Abstract
We show that HEXIM1 (hexamethylene bis-acetamide inducible 1) functions as an AR (androgen receptor) co-repressor as it physically interacts with the AR and is required for the ability of anti-androgens to inhibit androgen-induced target gene expression and cell proliferation. Oncomine™ database and IHC (immunohistochemistry) analyses of human prostate tissues revealed that expression of HEXIM1 mRNA and protein are down-regulated during the development and progression of prostate cancer. Enforced down-regulation of HEXIM1 in parental hormone-dependent LNCaP cells results in resistance to the inhibitory action of anti-androgens. Conversely, ectopic expression of HEXIM1 in the CRPC (castration-resistant prostate cancer) cell line, C4-2, enhances their sensitivity to the repressive effects of the anti-androgen bicalutamide. Novel insight into the mechanistic basis for HEXIM1 inhibition of AR activity is provided by the present studies showing that HEXIM1 induces expression of the histone demethylase KDM5B (lysine-specific demethylase 5B) and inhibits histone methylation, resulting in the inhibition of FOXA1 (forkhead box A1) licensing activity. This is a new mechanism of action attributed to HEXIM1, and distinct from what has been reported so far to be involved in HEXIM1 regulation of other nuclear hormone receptors, including the oestrogen receptor.
Keywords: androgen receptor (AR), castration-resistant prostate cancer (CRPC), forkhead box A1 (FOXA1), hexamethylene bis-acetamide inducible 1 (HEXIM1), positive transcription elongation factor b (P-TEFb), prostate-specific antigen (PSA), RNA polymerase II (RNAPII), RNA elongation
INTRODUCTION
The AR (androgen receptor) is a member of the nuclear steroid receptor family that is critical for the development of male reproductive tissues and skeletal muscle. AR mediates these effects by regulating gene transcription following androgen binding and receptor activation [1]. Deregulated expression and activation of AR is well recognized to be critical for prostate cancer progression [1]. AR transcriptional activity results in the regulation of genes responsible for promoting growth, inhibiting apoptosis and possibly enhancing metastasis of the prostate tumours [1]. Androgens have been shown to regulate numerous genes, including PSA (prostate-specific antigen), EGFR (epidermal growth factor receptor), CDK2 (cyclin-dependent kinase 2) and CDK4, p21WAF1 (presumed to play an anti-apoptotic role in prostate cancer), cyclin Ds, survivin, and the AR co-activator ARA70 [1].
AR plays a role in all the stages of prostate cancer, from initiation, development and resistance to hormone therapy [2]. Owing to the dependence of prostate cancer on the AR and androgens, the most common therapy for prostate cancer, in particular in advanced prostate cancer, is androgen ablation via orchidectomy or AR antagonists [2]. Although prostate tumours are typically first responsive to such treatment, they invariably become resistant to anti-androgen therapy. However, AR-silencing studies show that CRPC (castration-resistant prostate cancer) cells retain dependence on AR [2]. This supports the model that the progression to CRPC is functionally linked to the constitutive activation of the AR, which may result from some combination of the enhanced expression of the AR and AR co-activator(s), reduced expression of AR co-repressor(s), and production of androgens in tumours [2].
We have previously reported that HEXIM1 (hexamethylene bis-acetamide inducible 1) inhibits the activity of ERα (oestrogen receptor α) in vitro by intercepting an interaction between ERα and P-TEFb (positive transcription elongation factor b) [3], a protein complex comprised of cyclin T1 and CDK9. ERα directly binds to the cyclin T1 component of this complex (reviewed in [4]), recruiting P-TEFb to ER target genes whereby P-TEFb phosphorylates the C-terminal domain of RNAPII (RNA polymerase II) at Ser2 to promote productive phases of transcriptional elongation [4]. By associating with both ERα and the 7SK snRNP complex, HEXIM1 inhibits the co-recruitment of both ERα and cyclin T1 to the promoter region of ERα target genes. As a result, HEXIM1 inhibits the phosphorylation of the C-terminal domain of RNAPII and thereby prevents ERα-mediated transcriptional elongation in breast cells. HEXIM1 inhibits mammary tumorigenesis, partly by inhibiting ER-dependent transcription of cyclin D1 andVEGF (vascular endothelial growth factor) [5,6].
In the present study we now report on the role of HEXIM1 as a putative co-repressor of the AR that is required for the inhibition of the AR by anti-androgens. The mechanism for HEXIM1 inhibition of the AR involves decreases in the levels of active histone marks, notably histone 3 dimethylated at Lys4 (H3K4me2), that guide FOXA1 (forkhead box A1) recruitment to AR-regulated cell cycle genes. These M-phase cell cycle genes were previously shown to be critical for AR induction in CRPC [7]. Although HEXIM1 also inhibited recruitment of cyclin T1 to the AR target gene PSA, regulation of active histone marks also plays an important role in HEXIM1 regulation of AR activity. Thus the mechanism of HEXIM1 regulation of AR transcription is distinct from what we have reported so far for ERα.
MATERIALS AND METHODS
Cell culture and transfections
LNCaP cells were obtained from the A.T.C.C. (Manassas, VA, U.S.A.), C4-2 and C4-2B cells [8] were obtained from Dr Leland Chung (Department of Medicine, University of California Los Angeles, Los Angeles, CA, U.S.A.), and LAPC4 cells were obtained from Dr Robert Reiter (Department of Urology, University of California Los Angeles, Los Angeles, CA, U.S.A.). LNCaP and C4-2 cells were maintained as described previously [9], and LAPC4 cells were grown in Iscove’s medium (Invitrogen) supplemented with 10% FBS plus 1 nM R1881. To examine androgenic responses, cells were cultured in their respective basal medium containing 10%dextran-coated charcoal-extracted FBS (Invitrogen). Construction of expression vectors for control miRNA or HEXIM1 miRNAs (clones 35 and 609) has been described previously [5]. LNCaP cells were transfected with expression vectors containing either the HEXIM1 miRNA insert or a control miRNA insert as described previously [5]. Following blasticidin selection, cells expressing the highest level of GFP were flow-sorted and expanded. C4-2 cells were transfected with control or expression vector for FLAG–HEXIM1 [3] as described previously [5].
Co-immunoprecipitation
Endogenous proteins were co-immunoprecipitated and analysed as described previously [3].
In vitro translation and protein–protein interaction assays
In vitro transcription and translation of AR were performed using the Promega TNT kit according to the manufacturer’s recommendations. GST pull-down assays were performed as described previously [10].
Western blot analysis
Cell lysates were analysed by Western blotting as described previously [5]. The anti-HEXIM1 antibody was generated in the Montano laboratory [11]. The primary antibodies against UBE2C (ubiquitin-conjugating enzyme E2C; H-90; sc-99146), AR (441; sc-7305) and KDM5B (lysine-specific demethylase 5B; H-180; sc-67035) were obtained from Santa Cruz Biotechnology. The anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) antibody was obtained from Millipore.
Immunohistochemistry
Human prostate tissue samples were obtained from the CHTN (Cooperative Human Tissue Network) and a tissue microarray from US Biomax (# PR8011). All samples were confirmed to be AR-positive by IHC (immunohistochemistry). We carried out immunohistochemical staining to detect HEXIM1 levels as described previously [11,12].
Chromatin immunoprecipitation
ChIP assays were carried out as described previously [3]. The primers sequences used were: PSA ARE (androgen-response element) proximal forward, 5′-TCCTGAGTGCTGGTGTCTTAG- 3′, reverse, 5′-AGCCCTATAAAACCTTCATTCC-3′; PSA ARE enhancer forward, 5′-CATGTTCACATTAGTACACCTTG-3′, reverse, 5′-TCTCAGATCCAGGCTTGCTTAC-3′; PSA coding sequence forward, 5′-CACACCCGCTCTACGATATGAG-3′, reverse, 5′-GAGCTCGGCAGGCTCTGA-3′; UBE2C enhancer 1+ forward, 5′-TGCCTCTGAGTAGGAACAGGTAAGT-3′, reverse, 5′-TGCTTTTTCCATCATGGCAG-3′; UBE2C enhancer 2+ forward, 5′-CCACAAACTCTTCTCAGCTGGG- 3′, reverse, 5′-TTCTTTCCTTCCCTGTTACCCC-3′; CDK1 enhancer forward, 5′-GGGAAAGAGAAGCCCTACACTTG- 3′, reverse, 5′-GGGCTGTGCTACTTCTCTGGG-3′; CDC20 enhancer forward, 5′-GGAGTTGTGAGAACACCCGG-3′, reverse, 5′-AACACCCAGGTACACCCTCG-3′; KDM5B enhancer forward, 5′-GGCAACCCATGTCTATCACAAGAGG-3′, reverse, 5′-CTGGATACTTTGATACTCATCTG-3′.
RT (reverse transcription)–PCR analyses
Cells were subjected to RT–PCR analyses as described previously [5]. The primers sequences used were: PSA forward, 5′-TGTGTGCTGGACGCTGGA- 3′, reverse, 5′-CACTGCCCCATGACGTGAT- 3′; UBE2C forward, 5′-TGGTCTGCCCTGTATGATGT- 3′, reverse, 5′-AAAAGCTGTGGGGTTTTTCC-3′; CDK1 forward, 5′-CCTAGTACTGCAATTCGGGAAATT-3′, reverse, 5′-CCTGGAATCCTGCATAAGCAC-3′; CDC20 forward, 5′-CCTCTGGTCTCCCCATTAC-3′, reverse, 5′-ATGTGTGACCTTTGAGTTCAG- 3′; KDM5B forward, 5′-CATCACTGGCATGTTGTTCAAATTC- 3′, reverse, 5′-GAATGTAGTAAGCCACAAGAAGC- 3′.
Proliferation assay
LNCaP cells transfected with expression vector for control miRNA or HEXIM1 miRNA were plated on to 96-well plates. Cells were treated with 10 nM R1881, 10 μM bicalutamide or both for 7 days. Cell proliferation was assessed using the MTT-based Cell Growth Determination Kit from Sigma–Aldrich according to the manufacturer’s protocol.
Data analyses
Statistical significance was determined using the Student’s t test comparison for unpaired data.
RESULTS
Normal prostate epithelial cells express nuclear HEXIM1, and HEXIM1 expression is lower in prostate cancer cells
Our initial test for a role of HEXIM1 in prostate tumour progression was the examination of HEXIM1 expression in human prostate tissues by IHC using an anti-HEXIM1 antibody that we have carefully validated for IHC [11-14]. Strong nuclear HEXIM1 expression was evident in normal prostate tissues, and statistically significant lower levels of HEXIM1 were observed in BPH (benign prostatic hyperplasia) and tumours. Moreover, HEXIM1 expression was higher in well or moderately differentiated tumours (Gleason grade <7) when compared with poorly or undifferentiated tumours (Gleason grade >7, Figure 1A and Supplementary Figure S1 at http://www.biochemj.org/bj/462/bj4620315add.htm, P=0.0096). Human prostate tissue samples with Gleason score ≥7 are associated with a shorter duration to CRPC among patients with metastatic disease [15,16]. Our analyses of HEXIM1 expression in human tissues were also consistent with other studies as analysed by Oncomine™ [17-19]. Microarray gene expression data from another laboratory (also analysed by Oncomine™) showed a statistically significant decrease in the expression of HEXIM1 in CRPC when compared with benign and prostate carcinomas (Figure 1B) [18]. We also observed lower levels of HEXIM1 expression in the CRPC cell lines C4-2 and C4-2B relative to the androgen-dependent cell lines LNCaP and LAPC4 (Figure 1C). These cancer cell lines were selected for this comparison because they also express AR, a characteristic of the vast majority of prostate cancers.
Figure 1. Expressions levels of HEXIM1 in normal prostate and prostate cancer tissue.
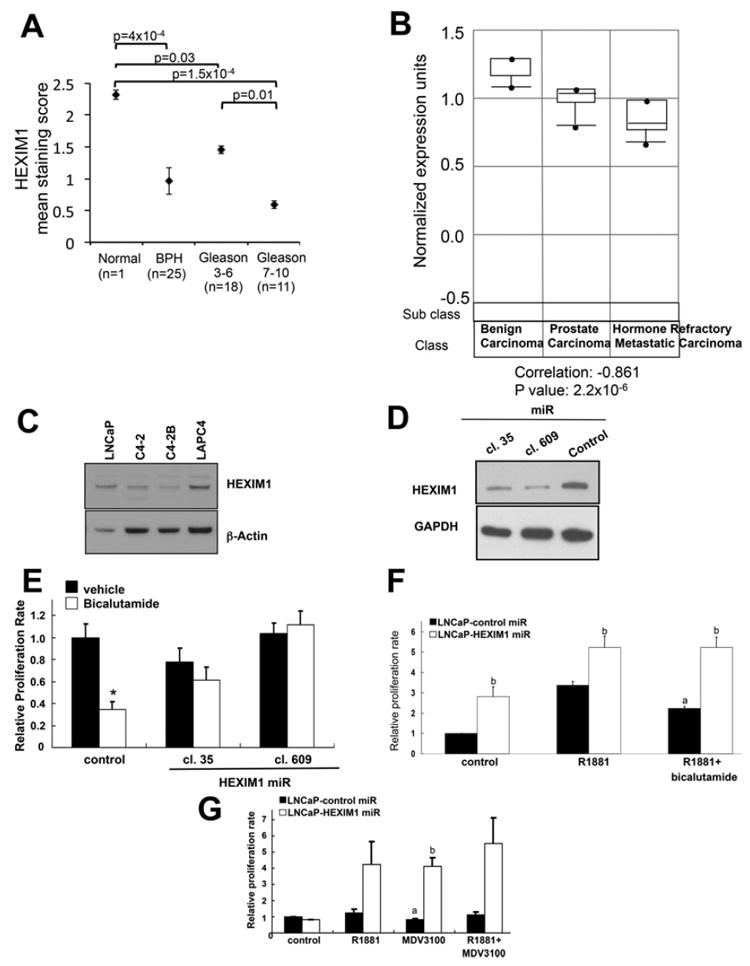
(A) Sections obtained from normal prostate tissues, BPH tissues and prostate tumour were stained for endogenous HEXIM1. The staining score was the product of the intensity of HEXIM1 nuclear staining and percentage of HEXIM1-positive cells. (B) Oncomine™ analyses of microarray gene expression of HEXIM1 in human benign carcinomas (n=6), prostate carcinomas (n=7) and hormone refractory metastatic carcinomas (n=6) [18]. For (A) and (B), P values were calculated using the Student’s t test. (C) Western blot analyses of HEXIM1 expression in LNCaP, LAPC4, C4-2 and C4-2B cells. LNCaP cells were stably transfected with control or two different HEXIM1miR clones separately (D and E) or together (F and G) and plated on to 94-well plates, treated as indicated for 6 days, then processed for MTT assays to assess proliferation. Figures are representative of at least three independent experiments. aP <0.05 relative to R1881 alone, bP <0.05 relative to control transfected cells with the same treatment and *P <0.05 relative to vehicle.
Owing to the decreased expression of HEXIM1 in CRPC, we examined whether HEXIM1 can regulate the response to antiandrogens. We used two different miRNA clones to down-regulate HEXIM1 expression in LNCaP cells (Figure 1D). Decreased HEXIM1 expression resulted in increased cell growth and a complete loss of the ability of bicalutamide to repress basal or R1881-stimulated growth, although these cells retained some sensitivity to growth stimulation by R1881 (Figures 1E and 1F). Down-regulation of HEXIM1 also resulted in attenuation of repressive effects of the anti-androgen MDV3100 on cell proliferation (Figure 1G).
HEXIM1 interacts with the AR
Using co-immunoprecipitation assays we show that endogenous AR interacts with endogenous HEXIM1 (Figure 2A). The interaction was attenuated in cells transfected with HEXIM1miR (Figure 2B). This interaction occurred in the absence or presence of AR agonists and antagonists (Figures 2A and 2B). In vitro GST pull-down assays were used to determine whether HEXIM1 and AR directly interact. GST–HEXIM1 bound to Sepharose beads pulled down in vitro translated AR. In vitro translated AR did not interact with GST alone. We have previously reported that amino acids 150–177 of HEXIM1 were required for the interaction with ERα [3]. Our data indicate that interaction of HEXIM1150–177 with AR occurs despite the lower expression levels of HEXIM1150–177 (Figure 2C), suggesting that amino acids 150–177 were also involved in the interaction of AR with HEXIM1.
Figure 2. Physical interaction between HEXIM1 and AR.
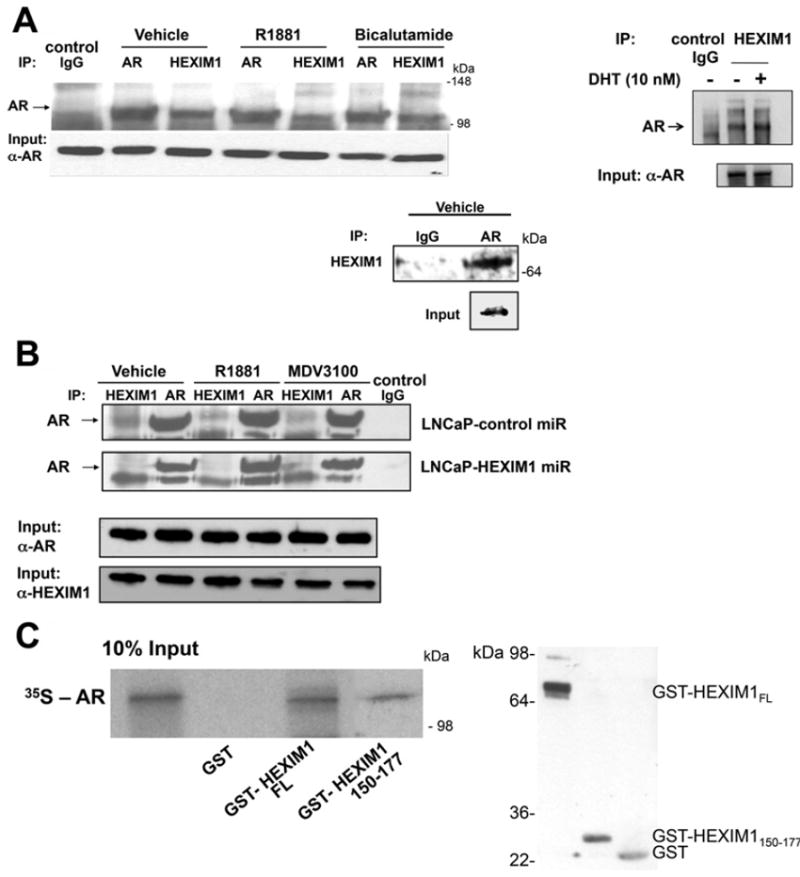
(A) LNCaP cells were treated with either vehicle, 10 nM R1881, 10 nM DHT (dihydrotestosterone) or 10 μM bicalutamide for 90 min. (B) LNCaP cells stably transfected with control or HEXIM1miR were treated with vehicle, 10 nm R1881 or 10 μM MDV3100 for 90 min. In (A) and (B) lysates were immunoprecipitated using antibodies against HEXIM1 or AR and analysed for co-immunoprecipitation of HEXIM1 or AR by Western blotting. Normal rabbit IgG was used as a specificity control. Input lanes represent 25% of the total protein. (C) Left-hand panel: in vitro translated and [35S]methionine-labelled AR was incubated with GST alone, GST–HEXIM1 or GST–HEXIM1150–177 bound to Sepharose. Bound protein was eluted and analysed by SDS/PAGE (12.5% gel). The Input lane represents 10% in vitro translated product added to the samples. Right-hand panel, Western blot analyses of GST, GST–HEXIM1 or GST–HEXIM1150–177 expression. Figures are representative of at least three independent experiments.
HEXIM1 regulates recruitment of transcriptional elongation factors to AR target genes
To test the potential for HEXIM1 regulation of AR-mediated transcription, we conducted ChIP analyses to examine the recruitment of HEXIM1 to the promoter of AR target genes in response to R1881 or bicalutamide and concomitantly assessed whether down-regulating the expression of HEXIM1 would alter the recruitment of transcriptional elongation factors to those promoters. Our data supported that HEXIM1 was recruited to the ARE-containing regions of the PSA promoter in LNCaP cells, and that such recruitment was increased following treatment with the anti-androgens bicalutamide, nilutamide and MDV3100 (Figures 3B and 3C). Down-regulation of HEXIM1 in LNCaP cells by synthetic HEXIM1miRs (Figure 3A) resulted in decreased recruitment of HEXIM1 to this promoter region as expected (Figure 3B), and enhanced recruitment of cyclin T1 (Figure 3D) and RNAPII phosphorylated at Ser2 (S2P RNAPII, Figure 3E) to the coding region of the PSA gene following treatment with bicalutamide. These results support that recruitment of HEXIM1 by treatment with bicalutamide inhibits the recruitment of both cyclin T1 and RNAPII to the coding region of PSA. Bicalutamide did not induce HEXIM1 recruitment to the non-ARE-containing region of the PSA gene (Supplementary Figure S2 at http://www.biochemj.org/bj/462/bj4620315add.htm).
Figure 3. Recruitment patterns of HEXIM1 and transcriptional elongation factors in LNCaP cells.
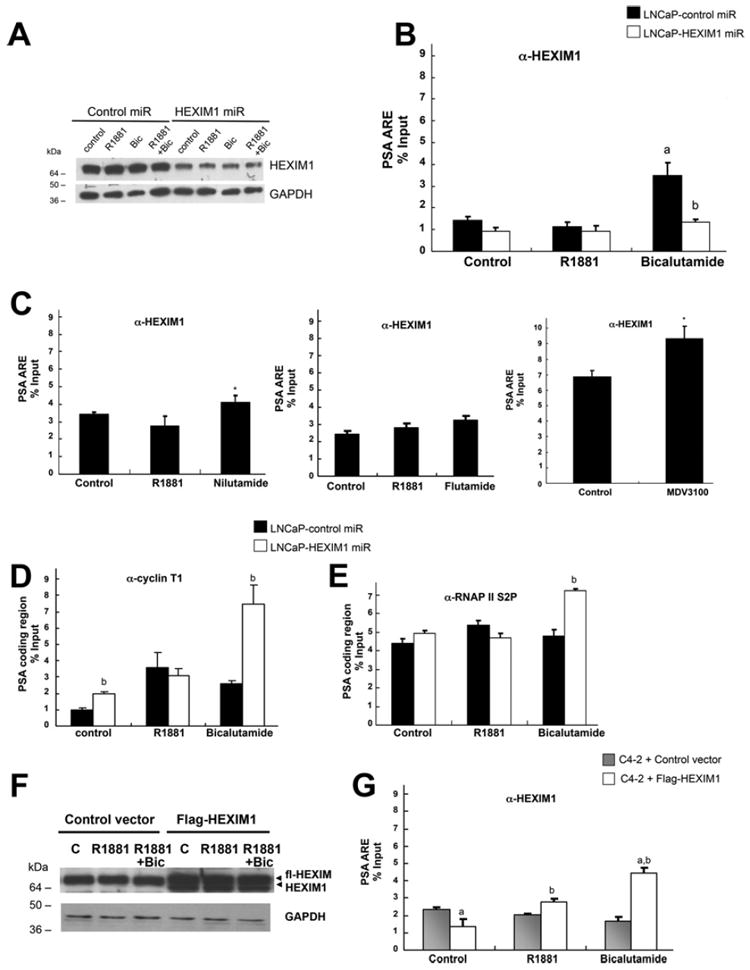
LNCaP cells stably transfected with control or HEXIM1miR were treated with vehicle, 10 nm R1881 or 10 μM bicalutamide (Bic), nulatimide, flutamide or MDV3100 for 90 min. (A) Cells were processed for Western blot analyses of HEXIM1 relative to the GAPDH loading control. Also shown are ChIP analyses of lysates immunoprecipitated with antibodies against HEXIM1 (B and C), cyclin T1 (D), RNAPII phosphorylated at Ser2 (E) or control non-specific rabbit Ig, followed by PCR amplification of the proximal ARE-containing region or the coding region of the PSA promoter. C4-2 cells transfected with control vector or expression vector for FLAG-tagged HEXIM1 (fl-HEXIM1) were treated with vehicle, 10 nm R1881 or 10 μM bicalutamide for 90 min. (F) Cells were processed for Western blot analyses of HEXIM1 relative to the GAPDH loading control. (G) ChIP analyses of lysates immunoprecipitated with antibodies against HEXIM1. The results are means±S.E.M. of three independent experiments. *P <0.05 relative to vehicle treated cells, aP <0.05 relative to R1881 alone and bP <0.05 relative to control transfected cells with the same treatment.
In contrast with LNCaP cells, bicalutamide did not induce recruitment of endogenous HEXIM1 to the PSA promoter in C4-2 cells, an androgen-independent derivative of the LNCaP cell line (Figure 3G). Introduction of a transfection vector for FLAG-tagged HEXIM1 resulted in enhanced HEXIM1 recruitment (Figure 3F), the consequences of which are described below.
HEXIM1 modifies histone marks on the enhancer regions of AR target genes
It has been proposed that the role of AR in androgen-independent cancer cells is not to direct the androgen-dependent gene expression programme without androgens, but rather to execute a distinct programme resulting in androgen-independent growth [7]. In that study, AR selectively up-regulated M-phase cell-cycle genes, including CDC20, CDK1 and UBE2C in an androgen-independent variant of LNCaP cells, abl. FOXA1 was shown to direct AR–enhancer binding and activation of CDC20, CDK1 and UBE2C [7]. FOXA1 recruitment occurred primarily on H3K9me2-poor but H3K4me1/2-rich regions, with H3K4me1/2 guiding FOXA1 cell type-specific recruitment through direct physical interactions.
ChIP assays for AR, FOXA1 and H3K4me2 were performed in control and HEXIM1miR LNCaP cells after their treatment with vehicle, R1881 or bicalutamide. We observed that the down-regulation of HEXIM1 resulted in increased recruitment of AR and FOXA1, and increased levels of H3K4me2 in the enhancer regions of CDC20 and CDK1 in the bicalutamide-treated groups (Figure 4). Enhancement of FOXA1 and the levels of H3K4me2 also occurred on the PSA promoter upon enforced down-regulation of HEXIM1 in bicalutamide-treated cells.
Figure 4. HEXIM1 inhibits FOXA1 recruitment and H3K4me2 enrichment.
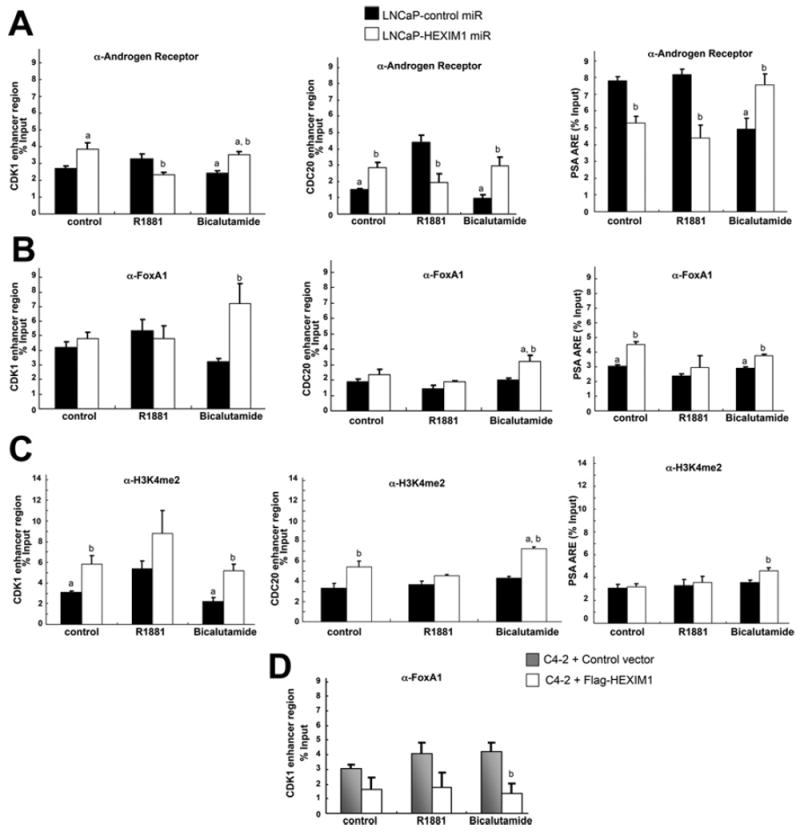
LNCaP cells stably transfected with control or HEXIM1miR were treated with vehicle, 10 nm R1881 or 10 μM bicalutamide for 90 min. Results show ChIP analyses of lysates immunoprecipitated with antibodies against AR (A), FOXA1 (B) or H3K4me2 (C) or control non-specific rabbit immunoglobin and PCR amplification of the enhancer regions of CDC20 (left-hand panels), CDK1 (middle panels) or PSA (right-hand panels). (D) C4-2 cells transfected with control vector or expression vector for FLAG-tagged HEXIM1 (fl-HEXIM1) were treated with vehicle, 10 nM R1881 or 10 μM bicalutamide for 90 min. Shown are ChIP analyses of lysates immunoprecipitated with antibodies against FOXA1 or control non-specific rabbit immunoglobin and PCR amplification of the enhancer region of CDK1. The results are means±S.E.M. of three independent experiments. aP <0.05 relative to R1881 alone, bP <0.05 relative to control transfected cells with the same treatment.
Although AR occupancy in bicalutamide-treated LNCaP cells was lower relative to R1881-treated cells, down-regulation of HEXIM1 resulted in increased AR occupancy in bicalutamide-treated cells relative to R1881-treated cells (Figure 4A). However, recruitment of R1881-liganded AR was decreased upon down-regulation of HEXIM1, suggesting that, although HEXIM1 attenuated recruitment of bicalutamide-liganded AR, it enhanced recruitment of R1881-liganded AR.
We observed similar recruitment patterns of FOXA1 in a CRPC variant of LNCaP, C4-2 cells, as those observed in LNCaP HEXIM1miR cells (Figure 4D). Increased expression of HEXIM1 inC4-2 cells (Figure 3F) attenuated FOXA1recruitment (Figure 4D). These results suggested that decreased levels of HEXIM1 in CRPC may contribute to dysregulated AR activation or activation of a distinct AR transcriptional program.
HEXIM1 regulates KDM5B expression and recruitment to AR target genes
Although we observed significantly increased recruitment of HEXIM1 to AR-binding sites in the CDK1 or CDC20 genes in the presence of bicalutamide (Figure 5A), the increase was not particularly impressive. We thus searched for other factors involved in HEXIM1 down-regulation of H3K4me2 on AR target genes. We screened known H3K4me2 demethylases for regulation by HEXIM1. KDM5B has been reported to be up-regulated in prostate cancers and to enhance AR transcriptional activity [20]. Conversely, KDM5B was shown to be part of a repressive complex on PR target genes [21]. Progestin-induced displacement of KDM5B and MLL2/MLL3-mediated H3K4 trimethylation during the initial chromatin remodelling events was required for progesterone gene activation [21]. Moreover, an Oncomine™ dataset indicated down-regulation of KDM5B in CRPC (fold change: −2.7, P=6.6×10−5 [17]). It is possible that KDM5B interaction with HEXIM1 resulted in a different function depending on gene context.
Figure 5. HEXIM regulates KDM5B expression.
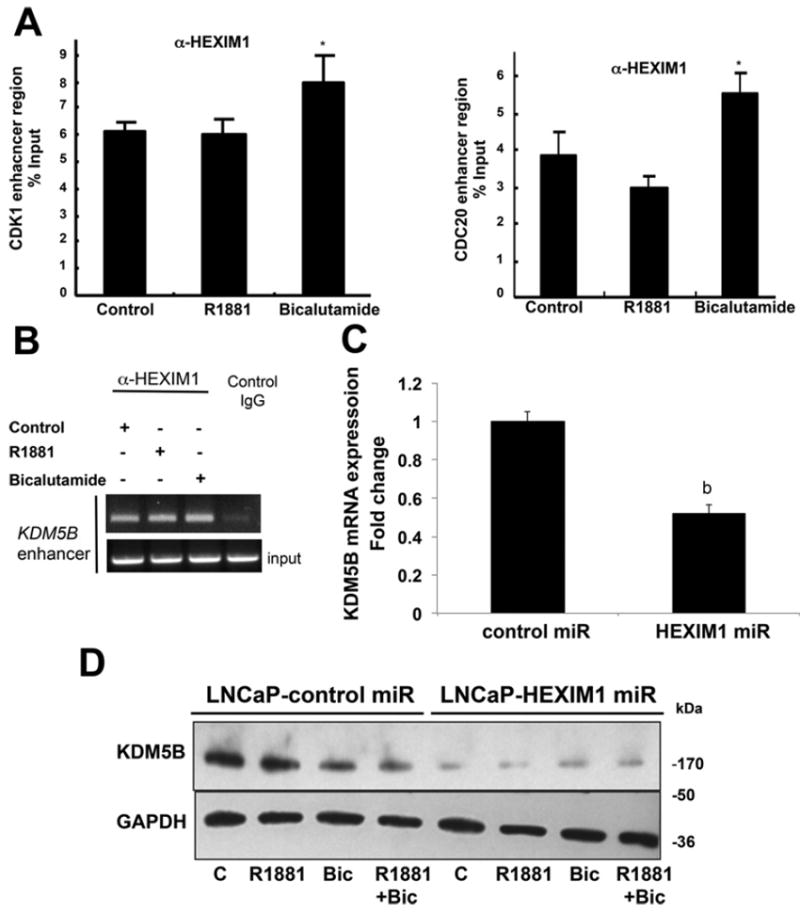
(A) LNCaP cells stably transfected with control or HEXIM1miR were treated with vehicle, 10 nMR1881 or 10 μMbicalutamide (Bic) for 90 min. Results show ChIP analyses of lysates immunoprecipitated with antibodies against HEXIM1 or control non-specific rabbit immunoglobin and PCR amplification of the enhancer regions of CDK1 or CDC20. *P <0.05 relative to vehicle treated cells. (B) ChIP analyses of lysates from LNCaP cells immunoprecipitated with antibodies against HEXIM1 or control non-specific rabbit immunoglobin and PCR amplification of the −73339/−72922 region of KDM5B. (C) RNA was harvested from LNCaP cells stably transfected with control or HEXIM1miR and subjected to RT–PCR to assess KDM5B mRNA levels using GAPDH as control. bP <0.05 relative to control transfected cells with the same treatment. (D) Western blot analyses of endogenous KDM5B relative to GAPDH loading control in LNCaP cells stably transfected with control or HEXIM1miR. Figures are representative of at least three independent experiments. C, control.
Our ChIP-seq analyses and validation by standard ChIP indicated that HEXIM1 is recruited to the enhancer region of the KDM5B gene (Figure 5B). Down-regulation of HEXIM1 resulted in decreased expression of KDM5B (Figures 5C and 5D). These results suggest direct transcriptional regulation of the KDM5B gene by HEXIM1.
We then determined whether KDM5B can modulate H3K4me2 on AR target genes, and the relative role of HEXIM1 in regulating KDM5B. Enforced expression of KDM5B (Figure 6A) resulted in decreased levels of H3K4me2 on the promoter of AR target genes, and down-regulation of HEXIM1 resulted in attenuation of the ability of KDM5B to down-regulate levels of H3K4me2 on AR target genes, but not on the control GAPDH gene (Figure 6B). The basis for the regulation of KDM5B by HEXIM1 was revealed by our observation that down-regulation of HEXIM1 resulted in decreased recruitment of KDM5B (Figure 6C). Thus our studies implicated HEXIM1 not only in the up-regulation of expression of a H3K4me2 demethylase, but also the recruitment of KDM5B to AR target genes as a mechanism for HEXIM1 down-regulation of H3K4 methylation on AR target genes. The regulation of KDM5B recruitment may not just be due to HEXIM1 regulation of KDM5B expression, because co-immunoprecipitation assays indicated that endogenous KDM5B interacts with endogenous HEXIM1 (Figure 6D).
Figure 6. HEXIM1 regulates KDM5B recruitment.
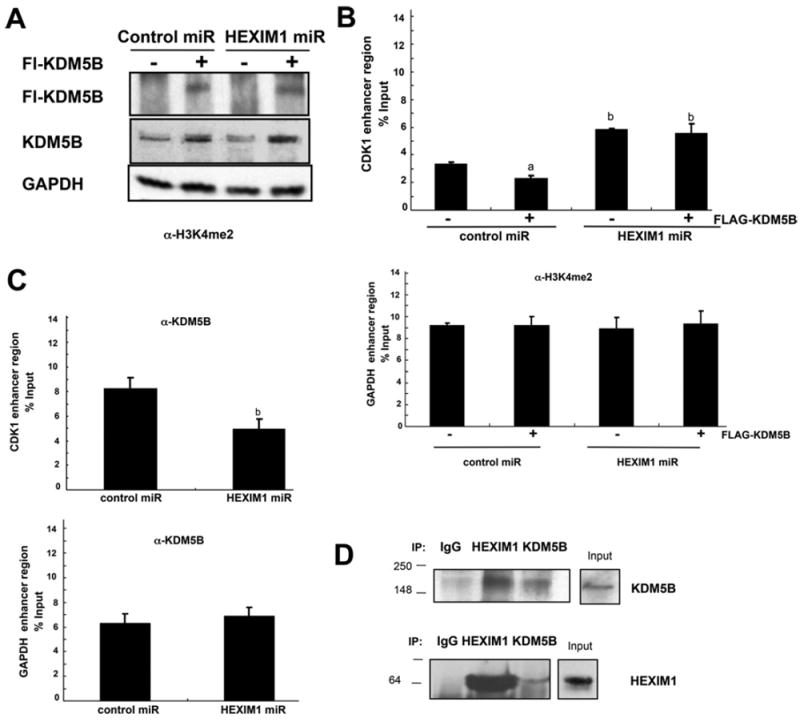
LNCaP cells stably transfected with control or HEXIM1miR were treated with 10 μM bicalutamide for 90 min. (A) Western blot analyses of endogenous KDM5B or FLAG–KDM5B relative to the GAPDH loading control. ChIP analyses of lysates immunoprecipitated with antibodies against (B) H3K4me2 or (C) KDM5B or control non-specific rabbit immunoglobin and PCR amplification of the enhancer regions of CDK1 or GAPDH. The results are means±S.E.M. of three independent experiments. aP <0.05 relative to non-FLAG–KDM5B transfected cells and bP <0.05 relative to control miRNA transfected cells. (D) Lysates from LNCaP cells were immunoprecipitated (IP) using antibodies against HEXIM1 or KDM5B and analysed for co-immunoprecipitation of HEXIM1 or KDM5B by Western blotting. Normal rabbit IgG was used as a specificity control. Input lanes represent 25% of the total protein. Figures are representative of at least three independent experiments.
Modulation of HEXIM1 expression results in altered gene expression in response to R1881 and bicalutamide
We next examined the functional consequences of HEXIM1 expression on AR-dependent gene expression. Down-regulation of HEXIM1 repressed CDK1, CDC20 and UBE2C expression in R1881-treated cells, but enhanced expression of CDC20, CDK1 and UBE2C mRNAs in vehicle- or bicalutamide-treated cells (Figure 7A). The attenuation of R1881-induced gene expression when HEXIM1 expression was down-regulated reflected the down-regulation of AR recruitment to AR target genes upon decreased HEXIM1 expression. Due to the emerging role of UBE2C in CRPC [22], we also examined regulation of UBE2C protein levels by HEXIM1. The effects of HEXIM1 downregulation on the expression of UBE2C mRNA in vehicle- and bicalutamide-treated cells were reflected at the protein level (Figure 7B). Down-regulation of HEXIM1 enhanced expression of PSA mRNA and protein in vehicle-, R1881- or bicalutamide-treated cells.
Figure 7. Altered HEXIM1 expression results in altered cellular response to an anti-androgen.
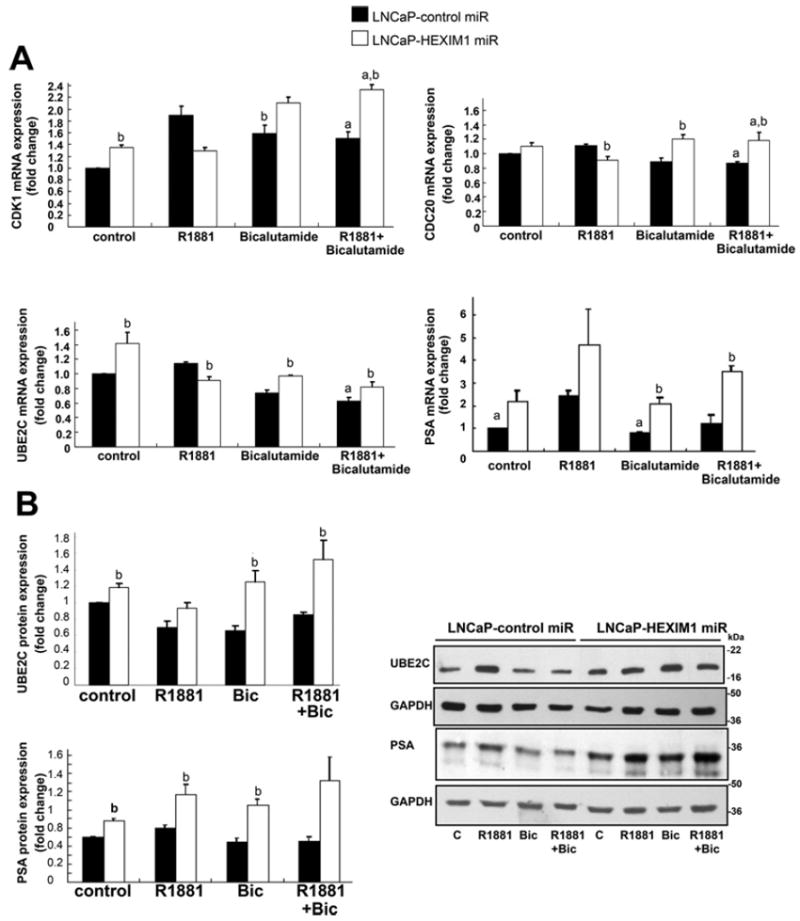
(A) LNCaP cells stably transfected with control or HEXIM1miR were treated with vehicle, 1 nM R1881 and/or 1 μM bicalutamide for 4 h. RNA was harvested and subjected to RT–PCR to assess CDK1, CDC20, UBE2C and PSA mRNA levels using GAPDH as a control. (B) LNCaP cells were stably transfected with control or HEXIM1miR and processed for Western blot analyses of UBE2C and PSA levels. Figures are representative of at least three independent experiments. aP <0.05 relative to R1881 alone and bP <0.05 relative to control transfected cells with the same treatment.
DISCUSSION
Although androgen ablation therapy is initially effective for the majority of men with metastatic prostate cancer, resistance to such treatment invariably develops through mechanisms that remain incompletely understood [23,24]. Most CRPCs express AR and are dependent on AR for growth and survival, through mechanisms reported to involve constitutive activation of AR [25,26]. Comparative gene expression profiling of androgen-regulated gene changes in LNCaP cells with AR-regulated genes in the LNCaP androgen-independent variant, abl, strongly suggest that AR controls the expression of a set of genes in CRPC that is unique from those regulated by androgens in ADPC (androgen-dependent prostate cancer) [7]. The latter study provides compelling support that CPRC represents an alteration in the function of the AR rather than just its constitutive activation.
Although studies on AR co-regulatory factors in prostate cancer are not novel, a role for HEXIM1 in the inhibitory actions of anti-androgens has not been previously reported. We also identified a new mechanism of action of HEXIM1 that is distinct from what we have reported for HEXIM1 regulation of ERα. Our new mechanism involves epigenetic regulation through the H3K4 demethylase KDM5B, that subsequently inhibits FOXA1 licensing activity. On a related note, H3K4me1, H3K4me2 and H3K4me3 levels were reported to be significantly increased in CRPC [27].
Previous studies demonstrated that P-TEFb binds to liganded AR, which recruits P-TEFb to the promoters of androgen target genes to facilitate their transcriptional elongation [28,29]. The co-immunoprecipitation and GST pull-down experiments of the present study indicated that HEXIM1 directly interacts with and prevents the AR from recruiting P-TEFb to AR target genes, similar to the action of HEXIM1 on ER target genes [3,5]. HEXIM1 may repress androgenic responses by inhibiting the release of P-TEFb from the 7SK small nuclear RNA protein complex (snRNP) [30,31], sequestering P-TEFb in a transcriptionally inactive complex that is unable to promote transcriptional elongation of AR target genes through phosphorylation of RNAPII [32,33]. We observed regulation of cyclin T1 recruitment by HEXIM1 on the PSA promoter, and ensuing significant change in PSA mRNA levels. HEXIM1 also regulated H3K4me2 levels and FOXA1 recruitment on AR target genes, CDK1 and CDC20, with ensuing changes in mRNA levels. Thus our data suggest that HEXIM1 regulation of H3K4 methylation and FOXA1 recruitment may be more critical in HEXIM1 regulation of AR target gene expression.
There is an established link between transcriptional elongation and histone methylation [34-38]. H2B monoubiquitination (ubH2B) plays a critical role in H3K4 methylation [39,40]. Conversely, H2B monoubiquitination depends upon the early steps of transcriptional elongation, and CDK9 activity is essential for maintaining global and gene-associated levels of histone H2B monoubiquitination [36]. However, HEXIM1 may act independently of P-TEFb. Results showing P-TEFb-independent actions of HEXIM1 would not be unexpected as our studies indicate that HEXIM1 induced phenotypic effects that are independent of its ability to inhibit P-TEFb [6,14]. In particular, inhibition of ER-mediated activation of VEGF gene transcription by HEXIM1 is independent of the ability of HEXIM1 to inhibit P-TEFb.
It is important to note thatHEXIM1regulation of transcriptional elongation and histone modifications do not necessarily translate to global regulation of gene expression. HEXIM1 does not bind directly to DNA, but is recruited through its interaction with other transcription factors, allowing for its selective regulation. Moreover, the genomic targeting of KDM5B is mediated by sequence-specific DNA binding and by binding to post-translationally modified histones [41].
In addition to its overexpression in breast cancers, KDM5B dysregulation has been reported in several types of solid tumours. It has been reported that, functionally, KDM5B plays an important role in the proliferative capacity of breast cancer cells through repression of tumour suppressor genes, including BRCA1 (breast cancer 1, early onset) [42]. However, KDM5B was shown to be part of a repressive complex on PR (progesterone receptor) target genes [21]. Moreover, KDM5B has putative tumour-suppressive activity, partly due to its ability to bind and stabilize hypophosphorylated pRb, leading to maintenance of pRb-mediated cell-cycle control [43]. In agreement with this, it has been observed that the expression of KDM5B is lost in the majority of advanced and metastatic melanomas [44,45]. Importantly, KDM5B suppresses mammary angiogenesis and metastasis [46]. It is likely that KDM5B-interacting partners, such as HEXIM1, influence its function.
Although we observed overall repression of the recruitment of bicalutamide-liganded AR and associated factors, HEXIM1 enhanced recruitment of R1881-liganded AR and R1881-induced mRNA expression. However, these ligand-specific effects of HEXIM1 did not solely dictate HEXIM1’s effects on cell proliferation. HEXIM1 inhibited R1881-induced proliferation, implicating that the overall effect of HEXIM1 on R1881-induced proliferation can be attributed to regulation of other R1881- regulated genes. Decreased recruitment of R1881-liganded AR upon down-regulation of HEXIM1 (Figure 4A) is reminiscent of the impact of PTEN (phosphatase and tensin homologue deleted on chromosome 10) loss on AR activity. The up-regulation of AR activity by PTEN was attributed to down-regulation of EZH2 expression that resulted in increased dependence on androgens [47]. It has also been reported that the AR induces transcription of genes that promote differentiation (e.g. CDK1 [48]) and inhibits those that promote metastasis [49].
The role of HEXIM1 as a tumour suppressor in prostate cancer was supported by high nuclear HEXIM1 expression in normal prostate tissue and decreased expression of HEXIM1 in BPH and tumours. Further decreased expression was observed during the progression from well-differentiated tumours to poorly differentiated tumours. Further validation was provided by datasets in the Oncomine™ database that indicated decreased expression of HEXIM1 in CRPC relative to hormone-dependent prostate carcinomas. COPA (cancer outlier profile analyses), the algorithm that led to the discovery of TMPRSS2 (transmembrane protease, serine 2) and ETS family gene fusion events in prostate cancer, was recently modified to allow for the identification of down-regulated outliers [50]. As a result HEXIM1 was identified as a potential tumour suppressor in prostate cancer [50]. These sets of data are in sharp contrast with a recent report that HEXIM1 expression is absent in normal prostate, but highly expressed in adenocarcinoma of the prostate [51]. Moreover, there is incongruence between that group’s expression data and animal data showing heterozygosity for HEXIM1 accelerated tumour progression in the TRAMP model of prostate cancer. Although the group reported that heterozygosity for HEXIM1 resulted in increased phosphorylation of the AR, an interaction between the AR and HEXIM1 and the functional consequences of increased AR phosphorylation at Ser81 was not reported. The phosphorylation of AR at Ser81 by CDK9 has been reported previously [52] and the up-regulation of AR phosphorylation in heterozygote HEXIM1 mice was probably through its inhibition of CDK9, rather than direct effects of HEXIM1 on AR transcriptional activity as we reported in the present study. Moreover, there are conflicting reports on the functional relevance of the phosphorylation at Ser81 in AR transcriptional activity, and that perhaps its role in AR transcription is gene-context-dependent [53]. Finally, Mascareno et al. [51] did not report on HEXIM1 regulation of anti-androgenic responses.
Supplementary Material
Acknowledgments
We are grateful to Dr Ralf Janknecht (University of Oklahoma, Norman, OK, U.S.A.) for providing the KDM5B expression vector.
FUNDING
This work was supported by the National Institutes of Health [grant numbers CA092440 (to M.M.M.) and CA134878 (to D.D.)].
Abbreviations
- AR
androgen receptor
- ARE
androgen-response element
- BPH
benign prostatic hyperplasia
- CDK
cyclin-dependent kinase
- CRPC
castration-resistant prostate cancer
- ER
oestrogen receptor
- FOXA1
forkhead box A1
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- H3K4me2
histone 3 dimethylated at Lys4
- HEXIM1
hexamethylene bis-acetamide inducible 1
- IHC
immunohistochemistry
- KDM5B
lysine-specific demethylase 5B
- P-TEFb
positive transcription elongation factor b
- PSA
prostate-specific antigen
- PTEN
phosphatase and tensin homologue deleted on chromosome 10
- RNAPII
RNA polymerase II
- RT
reverse transcription
- UBE2C
ubiquitin-conjugating enzyme E2C
- VEGF
vascular endothelial growth factor
Footnotes
AUTHOR CONTRIBUTION
I-Ju Yeh, Kyung Song, Bryan Wittmann, David Danielpour and Monica Montano designed the research. I-Ju Yeh, Kyung Song, Bryan Wittmann and Xiaodong Bai performed the research. I-Ju Yeh analysed the data. I-Ju Yeh, Kyung Song, Bryan Wittmann, Xiaodong Bai, David Danielpour and Monica Montano wrote the paper.
References
- 1.Dehm SM, Tindall DJ. Androgen receptor structural and functional elements: role and regulation in prostate cancer. Mol Endocrinol. 2007;21:2855–2863. doi: 10.1210/me.2007-0223. [DOI] [PubMed] [Google Scholar]
- 2.Devlin HL, Mudryj M. Progression of prostate cancer: multiple pathways to androgen independence. Cancer Lett. 2009;274:177–186. doi: 10.1016/j.canlet.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Wittmann BM, Fujinaga K, Deng H, Ogba N, Montano MM. The breast cell growth inhibitor, estrogen down regulated gene 1, modulates a novel functional interaction between estrogen receptor alpha and transcriptional elongation factor cyclin T1. Oncogene. 2005;24:5576–5588. doi: 10.1038/sj.onc.1208728. [DOI] [PubMed] [Google Scholar]
- 4.Garriga J, Grana X. Cellular control of gene expression by T-type cyclin/CDK9 complexes. Gene. 2004;337:15–23. doi: 10.1016/j.gene.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Ogba N, Chaplin L, Doughman YQ, Fujinaga K, Montano MM. HEXIM1 regulates E2/ERα-mediated expression of cyclin D1 in mammary cells via modulation of P-TEFb. Cancer Res. 2008;68:7015–7024. doi: 10.1158/0008-5472.CAN-08-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogba N, Doughman YQ, Chaplin LJ, Hu Y, Gargesha M, Watanabe M, Montano MM. HEXIM1 modulates vascular endothelial growth factor expression and function in breast epithelial cells and mammary gland. Oncogene. 2010;29:3639–3649. doi: 10.1038/onc.2010.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thalmann GN, Anezinis PE, Chang SM, Zhau HE, Kim EE, Hopwood VL, Pathak S, von Eschenbach AC, Chung LW. Androgen-independent cancer progression and bone metastasis in the LNCaP model of human prostate cancer. Cancer Res. 1994;54:2577–2581. [PubMed] [Google Scholar]
- 9.Song K, Wang H, Krebs TL, Wang B, Kelley TJ, Danielpour D. DHT selectively reverses Smad3-mediated/TGF-β-induced responses through transcriptional down-regulation of Smad3 in prostate epithelial cells. Mol Endocrinol. 2010;24:2019–2029. doi: 10.1210/me.2010-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montano MM, Ekena K, Delage-Mourroux R, Chang W, Martini P, Katzenellenbogen BS. An estrogen receptor-selective coregulator that potentiates the effectiveness of antiestrogens and represses the activity of estrogens. Proc Natl Acad Sci U S A. 1999;96:6947–6952. doi: 10.1073/pnas.96.12.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wittmann BM, Wang N, Montano MM. Identification of a novel inhibitor of cell growth that is down-regulated by estrogens and decreased in breast tumors. Cancer Res. 2003;63:5151–5158. [PubMed] [Google Scholar]
- 12.Ketchart W, Ogba N, Kresak A, Albert JM, Pink JJ, Montano MM. HEXIM1 is a critical determinant of the response to tamoxifen. Oncogene. 2011;30:3563–3569. doi: 10.1038/onc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ketchart W, Smith KM, Krupka T, Wittmann BM, Hu Y, Rayman PA, Doughman YQ, Albert JM, Bai X, Finke JH, et al. Inhibition of metastasis by HEXIM1 through effects on cell invasion and angiogenesis. Oncogene. 2013;32:3829–3839. doi: 10.1038/onc.2012.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montano MM, Doughman YQ, Deng H, Chaplin L, Yang J, Wang N, Zhou Q, Ward NL, Watanabe M. Mutation of the HEXIM1 gene results in defects during heart and vascular development partly through downregulation of vascular endothelial growth factor. Circ Res. 2008;102:415–422. doi: 10.1161/CIRCRESAHA.107.157859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross RW, Xie W, Regan MM, Pomerantz M, Nakabayashi M, Daskivich TJ, Sartor O, Taplin ME, Kantoff PW, Oh WK. Efficacy of androgen deprivation therapy (ADT) in patients with advanced prostate cancer: association between Gleason score, prostate-specific antigen level, and prior ADT exposure with duration of ADT effect. Cancer. 2008;112:1247–1253. doi: 10.1002/cncr.23304. [DOI] [PubMed] [Google Scholar]
- 16.Shibata Y, Suzuki K, Arai S, Miyoshi Y, Umemoto S, Masumori N, Kamiya N, Ichikawa T, Kitagawa Y, Mizokami A, et al. Impact of pre-treatment prostate tissue androgen content on the prediction of castration-resistant prostate cancer development in patients treated with primary androgen deprivation therapy. Andrology. 2013;1:505–511. doi: 10.1111/j.2047-2927.2013.00068.x. [DOI] [PubMed] [Google Scholar]
- 17.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 18.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, Wei JT, Pienta KJ, Ghosh D, Rubin MA, Chinnaiyan AM. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 20.Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z, Ma Y, Yu Y, Lin H, Chen AP, Chen CD. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc Natl Acad Sci U S A. 2007;104:19226–19231. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vicent GP, Nacht AS, Font-Mateu J, Castellano G, Gaveglia L, Ballare C, Beato M. Four enzymes cooperate to displace histone H1 during the first minute of hormonal gene activation. Genes Dev. 2011;25:845–862. doi: 10.1101/gad.621811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z, Zhang C, Wu D, Chen H, Rorick A, Zhang X, Wang Q. Phospho-MED1-enhanced UBE2C locus looping drives castration-resistant prostate cancer growth. EMBO J. 2011;30:2405–2419. doi: 10.1038/emboj.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cannata DH, Kirschenbaum A, Levine AC. Androgen deprivation therapy as primary treatment for prostate cancer. J Clin Endocrinol Metab. 2012;97:360–365. doi: 10.1210/jc.2011-2353. [DOI] [PubMed] [Google Scholar]
- 24.Kirby M, Hirst C, Crawford ED. Characterising the castration-resistant prostate cancer population: a systematic review. Int J Clin Pract. 2011;65:1180–1192. doi: 10.1111/j.1742-1241.2011.02799.x. [DOI] [PubMed] [Google Scholar]
- 25.Shiota M, Yokomizo A, Fujimoto N, Naito S. Androgen receptor cofactors in prostate cancer: potential therapeutic targets of castration-resistant prostate cancer. Curr Cancer Drug Targets. 2011;11:870–881. doi: 10.2174/156800911796798904. [DOI] [PubMed] [Google Scholar]
- 26.Basu S, Tindall DJ. Androgen action in prostate cancer. Horm Cancer. 2010;1:223–228. doi: 10.1007/s12672-010-0044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellinger J, Kahl P, von der Gathen J, Rogenhofer S, Heukamp LC, Gutgemann I, Walter B, Hofstadter F, Buttner R, Muller SC, et al. Global levels of histone modifications predict prostate cancer recurrence. Prostate. 2010;70:61–69. doi: 10.1002/pros.21038. [DOI] [PubMed] [Google Scholar]
- 28.Lee DK, Chang C. Molecular communication between androgen receptor and general transcription machinery. J Steroid Biochem Mol Biol. 2003;84:41–49. doi: 10.1016/s0960-0760(03)00005-0. [DOI] [PubMed] [Google Scholar]
- 29.Lee DK, Duan HO, Chang C. Androgen receptor interacts with the positive elongation factor P-TEFb and enhances the efficiency of transcriptional elongation. J Biol Chem. 2001;276:9978–9984. doi: 10.1074/jbc.M002285200. [DOI] [PubMed] [Google Scholar]
- 30.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 31.Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, Sedore SC, Price JP, Price DH, Lania L, Bensaude O. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23:2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Byers SA, Price JP, Cooper JJ, Li Q, Price DH. HEXIM2, a HEXIM1-related protein, regulates positive transcription elongation factor b through association with 7SK. J Biol Chem. 2005;180:16360–16367. doi: 10.1074/jbc.M500424200. [DOI] [PubMed] [Google Scholar]
- 33.Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J Biol Chem. 2005;280:28819–28826. doi: 10.1074/jbc.M502712200. [DOI] [PubMed] [Google Scholar]
- 34.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 35.Pekowska A, Benoukraf T, Zacarias-Cabeza J, Belhocine M, Koch F, Holota H, Imbert J, Andrau JC, Ferrier P, Spicuglia S. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011;30:4198–4210. doi: 10.1038/emboj.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pirngruber J, Shchebet A, Schreiber L, Shema E, Minsky N, Chapman RD, Eick D, Aylon Y, Oren M, Johnsen SA. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3′-end processing. EMBO Rep. 2009;10:894–900. doi: 10.1038/embor.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pirngruber J, Shchebet A, Johnsen SA. Insights into the function of the human P-TEFb component CDK9 in the regulation of chromatin modifications and co-transcriptional mRNA processing. Cell Cycle. 2009;8:3636–3642. doi: 10.4161/cc.8.22.9890. [DOI] [PubMed] [Google Scholar]
- 38.Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138:1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 39.Chandrasekharan MB, Huang F, Sun ZW. Histone H2B ubiquitination and beyond: regulation of nucleosome stability, chromatin dynamics and the trans-histone H3 methylation. Epigenetics. 2010;5:460–468. doi: 10.4161/epi.5.6.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakanishi S, Lee JS, Gardner KE, Gardner JM, Takahashi YH, Chandrasekharan MB, Sun ZW, Osley MA, Strahl BD, Jaspersen SL, Shilatifard A. Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J Cell Biol. 2009;186:371–377. doi: 10.1083/jcb.200906005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scibetta AG, Santangelo S, Coleman J, Hall D, Chaplin T, Copier J, Catchpole S, Burchell J, Taylor-Papadimitriou J. Functional analysis of the transcription repressor PLU-1/JARID1B. Mol Cell Biol. 2007;27:7220–7235. doi: 10.1128/MCB.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Roesch A, Becker B, Schneider-Brachert W, Hagen I, Landthaler M, Vogt T. Re-expression of the retinoblastoma-binding protein 2-homolog 1 reveals tumor-suppressive functions in highly metastatic melanoma cells. J Invest Dermatol. 2006;126:1850–1859. doi: 10.1038/sj.jid.5700324. [DOI] [PubMed] [Google Scholar]
- 44.Roesch A, Becker B, Meyer S, Wild P, Hafner C, Landthaler M, Vogt T. Retinoblastoma-binding protein 2-homolog 1: a retinoblastoma-binding protein downregulated in malignant melanomas. Mod Pathol. 2005;18:1249–1257. doi: 10.1038/modpathol.3800413. [DOI] [PubMed] [Google Scholar]
- 45.Roesch A, Mueller AM, Stempfl T, Moehle C, Landthaler M, Vogt T. RBP2-H1/JARID1B is a transcriptional regulator with a tumor suppressive potential in melanoma cells. Int J Cancer. 2008;122:1047–1057. doi: 10.1002/ijc.23211. [DOI] [PubMed] [Google Scholar]
- 46.Li Q, Shi L, Gui B, Yu W, Wang J, Zhang D, Han X, Yao Z, Shang Y. Binding of the JmjC demethylase JARID1B to LSD1/NuRD suppresses angiogenesis and metastasis in breast cancer cells by repressing chemokine CCL14. Cancer Res. 2011;71:6899–6908. doi: 10.1158/0008-5472.CAN-11-1523. [DOI] [PubMed] [Google Scholar]
- 47.Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, Wang S, Plaisier S, Garraway IP, Huang J, Graeber TG, Wu H. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell. 2011;19:792–804. doi: 10.1016/j.ccr.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B, Yang CC, Yang JY, Lin CY, Lai CC, Hung MC. CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat Cell Biol. 2011;13:87–94. doi: 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chng KR, Chang CW, Tan SK, Yang C, Hong SZ, Sng NY, Cheung E. A transcriptional repressor co-regulatory network governing androgen response in prostate cancers. EMBO J. 2012;31:2810–2823. doi: 10.1038/emboj.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang C, Taciroglu A, Maetschke SR, Nelson CC, Ragan MA, Davis MJ. mCOPA: analysis of heterogeneous features in cancer expression data. J Clin Bioinforma. 2012;2:22. doi: 10.1186/2043-9113-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mascareno EJ, Belashov I, Siddiqui MA, Liu F, Dhar-Mascareno M. Hexim-1 modulates androgen receptor and the TGF-β signaling during the progression of prostate cancer. Prostate. 2012;72:1035–1044. doi: 10.1002/pros.21510. [DOI] [PubMed] [Google Scholar]
- 52.Gordon V, Bhadel S, Wunderlich W, Zhang J, Ficarro SB, Mollah SA, Shabanowitz J, Hunt DF, Xenarios I, Hahn WC, et al. CDK9 regulates AR promoter selectivity and cell growth through serine 81 phosphorylation. Mol Endocrinol. 2010;24:2267–2280. doi: 10.1210/me.2010-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gioeli D, Ficarro SB, Kwiek JJ, Aaronson D, Hancock M, Catling AD, White FM, Christian RE, Settlage RE, Shabanowitz J, et al. Androgen receptor phosphorylation. Regulation and identification of the phosphorylation sites. J Biol Chem. 2002;277:29304–29314. doi: 10.1074/jbc.M204131200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


