Abstract
Autophagy activated after DNA damage or other stresses mitigates cellular damage by removing damaged proteins, lipids and organelles. Activation of the master metabolic kinase AMPK enhances autophagy. Here we report that cyclin D1 restrains autophagy by modulating the activation of AMPK. In cell models of human breast cancer or in a cyclinD1-deficient model, we observed a cyclin D1-mediated reduction in AMPK activation. Mechanistic investigations showed that Cyclin D1 inhibited mitochondrial function, promoted glycolysis and reduced activation of AMPK (pT172), possibly through a mechanism that involves cyclin D1-Cdk4/Cdk6 phosphorylation of LKB1. Our findings suggest how AMPK activation by cyclin D1 may couple cell proliferation to energy homeostasis.
Keywords: Cell cycle, breast cancer, autophagy, cyclin D1, AMPK, mitochondria
Introduction
Autophagy is an evolutionarily conserved catabolic process that degrades damaged proteins and organelles, recycling their biochemical components for energy production and other biosynthetic reactions. As a protective mechanism, autophagy allows the cell to survive in response to different stressors such as nutrient deprivation, DNA damage and low energy conditions (1). The core pathway governing autophagy is highly conserved (1). 5′ AMP-activated protein kinase (AMPK) is a sensor of cellular energy homeostasis and is activated by increased cellular adenosine mono- and diphosphate (AMP and ADP) (2). Glucose deprivation induces phosphorylation of AMPK at Threonine 172 (T172), which restrains growth and switches cellular metabolism toward both catabolic and oxidative processes. The ability of AMPK to respond to altered AMP levels is dependent upon liver kinase B1 (LKB1), which phosphorylates AMPK (3). LKB1 was originally identified as a mutated tumor suppressor in Peutz-Jeghers syndrome. The discovery that LKB1 was the major kinase required for AMPK activation in response to metabolic stress provided a direct link between AMPK and cancer (4). LKB1 is constitutively active in cells (5) and its phosphorylation in response to oncogenic B-RAF compromises the ability of LKB1 to bind and activate AMPK (6).
The mitigation of cellular damage by autophagic removal of damaged proteins and lipidated organelles is important for tumor suppression in some circumstances but promotes proliferation and maintains tumorigenesis in other circumstances. Thus, autophagy inhibits progression and delays tumor initiation, whereas it is required for tumor maintenance (7). The promotion of tumor growth by autophagy involves provision of energy substrates required for cellular proliferation. In contrast, there is a strong correlation between mitogenic signaling and the inhibition of autophagy (8). In a genome wide screen, a large percentage of genes that negatively regulate autophagy were also involved in cellular growth and proliferation (9). Despite strong evidence for an association between mitogenic signaling in the restraint of autophagy, the molecular mechanisms remain poorly defined. In this regard LKB1-AMPK activates p27KIP1, a cyclin-dependent kinase inhibitor that leads to cell cycle arrest (10). Furthermore, the induction of RAF-MEK-ERK negatively regulates AMPK, as ERK and Rsk phosphorylate LKB1, reducing AMPK activity (6). The phosphorylation of LKB1 at Serine 325 (S325) occurs within a motif that is predicted to serve as a substrate for proline-directed serine kinases, including the cell-cycle regulators belonging to the cyclin-dependent kinase family (11).
The CCND1 gene encodes cyclin D1, the regulatory subunit of the holoenzyme that phosphorylates and inactivates the retinoblastoma protein (Rb). CCND1 is overexpressed in a variety of malignancies and is increased in the early phase of breast cancer malignancy, including ductal carcinoma in situ (12). In recent studies of transgenic mice, cyclin D1 mediated promotion of breast tumorigenesis was kinase-independent, but autophagy restraint was kinase-dependent raising the question of the molecular mechanisms governing autophagy restraint (13,14). The current studies confirmed these prior findings and extended these observations to identify the molecular mechanisms. The phosphorylation of LKB1 and consequent reduction in AMPK by cyclin D1 may serve to couple cellular proliferation to energy homeostasis.
Materials and Methods
Cell culture, vectors and reagents
Cell lines were authenticated by examination of morphology, growth profile and are mycoplasma free. MCF10A cells transformed with the oncogene NeuT (MCF10A-NeuT), Ha-Ras (MCF10-Ha-Ras) or v-Src (MCF10A-Src) (MCF10A lines generated 2009, Thomas Jefferson University), Human Embryonic Kidney (HEK293T), cyclin D1 wild type (cyclin D1+/+) and cyclin D1 knockout (cyclin D1−/−) mouse embryonic fibroblasts (MEF) and immortalized mouse embryonic fibroblasts (3T3) were previously described (15) (MEFs and 3T3 cells generated 2011 at Thomas Jefferson University).
The expression vectors encoding LKB1 with an N-terminal FLAG epitope (FLAG-LKB1) and the point mutant S325A (FLAG-LKB1-S325A) were previously described (6). The pSM2 vector containing shRNA for cyclin D1 and control vector were used as described (16). The pCDNA3.1-3X-FLAG expression vector encoding human cyclin D1 wild type (cyclin D1WT), human cyclin D1 kinase dead mutant (cyclin D1KE) and control vector were used as described (17).
MCF10A-NeuT cells were incubated with the Cdk4/6 inhibitor PD0332991 (Cat. No. PZ0199, Sigma-Aldrich, St. Louis, MO, USA) at a working concentration of 500 nM for 24 hours. Autophagy was induced by treating the cells either with 10 μM of Rapamycin (Cat. No. R0395, Sigma-Aldrich, St. Louis, MO, USA) or Hanks Buffered Salt Solution (HBSS) (Cat. No. 21-020, Corning, Corning, NY, USA) for 2 hours. In order to activate the AMPK signaling pathway, MEF were treated using metabolic stressors including ultraviolet (UV) or hydrogen peroxide (H2O2, 100 μM).
Transgenic mice
Conditional cyclin D1fl/fl mice were a generous gift from Dr. Sicinski. C57BL/6J ROSA26-Cre-ERT2 mice were a gift from Dr. Streamson C. Chua Jr. The appropriate Thomas Jefferson University institutional committee approved protocols were followed when working with these mice. ROSA26-Cre-ERT2-cyclin D1fl/fl mice were generated by crossing cyclin D1fl/fl and ROSA26-Cre-ERT2 mice. Ccnd1 knock-out mice were generated by a daily intra-peritoneal injection of Tamoxifen (1 mg/200 μl) for 5 days, followed by a wash out period of four weeks. Mammary gland tissues were then collected. Wild type Cre-ERT2 mice were used as control.
Immunofluorescence
Autophagy activation was measured by immunodetection of microtubule-associated protein light-chain 3 (LC3) puncta using LC3-A/B antibody (Cat. No. 4108, Cell Signaling, Beverly, MA, USA). Alpha tubulin (Cat. No. T6199, Sigma-Aldrich, St. Louis, USA) was used as a reference marker for cellular area. The amount of LC3 puncta was measured with ImageJ and normalized by cell area. The autofluorescent agent monodansylcadaverine (Sigma) (18) was used to measure autophagy activation in 96 wells plates. Cells were plated at a density of 2×104 cells/cm2 one day before treatment and were incubated overnight at 37°C. After treatment, as described above, cells were incubated for 10 minutes with 0.05 mmol/L monodansylcadaverine at 37°C and were then observed with a fluorescence plate reader using a UV filter. Autophagic flux was assessed using LC3-GFP in the presence of chloroquine.
In order to assess autophagy flux, cyclin D1+/+ and cyclin D1−/− 3T3 cells (1 × 106) were transfected by electroporation (Nucleofector 2b Device, Program NIH 3T3 U-030; Lonza, Allendale, NJ) with the mCherry-EGFP-LC3 dual reporter (Addgene, #22418). Analysis of the labeled LC3 puncta was conducted by confocal microscopy (Nikon Cl PLUS) and quantitated using ImageJ (19).
Western blot and Immunoprecipitation Western blot
Whole cell lysates (50 μg) were separated by SDS-PAGE and the proteins transferred to a nitrocellulose membrane for Western blotting as previously described (20). Specific primary antibodies were prepared according to the manufacturer’s instructions; Cyclin D1 (Cat. No. sc-753, clone H295, Santa Cruz, Santa Cruz, CA, USA), phospho Rb S780 (Cat. No. 9307S, Cell Signaling, Beverly, MA, USA), LC3-A/B (Cat. No. 4108, Cell Signaling, Danvers, MA, USA), AMPK (Cat. No. 2532S, Cell Signaling, Beverly, MA, USA), phospho AMPK T172 (Cat. No. 2531S, Cell Signaling, Beverly, MA, USA), Bnip3 (Cat. No. 3485-1, Epitomics, Burlingame, CA, USA) and Vinculin (Cat. No. V9131, Sigma-Aldrich, St. Louis, USA), which was used as an internal protein loading control. The LKB1 phospho-serine 325 (pS325) antibody was previously described (6). Anti rabbit (Cat. No. sc-2004, Santa Cruz, Santa Cruz, CA, USA) or mouse (Cat. No. sc-2005, Santa Cruz, Santa Cruz, CA, USA) secondary HRP-conjugated antibodies were diluted at 1:3000.
Immunoprecipitation Kinase assays
HEK293T cells were co-transfected with expression vectors encoding FLAG-LKB1 and either cyclin D1WT or cyclin D1KE. The lysates were prepared using cellular lysis buffer (HEPES pH=7.0, 150mM NaCl 1% NP-40, 1mM Na3VO4 and protease inhibitor cocktail). Protein concentrations were normalized using the Bradford assay. For immuno-precipitation, cell lysates were incubated with primary antibodies of anti-FLAG M2 (Cat. No. F3165, Sigma-Aldrich, St. Louis, USA) affinity gel overnight at 4°C, followed by incubation with protein A/G sepharose for 1 hour at 4°C. Immunoprecipitated samples were incubated in kinase assay buffer in the presence of γ-32P dATP for 20 minutes at 37°C. After labeling, samples were run on a 10% SDS-PAGE gel. The gel was dried and exposed to an autoradiography film (21).
Statistical analysis
The statistical analysis was performed using Prisma 4 software (GraphPad Software, San Diego, CA). Differences were considered statistically significant with P-values < 0.05.
Measurements of oxygen consumption rate and extracellular acidification rate
A Seahorse Bioscience XF24-3 Extracellular Flux Analyzer was used to measure the rate change of dissolved O2 and pH of medium immediately surrounding adherent cells cultured in a XF24 V7 cell culture microplate (Seahorse Bioscience). Measurements of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were performed after equilibration in assay medium for 1 h. Measurements of O2 concentration and pH were made periodically over 4 min, and OCR and ECAR were obtained from the slopes of concentration change in these parameters vs. time.
Lactate assay, ATP assay and NAD+/NADH Ratio
Lactate concentration in cell culture media was assayed using BioAssay Systems’ EnzyChromTM lactate assay kit. Total cellular ATP was assayed based on the manufacturers’ guidelines (Molecular Probes, Invitrogen). The NAD+/NADH ratio was quantitated per manufacturers guidelines (BioVision) through a reaction that specifically detects NADH and NAD+, but not NADP nor NADPH.
Oxygen Consumption Assay
Oxygen consumption rates in cells were conducted in an Oxygen Biosensor System (BD). Oxygen consumption rates were measured over 3 hours every 15 minutes using a fluorescent plate reader.
Quantitation of Mitochondrial Morphology
The morphological analysis of mitochondria in live MEF was performed as described (22), by using confocal Z-stack maximal projections of TMRE loaded mitochondria and running the analysis using ImageJ.
FACS analysis
For FACS analysis of mitochondria, MEF were loaded with Vybrant DyeCycle Violet according to manufacturer’s protocol (LifeScience) and 10 μM JC-1 dye (Molecular Probes) for 30 min at 37°C. Green and red fluorescence components from JC-1 staining were measured and segregated based on the cell cycle phase.
Mitochondrial FRAP Assay
Live MEF were stained with tetramethylrhodamine methyl ester (TMRM) (50 nM) in DMEM for 30 min at 37°C and mitochondrial dynamics were analyzed by FRAP as previously published (23) using a Zeiss inverted confocal microscope equipped with a 63x Oil objective. 50μg/mL ethacrinic acid and 5μM FCCP were used as positive and negative controls for the analysis. Recovery of fluorescence was monitored for 120 seconds. 3 different clones were used for each genotype, and for each of them 3 to 5 cells were analyzed. High resolution images of TMRM loaded mitochondria were used for quantification of mitochondrial morphology using ImageJ.
Results
Endogenous cyclin D1 restrains oncogene-induced autophagy
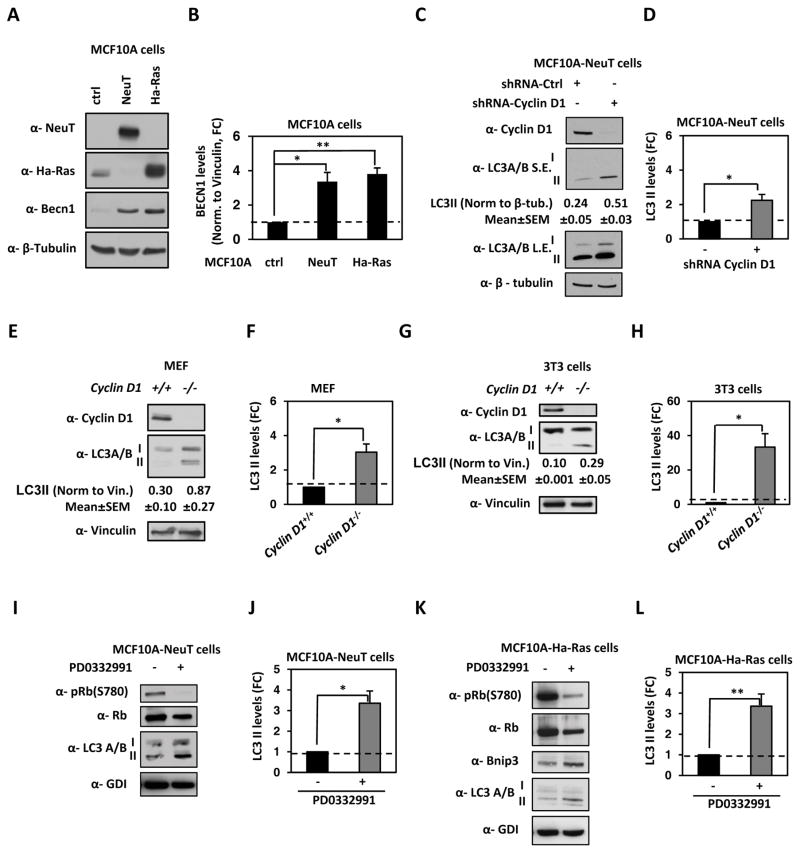
Cyclin D1 abundance is induced during oncogenic transduction of immortalized human breast MCF10A epithelial cells and is a direct transcriptional target of NeuT, v-Src and Ha-Ras (24–26). To determine whether cyclin D1 regulates autophagy induced by distinct oncogenes, isogenic human breast cancer cell lines were assessed. We first determined the role of endogenous cyclin D1 in regulating basal autophagy by using short hairpin RNA (shRNA) against cyclin D1. We verified that oncogene transformation of MCF10A-ErbB2 (NeuT) and MCF10A-Ha-Ras induced autophagy compared with control (ctrl) parental MCF10A cells (Figure 1A). Both NeuT (P=0.012) and Ha-Ras (P=0.002) induced autophagy based on Becn1 abundance (Figure 1B). In MCF10A-NeuT cells, cyclin D1 shRNA reduced the abundance of cyclin D1 by >80% (Figure 1C). We determined the basal autophagy status by detecting both isoforms of microtubule-associated protein 1 light-chain 3 (LC3) I and II, as the lipidated form (LC3-II), stably associates with the autophagosomes, serving as a marker of autophagy induction (Figure 1C and 1D) (P=0.021) (27). Next, we deployed MEF and 3T3 cells derived from cyclin D1−/− and cyclin D1+/+ mice. LC3-II was increased in cyclin D1−/− MEF (P=0.013) and 3T3 cells (P=0.014) compared with cyclin D1+/+ cells (Figure 1E–H). We verified that the mechanism of cyclin D1 inhibition of autophagy involved cyclin-dependent kinase activity by treating cells with a cdk4/6 inhibitor (PD0332991-Pablociclib). PD0332991 treatment of MCF10A-NeuT (Figure 1I and 1J) and MCF10A-Ha-Ras (Figure 1K and 1L) significantly reduced the phosphorylation of Rb at serine 780 (pRb S780) (cdk4/6 phosphorylation site) with a commensurate increase in LC3-II form (P=0.016 and P=0.006, respectively) and Bnip3.
Figure 1. Endogenous cyclin D1 restrains stress-induced autophagy in oncogene transformed human breast cancer cells.
(A) MCF10A cells stably transduced with oncogenes ErbB2 (NeuT) and Ha-Ras were immunoblotted for NeuT, Ha-Ras, Becn1 and β-Tubulin. (B) Quantification of Becn1 in MCF10A-Ha-Ras cells and MCF10A-NeuT vs control vector showed increased Becn1 abundance in the isogenic MCF10A cell lines. (C) MCF10A-NeuT cells stably transduced with cyclin D1 shRNA or shScramble as control vector and immunoblotted for cyclin D1, LC3A/B and β-Tubulin. (D) Quantification of the lipidated form of LC3 in MCF10A/NeuT cells treated with shCCND1 or control vector. (E) Western blot assay of cyclin D1+/+ and cyclin D1−/− mouse embryonic fibroblasts (MEF) for the lipidated form of LC3. (F) Quantitation of the lipidated form of LC3 in MEF. (G) Western blot assay of immortalized cyclin D1+/+ and cyclin D1−/− mouse embryonic fibroblasts (3T3) for LC3A/B. (H) Quantitation of the lipidated form of LC3 in 3T3 cells. (I) MCF10A-NeuT and (K) MCF10A-Ha-Ras cells cultured in presence/absence of the cdk4/6 inhibitor PD0332991(0.5μM), immunoblotted for phosphorylated pRB (S780), total pRB, LC3A/B and GDI. Quantitation of the lipidated form of LC3 in (J) MCF10A-NeuT and (L) MCF10A-Ha-Ras cells. (* P<0.05, ** P<0.01).
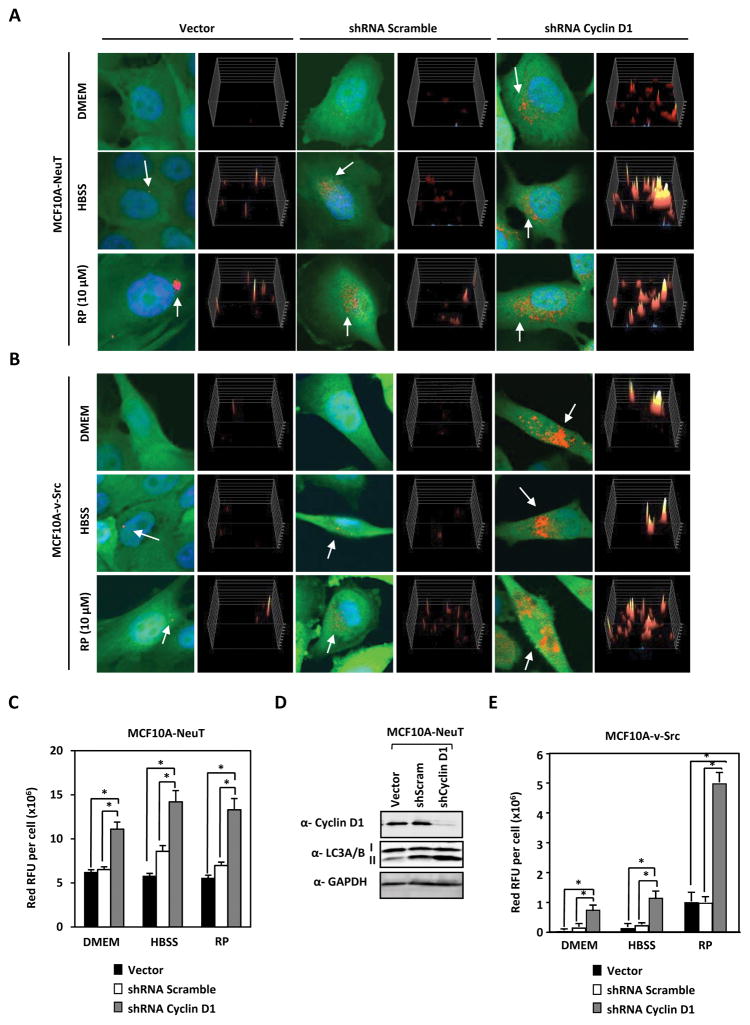
MCF10A-NeuT and MCF10A-v-Src cells were treated with distinct autophagy inducers (Figure 2A and 2B). Cyclin D1 shRNA enhanced basal autophagy in MCF10A-NeuT cells approximately 2-fold and in MCF10A-v-Src cells approximately 5-fold (P<0.001) (Figure 2C, 2D and 2E). Cyclin D1 shRNA enhanced autophagy in HBSS treated MCF10A-NeuT cells approximately 1.7-fold and in MCF10A-v-Src cells approximately 3.5-fold (P<0.001) (Figure 2C, 2D and 2E) and cyclin D1 shRNA enhanced autophagy in Rapamycin (RP) treated MCF10A-NeuT cells approximately 1.8-fold and in MCF10A-v-Src cells approximately 5-fold (<0.001) (Figure 2C and 2E). Collectively, these data demonstrate that endogenous cyclin D1 restrains autophagy in breast cancer cells transformed by ErbB2, Ha-Ras or oncogenic v-Src.
Figure 2. Endogenous cyclin D1 restrains stress-induced autophagy in oncogene transformed human breast cancer cells.
(A) MCF10A-NeuT and (B) MCF10A-v-Src cells expressing RFP-LC3 were treated with shRNA against cyclin D1 or control shRNA in the presence of either DMEM, HBSS or Rapamycin (RP) (10 μM) for 2 hours. Immunofluorescence detection of RFP-LC3 shows increased LC3 redistribution to autophagosomes (yellow/red perinuclear dots) in cells transduced with shCCND1. (C) Quantitation of LC3 puncta/cell area in MCF10A-NeuT and (D) Western blotting on the same cells demonstrates increased LC3-II in MCF10A-NeuT. (E) Quantification of LC3 puncta/cell area in MCF10A-c-Src (expressed in square pixels, n=50). Cyclin D1 reduces autophagosome formation (* P<0.05 and ** P<0.01).
Cyclin D1 restrains stress induced AMPK activation
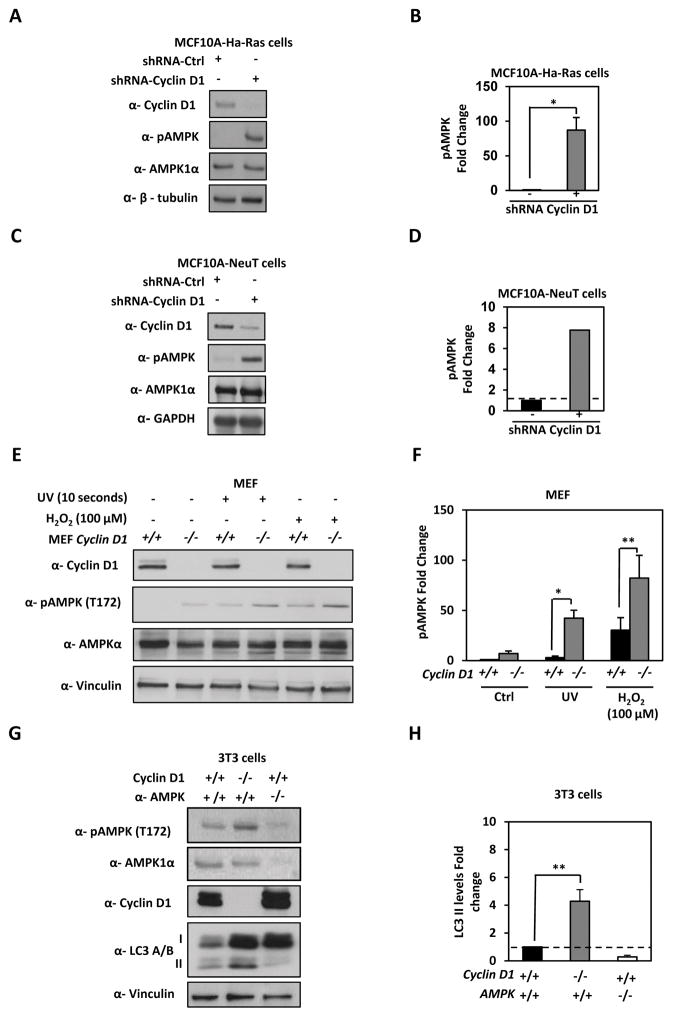
AMPK is a central mediator of cellular adaption to metabolic flux and promotes autophagy. AMPK is activated upon depletion of nutrients by binding AMP and by LKB1 inducing phosphorylation at threonine 172 (T172). To determine if cyclin D1-mediated restraint of autophagy involves the inhibition of AMPK, we examined AMPK signaling in tissue culture and in vivo upon cyclin D1 depletion. We deployed MCF10A-NeuT and MCF10A-Ha-Ras cells treated with cyclin D1 shRNA. Cyclin D1 depletion correlated with a significant increase in the phosphorylation level of AMPK at T172 in both MCF10A-Ha-Ras (P<0.001) (Figure 3A and 3B) and MCF10A-NeuT (P<0.001) (Figure 3C and 3D).
Figure 3. Endogenous cyclin D1 restrains stress induced AMPK.
(A) MCF10A-Ha-Ras and (C) MCF10A-NeuT expressing cyclin D1 shRNA were analyzed by Western blotting for LC3A/B-II, AMPK and pAMPK. Quantitation of pAMPK levels in (B) MCF10A-Ha-Ras and (D) MCF10A-NeuT, fold enrichment of pAMPK is shown. (E) cyclin D1+/+ and cyclin D1−/− MEF were treated with cellular stressors (UV and H2O2) and cell lysates analyzed by Western blot as shown. (F) Fold enrichment of pAMPK were quantified and shown. (G) cyclin D1+/+ and cyclin D1−/− 3T3 cells compared to Ampk1α2α−/− 3T3 by Western blot to analyze phosphorylated Ampk (T172), lipidated LC3-II levels, cyclin D1 and normalized to vinculin to calculate LC3-II levels (H)(* P<0.05; ** P<0.01 and *** P<0.001).
To determine whether cyclin D1 restrains AMPK activation by cellular stressors, cyclin D1−/− and cyclin D1+/+ MEF were treated with cellular stressors such as H2O2 and UV. Cellular stress activated pAMPK (T172), which was further enhanced by cyclin D1 knockdown in both treatments, UV and H2O2 (P=0.023 and P=0.004, respectively) (Figure 3E and 3F). Total AMPK kinase was unchanged by cyclin D1 in MCF10A-NeuT, MCF10A-Ha-Ras and MEF. Total AMPK remained unchanged upon bioenergetic stress (Figure 3). In addition, we determined that AMPK phosphorylation at T172 was inhibited by cyclin D1 in 3T3 cells (Fig. 3G and 3H) commensurate with a reduction in lipidated LC3 with AMPK1α2α−/− 3T3 cells used as a control.
AMPK is activated following cell stress and is required under stress to promote cell viability (28). We therefore tested the role of cyclin D1 in glucose stress. Cyclin D1 sensitizes MEF to glucose stress, growth rates of cyclin D1+/+ MEF vs. cyclin D1−/− MEF was compromised at glucose concentrations below 4.5g/L shown in Supplemental Fig. 1A (P=1E-05 and P=0.05 respectively) and 1B (cyclin D1−/− vs cyclin D1+/+ at 2g/L of glucose P=3.5E-06, at 1g/L of glucose P=4.2E-08 and minus glucose P=8E-05).
Endogenous cyclin D1 restrains autophagy in the mammary gland
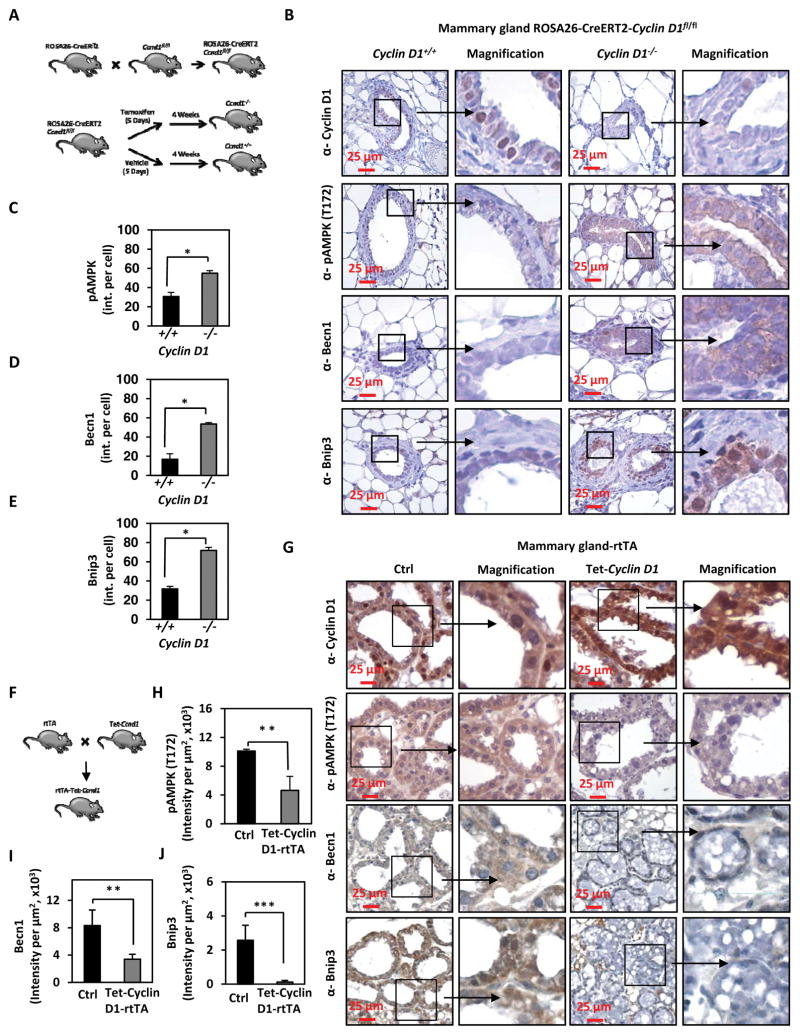
To determine whether cyclin D1 restrains autophagy in vivo, we examined transgenic mice that delete the Ccnd1 gene in the adult mouse (ROSA26-Cre-ERT2-Ccnd1fl/fl mice) (Figure 4A). Animals were treated with Tamoxifen to induce Cre expression and analysis was conducted 4 weeks later to ensure washout of Tamoxifen. Mammary gland tissues of these mice were assessed using immunohistochemistry for autophagy (Becn1 and Bnip3) and AMPK activity (pAMPK T172) (Figure 4B). Cyclin D1 was detected in the mammary epithelium and fibroblasts of the vehicle treated bi-transgenic mice but was not detectable in animals that had been treated with Tamoxifen (Figure 4B). pAMPK at T172 was increased in the cyclin D1−/− mammary epithelial cells (P< 0.05) (Figure 4C). The autophagy markers, Becn1 and Bnip3 were increased in the cyclin D1−/− mammary gland tissue (P<0.05 and P<0.001, respectively) (Figure 4D and 4E). Immunohistochemistry was conducted on murine mammary gland-targeted cyclin D1 transgenic mice (rtTA/CCND1WT) (Figure 4F and 4G). Transient expression of cyclin D1 (17 days), through doxycycline induction of a tet-regulated cyclin D1 cDNA cassette directed to the murine mammary gland, induced cyclin D1 abundance, decreased pAMPK T172 (a correlate of AMPK activity), and reduced autophagy as attested by IHC showing reduced Becn1 and Bnip3 (P<0.001 and P<0.01, respectively) (Figure 4H–I).
Figure 4. Endogenous cyclin D1 restrains autophagy in vivo.
(A) Schematic representation of the ROSA26-Cre-ERT2-cyclin D1fl/fl generated mice. B) Immunohistochemistry for cyclin D1, pAMPK (T172), BECN1 and Bnip3 in cyclin D1+/+ vs cyclin D1−/− mouse mammary gland tissue. (C–E) Quantitation of immunoreactivity for pAMPK (T172), BECN1 and Bnip3 in cyclin D11−/− mouse mammary gland tissue, shown as mean SEM. (F) Schematic representation of the rtTA Tet-cyclin D1 mice. (G) Immunohistochemistry for cyclin D1, BECN1, Bnip3 and pAMPK (T172) in tetracycline inducible cyclin D1 mammary gland tissue. (H, I and J) Quantitation of immunoreactivity for Bnip3, Becn1 and pAMPK (T172) in tetracycline induced rtTA-cyclin D1 mouse mammary gland compared to control mammary gland are shown as mean SEM (* P<0.05 ** P<0.01 and *** P<0.001).
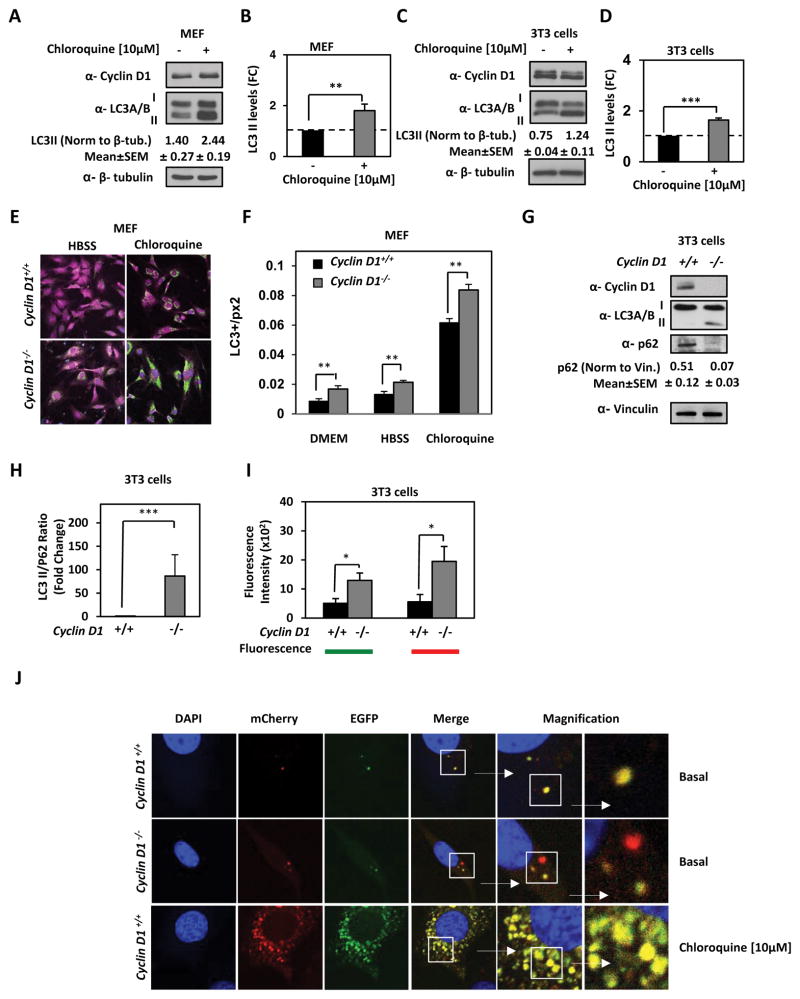
Cyclin D1 restrains chloroquine induced autophagic cargo flux
We next conducted analysis of autophagy cargo flux using endogenous LC3. Prior to engaging in the assessment of cyclin D1 restraint of autophagic cargo flux in the model cell lines we examined the autophagic response via treatment with chloroquine. Chloroquine inhibition of autophagy measured by lipidated LC3 in MCF10A-NeuT (Supplemental Fig. 2A and 2B), cyclin D1+/+ MEF and cyclin D1+/+ 3T3 cells was induced by 5-fold (P=<0.001), 1.7-fold (P=0.007) and 1.7-fold (P<0.001) respectively (Fig. 5A–D). We had previously confirmed that retroviral transduction of fluorescent protein-tagged LC3 relocates to the autophagosome membrane upon induction of autophagy. The induction of autophagy by culturing cells in nutrition free balanced salt solution (Hank’s Balanced Salt Solution) (HBSS) (Fig. 5E) was quantitated using confocal images and the corresponding general intensity surface spots were quantitated as mean data ± SEM (Fig. 5F). A comparison was made between cyclin D1−/− and cyclin D1+/+ MEF. The redistribution of the LC3-RFP to autophagosomes to form puncta was quantitated. In response to HBSS, the relative abundance of LC3 in autophagosomes characterized by puncta was increased, with a further increase in cyclin D1−/− compared to cyclin D1+/+ MEF (P=0.004). An increase in the relative amount of LC3-II can be due to either an increase in autophagy or a decrease in the degradation of autophagic intermediates. Autophagic activity may therefore reflect changes in autophagic flux. Flux can also be measured by blocking lysosomal degradation of autophagosomes using chloroquine, resulting in a buildup of autophagic intermediates. After chloroquine treatment GFP-LC3 puncta increased 7-fold, which was increased 10-fold in cyclin D1−/− cells (Figure 5F) (P=3.7E-06). In addition, we assayed autophagic flux by analyzing the ratio of LC3-II to p62. Since p62 levels increase when autophagy is inhibited, and decreased when autophagy is induced, p62 can be used as a marker to study autophagic flux. Autophagic flux in 3T3 cells derived from cyclin D1−/− (p62/LC3II ratio=15.7) compared cyclin D1+/+ mice (p62/LC3II ratio=0.41) (P=0.0014) showed a significantly greater LC3-II/p62 ratio (Fig. 5G and 5H). These, findings are consistent with cyclin D1 restraining autophagic cargo flux.
Figure 5. Cyclin D1 inhibits autophagic cargo flux by reducing AMPK phosphorylation.
Chloroquine induction of autophagy in (A) MEF and (C) 3T3 fibroblast cells. Protein lysates immunoblotted for cyclin D1, LC3A/B and β-tubulin. Relative abundance of LC3-II was quantitated for (B) MEF and (D) 3T3 fibroblast cells. (E) cyclin D1+/+ and cyclin D1−/− MEF were immunostained for LC3 (green) and actin (purple). (F) Quantitation of LC3 levels. The amounts of LC3 puncta were normalized by the total cell area expressed in square pixels. LC3 levels are increased in cyclin D1−/− MEF (n=12) vs cyclin D1+/+ MEF (n=6) cultured with regular media, in cyclin D1−/− MEF (n=6) vs cyclin D1+/+ MEF (n=7) in HBSS and in cyclin D1−/− MEF (n=23) vs cyclin D1+/+ MEF (n=24) treated with chloroquine (**P<0.01 for number of stated cells, and average LC3 puncta depicted ±SEM). (G) Autophagic flux assayed by Western blot for p62 and LC3A/B lapidated form. Abundance of autophagic flux markers were normalized to vinculin and graphically represented as (H) LC3II/p62 ratio. (**; P<0.01 and ***; P<0.001).
In order to define further the role of cyclin D1 in autophagic cargo flux, we deployed a Autophagy Tandem Sensor to visualize maturation of the autophagasome to the lysosome. By combining an acid-sensitive GFP with an acid-insensitive RFP, the change from autophagosome to autolysosome can be visualized by imaging the specific loss of the GFP fluorescence, leaving only red fluorescence allowing for dynamic visualization of the autophagic process. The autophagy flux in both 3T3 cyclin D1+/+ and cyclin D1−/− cells was assessed using a dual report mCherry-EGFP-LC3. At basal condition the knocking down of cyclin D1 in 3T3 cells shows an increase of both signals, EGFP (P<0.037) and mCherry (P<0.025) compared to 3T3 cyclin D1+/+ (Figure 5I). Moreover, in 3T3 cells cyclin D1−/− there is an increase of autophagosomes labeled exclusively with mCherry signal (Figure 5J). These data suggest that autophagy flux is restrained by cyclin D1.
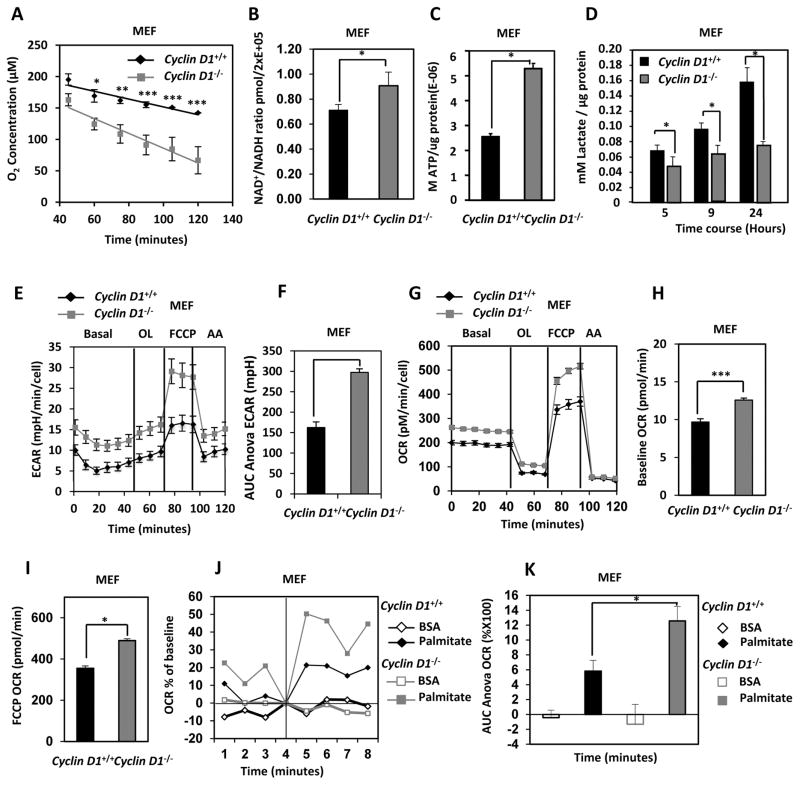
Deletion of cyclin D1 leads to a gain-of-function function phenotype resulting in increased mitochondrial respiration
AMPK is a central cellular mediator of the nutrient stress response. Under conditions of nutrient flux, AMPK inhibits anabolic processes including lipogenesis, protein metabolism and cell growth and activates catabolic processes that increase cellular ATP (β-fatty acid oxidation, glycolysis, β-fatty acid oxidation, mitochondrial biogenesis and autophagy) (29). A metabolic analysis was conducted to determine whether cyclin D1 inhibition of AMPK recapitulated the metabolic profile of AMPK inhibition. Oxygen consumption rates (OCR) were determined using an oxygen biosensor system (Fig. 6A). Endogenous cyclin D1 blunted oxygen consumption rates (cyclin D1+/+ vs. cyclin D1−/− MEF, P<0.001). Reflecting increased mitochondrial oxidation rates the NAD/NADH ratio was increased in cyclin D1−/− MEF (Fig. 6B) (P=0.04). Cellular ATP levels (Fig. 6C) (P=3.7E-14) and increased lactate secretion was observed in cyclin D1−/− MEF (Fig. 6D) (P=0.039).
Figure 6. Cyclin D1 reduces basal and palmitate dependent cellular oxygen consumption.
(A) Measurement of O2 consumption rates (OCR) in cyclin D1+/+ vs. cyclin D1−/− MEF. Average oxygen consumption rates for cyclin D1+/+ vs. cyclin D1−/− MEF (P<0.001 for n=10). (B) NAD+/NADH ratio in cyclin D1+/+ vs. cyclin D1−/− MEF (*P<0.05 data are mean of n=2 cyclin D1+/+ and n=3 cyclin D1−/− MEF lines ±SEM) (C) Concentration of ATP in cyclin D1+/+ vs. cyclin D1−/− MEF normalized to total cellular protein concentration (*P<0.001 data are mean of n=3 MEF lines ±SEM) (D) Time course of concentration of lactate in the media from cyclin D1+/+ vs. cyclin D1−/− MEF normalized to total cellular protein concentration (*P<0.05 mean of n=3 samples ±STDV) (E & F). Glucose flux ECAR and (G & H) Seahorse FX bioanalyzer of OCR profiles for cyclin D1+/+ vs. cyclin D1−/− MEF, under a BOFA protocol and (F) ECAR measurement as AUR during FCCP treatment and (H) baseline OCR rates and (I) OCR under FFCP (3μM) stress. (J) Seahorse FX bioanalyzer of OCR profiles for cyclin D1+/+ vs. cyclin D1−/− MEF under a palmitate utilization protocol and (K) quantitation of palmitate utilization for cyclin D1+/+ vs. cyclin D1−/− MEF (BSA control) (* P<0.001, ±SEM, calculations done using AUC ANOVA).
Next we deployed the Seahorse Extracellular Flux Analyzer (XF) that measures oxygen consumption (OCR) and glycolytic flux (ECAR) in cyclin D1+/+ MEF and cyclin D1−/− MEF. Multiple independent MEF preparations were analyzed using a standard bioenergetics protocol (Basal, Oligomycin, FCCP and Antimycin A; BOFA). Glycolytic flux measured through external acidification rates (ECAR) were decreased by cyclin D1 (Fig. 5E and 5F). Oxygen consumption rates (OCR) were significantly increased in cyclin D1−/− MEF under basal conditions indicating a gain-of-function phenotype in cyclin D1−/− MEF ((Fig. 6G and 6H (P=9.1E-15)).
Following oligomycin addition, the maximal respiratory rate was determined following uncoupling of mitochondrial metabolism by FCCP. Cyclin D1−/− MEF showed increased mitochondrial oxygen consumption compared to cyclin D1+/+ MEF cells and significantly increased OCR following mitochondrial uncoupling with FCCP (Fig. 6I) (P=3.67E-06), indicative of greater mitochondrial reserve in the cyclin D1−/− MEF. The OCR due to non-mitochondrial sources, determined as OCR in the presence of antinomycin A, was unchanged.
We next determined the ability of cyclin D1 to oxidize exogenously added fatty acids (FAs). Substrate utilization of cyclin D1−/− vs cyclin D1+/+ MEF was determined with palmitate-BSA in a FA oxidation protocol on the Seahorse Extracellular Flux (XF) Analyzer. OCR rates were 2-fold higher in the cyclin D1−/− MEF compared to cyclin D1+/+ MEF (Figure 6J and 6K) (P=9.3E-05).
Cyclin D1 inactivation reduces mitochondrial interconnectivity and mitochondrial membrane potential
Mitochondrial interconnectivity can be measured by Fluorescent Recovery After Photobleaching (FRAP). To determine if the mitochondrial network of cyclin D1−/− MEF demonstrated increased connectivity and therefore a more efficient capacity for substrate oxidation, we conducted FRAP on cyclin D1−/− vs. cyclin D1+/+ MEF cells alongside AMPK1α−/− fibroblasts as a control (Supplemental Fig. 3). We treated cells with FCCP, a protonophore (H+ ionophore) and uncoupler of oxidative phosphorylation in mitochondria (fragmented mitochondrion; negative control), or ethacrynic acid, a potent inducer of mitochondrial fusion (interconnected mitochondrion; positive control) as controls, and compared cyclin D1−/− vs. cyclin D1+/+ MEF (Supplemental Fig. 3A and 3B). Cells were loaded with the fluorescent tracer, TMRM, then a mask was used to photobleach a small region of the mitochondrion. The fluorescent recovery rate was timed (Supplemental Fig. 3A and 3B). In cyclin D1−/− MEF the fluorescent recovery was significantly faster indicating increased mitochondrial connectivity. The AMPK1α2α−/− fibroblasts demonstrated increased recovery compared to cyclin D1−/− MEF, implying that in AMPK1α2α−/− fibroblasts the mitochondria have increased connectivity (Supplemental Fig. 3C)
To determine whether cyclin D1 expression reduces mitochondrial area and number, quantification was conducted on MEF. Endogenous cyclin D1 inhibited both mitochondrial area and number (Supplemental Fig. 4A and 4B) (P=0.015 and P=0.003 respectively). Reduced AMPK levels correlates with a reduction in mitochondrial membrane potential and mass (30). Mitochondrial membrane potential (red component of JC1) and mitochondrial mass (green component of JC1) were inhibited by cyclin D1 at each phase of the cell cycle (Supplemental Fig. 4C) (mitochondrial mass at G0-G1 P=8.4E-04, S P=3.4E-05 and G2-M P=0.003) and (Supplemental Fig. 4D) (mitochondrial membrane potential at G0-G1 P=0.001, S P=0.0003 and G2-M P=0.0039).
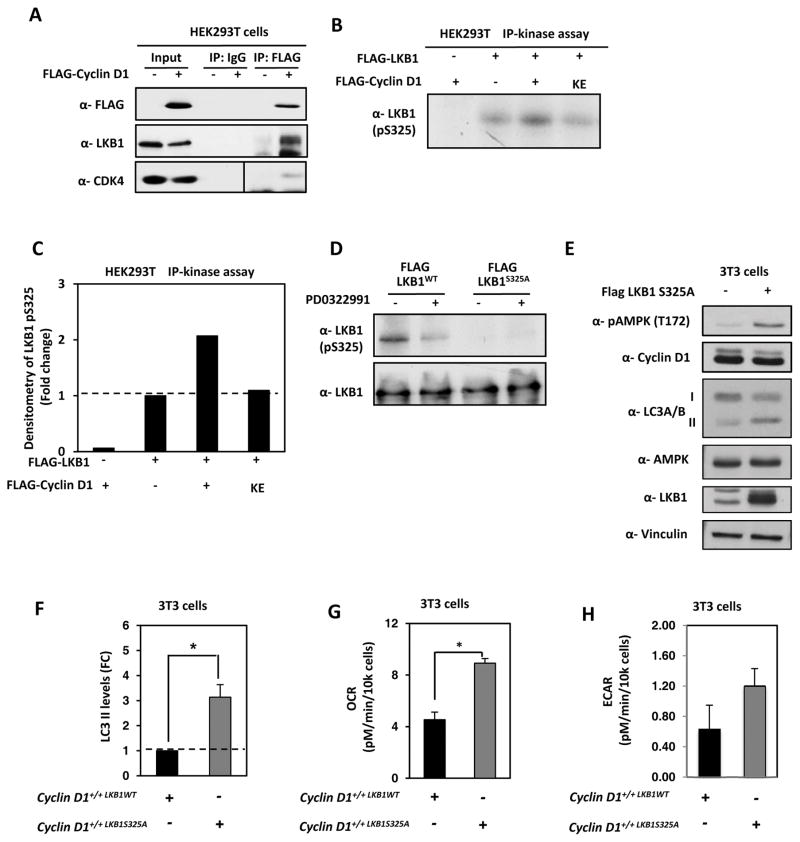
Cyclin D1-associated kinase phosphorylates LKB1 at S325A
Herein, cyclin D1 restrained autophagy and inhibited AMPK phosphorylation. LKB1 binds to phosphorylated AMPK at threonine 172 (T172). Phosphorylation of LKB1 at Serine 325 by ERK and Rsk compromises its ability to bind and activate AMPK (6). As the LKB1 phosphorylation site resembles a consensus for a proline-directed serine/threonine kinase, we considered the possibility that LKB1 served as a substrate for cyclin D1/cdk4/6. HEK293T cells were transfected with a mammalian expression vector encoding cyclin D1 with an N-terminal FLAG epitope. Immunoprecipitation of cell extracts using the anti-FLAG antibody to precipitate cyclin D1, co-precipitated cdk4 and LKB1 (Fig. 7A). To determine whether LKB1 serves as a substrate for cyclin D1/cdk4, using GST-LKB1 as a substrate we conducted a cyclin D1-IP kinase assay. Cells were transfected with an expression vector encoding cyclin D1, or a kinase-defective cyclin D1 cDNA (cyclin D1KE) (Fig. 7B). Expression of cyclin D1 enhanced LKB1 phosphorylation as assessed by incorporation of γ-32P into LKB1. Cells transduced with a kinase-defective mutant of cyclin D1 did not significantly enhance LKB1 phosphorylation (Fig. 7B and 7C). Western blot analysis of cells transfected with an expression vector encoding LKB1WT or a point mutant at Serine 325 (LKB1S325A) were immunoblotted with an antibody to phosphorylated LKB1 (S325A). We demonstrated the presence of the phosphorylated form of LKB1, which was reduced by the addition of the cdk4/6 inhibitor PD0322991 and abolished by point mutation of LKB1 at S325A (Fig. 7D).
Figure 7. Cyclin D1/cdk4 complex phosphorylates LKB1 at Serine 325 (S325).
(A) Immunoprecipitated FLAG-cyclin D1 was immunoblotted for LKB1 and FLAG-cyclin D1. (B) HEK293T cells were transfected with expression vectors encoding FLAG-LKB1 and with expression vectors encoding either FLAG-cyclin D1WT or FLAG-cyclin D1KE. Total cell lysates were immunoprecipitated with an antibody directed to the FLAG epitope, then incubated in the presence of [γ-32P] ATP and subjected to autoradiography. (C) LKB1 phosphorylation was increased in the presence of cyclin D1WT, but not the kinase dead mutant (cyclin D1KE). (D) MCF10A cells were transfected with FLAG-LKB1 or FLAG-LKB1 S325A and treated for 24h with the cdk4/6 inhibitor PD0322991 at a concentration of 0.5 μM. Total lysates were run on SDS-PAGE gel and immunoblotted for LKB1 and phosphorylated S325 LKB1. The inhibition of cdk4/6 activity reduced the phosphorylation of S325A. (E) 3T3 WT cells were infected with LKB1 S325A and protein lysates subjected to immunoblotting for pAMPK (T172), cyclin D1, LC3A/B, LKB1 and β-tubulin used as a loading control. (F) Quantitation of the abundance of lipidated LC3-II was normalized to vinculin and graphically represented. Seahorse FX bioanalyzer of basal OCR (G) and (H) basal ECAR using a Seahorse FX bioanalyzer of cyclin D1+/+, cyclin D1−/− and cyclin D1+/+LKB1S325A cells (*; P<0.05 and ***; P<0.001).
To determine whether phosphorylation of LKB1 at S325 by cyclin D1 cdk4/6 contributes to autophagic restraint, we deployed a point mutation of LKB1 S325A and conducted Western blots for activated AMPK and autophagic markers. Expression of LKB1 S325A in wild type MEF abrogated cyclin D1 restraint of autophagy compared to MEF expressing LKB1 WT (Fig. 7E and 7F). To determine whether cyclin D1/cdk4/6 phosphorylation of LKB1 at S325 also contributed to mitochondrial function we deployed the point mutation of LKB1 S325A and conducted ECAR and OCR analysis using the Seahorse Extracellular Flux Analyzer (XF). The expression of LKB1 S325A in cyclin D1+/+ MEF significantly increased OCR (2-fold; P=0.02) (Fig. 7G) and increased ECAR (2 fold; P=0.052) (Fig. 7H).
Discussion
Cyclin D1 restrains autophagy
The current studies demonstrated endogenous cyclin D1 restrains autophagy demonstrated by using shRNA to cyclin D1, cyclin D1 kinase inhibition, or cyclin D1 gene deletion. The restraint of autophagy was demonstrated by the reduction in LC3-II/LC3-I by Western blot and the formation of autophagic puncta visualized using an LC3-GFP fusion protein. Endogenous cyclin D1 restrained the induction of autophagy induced by either ErbB2, Ha-Ras or v-Src, or by the cellular stress of nutrient deprivation or by Rapamycin treatment. The current findings that cyclin D1, which is induced by a variety of mitogenic stimuli, restrains autophagy is consistent with prior studies on mitogenic stimuli and autophagy, as there is a strong correlation between mitogenic signaling and the inhibition of autophagy (8). In prior studies the mammary gland of MMTV-ErbB2-cyclin D1KE/KE knock-in mice showed enhanced mammary epithelial cell cellular proliferation similar to MMTV-ErbB2-cyclin D1WT/WT knock-in mice, but increased autophagy, suggesting cyclin D1 restrained basal autophagy in a kinase-dependent manner (14). The current studies extend these findings by demonstrating the mechanism in which cyclin D1 phosphorylates LKB1 at S325 to inactivate AMPK and thereby restrain autophagy. The demonstration that the cell cycle control protein cyclin D1 controls autophagy via phosphorylation of LKB1 brings together two of the important hallmarks of cancer, deregulation of cellular energetics and sustained proliferative signaling (31).
Cyclin D1 inhibits AMPK
Cyclin D1 reduced AMPK activity in tissue culture and in the mammary gland in vivo. Increased phosphorylation of AMPK (T172) was demonstrated upon reduction of cyclin D1 in human breast cancer cell lines, MEF, and in the tissue of mammary epithelial cell targeted inducible cyclin D1 deletion mice. Endogenous cyclin D1 restrained AMPK induction by the cellular stresses of nutrient deprivation, reactive oxygen species (H2O2) and ultraviolet irradiation. AMPK is down-regulated in tumors compared with normal epithelium through an unknown mechanism (29). Cyclin D1 is overexpressed in the majority of human breast tumors and may contribute to the down-regulation of AMPK in tumors. AMPK is known to induce (2) and cyclin D1 is known to restrain mitochondrial biogenesis and thereby increase cytosolic glycolysis (21,32).
In the current studies, endogenous cyclin D1 was shown to enhance glycolytic flux, blunt OCR, and to reduce OCR when MEF were supplemented with palmitate. Cyclin D1 reduced mitochondrial area and number and reduced mitochondrial connectivity. Cyclin D1 increased lactate secretion and reduced oxidative phosphorylation in the mitochondria. AMPK activation increases shuttling of glycolytic pyruvate into the mitochondria. The decrease in lactate secretion in the cyclin D1−/− MEF is indicative of elevated shuttling of glycolytic pyruvate into the mitochondria. Introducing the LKB1 325A mutant abrogates the suppression of glycolysis and oxygen consumption evidenced by increased ECAR and OCR in cyclin D1+/+LKB1S325A cells compared to cyclin D1+/+LKB1WT.
AMPK is known to be involved in mitochondrial biogenesis in hepatocytes (33,34). Relevant to the finding here in both mitochondrial respiration and basal level of ATP were significantly lower in hepatocytes isolated from a liver specific AMPKα1α−/− mouse compared with control mice (35). Reduced AMPK activity may be an important factor in the reduced mitochondrial function. AMPK has been reported to induce cellular glycolysis under a number of cellular stress events (36–38). These findings are consistent with our findings that cyclin D1 inhibits mitochondrial function and reduces glycolytic flux through regulating the LKB1/AMPK signaling axis.
Cyclin D1 phosphorylates LKB1 requiring S325A
To examine the mechanism by which cyclin D1 inhibited AMPK, we considered the role of the upstream kinase LKB1. LKB1 itself is constitutively active in cells (5,6), however, phosphorylation by Raf reduced AMPK phosphorylation (6). The MAPKs, ERK and Rsk phosphorylate Ser325 inactivating LKB1-dependent AMPK activity (6). The phosphorylation of LKB1 interferes with the ability of LKB1 to phosphorylate and activate AMPK (6). Herein, a cyclin D1 complex, including cdk4/6, associated with LKB1. Transfection of HEK293T cells with cyclin D1 enhanced LKB1 phosphorylation 3-fold, however, a kinase-dead mutant of cyclin D1 failed to induce LKB1 phosphorylation. By Scan Site analysis, LKB1 Ser325 was a candidate site for proline-dependent serine/threonine kinases and was required for phosphorylation in cyclin D1-immune kinase assays. Finally, treatment of cells with the cdk inhibitor PD0322991 reduced LKB1 phosphorylation. The binding and inactivation of the LKB1 tumor suppressor by cyclin D1 (Supplemental Fig. 5) is consistent with the known role of cyclin D1 to bind and inactivate other tumor suppressors (pRb, p130, p107, BRCA1) (39,40). Endogenous cyclin D1 phosphorylated LKB1 and repressed the induction of AMPK associated with cellular stress. It has been suggested that activation of AMPK may explain the tumor suppressor effects of LKB1, as AMPK is the only downstream target of LKB1 known to cause inhibition of cell growth (2).
What might be the clinical significance of these findings?
The finding that cyclin D1 restrains autophagy may be important in therapeutic strategies depending upon the role of autophagy in tumor progression and therapy resistance. The removal of damaged proteins by autophagy may be important for both cell survival and tumor-suppression (8). Autophagy has a cellular context-dependent role in cancer and may also vary depending on the stage of tumorigenesis. Evidence exists for both a tumor suppressor and for a tumor promoting role of autophagy. Evidence for a tumor suppressive role includes findings that, autophagy may inhibit tumorigenesis by limiting necrosis and inflammation, which are thought to be sites that generate cancer-causing mutations (41). Defects in autophagy are associated with increased ROS production (18) and tumor development and several oncogenic pathways (PI3K mutations, AKT amplification, Pten loss) decrease autophagy (42). The tumor suppressor p53 induces autophagy (43). Furthermore, Beclin 1, the mammalian homologue of yeast ATG6, functions as a tumor suppressor with haploinsufficiency (44).
In some circumstances autophagy functions to maintain tumors, providing energy substrates required for cellular growth (45). Autophagy may promote tumor development later in the tumorigenic process by providing energy substrates during periods of nutrient limitation (46). Autophagy is required for the survival of tumor cells in hypoxic tumor regions. Oncogenic RAS transformation upregulates basal autophagy required for the maintenance and progression of tumorigenesis (46). Engineered mouse models have also demonstrated a pro-tumorigenic role for autophagy. Deletion of Atg7 in KrasG12D or BrafV600E non-small cell lung cancer (NSCLC) in adult mice suppresses tumor cell proliferation. Autophagy promotes tumorigenesis by mitigating Trp53 tumor suppression (47,48). Using temporally controlled genetic ablation of the autophagy gene Atg7, recent studies suggest the function of autophagy may change with time (7). In the setting of cancer, 5 weeks of acute autophagy ablation converted established lung adenocarcinomas to oncocytomas and blocked cell proliferation.
The current studies are consistent with, and extend, the repertoire of cyclin D1-mediated functions. Autophagy limits genomic instability and the inhibition of this autophagic function increases genomic instability (49). The inhibition of autophagy by cyclin D1 is therefore consistent with the known effects of cyclin D1 to induce chromosomal instability (13,20). The current studies extend the reported cyclin D1 functions that may contribute to the collaborative oncogenic function of cyclin D1.
Supplementary Material
Acknowledgments
Financial Support
This work was supported in part by NIH grants (1R01CA137494, R01CA132115, R01CA086072 to R.G. Pestell), the Sidney Kimmel Cancer Center NIH Cancer Center Core grant P30CA056036 (R.G. Pestell), generous grants from The Breast Cancer and Research Foundation (R.G. Pestell) and the Dr. Ralph and Marian C. Falk Medical Research Trust (R.G. Pestell), a grant from the Pennsylvania Department of Health (R.G. Pestell) and an American-Italian Cancer Foundation Post-doctoral Research Fellowship (G. Di Sante).
This work was supported in part by NIH grants (1R01CA137494, R01CA132115, R01CA086072 to R.G.P.), the Sidney Kimmel Cancer Center NIH Cancer Center Core grant P30CA056036 (R.G.P.), generous grants from The Breast Cancer and Research Foundation (R.G.P.) and the Dr. Ralph and Marian C. Falk Medical Research Trust (R.G.P.), a grant from the Pennsylvania Department of Health (R.G.P.) and an American-Italian Cancer Foundation Post-doctoral Research Fellowship (G.D.). The Department specifically disclaims responsibility for an analysis, interpretations or conclusions. There are no conflicts of interest associated with this manuscript.
Abreviations
- AMPK
5′-prime-AMP-activated protein kinase
- LKB1
Liver Kinase B1
- LC3
microtubule-associated protein 1 light-chain 3
- Rb
Retinoblastoma protein
- HBSS
Hank’s Balanced Salt Solution
- MEF
Mouse Embryonic Fibroblasts
- Tet
Tetracycline
- rtTA
reverse tetracycline-controlled transactivator
- FCCP
Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone
Footnotes
Competing financial interests: The authors disclose no potential conflicts of interest.
References
- 1.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Molecular cell. 2010;40:280–93. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25:1895–908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Makela TP, et al. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. Journal of biology. 2003;2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canto C, Auwerx J. AMP-activated protein kinase and its downstream transcriptional pathways. Cell Mol Life Sci. 2010;67:3407–23. doi: 10.1007/s00018-010-0454-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakamoto K, Goransson O, Hardie DG, Alessi DR. Activity of LKB1 and AMPK-related kinases in skeletal muscle: effects of contraction, phenformin, and AICAR. Am J Physiol Endocrinol Metab. 2004;287:E310–7. doi: 10.1152/ajpendo.00074.2004. [DOI] [PubMed] [Google Scholar]
- 6.Zheng B, Jeong JH, Asara JM, Yuan YY, Granter SR, Chin L, et al. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Molecular cell. 2009;33:237–47. doi: 10.1016/j.molcel.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014;4:914–27. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipinski MM, Hoffman G, Ng A, Zhou W, Py BF, Hsu E, et al. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Developmental cell. 2010;18:1041–52. doi: 10.1016/j.devcel.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nature cell biology. 2007;9:218–24. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- 11.Sapkota GP, Boudeau J, Deak M, Kieloch A, Morrice N, Alessi DR. Identification and characterization of four novel phosphorylation sites (Ser31, Ser325, Thr336 and Thr366) on LKB1/STK11, the protein kinase mutated in Peutz-Jeghers cancer syndrome. Biochem J. 2002;362:481–90. doi: 10.1042/0264-6021:3620481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: Normal and Abnormal Functions. Endocrinology. 2004;145:5439–47. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- 13.Casimiro MC, Di Sante G, Crosariol M, Loro E, Dampier W, Ertel A, et al. Kinase-independent role of cyclin D1 in chromosomal instability and mammary tumorigenesis. Oncotarget. 2015;6:8525–38. doi: 10.18632/oncotarget.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown NE, Jeselsohn R, Bihani T, Hu MG, Foltopoulou P, Kuperwasser C, et al. Cyclin D1 activity regulates autophagy and senescence in the mammary epithelium. Cancer Res. 2012;72:6477–89. doi: 10.1158/0008-5472.CAN-11-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albanese C, D’Amico M, Reutens AT, Fu M, Watanabe G, Lee RJ, et al. Activation of the cyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J Biol Chem. 1999;274:34186–95. doi: 10.1074/jbc.274.48.34186. [DOI] [PubMed] [Google Scholar]
- 16.Casimiro MC, Di Sante G, Ju X, Li Z, Chen K, Crosariol M, et al. Cyclin D1 Promotes Androgen-Dependent DNA Damage Repair in Prostate Cancer Cells. Cancer Res. 2016;76:329–38. doi: 10.1158/0008-5472.CAN-15-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Pattabiraman N, Zhou JN, Fu M, Sakamaki T, Albanese C, et al. Cyclin D1 repression of peroxisome proliferator-activated receptor gamma expression and transactivation. Molecular and Cell Biology. 2003;23:6159–73. doi: 10.1128/MCB.23.17.6159-6173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Sante G, Pestell TG, Casimiro MC, Bisetto S, Powell MJ, Lisanti MP, et al. Loss of Sirt1 promotes prostatic intraepithelial neoplasia, reduces mitophagy, and delays PARK2 translocation to mitochondria. Am J Pathol. 2015;185:266–79. doi: 10.1016/j.ajpath.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casimiro MC, Crosariol M, Loro E, Ertel A, Yu Z, Dampier W, et al. ChIP sequencing of cyclin D1 reveals a transcriptional role in chromosomal instability in mice. The Journal of Clinical Investigation. 2012;122:833–43. doi: 10.1172/JCI60256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C, Li Z, Lu Y, Du R, Katiyar S, Yang J, et al. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc Natl Acad Sci U S A. 2006;103:11567–72. doi: 10.1073/pnas.0603363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koopman WJ, Visch HJ, Verkaart S, van den Heuvel LW, Smeitink JA, Willems PH. Mitochondrial network complexity and pathological decrease in complex I activity are tightly correlated in isolated human complex I deficiency. Am J Physiol Cell Physiol. 2005;289:C881–90. doi: 10.1152/ajpcell.00104.2005. [DOI] [PubMed] [Google Scholar]
- 23.Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc Natl Acad Sci U S A. 2009;106:11960–5. doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee RJ, Albanese C, Fu M, D’Amico M, Lin B, Watanabe G, et al. Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Molecular and Cellular Biology. 2000;20:672–83. doi: 10.1128/mcb.20.2.672-683.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee RJ, Albanese C, Stenger RJ, Watanabe G, Inghirami G, Haines GK, 3rd, et al. pp60(v-src) induction of cyclin D1 requires collaborative interactions between the extracellular signal-regulated kinase, p38, and Jun kinase pathways. A role for cAMP response element-binding protein and activating transcription factor-2 in pp60(v-src) signaling in breast cancer cells. The Journal of Biological Chemistry. 1999;274:7341–50. doi: 10.1074/jbc.274.11.7341. [DOI] [PubMed] [Google Scholar]
- 26.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, et al. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–97. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 27.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–92. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 28.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Molecular cell. 2005;18:283–93. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 29.Liang J, Mills GB. AMPK: a contextual oncogene or tumor suppressor? Cancer Res. 2013;73:2929–35. doi: 10.1158/0008-5472.CAN-12-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Neill HM, Maarbjerg SJ, Crane JD, Jeppesen J, Jorgensen SB, Schertzer JD, et al. AMP-activated protein kinase (AMPK) beta1beta2 muscle null mice reveal an essential role for AMPK in maintaining mitochondrial content and glucose uptake during exercise. Proc Natl Acad Sci U S A. 2011;108:16092–7. doi: 10.1073/pnas.1105062108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 32.Sakamaki T, Casimiro MC, Ju X, Quong AA, Katiyar S, Liu M, et al. Cyclin D1 determines mitochondrial function in vivo. Mol Cell Biol. 2006;26:5449–69. doi: 10.1128/MCB.02074-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guigas B, Taleux N, Foretz M, Detaille D, Andreelli F, Viollet B, et al. AMP-activated protein kinase-independent inhibition of hepatic mitochondrial oxidative phosphorylation by AICA riboside. Biochem J. 2007;404:499–507. doi: 10.1042/BJ20070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guigas B, Bertrand L, Taleux N, Foretz M, Wiernsperger N, Vertommen D, et al. 5-Aminoimidazole-4-carboxamide-1-beta-D-ribofuranoside and metformin inhibit hepatic glucose phosphorylation by an AMP-activated protein kinase-independent effect on glucokinase translocation. Diabetes. 2006;55:865–74. doi: 10.2337/diabetes.55.04.06.db05-1178. [DOI] [PubMed] [Google Scholar]
- 36.Wu SB, Wei YH. AMPK-mediated increase of glycolysis as an adaptive response to oxidative stress in human cells: implication of the cell survival in mitochondrial diseases. Biochim Biophys Acta. 2012;1822:233–47. doi: 10.1016/j.bbadis.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 37.Marsin AS, Bouzin C, Bertrand L, Hue L. The stimulation of glycolysis by hypoxia in activated monocytes is mediated by AMP-activated protein kinase and inducible 6-phosphofructo-2-kinase. J Biol Chem. 2002;277:30778–83. doi: 10.1074/jbc.M205213200. [DOI] [PubMed] [Google Scholar]
- 38.Almeida A, Moncada S, Bolanos JP. Nitric oxide switches on glycolysis through the AMP protein kinase and 6-phosphofructo-2-kinase pathway. Nature cell biology. 2004;6:45–51. doi: 10.1038/ncb1080. [DOI] [PubMed] [Google Scholar]
- 39.Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes and Development. 1993;7:331–42. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 40.Wang C, Fan S, Li Z, Fu M, Rao M, Ma Y, et al. Cyclin D1 antagonizes BRCA1 repression of estrogen receptor alpha activity. Cancer Res. 2005;65:6557–67. doi: 10.1158/0008-5472.CAN-05-0486. [DOI] [PubMed] [Google Scholar]
- 41.Mathew R, White E. Autophagy in tumorigenesis and energy metabolism: friend by day, foe by night. Curr Opin Genet Dev. 2011;21:113–9. doi: 10.1016/j.gde.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diaz-Troya S, Perez-Perez ME, Florencio FJ, Crespo JL. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy. 2008;4:851–65. doi: 10.4161/auto.6555. [DOI] [PubMed] [Google Scholar]
- 43.Balaburski GM, Hontz RD, Murphy ME. p53 and ARF: unexpected players in autophagy. Trends in cell biology. 2010;20:363–9. doi: 10.1016/j.tcb.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 45.Martinez-Outschoorn UE, Trimmer C, Lin Z, Whitaker-Menezes D, Chiavarina B, Zhou J, et al. Autophagy in cancer associated fibroblasts promotes tumor cell survival: Role of hypoxia, HIF1 induction and NFkappaB activation in the tumor stromal microenvironment. Cell cycle. 2010:9. doi: 10.4161/cc.9.17.12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 48.Rao S, Tortola L, Perlot T, Wirnsberger G, Novatchkova M, Nitsch R, et al. A dual role for autophagy in a murine model of lung cancer. Nat Commun. 2014;5:3056. doi: 10.1038/ncomms4056. [DOI] [PubMed] [Google Scholar]
- 49.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.