Abstract
Introduction
There remains a significant therapeutic need for small-cell lung cancer (SCLC). We and others have reported high frequency of copy number gains in cytogenetic bands encoding fibroblast growth factor receptor 1 (FGFR1) in SCLC tumors and cell lines.
Methods
Thirteen SCLC cell lines and 68 SCLC patient tumor samples were studied for FGFR1 amplification. Growth inhibition assays were performed using PD173074, a pan-FGFR inhibitor to determine the correlation between FGFR1 expression and drug sensitivity.
Results
We did not detect FGFR1 mutations in SCLC cell lines. Focal amplification of FGFR1 gene was found in five tumor samples (7%), with high-level focal amplification in only one tumor sample (1%). Amplification owing to polysomy of chromosome 8, where FGFR1 locates, was observed in 22 tumor samples (32%). There was no correlation between FGFR1 gene copy number and messenger RNA expression or protein expression in SCLC cells. FGFR inhibitor sensitivity correlated with FGFR1 copy number determined by real-time polymerase chain reaction assay (r= −0.79; p = 0.01).
Conclusion
FGFR1 gene mutations and focal amplification are rare in SCLC, but polysomy of chromosome 8 is relatively common. FGFR1 copy number gain predicts sensitivity to FGFR inhibition, and FGFR expression correlates inversely with chemosensitivity.
Keywords: Small-cell lung cancer, FGFR1, Focal amplification, Copy number gain
Small-cell lung cancer (SCLC) comprises 13% of all lung cancers.1 Despite extensive research over the last two decades, treatment for SCLC has not improved significantly and prognosis remains poor.2 Genetic characterization of driver mutations could potentially offer effective drug targets and novel therapies in SCLC. Although previous studies have reported recurrent genetic alterations in SCLC,3–5 therapeutically tractable targets have so far been elusive.
Fibroblast growth factors and fibroblast growth factor receptors (FGFR) play essential roles in regulation of normal cell proliferation, survival, migration, and differentiation.6 The FGFR tyrosine kinase family comprises four kinases: FGFR1, FGFR2, FGFR3, and FGFR4. FGFR1 amplification has been reported in 22% of squamous cell lung carcinoma and may predict sensitivity to FGFR inhibition.7 The fibroblast growth factor signaling pathway may influence growth of SCLC. FGFR inhibition impairs SCLC growth in vitro and in vivo, reduces intratumor proliferation, and increases apoptosis.8 We and others have reported high copy number gains (CNGs) in cytogenetic bands encoding FGFR1 in SCLC cell lines and tumors.4,5 High CNGs in some of the samples suggest a role for FGFR1 as a potential therapeutic target in a subgroup of SCLC. In this study, we sought to further characterize the role of FGFR1 in SCLC.
PATIENTS AND METHODS
Tumor Samples, Tissue Microarray, and Cancer Cell Lines
Thirteen SCLC cell lines and 68 SCLC tumor samples were studied. Use of human samples was approved by the National Cancer Institute (NCI) Institutional Review Board. Details of cells and tumors are described in the Supplementary Materials and Methods (Supplementary Digital Content 1, http://links.lww.com/JTO/A534).
Array Comparative Genomic Hybridization Analysis
We performed array comparative genomic hybridization (aCGH) analysis on five patient samples from the NCI cohort. aCGH was performed and analyzed as described previously.5 A reference genomic DNA was used for hybridization. Data analysis was performed using Nexus 4.0 software (Biodiscovery Inc., Hawthorne, CA). CNG and loss of the FGFR1 gene were defined if the log2 ratio values of sample DNA/reference genomic DNA were more than 0.2 and less than −0.2, respectively.
Sequencing
Exons 2 to 18 of FGFR1 gene were sequenced in 13 SCLC cell lines as described in the Supplementary Materials and Methods (Supplementary Digital Content 1, http://links.lww.com/JTO/A534).
Real-Time Polymerase Chain Reaction
The messenger RNA (mRNA) expression of FGFR1 gene and the copy numbers of the FGFR1 gene in SCLC cells were evaluated using the Taqman gene expression assay and Taqman copy number assay, respectively (Applied Biosystems, Foster City, CA), following manufacturer’s instruction. Details of the method are described in the Supplementary Materials and Methods (Supplementary Digital Content 1, http://links.lww.com/JTO/A534).
Western Blot
Western blotting was performed as previously described.9 Anti-FGFR1 antibody was obtained from Abcam (Cambridge, United Kingdom), anti-p-FGFR antibody was from Cell Signaling Technology (Danvers, MA), and anti-Actin antibody was from Sigma-Aldrich (St. Louis, MO). Semiquantification of signals of Western blot was analyzed by Image J software (National Institutes of Health, Bethesda, MD). FGFR1 protein expression was normalized by actin protein expression and then calibrated by the expression level of the H69 cells before further analysis.
Growth Inhibition Assays
The concentration that inhibits 50% to drugs in SCLC cells was determined as described in the Supplementary Materials and Methods (Supplementary Digital Content 1, http://links.lww.com/JTO/A534).
Fluorescence In Situ Hybridization for the FGFR1 Gene
Fluorescence in situ hybridization (FISH) assays were performed on 5 μm formalin-fixed paraffin-embedded tumor sections using a laboratory standardized protocol with slight modifications.10 Details of the method are described in the Supplementary Materials and Methods (Supplementary Digital Content 1, http://links.lww.com/JTO/A534). CNG was defined as red FGFR1 signals with the corresponding control chromosome enumeration probes (CEP8) more than or equal to four copies per cell in more than 20% of cells. Focal amplification was defined if the ratio of the FGFR1 gene to the CEP8 was more than 2 (low-level amplification 2–4; high-level amplification >4).
Statistical Analysis
Correlation between variables was analyzed using Spearman’s method, and p values less than 0.05 were regarded as significant.
RESULTS
Expression of FGFR1 Is Unrelated to Gene Copy Number
We previously reported genetic alterations of 13 SCLC cell lines using aCGH assay.5 We further determined the copy number of the FGFR1 gene using real-time polymerase chain reaction assay and evaluated whether the copy number alteration of the FGFR1 gene is associated with its protein expression (Fig. 1A). Whereas the protein expression of the FGFR1 gene was related to its mRNA expression, protein or mRNA expression of the FGFR1 gene in SCLC cells was unrelated to its gene copy number (Fig. 1B). As the anti-p-FGFR antibody is not FGFR1 specific, we failed to observe correlation between expression of p-FGFR and copy number of the FGFR1 gene (r = −0.19, p = 0.66), FGFR1 mRNA expression (r = −0.31, p = 0.45), and FGFR1 protein expression (r = −0.34, p = 0.42). We sequenced the exons 2 to 18 of the FGFR1 gene in 13 SCLC cell lines, and no mutation was detected.
FIGURE 1.
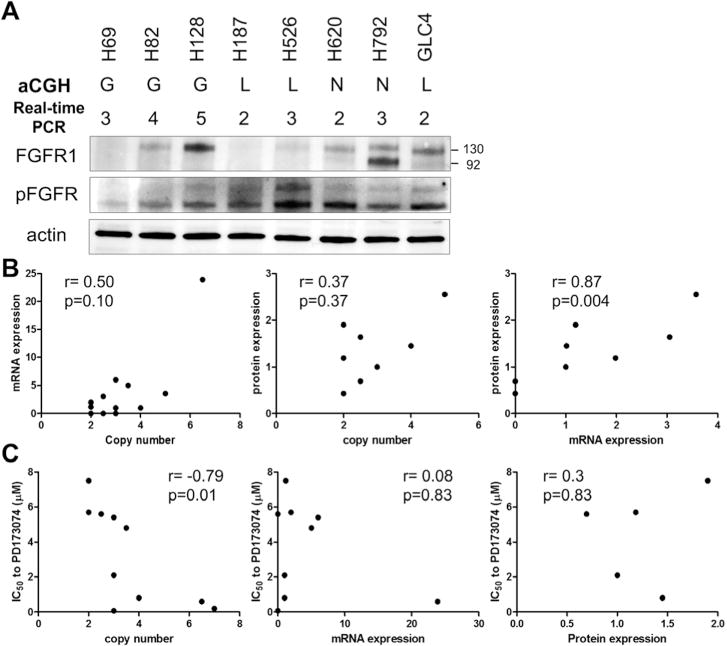
FGFR1 in SCLC cells. A, FGFR1 protein expression in SCLC cells. G, copy number gain; N, copy number normal; L, copy number loss. The numbers indicate the copy number of the FGFR1 gene determined by the real-time PCR assay. B, Correlation of FGFR1 copy number assessed by real-time PCR and mRNA expression and protein expression. C, Correlation of concentration that inhibits 50% to PD173074 and FGFR1 copy number assessed by real-time PCR assay, FGFR1 mRNA expression, or FGFR1 protein expression. FGFR1, fibroblast growth factor receptor 1; SCLC, small-cell lung cancer; PCR, polymerase chain reaction; mRNA, messenger RNA.
FGFR1 Inhibitor Sensitivity Is Related to CNG of the Gene
SCLC cells with higher copies of the FGFR1 gene, detected by real-time polymerase chain reaction assay, were more sensitive to PD173074, a pan-FGFR inhibitor (r = −0.79; p = 0.01) (Fig. 1C). Nevertheless, we failed to observe a correlation between the sensitivity to PD173074 and FGFR1 mRNA expression (r = 0.45; p = 0.13) or FGFR1 protein expression (Fig. 1C). The expression of p-FGFR was unrelated to the sensitivity to PD173074 (r = 0.31, p = 0.61). We studied the correlation between FGFR1 copy number (Supplementary Fig. S1A and B, Supplementary Digital Content 1, http://links.lww.com/JTO/A534) and FGFR1 mRNA expression (Supplementary Fig. S1C and D, Supplementary Digital Content 1, http://links.lww.com/JTO/A534) and sensitivity to the cytotoxic agents cisplatin and etoposide, both of which are standard therapies for SCLC. We observed that SCLC cells expressing higher FGFR1 mRNA were more resistant to etoposide (r = 0.68; p = 0.01) (Supplementary Fig. S1D, Supplementary Digital Content 1, http://links.lww.com/JTO/A534). Nevertheless, the protein expression of the FGFR1 gene was unrelated to the sensitivities to etoposide (r = 0.43, p = 0.29).
Focal Amplification of FGFR1 Gene Was Infrequent, But CNG Was Common
We studied, using FISH, whether the CNG was due to focal amplification or to polysomy of chromosome 8. Focal amplification was observed in only five of 68 tumor samples (7%) and among those high-level focal amplification was detected only in 1% (Fig. 2A).
FIGURE 2.
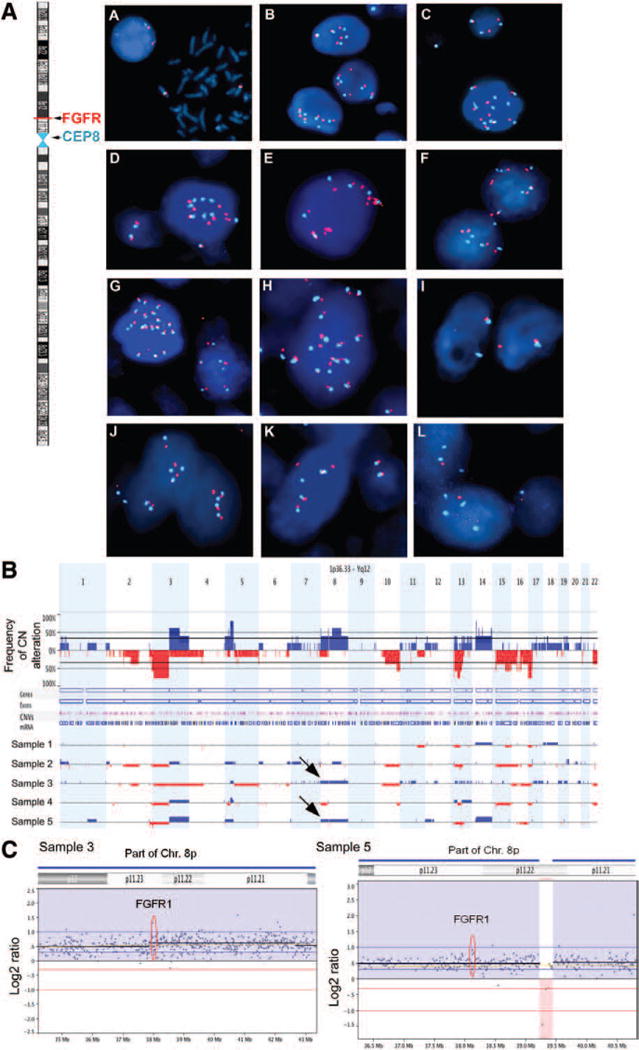
FGFR1 in SCLC tumors. A, Dual-color fluorescence in situ hybridization showing FGFR1 (red fluorescence signal) gene copy number variations along with the reference chromosome 8 centromeric probe (aqua fluorescence signal) in SCLC cell lines and primary tumors. Left: Ideogram of chromosome 8 with FGFR1 gene mapped to 8p12–p11.23 chromosomal region. Normal peripheral blood lymphocytes showing two copies of FGFR1 gene on interphase and metaphase cells. B, Genome-wide frequency of copy number alterations. Gains and losses are shown in blue and red, respectively. C, High-resolution analysis showing copy number alterations in chromosome 8p where FGFR1 is located of patient samples 3 and 5 which had chromosome 8p polysomy. FGFR1, fibroblast growth factor receptor 1; SCLC, small-cell lung cancer.
Nevertheless, 22 of the 68 tumor samples (32%) displayed moderate amplification ranging from 4 to 9 FGFR1 gene copy numbers per cell, which includes the entire chromosome 8 (Table 1). The same pattern, that is, polysomy of chromosome 8, was seen in the SCLC cell lines H678 and DMS114. We performed aCGH in five samples from the NCI cohort and observed CNG of the whole chromosome 8p, implying polysomy in two samples (Fig. 2B and C). The results were consistent with what was observed by using FISH.
TABLE 1.
FGFR1 Fluorescence In Situ Hybridization in the Small-Cell Lung Cancer Tumor Samples and TMA Specimens
| Tumor Source | No. of Patients | FGFR1 Copy Number Gain (%) | FGFR1 Focal Amplification (%) |
|---|---|---|---|
| National Cancer Institute cohort | 20 | 12 (60) | 1 (5) |
| TMA | 14 | 5 (36) | 0 |
| Multitumor block | 34 | 5 (15) | 4 (12) |
FGFR1, Fibroblast growth factor receptor 1; TMA, tissue microarray.
DISCUSSION
Recent large-scale sequencing efforts have identified FGFR1 as a potentially therapeutically tractable target in a subgroup of SCLC. Consistent with previous reports, we found no mutations in FGFR1 exons.11 Peifer et al.4 detected focal amplification of FGFR1 locus in SCLC by copy number analysis (4 of 63, 6%) and validated the results by FISH in an independent cohort (3 of 51, 6%). Nevertheless, no amplification of the FGFR1 locus was identified in a similar analysis of 56 cases.3 Among 68 SCLC tumor specimens, we found focal FGFR1 amplification in 7% of the samples and true high-level amplification in only 1% of the sample. Our findings are comparable to results of large-scale integrated genomics studies where the frequency of FGFR1 amplification ranged from 0% to 6%,3,4 and mutation was absent.
We found no correlation between FGFR1 copy number and mRNA expression and between FGFR1 copy number and protein expression. A similar lack of correlation between FGFR1 mRNA expression and copy number of the gene was also demonstrated in squamous cell carcinomas of lung.12 In SCLC cells, the lack of correlation between FGFR1 copy number, mRNA expression, and protein expression suggests that chromosome 8 polysomy may have little impact on FGFR1 expression. Despite SCLC cells with higher copies of the FGFR1 gene were more sensitive to PD173074, a non-isoform-specific FGFR inhibitor, we cannot exclude the possibility that other genes locating in the chromosome 8 contribute to the sensitivity to PD173074 in SCLC cells.
In conclusion, mutations or focal amplification of FGFR1 gene are rare in SCLC. Nevertheless, FGFR1 gene CNG is common and is related to sensitivity to FGFR inhibition. FGFR1 inhibitors should be explored in patients with SCLC with high copy numbers of the gene.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health.
Footnotes
Disclosure: The authors declare no conflict of interest.
References
- 1.van Meerbeeck JP, Fennell DA, De Ruysscher DK. Small-cell lung cancer. Lancet. 2011;378:1741–1755. doi: 10.1016/S0140-6736(11)60165-7. [DOI] [PubMed] [Google Scholar]
- 2.Oze I, Hotta K, Kiura K, et al. Twenty-seven years of phase III trials for patients with extensive disease small-cell lung cancer: disappointing results. PLoS One. 2009;4:e7835. doi: 10.1371/journal.pone.0007835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111–1116. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peifer M, Fernández-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voortman J, Lee JH, Killian JK, et al. Array comparative genomic hybridization-based characterization of genetic alterations in pulmonary neuroendocrine tumors. Proc Natl Acad Sci U S A. 2010;107:13040–13045. doi: 10.1073/pnas.1008132107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 7.Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pardo OE, Latigo J, Jeffery RE, et al. The fibroblast growth factor receptor inhibitor PD173074 blocks small cell lung cancer growth in vitro and in vivo. Cancer Res. 2009;69:8645–8651. doi: 10.1158/0008-5472.CAN-09-1576. [DOI] [PubMed] [Google Scholar]
- 9.Lee JH, Voortman J, Dingemans AM, et al. MicroRNA expression and clinical outcome of small cell lung cancer. PLoS One. 2011;6:e21300. doi: 10.1371/journal.pone.0021300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pack SD, Zhuang Z. Fluorescence in situ hybridization: application in cancer research and clinical diagnostics. Methods Mol Med. 2001;50:35–50. doi: 10.1385/1-59259-084-5:35. [DOI] [PubMed] [Google Scholar]
- 11.Pleasance ED, Stephens PJ, O’Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammerman PS, Lawrence MS, Voet D, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


