 (9 min) | 126.3 ± 2.05 (90 min) | 1.10 | 325 |
| Sodium Chloride (NaCl) | 100 NaCl | monomodal | 49.1 ± 6.41 (27 min) | 228.8 ± 29.3 (90 min) | 2.42 | 465 |
| Potassium (K-buffer) | 4.02 KH2PO4 155 KCl |
bimodal (15 min)c | 177.1 ± 23.9 (0 min) | 138.57 ± 4.45 1042.4 ± 64.6 (39 min) |
4.94 (0–15) 17.6 (15–39) −3.41 (15–39)d |
588 |
Numbers in parenthesis indicate aging time when particles become measurable using DLS.
Best fit lines and R2-values reported in the Supporting Information (Figure S1).
Aging time when PSD becomes bimodal
The numbers in parenthesis indicate the aging time interval (in min.) for which the particle growth rate is calculated. The first growth rate is associated with the particle population before transition to a bimodal distribution. The second growth rate is associated with the large particle population of the bimodal particle distribution and the third (negative) growth rate is associated with the small particle population. See Figure S1 for explanation of negative growth curve.
Comparing the growth curves presented in Figure 2A, and the data in Table 1, buffer composition has a clear effect on particle growth dynamics in SG-CViL silica sols. In K-buffer, particles are measurable at 0 min, compared to 6 or more minutes for Na-based buffer systems. Additionally, in sodium containing sols, while initial particle size values are within 20 nm of each other (~39–61 nm), the aging time required to achieve measurable particles decreases in the order of PBS < NaAc < NaPB < NaCl. Together these trends suggest the presence of even a small amount of potassium in buffers results in faster particle generation. A detailed discussion of the mechanisms governing particle growth changes with differing buffer constituents can be found in the supporting information. Further particle growth studies were conducted using PBS due to its known biocompatibility, fast generation of DLS observable particles (low monomer content), highest particle growth rate (2 nm/min), and widest range of particle sizes (60–255 nm).
3.2.2. Increasing Salinity Increases Particle Growth Rate
Particle growth as a function of aging time is plotted in Figure 2B for 0.5× (79.5 mM), 1× (159 mM), 1.5× (238.5 mM), and 2× (318 mM) PBS solutions. In general, as PBS concentrations increase, there is faster generation of measurable particles and an increase in particle growth rate (data summarized in Table 2). These trends are likely a function of an overall increased electrolyte concentration.9–10 In 2× PBS, the high concentration of cationic species promotes rapid particle formation and aggregation through the bridging mechanism discussed earlier. As particles grow, more sodium ions bind to the surface of larger particles, increasing the electrostatic bonding of phosphate anions. The increased anionic layer on larger silica particles potentially shields smaller silica particles from binding with larger particles. This generates two particle populations (note the bimodal particle distribution in Figure 2B) as smaller particles do not have enough surface associated cations to overcome the anionic repulsion imparted by negatively charged silica particles, resulting in a lower frequency of particle-particle interactions
Table 2.
Impact of Salinity on SG-CViL Silica Sol Particle Size Dynamics
| PBS Buffer | Salt concentration (mM) | Particle size distribution | Initial particle size (nm)a | Final particle size (nm) | Particle growth rate (nm/min)b | Particle Size Increase (%) |
|---|---|---|---|---|---|---|
| 0.5× | 1.49 Na2PO4 0.525KH2PO4 77.5 NaCl |
monomodal | 39.4 ± 2.63 (18 min) | 98.0 ± 4.41 (90 min) | 0.88 | 248 |
| 1.0× | 2.97 Na2PO4 1.05 KH2PO4 155 NaCl |
monomodal | 59.3 ± 3.44 (6 min) | 255.7 ± 78.2 (90 min) | 2.03 | 431 |
| 1.5× | 4.46 Na2PO4 1.575 KH2PO4 232.5 NaCl |
monomodal | 105.1 ± 5.38 (0 min) | 155.8 ± 5.95 ( 21 min) | 2.60 | 148 |
| 2.0× | 5.94 Na2PO4 2.10 KH2PO4 310 NaCl |
bimodal (12 min)c | 49.1 ± 6.41 (0 min) | 109.5 ± 4.38 (36 min) 907.1 ± 33.8 (36 min) |
8.22 (0–9) 14.4 (12–36) −2.79 (12–36)d |
1,847 |
Numbers in parenthesis indicate aging time when particles become measurable using DLS.
Best fit lines and R2-values reported in the Supporting Information (Figure S1).
Aging time when PSD becomes bimodal
The numbers in parenthesis indicate the aging time interval (in min.) for which the particle growth rate is calculated. The first growth rate is associated with the particle population before transition to a bimodal distribution. The second growth rate is associated with the large particle population of the bimodal particle distribution and the third (negative) growth rate is associated with the small particle population. See Figure S1 for explanation of negative growth curve
3.2.3. Increasing pH Decreases Particle Growth
In order to assess the effect of pH on particle growth, SG-CViL was performed in buffers with pH varied between slightly acidic (pH 6.4), neutral (pH 7.4) or basic (pH 8.4, 9.8). Distinct differences in particle size distributions and particle growth behavior were observed. At pH 6.4 (Figure 2C, blue diamonds), the particle size distribution comprises three regimes. From t = 0–33 min, the particle size distribution was monomodal, showing particle growth from approximately 94.7 ± 10.9 nm to approximately 225.9 ± 68.2 nm. From t = 33–45 min, the particle size distribution increased (note larger error bars). This increased distribution likely results from the development of large particle aggregates which are hydrolytically unstable, assembling and disassembling. At 45 minutes and longer, the larger particle aggregates become stable, leading to bimodal silica sols composed of a small particle population and a large particle population (see inset Figure 2C). As aging progresses, the large particles continue to increase in size; the size of the small particles decreases slightly. This is thought to be due to both large particles cannibalizing silicic acid monomers that disassociate from smaller silica particle surfaces, and condensation of small particles with larger silica particles. The overall shape of the distribution remains bimodal through 75 minutes of aging, at which point the sols become un-measurable using DLS, implying that silica species in the sol have condensed and formed large, polymeric structures.
In contrast to sols generated at pH 6.4, sols generated at pH 7.4, 8.4, and 9.8 display linear particle growth with particle growth rates of 2.03 nm/min (pH 7.4), 1.29 nm/min (pH 8.4), and 0.42 nm/min (pH 9.8). The sols further maintain monodisperse particle sizes of 255.7 ± 78.22 nm (pH 7.4), 156.2 ± 23.31 nm (pH 8.4), 68.1 ± 2.24 nm after 90 minutes of aging, showing that both particle growth rate and overall particle size decrease with increasing sol pH. These data are summarized in Table 3. See the supporting information for detailed mechanistic discussion of particle growth with varying pH.
Table 3.
Impact of pH on SG-CViL Silica Sol Particle Size Dynamics
| pH | Particle size distribution | Initial particle sizea | Final particle size | Particle growth rate (nm/min)b | Particle size increase (%) |
|---|---|---|---|---|---|
| 6.4 | bimodal (42 min)c | 94.8 ± 11.0 (15 min) | 143.3 ± 1.88 (66 min) 1,225.3 ± 180.2 (66 min) |
6.01 (15–42) 24.95 (42–66) −3.89 (42–66)d |
1,292 |
| 7.4 | monomodal | 59.3 ± 3.44 (6 min) | 255.7 ± 78.2 (90 min) | 2.03 | 431 |
| 8.4 | monomodal | 59.4 ± 8.56 (12 min) | 156.2 ± 23.3 (90 min) | 1.29 | 262 |
| 9.8 | monomodal | 48.4 ± 2.41 (39 min) | 68.1 ± 2.24 (90 min) | 0.42 | 142 |
Numbers in parenthesis indicate aging time when particles become measurable using DLS.
Best fit lines and R2-values reported in the Supporting Information (Figure S1).
Aging time when PSD becomes bimodal
The numbers in parenthesis indicate the aging time interval (in min.) for which the particle growth rate is calculated. The first growth rate is associated with the particle population before transition to a bimodal distribution. The second growth rate is associated with the large particle population of the bimodal particle distribution and the third (negative) growth rate is associated with the small particle population. See Figure S1 for explanation of negative growth curve
3.2.4. Increasing Temperature Increases Particle Growth Rate
The average particle size for silica sols prepared in 1× PBS, pH 7.4, at various aging temperatures is shown in Figure 2D, and summarized in Table 4. Of note is the decrease in aging time at which particles become measurable with increasing aging temperature. At 25°C, measurable particle sizes are not achieved until 48 min post SG-CViL deposition; whereas at 40°C measurable particles are achieved 9 min post SG-CViL. Knowing the time needed to achieve measurable particles is important as when particles are measurable, it is likely that the presence of potentially cytotoxic silica monomers is minimized, maximizing silica sol biocompatibility.
Table 4.
Impact of Temperature on SG-CViL Silica Sol Particle Size Dynamics.a
| Aging Temperature | Particle size distribution | Initial particle sizeb | Final particle size | Particle growth rate (nm/min)c | Particle size increase (%) |
|---|---|---|---|---|---|
| 40°C | monomodal | 59.3 ± 3.44 (6 min) | 255.7 ± 78.2 (90 min) | 2.03 | 431 |
| 35°C | monomodal | 56.4 ± 6.50 (15 min) | 138.7 ± 9.16 (90 min) | 1.16 | 245 |
| 30°C | monomodal | 63.6 ± 3.64 (33 min) | 99.1 ± 5.58 (84 min) | 0.73 | 155 |
| 25°C | monomodal | 64.7 ± 6.63 (48 min) | 89.1 ± 0.6 (90 min) | 0.6 | 137 |
SG-CViL performed for 30 min at 40°C in 1× PBS.
Numbers in parenthesis indicate aging time when particles become measurable using DLS.
Best fit lines and R2-values reported in the Supporting Information (Figure S1).
An increase in particle growth rate with increasing aging temperature was also observed in Figure 2D and Table 4. The higher silica particle growth rate in sols aged at higher temperatures is likely due to temperature effects on silica solubility and enhanced reaction kinetics. Above pH 7, the solubility of amorphous silica increases with increasing temperature.9 Therefore, at the pH of 7.4 in these experiments, silica solubility increases with temperature. This leads to greater accessibility to hydrolyzed silica monomers needed for nucleation and particle growth. Additionally, higher aging temperatures result in relatively greater particle movement, increasing the frequency of particle-particle interaction, and in higher condensation reaction kinetics.10
3.3. Parameter Tuning for Liposome Encapsulation
Having examined the effect of SG-CViL reaction parameters on particle growth in silica sols in the absence of biomolecules, mechanistic studies were next performed to gain understanding on how reaction parameters affect the interaction of silica sols with a model phospholipid assembly (Figure 1, path II). For these mechanistic studies, a liposome system was utilized to model silica interaction with cell membranes. Insights from this model were applied to advance the ability to control SG-CViL parameters that facilitate integration of cells within SG-CViL generated silica materials.. Liposomes formulated with a polyethylene glycol (PEG) polymer covalently attached to the hydrophilic head group of the lipid were used for hydrogen bonding studies. Pegylated liposomes were utilized in order to increase liposome colloidal stability. Liposome formulations lacking PEG, or other hydrophilic coatings, rapidly fused together before TMOS exposure could be completed (data not shown). These studies reveal that controlling hydrogen bonding dynamics via pH variation is necessary for stable silica deposition on PEG liposomes. Subsequent studies substituting PEG conjugated lipids for cationic lipids demonstrate cationic based electrostatic interactions facilitate greater control over silica-biomolecule interactions
3.3.1. SG-CViL Liposome Encapsulation is Time Sensitive
Initial liposome experiments focused on varying deposition time and are presented in Figure S2. Liposomes showed no significant size increase upon addition to silica sols generated by depositing TMOS for 5, 10, or 12 min. However, liposomes added to sols generated by depositing TMOS for 15 min increased in size to greater than 1 μm, with samples containing clearly visible aggregates (See inset Figure S2A). Aggregation was also observed in 5, 10, and 12 min samples approximately 15 min post liposome addition to sols. SEM-EDS analysis of 12 min and 15 min samples (Figure S2B) confirms aggregates are micron sized and contain silica, suggesting silica-liposome interaction results in aggregate formation, Liposome aggregation was also observed when experiments were performed using PEG 350 and PEG 5000 conjugated lipids, lower PBS concentrations (0.1×), and longer TMOS deposition times (30 min). (data not shown). The results indicate liposome aggregation is independent of PEG chain length, buffer salt concentration, or hydrolyzed silica content.
3.3.2. Increasing pH Decreases Liposome Aggregation
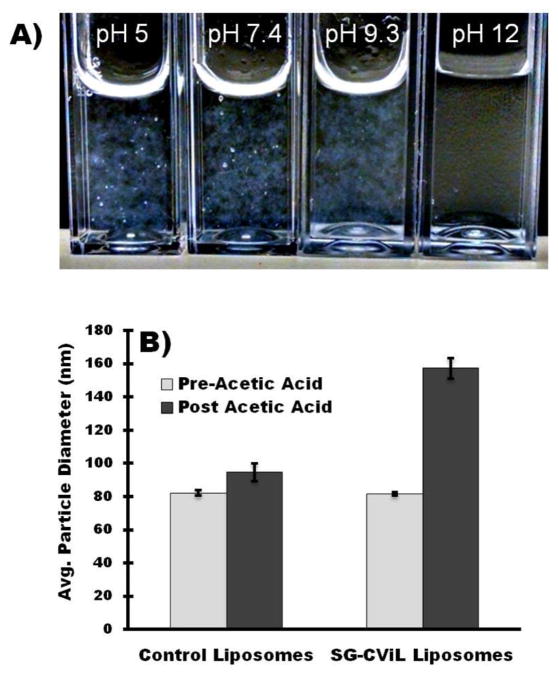
Numerous reports have shown silica interacts with PEG via hydrogen bonding of surface silanols with polyether oxygens in PEG, leading to silica particle formation and aggregation.25–27 Given this, we hypothesized that silica induced liposome aggregation results from hydrogen bonding of SG-CViL generated silica with PEG liposomes. To examine this interaction, SG-CViL deposition was performed at differing pHs, as shown in Figure 3. When liposomes were added to silica sols generated at pH < 12 (Figure 3A), aggregation similar to that shown in Figure S2A occurred within one minute of liposome addition to sols. However, no aggregation was observed when liposomes were added to pH 12 silica sols. Further, liposomes increased in size by less than 4 nm (83.8 nm to 87.1 nm) 3 days post liposome addition to pH 12 sols (Figure S3) indicating that highly basic conditions inhibit silica induced liposome aggregation, likely due to deprotonation of surface silanols at basic pH inhibiting silica-PEG hydrogen bonding interactions.10, 28
Figure 3.
(A) Optical image of SG-CViL silica sols generated at varying pH post liposome addition to sols. Images show silica induced liposome aggregation is inhibited at high pH. (B) Control of liposome-silica interaction via pH and ion parameters. Post-acetic acid addition, SG-CViL exposed liposomes increase in size significantly (p<0.05, student’s t-test), whereas control liposomes show insignificant increase in size.
The lack of liposome aggregation in pH 12 sols, in addition to data suggesting that the acetate ion stabilizes silica particle growth (Figure 2A), suggest an avenue for controlling liposome-silica interaction through pH variation using acetic acid. To test this hypothesis liposomes were first added to pH 12 silica sols (SG-CViL silica-liposomes), or to 1× PBS, pH 12, without silica (control liposomes). The initial particle size was determined to be approximately 82 nm for both conditions (Figure 3B, light grey bars). Following analysis, acetic acid (1.2% final concentration in sols, pH ~ 5.0) was added to the samples, and the particle size was again measured (Figure 3B, black bars). When acetic acid was added to the control liposome sample, there was a small increase in the average particle size and size distribution (82.2 ± 1.6 nm to 94.7 ± 5.5 nm, N=3 p>0.05), likely due to some hydrogen bonding of acetate ions with PEG. When acetic acid was added to SG-CViL silica-liposome solutions, there was a substantial increase in the average particle size from 82.6 ± 1.2 nm to 123.3 ± 6.2 nm (N = 3, p<0.05). The SG-CViL-liposome suspension does not aggregate after addition of acetic acid, as the particle suspension remained monodisperse 16 days post acetic acid addition (Figure S4).
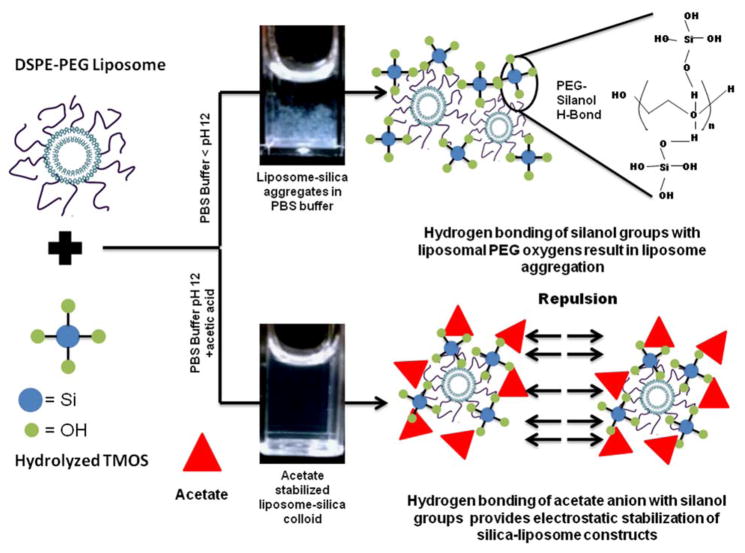
We propose that the liposome size increase was likely a result of the added acetic acid reducing the sol pH and protonating some of the silanol groups. This facilitated hydrogen bonding between silica and PEG, coating the liposomes in silica. The lack of aggregation indicates that the acetate ion stabilized the particle suspension against aggregation via electrostatic particle repulsion and shielding of surface silanols by the acetate ion. This mechanism is represented schematically in Figure 4.
Figure 4.
Schematic illustration of liposome-silica hydrogen bonding interactions in PBS, and control over aggregation via electrostatic stabilization.
3.3.3. Cationic Liposome Surface Increases Stability
Cationic liposomes were prepared by replacing DSPE-PEG 5000 lipids in lipid formulations with either 10 mol% or 20 mol% of the positively charged lipid, 1,2-dipalmitoyl-3-trimethylammonium-propane (TAP). To interrogate potential differences in cationic liposome-silica interaction at different stages of silica generation, two silica exposure methods were utilized: an in-situ method, where cationic liposomes were present in 1× PBS during TMOS deposition (Figure 1, step 2), and the SG-CViL method, where cationic liposomes were added to silica sols post TMOS deposition (Figure 1, step 3).
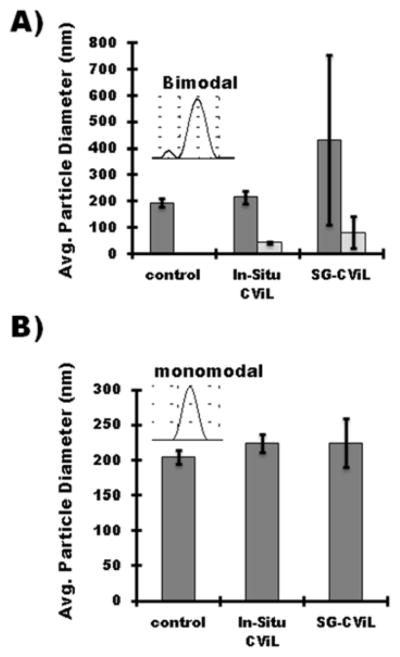
Figure 5 plots the particle diameter of liposomes containing 10 mol% (10% TAP-liposomes, Figure 5A), or 20 mol% TAP (20% TAP-liposomes, Figure 5B), using both in-situ and SG-CViL exposure methods. For 10% TAP-liposomes, exposed to silica via the in-situ approach, liposome size increased roughly 10% from 195 ± 15.2 to 216.6 ± 12.9 nm (p > 0.05 N=3). Treatment with silica via SG-CViL resulted in liposome size increase of approximately 120% (432.2 ± 320. 9; p > 0.05 N=3 nm), indicating generation of nanoscale liposome-silica aggregates. Additionally, a small peak (43.3 ± 3.4 for the in-situ approach and 81.5 ± 59.6 nm for SG-CViL), not present in control liposomes is evident in liposomes with 10 mol% TAP, indicating generation of small silica particles in the liposome suspension.
Figure 5.
Cation mediated interaction of SG-CViL generated silica with cationic liposomes Graphs represent particle size of (A) 10 mol% TAP-liposomes and (B) 20 mol% TAP-liposomes in buffer without TMOS exposure (control), present in buffer during TMOS deposition (in-Situ), or added to buffer post TMOS deposition (SG-CViL). Light grey bars in (A) correspond to small particle population peak in particle size distribution (see inset). TMOS deposition was performed for 10 min at 30C in 1X pH 7.4 PBS buffer. Error Bars represent the standard deviation of N=3 experiments
Compared to 10 mol% TAP-liposomes, 20 mol%-TAP liposomes exposed to silica increased in size by approximately 10%, from 203.7 ± 9.8 nm to 224.3 ± 12.9 nm (in-situ CViL; p < 0.05 N=3) and 224.4 ± 34.8 nm (SG-CViL; p > 0.05, N=3) nm. Zeta potential measurements of liposome suspensions showed greater positive zeta-potential for 20 mol% TAP-liposomes (34.3 ± 5.16 mV) compared to 10 mol% TAP-liposomes (12.6 ± 0.96 mV) with a decrease of approximately −15 mV upon liposome exposure to silica, confirming silica association with the liposome surface (see Figure S5 for zeta potential values).
While only one experimental condition resulted in a statistically significant increase in liposome size, the lack of small particle, and reduced size variability observed for 20 mol% TAP liposomes compared to 10 mol%-TAP, suggests increasing TAP concentration provides a sufficiently high positive surface charge to the liposomes to stabilize silica at the liposome surface through electrostatic interactions.29–30 However, when silica deposition time is increased to 15 minutes for 20 mol%-TAP liposomes, liposomes aggregate(data not shown). This indicates the stabilization is dependent on silica concentration as well as charge. Importantly, the nano-scale liposome-silica interaction observed with 20 mol% TAP liposomes, compared to the aggregates observed using PEG containing liposomes (Figure S2), shows that cation mediated electrostatic interactions provide an avenue for controlled silica deposition on lipid membranes using SG-CViL under physiologic conditions that are relevant for living cell encapsulation.
3.4. Electrostatic Interactions and SG-CViL Particle Size Variation Facilitate Whole Cell Encapsulation
3.4.1. SG-CViL Induces Aggregation of Cation Coated Suspension Cells
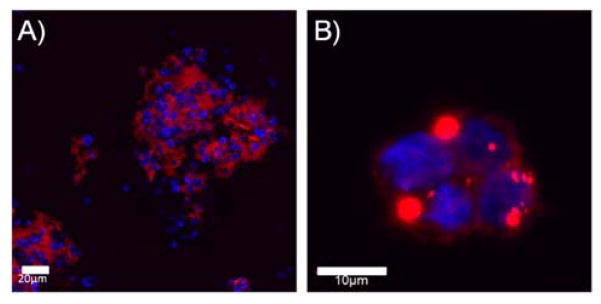
To examine silica deposition on cation coated suspensions cells, S. cerevisiae and Jurkat cells were coated with the natural polycation, spermidine, and exposed to Rhodamine B-labeled SG-CViL silica sols (20 min deposition time, 1× PBS, pH 7.4, no aging). When cells are exposed to silica sol, they become surrounded by red fluorescence, as shown in Figure 6A and 7B, indicating silica deposition on the cell surface of cation coated cells. However, there is pronounced aggregation of cells similar to the liposome aggregation observed in Figure 3. This suggests that the cell associated silica remains reactive and capable of condensing with the silica on neighboring cells despite cation mediated electrostatic cell-silica interactions, likely because there are insufficient positive charge sites to completely shield hydrolyzed silanols. Pelleting cells likely facilitates condensation of unshielded silanols by bringing these species into intimate contact, leading to cell aggregates even after resuspension. While silica induced aggregation of suspension cells could be useful for generating multi-cellular constructs for investigating proximity based cell-cell communication phenomena such as flocculation,31 it highlights the need for cell isolation in order to access and maintain single cell-silica geometries using SG-CViL.
Figure 6.
Florescence microscopy images of spermidine coated S. cerevisiae (A) and Jurkat (B) cells exposed to SG-CViL generated silica. Images show cell clusters (blue fluorescence) surrounded by red fluorescence (silica), indicating silica is deposited on cells and condenses with silica on neighboring cells, generating cells aggregates. S. cerevisiae stained with calcofluor white, Jurkat with DAPI, and silica with Rhodamine B
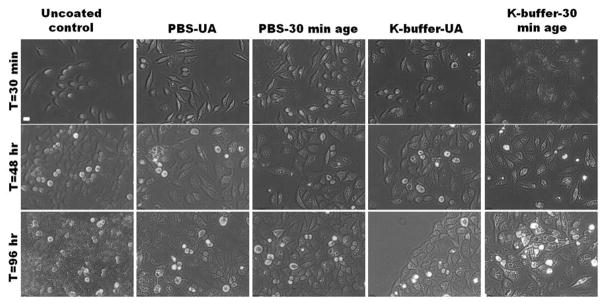
Figure 7.
SG-CViL generated silica sol buffer composition and aging time can be tuned to provide control over silica particle size and silica deposition thickness and morphology on spermidine coated HeLa cells. (A, B) Representative images of HeLa cells treated with un-aged PBS silica sol (A, PBS-UA), or PBS silica sol aged for 30 min (B, PBS-30 min age). (C, D) Representative images of HeLa cells treated with un-aged K-buffer silica sol (C, K-buffer-UA) or K-buffer silica sol aged for 30 min (D, K-buffer-30 min age). Greater particle size in silica sol results in thicker and less conformal silica deposition on cells. Red fluorescence from Rhodamine B stained silica; blue fluorescence from DAPI stained cell nuclei. Scale = 10 μm.
3.4.2. Tunable Silica Deposition on Adherent HeLa Cells
Given the liposome and polycation modified cells in suspension results in Figure 5, along with previous work showing that polycation layers promote silica condensation on cell surfaces through electrostatic interaction of silica with the polycation,12, 19–21 studies were performed to determine if controlling silica particle size via deposition buffer and sol aging (Section 2.2) facilitated tunable silica deposition on spermidine coated HeLa cells. The adherent nature of HeLa cells allows cell exposure to SG-CViL silica sols while preventing major cell-cell physical interactions, reducing or eliminating the silica induced aggregation seen with cells in suspension (Figure 6).
Figure 7 contains representative images of spermidine coated HeLa cells exposed to unaged silica sols (abbreviated UA) or silica sols aged for 30 minutes (abbreviated 30 min-age) generated using 1× PBS, pH 7.4 (Figure 7A, B), or K-buffer (Figure 7C, D). When spermidine coated HeLa cells were treated with PBS generated silica sols aged for 0 minutes (Figure 7A, PBS-UA) a thin, conformal red fluorescence (Rhodamine B labeled silica, see Experimental Section 2.5.2) was visible on the cell surface, indicating deposition of a thin silica layer on the HeLa cell surface, measured at approximately 1 μm in thickness using the measurement tool in Fluoview imaging software. Increasing sol aging time to 30 minutes resulted in an increase in the silica layer deposited on HeLa cell surfaces to approximately 5 μm (Figure 7B, PBS-30 min age). Additionally, the presence of areas lacking red fluorescence within the deposited silica layer indicates a certain level of porosity on part of the silica layer. When spermidine coated HeLa were exposed to K-buffer generated silica sols aged for 0 min (Figure 7C, K-buffer-UA), the amount of cell associated silica increased substantially compared to cells treated with PBS-UA cells, resulting in an approximately 15 μm thick silica layer deposited on the cells. A further increase in silica deposition to a thickness of greater than 20 μm was observed on cells exposed to K-buffer generated sols aged for 30 min (Figure 7D, K-buffer-30 min age). Additionally, areas lacking red fluorescence are limited in Figure 7D, indicating, perhaps, a more condensed silica layer on cells in Figure 7D when compared to cells in Figure 7B.
The change in thickness of the deposited silica layer on HeLa cells with different sol generating parameters (i.e. deposition buffer and aging) likely results from the effect of these parameters on silica particle size in sols. At pH 7.4, as particle sizes increase, particle surface charge becomes more negative.32 The particle size in unaged K-buffer sols was nearly 4 times larger than unaged PBS sols (177.1 ± 23.9 nm compared to 47.8 ± 8.22 nm). Given the much larger particles in K-buffer-UA sols compared to PBS-UA sols, the increased silica deposition observed on K-buffer UA sols treated cells may also be due to the larger particles having more electrostatic attraction to the positively charged spermidine on the HeLa cell surface.33 Additionally, the larger particles that initially bind to the HeLa cell surface could provide more sites for particle condensation than the smaller particles generated using PBS. The increase in potential condensation sites could increase the frequency of particle-particle interactions such that more silica is deposited from sols with larger particles than from sols containing smaller particles (see Figure 7A, C). A similar mechanism may explain the increased coating of cells when sols are aged for 30 min (compare Figure 7B & D), as sol aging increases silica particle size in both buffer systems, increasing both electrostatic binding of particles with the HeLa cell surface and subsequent particle condensation.
3.4.3. Morphology and Viability of SG-CViL Encapsulated HeLa
To characterize HeLa cell response to encapsulation using different sol generation parameters, cells were encapsulated using sols generated in either PBS or K-buffer that were unaged or aged for 30 min (section 2.5.3). Post encapsulation, growth media was added to cells and cells were incubated under standard culture conditions. Phase contrast images of cells 30 min, 48 hours, and 96 hours post SG-CViL are presented in Figure 8. When examined over time, encapsulated cells density increases over the 96 hour analysis time frame, indicating cells remained capable of growth and division. Taken together, data in Figure 7 and 8 suggest the possibility that silica deposited on cells exposed to SG-CViL silica sols was not deposited as a fully condensed, cross-linked layer, but as a loosely cross-linked polymeric structure, or particle assemblage. Under these conditions, the less cross-linked structure of the deposited silica layers would be mechanically flexible enough to allow cells the freedom to grow and divide 11, though more detailed time lapse fluorescence microscopy studies are needed to confirm this.
Figure 8.
Morphology of silica encapsulated HeLa cells over time. HeLa cells exposed to unaged (PBS-UA, K-buffer-UA) silica sols or silica sols aged for 30 min (PBS-30 min age, K-buffer 30 min age) visualized 0.5, 48, and 96 hours post encapsulation show minimal signs of cell death (i.e. detachment of cells from culture dishes) despite development of vacuole like structures on the cell interior. Scale = 20 μm.
In addition to cell growth, phase contrast images taken 48 and 96 hours post encapsulation show silica encapsulated cells contain vacuole-like structures that are absent in uncoated controls regardless of the sol generation parameters (see Figure S6 A & B for larger sized images from Figure 8 to more easily visualize vacuoles). 96 hours post encapsulation the vacuoles are less abundant in cells treated with PBS-UA, PBS-30 min age, and K-buffer-UA sols, with cells in these three conditions having similar morphology to uncoated control cells. In contrast, cells treated with K-buffer-30 min sols are larger in size than uncoated controls and contain prominent vacuoles.
Based on previous work showing that silica particles can be internalized by cells via formation of micropinosomes, we hypothesize that the vacuole structures observed in cells treated with SG-CViL generated silica sols could arise from cell internalization of silica after deposition.34–36 Importantly, when cells were examined using vital fluorescent dyes,12 cell viability was determined to be 99.0 ± 0.27 % (uncoated control), 98.3 ± 2.7 % (PBS-UA), 98.0 ± 2.1 % (PBS-30 min age), 95.9 ± 7.2 % (K-buffer-UA), and 89.8 ± 12.4 % (K-buffer-30 min age) 96 hours post encapsulation. (For representative fluorescent viability images, see Figure S7). This, combined with growth and division of cells exposed to SG-CViL, indicates that the silica coating treatment, encapsulation within silica, and the development of vacuoles in response to silica encapsulation, did not compromise cell viability under the time frame investigated. However, it cannot be discounted that vacuole formation in SG-CViL treated cells results from induction of non-apoptotic cell death pathways such as oncosis that would result in viability loss for cells at extended storage times 37. Given vital dye staining only provides information on cellular esterase activity and membrane permability, further experiments to assess ATP synthesis and measure intracellular ROS generation are needed to develop a more complete understanding of cellular behavior post silica encapsulation.
To our knowledge, this is one of the first reports to show silica deposition on adherent mammalian cells with micron scale thickness control using a natural polycationic polymer, minimal cell washing steps, with potential processing and internalization of the deposited silica layer by cells.19–20. The data suggest SG-CViL does not lead to highly cross-linked, mechanically rigid silica layers, but layers that are lightly cross-linked, and mechanically malleable. These layers are difficult to control when deposited on colloidal phospholipid assemblies (liposomes and cells, see Figure 3, 4 and 7), suggesting that the SG-CViL technique requires further adaptation for coating of these assemblies. However, in adherent cells systems SG-CViL facilitates well-defined silica deposition by controlling deposition buffer and sol aging to precisely generate a desired silica particle size (Figure 7). The reactivity of SG-CViL generated silica, while a limiting factor in coating colloidal phospholipid assemblies, could provide an avenue for applications such as cell transfection in adherent cells by complexing the deposited silica layers with charged molecules such as DNA or siRNA using cationic polymers as binding intermediates. Furthermore, the flexibility of the silica deposited using SG-CViL may also facilitate integration of fragile eukaryotic cells in silica gel monoliths by shielding cells from the compressive stresses imparted on cells by condensation of the bulk gel.
4. SUMMARY AND CONCLUSIONS
Data presented herein provide mechanistic insights into particle growth dynamics in chemical vapor deposition generated silica sols and the interaction of generated silica particles with phospholipid assemblies (liposomes and cells). Studies utilizing different buffer systems and buffers of varying salinity showed differing trends in particle growth in silica sols. Investigations varying buffer pH demonstrated a pH dependent transition in silica particle growth from a cluster-cluster mechanism to a monomer-cluster growth mechanism. Additional DLS experiments show that by adjusting aging temperature, silica particle growth in SG-CViL silica sols can be controlled at the nanoscale. Employing a model liposome system for examination of SG-CViL silica interaction with phospholipid assemblies, we suggest silica induces liposome aggregation through a hydrogen bonding mechanism when PEG is present in liposome formulations, and that this aggregation can be ameliorated through control of hydrogen bonding or substitution of PEG lipids with cationic lipids to promote electrostatic interaction. Ultimately, we utilize insights gained from mechanism studies and previous work to define a SG-CViL protocol to deposit silica on the surface of HeLa cells with micron scale tunability, while maintaining cell viability.
Supplementary Material
Acknowledgments
The authors thank Dr. Snezna Rogelj and Dr. Lillya Frolova for helpful technical discussion and cell culture assistance, as well as Gary Chandler for assisting in SEM prep and analysis. We also wish to thank Mendi Marquez, Eligio Madrid, Jayla Nicole Irving and Anna Wermer for assistance with liposome preparation and DLS measurements. This project was supported by grants from the Sandia Lab Directed Research and Development (LDRD) Campus Executive Program (151375), the National Center for Research Resources (5P20RR016480-12), and the National Institute of General Medical Sciences (8 P20 GM103451-12) from the National Institutes of Health (NIH). Sandia National Laboratories is a multi-mission laboratory managed and operated by National Technology and Engineering Solutions of Sandia, LLC., a wholly owned subsidiary of Honeywell International, Inc., for the U.S. Department of Energy’s National Nuclear Security Administration under contract DE-NA-0003525
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Supporting Information. Figures S1–S7, containing analysis of particle growth curves, dynamic light scattering and zeta-potential measurements, as well as representative viability images are provided in the supplementary information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Srinivasan G, Chen J, Parisi J, Brückner C, Yao X, Lei Y. An Injectable PEG-BSA-Coumarin-GOx Hydrogel for Fluorescence Turn-on Glucose Detection. Applied Biochemistry and Biotechnology. 2015;177(5):1115–1126. doi: 10.1007/s12010-015-1800-2. [DOI] [PubMed] [Google Scholar]
- 2.Cui L, Wu J, Ju H. Electrochemical Sensing Of Heavy Metal Ions With Inorganic, Organic And Bio-Materials. Biosensors and Bioelectronics. 2015;63:276–286. doi: 10.1016/j.bios.2014.07.052. DOI: http://dx.doi.org/10.1016/j.bios.2014.07.052. [DOI] [PubMed] [Google Scholar]
- 3.Coradin T, Allouche J, Boissière M, Livage J. Sol-Gel Biopolymer/Silica Nanocomposites In Biotechnology. Current Nanoscience. 2006;2(3):219–230. DOI: https://doi.org/10.2174/1573413710602030219. [Google Scholar]
- 4.Coradin T, Mercey E, Lisnard L, Livage J. Design Of Silica-Coated Microcapsules For Bioencapsulation. Chem Commun. 2001;(23):2496–2497. doi: 10.1039/b108324b. DOI: https://doi.org/10.2174/1573413710602030219. [DOI] [PubMed]
- 5.Nassif N, Bouvet O, Rager MN, Roux C, Coradin T, Livage J. Living Bacteria In Silica Gels. Nature Materials. 2002;1(1):42–44. doi: 10.1038/nmat709. [DOI] [PubMed] [Google Scholar]
- 6.Nassif N, Roux C, Coradin T, Rager MN, Bouvet OMM, Livage J. A Sol-Gel Matrix To Preserve The Viability Of Encapsulated Bacteria. Journal of Materials Chemistry. 2003;13(2):203–208. DOI: https://doi.org/10.1039/b210167j. [Google Scholar]
- 7.Harper JC, Lopez DM, Larkin EC, Economides MK, McIntyre SK, Alam TM, Tartis MS, Werner-Washburne M, Brinker CJ, Brozik SM, Wheeler DR. Encapsulation of S. cerevisiae in Poly(glycerol) Silicate Derived Matrices: Effect of Matrix Additives and Cell Metabolic Phase on Long-Term Viability and Rate of Gene Expression. Chemistry of Materials. 2011;23(10):2555–2564. doi: 10.1021/cm103525u. [DOI] [Google Scholar]
- 8.Harper JC, Edwards TL, Savage T, Harbaugh S, Kelley-Loughnane N, Stone MO, Brinker CJ, Brozik SM. Orthogonal Cell-Based Biosensing: Fluorescent, Electrochemical, and Colorimetric Detection with Silica-Immobilized Cellular Communities Integrated with an ITO–Glass/Plastic Laminate Cartridge. Small. 2012;8(17):2743–2751. doi: 10.1002/smll.201200343. [DOI] [PubMed] [Google Scholar]
- 9.Brinker CJ, Scherer GW. Sol-Gel Science: The Physics And Chemistry Of Sol-Gel Processing. Academic press; 2013. [Google Scholar]
- 10.Iler RK. The Chemistry Of Silica: Solubility, Polymerization, Colloid And Surface Pro Perties, And Biochemistry. Wiley; 1979. [Google Scholar]
- 11.Eleftheriou NM, Ge X, Kolesnik J, Falconer SB, Harris RJ, Khursigara C, Brown ED, Brennan JD. Entrapment of Living Bacterial Cells in Low-Concentration Silica Materials Preserves Cell Division and Promoter Regulation. Chemistry of Materials. 2013;25(23):4798–4805. doi: 10.1021/cm403198z. [DOI] [Google Scholar]
- 12.Harper JC, Khirpin CY, Carnes EC, Ashley CE, Lopez DM, Savage T, Jones HD, Davis RW, Nunez DE, Brinker LM. Cell-Directed Integration into Three-Dimensional Lipid− Silica Nanostructured Matrices. ACS nano. 2010;4(10):5539–5550. doi: 10.1021/nn101793u. [DOI] [PubMed] [Google Scholar]
- 13.Baca HK, Ashley C, Carnes E, Lopez D, Flemming J, Dunphy D, Singh S, Chen Z, Liu N, Fan H. Cell-Directed Assembly Of Lipid-Silica Nanostructures Providing Extended Cell Viability. Science. 2006;313(5785):337–341. doi: 10.1126/science.1126590. http://dx.doi.org/10.1126/science.1126590. [DOI] [PubMed] [Google Scholar]
- 14.Carturan G, Dal Toso R, Boninsegna S, Dal Monte R. Encapsulation Of Functional Cells By Sol-Gel Silica: Actual Progress And Perspectives For Cell Therapy. Journal of Materials Chemistry. 2004;14(14):2087–2098. doi: 10.1039/B401450B. [DOI] [Google Scholar]
- 15.Sglavo VM, Carturan G, Dal Monte R, Muraca M. SiO2 Entrapment Of Animal Cells Part I Mechanical Featurs Of Sol-Gel Sio2 Coatings. Journal of Materials Science. 1999;34(15):3587–3590. doi: 10.1023/a:1004626632730. [DOI] [Google Scholar]
- 16.Avnir D, Coradin T, Lev O, Livage J. Recent Bio-Applications Of Sol-Gel Materials. Journal of Materials Chemistry. 2006;16(11):1013–1030. doi: 10.1039/B512706H. [DOI] [Google Scholar]
- 17.Baca HK, Carnes E, Singh S, Ashley C, Lopez D, Brinker CJ. Cell-directed assembly of bio/nano interfaces—a new scheme for cell immobilization. Accounts of chemical research. 2007;40(9):836–845. doi: 10.1021/ar600027u. [DOI] [PubMed] [Google Scholar]
- 18.Baca HK, Carnes EC, Ashley CE, Lopez DM, Douthit C, Karlin S, Brinker CJ. Cell-directed-assembly: directing the formation of nano/bio interfaces and architectures with living cells. Biochimica et Biophysica Acta (BBA)-General Subjects. 2011;1810(3):259–267. doi: 10.1016/j.bbagen.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H, Hong D, Choi JY, Kim JY, Lee SH, Kim HM, Yang SH, Choi IS. Layer-by-Layer-Based Silica Encapsulation of Individual Yeast with Thickness Control. Chemistry–An Asian Journal. 2015;10(1):129–132. doi: 10.1002/asia.201402993. https://doi.org/10.1002/asia.201402993. [DOI] [PubMed] [Google Scholar]
- 20.Park JH, Hong D, Lee J, Choi IS. Cell-In-Shell Hybrids: Chemical Nanoencapsulation Of Individual Cells. Accounts of chemical research. 2016 doi: 10.1021/acs.accounts.6b00087. https://doi.org/10.1021/acs.accounts.6b00087. [DOI] [PubMed]
- 21.Yang SH, Lee KB, Kong B, Kim JH, Kim HS, Choi IS. Biomimetic Encapsulation of Individual Cells with Silica. Angewandte Chemie-International Edition. 2009;48(48):9160–9163. doi: 10.1002/anie.200903010. [DOI] [PubMed] [Google Scholar]
- 22.Rider TH, Petrovick MS, Nargi FE, Harper JD, Schwoebel ED, Mathews RH, Blanchard DJ, Bortolin LT, Young AM, Chen JZ, Hollis MA. A B Cell-Based Sensor For Rapid Identification Of Pathogens. Science. 2003;301(5630):213–215. doi: 10.1126/science.1084920. https://doi.org/10.1126/science.1084920. [DOI] [PubMed] [Google Scholar]
- 23.Johnston R, Rogelj S, Harper JC, Tartis M. Sol-Generating Chemical Vapor Into Liquid (SG-Cvil) Deposition–A Facile Method For Encapsulation Of Diverse Cell Types In Silica Matrices. Journal of Materials Chemistry B. 2015;3(6):1032–1041. doi: 10.1039/C4TB01349B. https://doi.org/10.1039/C4TB01349B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta G, Rathod SB, Staggs KW, Ista LK, Oucherif KA, Atanassov PB, Tartis MS, Montano GA, Lopez GP. CVD for the Facile Synthesis of Hybrid Nanobiomaterials Integrating Functional Supramolecular Assemblies. Langmuir. 2009;25(23):13322–13327. doi: 10.1021/la903475d. [DOI] [PubMed] [Google Scholar]
- 25.Preari M, Spinde K, Lazic J, Brunner E, Demadis KD. Bioinspired Insights into Silicic Acid Stabilization Mechanisms: The Dominant Role of Polyethylene Glycol-Induced Hydrogen Bonding. Journal of the American Chemical Society. 2014;136(11):4236–4244. doi: 10.1021/ja411822s. [DOI] [PubMed] [Google Scholar]
- 26.Vong MSW, Bazin N, Sermon PA. Chemical Modification Of Silica Gels. Journal of Sol-Gel Science and Technology. 1997;8(1):499–505. doi: 10.1007/bf02436889. [DOI] [Google Scholar]
- 27.Kuraoka K, Ueda T, Sato M. Preparation And Properties Of Organic-Inorganic Hybrid Flexible Hardcoat Films. Journal of Materials Science. 2005;40(13):3577–3579. doi: 10.1007/s10853-005-2880-0. [DOI] [Google Scholar]
- 28.Brinker CJaS. Sol-Gel Science. Academic Press; San Diego: 1990. [Google Scholar]
- 29.Mornet S, Lambert O, Duguet E, Brisson A. The Formation of Supported Lipid Bilayers on Silica Nanoparticles Revealed by Cryoelectron Microscopy. Nano Letters. 2005;5(2):281–285. doi: 10.1021/nl048153y. [DOI] [PubMed] [Google Scholar]
- 30.Richter R, Mukhopadhyay A, Brisson A. Pathways of Lipid Vesicle Deposition on Solid Surfaces: A Combined QCM-D and AFM Study. Biophysical Journal. 2003;85(5):3035–3047. doi: 10.1016/S0006-3495(03)74722-5. DOI: http://dx.doi.org/10.1016/S0006-3495(03)74722-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng C, Zhang M, Xue C, Bai F, Zhao X. Development Of Stress Tolerant Saccharomyces Cerevisiae Strains By Metabolic Engineering: New Aspects From Cell Flocculation And Zinc Supplementation. Journal of Bioscience and Bioengineering. 2016 doi: 10.1016/j.jbiosc.2016.07.021. https://doi.org/10.1016/j.jbiosc.2016.07.021. [DOI] [PubMed]
- 32.Puddu V, Perry CC. Interactions at the Silica–Peptide Interface: The Influence of Particle Size and Surface Functionality. Langmuir. 2014;30(1):227–233. doi: 10.1021/la403242f. [DOI] [PubMed] [Google Scholar]
- 33.Belton DJ, Patwardhan SV, Perry CC. Spermine, Spermidine And Their Analogues Generate Tailored Silicas. Journal of Materials Chemistry. 2005;15(43):4629–4638. https://doi.org/10.1039/b509683a. [Google Scholar]
- 34.Meng H, Yang S, Li Z, Xia T, Chen J, Ji Z, Zhang H, Wang X, Lin S, Huang C, Zhou ZH, Zink JI, Nel AE. Aspect Ratio Determines the Quantity of Mesoporous Silica Nanoparticle Uptake by a Small GTPase-Dependent Macropinocytosis Mechanism. ACS Nano. 2011;5(6):4434–4447. doi: 10.1021/nn103344k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Hu L, Yu D, Gao C. Influence Of Silica Particle Internalization On Adhesion And Migration Of Human Dermal Fibroblasts. Biomaterials. 2010;31(32):8465–8474. doi: 10.1016/j.biomaterials.2010.07.060. https://doi.org/10.1016/j.biomaterials.2010.07.060. [DOI] [PubMed] [Google Scholar]
- 36.Pritz CO, Bitsche M, Salvenmoser W, Dudás J, Schrott-Fischer A, Glueckert R. Endocytic Trafficking Of Silica Nanoparticles In A Cell Line Derived From The Organ Of Corti. Nanomedicine. 2013;8(2):239–252. doi: 10.2217/nnm.12.91. https://doi.org/10.2217/nnm.12.91. [DOI] [PubMed] [Google Scholar]
- 37.Majno G, Joris I. Apoptosis, Oncosis, And Necrosis. An Overview Of Cell Death. The American journal of pathology. 1995;146(1):3. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.