Abstract
The lack of therapies for neurodegenerative diseases arises from our incomplete understanding of their underlying cellular toxicities and the limited number of predictive model systems. It is critical that we develop approaches to identify novel targets and lead compounds. Here, a phenotypic screen of yeast proteinopathy models identified dihydropyrimidine-thiones (DHPM-thiones) that selectively rescued the toxicity caused by β-amyloid (Aβ), the peptide implicated in Alzheimer’s disease. Rescue of Aβ toxicity by DHPM-thiones occurred through a metal-dependent mechanism of action. The bioactivity was distinct, however, from that of the 8-hydroxyquinoline clioquinol (CQ). These structurally dissimilar compounds strongly synergized at concentrations otherwise not competent to reduce toxicity. Cotreatment ameliorated Aβ toxicity by reducing Aβ levels and restoring functional vesicle trafficking. Notably, these low doses significantly reduced deleterious off-target effects caused by CQ on mitochondria at higher concentrations. Both single and combinatorial treatments also reduced death of neurons expressing Aβ in a nematode, indicating that DHPM-thiones target a conserved protective mechanism. Furthermore, this conserved activity suggests that expression of the Aβ peptide causes similar cellular pathologies from yeast to neurons. Our identification of a new cytoprotective scaffold that requires metal-binding underscores the critical role of metal phenomenology in mediating Aβ toxicity. Additionally, our findings demonstrate the valuable potential of synergistic compounds to enhance on-target activities, while mitigating deleterious off-target effects. The identification and prosecution of synergistic compounds could prove useful for developing AD therapeutics where combination therapies may be required to antagonize diverse pathologies.
Keywords: Alzheimer’s disease, β-amyloid, metal homeostasis, dihydropyrimidine-thione, clioquinol, phenotypic screen, yeast, C. elegans
Graphical Abstract

INTRODUCTION
Protein misfolding causes a diverse set of neurodegenerative diseases (ND) that exact a tremendous personal and socioeconomic burden. Each disease is characterized by the stereotypic insult of a disease-specific protein on distinct neuronal subpopulations. The dysfunction and death of neurons manifest in diverse clinical pathologies, ranging from motor defects to dementia. Our lack of effective disease-modifying therapies for these disorders has obvious and significant ramifications as the number of aged individuals continues to increase.
To elucidate the relationship between neurotoxic proteins and cell pathology, we and others have exploited the unique advantages of budding yeast. Yeast harbor nearly all the same conserved cellular pathways as human neurons, including vesicle trafficking, mitochondrial function, lipid biology, and most aspects of protein homeostasis.1–3 Moreover, the genetic tractability of yeast and its ease of growth enable unparalleled genetic and chemical screens in defined, genetically homogeneous cell populations. Several disease proteins, including α-synuclein (α-syn, Parkinson’s disease, PD), amyloid-β peptide (Aβ, Alzheimer’s disease, AD), and TDP-43 (amyotrophic lateral sclerosis/ALS and frontotemporal lobar degeneration/FTLD) elicit specific perturbations to cellular pathways in yeast that lead to overt cytotoxicity.4–6
Using the yeast Aβ model, critical links have been established between cellular pathologies caused by intracellular Aβ and characteristics specific to the human disease. In humans, Aβ is produced throughout life as a cleavage product from the amyloid precursor protein (APP), and, in AD patients, it accumulates in the form of extracellular amyloid plaques.7,8 Although these plaques are conspicuous disease hallmarks, healthy, nondemented individuals also exhibit Aβ plaque burden,9 suggesting that it is actually smaller assemblies of Aβ oligomers that poison neuronal function and cause disease. In the yeast model, a signal sequence is added to the Aβ expression construct so that the Aβ peptide is cotranslationally imported into endoplasmic reticulum (ER). The signal sequence is then cleaved and native Aβ forms oligomers within the secretory and endosomal compartment that elicit cellular toxicity. Importantly, the yeast model recapitulates the enhanced pathogenicity and oligomer formation of the Aβ1–42 peptide (as compared to Aβ1–40) observed in AD.10,11 Related yeast models have used a non-native Aβ-GFP fusion protein expressed directly in the cytoplasm, where it causes cellular toxicity and aggregates.12
The robust growth inhibition caused by secretory Aβ expression in yeast enabled two unbiased phenotypic screens for modulators of Aβ toxicity, one genetic4 and one chemical.13 From the genetic screen, we identified several genes that function in endocytosis, suggesting that Aβ poisons this process. Indeed, endosomal transport was largely abrogated in Aβ-expressing yeast. Even more strikingly, multiple genetic modifiers were shown to be yeast homologues of, or to interact with, known human AD risk alleles.14–18 One example, PICALM, is one of the most highly validated risk factors for AD, aside from APOe4.14,19 A conserved mechanism of toxicity with relevance to neurons was further supported by the demonstration that homologues of the yeast genetic modifiers reversed Aβ toxicity in both nematode and mammalian neurons. A genetic screen of an Aβ-GFP model revealed related pathways, where phospholipid metabolism was implicated in mediating toxicity.20 Thus, unbiased screens using the yeast Aβ model can identify genes that are directly related to the human disease.
A large-scale Aβ chemical screen of the yeast Aβ model was used to identify compounds that restored growth with no preconception of potential protective pathways or targets.13 Once uncovered, the cytoprotective small molecule itself can be used to reveal underlying pathological mechanisms and potential drug targets.21 For example, our chemical screen hits included several 8-hydroxyquinolines, a class of metal-binding compounds previously shown to robustly antagonize Aβ pathology in mouse models of Aβ toxicity. Strikingly, this scaffold has been tested through phase II clinical trials22–25 and, in yeast, operates through a metal-dependent stimulation of Aβ degradation in yeast.13 This work indicates that the role of metals in Aβ aggregation in AD is recapitulated in the Aβ yeast model.26,27 Specifically, it appears that the aggregation-promoting interaction of the Aβ peptide with metals can be targeted with metal-binding compounds to promote peptide degradation and cellular rescue. Other mechanisms of action targeting misfolded Aβ oligomers can likely be targeted as well. Dimebon, for example, enhances autophagy and reduces intracellular levels of GFP-Aβ42 in yeast.28 And an Aβ oligomerization yeast model has identified small molecules that target toxic Aβ structures directly.29 Thus, multiple different models have revealed a path to identifying cytoprotective compounds targeting key aspects of Aβ cellular pathology that provide starting points for lead compound discovery.
The ability to robustly identify new chemical matter that suppresses a cellular growth defect provides a powerful and unbiased approach to elucidate relevant druggable pathways. Here, we expand on our previous screens by reporting screening results from a small, yet diverse, chemical library. One series of compounds, dihydropyrimidine-thiones (DHPM-thiones), protected cells against Aβ toxicity through a metal-dependent mechanism and strongly synergized with a known metal-binding compound in both yeast and nematode models. The ability to identify and evaluate synergy between compounds may prove powerful given that treating human neurodegenerative disease may require complementary treatment modalities.
RESULTS AND DISCUSSION
Phenotypic Small Molecule Screen Identifies Cytoprotective DHPM-Thiones
To identify new compounds and potential drug targets using the Aβ yeast model, we expanded the chemical space that we had previously surveyed by screening a small chemical library of novel chemistry (assembled by the Boston University Center for Molecular Discovery; BU-CMD). We screened the secretory Aβ model against ~3000 compounds and monitored growth using optical density at 24 and 48 h after inducing toxic protein expression with galactose and administering screening compounds. Hits were identified by calculating Z-scores relative to the plate mean and using a 3σ cutoff.
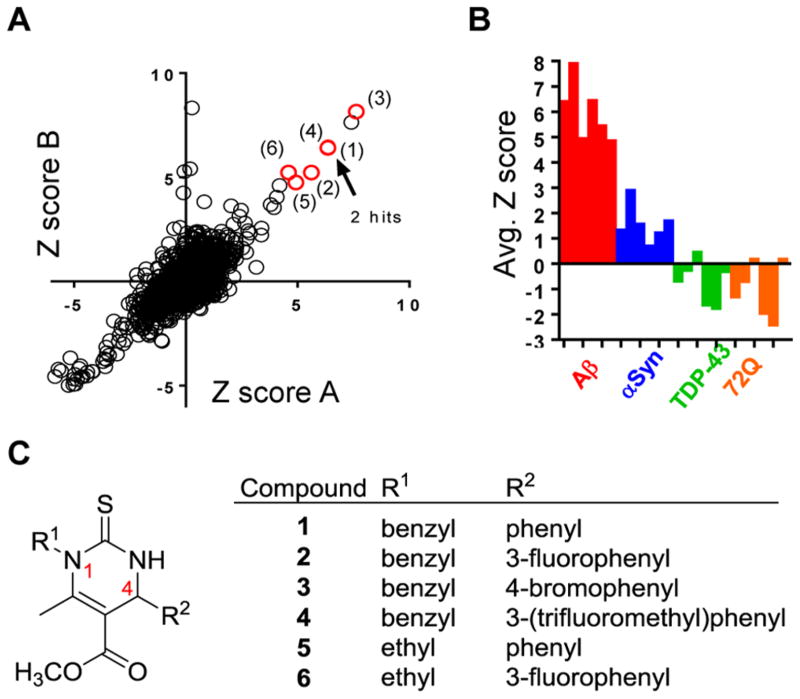
This screen identified a small number of compounds that partially rescued cells from Aβ toxicity (Figure 1A). These initial hits were selective for Aβ toxicity, as yeast models expressing α–synuclein, TDP-43, and htt72Q were not rescued by the hits in parallel analyses (Figure 1B). This selectivity profile suggests that Aβ elicits cellular pathologies distinct from those of the other models, and that the target of this hit series specifically ameliorates this pathology.4–6,30
Figure 1.
Dihydropyrimidine-thiones protect yeast cells against Aβ toxicity. (A) Z-Scores of a small molecule screen of the BU-CMD compound collection against our yeast Aβ toxicity model. The screen was performed in duplicate (replicates A and B) with the six DHPM-thione hits indicated by red circles and numbers. Hits (1) and (4) have overlapping Z-scores. (B) Compound plates were also screened against other yeast toxicity models, including α-syn, TDP-43, and polyglutamine (72Q). Z-Scores under 3.0 for six DHPM-thione hits indicate that the series was selective for the Aβ model. (C) General structure of DHPM-thione hits with table indicating R1 and R2 group substituents.
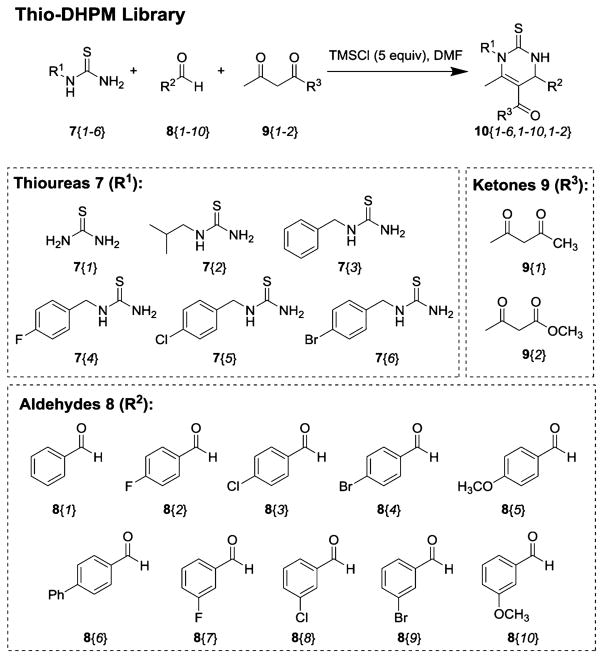
Analysis showed that the chemical structure of the six hit compounds (1–6) comprised a dihydropyrimidine-thione scaffold (DHPM-thiones; Figure 1C). These heterocyclic compounds had either ethyl or benzyl substituents at N1 (R1 indicated in Figure 1C) and a range of substituted phenyl rings at C4. This particular scaffold has been extensively studied based on its robust, accessible synthesis via the Bignelli reaction, a three-component, one-pot reaction that is amenable to library-type synthesis. We took advantage of the flexible nature of DHPM-thione synthesis to construct 82 additional analogues (10) (Figure 2 and Supporting Information Table S1, Methods S1) for further evaluation of structure–activity relationships (SAR). We incorporated various alkyl and benzylic substituents at R1, alterations in phenyl ring substitution at R2, and either methyl esters or methyl ketones at the 5-position (only the methyl ester is indicated in Figure 1C). The resulting DHPM-library was then evaluated for efficacy against Aβ toxicity in yeast.
Figure 2.
DHPM-thione library generation. A schematic of the general three-component, one-pot Biginelli reaction is shown. The 82 compound member library was generated using this reaction and combinations of the three components, including (7) thioureas (R1), (8) aldehydes (R2), and (9) ketones (R3).
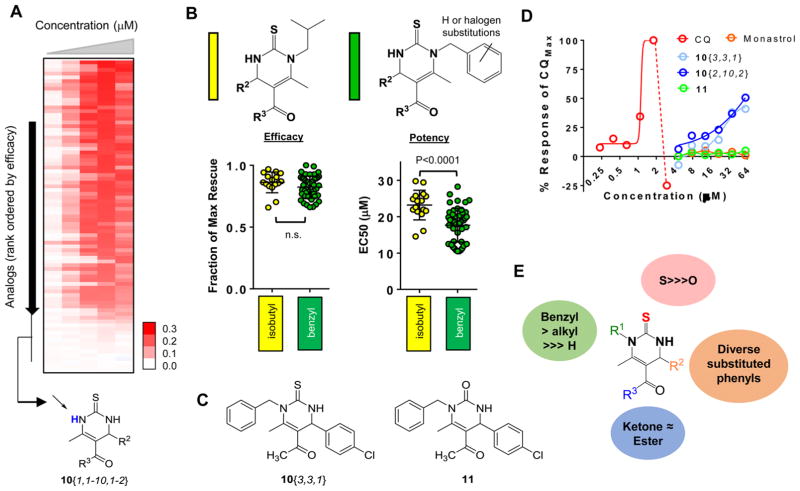
Dose-response curves of each analogue using the Aβ yeast strain revealed that many compounds retained significant efficacy (Figure 3A). All compounds synthesized lacking an R1 substituent (derived from thiourea 7{1}) were inactive, indicating a strict requirement for substitution at this position. Within R1-substituted analogues, we compared the isobutyl and substituted benzyl analogues. Active compounds within each of these groups achieved comparable levels of Aβ rescue (Figure 3B, left plot). Substituted benzyl derivatives, however, exhibited overall increased potency as compared to isobutyl analogues (P < 0.0001; Figure 3B, right plot). While aromatic halides contribute to increased hydrophobicity (Supporting Information Table S1), it is likely that these structural changes enable the compounds to more efficiently engage a target. For example, if these compounds bind a hydrophobic pocket, increasing this character with an associated increase in target affinity may improve potency. We also evaluated the impact of R3 as a ketone or an ester within the context of isobutyl or benzyl R1 substituents. These two modifications did not significantly impact efficacy or potency of either subseries of DHPM-thione (Supporting Information Figure 1); thus, this position appeared less important for target engagement.
Figure 3.
Structure–activity relationships (SAR) of DHPM-thione series. (A) Heat map representation of the dose–response curves for 82 DHPM-thiones. Compounds are sorted from highest efficacy to lowest efficacy. Red indicates rescue (OD600 values), white indicates no rescue. Doses of compounds increased from left to right and range from 2 to 40 μM. The inactive cluster of R1-unsubstituted DHPM-thiones is indicated at the bottom of the heat map. (B) General structures of isobutyl and substituted/unsubstituted benzyl DHPM-thione analogues indicated in yellow and green, respectively, in the following plots. The left plot is efficacy and right plot is potency of all analogues falling within that substitution pattern. The y-axis for efficacy is fraction of maximal rescue by the series and the y-axis for potency is EC50 (μM). The indicated P-values were calculated according using a Student’s t test. (C) Structures of the thiourea analogue, 10{3,3,1}, and the urea variant, 11. (D) Dose–response curves of 10{3,3,1}, 11, CQ, and monastrol. X-axis is concentration (μM) on a log2 scale, and y-axis is % of the maximal CQ response. (E) Summary of SAR for R1, R2, R3, and S/O urea. While significant tolerability existed within the R1 and R2 groups, a benzylic substitution at the R1 group was preferred. The thiourea was required, as was at least an alkyl group at R1. Both ketone and ester groups were well-tolerated at R3.
One of the more potent compounds from this series was 10{3,3,1} (Figure 3C), an N-benzyl, methyl ketone DHPM-thione with 4-chlorophenyl substitution at R2. To examine whether the thiourea function was important to activity, we made the corresponding urea substitution (11, Figure 3C). Along with 10{3,3,1} and 11, we also tested monastrol (a characterized DHPM-thione that targets kinesin-531) and clioquinol (CQ), an 8-hydroxyquinoline with demonstrated efficacy in both yeast and neuronal models of Aβ toxicity.13,22,23 Neither the oxygenated analogue 11 nor monastrol rescued Aβ toxicity (Figure 3D). These results indicated the requirement for a thiourea and were consistent with our earlier observations that R1-unsubstituted DHPM-thiones did not exhibit rescue. In addition to the thiourea and R1 substitutions, diverse substituted phenyl R2 substituents were tolerated, as were ketone or ester derivatives. Thus, while multiple substitutions were at least partially tolerated, the range of effects on potency and efficacy suggest that this series engages a protein target. These data are summarized in Figure 3E.
DHPM-Thiones Protect against Aβ Toxicity in a Nematode Model
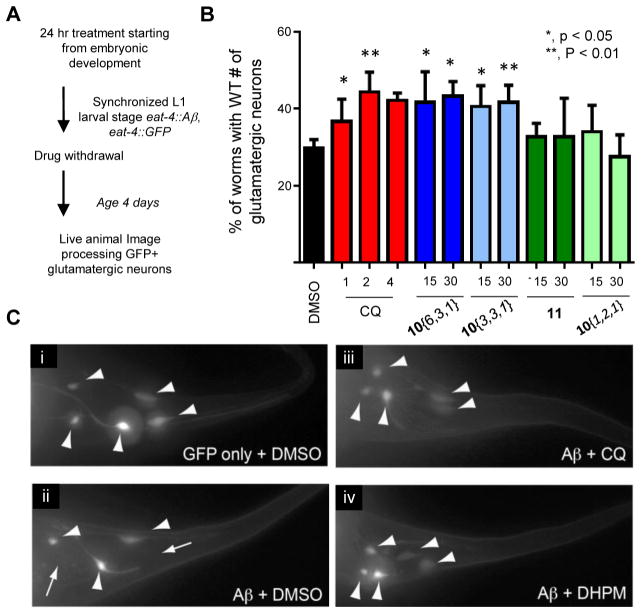
While the yeast advantages of genetic tractability and ease of growth are clear, they of course, lack the cell-to-cell contacts and communication inherent to a functioning nervous system.32 We therefore used a nematode model to test if DHPM-thiones could ameliorate Aβ toxicity in a fully intact nervous system. In our Caenorhabditis elegans model, Aβ is targeted to the secretory pathway in the same manner as in our yeast model.4 Using a glutamatergic neuron-specific promoter, we monitored Aβ proteotoxicity in six tail glutamatergic neurons in the nematode and scored the number of viable neurons over the course of 5 days. Importantly, the progressive neurotoxicity elicited by Aβ in these neurons is attenuated by the same genetic and small molecule modulators that were identified in unbiased yeast screens,33 indicating a conserved mechanism of Aβ toxicity from yeast to neurons.
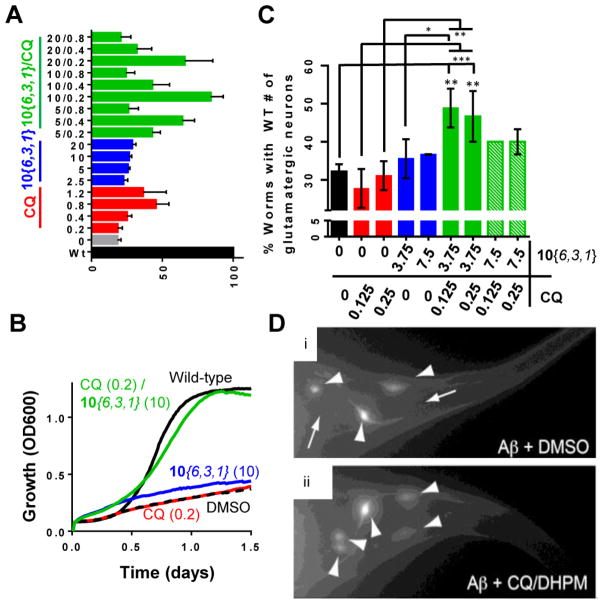
Early larval stage worms were treated with compound for 24 h, after which, worms were allowed to develop and age in the absence of compound for 4 days (Figure 4A). Toxicity was then assessed by counting the remaining GFP-positive glutamatergic neurons. Consistent with previous results,13,34 CQ increased the survival of Aβ-expressing glutamatergic neurons (Figure 4B, red). Two active DHPM-thiones (10{6,3,1} and 10{3,3,1}) partially reduced Aβ toxicity at 15 and 30 μM (Figure 4B, blue), while inactive compounds 10{1,2,1} and 11 failed to confer protection (Figure 4B, green). Representative fluorescent microscopy images of individual worms treated with solvent DMSO alone or compounds shows the position and viability of Aβ-expressing neurons (Figure 4C). The efficacy of the DHPM-thiones in yeast, as well as neurons, suggests that both the mechanism of Aβ toxicity and the mechanism of DHPM-thione rescue are fundamentally conserved across significant evolutionary distance.
Figure 4.
DHPM-thiones rescue nematode model of Aβ toxicity. (A) Schematic of the compound treatment regimen for nematode Aβ model. (B) Percent of worms with wild-type number of glutamatergic neurons labeled with eat-4::GFP. Three independent experiments were performed for each compound (concentration tested in μM) where 30 worms were scored in each trial (for a total of 90 worms). P-Values comparing DMSO to each compound were calculated using a one-way ANOVA and Tukey’s test. (C) Representative fluorescent images of GFP-labeled tail-region glutamatergic neurons in the Aβ nematode model for GFP only (i), Aβ plus DMSO (ii), Aβ plus CQ (iii), and Aβ plus compound 10{6,3,1} (iv). Large arrowheads indicate neurons. Small arrows with a line indicate a missing neuron.
DHPM-Thiones Impinge on Metal Homeostasis
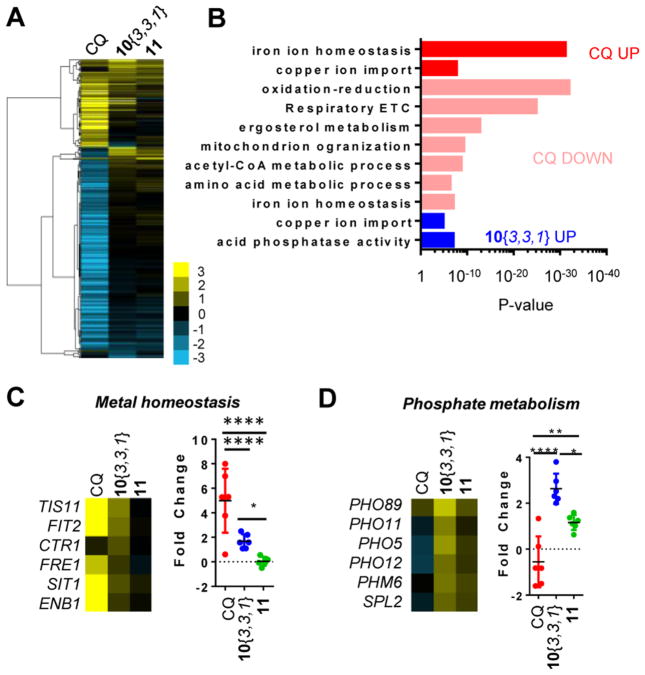
To begin our investigation into the mechanism of action (MOA) for the cytoprotective DHPM-thiones, we assessed changes in gene expression that were specific to the active compound 10{3,3,1} as compared to both the oxygenated analogue 11 and CQ. Wild-type yeast were treated with CQ, 10{3,3,1}, or 11 at each compound’s effective dose for 6 h, and then the cells were harvested for mRNA sequencing analysis.
Wild-type cells treated with each compound yielded distinct gene expression profiles, with CQ most dramatically inducing changes in mRNA abundance (Figure 5A, Table S2). As anticipated based on the known metal-binding capacity of CQ, gene ontology analysis of CQ-treated cultures revealed a clear metal-depletion signature as transcripts involved in copper and iron import increased in abundance, while those encoding proteins functioning in mitochondria (including electron transport chain, ETC) and metabolism (e.g., TCA cycle) dramatically decreased in abundance (Figure 5B). Interestingly, the active analogue 10{3,3,1} specifically increased metal-related genes compared to the inactive analogue 11 (Figure 5A and B, Table S2), although the magnitude of change was significantly greater for CQ than 10{3,3,1}. Importantly, as compared to 10{3,3,1}, metal-responsive genes remained unchanged upon treatment with the urea analogue, 11 (Figure 5C). Evaluation of a subset of genes from the RNA-seq data set reveals this more clearly (Figure 5C and D). Comparison of the entire group of metal-responsive genes revealed a highly significant difference between the active and inactive analogue (Figure 5C, right panel).
Figure 5.
DHPM-thiones bind copper and alter cellular metal ion homeostasis. (A) Heat map of gene expression changes in wild-type yeast cells treated with CQ, 10{3,3,1}, or 11. Cells were treated with compound and RNA levels analyzed by RNA-Seq relative to DMSO vehicle control. Thresholds were set at a fold change of 2 and a P-value of 10−5 for at least one of the treatments. Scale indicates log2 fold change. (B) Gene ontology enrichment for each compound treatment. P-Values are indicated on the x-axis. Changes are color-coded according to the indicated treatment and direction of effect. (C) Expression data for select metal homeostasis genes were extracted from the larger data set and analyzed as a group. Scale is as in (A). One-way ANOVA with Tukey’s statistical tests revealed highly significant P-values between groups where gene expression changed in response to compound treatment (*P < 0.05; ***P < 0.0001). CTR1 was removed from the statistical analysis for CQ because it was not significantly altered. (D) Expression data for select phosphate metabolism genes were extracted from the larger data set and analyzed as a group. Scale is as in (A). One-way ANOVA and Tukey’s statistical tests revealed highly significant p-values between groups (*P < 0.05; ***P < 0.001; ****P < 0.0001).
Phosphate metabolism genes were also induced by 10{3,3,1} (Figure 5B and D). However, the inactive 11 also induced this gene set, albeit to a lesser extent (Figure 5D). This data was insufficient to discern whether the metal or phosphate signatures were relevant to Aβ rescue. Regardless, it is likely that the signatures most strongly correlating with Aβ rescue were relevant to on-target activity. In this case, this suggested an interaction with metal homeostasis may be most related to Aβ rescue. After obtaining these results, we further confirmed that 10{6,3,1} and 10{3,3,1} interacted with cellular metal homeostasis: compromising cellular metal levels either through genetic or pharmacological means led to an enhanced sensitivity of cells to both CQ and DHPM-thiones, while the unrelated Rsp5-activating compounds (NAB2)21 had no effect (Supporting Information Figure 2).
DHPM-Thiones Rescue Aβ Toxicity in a Metal-Dependent Manner
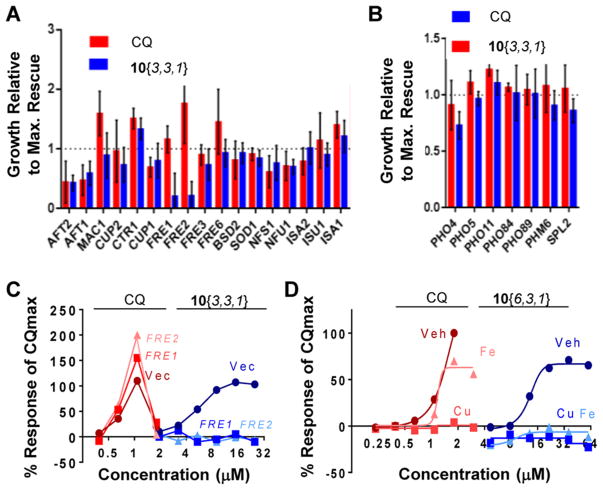
Both metal and phosphate metabolism genes were altered in response to 10{3,3,1} (Figure 5C and D). We therefore tested whether modifying these pathways influenced rescue by either CQ or 10{3,3,1}. A set of genes encompassing both metal homeostasis and phosphate metabolism functions were evaluated for modulatory effects on rescue by CQ and 10{3,3,1} (Figure 6A and B). Expression of iron-responsive transcription factors AFT1 and AFT2 reduced rescue by both CQ and 10{3,3,1}. However, this was due to overall reduced growth of the Aβ strain caused by AFT1/AFT2 overexpression (Supporting Information Figure 3). Two genes, FRE1 and FRE2, specifically mitigated rescue by 10{3,3,1} as compared to CQ (Figure 6A), without having any effects on Aβ growth on their own (Supporting Information Figure 3). The products of these two genes are the major cell surface copper and iron reductases that reduce Cu(II) and Fe(III) to the bioavailable Cu(I) and Fe(II) forms.35,36 FRE1/FRE2 overexpression may increase intracellular copper and/or iron levels in a manner that antagonized the protective effect of 10{3,3,1} on Aβ toxicity.
Figure 6.
Phenotypic confirmation of a metal-dependent activity for DHPM-thiones and CQ. (A, B) Rescue of Aβ by CQ or 10{3,3,1} was examined in the presence of overexpression plasmids functioning in (A) metal homeostasis or (B) phosphate metabolism. Maximal rescue is reported relative to the response of the maximal rescue of the vector-only control. Three independent experiments were performed. (C) Dose–response curves for CQ and 10{3,3,1} in Aβ strains harboring empty vector or FRE1/FRE2 overexpression plasmids. X-Axis is concentration (μM). Y-Axis is the response of compound as a percent of the maximal rescue afforded by CQ. (D) Rescue of Aβ toxicity by both CQ and 10{6,3,1} was reversed by exogenous copper (blue) and iron (orange). X-Axis is log2 concentration (μM). Y-Axis is the response of compound as a percent of the maximal rescue afforded by CQ.
In contrast to metal-related genes, overexpression of phosphate metabolism genes had no effect on rescue by neither CQ nor 10{3,3,1} (Figure 6B). A trend toward an effect by PHO4 overexpression on 10{3,3,1} was not statistically significant. We therefore posited that the metal homeostasis signature was more relevant in ameliorating Aβ toxicity than the phosphate metabolism genes. Full dose–response curves show that the reversal of Aβ rescue conferred by FRE1/FRE2 overexpression was distinct between CQ and 10{3,3,1}, indicating these two compounds likely work through different mechanisms of action (Figure 6C). To reconcile the partial effect of 11 on phosphate metabolism genes, it is possible that there are multiple targets of this scaffold. Some targets are shared between the active and inactive analogues independent of the urea or thiourea moiety, but only the metal-dependent target mediates rescue of Aβ toxicity. Alternatively, the inactive compound may inefficiently engage the target, which was sufficient to elicit a partial response that was insufficient to elicit the metal response or rescue Aβ toxicity.
To further examine whether the rescue of Aβ toxicity was due to a metal-dependent target, we added exogenous copper or iron to determine if they could compete with the compound for the target. As shown previously,13 copper prevented rescue by CQ, while iron shifted the dose–response and reduced toxicity at higher concentrations (Figure 6D). Consistent with the FRE1/FRE2 overexpression result, exogenous copper and iron fully blocked rescue by 10{6,3,1} (Figure 6D). These results further corroborate the gene expression data and overexpression studies, indicating the rescue by DHPM-thiones likely operates through a target that binds to copper or iron.
DHPM-Thiones Directly Bind Copper Ions
Thus far, data indicate that rescue of Aβ by both CQ and DHPM-thiones operate through a mechanism that operates through metal homeostasis or a specific metalloprotein. There are two possible mechanisms by which DHPM-thiones could elicit these phenotypes. Either DHPM-thiones rescue Aβ through a metal-dependent process that can be antagonized by genetic or pharmacological alterations in metal bioavailability (Figure 6C and D), or DHPM-thiones directly bind metal–either freely or as part of metalloprotein–to elicit a protective response.
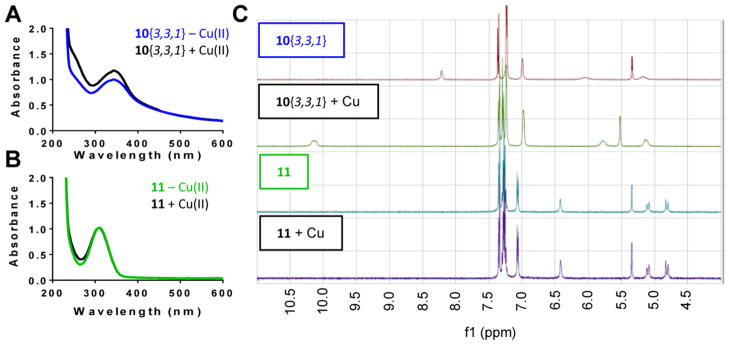
To test whether DHPM-thiones bind metal directly, we first used UV/vis absorbance profiles to monitor copper. As expected, the addition of Cu(II) specifically altered the UV/vis spectra of CQ (Supporting Information Figure 4). For 10{3,3,1}, there was a modest change to the absorbance profile when treated with Cu(II) (Figure 7A); however, 11 exhibited no change in the UV/vis absorbance profile (Figure 7B), suggesting the difference in absorbance was due to Cu(II) binding. To further substantiate this result, we used nuclear magnetic resonance (NMR) with 10{3,3,1} and 11 in combination with Cu(I), as Cu(II) is incompatible with NMR. Chemical shifts indicative of metal binding were detected with 10{3,3,1} in the presence of copper (Figure 7C, top). In contrast, 11 showed no evidence of metal binding (Figure 7C, bottom), consistent with the UV/vis result. Together with the genetic modulation (Figure 6C) and metal competition (Figure 6D) experiments, these data strongly suggest that DHPM-thione rescue of Aβ toxicity occurs through a mechanism and target whose activity is impacted by metal binding.
Figure 7.
DHPM-thiones bind copper directly. (A, B) UV/vis absorbance scan of (A) 10{3,3,1} and (B) 11 in the presence or absence of equimolar copper(II). Y-Axis is absorbance, and x-axis is wavelength (nM). Scans were performed independently three times. (C) NMR spectra of 10{3,3,1} and 11 in the presence or absence of Cu(I). X-Axis is proton chemical shift (ppm).
Although not as prevalent as for 8-hydroxyquinoline metal ligands, thioureas can complex with metals to target metalloproteins. Examples of bioactive thionyl-containing compounds that require metal-binding for their activity include hydroxypyrothione and hydroxypyridinethione inhibitors of metalloproteinases,37 as well as diisopropylthiourea-based antimicrobial agents.38 More recently, a multifunctional pyrimidinylthiourea with both inhibitory acetylcholine esterase activity and metal-binding potential were effective in vitro and in vivo.39
The data presented here thus provides a direct link between bioactive thiourea-based metal ligands and the toxicity caused by the Aβ peptide implicated in AD. This identification of another class of metal-dependent cytoprotective compounds underscores the significant evidence linking Aβ and metal-dependent phenomenology in AD. Metals bind and promote Aβ aggregation, facilitate formation of reactive oxygen species (ROS), accumulate in extracellular plaques, and are depleted in the surrounding areas.26,27 Thus, the ability to mitigate these effects and potentially redistribute bioavailable metal is a viable therapeutic strategy that has been pursued, especially with the 8-hydroxyquinoline, PBT2.19,23
Mechanism of Action: Evaluation of the Effects of CQ and DHPM-Thiones on Aβ Levels and Vesicle Trafficking
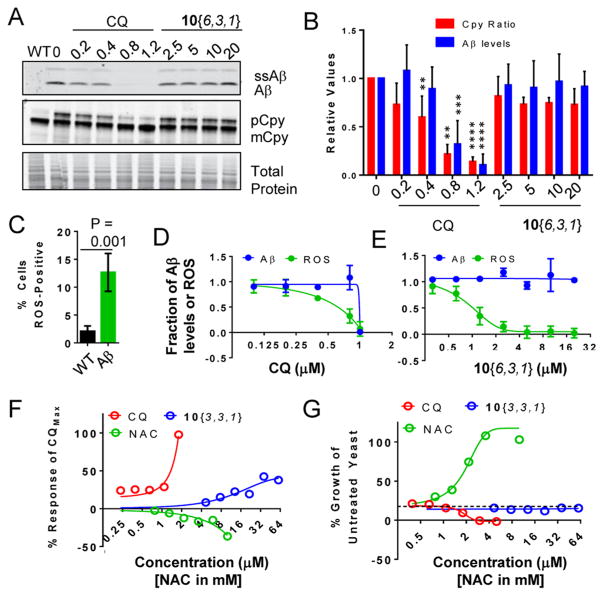
To evaluate the mechanism of action for DHPM-thiones, we first examined the effects of 10{6,3,1} on steady-state Aβ peptide levels. Confirming previous observations,13 immunoblot analysis revealed that CQ reduced Aβ peptide levels by ~80% at its most effective concentration of 0.8 μM (Figure 8A, top panel). This reduction was consistent with CQ preventing and/or reversing the metal-dependent oligomer formation and promoting Aβ degradation. Compound 10{6,3,1}, in contrast, did not reduce Aβ levels under these conditions (Figure 8A, top panel).
Figure 8.
Effects of compounds on Aβ levels, vesicle trafficking, and ROS generation. (A) Western blot analysis of Aβ-expressing yeast treated with increasing doses of CQ or 10{6,3,1}. Top panel shows Aβ peptide (higher molecular band is the signal sequence-Aβ fusion prior to cleavage). The middle panel is Cpy, where the larger immature Cpy band reflects a block in trafficking to the vacuole. Bottom panel is a Coomassie stain of the top portion of the same gel used for blotting Aβ and Cpy to show equal protein load. (B) Quantitation of three independent experiments of (A). Data was normalized to total protein (quantified by total protein) and set to 1.0 for wild-type (Cpy) or Aβ/untreated (Aβ levels). P-Values according to a one-way ANOVA and Tukey’s test where P-values are **<0.01 and ***<0.001; ****<0.0001. (C) Detection of increased reactive oxygen species (ROS) in Aβ-expressing yeast compared to wild-type. Aβ was induced for 7 h and ROS detected by flow cytometry using a ROS-sensing dye (CM-H2DCFDA). Data are expressed as percent of cells that were ROS-positive after 7 h of Aβ expression. (D, E) Dose–response of CQ (D) and 10{6,3,1} (E) for both Aβ protein levels (blue) and ROS levels (green). Y-Axis is fraction of Aβ or ROS in the untreated condition. X-Axes are concentration (μM). (F) Dose–response curves for rescue of Aβ toxicity by CQ, 10{3,3,1}, or the antioxidant N-acetylcysteine (NAC). The y-axis is percent of maximal rescue by CQ. X-Axis is as in (D). Data indicate that a canonical antioxidant could not rescue Aβ toxicity. (G) Dose–response curves for growth of wild-type yeast treated with an ~80% lethal concentration of H2O2 in the presence of CQ (red), 10{3,3,1} (blue), or NAC (green). The y-axis is percent of growth of untreated wild-type yeast. X-Axis is as in (D).
We simultaneously assessed the effects of CQ and 10{6,3,1} on restoration of vesicle trafficking using Cpy as representative trafficking protein substrate. Cpy is a vacuolar protease that is trafficked from the ER to the Golgi, where it is post-translationally modified through glycosylation.40 Cpy then traffics through multivesicular bodies to the vacuole and is cleaved to yield the active Cpy protease. Defects in trafficking from ER-to-Golgi and from the Golgi-to-vacuole can lead to accumulation of immature Cpy. Thus, monitoring Cpy trafficking can illuminate the trafficking defects caused by α-syn in yeast.5 Additionally, defects in recycling of its receptor from the vacuole back to the Golgi (via the retromer) can indirectly influence Cpy trafficking. We therefore performed immunoblot analysis of Cpy in both WT and Aβ-expressing yeast to monitor changes in the ratio of immature to fully processed Cpy.
After inducing Aβ expression, improperly trafficked Cpy accumulated, indicating Aβ inhibits features of secretory or endosomal trafficking (Figure 8A, middle panel; quantitation in Figure 8B). Given our previous demonstration that secretion was not inhibited by Aβ expression,4 this defect is likely due to dysfunctional endosomal transport consistent with the Mup1-GFP, Ste3-GFP, and clathrin defects we previously observed.4,13 The Cpy defect was fully reversed by CQ; compound 10{6,3,1} did not significantly reduce the Cpy trafficking defect (Figure 8A, middle panel and Figure 8B). It thus does not appear that DHPM-thiones confer cytoprotection against Aβ toxicity by reversing trafficking defects.
Mechanism of Action: Evaluation of CQ and DHPM-Thiones Effects on Aβ-Induced ROS Generation
The generation of reactive oxygen species (ROS) is a toxic common feature of many neurodegenerative disease models. ROS may be generated by dysfunctional mitochondria either when transition metals, such as copper and iron, directly catalyze the conversion of H2O2 to highly reactive hydroxyl radicals, or when the disease proteins themselves catalyze this conversion. Aβ-metal complexes, for example, can directly facilitate generation of ROS.41 In yeast, targeting Aβ to the secretory pathway has also been reported to facilitate ROS generation and mitochondrial dysfunction.42 We therefore tested whether the yeast Aβ model generates ROS, and, if so, was this phenotype reversed by CQ or DHPM-thiones.
Wild-type and Aβ-expressing yeast were shifted to galactose for 7 h and ROS detected using CM-H2DCFDA, which fluoresces green when oxidized. After 7 h, the percent of ROS-positive cells modestly increased (Figure 8C), consistent with previous results.42 We next tested whether CQ and/or 10{3,3,1} could prevent this accumulation. Aβ was induced with simultaneous compound administration and ROS levels evaluated after 7 h. For comparison with Aβ levels and trafficking defects, the same samples were also assessed for by immunoblotting against Aβ peptide and Cpy. CQ reduced ROS generation with a dose–response that was slightly shifted toward lower concentrations as compared to the reduction in protein level (Figure 8D). This shift suggests that CQ can reduce ROS before the dramatic reduction of protein levels. Treatment with 10{3,3,1} also reduced ROS generation, albeit without reducing Aβ levels (Figure 8E). Interestingly, the doses at which ROS reduction occurred were significantly lower than those required for rescue. This may suggest that the compound was not stable during the duration of a growth assay (48 h), or, that the compound may be modified while exerting its protective activity. In addition, the results with 10{3,3,1} indicate that reversal of ROS could be accomplished without reducing Aβ levels or restoring vesicle trafficking.
To test whether 10{3,3,1} rescued Aβ toxicity by directly reducing ROS, we examined (1) whether antioxidants could rescue Aβ toxicity and (2) if 10{3,3,1} could rescue H2O2 toxicity. We first evaluated the ability of a robust antioxidant, N-acetylcysteine (NAC), to rescue Aβ toxicity. If simple radical scavenging was a productive mode of action, then NAC should at least partially reduce Aβ toxicity. Dose–response analysis showed that while CQ and 10{3,3,1} rescued toxicity, NAC at concentrations from 0.5 to 10 mM had no effect on Aβ toxicity in yeast (Figure 8F). Note that these concentrations of NAC were ~1000-fold higher (mM vs μM) than that required for CQ or 10{3,3,1} efficacy. The opposite question was then asked: could CQ or 10{3,3,1} reduce ROS toxicity? We tested a concentration of H2O2 that reduced the growth of wild-type yeast by ~80% over a 24 h period and examined dose–response curves for each compound. While NAC strongly reduced H2O2 toxicity with an EC50 of ~1.5 mM, CQ and 10{3,3,1} conferred no protection (Figure 8G).
These data suggest that ROS itself is not the primary source of toxicity in the Aβ yeast model and that neither CQ nor 10{3,3,1} rescue Aβ toxicity by simply scavenging radicals or reducing metal-catalyzed H2O2 production. However, this result does suggest that the dysfunctional process leading to ROS generation–not the ROS itself–is likely contributing to toxicity. For example, if Aβ-influenced mitochondrial function is leading to ROS generation, then CQ and DHPM-thiones could reduce ROS because of their main function in rectifying a dysfunctional process. In such a scenario, NAC would not be expected to confer cytoprotection as the underlying process would remain dysfunctional. For CQ, the reduction of ROS-generating dysfunction closely follows a reduction in Aβ levels. 10{3,3,1}, in contrast, achieved a reduction in ROS without appreciably affecting Aβ or vesicle trafficking, thereby indicating these processes can be uncoupled and independently targeted pharmacologically.
CQ and 10{6,3,1} Synergize To Rescue Aβ Toxicity in Yeast and Nematode Aβ Models
CQ displayed distinct rescuing activities and phenotypic responses in metal-related experiments as compared to the DHPM-thiones 10{6,3,1} and 10{3,3,1}. While both bound metals to rescue Aβ toxicity, they appear to have variable effects on cell physiology, such as gene expression profile differences and variable responses to FRE1/FRE2 overexpression (Figures 5 and 6). These chemicals also have distinct abilities to facilitate Aβ turnover and rescue Cpy trafficking defects (Figure 8). These results and previous studies clearly link Aβ rescue to metal-dependent Aβ turnover for CQ (Figure 8).13 However, for the DHPM-thiones, the metal-responsive protective activity is less clear. Because of these distinct activities, we posited that they do indeed work through distinct mechanisms of action. These differences provide an opportunity to evaluate any synergistic activity. Compounds that work by the same mechanism of action show additive effects, while those working through distinct mechanisms can exhibit synergy. We therefore tested how cells expressing Aβ responded to the combination of CQ and 10{6,3,1} in both yeast and worm models.
We examined various combinations of CQ and 10{6,3,1} and compared each combination to the single agent dose for their ability rescue Aβ toxicity (Figure 9A). An exceptionally strong rescue of Aβ toxicity was achieved with a combined dose of 0.2 μM CQ and 10 μM 10{6,3,1}, bringing growth almost to the level of WT cells (Figure 9B). At this concentration of each compound alone, or even at twice the single agent concentration, there was no rescue of Aβ toxicity. Taken together with the distinct behaviors observed in various other assays, the synergy between 10{6,3,1} and CQ suggest that they work through distinct, complementary mechanisms.
Figure 9.
CQ and DHPM-thione synergize to rescue Aβ toxicity in both yeast and nematode. (A) Quantitation of maximal growth as a percent of wild-type growth over a 48-h time course for single or combined doses (as indicated on y-axis) of CQ and 10{6,3,1}. (B) Growth curves of wild-type or Aβ-expressing yeast treated with subprotective doses of CQ (0.2 μM) and 10{6,3,1} (10 μM), as well as their synergistic rescue of combined doses. (C) Synergy of CQ and 10{6,3,1} in the nematode model of Aβ toxicity. The y-axis is percent of worms with a wild-type number of tail glutamatergic neurons. Black is DMSO solvent control, red is CQ, and blue is 10{6,3,1} at subeffective doses. Green is the combination. For each treatment condition, three independent experiments were performed with 30 worms scored in each trial (a total of 90 worms examined for each condition tested in μM). One-way ANOVA with Tukey’s test was used to compare the neurodegeneration among various treatment groups. *P < 0.05; **P < 0.01; ***P < 0.001. (D) Representative fluorescent images of nematodes expressing Aβ and treated with DMSO alone (i) or the combination of CQ/10{6,3,1} (ii). Large arrowheads indicate posterior glutamatergic neurons. Small arrows with line indicate a missing neuron.
Since both CQ and the DHPM-thiones 10{3,3,1} and 10{6,3,1} individually rescued nematode neurons from Aβ toxicity (Figure 4), we next tested if the two compounds also synergized in a model with a fully integrated nervous system. Larval stage worms were treated with doses of CQ or 10{6,3,1} that were at least 4-fold lower than the active CQ and 10{6,3,1} doses. The low doses of each compound failed to restore neuronal viability, consistent with these doses being sub-effective as single agents (Figure 9C). However, combined treatment with lower doses (3.75 μM 10{6,3,1} and 0.125/0.25 μM CQ), conferred a robust protection of glutamatergic neurons expressing Aβ (Figure 9C and D). Thus, the ability of a combination treatment with DHPM-thiones and CQ to synergistically protect against Aβ toxicity is conserved from unicellular yeast through the intact nervous system of nematodes, indicating that these distinct mechanisms of action for each compound operate within each system.
DHPM-Thione/CQ Synergy Mitigates Off-Target Effects
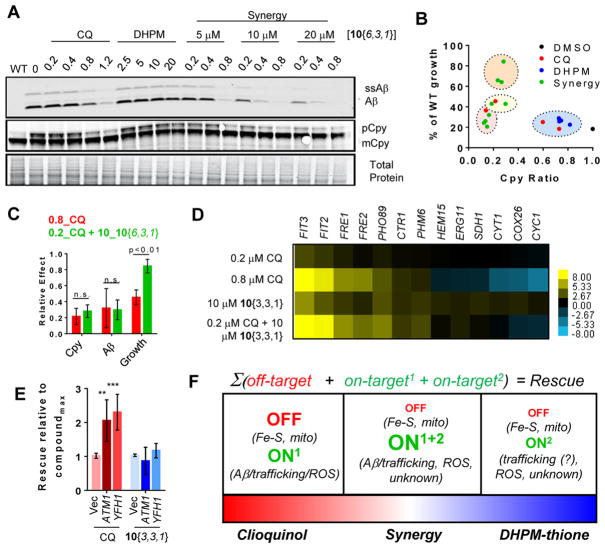
The supra-additive rescue of the combination treatment suggested that 10{6,3,1} and CQ function through distinct mechanisms. To determine if these mechanisms converge on altering the levels of Aβ and the associated trafficking defects, we examined the effects of synergistic doses by immunoblot as done previously (Figure 10A). Indeed, combined treatments of synergistic doses revealed a strong reduction of Aβ levels with concomitant rescue of Cpy trafficking (Figure 10A). The most effective combination (0.2 μM CQ and 10 μM 10{6,3,1}) reduced Aβ and rescued Cpy to a level comparable to treatment with 0.8 μM CQ alone. Across all compound treatments, both single and combined, there was a very strong correlation between the reduction in Aβ levels and the restoration of functional Cpy trafficking (Figure 10B, Supporting Information Figure 5). Thus, the ability of DHPM-thiones to enhance Aβ turnover and restore Cpy trafficking suggests these compounds may contribute to this pathway in a manner not robustly detectable in earlier experiments (Figure 8). Intriguingly, there were combinations of CQ and 10{6,3,1} that restored trafficking of Cpy to a similar extent as the maximal effective dose of CQ, yet the combined treatment restored growth to a greater extent than CQ alone (Figure 10B). In particular, the combined dose of 0.2 μM CQ and 10 μM 10{6,3,1} led to the same effects on both Aβ rescue and Cpy trafficking correction as 0.8 μM CQ alone, yet restored growth to nearly wild-type levels, unlike CQ alone (Figure 10C).
Figure 10.
CQ and DHPM-thiones synergize to reduce Aβ levels, reverse Cpy trafficking deficits, and mitigate deleterious off-target effects of CQ. (A) Western blot analysis of Aβ-expressing yeast treated with CQ, 10{6,3,1}, or the indicated combinations. (B) Comparison of rescue (y-axis) and rescue of Cpy restoration ratio. Compounds are color coded as follows: blue, subeffective; red, too high/toxic; orange, synergistic rescue; yellow; subeffective dose. (C) Comparison of Cpy trafficking, Aβ levels, and growth for the most synergistic dose of CQ/10{3,3,1} (0.2 μM CQ and 10 μM 10{3,3,1}) and CQ alone. P-Values were generated with one-way ANOVA with Tukey’s test and are indicated on the graph. (D) Heat map of RT-PCR quantitation of transcripts after treatment with single or combined synergistic doses of CQ and 10{3,3,1}. Genes were selected based on changes demonstrated by RNA-seq experiments (Figure 5). Scale indicates log2 fold change of transcripts. (E) Quantitation of the maximal rescue for Aβ strains with or without overexpression of mitochondrial iron–sulfur cluster regulating proteins, ATM1 and YFH1. Y-Axis is rescue relative to the maximal effect for that compound. One-way ANOVA Tukey’s statistical tests revealed highly significant p-values between groups (**P < 0.01; ***P < 0.001). (F) Model of synergy between CQ and DHPM-thiones. Red text indicates deleterious effects. Green indicates protective effects. The size of the text in the right box reflects their increased/decreased activities under synergistic combinations.
One possible explanation for the enhanced rescue of synergistic doses is that there are targets modulated by DHPM-thiones that are independent of Aβ and contribute to additional growth rescue, e.g., via the reduction of ROS (Figure 8). Another nonmutually exclusive possibility is that the dose-lowering effect of 10{6,3,1} was accomplished by 1) enhancing degradation of Aβ, 2) reducing ROS, and, in addition, 3) reducing spurious off-target effects on metal-homeostasis caused by CQ. This second possibility was supported by the RNA-seq data (Figure 5C), where 10{3,3,1} had modest effects on metal-regulated genes as compared to the dramatic effects of CQ, especially on mitochondrial proteins.
To examine how combined treatment generally impacted cellular metal availability, we examined the expression of a set of genes whose transcript levels were altered by either CQ or 10{3,3,1} (Figure 5C and D). Both 10{6,3,1} and 10{3,3,1} exhibited nearly identical synergy patterns, so we used 10{3,3,1} to be consistent with the previous RNA-seq experiments. Quantitative RT-PCR analysis was performed on RNA isolated from cells treated with the optimal dose of CQ alone (0.8 μM), the optimal synergistic doses (0.2 μM CQ, 10 μM 10{3,3,1}), and the single agent doses of the 10{3,3,1} compounds. We first examined genes that were upregulated in response to CQ, including iron-(FIT3, FIT2, FRE1, FRE2), copper-(FRE1, CTR1), and phosphate-regulated genes (PHO89, PHM6). Consistent with the RNA-seq data, 0.8 μM CQ led to a strong up-regulation of metal-regulated genes, while 10{3,3,1} had a moderate effect (Figure 10D). The combined, synergistic dose of two compounds induced upregulation of these genes to levels comparable to 0.8 μM CQ (Figure 10D), suggesting the combination promoted a broad metal response. In these experiments, phosphate-regulated genes were modestly upregulated by CQ, where they were not in the RNA-seq experiments. Combined treatment had little added effect on phosphate genes beyond that of 10{3,3,1} alone.
Downregulated genes were affected in a very different manner. Several mitochondrial genes functioning in the electron transport chain, some of which are Fe–S-containing proteins (e.g., SDH1, CYC1, CYT1, COX26), as well as ERG11 (ergosterol biosynthesis) and HEM15 (heme biosynthesis) were significantly down-regulated in response to CQ, but not affected by 10{3,3,1} (Figure 10D). Notably, the combined dose, which afforded supra-additive rescue, did not cause a strong reduction in expression of transcripts encoding mitochondrial proteins (Figure 10D). Thus, it appeared that the combination of CQ and 10{3,3,1} was significantly less impactful on mitochondrial function than CQ alone. This difference was likely due to a less pronounced effect on Fe–S cluster-containing proteins.
To test the notion that a competing, deleterious effect on mitochondrial function by the 0.8 μM dose of CQ limited its rescuing capacity, we overexpressed two proteins that promote iron–sulfur cluster generation (ATM1 and YFH1) and monitored rescue of Aβ toxicity. Indeed, the overexpression of ATM1 and YFH1 increased Aβ rescue by CQ, yet had no impact on Aβ rescue by 10{3,3,1} (Figure 10E). This result at least partially explained how maximally effective CQ had the same impact on Aβ and Cpy as the combined dose, yet an inferior rescue (Figure 10B): the deleterious off-target cost of maximally effective CQ out-weighed its effects on Aβ levels and restoration of trafficking. Thus, synergy likely was achieved by multiple mechanisms: (1) increasing Aβ turnover (Figure 10B), (2) restoring vesicle trafficking, (3) providing additional anti-ROS generating activity (Figure 8E), and (4) simultaneously reducing off-target consequences of CQ on mitochondrial function (Figure 10D). We have summarized these effects in Figure 10F.
The data presented herein underscores the importance of metal phenomenology in mediating Aβ toxicity and adds to the established set of compounds with metal-targeting mechanisms. Numerous efforts have led to both the identification and development of compounds with the capacity to bind metals with multiple mechanisms of action. Such compounds frequently reduce both Aβ levels and metal-dependent H2O2 generation and promote redistribution of bioavailable metal ions. Here, DHPM-thiones (herein referred to as a collective class of protective compounds unless otherwise noted) exerted protective activity in both yeast and nematode models through a metal-dependent mechanism of action that robustly synergized with CQ to target Aβ-dependent cellular toxicities.
Although the capacity to bind metals was essential for activity, the identity of the proximal metalloprotein target remains unknown. While Aβ itself is a likely candidate as a direct target, we did not observe robust effects of DHPM-thiones on Aβ levels or Cpy-trafficking reversal. It is possible that our assays could not reveal subtle, yet significant, effects on Aβ levels or conformation. However, based on estimates that one-third to one-half of all proteins bind a metal ion, there are several hundred proteins that could be targeted.43,44 Thus, navigating this expansive set of metalloproteins for one specific activity poses a significant challenge. Future efforts will attempt to identify the precise target for DHPM-thiones.
Both individual CQ and CQ/DHPM-thione synergy experiments demonstrated that the rescue of Aβ toxicity closely tracked with an ability to reverse vesicle trafficking defects (Figure 10). In this case, the rescue of trafficking occurred concomitant with a dramatic reduction of Aβ peptide. Based on the data herein, it is not possible to distinguish whether Aβ reduction led to a reversal of trafficking, or, if rescuing trafficking led to enhanced degradation of Aβ. Regardless, the finding of a tight correlation between rescue and correcting an underlying trafficking defect supports accumulating evidence for endosomal dysfunction as a prominent molecular feature of AD pathogenesis. In our system, we first connected endosomal trafficking defects through the identification of PICALM, a protein functioning in endocytosis and a well validated risk factor for AD,14,16,19 in our yeast genetic screens.4 Elevating PICALM levels both protected yeast cells from intravesicular Aβ, while also mitigating the toxicity of exogenously applied Aβ oligomers to rat primary cortical neurons.4 Furthermore, additional AD risk factors BIN1 and CD2AP function in vesicle trafficking as well.16,18 These genetic links, along with reports of endosomal abnormalities caused by intracellular Aβ,45,46 suggest this may be a key pathway to target pharmacologically for AD.
The inability to identify stand-alone therapies for AD partially derives from the fact that disease onset and progression are multifactorial processes in which numerous cellular pathways are perturbed. For this reason, and given the central role of Aβ in the amyloid cascade hypothesis, some efforts have focused on reducing Aβ levels directly. Positive indications for potential success of this approach have recently come to light with the early results of Adjucanimab, a human-derived antibody that reduced Aβ plaques and slowed clinical decline in phase II clinical trials.47 This promise comes in the wake of other less successful attempts at Aβ-targeted antibody therapies.48 Metal-targeting compounds may have the added benefit of targeting Aβ via metal-dependent destabilization in addition to redistributing bioavailable metals from plaques to surrounding neurons.23 PBT2, an 8-hydroxyquinoline, also has shown some therapeutic promise, though more recent results make its future unclear.24,25 Considering the multifactorial nature of disease progression and the limitation of any one given approach, it is plausible that combined therapies may ultimately prove important for successful treatment of AD.
While we focused on synergy and dose-lowering capacity here, others have taken a different approach to this problem by synthesizing multifunctional compounds. Hybrid compounds, many of which are based on 8-hydroxyquinolines, have been rationally designed to target multiple toxic attributes of neurodegenerative diseases.49 This approach could have particular importance in indications such as AD where multiple metal-dependent mechanisms could afford neuroprotection. Numerous other hybrid molecules have been developed as potential multifunctional anti-Alzheimer’s agents: flavonoids,50 a benzofuran hybrid that inhibits cholinesterase activity and Aβ aggregation,51 and memoquin, which combines the acetylcholinesterase inhibitor activity of caproctamine with the antioxidant activity of 1,4-benzoquinone,52,53 have demonstrated efficacy in numerous model systems. Of particular relevance to the current study, a pyrimidinylthiourea inhibited acetylcholinesterase, along with exhibiting activity against multiple Aβ-metal-dependent phenomena.39 All activities of this scaffold were enhanced by the thiourea moiety inherent to the inhibitory ACh esterase pharmacophore.
Combination therapy is not a new concept and may ultimately be critical for attaining maximal patient benefit. Symptom management approaches of cholinesterase inhibitors and memantine, for example, slow cognitive decline more effectively than single agent treatments.54 While these drugs attempt to maintain levels of acetylcholine and regulate glutamate, which have different functions in neurotransmission, they do not themselves address the underlying pathologies contributing to AD. Combinations of potential disease-modifying agents, like those described here or new chemical entities identified from phenotypic screens, may be effective in treating disease when combined with symptom management drugs that target neuronal function. Cell-based systems amenable to phenotypic screening, such as budding yeast, thus provide a path forward to both identify and evaluate novel mechanisms for single agents and synergistic combinations of small molecules protective against neurodegenerative disease proteins.
METHODS
Yeast Strains
All yeast strains were grown using standard methods. Strains were propagated on solid plates composed of complete synthetic media (CSM) containing 2% (w/v) glucose and 2% (w/v) agar. Most liquid growth was carried out in complete synthetic media (CSM) containing 2% (w/v) of glucose, raffinose, or galactose in 30 °C shaking incubators or rotating wheels. Strains harboring plasmids were maintained in media lacking uracil (SX-Ura, where “X” are sugars) was used to maintain the plasmid. All yeast model strains have been described previously4,6,30,60 The af t1Δ strain was generated using pAG3255 (pAG32 was a gift from John McCusker (Addgene plasmid # 35122) as a template to PCR amplify the hygromycin cassette to knock out AFT1. Transformations were carried out using a lithium acetate-based transformations protocol.56 For deletions, following overnight recovery in CSM-glucose, cells transformed with the AFT1-targeted hygromycin cassette were plated on YPD (yeast extract-peptone-dextrose) plates containing hygromycin (300 μg/mL). Individual transformants were analyzed by diagnostic PCR to verify deletion of AFT1. For experiments using overexpression of metal homeostasis-related genes, Flexgene plasmids57 were transformed into the indicated strains and plated on SD-Ura plates to select for plasmids harboring the URA3 selectable prototrophic marker.
Compound Screening and Treatments
A library of ~3000 compounds from the Boston University Center for Molecular Discovery (BU-CMD) was screened in the α-synuclein, Aβ, TDP-43, and 72Q yeast models. For the screen and most compound experiments, strains were grown overnight in CSM-glucose. Stationary overnight cultures were then diluted 1:20 in CSM-raffinose for roughly 6 h. Strains were then diluted to an OD600 of 0.01 in CSM-galactose to induce expression of the toxic genes. For the screen, culture was dispensed to 384-well clear plastic plates and compounds pinned using a robotic workstation outfitted with a 100 nL pin tool. Plates were then grown under humidified conditions at 30 °C and OD600 read using an Envision (PerkinElmer) plate reader after 24 or 48 h. z-Scores were then calculated using the formula (OD600 exp well – OD600 plate avg)/(OD600 plate stdev). A z-score of 3.0 was used as a cutoff to call screen hits.
The same general growth strategy was used for all compound treatment experiments. Compound dilutions for dose–response curves were carried out at a 2× concentration in the appropriate media (typically CSM-galactose) in 96-well plates. A 2× concentration of cells was then added to the entire plate to achieve the final desired compound concentration and cell density. Next, a Tecan Freedom EVO liquid handling robot was used to transfer culture (30 μL per well) from the 96-well plate to the four quadrants of a 384-well plate. Following a 24 to 48 h incubation and OD600 reading on the Envision plate reader, the average of the four wells was used as a single data point and averaged across multiple experiments. Data was typically normalized to the maximal rescue conferred by clioquinol (CQ). Raw data was first corrected for background OD600 contributed by the plate, a fold-change calculated for compound-treated versus DMSO-treated wells, and the maximal rescue for CQ set to 100%. For wild-type cell experiments, data was normalized to 100% for the DMSO treated condition. For dose–response curves, a logistic regression curve was fit to the normalized data using Spotfire (TIBCO). For growth curve experiments, a Bioscreen-C instrument (Growth Curves USA) was used to both aerate cultures and read OD600 values at 15 min intervals.
Additional variations of the above treatment regimen were also carried out as indicated. Exogenous metal competition experiments were carried out by diluting copper chloride or iron sulfate to an equimolar concentration as the highest concentration in the dilution plate. The compound/metal mixture was then serially diluted along with the compound such that an equimolar compound-to-metal concentration was maintained across all points of the dose–response curve. For 1,10-phenanthroline experiments, a final uniform concentration of 10 μM chelator was used at all doses of CQ or DHMP-thione. For N-acetylcysteine (NAC) growth experiments, a dilution of NAC to achieve a final concentration in millimolar, not micromolar, was carried out as described above. For H2O2 experiments, a concentration was used that achieved ~80% reduction in cell growth after 24 h was used (~0.0015% (w/v)). Synergy experiments were carried out by combining CQ and DHPM-thione at indicated concentrations. Growth was carried out using the Bioscreen-C instrument, which was used for both the growth curves and when growing samples for protein analysis.
DHPM-Thione Synthesis
Synthesis and experimental details of all compounds are described in the Supporting Information (Methods S1). The full compound list, properties, and activities are described in Table S1.
Nematode Experiments
Nematodes were maintained using standard procedures.58 The transgenic strain, UA198 {baIn34[-Peat-4::Aβ42; Pmyo-2::mCherry]; adIs[Peat-4::gfp]} was generated as described in ref 4. For the analysis of CQ or DHPM effect on Aβ-induced glutamatergic neurodegeneration, embryo preparation of UA198 worms was performed and then embryos were soaked in CQ or DHPM solution for the following 24 h, during which embryos were hatched and worms were age-synchronized59 at L1 larvae stage. Larva worms were rinsed three times with M9 solution to be withdrawn from drug treatment, and then transferred onto OP50 regular food plates to grow for additional 4 days at 20 °C.
For scoring neurodegeneration, worms were mounted on a 2% agarose pad, immobilized with 2 mM levamisole and analyzed with a CCD camera (Photometrics CoolSnap HQ) on a Nikon E800 microscope at 40× magnification. Neurodegeneration was considered rescued when all five posterior glutamatergic neurons were intact and had no visible signs of degeneration. For each drug condition, three independent experiments were performed with 30 worms in each trial. Statistical analyses were performed using one-way ANOVA in GraphPad Prism.
Immunoblot Analysis of Aβ and Cpy
Analysis of Aβ levels and Cpy-trafficking was carried out from cells grown in the Bioscreen-C instrument. Treatments were carried out as described above where CSM-raffinose cultures were diluted into CSM-galactose with the indicated compound treatment conditions. For protein analysis, cells were diluted to an OD600 of 0.2 and grown for 8 h. Three replicate wells (each 300 μL) were then pooled, centrifuged, washed in cold water, and then prepared for protein gels. Given the small volumes, cell pellet size (and roughly number) was adjusted to account for differences in cell growth. Cell pellets were resuspended in 100 μL of 1× SDS loading buffering and boiled for 15 min. Denatured samples were then vortexed for ~30 s and tubes centrifuged for 1 min at 1,000xg to pellet cell debris. Samples were resolved by SDS-PAGE (4–12% gradient gels, Invitrogen) in 1X MES buffer at 150 V until the dye reached the bottom of the gel. Proteins were then transferred to PVDF using an iBlot apparatus (Invitrogen) with a 5 min, 30 s transfer time (reduced time due to small size of Aβ peptide). Prior to transfer, the top one-third of the gel was excised and stained for total protein using SimplyBlue SafeStain (Life Technologies). After transfer to PVDF, blots were blocked with 5% milk phosphate-buffered saline with 0.1% (v/v) tween (PBST). Primary antibodies recognizing Aβ (6E10, Covance) and Cpy (10A5B5, ThermoFisher) were then incubated overnight with the blots in PBST/5% milk at 4 °C at a 1:2,000 and 1:5000 dilution, respectively. Blots were washed for 30 min, and secondary antibodies added (IRDye800 Licor) for 2 h at room temperature. Blots were washed and scanned using a Licor Odyssey instrument to enable quantification. For Aβ levels, signal was normalized to the total protein from the stained one-third portion of the gel (scanned with Licor Odyssey at 600 nm). For Cpy, the larger molecular weight (ER/Golgi localized) band was normalized to the lower molecular weight (mature) Cpy band and expressed as a ratio. For both Aβ and Cpy, values were then normalized to the DMSO-treated condition to monitor effects of compound treatment. At least three independent biological replicates were carried out for each experiment.
ROS Experiments
ROS detection was carried out as described previously.60 Briefly, Aβ yeast cultures were induced with galactose in the presence or absence of compounds and grown for 7 h in the Bioscreen-C instrument. Samples were then taken for ROS detection using a Guava easyCyte flow cytometer to measure the oxidation of 5-(and-6)-chloromethyl-2′7′-dichlorodihydrofluorescein diacetate (CM-H2DCFDA).
Copper-Binding Analysis
UV/vis experiments were carried out by scanning samples from 200 to 600 nM wavelength. Compounds (40 mM DMSO stocks) were diluted to 50 μM with or without 50 μM copper(II) sulfate in solution containing 1× PBS (pH 7.4) and 50% methanol and incubated at room temperature for 15 min prior to UV/vis scanning. Three independent compound dilutions and scans were obtained and averaged.
For NMR Cu-binding experiments, A mixture of CuCl (1.0 mg, 0.01 mmol, 1.0 equiv) and 10{3,3,1} (3.7 mg, 0.01 mmol, 1.0 equiv) in CD3CN was sonicated for 5 min and analyzed by 1H NMR. An overlaid comparison of the resulting spectrum to that of 10{3,3,1} alone is shown in the figure (Figure 7C, top). In analogy to the above experiment, a mixture of CuCl (1.0 mg, 0.01 mmol, 1.0 equiv) and 11 (3.5 mg, 0.01 mmol, 1.0 equiv) in CD3CN was sonicated for 5 min and analyzed by 1H NMR. An overlaid comparison of the resulting spectrum to that of 11 alone is shown in the figure (Figure 7C, bottom).
Compound Treatment for RNA-seq Experiments
Wild-type yeast (5 mL cultures) grown to log-phase in CSM-galactose were treated with 0.8 μM CQ, 20 μM 10{3,3,1}, 11, or a DMSO solvent control for 6 h at 30 °C. Cells were centrifuged, washed once with ice-cold water, and cell pellets flash frozen in liquid nitrogen prior to storage at −80 °C. Cell treatments were carried out in duplicate on separate days. RNA was extracted from yeast pellets using a hot acid phenol protocol.61 RNA-seq libraries from yeast total RNA (1 μg) were prepared for sequencing with the PrepX RNA-Seq Kit from IntegenX according to the standard protocol and sequenced on a HiSeq 2500. Between 16 and 26 million 40nt long reads were obtained per sample. Reads were trimmed with FASTQ Quality Trimmer (http://hannonlab.cshl.edu/fastx_toolkit/index.html) with parameters “-v -t 20 -l 25”. Trimmed reads were mapped using TopHat262 using the following parameters: “--no-coverage-search --library-type = fr-secondstrand--solexa1.3-quals --segment-length 20 -I 2000 --no-novel-juncs” and a gtf file from ENSEMBL release 65 (Saccharomyces_cerevisiae. EF4.65.gtf). Reads were assigned to genes using htseq-count63 with parameters “--mode = intersection-strict --stranded = yes”. Read normalization and differential analysis was done with DEseq64 Data is found in Supporting Information Table S2.
RT-PCR of Compound Treatments
RNA expression was analyzed from wild-type yeast treated with CQ, 10{3,3,1}, or combined treatments. CSM-raffinose cultures were transferred to CSM-galactose to an OD600 of 0.1 and compounds added. Treated cultures were grown for 6 h at 30 °C. Cells were then centrifuged, washedd with cold water, flash frozen in liquid nitrogen, and stored at −80 °C. RNA was then isolated using the hot acid phenol method.61 cDNA was generated from 500 ng of total yeast RNA using reverse transcriptase (Invitrogen, Superscript III) primed with oligo dT (Thermofisher). Gene-specific primers were designed with “Primer3” to amplify an approximate 100–150 base pair PCR product localized within the 300 bp region of the 3′ end of each gene of interest. Reverse transcribed cDNA was diluted 1:100 in TE and used as a template in triplicate real-time PCR reactions (Applied Biosystems 7900HT real time PCR machine). Data was normalized relative to the housekeeping gene, ACT1. Four independent experiments were performed and averaged.
Supplementary Material
Acknowledgments
Funding
D.F.T was funded by NRSA Fellowship NIH 5F32NS061419. D.F.T. and S.L. were supported by WIBR funds in support of research on Regenerative Disease, the Picower/JPB Foundation, and the Edward N. and Della L. Thome Foundation. G.A.C. and S.L. were funded by a Howard Hughes Medical Institute (HHMI) Collaborative Innovation Award. L.E.B., R.T., and S.E.S. were funded by NIH GM086180, NIH GM067041, and NIH GM111625.
We gratefully acknowledge the late Susan Lindquist and dedicate this manuscript in her memory. We thank John A. Porco Jr. and the entire BU-CMD for contributing to and sharing their diversity library for screening. pAG32 was a gift from John McCusker (Addgene plasmid # 35122). We thank all funding sources, especially the Thome Foundation for their support, the WIBR Genome Technology Core for RNA-sequencing, Stephen Buchwald for advice, Priyanka Narayan, George Bell, and Linda Clayton for reading the manuscript.
ABBREVIATIONS
- ND
neurodegenerative disease
- AD
Alzheimer’s disease
- Aβ
β-amyloid peptide
- ER
endoplasmic reticulum
- WT
wild-type
- DHPM-thione
dihydropyrimidine-thione
- CQ
clioquinol
- SAR
structure activity relationship
- ROS
reactive oxygen species
Footnotes
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschemneuro.7b00187.
Full experimental details for compound synthesis and characterization; a table of the properties and structures for the 82-member DHPM-thione library; five supporting figures that describe (1) the effects of R3 on DHPM-thione activity, (2) functional interaction between CQ/DHPM-thiones and metals, (3) chemical-genetic interactions between CQ/DHPM-thiones and metal-related genes, (4) UV/vis trace of CQ +/− copper, and (5) the relationship between Aβ levels and Cpy trafficking after compound treatment; a data set for CQ/DHPM-thione RNA-seq experiment (PDF)
CQ and DHPM-thione RNA-seq data GEO accession number GSE99884 (XLSX)
Author Contributions
D.F.T. conceived of, conducted, and analyzed all yeast experiments and wrote the manuscript. L.E.B. synthesized the DHPM-thione library and analyzed SAR. X.Y. conducted C. elegans experiments. R.T. characterized the DHPM-thione library. N.J. conducted the NMR experiments. M.I.B analyzed RNA-seq data. K.A.C. and G.A.C analyzed the C. elegans experiments. S.E.S. designed DHPM-thione library and analyzed SAR. S.L. helped conceive experiments and analyze data. All authors edited the manuscript.
Notes
The authors declare the following competing financial interest(s): D.F.T. is a scientific co-founder and S.L. was the founder of Yumanity Therapeutics. No funds were received for this research and there are no competing or financial interests to disclose.
References
- 1.Khurana V, Lindquist S. Modelling neurodegeneration in Saccharomyces cerevisiae: why cook with baker’s yeast? Nat Rev Neurosci. 2010;11(6):436–49. doi: 10.1038/nrn2809. [DOI] [PubMed] [Google Scholar]
- 2.Tenreiro S, Munder MC, Alberti S, Outeiro TF. Harnessing the power of yeast to unravel the molecular basis of neurodegeneration. J Neurochem. 2013;127(4):438–52. doi: 10.1111/jnc.12271. [DOI] [PubMed] [Google Scholar]
- 3.Kachroo AH, Laurent JM, Yellman CM, Meyer AG, Wilke CO, Marcotte EM. Evolution. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science. 2015;348(6237):921–5. doi: 10.1126/science.aaa0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Treusch S, Hamamichi S, Goodman JL, Matlack KE, Chung CY, Baru V, Shulman JM, Parrado A, Bevis BJ, Valastyan JS, Han H, Lindhagen-Persson M, Reiman EM, Evans DA, Bennett DA, Olofsson A, DeJager PL, Tanzi RE, Caldwell KA, Caldwell GA, Lindquist S. Functional links between Abeta toxicity, endocytic trafficking, and Alzheimer’s disease risk factors in yeast. Science. 2011;334(6060):1241–5. doi: 10.1126/science.1213210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, Cao S, Caldwell KA, Caldwell GA, Marsischky G, Kolodner RD, Labaer J, Rochet JC, Bonini NM, Lindquist S. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science. 2006;313(5785):324–8. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson BS, McCaffery JM, Lindquist S, Gitler AD. A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci U S A. 2008;105(17):6439–44. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–6. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 8.McLean CA, Cherny RA, Fraser FW, Fuller SJ, Smith MJ, Beyreuther K, Bush AI, Masters CL. Soluble pool of Abeta amyloid as a determinant of severity of neurodegeneration in Alzheimer’s disease. Ann Neurol. 1999;46(6):860–6. doi: 10.1002/1531-8249(199912)46:6<860::aid-ana8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 9.Walsh DM, Klyubin I, Shankar GM, Townsend M, Fadeeva JV, Betts V, Podlisny MB, Cleary JP, Ashe KH, Rowan MJ, Selkoe DJ. The role of cell-derived oligomers of Abeta in Alzheimer’s disease and avenues for therapeutic intervention. Biochem Soc Trans. 2005;33(Pt 5):1087–90. doi: 10.1042/BST20051087. [DOI] [PubMed] [Google Scholar]
- 10.Iwatsubo T, Odaka A, Suzuki N, Mizusawa H, Nukina N, Ihara Y. Visualization of A beta 42(43) and A beta 40 in senile plaques with end-specific A beta monoclonals: evidence that an initially deposited species is A beta 42(43) Neuron. 1994;13(1):45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- 11.Jarrett JT, Berger EP, Lansbury PT., Jr The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: implications for the pathogenesis of Alzheimer’s disease. Biochemistry. 1993;32(18):4693–7. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 12.Caine J, Sankovich S, Antony H, Waddington L, Macreadie P, Varghese J, Macreadie I. Alzheimer’s Abeta fused to green fluorescent protein induces growth stress and a heat shock response. FEMS Yeast Res. 2007;7(8):1230–6. doi: 10.1111/j.1567-1364.2007.00285.x. [DOI] [PubMed] [Google Scholar]
- 13.Matlack KE, Tardiff DF, Narayan P, Hamamichi S, Caldwell KA, Caldwell GA, Lindquist S. Clioquinol promotes the degradation of metal-dependent amyloid-beta (Abeta) oligomers to restore endocytosis and ameliorate Abeta toxicity. Proc Natl Acad Sci U S A. 2014;111(11):4013–8. doi: 10.1073/pnas.1402228111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, Heun R, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1088–93. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ER, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Ruther E, Schurmann B, Heun R, Kolsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Gallacher J, Hull M, Rujescu D, Giegling I, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, van Duijn CM, Breteler MM, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, Berr C, Campion D, Epelbaum J, Dartigues JF, Tzourio C, Alperovitch A, Lathrop M, Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Bjornsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossu P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jonsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O’Donovan M, Amouyel P, Williams J. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43(5):429–35. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, Harold D, Schrijvers EM, Ramirez-Lorca R, Debette S, Longstreth WT, Jr, Janssens AC, Pankratz VS, Dartigues JF, Hollingworth P, Aspelund T, Hernandez I, Beiser A, Kuller LH, Koudstaal PJ, Dickson DW, Tzourio C, Abraham R, Antunez C, Du Y, Rotter JI, Aulchenko YS, Harris TB, Petersen RC, Berr C, Owen MJ, Lopez-Arrieta J, Varadarajan BN, Becker JT, Rivadeneira F, Nalls MA, Graff-Radford NR, Campion D, Auerbach S, Rice K, Hofman A, Jonsson PV, Schmidt H, Lathrop M, Mosley TH, Au R, Psaty BM, Uitterlinden AG, Farrer LA, Lumley T, Ruiz A, Williams J, Amouyel P, Younkin SG, Wolf PA, Launer LJ, Lopez OL, van Duijn CM, Breteler MM Consortium C; Consortium G; Consortium E. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303(18):1832–40. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramjaun AR, Micheva KD, Bouchelet I, McPherson PS. Identification and characterization of a nerve terminalenriched amphiphysin isoform. J Biol Chem. 1997;272(26):16700–6. doi: 10.1074/jbc.272.26.16700. [DOI] [PubMed] [Google Scholar]
- 18.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43(5):436–41. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossu P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Jr, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nothen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45(12):1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nair S, Traini M, Dawes IW, Perrone GG. Genome-wide analysis of Saccharomyces cerevisiae identifies cellular processes affecting intracellular aggregation of Alzheimer’s amyloid-beta42: importance of lipid homeostasis. Mol Biol Cell. 2014;25(15):2235–49. doi: 10.1091/mbc.E13-04-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tardiff DF, Jui NT, Khurana V, Tambe MA, Thompson ML, Chung CY, Kamadurai HB, Kim HT, Lancaster AK, Caldwell KA, Caldwell GA, Rochet JC, Buchwald SL, Lindquist S. Yeast reveal a “druggable” Rsp5/Nedd4 network that ameliorates alpha-synuclein toxicity in neurons. Science. 2013;342(6161):979–83. doi: 10.1126/science.1245321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cherny RA, Atwood CS, Xilinas ME, Gray DN, Jones WD, McLean CA, Barnham KJ, Volitakis I, Fraser FW, Kim Y, Huang X, Goldstein LE, Moir RD, Lim JT, Beyreuther K, Zheng H, Tanzi RE, Masters CL, Bush AI. Treatment with a copper-zinc chelator markedly and rapidly inhibits beta-amyloid accumulation in Alzheimer’s disease transgenic mice. Neuron. 2001;30(3):665–76. doi: 10.1016/s0896-6273(01)00317-8. [DOI] [PubMed] [Google Scholar]
- 23.Adlard PA, Cherny RA, Finkelstein DI, Gautier E, Robb E, Cortes M, Volitakis I, Liu X, Smith JP, Perez K, Laughton K, Li QX, Charman SA, Nicolazzo JA, Wilkins S, Deleva K, Lynch T, Kok G, Ritchie CW, Tanzi RE, Cappai R, Masters CL, Barnham KJ, Bush AI. Rapid restoration of cognition in Alzheimer’s transgenic mice with 8-hydroxy quinoline analogs is associated with decreased interstitial Abeta. Neuron. 2008;59(1):43–55. doi: 10.1016/j.neuron.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 24.Lannfelt L, Blennow K, Zetterberg H, Batsman S, Ames D, Harrison J, Masters CL, Targum S, Bush AI, Murdoch R, Wilson J, Ritchie CW. Safety, efficacy, and biomarker findings of PBT2 in targeting Abeta as a modifying therapy for Alzheimer’s disease: a phase IIa, double-blind, randomised, placebo-controlled trial. Lancet Neurol. 2008;7(9):779–86. doi: 10.1016/S1474-4422(08)70167-4. [DOI] [PubMed] [Google Scholar]
- 25.Faux NG, Ritchie CW, Gunn A, Rembach A, Tsatsanis A, Bedo J, Harrison J, Lannfelt L, Blennow K, Zetterberg H, Ingelsson M, Masters CL, Tanzi RE, Cummings JL, Herd CM, Bush AI. PBT2 rapidly improves cognition in Alzheimer’s Disease: additional phase II analyses. J Alzheimer’s Dis. 2010;20(2):509–16. doi: 10.3233/JAD-2010-1390. [DOI] [PubMed] [Google Scholar]
- 26.Faller P, Hureau C, Berthoumieu O. Role of metal ions in the self-assembly of the Alzheimer’s amyloid-beta peptide. Inorg Chem. 2013;52(21):12193–206. doi: 10.1021/ic4003059. [DOI] [PubMed] [Google Scholar]
- 27.Crouch PJ, Barnham KJ. Therapeutic redistribution of metal ions to treat Alzheimer’s disease. Acc Chem Res. 2012;45(9):1604–11. doi: 10.1021/ar300074t. [DOI] [PubMed] [Google Scholar]
- 28.Bharadwaj PR, Verdile G, Barr RK, Gupta V, Steele JW, Lachenmayer ML, Yue Z, Ehrlich ME, Petsko G, Ju S, Ringe D, Sankovich SE, Caine JM, Macreadie IG, Gandy S, Martins RN. Latrepirdine (dimebon) enhances autophagy and reduces intracellular GFP-Abeta42 levels in yeast. J Alzheimer’s Dis. 2012;32(4):949–67. doi: 10.3233/JAD-2012-120178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park SK, Pegan SD, Mesecar AD, Jungbauer LM, LaDu MJ, Liebman SW. Development and validation of a yeast high-throughput screen for inhibitors of Abeta(4)(2) oligomerization. Dis Models & Mech. 2011;4(6):822–31. doi: 10.1242/dmm.007963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci U S A. 2006;103(29):11045–50. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286(5441):971–4. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 32.Dexter PM, Caldwell KA, Caldwell GA. A predictable worm: application of Caenorhabditis elegans for mechanistic investigation of movement disorders. Neurotherapeutics. 2012;9(2):393–404. doi: 10.1007/s13311-012-0109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Douglas PM, Treusch S, Ren HY, Halfmann R, Duennwald ML, Lindquist S, Cyr DM. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc Natl Acad Sci U S A. 2008;105(20):7206–11. doi: 10.1073/pnas.0802593105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McColl G, Roberts BR, Pukala TL, Kenche VB, Roberts CM, Link CD, Ryan TM, Masters CL, Barnham KJ, Bush AI, Cherny RA European Alzheimer’s Disease I; Genetic; Environmental Risk in Alzheimer’s D; Alzheimer’s Disease Genetic C; Cohorts for H; Aging Research in Genomic E, Alzheimer’s Disease Neuroimaging I, consortium E, consortium C. Utility of an improved model of amyloid-beta (Abeta(1)(−)(4)(2)) toxicity in Caenorhabditis elegans for drug screening for Alzheimer’s disease. Mol Neurodegener. 2012;7:57. doi: 10.1186/1750-1326-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hassett R, Kosman DJ. Evidence for Cu(II) reduction as a component of copper uptake by Saccharomyces cerevisiae. J Biol Chem. 1995;270(1):128–34. doi: 10.1074/jbc.270.1.128. [DOI] [PubMed] [Google Scholar]
- 36.Dancis A, Roman DG, Anderson GJ, Hinnebusch AG, Klausner RD. Ferric reductase of Saccharomyces cerevisiae: molecular characterization, role in iron uptake, and transcriptional control by iron. Proc Natl Acad Sci U S A. 1992;89(9):3869–73. doi: 10.1073/pnas.89.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobsen JA, Fullagar JL, Miller MT, Cohen SM. Identifying chelators for metalloprotein inhibitors using a fragment-based approach. J Med Chem. 2011;54(2):591–602. doi: 10.1021/jm101266s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ajibade PA, Zulu NH. Metal complexes of diisopropylthiourea: synthesis, characterization and antibacterial studies. Int J Mol Sci. 2011;12(10):7186–98. doi: 10.3390/ijms12107186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Wang H, Lu Z, Zheng X, Ni W, Zhu J, Fu Y, Lian F, Zhang N, Li J, Zhang H, Mao F. Development of Multifunctional Pyrimidinylthiourea Derivatives as Potential Anti-Alzheimer Agents. J Med Chem. 2016;59(18):8326–44. doi: 10.1021/acs.jmedchem.6b00636. [DOI] [PubMed] [Google Scholar]
- 40.Bryant NJ, Stevens TH. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol Mol Biol Rev. 1998;62(1):230–47. doi: 10.1128/mmbr.62.1.230-247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith DG, Cappai R, Barnham KJ. The redox chemistry of the Alzheimer’s disease amyloid beta peptide. Biochim Biophys Acta, Biomembr. 2007;1768(8):1976–90. doi: 10.1016/j.bbamem.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 42.Chen X, Petranovic D. Amyloid-beta peptide-induced cytotoxicity and mitochondrial dysfunction in yeast. FEMS Yeast Res. 2015;15(6):fov061. doi: 10.1093/femsyr/fov061. [DOI] [PubMed] [Google Scholar]
- 43.Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol. 2009;7(1):25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- 44.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460(7257):823–30. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- 45.Cataldo AM, Peterhoff CM, Troncoso JC, Gomez-Isla T, Hyman BT, Nixon RA. Endocytic pathway abnormalities precede amyloid beta deposition in sporadic Alzheimer’s disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. Am J Pathol. 2000;157(1):277–86. doi: 10.1016/s0002-9440(10)64538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cataldo AM, Barnett JL, Pieroni C, Nixon RA. Increased neuronal endocytosis and protease delivery to early endosomes in sporadic Alzheimer’s disease: neuropathologic evidence for a mechanism of increased beta-amyloidogenesis. J Neurosci. 1997;17(16):6142–51. doi: 10.1523/JNEUROSCI.17-16-06142.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sevigny J, Chiao P, Bussiere T, Weinreb PH, Williams L, Maier M, Dunstan R, Salloway S, Chen T, Ling Y, O’Gorman J, Qian F, Arastu M, Li M, Chollate S, Brennan MS, Quintero-Monzon O, Scannevin RH, Arnold HM, Engber T, Rhodes K, Ferrero J, Hang Y, Mikulskis A, Grimm J, Hock C, Nitsch RM, Sandrock A. The antibody aducanumab reduces Abeta plaques in Alzheimer’s disease. Nature. 2016;537(7618):50–6. doi: 10.1038/nature19323. [DOI] [PubMed] [Google Scholar]
- 48.Castello MA, Jeppson JD, Soriano S. Moving beyond anti-amyloid therapy for the prevention and treatment of Alzheimer’s disease. BMC Neurol. 2014;14:169. doi: 10.1186/s12883-014-0169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng H, Fridkin M, Youdim MB. Novel chelators targeting cell cycle arrest, acetylcholinesterase, and monoamine oxidase for Alzheimer’s therapy. Curr Drug Targets. 2012;13(8):1096–1113. doi: 10.2174/138945012802009026. [DOI] [PubMed] [Google Scholar]
- 50.Lee S, Zheng X, Krishnamoorthy J, Savelieff MG, Park HM, Brender JR, Kim JH, Derrick JS, Kochi A, Lee HJ, Kim C, Ramamoorthy A, Bowers MT, Lim MH. Rational design of a structural framework with potential use to develop chemical reagents that target and modulate multiple facets of Alzheimer’s disease. J Am Chem Soc. 2014;136(1):299–310. doi: 10.1021/ja409801p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rizzo S, Tarozzi A, Bartolini M, Da Costa G, Bisi A, Gobbi S, Belluti F, Ligresti A, Allara M, Monti JP, Andrisano V, Di Marzo V, Hrelia P, Rampa A. 2-Arylbenzofuran-based molecules as multipotent Alzheimer’s disease modifying agents. Eur J Med Chem. 2012;58:519–32. doi: 10.1016/j.ejmech.2012.10.045. [DOI] [PubMed] [Google Scholar]
- 52.Capurro V, Busquet P, Lopes JP, Bertorelli R, Tarozzo G, Bolognesi ML, Piomelli D, Reggiani A, Cavalli A. Pharmacological characterization of memoquin, a multi-target compound for the treatment of Alzheimer’s disease. PLoS One. 2013;8(2):e56870. doi: 10.1371/journal.pone.0056870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bolognesi ML, Cavalli A, Melchiorre C. Memoquin: a multi-target-directed ligand as an innovative therapeutic opportunity for Alzheimer’s disease. Neurotherapeutics. 2009;6(1):152–62. doi: 10.1016/j.nurt.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gauthier S, Molinuevo JL. Benefits of combined cholinesterase inhibitor and memantine treatment in moderate-severe Alzheimer’s disease. Alzheimer’s Dementia. 2013;9(3):326–31. doi: 10.1016/j.jalz.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 55.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15(14):1541–53. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 56.Gietz RD. Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol Biol. 2014;1205:1–12. doi: 10.1007/978-1-4939-1363-3_1. [DOI] [PubMed] [Google Scholar]
- 57.Hu Y, Rolfs A, Bhullar B, Murthy TV, Zhu C, Berger MF, Camargo AA, Kelley F, McCarron S, Jepson D, Richardson A, Raphael J, Moreira D, Taycher E, Zuo D, Mohr S, Kane MF, Williamson J, Simpson A, Bulyk ML, Harlow E, Marsischky G, Kolodner RD, LaBaer J. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Res. 2007;17(4):536–43. doi: 10.1101/gr.6037607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lewis JA, Fleming JT. Basic culture methods. Methods Cell Biol. 1995;48:3–29. [PubMed] [Google Scholar]
- 60.Su LJ, Auluck PK, Outeiro TF, Yeger-Lotem E, Kritzer JA, Tardiff DF, Strathearn KE, Liu F, Cao S, Hamamichi S, Hill KJ, Caldwell KA, Bell GW, Fraenkel E, Cooper AA, Caldwell GA, McCaffery JM, Rochet JC, Lindquist S. Compounds from an unbiased chemical screen reverse both ER-to-Golgi trafficking defects and mitochondrial dysfunction in Parkinson’s disease models. Dis Models & Mech. 2010;3(3–4):194–208. doi: 10.1242/dmm.004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Collart MA, Oliviero S. Preparation of yeast RNA. Curr Protoc Mol Biol. 2001:13.12.1–13.12.5. doi: 10.1002/0471142727.mb1312s23. [DOI] [PubMed] [Google Scholar]
- 62.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.