Abstract
Purpose
Mammalian target of rapamycin (mTOR) inhibition activates compensatory insulin–like growth factor receptor (IGFR) signaling. We evaluated the ridaforolimus (mTOR inhibitor) and dalotuzumab (anti-IGF1R antibody) combination.
Experimental Design
In vitro and in vivo models, and a phase I study in which patients with advanced cancer received ridaforolimus (10–40 mg/day every day × 5/week) and dalotuzumab (10 mg/kg/week or 7.5 mg/kg/every other week) were explored.
Results
Preclinical studies demonstrated enhanced pathway inhibition with ridaforolimus and dalotuzumab. With 87 patients treated in the phase I study, main dose-limiting toxicities (DLT) of the combination were primarily mTOR-related stomatitis and asthenia at doses of ridaforolimus lower than expected, suggesting blockade of compensatory pathways in normal tissues. Six confirmed partial responses were reported (3 patients with breast cancer); 10 of 23 patients with breast cancer and 6 of 11 patients with ER+/high-proliferative breast cancer showed anti-tumor activity.
Conclusions
Our study provides proof-of-concept that inhibiting the IGF1R compensatory response to mTOR inhibition is feasible with promising clinical activity in heavily pretreated advanced cancer, particularly in ER+/high-proliferative breast cancer (ClinicalTrials.gov identifier: NCT00730379).
Introduction
The phosphatidylinositol 3-kinase (PI3K)–AKT–mammalian target of rapamycin (mTOR) pathway plays a critical role in cell growth, proliferation, and survival (1). Multiple receptor tyrosine kinases [e.g., insulin-like growth factor-1 receptor (IGF1R) and human epidermal growth factor (HER) receptor family] activate this pathway via adapter proteins or binding of the p85–PI3K regulatory subunit (2). In breast cancer, aberrant activation of the PI3K pathway has been reported in approximately 50% of primary tumors and has been associated with the HER receptor family, estrogen receptor (ER), and IGF1R signaling pathways (3–6), and with resistance to endocrine therapy and anti-HER2 therapy (7, 8).
Because of its central role in cancer development and progression, various therapeutic strategies have focused on blocking specific signaling molecules of the PI3K pathway. Perhaps the most extensively tested druggable component of the pathway has been the kinase mTOR (composed of mTORC1 and mTORC2) due to a better understanding of and discovery of its modulation by rapamycin and its analogues (9, 10).
However, rapamycin analogues have shown modest and variable antitumor activity as single-agent therapies for most tumor types, possibly due to the lack of identification of the most suitable patient population and/or optimal dose (10, 11). One mechanism of suboptimal response to mTOR inhibitors is related to the activation of compensatory pathways. When mTORC1 is active, S6K1 directly phosphorylates the adapter protein of IGF1R insulin receptor substrate-1 (IRS1), leading to its degradation. A decrease in IRS1 levels reduces IGF1R signaling and activation of the PI3K–AKT pathway. Conversely, mTORC1 inhibition relieves this negative feedback loop, resulting in sustained IGF1R/IRS1 signaling and activation of AKT (12, 13).
Inhibition of mTOR in cancer cell lines and in patient tumor biopsies causes activation of AKT kinase, which is associated with induction of IRS1 but could be prevented by IGF1R inhibition (13, 14). Furthermore, activation of compensatory pathways has also been observed with other inhibitors of the PI3K–AKT–mTOR pathway, suggesting that it is a general mechanism of response to inhibition of this pathway and that concomitant blockade of these compensatory responses may be required for optimal therapeutic efficacy (15, 16).
We have tested the hypothesis of preventing activation of IGF1R signaling by exploring the combination of the mTOR inhibitor ridaforolimus with the anti-IGF1R monoclonal antibody dalotuzumab in preclinical models and in a phase I study in patients with advanced solid tumors, including patients with ER+ breast cancer.
Materials and Methods
Preclinical assessment of combination effects between ridaforolimus and dalotuzumab
Cell lines
293FT producer cells were obtained from Invitrogen. Other cell lines were purchased from cell line banks the American Type Culture Collection (ATCC), Japanese Collection of Research Bioresources Cell Bank (RIKEN), or Deutsche Sammlung von Mikroorganismen und Zellkulturen (DKMZ). The cells were grown under culture conditions recommended by the vendors. Cells were expanded and immediately frozen for experimentation. The authenticity of the cell lines were verified by short tandem repeat (STR) profiling analysis or similar methodologies by the banks. In addition, the mutation and gene expression levels from the targeted exome sequencing (TES) data were compared with the published mutation (COSMIC; Sanger data base) and gene expression data.
In vitro studies
A short hairpin RNA (shRNA) enhancer screen was performed in a colorectal cancer cell line. Details are available in the Supplementary Appendix.
In vivo xenograft studies
Immunodeficient female mice (HsdCpb:NMRI-Foxn1nu; Harlan Laboratories) were implanted with tumor fragments from the lung adenocarcinoma LXFA629. The control group was dosed with dalotuzumab vehicle once per week by intraperitoneal (i.p.) injection for 4 weeks (once per week × 4). Ridaforolimus vehicle was dosed five times per week i.p. for 4 weeks. The experimental groups were dosed with dalotuzumab once per week (20 mg/kg, i.p.), ridaforolimus five times per week (1 mg/kg, i.p.), or the combination of dalotuzumab and ridaforolimus at the same dose/schedules as the monotherapy arms. Details are available in the Supplementary Appendix.
Phase I clinical study of the combination of dalotuzumab and ridaforolimus in patients with solid tumors
Patients of ages 18 years or older with histologically confirmed advanced tumors unresponsive to standard therapy—or for whom standard therapy does not exist—were enrolled into the phase I, international, multicenter, open-label, single-arm, non-randomized trial (ClinicalTrials.gov identifier: NCT00730379; http://clinicaltrials.gov/ct2/show/NCT00730379; Protocol 004). Eligible patients had Eastern Cooperative Oncology Group (ECOG) performance status of 0 or 1 and adequate bone marrow, hepatic, and renal function. Patients with uncontrolled diabetes and uncontrolled hyperlipidemia at screening were excluded in the dose escalation portion. Diabetic patients requiring insulin were excluded from the dose-escalation portion; however, well-controlled insulin-using diabetic patients were allowed in the dose-expansion cohorts. The study was conducted in accordance with Good Clinical Practice guidelines and the Declaration of Helsinki. All patients gave informed consent, and approval was obtained from the ethics committees at each participating institution.
In the dose-escalation phase, the main objective was to determine the maximum-tolerated dose (MTD) of the two drugs in combination. Enrollment in dose escalation followed a variation of the traditional “3 + 3” dose-escalation scheme, called the “rolling 6” trial design (17). Enrollment of a minimum of 3 patients per dose cohort was required, but enrollment of up to 6 patients per cohort was allowed (no intrapatient dose escalation was permitted). In dose expansion, at least 12 additional patients were to be enrolled at the recommended phase II dose (RP2D). Antitumor activity was analyzed in the dose-escalation and dose-expansion phases. A third part of the study was added as an amendment to seek preliminary evidence of antitumor activity in specific tumor types [advanced colorectal and non–small cell lung cancer (NSCLC)], and to gain additional safety and tolerability experience with the combination at the RP2D to better describe the adverse event (AE) profile. Secondary and tertiary objectives included pharmacodynamic (PD) and pharmacokinetic (PK) analyses, as well as measurements of antitumor activity using various endpoints [objective response, metabolic response, tumor markers, 4-month progression-free survival (PFS) rate, and time on treatment].
Ridaforolimus was administered per os at a starting dose of 10 mg/day, once daily for 5 days (10 mg/day every day × 5; dose level 1); ridaforolimus doses were escalated in sequential cohorts of patients in increments of 10 mg/day up to 40 mg/day (dose levels, 2–4). Dalotuzumab was administered intravenously at 10 mg/kg/week for the first dose level; subsequent dose levels tested sequential escalating doses of ridaforolimus in combination with dalotuzumab at either 10 mg/kg/week (dose levels, 2–4) or 7.5 mg/kg every other week (every 14 days; dose levels, 1.5–3.5). Ridaforolimus was given as monotherapy for the first week (days, 1–5); this period was referred to as cycle 0. Cycle 1 began on the first day of dosing with the combination of ridaforolimus and dalotuzumab and lasted for 4 weeks (Supplementary Fig. S1). The first 4 weeks of dosing were used as the toxicity evaluation period for the purposes of dose escalation. Dose-limiting toxicities (DLT) were defined as any grade 4 neutropenia persisting for at least 5 days; grade 3 or 4 neutropenia associated with fever, antibiotics, or hospitalization for infection; grade 4 thrombocytopenia persisting for at least 5 days; grade 3 or greater hyperglycemia persisting for at least 5 days, grade 3 or greater diarrhea persisting for more than 24 hours, or grade 3 or greater nausea or vomiting (despite optimal medical management); and any other grade 3 or higher nonhematologic toxicity—except for alopecia and transient electrolyte abnormalities—persisting despite treatment. Any toxicities that were at least possibly related to study drug that resulted in an inability to complete the DLT assessment period, interruption in dosing for more than 10 dosing days during the DLT assessment period, or a delay in the initiation of the next cycle for more than 10 dosing days were also considered DLTs. AEs were graded according to the National Cancer Institute Common Terminology Criteria for Adverse Events (NCI-CTCAE version 3.0).
Safety and efficacy assessments
Patients were evaluated predose on day 1 of each week to assess ECOG performance status, vital signs, body weight, and AEs. Twelve-lead electrocardiogram measurements were taken at screening and predose for all cycles after cycle 1. Tumor assessment by Response Evaluation Criteria in Solid Tumors (RECIST v1.0) was performed by computed tomography scan or magnetic resonance imaging predose and every 8 weeks thereafter for the remainder of the study. Measurable disease was not required for eligibility for efficacy analysis. [18F]Fluorodeoxyglucose-positron emission tomography (18FDG-PET) was used to assess tumor metabolic rates during dose expansion within 14 days of starting therapy, and at the end of cycle 1, within 7 days before starting cycle 2. Metabolic response was defined in accordance with the European Organization for Research and Treatment of Cancer (EORTC) guidelines on 18FDG-PET imaging as a 20% decrease in tumor standardized uptake values (SUV) compared with baseline.
Pharmacodynamics assessments and biomarker evaluation
Pharmacodynamic effects of the combination of ridaforolimus and dalotuzumab in skin and tumor samples were determined in patients enrolled in the dose-expansion phase for whom paired specimens were available. Processing of the samples, immunohistochemistry, and statistical analysis were performed as described previously (18). Immunohistochemical analysis of skin and tumor biopsies was performed in formalin-fixed paraffin-embedded sections. Pharmacodynamic effects were also assessed in patients with ER+ breast cancer. On the basis of the gene expression signature of a set of 97 genes that define the genomic grade index (GGI), ER+ breast cancer can be subdivided into highly proliferative poor prognosis [Luminal B (high GGI)] and low proliferative [Luminal A (low GGI)] categories.
To identify potential biomarkers that may help determine patient populations responsive to the combination of ridaforolimus and dalotuzumab, breast cancer cell lines and human breast tumor were used to evaluate correlations between mTOR inhibition and the RAS pathway signature, which is a transcription readout of RAS pathway activation (19). The RAS signature score was determined as the log(10) ratio relative to the mean of all samples in the respective experiment. To assess the expression of IGF1R signaling pathway components in human breast cancers, we evaluated gene expression profiling datasets from The Cancer Genome Atlas (TCGA).
Results
Preclinical assessment of the combination of ridaforolimus and dalotuzumab
IGF1R signaling mediates feedback activation of AKT upon mTOR inhibition
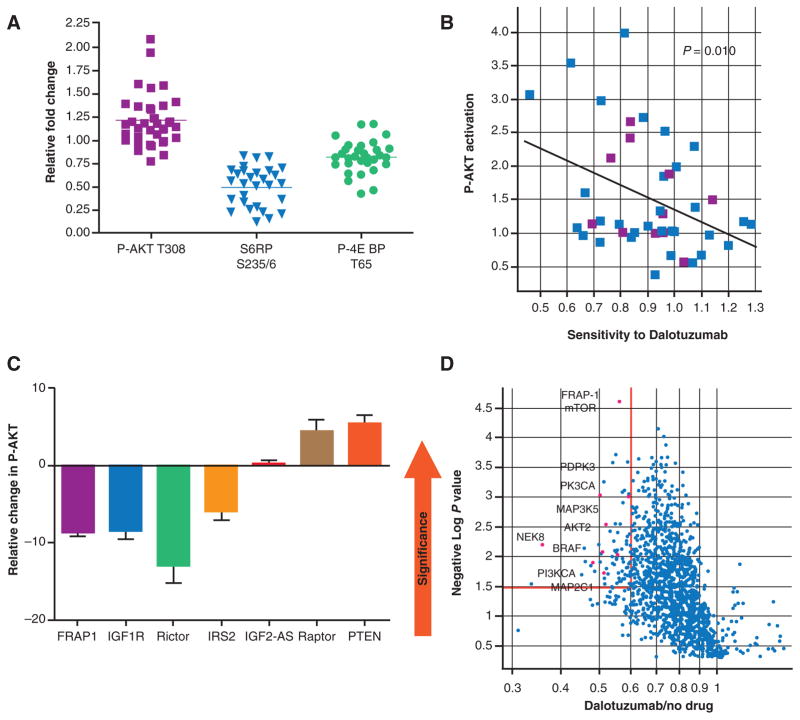
Treatment with ridaforolimus resulted in inhibition of phosphorylation of downstream pathway components, p-S6 ribosomal protein, and p-4EBP, and in compensatory activation of AKT (Fig. 1A). Cell lines exhibiting high levels of feedback induction by ridaforolimus were more sensitive to dalotuzumab (Fig. 1B). Inhibition of IGF1R and IRS2, as well as components of mTORC2 complex, blocked the activation of AKT (Fig. 1C), suggesting a causal link between the feedback activation of AKT following ridaforolimus exposure and IGF1R signaling.
Figure 1.
Combination therapy with mTOR and IGF1R inhibitors block feedback activation of AKT. A, diversity in feedback activation of AKT by ridaforolimus was observed in a panel of breast cancer cell lines. B, cell lines exhibiting feedback activation of AKT following ridaforolimus treatment showed sensitivity to anti-IGF1R therapy. C, knockdown of IGF1R and its signaling pathway components blocked feedback activation of AKT. A lentiviral shRNA screen identified mTOR as an enhancer of the activity of dalotuzumab in tumor cell lines, and AKT activation by mTOR was confirmed to be mediated via the IGF1R signaling pathway. D, representative results from a shRNA screen targeting various PI3K and MAPK kinases in HT-29 CRC cells. mTOR, mammalian target of rapamycin; IGF1R, insulin-like growth factor 1 receptor; shRNA, short hairpin RNA; PI3K, phosphatidylinositol 3-kinase; CRC, colorectal cancer; PTEN, phosphatase and tensin homolog.
Enhancers of sensitivity to dalotuzumab
The lentiviral shRNA enhancer screen targeting 480 distinct human kinases revealed that the strongest enhancers of dalotuzumab (top 3%) included several members of the PI3K pathway (PIK3CA, PDPK1, AKT2, and FRAP1/mTOR; Fig. 1D and Supplementary Table S1). In addition, key members of the RAS–MAPK pathway, BRAF and MAP2K1, were also identified. To confirm the screening hits, we performed colony formation assays using HT29 cells. Seventeen of 31 shRNA hits inhibited HT29 cells growth in the absence of drug, preventing an assessment of dalotuzumab sensitization. Eleven out of 20 shRNAs screening hits resulted in 2-fold or greater enhancement of dalotuzumab mediated cell proliferation—shRNAs targeting both PI3KCA and mTOR (FRAP1) significantly enhanced dalotuzumab-mediated colony growth inhibition (Fig. 1F). When we tested a shRNA vector that efficiently silences lipid phosphatase PTEN, the primary negative regulator of PI3K signaling, we found that dalotuzumab-mediated growth inhibition was abrogated by the PTEN shRNA (Fig. 1F), further confirming the role of PI3K pathway components (e.g., mTOR and PI3KCA) in enhancing the antitumor activity of dalotuzumab.
Combination therapy with ridaforolimus and dalotuzumab potentiates antitumor activity
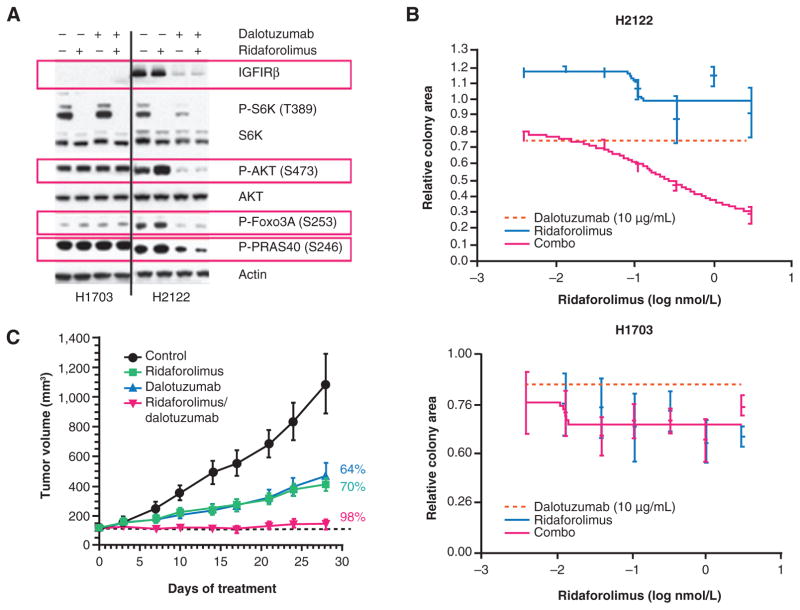
Western blot analysis showed that in the IGF1R-expressing H2122 NSCLC and MCF7 breast cancer cell line, ridaforolimus inhibited mTOR signaling (decreased levels of p-S6K) and activated AKT signaling (increased levels of p-AKT and its downstream targets, Foxo3A and PRAS40). Despite inhibiting mTOR signaling, AKT activation was not observed following treatment with ridaforolimus in H1703 cells, which express low levels of IGF1R (Fig. 2A and Supplementary Fig. S2). These results suggest that active IGF1R signaling is associated with feedback activation of AKT following ridaforolimus treatment. Treatment of H2122 cells with dalotuzumab alone significantly inhibited IGF1R signaling by blocking ligand binding and promoting receptor internalization and degradation while combination treatment with ridaforolimus and dalotuzumab prevented AKT activation and blocked phosphorylation of Foxo3A and PRAS40, resulting in effective targeting of both upstream and downstream components of the PI3K pathway (Fig. 2A). In MCF7 cells, combination therapy blocked feedback activation of AKT and potentiated PI3K pathway inhibition (Supplementary Fig. S2).
Figure 2.
Cotreatment with dalotuzumab and ridaforolimus enhances antitumor activity in IGF1R-expressing tumors. A, Western blot analysis of PI3K pathway components in cancer cells treated with dalotuzumab, ridaforolimus, or the combination. In IGF1R-expressing cancer cell lines (H2122), combination treatment suppresses the activation of AKT demonstrated by decreased phosphorylation of AKT and downstream targets FOXO3A and PRAS40. B, combination therapy with ridaforolimus and dalotuzumab results in increased cell death in anchorage-independent growth inhibition assays in IGF1R-expressing cells (H2122). No such combination benefit was observed in cancer cells expressing low levels of IGF1R (H1703). C, In vivo, combination therapy with ridaforolimus and dalotuzumab significantly inhibits growth over monotherapies. Tumor growth of primary human tumor–derived lung adenocarcinoma xenograft model LXFA629 in control (vehicle) or experimental agent–treated mice (n =10/group) were plotted. Combination therapy with ridaforolimus and dalotuzumab significantly enhanced tumor growth inhibition compared to vehicle-treated (P < 0.0001) or single-agent treated groups (P < 0.05).
Dalotuzumab alone or in combination did not alter PI3K signaling in the low IGF1R H1703 cells (Fig. 2A). The efficacy of combined treatment was further evaluated using an anchorage-independent growth assay. Although single-agent treatment with dalotuzumab or ridaforolimus showed minimal to modest growth inhibition in the high IGF1R H2122 cell line, significant growth inhibition was observed in the combination-treated group (Fig. 2B). In contrast, combination therapy did not enhance growth inhibition in the low IGF1R H1703 cell line compared with monotherapy alone. Treatment with ridaforolimus resulted in G1 cell-cycle arrest in H1703 cells, but not in H2122 cells. The addition of dalotuzumab to ridaforolimus had minimal effect in H1703 cells, while in H2122 cells, the combination decreased the percentage of actively cycling cells and increased the proportion of dead or dying cells (Fig. 2B). Combination therapy also induced apoptosis markers assessed by PARP cleavage in IGF1R-positive breast cancer cells (Supplementary Fig. S2). These results were also observed in additional cell lines and are consistent with enhanced inhibition of PI3K signaling by combination therapy in high IGF1R-expressing cancer cells.
Combination treatment was further evaluated in vivo using a primary human tumor–derived NSCLC xenograft model. Significant antitumor activity over single agents was observed (Fig. 2C).
Biomarker analysis of response to ridaforolimus and dalotuzumab therapy
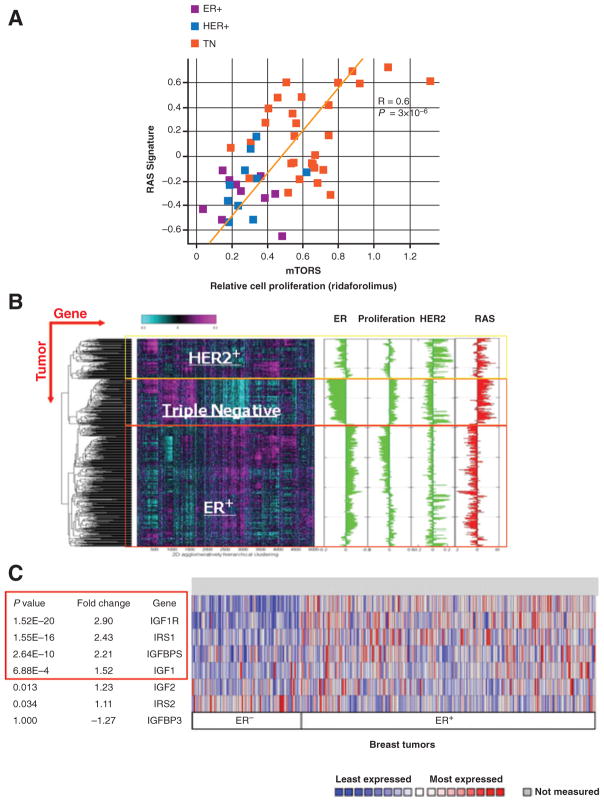
Sensitivity of breast cancer cell lines to mTORC1 inhibition significantly correlated with the RAS pathway signature (Fig. 3A; ref. 19). Notably, ER+ breast cancer cells expressed low RAS signature and were sensitive to ridaforolimus treatment. A previous study found that IGF1R expression and activation was associated with response to anti-IGF1R monotherapy in a panel of breast cancer cell lines (20). We evaluated these biomarkers in 800 human breast tumors obtained from the Karolinska Institutet (Solna, Sweden). Two-dimensional hierarchical clustering of 5,000 differentially expressed genes classified the tumors into three major subtypes: ER+, HER2+, and triple negative. The RAS gene expression signature was high in the triple-negative subtype and low in the ER+ subtype (Fig. 3B), consistent with the distribution of the RAS signature in the cell line panel. Analysis of human breast cancer gene expression profiling data from the TCGA demonstrated that IGF1R, its ligand IGF1, and its adaptor protein IRS1 are significantly overexpressed (P <1 × 10−10) in ER+ breast cancer (Fig. 3C). Analysis of two independent breast cancer gene expression datasets suggests that within ER+ breast tumors, those that have high levels of proliferation have the highest levels of IGF1R (Fig. 3C). On the basis of these biomarkers, we developed a hypothesis for defining the responder population to ridaforolimus and dalotuzumab combination therapy (Supplementary Fig. S4). Containing all of the features proved to predict sensitivity to the combination at the preclinical level, the luminal B breast cancer subtype represents the ideal model for the development of the combined treatment with ridaforolimus and dalotuzumab. This is why the combination is being evaluated in patients with luminal B breast cancer, although we cannot exclude that other types of tumors with high levels of IGF1R or lGF1 ligand, high PI3K signature, and low RAS signature may similarly benefit from the combination.
Figure 3.
RAS pathway activation and sensitivity to mTOR inhibition in ER+ breast cancer cell lines. A, RAS signature is associated with sensitivity to ridaforolimus in a panel of breast cancer cell lines. B, ER+ tumors have low levels of RAS pathway activation—unsupervised 2D clustering of the 5,000 most variable genes segregate breast tumors into ER+, HER2+, or triple negative (ER−, PR−, and HER2−) subtypes. RAS pathway activation is low in the ER+ tumors. C, IGF1R signaling pathway components are overexpressed in ER+ breast tumors. Differential expression of selected genes in the IGF1R receptor signaling pathway was compared between ER+(n =228) and ER−(n =87) breast tumors profiled by TCGA. The fold overexpression and P value for differential expression are indicated.
Phase I clinical evaluation of combined treatment with ridaforolimus and dalotuzumab
Between July 11, 2008 and March 10, 2010, 87 patients were enrolled and treated in the dose-escalation and dose-expansion phases. All patients receiving at least one dose of study medication were included in the safety and antitumor activity analyses. The median age was 56 years (range, 18–89), the median number of prior therapies was three (range, 1–17), and the most frequent malignancies were breast cancer (n = 23; excludes 2 patients with phylloides-type not included in breast cancer subtype efficacy analyses), colorectal cancer (n = 19), NSCLC (n = 16), and sarcoma (n = 14; Supplementary Table S2). The most common breast cancer subtype was ER+ (n = 18), of which 11 were ER+/high proliferative (Ki67 ≥ 15%). Sarcoma histology subtypes included Ewing sarcoma (n = 5), leiomyosarcoma (n = 3), osteosarcoma (n = 2), and chondrosarcoma, liposarcoma, synovial cell sarcoma, and desmoplastic round cell tumor (n = 1 for each).
Dose-limiting toxicities and maximum-tolerated dose
Thirty-seven patients were enrolled across five dose levels in the dose-escalation phase: ridaforolimus 10, 20, 30, and 40 mg/day every day × 5 plus dalotuzumab at 10 mg/kg/week, and ridaforolimus 20 mg/day every day × 5 plus dalotuzumab at 7.5 mg/kg every 14 days. Thirty-two patients completed the first cycle of treatment (DLT observation period), and were therefore considered DLT-evaluable. Only 1 patient experienced a DLT (prolonged grade 2 stomatitis), which occurred at dose level 1 (ridaforolimus 10 mg/day every day × 5/dalotuzumab 10 mg/kg/week). Dose level 4 (ridaforolimus 40 mg/day every day × 5/dalotuzumab 10 mg/kg/week) was determined to be the preliminary MTD because none of the 6 evaluable patients experienced a DLT. Subsequently, dose expansion was initiated and 12 additional patients were enrolled at dose level 4. The combined 15 patients from dose-escalation and dose-expansion were evaluable for DLTs at dose level 4. Four patients experienced a DLT—one with grade 4 fatigue and 3 with grade 3 mucosal inflammation/stomatitis. Because the cumulative rate of observed DLTs at dose level 4 (4 of 15; 27%) exceeded the prespecified goal of an observed rate of DLTs of less than 25% at the RP2D, dose level 3 was opened for expansion with 12 additional patients (permitted by the protocol). Initially, 19 DLT-evaluable patients were treated at dose level 3 (dose escalation and expansion), and only 1 patient experienced a DLT of grade 4 mucosal inflammation (5%). Therefore, ridaforolimus 30 mg/day every day × 5 and dalotuzumab 10 mg/kg/week were established as the RP2D. Considering all 41 DLT-evaluable patients treated at dose level 3 (all patients including those enrolled following protocol amendment), only 3 DLTs (grade 3 or 4 mucosal inflammation) were reported, for an overall cumulative DLT rate of 8% (see Supplementary Table S3 for a summary of all DLTs).
Safety
All treatment-related AEs reported in at least 10% of patients across all dose levels including the RP2D are listed in Table 1. Treatment-related AEs were mostly grade 1 to 2; grade 3 or worse AEs included hyperglycemia (12%), mucosal inflammation (9%), thrombocytopenia (9%), and asthenia (7%). Management of hyperglycemia included the use of oral antidiabetic agents or insulin in 26 of 50 patients (46%), with 21 patients receiving an oral agent and 5 patients receiving insulin. No patients required treatment discontinuation or dose reduction, but 6 patients required dose interruption due to hyperglycemic AEs; all restarted therapy after a period of interruption (dose level 2, n = 1; dose level 3, n = 4; dose level 4, n = 1). Treatment discontinuation and dose reductions due to stomatitis were required in 3 (dose level 1, n = 1; dose level 4, n = 2) and 9 patients (dose level 2, n = 1; dose level 3, n =6; dose level 4, n =2), respectively. The median time to onset of stomatitis and mucosal inflammation across all dose levels ranged from 9 to 15.5 days; at the RP2D (dose level 3), the median time to onset was 9 days. Treatment-related AEs in dose level 3 (RP2D) were similar to all dose levels with the exception of slightly higher levels of diarrhea and thrombocytopenia (Table 1). Patients received a mean dose of ridaforolimus ranging from 77% to 89% of the expected total dose across dose levels—notably, the mean total dose received (relative to planned) was the lowest in the dose level 4 cohort at 77%, reflecting a higher frequency of dose interruptions and dose reductions for the management of AEs.
Table 1.
Summary of treatment-related AEs occurring in at least 10% of patients across all dose levels or at dose level 3 (recommended phase II dose)
| Type of toxicity | All dose levels (N = 87) | Dose level 3 (N = 45) | ||
|---|---|---|---|---|
|
|
|
|||
| All grades n (%) | Grade >3 n (%) | All grades n (%) | Grade >3 n (%) | |
| Patients with ≥1 AE | 83 (95) | 40 (46) | 42 (93) | 23 (51) |
| Hyperglycemia | 49 (56) | 10 (12) | 29 (64) | 6 (13) |
| Mucosal inflammation | 40 (46) | 8 (9) | 18 (40) | 5 (11) |
| Hypercholesterolemia | 36 (41) | 1 (1) | 22 (49) | 1 (2) |
| Asthenia | 34 (39) | 6 (7) | 18 (40) | 4 (9) |
| Hypertriglyceridemia | 26 (30) | 0 | 18 (40) | 0 |
| Stomatitis | 25 (29) | 1 (1) | 14 (31) | 0 |
| Decreased appetite | 22 (25) | 2 (2) | 12 (27) | 1 (2) |
| Diarrhea | 20 (23) | 4 (5) | 16 (36) | 4 (9) |
| Thrombocytopenia | 20 (23) | 8 (9) | 15 (33) | 7 (16) |
| Nausea | 18 (21) | 1 (1) | 9 (20) | 1 (2) |
| Fatigue | 17 (20) | 3 (3) | 7 (16) | 1 (2) |
| Rash | 12 (14) | 0 | 6 (13) | 0 |
| Epistaxis | 10 (12) | 0 | 5 (11) | 0 |
| Anemia | 10 (12) | 1 (1) | 4 (9) | 0 |
| Vomiting | 9 (10) | 1 (1) | 6 (13) | 1 (2) |
NOTE: Only the highest reported grade of a given AE is counted for the individual subject.
Pharmacodynamic effects
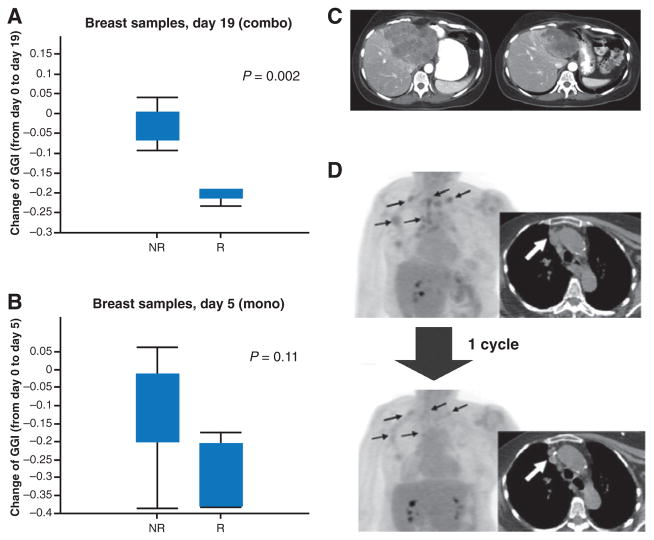
Twelve tumor-matched biopsy specimens were available for analysis. Biopsies were obtained at baseline (n = 22), day 5 of ridaforolimus monotherapy (n = 22), and day 19 after 2 doses of dalotuzumab (n = 19; Supplementary Fig. S1) and stained using hematoxylin and eosin, and for Ki67, p-S6K, and p-PRAS40. Gene expression profiling was also performed in available matched tumor samples. A marked inhibition of p-S6K was observed with ridaforolimus treatment that persisted with the addition of dalotuzumab (Fig. 4A; representative images are shown in Supplementary Fig. S2), indicating early and sustained inhibition of the mTOR signaling pathway. Phosphorylated PRAS40, an AKT substrate, was used as a surrogate marker to determine the level of AKT activation. The p-PRAS40 staining had a high background that was unchanged upon ridaforolimus or combination treatment with dalotuzumab; therefore, no conclusion could be made regarding the ability of the combination treatment to block feedback activation of AKT in tumor biopsy samples. However, we had previously shown that the dalotuzumab doses tested in this study efficiently blocked IG1R signaling (18). Analysis of changes in GGI in patients with ER+ breast cancer showed that by day 19 of combined treatment, patients responding to therapy demonstrated a significant reduction in the expression of GGI signature compared with nonresponders (no response by RECIST, PFS ≥6 months, metabolic response, or tumor markers decline; Fig. 4B). In contrast, single-agent treatment with ridaforolimus had a modest decrease in the GGI by day 5 that did not reach statistical significance (Fig. 3B). Staining for Ki67 also showed a corresponding decrease in proliferation in patients that responded to combination therapy or single-agent ridaforolimus; however, the change compared with nonresponders was not significant (Supplementary Fig. S3).
Figure 4.
Effect of ridaforolimus and dalotuzumab treatment on tumor markers. A, ridaforolimus and dalotuzumab treatment results in a significant decrease in the GGI at day 19 compared with day 0 in patients who responded to the combination therapy. B, patients responding to treatment with ridaforolimus alone had less pronounced changes in GGI by day 5 compared with day 0. C, example of tumor response (PR; RECIST) in a breast cancer patient with liver metastases treated at dose level 4 (ridaforolimus 40 mg/day and dalotuzumab 10 mg/kg/week). D, PET-CT evaluation of a patient with breast cancer after one cycle of treatment at dose level 4: CT scans revealed a 28% decrease in the sum of the longer diameters of the target lesions, which corresponded to 48% decrease in 18FDG uptake. Arrows indicate target lesions of interest. p-S6RP, phosphorylated S6 ribosomal protein; NR, nonresponders; R, responders.
Antitumor activity
All 87 patients enrolled were evaluable for antitumor activity; 6 partial responses (PR) were observed (breast cancer, n = 3; NSCLC, n = 1; ovarian cancer, n = 1; salivary gland cancer, n = 1) corresponding to an objective response rate (ORR) of 7% (95% confidence interval, 2.6, 14.4). The 3 patients with breast cancer with PR were treated at dose level 2 (n =1) and dose level 4 (n = 2), and the other 3 patients with a PR were treated at dose levels 1.5, 3, and 4 (n = 1 each). An additional 40 (46%) patients achieved stable disease (SD), and 27 (31%) had progressive disease (PD). Of note, out of 23 patients with breast cancer, 3 (13%) patients—all with ER+/high proliferative tumors—achieved a PR; another 11 (48%) were reported to have SD [5 (46%) with ER+/high-proliferative tumors] and 7 (30%) had PD [3 (27%) with ER+/high-proliferative tumors]. Among patients with ER+/high-proliferative breast cancer, the ORR was 27% (3 of 11 patients; Table 2; Supplementary Table S4; Fig. 4D and E). A total of 15 patients (17%; breast cancer, n = 5; NSCLC, n = 3; ovarian cancer, n =2; and 1 each with colon cancer, salivary gland cancer, endometrial cancer, sarcoma, rectal cancer carcinoid) had PFS >4 months. All patients with breast cancer with PR, and an additional patient with breast cancer with SD who also displayed a decline in tumor biomarker, had PFS >6 months. Time on treatment with ridaforolimus and dalotuzumab was longer than the most recent prior treatment in 23 of 81 (28%) evaluable patients and was at least doubled in 13 (15%) patients.
Table 2.
Summary of clinical efficacy in breast cancer patientsa
| Breast cancer type | n | RECIST (PR) | PFS ≥6 months | 18FDG-PET response | Tumor marker decline |
|---|---|---|---|---|---|
| Overall | 23 | 3 | 4 | 5 | 8 |
| Triple-negative breast cancer | 2 | 0 | 0 | 0 | 0 |
| HER2+ | 3 | 0 | 0 | 3 | 2 |
| ER+ | 18 | 3 | 4 | 2 | 6 |
| High proliferation (Ki67 ≤15%) | 11 | 3 | 4 | 2 | 5 |
| Low proliferation (Ki67 < 15%) | 5 | 0 | 0 | 0 | 0 |
| Unknown proliferation | 2 | 0 | 0 | 0 | 1 |
Overall, 10 of 23 (43%) breast cancer patients and 6 of 11 (54%) patients with ER+/high-proliferative breast cancer showed some evidence of antitumor activity (either response by RECIST, PFS ≥6 months, metabolic response, or tumor markers decline).
Seventeen patients had baseline and follow-up PET scans that could be analyzed for metabolic response. On the basis of changes in SUVmean, 9 (53%) of 17 evaluable patients were classified as metabolic responders (breast cancer, n = 5; ovarian cancer, n = 2; melanoma, n =1; rectal cancer, n =1). Forty-two patients also had a measurable serum tumor marker (e.g., CA 15-3 for breast cancer) at baseline, and at least one posttreatment evaluation. Eleven (41%) of 27 patients (breast cancer, n = 8; ovarian cancer, n = 2; endometrial cancer, n = 1) had a 25% to 97% decrease in serum tumor markers and were considered to have a biomarker response. Ten patients with breast cancer had evidence of anti-tumor activity following treatment with the combination of ridaforolimus and dalotuzumab, based on objective response by RECIST, PFS greater than 6 months, PET response, and decrease of tumor markers during treatment.
Discussion
Inhibition of mTOR induces feedback activation of AKT, which may potentially counteract antitumor effects of mTOR inhibitors (12, 13). Here, we have confirmed that mTORC1 inhibition alone led to activation of AKT, both in cell lines and in a primary human tumor-derived xenograft model. We observed that the sensitivity to dalotuzumab correlated with the level of induced AKT activation, which could therefore become a potential biomarker of sensitivity to the combination. In addition, knockdown of IGF1R and its signaling pathway also blocked AKT activation, providing further evidence for the role of the IGF1R pathway in this setting. Conversely, an RNAi enhancer screen indicated that silencing components of the PI3K pathway, notably mTOR, resulted in enhanced sensitivity to the IGF1R inhibitor dalotuzumab. In addition, shRNAs targeting PI3KCA and mTOR (FRAP1) enhanced inhibition of colony growth by dalotuzumab. Taken together, our preclinical studies strongly supported the rationale for targeting both mTOR and IGF1R signaling pathways by combined ridaforolimus and dalotuzumab.
The results from our phase I study demonstrate that the combination of ridaforolimus and dalotuzumab is feasible, with encouraging activity in patients with breast cancer with ER+/high-proliferative tumors. The observed AEs were generally mild to moderate in severity and manageable, allowing patients to remain on treatment for long periods. However, as noted below, the grade of oral mucositis was higher than the one observed with single-agent ridaforolimus, suggesting that the inhibition of compensatory pathways may also be at play in tissues sensitive to mTOR inhibition. There was no evident pharmacologic interaction between study drugs, and drug levels were comparable with expected levels (based on single-agent trial reports). On the basis of our results, we determined that the RP2D of ridaforolimus to combine with dalotuzumab 10 mg/kg/week for further clinical development is 30 mg every day × 5. The RP2D for ridaforolimus is lower than the dose that has been evaluated for therapy as a single agent; this is due to the higher rates of stomatitis and mucositis observed with the combination, which is probably an indication that the combination also inhibits the feedback loop in normal tissues. Stomatitis/mucositis are well-known class effects of mTOR inhibitors and have been previously reported with similar incidence and grade with single-agent ridaforolimus (21) or other rapamycin analogues (22). In the present study, stomatitis/mucositis were manageable with local treatment and temporary dose interruption in most cases. Although concerns regarding metabolic toxicity have been raised with both mTOR (23) and IGF1R inhibitors (24), all the episodes of hyperglycemia and hypercholesterolemia reported with ridaforolimus and dalotuzumab occurred outside of the DLT period and did not interfere with dosing.
Although ridaforolimus has not been formally tested as a single agent in breast cancer, in the initial phase I trial of single-agent oral ridaforolimus, 1 of 8 treated patients with metastatic breast cancer reported SD for about 5 months (21). Single-agent therapy with other mTOR inhibitors has shown modest activity in patients with breast cancer (25, 26), while no signal of antitumor activity was observed with dalotuzumab in a recently reported phase I study that included 17 patients with breast cancer (18). In our phase I study, clinical evidence of antitumor activity was observed in several patients; the combination of ridaforolimus and dalotuzumab produced an overall ORR of 7% and a 4-month SD rate of 17%. Notably, the most prominent results were observed in patients with breast cancer (ORR of 13% and a SD of 48%); especially among patients with ER+/high-proliferation breast cancer who achieved an ORR of 27% (3 out of 11 patients), which was 20% higher than the overall population. With the limits of a phase I study, the clinical data with ridaforolimus and dalotuzumab support the antitumor activity seen in preclinical models, and provides evidence that the combination is safe and active in patients with advanced cancer, and appears to be particularly promising in patients with breast cancer.
Until recently, there has been little overall success in identifying biologic markers that may identify a population responsive to either mTOR or IGF1R inhibitors. In this study, most patients with breast cancer deriving benefit from the combination had ER+/high-proliferative tumors with 15% or more Ki67-positive staining. A Ki67 labeling index (LI) of greater than 13.25% is typically used to distinguish luminal B from luminal A tumors (27). Dysregulation of IGF1R or PI3K pathways has been associated with luminal B tumors; it has been recently reported that luminal B tumors have hyperactive PI3K signaling and are more likely to express IGF gene signature as compared with luminal A breast cancer (3, 4). The high-proliferative luminal B subtype is associated with poorer prognosis (6), and long-term outcomes following adjuvant tamoxifen with a 10-year disease-free survival (DFS) of approximately 50% (28). In addition, luminal B breast tumors are poorly responsive to primary chemotherapy, with pathologic complete response rates that are worse than those of other molecular subtypes (29).
We assessed potential biomarkers that would correlate with response to ridaforolimus and dalotuzumab. Breast cancer cell line sensitivity to ridaforolimus was highly correlated with activation of the RAS pathway, a finding that is consistent with resistance to other PI3K pathway inhibitors via activation of the parallel MAPK pathway. Activation of the RAS pathway has also been correlated with resistance to AKT inhibition (19). Conversely, sensitivity to anti-IGF1R therapy has been associated with expression levels of IGF1R and activation of the IGF1R pathway (20). The ER has been shown to regulate the expression of IGF1R (30). Moreover, primary or acquired resistance to endocrine therapy, a major cause of mortality in patients with ER+ breast cancer, is associated with reactivation of IGF1R signaling (31). These results suggest that the IGF1R pathway may play an important role in the progression of ER+breast cancer. Our analysis of the RAS signature in human breast cancer tumors showed high levels expressed in triple-negative breast cancer and low levels in the ER+ subtype. Gene expression profiling based on the TCGA dataset revealed overexpression of IGF1R, its ligand IGF1, and the adaptor protein IRS1 in ER+breast cancer. Although indirect, these findings suggest that patients with breast cancer with the luminal B subtype expressing high levels of the IGF1R pathway and low levels of RAS pathway activation would be responsive to ridaforolimus and dalotuzumab combination therapy (Supplementary Fig. S4).
Results from the Breast Cancer Trials of Oral Everolimus-2 (BOLERO-2) trial demonstrated significant increases in PFS and ORR with everolimus (10 mg every day) and the aromatase inhibitor exemestane (25 mg every day) compared with exemestane alone, strongly supporting the value of mTOR inhibition in patients with hormone receptor–positive breast cancer (32). Our findings reinforce the value of mTOR inhibition in the treatment of ER+ breast cancer and support the additional value of dalotuzumab to abrogate the feedback activation of AKT by mTOR inhibitors. On the basis of the results of this phase I study, several studies are exploring the combination of ridaforolimus and dalotuzumab in breast cancer and other cancers. A phase II trial to compare the combination of ridaforolimus and dalotuzumab with endocrine therapy in patients with advanced luminal B breast cancer has been recently completed (NCT01234857).
Supplementary Material
Translational Relevance.
There is a strong rationale for targeting the phosphatidylinositol 3-kinase (PI3K)–AKT–mTOR pathway in patients with advanced cancer, with potential to improve clinical outcomes in subsets of patients. Preclinical and early clinical results support the combination of ridaforolimus and dalotuzumab to abrogate feedback activation of AKT following mTOR inhibition. In particular, our findings reinforce the value of mTOR inhibition in the treatment of estrogen receptor–positive (ER+) breast cancer.
Acknowledgments
The authors thank Meritxell Soler, Gemma Sala, Javier Cortes, Rafael Simó, and Ryan Geschwindt for their assistance in data collection and patient management, and for dalotuzumab pharmacokinetic analyses. The authors also wish to thank all the patients, nurses, and site staff who participated in the study. Editorial assistance was provided by Jennifer Granit of Integrus Scientific, a division of Medicus International New York (New York, NY). This assistance was funded by Merck & Co., Inc., Whitehouse Station, NJ, USA. Editorial assistance was also provided by Amy O. Johnson-Levonas, Ph.D., and Kristen Lewis of Merck & Co., Inc., Whitehouse Station, NJ, USA.
Grant Support
This study was funded by Merck & Co., Inc., Whitehouse Station, NJ, USA.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Note: Portions of this work were presented at the 2010 Annual Meeting of the American Society of Clinical Oncology (ASCO), Chicago, IL.
Disclosure of Potential Conflicts of Interest
B.B. Haines, S. Jha, Y. Song, Y. Xu, and T. Zhang are employees of Merck & Co., Inc. S. Ebbinghaus, S. Sathyanarayanan, andA. Leighton-Swayze are employees of and have ownership interest in Merck & Co., Inc. J. Frazier was an employee of and has ownership interest in Merck and Co. R.A. Klinghoffer and C.G. Winter were employees of Merck & Co., Inc. S. Ebbinghaus, R.A. Klinghoffer, S. Sathyanarayanan, and C.G. Winter are listed on a patent application for ridaforolimus owned by Merck & Co., Inc. No other potential conflicts of interest were disclosed by the other authors.
Disclaimer
The authors are fully responsible for all content and editorial decisions and received no financial support or other compensation related to the development of the article.
Authors’ Contributions
Conception and design: S. Di Cosimo, S. Sathyanarayanan, M.N. Stein, D. Roda, C.G. Winter, R.A. Klinghoffer, A. Leighton-Swayze, Y. Song, S. Ebbinghaus, J. Baselga
Development of methodology: S. Di Cosimo, S. Sathyanarayanan, D. Roda, S. Jha, R.A. Klinghoffer, A. Leighton-Swayze, S. Ebbinghaus, J. Baselga
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): S. Di Cosimo, S. Sathyanarayanan, J.C. Bendell, A. Cervantes, M.N. Stein, I. Braña, D. Roda, B.B. Haines, S. Jha, Y. Xu, J. Frazier, R.A. Klinghoffer, J. Baselga
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): S. Di Cosimo, S. Sathyanarayanan, J.C. Bendell, M.N. Stein, D. Roda, T. Zhang, C.G. Winter, S. Jha, Y. Xu, Y. Song, S. Ebbinghaus, J. Baselga
Writing, review, and/or revision of the manuscript: S. Di Cosimo, S. Sathyanarayanan, J.C. Bendell, A. Cervantes, M.N. Stein, I. Braña, D. Roda, T. Zhang, C.G. Winter, R.A. Klinghoffer, A. Leighton-Swayze, Y. Song, S. Ebbinghaus, J. Baselga
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): S. Jha, A. Leighton-Swayze
Study supervision: S. Sathyanarayanan, S. Ebbinghaus, J. Baselga
References
- 1.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 2.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–7. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 3.Creighton CJ, Casa A, Lazard Z, Huang S, Tsimelzon A, Hilsenbeck SG, et al. Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol. 2008;26:4078–85. doi: 10.1200/JCO.2007.13.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Creighton CJ, Fu X, Hennessy BT, Casa AJ, Zhang Y, Gonzalez-Angulo AM, et al. Proteomic and transcriptomic profiling reveals a link between the PI3K pathway and lower estrogen-receptor (ER) levels and activity in ER+ breast cancer. Breast Cancer Res. 2010;12:R40. doi: 10.1186/bcr2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loi S, Haibe-Kains B, Desmedt C, Wirapati P, Lallemand F, Tutt AM, et al. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics. 2008;9:239. doi: 10.1186/1471-2164-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–91. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox EM, Miller TW, Balko JM, Kuba MG, Sánchez V, Smith RA, et al. A kinome-wide screen identifies the insulin/IGF-I receptor pathway as a mechanism of escape from hormone dependence in breast cancer. Cancer Res. 2011;71:6773–84. doi: 10.1158/0008-5472.CAN-11-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller TW, Balko JM, Arteaga CL. Phosphatidylinositol 3-kinase and antiestrogen resistance in breast cancer. J Clin Oncol. 2011;29:4452–61. doi: 10.1200/JCO.2010.34.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Meric-Bernstam F, Gonzalez-Angulo AM. Targeting the mTOR signaling network for cancer therapy. J Clin Oncol. 2009;27:2278–87. doi: 10.1200/JCO.2008.20.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Cosimo S. Controversies in breast cancer: the mammalian target of rapamycin as a target for breast cancer therapy. Breast Cancer Res. 2009;11(Suppl 3):S25. doi: 10.1186/bcr2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Cosimo S, Seoane J, Guzman M, Rojo F, Jimenez J, Anido J, et al. Combination of the mammalian target of rapamycin (mTOR) inhibitor everolimus (E) with the insulin like growth factor-1-receptor (IGF-1-R) inhibitor NVP-AEW-541: A mechanistic based anti-tumor strategy. J Clin Oncol. 2005;23(Suppl):3112. [Google Scholar]
- 13.O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–8. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabernero J, Rojo F, Calvo E, Burris H, Judson I, Hazell K, et al. Dose- and schedule-dependent inhibition of the mammalian target of rapamycin pathway with everolimus: a phase I tumor pharmacodynamic study in patients with advanced solid tumors. J Clin Oncol. 2008;26:1603–10. doi: 10.1200/JCO.2007.14.5482. [DOI] [PubMed] [Google Scholar]
- 15.Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serra V, Scaltriti M, Prudkin L, Eichhorn PJ, Ibrahim YH, Chandarlapaty S, et al. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene. 2011;30:2547–57. doi: 10.1038/onc.2010.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skolnik JM, Barrett JS, Jayaraman B, Patel D, Adamson PC. Shortening the timeline of pediatric phase I trials: the rolling six design. J Clin Oncol. 2008;26:190–5. doi: 10.1200/JCO.2007.12.7712. [DOI] [PubMed] [Google Scholar]
- 18.Atzori F, Tabernero J, Cervantes A, Prudkin L, Andreu J, Rodríguez-Braun E, et al. A phase I pharmacokinetic and pharmacodynamic study of dalotuzumab (MK-0646), an anti-insulin-like growth factor-1 receptor monoclonal antibody, in patients with advanced solid tumors. Clin Cancer Res. 2011;17:6304–12. doi: 10.1158/1078-0432.CCR-10-3336. [DOI] [PubMed] [Google Scholar]
- 19.Loboda A, Nebozhyn M, Klinghoffer R, Frazier J, Chastain M, Arthur W, et al. A gene expression signature of RAS pathway dependence predicts response to PI3K and RAS pathway inhibitors and expands the population of RAS pathway activated tumors. BMC Med Genomics. 2010;3:26. doi: 10.1186/1755-8794-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zha J, O’Brien C, Savage H, Huw LY, Zhong F, Berry L, et al. Molecular predictors of response to a humanized anti-insulin-like growth factor-I receptor monoclonal antibody in breast and colorectal cancer. Mol Cancer Ther. 2009;8:2110–21. doi: 10.1158/1535-7163.MCT-09-0381. [DOI] [PubMed] [Google Scholar]
- 21.Mita MM, Britten CD, Poplin E, et al. Deforolimus trial 106—a phase I trial evaluating 7 regimens of oral deforolimus (AP23573, MK-8669) J Clin Oncol. 2008;26(Suppl):3509. [Google Scholar]
- 22.Sankhala K, Mita A, Kelly K, Mahalingam D, Giles F, Mita M. The emerging safety profile of mTOR inhibitors, a novel class of anticancer agents. Target Oncol. 2009;4:135–42. doi: 10.1007/s11523-009-0107-z. [DOI] [PubMed] [Google Scholar]
- 23.Motzer RJ, Escudier B, Oudard S, Hutson TE, Porta C, Bracarda S, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008;372:449–56. doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- 24.Gualberto A, Pollak M. Emerging role of insulin-like growth factor receptor inhibitors in oncology: early clinical trial results and future directions. Oncogene. 2009;28:3009–21. doi: 10.1038/onc.2009.172. [DOI] [PubMed] [Google Scholar]
- 25.Chan S, Scheulen ME, Johnston S, Mross K, Cardoso F, Dittrich C, et al. Phase II study of temsirolimus (CCI-779), a novel inhibitor of mTOR, in heavily pretreated patients with locally advanced or metastatic breast cancer. J Clin Oncol. 2005;23:5314–22. doi: 10.1200/JCO.2005.66.130. [DOI] [PubMed] [Google Scholar]
- 26.Ellard SL, Clemons M, Gelmon KA, Norris B, Kennecke H, Chia S, et al. Randomized phase II study comparing two schedules of everolimus in patients with recurrent/metastatic breast cancer: NCIC Clinical Trials Group IND. 163. J Clin Oncol. 2009;27:4536–41. doi: 10.1200/JCO.2008.21.3033. [DOI] [PubMed] [Google Scholar]
- 27.Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–50. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Ronde JJ, Hannemann J, Halfwerk H, Mulder L, Straver ME, Vrancken Peeters MJ, et al. Concordance of clinical and molecular breast cancer subtyping in the context of preoperative chemotherapy response. Breast Cancer Res Treat. 2010;119:119–26. doi: 10.1007/s10549-009-0499-6. [DOI] [PubMed] [Google Scholar]
- 30.Massarweh S, Osborne CK, Creighton CJ, Qin L, Tsimelzon A, Huang S, et al. Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res. 2008;68:826–33. doi: 10.1158/0008-5472.CAN-07-2707. [DOI] [PubMed] [Google Scholar]
- 31.Casa AJ, Dearth RK, Litzenburger BC, Lee AV, Cui X. The type I insulin-like growth factor receptor pathway: a key player in cancer therapeutic resistance. Front Biosci. 2008;13:3273–87. doi: 10.2741/2925. [DOI] [PubMed] [Google Scholar]
- 32.Baselga J, Campone M, Piccart M, Burris HA, III, Rugo HS, Sahmoud T, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366:520–9. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.