Abstract
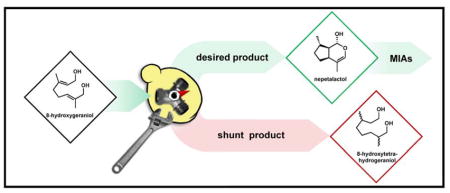
Monoterpene indole alkaloids (MIAs) represent a structurally diverse, medicinally essential class of plant derived natural products. The universal MIA building block strictosidine was recently produced in the yeast Saccharomyces cerevisiae, setting the stage for optimization of microbial production. However, the irreversible reduction of pathway intermediates by yeast enzymes results in a non-recoverable loss of carbon, which has a strong negative impact on metabolic flux. In this study, we identified and engineered the determinants of biocatalytic selectivity which control flux towards the iridoid scaffold from which all MIAs are derived. Development of a bioconversion based production platform enabled analysis of the metabolic flux and interference around two critical steps in generating the iridoid scaffold: oxidation of 8-hydroxygeraniol to the dialdehyde 8-oxogeranial followed by reductive cyclization to form nepetalactol. In vitro reconstitution of previously uncharacterized shunt pathways enabled the identification of two distinct routes to a reduced shunt product including endogenous ‘ene’-reduction and non-productive reduction by iridoid synthase when interfaced with endogenous alcohol dehydrogenases. Deletion of five genes involved in α,β-unsaturated carbonyl metabolism resulted in a 5.2-fold increase in biocatalytic selectivity of the desired iridoid over reduced shunt product. We anticipate that our engineering strategies will play an important role in the development of S. cerevisiae for sustainable production of iridoids and MIAs.
Keywords: Saccharomyces cerevisiae, Iridoids, Monoterpene indole alkaloids, Old Yellow Enzyme
Graphical Abstract
1. INTRODUCTION
Natural products and their derivatives comprise a pharmaceutically indispensable category of molecules. The monoterpene indole alkaloids (MIAs) are a class of plant natural products with structural complexities matched by potent biological activities (O’Connor and Maresh, 2006). The functional diversity of MIAs ranges from anticancer to antimalarial to antiaddiction, resulting in immense clinical relevance for this class of natural products. Of particular importance is the microtubule inhibitor vinblastine, which is a chemotherapeutic on the World Health Organization’s List of Essential Medicines. However, the native producer Catharanthus roseus yields just 0.001 percentage dry weight vinblastine (Datta and Srivastava, 1997), making it one of the most expensive small molecules on the market. Increasing clinical demand coupled with the environmental impact of isolation make MIAs an ideal target for heterologous microbial production (Leonard et al., 2009). Strictosidine is a universal biosynthetic precursor to ~3,000 plant MIAs. Strictosidine itself is biosynthesized from the monoterpene geraniol via an iridoid intermediate nepetalactol 3 (Fig. 1), which is further decorated through a series of oxidations, methylation, and glucosylation before ultimately being condensed with tryptamine (Fig. S1). The biosynthesis of strictosidine was recently reconstituted in yeast, albeit with low yields largely attributed to suboptimal flux through the early seco-iridoid pathway (Brown et al., 2015; Miettinen et al., 2014). In addition to serving as the monoterpene scaffold in MIAs, a number of iridoid natural products are gaining clinical interest due to their anticancer, antibacterial, and anti-inflammatory properties (Tundis et al., 2008). Optimizing nepetalactol production in yeast is therefore a crucial requisite for realizing microbial production of a wide array of high value compounds including iridoids, strictosidine, as well as all downstream MIA natural products. Accordingly, the recent surge in metabolic engineering technologies in yeast has invigorated natural product pathway engineering efforts (Billingsley et al., 2016; Krivoruchko and Nielsen, 2015).
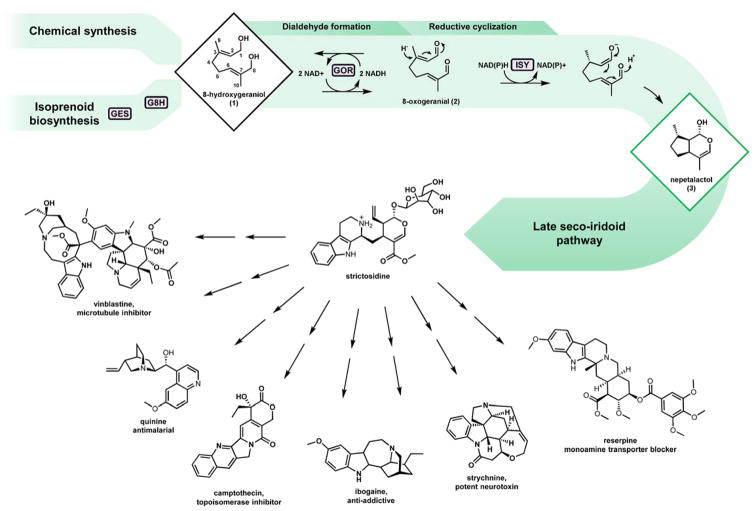
Fig. 1. Monoterpene indole alkaloid biosynthesis.
The pathway intermediate 8-hydroxygeraniol 1 may be produced via chemical synthesis or de novo via isoprenoid biosynthesis. De novo production proceeds via the monoterpene precursor geranyl pyrophosphate, which may be hydrolyzed and ω-hydroxylated by GES and G8H respectively. The resulting diol 1 is converted by GOR to the dialdehyde 8-oxogeranial 2, which undergoes reductive cyclization by ISY. The iridoid 3 scaffold is then subject to a number of oxidations, methylation, glucosylation, and condensation with tryptamine to generate the universal monoterpene indole alkaloid (MIA) precursor strictosidine. Several MIAs with important biological activities are shown.
Biosynthesis of nepetalactol in yeast requires heterologous expression of four enzymes. The terpene synthase geraniol synthase (GES) hydrolyzes geranyl diphosphate (GPP) into geraniol, which is then oxidized by the P450 geraniol 8-hydroxylase (G8H) into 8-hydroxygeraniol 1. Double two e− oxidation catalyzed by geraniol oxidoreductase (GOR) afford the dialdehyde 8-oxogeranial 2, which then undergoes reductive cyclization by iridoid synthase (ISY) to yield the bicyclic nepetalactol 3 (Geu-Flores et al., 2012). Production of the iridoid scaffold appears concise compared to other plant pathways that have recently been reconstituted in yeast, such as those producing sesquiterpenoids (Paddon et al., 2013) and benzylisoquinoline alkaloids (Galanie et al., 2015; Li and Smolke, 2016). However, building iridoid (and in turn MIA) “platform strains” (Nielsen, 2015) has proven to be challenging with each of the four enzymatic steps posing its own metabolic engineering obstacles. Compared to the sesquiterpene precursor FPP, the monoterpene precursor GPP exists only transiently in native yeast metabolism and readily undergoes isoprenoid chain elongation by ERG20 (Fischer et al., 2011). Strain engineering to increase monoterpene precursor production has recently led to significant improvements in geraniol titers. (Jiang et al., 2017; Zhao et al., 2017). The G8H-mediated ω-hydroxylation of geraniol at C-8 was described as a major pathway bottleneck in S. cerevisiae due to poor activity of the P450 (Brown et al., 2015). Engineering strategies aimed at optimizing P450 performance are expected to overcome this limitation (Jung et al., 2011; Renault et al., 2014). As substrates of GOR and ISY, the α,β-unsaturated alcohol 1 and aldehyde 2, respectively, are subject to significant crosstalk and interference with the endogenous yeast redox metabolism. For example, efforts in de novo iridoid production were derailed when it was discovered that saturated shunt products accumulate when the iridoid pathway is expressed in yeast (Campbell et al., 2016). The α,β-unsaturated aldehydes present in the linear iridoid precursor 2 are required in the cyclization mechanism of ISY towards the formation of nepetalactol 3 (Lindner et al., 2014). Thus, saturated shunt products represent a non-recoverable loss in carbon which severely limits overall iridoid yields. While several reports have described the reduction of the double bonds in monoterpene pathway intermediates by yeast (Gramatica et al., 1982; King and Richard Dickinson, 2000; Steyer et al., 2013), the exact mechanism has remained unresolved particularly with regard to crosstalk between exogenous biosynthetic enzymes (GOR and ISY) and endogenous yeast enzymes. However the low benchmark titers of 8-hydroxygeraniol 2 achieved to date (5.3 mg/L) (Campbell et al., 2016), in combination with the obscured mechanistic details of shunt product formation have significantly limited the reconstitution of nepetalactol 3 production and overall MIA pathway engineering in yeast.
Towards gaining deeper understanding of the enzymatic basis of shunt product formation, we designed a yeast-based bioconversion platform to assess activities of GOR and ISY. Our strategy leverages the facile synthesis of the substrate 8-hydroxygeraniol 1 (Bogazkaya et al., 2014), followed by bioconversion of this pathway intermediate by engineered S. cerevisiae. Characterization of the major shunt products followed by in vitro pathway reconstitution allowed for the clarification of two distinct mechanisms of shunt product formation en route to iridoid 3 production: endogenous ‘ene’-reduction and reduction by iridoid synthase in a non-productive redox cascade with yeast alcohol dehydrogenases. Our study provides strategies that can be directly employed to improve the performance of iridoid producing strains.
2. MATERIALS AND METHODS
2.1. Strains and plasmids
The strains and plasmids used in this study are listed in Table 1. JHY651 (S1) was constructed from BY4742 (Brachmann et al., 1998) to correct previously identified issues associated with sporulation, mitochondrial stability, and heterologous protein degradation. (Method S1). Plasmids pJB046, pJB047, pJB053, pJB057, and pJB097 were utilized for subsequent CRISPR/Cas9 mediated strain modifications of JHY651. Unless otherwise stated, HICRISPR (Bao et al., 2015) was successfully employed for subsequent gene deletions using gRNAs designed with CRISPy Cas9 target finder (Jakounas et al., 2015), pCRCT was a gift from Huimin Zhao (Addgene plasmid # 60621). Knockout of ADH7 and ARI1 required a two-stage deletion strategy involving integration of a LEU2 marker followed CRISPR-mediated pop-out of the marker using pJB097. Plasmids were maintained and propagated in XL1-Blue E. coli. The pathway genes GOR and ISY were codon optimized for expression in S. cerevisiae and synthesized by Gen9 (Fig. S2). Late-stage inducible promoters (ADH2p (Lee and DaSilva, 2005) and PCK1p (Proft et al., 1995)) and high-capacity terminators (PRM9t and CPS1t (Curran et al., 2013)) were amplified from yeast gDNA. Expression cassettes were first generated via SOE PCR and then cloned into PCR blunt. Yeast expression plasmids pJB031, pJB033, and pJB034 were assembled using yeast homologous recombination. All yeast transformations were carried out using the LiOAc/ssDNA method (Gietz and Schiestl, 2007). Transformants were selected using uracil dropout media or YPD supplemented with 200 μg/mL hygromycin. Protein expression plasmids were assembled using digestion ligation (pJB042, pJB043, pJB044, pJB045, pJB071, pJB072) or yeast homologous recombination (pJB095). Primers used for strain and plasmid construction are listed in Table S1.
Table 1.
Strains and plasmids used in this study.
| Strain | Parent | Genotype | Reference | |
|---|---|---|---|---|
| BY4742 | S288C | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | Brachmann et al., Yeast, 1998 | |
| DHY214 | BY4742 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 SAL1+ CAT5(91M) MIP1(661T) MKT1(30G) RME1(INS-308A) TAO3(1493Q) HAP1+ | Chu and Horecka, Stanford Genome Technology Center | |
| S1 | JHY651* | DHY214 | MATα prb1Δ pep4Δ his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | ibid. |
| S2 | YJB011* | JHY651 | MATα prb1Δ pep4Δ his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 oye2Δ1 | This study |
| S3 | YJB012* | JHY651 | MATα prb1Δ pep4Δ his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 oye3Δ1 | This study |
| S4 | YJB013* | YJB011 | MATα prb1Δ pep4Δ his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 oye2Δ1 oye3Δ1 | This study |
| S5 | YJB023* | YJB013 | MATα prb1Δ pep4Δ his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 oye2Δ1 oye3Δ1 gre2Δ1 | This study |
| YJB025* | YJB013 | MATα prb1Δ pep4Δ his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 oye2Δ1 oye3Δ1 ari1::LEU2 | This study | |
| S6 | YJB027* | YJB025 | MATα prb1Δ pep4Δ his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 oye2Δ1 oye3Δ1 ari1Δ0 | This study |
| S7 | YJB018* | YJB013 | MATα prb1Δ pep4Δ his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 oye2Δ1 oye3Δ1 adh6Δ1 | This study |
| YJB024* | YJB013 | MATα prb1Δ pep4Δ his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 oye2Δ1 oye3Δ1 adh7::LEU2 | This study | |
| S8 | YJB052* | YJB024 | MATα prb1Δ pep4Δ his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 oye2Δ1 oye3Δ1 adh7Δ0 | This study |
| YJB030* | YJB027 | MATα prb1Δ pep4Δ his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 oye2Δ1 oye3Δ1 ari1Δ0 adh7::LEU2 | This study | |
| YJB033* | YJB030 | MATα prb1Δ pep4Δ his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 oye2Δ1 oye3Δ1 ari1Δ0 adh7Δ0 | This study | |
| S9 | YJB051* | YJB033 | MATα prb1Δ pep4Δ his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 oye2Δ1 oye3Δ1 ari1Δ0 adh7Δ0 adh6Δ1 | This study |
| Plasmid | Description | Reference |
|---|---|---|
| pJB031 | 2μ; URA3; AmpR | This study |
| pJB033 | 2μ; URA3; AmpR; ADH2p-GOR-PRM9t | This study |
| pJB034 | 2μ; URA3; AmpR; ADH2p-GOR-PRM9t; PCK1p-ISY-CPS1t | This study |
| pJB046 | pCRCT-Δoye2 | This study |
| pJB047 | pCRCT-Δoye3 | This study |
| pJB057 | pCRCT-Δgre2 | This study |
| pJB053 | pCRCT-Δadh6 | This study |
| pJB097 | 2μ; kanMX; AmpR; iCas9; SNR52p-LEU2crRNA-sgRNA-SUP4t | This study |
| pJB042 | pET-28a-6×HIS-GOR | This study |
| pJB043 | pET-28a-6×HIS-ISY | This study |
| pJB044 | pET-28a-6×HIS-OYE2 | This study |
| pJB045 | pET-28a-6×HIS-OYE3 | This study |
| pJB072 | pET-28a-6×HIS-ADH6 | This study |
| pJB071 | pET-28a-6×HIS-ADH7 | This study |
| pJB095 | 2μ; URA3; AmpR; ADH2p-6xHIS-ARI1-PRM9t | This study |
Notes:
strain is also: SAL1+ CAT5(91M) MIP1(661T) MKT1(30G) RME1(INS-308A) TAO3(1493Q) HAP1+
2.2. Single fed-batch and repeated fed-batch bioconversion assay
Single colony transformants were repatched onto uracil dropout plates. A starter culture was prepared by stabbing through a patch and inoculating 1 mL of liquid uracil dropout media in a culture tube. This starter culture was shaken at 28 °C and 250 rpm for 24 hours. Next, three culture tubes containing 3 mL YPD were each inoculated with 100 μL of the starter culture and shaken at 28 °C and 250 rpm for 24 hours. The sub-cultures were pooled and centrifuged at 4,000 g for 5 minutes, after which 6 mL of the supernatant was removed. Concentrated culture was split into two 1.5 mL volumes, one sample to which substrate was added and a negative control to which no substrate was added. For the single fed-batch bioconversion assay, 15 μL of 200 mM 8-hydroxygeraniol dissolved in ethanol was added to the sample, and 15 μL of ethanol was added to the negative control. For the repeated fed-batch bioconversion assay, 0.75 μL of 200 mM 8-hydroxygeraniol dissolved in ethanol was added to the sample every hour for 20 hours. During the 8-hydroxygeraniol bioconversion phase, cultures were shaken at 28 °C and 250 rpm. 24 hours after the initial addition of substrate, biocatalytic selectivity was analyzed.
2.3. Culture extraction and quantification
Samples were analyzed by extracting 700 μL culture with 300 μL of an organic phase consisting of 75% ethyl acetate and 25% acetone. The samples were vortexed for 1 minute and then centrifuged for 10 minutes. The organic layer was analyzed on an Agilent Technologies GC-MS 6890/5973 equipped with a DB-FFAP column. An inlet temperature of 220 °C and constant pressure of 4.2 psi were used. The oven temperature was held at 60 °C for 5 minutes and then ramped at 60 °C/min for 1.5 min, followed by a ramp of 15 °C/min for 16 minutes and a hold for 10 minutes. A monoterpene standard curve was generated using 8-hydroxygeraniol and used to quantify all analytes by assuming an equivalent mass response factor (Fig. S3). Biocatalytic selectivity is defined as molar production rate of iridoid relative to molar production rate of reduced shunt product, and was calculated based on end-point concentrations of monoterpenes. For each strain, the biocatalytic selectivity of three biological isolates was measured. Means and standard errors are reported.
2.4. Compound isolation and NMR analysis
Large scale shake flask culture was utilized for compound isolation. A starter culture of S1 transformed with pJB034 was prepared by stabbing through a patch and inoculating 2 mL of liquid uracil dropout media in a culture tube. This starter culture was shaken at 28 °C and 250 rpm for 24 hours and then transferred to 50 mL liquid uracil dropout. After 24 hours, 2 L culture was inoculated. 24 hours later 340 mg 8-hydroxygeraniol was fed to 2 L culture to a final concentration of 1 mM. After 24 hours, the culture was spun down. The supernatant was extracted with an equal volume of ethyl acetate and the pellet was extracted with an equal volume of 50:50 mixture of ethyl acetate:acetone. The organic fractions were combined and dried. The resulting crude extract was separated using ISCO-CombiFlash Rf 200 (Teledyne Isco) with a gradient of hexane and acetone. The fractions containing the reduced shunt product were pooled and purified with HPLC using a Shimadzu Prominence HPLC (Phenonenex Kinetex, 5μ, 10.0 × 250 mm, C-18 column). The elution method was a linear gradient of 35–55% (v/v) CH3CN/H2O in 20.5 minutes, followed by a linear gradient of 55–95% in 2 minutes, followed by 95% CH3CN/H2O for 3 minutes with a flow rate of 3.0 mL/min. Structures were solved by NMR (Fig. S13 – S22, Table S3 – S4).
2.5. Protein purification and in vitro assays
The expression plasmids (pJB042, pJB043, pJB044, pJB045, pJB072, and pJB071 were each transformed into the E. coli strain BL21 for expression of GOR, ISY, OYE2, OYE3, ADH6, and ADH7, respectively. Cells were inoculated into 1 L LB containing 100 μg/mL kanamycin and cultured at 37 °C and 250 rpm to an OD600 of 0.6. Protein expression was induced via addition of 0.1 mM IPTG, followed 16 hours of continued shaking at 16 °C and 250 rpm. The expression plasmid pJB095 was transformed into JHY651 for expression of ARI1. Cells were inoculated into 2 mL liquid uracil dropout media and cultured at 28 °C and 250 rpm for 24 hours. This culture was used to inoculate YPD (1L) and cultured at 28 °C and 250 rpm for 3 days. Protein purification was carried out at 4 °C following standard Nickel-NTA affinity chromatography protocols. Purified enzymes were concentrated and exchanged into protein storage buffer, then visualized using SDS-PAGE (Fig. S5). Concentration was determined by Bradford assay and protein was stored at −80 °C. In vitro reactions were performed in 250 μL reactions. Unless otherwise stated, reactions consisted of 200 μM 8-hydroxygeraniol, 400 μM NAD+, 400 μM NADPH, and 1 μM of the specified enzyme(s) in 50 mM bis-tris propane buffer, pH = 9. After the specified amount of time, the assay was extracted with 100 μL ethyl acetate and analyzed using GC-MS as described for metabolite quantification.
2.6. Fermentation, growth rate, and plasmid stability
A New Brunswick BioFlo/CelliGen 115 fermenter equipped with a 2 L vessel was utilized for the bioconversion-based continuous fed-batch fermentation. A starter culture was prepared by stabbing through a patch and inoculating 2 mL of liquid uracil dropout media in a culture tube. This starter culture was shaken at 28 °C and 250 rpm for 24 hours and then transferred to 50 mL liquid uracil dropout. After 24 hours, the culture was centrifuged at 800 g for 5 minutes and resuspended in 20 mL YPD. The fermenter working volume was 1 L; the media consisted of YPD. Following inoculation to an OD600 of 0.1, the fermenter pH, dissolved oxygen, and temperature were automatically regulated. pH was maintained at pH 5 by addition of 2 M NaOH or 2 M HCl. Dissolved oxygen was maintained at 25% by adjustment of the agitation rate. Temperature was maintained at 28 °C using a heating jacket and recirculated chilled water. Recirculated chilled water was also used to cool the exhaust condenser. Following 18 hours of culture, 30 mL ethanol was added to the fermenter using a syringe pump at a flow rate of 2 mL/minute for increased induction of pathway genes. Six hours later, continuous substrate addition was carried out; 30 mL of 67 mM 8-hydroxygeraniol was added to the fermenter using a syringe pump at a flow rate of 2.5 mL/hr. Growth rates on glucose and ethanol were calculated using data points falling within the linear portions of the log plot. Plasmid stability was calculated by diluting fermentation samples 105 fold, plating 10 μL on YPD and uracil dropout plates, and comparing total CFU.
2.7 Synthesis of 8-hydroxygeraniol
8-Hydroxygeraniol 1 was prepared using minor modifications of the known route (Bogazkaya et al., 2014). Beginning with geranyl acetate, treatment with SeO2 and tert-butyl hydroperoxide led to regioselective allylic hydroxylation. Subsequent cleavage of the acetate group using methanolysis conditions afforded 8-hydroxygeraniol 1 in 49% yield over 2 steps (Method S2, Fig. S11 – S12).
3. RESULTS AND DISCUSSION
3.1. Purification of the major shunt product reveals complete reduction of 8-hydroxygeraniol to 8-hydroxytetrahydrogeraniol
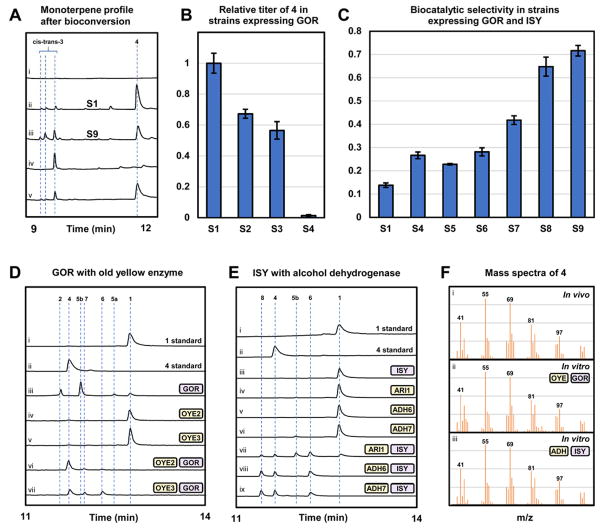
To probe the metabolic fate of 8-hydroxygeraniol 1 in yeast both in the presence and absence of the downstream enzymes GOR and ISY, we performed bioconversion assays followed by metabolite analysis. To obtain sufficient amounts of the substrate for thorough analysis, we chemically synthesized 8-hydroxygeraniol 1 following reported methods (Bogazkaya et al., 2014). Both single fed-batch and repeated fed-batch methods of substrate delivery were explored. We began by performing the single fed-batch bioconversion assay on the parent strain JHY651 (S1) transformed with a control vector (pJB031), a plasmid encoding GOR (pJB033), and a plasmid encoding both GOR and ISY (pJB034). The ethanol inducible, late-stage promoters ADH2p and PCK1p were employed to drive expression of GOR and ISY respectively (Lee and DaSilva, 2005; Proft et al., 1995). The use of these late-stage promoters decoupled the growth phase from protein production phase as well as eliminated the need for addition of an inducer for transcriptional activation (Peng et al., 2015; Weinhandl et al., 2014). 2 mM of 1 was added at the onset of yeast diauxic shift and the biotransformation was allowed to proceed for 24 hours followed by metabolite analysis. While a small amount of nepetalactol 3 was detected in S1 expressing both GOR and ISY (Fig. 3A, trace ii), most of the fed substrate 1 was converted to shunt product 4, which was absent from cultures not supplemented with 8-hydroxygeraniol (Fig. 3A, trace i). Large-scale purification followed by NMR analysis (Fig. S13 – S17, Table S3) allowed us to identify 4 as the saturated 8-hydroxytetrahydrogeraniol (Fig. 2). We also observed oxidation products of 4 and were able to isolate a mixture of the saturated 8-hydroxy-2,6-dimethyloctanoic and 8-hydroxy-3,7-dimethyloctanoic acids (Fig. S18 – 22, Table S4). These reduced shunt compounds also dominated the metabolic profiles of S1 expressing GOR (Fig. S5, trace ii). The reduction of 1 to 4 in the parent strain transformed with the control vector pJB031, however, is greatly attenuated (Fig. S5, trace i). In strains not expressing GOR, 1 remained present after 24 hours, with a small amount converted to 4 and the partially reduced 8-hydroxy-2,3-dihydrogeraniol 6. This result therefore suggested that GOR expression plays a key role in the formation of the saturated shunt product 4. To establish a starting point for strain improvement, we performed a repeated fed-batch bioconversion assay during which 0.1 mM 1 was added every hour for 20 hours. In the parent strain expressing GOR and ISY, biocatalytic selectivity of iridoid 3 over the primary shunt product 4 was determined to be 0.14 ± 0.01.
Fig. 3. Engineering and elucidation of 8-hydroxygeraniol reduction.
(A) GC-MS analysis of monoterpene profile after in vivo and in vitro bioconversion, the identity of 3 was confirmed by comparison to reported mass spectra, (i) S1 + pJB034 repeated fed-batch bioconversion with no substrate, (ii) S1 + pJB034 repeated fed-batch bioconversion with substrate, (iii) S9 + pJB034 repeated fed-batch bioconversion with substrate, (iv) product profile for in vitro assay with 200 μM 8-hydroxygeraniol, 400 μM NAD+, GOR, ISY (v) product profile for in vitro assay with 200 μM 8-hydroxygeraniol, physiological concentration of cofactors, GOR, ISY, OYE2, OYE3, ARI1, ADH6, ADH7. (B) Relative shunt product titer in strains expressing GOR. Deletion of OYE2 and OYE3 (S4) resulted in elimination of endogenous ‘ene’-reduction. Means and standard errors are reported. (C) Deletion of yeast ADHs resulted in an increase in the production of iridoid relative to reduced shunt product, corresponding to an increase in the biocatalytic selectivity. Means and standard errors are reported. (D) Reconstitution of endogenous ‘ene’-reduction in vitro, (i) 8-hydroxygeraniol standard, (ii) 8-hydroxytetrahydrogeraniol standard, (iii) 8-hydroxygeraniol + GOR, (iv) 8-hydroxygeraniol + OYE2, (v) 8-hydroxygeraniol + OYE3, (vi) 8-hydroxygeraniol + GOR + OYE2, (vii) 8-hydroxygeraniol + GOR + OYE3. (E) Reconstitution of non-productive reduction by ISY in vitro, (i) 8-hydroxygeraniol standard (ii) 8-hydroxytetrahydrogeraniol standard, (iii) 8-hydroxygeraniol + ISY, (iv) 8-hydroxygeraniol + ARI1, (v) 8-hydroxygeraniol + ADH6, (vi) 8-hydroxygeraniol + ADH7, (vii) 8-hydroxygeraniol + ARI1 + ISY, (viii) 8-hydroxygeraniol + ADH6 + ISY, (ix) 8-hydroxygeraniol + ADH7 + ISY. (F) Comparison of mass spectra of (i) in vivo shunt product, (ii) GOR + OYE in vitro reaction product, and (iii) ADH + ISY in vitro reaction product.
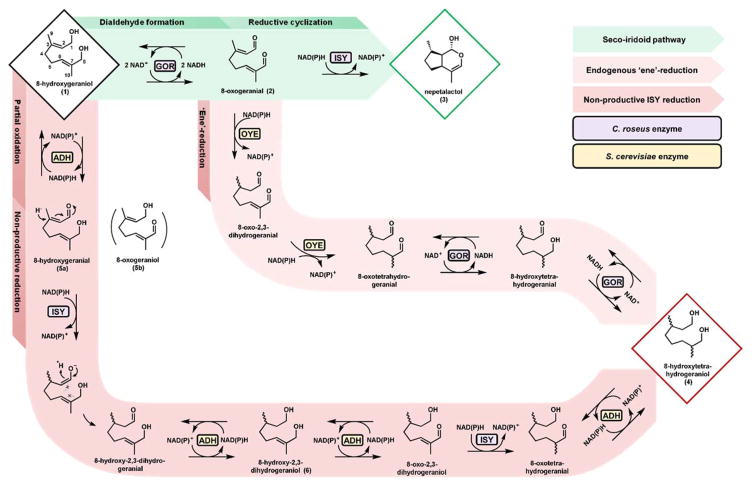
Fig. 2. Two distinct routes to a reduced shunt product.
Endogenous ‘ene’-reduction (top shunt pathway) by yeast OYEs occurs after oxidation of the fed substrate to an α,β-unsaturated carbonyl by GOR. The resulting saturated dialdehyde can then be reduced by an endogenous or exogenous alcohol dehydrogenase, as substrate oxidation has been shown to be reversible and cofactor driven. Alternatively, non-productive reduction (bottom pathway) by ISY occurs when the enzyme acts on a partially oxidized substrate. Fed substrate is incompletely oxidized by a yeast ADH, which enables premature hydride addition. The resulting enolate is quenched by a proton instead of the anticipated Michael acceptor, and the intended cyclization is interrupted. For each of the shunt pathways, one possible route to the reduced shunt product is highlighted, whereas multiple permutations of enzymatic sequence contribute to shunt product formation in practice.
3.2. Knockout of OYE2 and OYE3 results in elimination of endogenous α,β-unsaturated carbonyl reductase activity
We hypothesized that the product of GOR, 8-oxogeranial 2, may be prone to ‘ene’-reduction (reduction of the α,β-double bond). S. cerevisiae harbors two old yellow enzymes (OYE2 and OYE3) which catalyze the flavin-dependent, stereospecific reduction of α,β-unsaturated double bonds (Williams and Bruce, 2002). Previous studies concluded that OYE2 (but not OYE3) is involved in direct α,β-unsaturated monoterpenol reduction and is a target for increasing flux through the strictosidine pathway (Steyer et al., 2013). Other reports however, indicated that OYEs cannot reduce α,β-unsaturated alcohols, and that substrate reduction must proceed through activated α,β-unsaturated carbonyl intermediates (Brenna et al., 2012; Lonsdale and Reetz, 2015). In order to ascertain the role of yeast OYEs in shunt product 4 formation, we deleted these enzymes in the parent strain using CRISPR/Cas9 (Fig. 3B). We first deleted OYE2 to generate strain S2. This strain was transformed with pJB033 expressing GOR and the single fed-batch bioconversion assay was performed. The final amount of saturated substrate 4 in S2 decreased by 33% compared to S1/pJB033, validating the role of OYE2 in reduction of 8-hydroxygeraniol 1. We then deleted OYE3 in the parent strain to yield S3, which was transformed with pJB033 and similarly assayed for formation of 4. Curiously, the OYE3 knockout provided a greater decrease in shunt product 4 level of 44%, indicating that OYE3 is also involved in shunt product 4 formation and suggesting degenerate roles of the enzymes in ‘ene’-reduction. A combined knockout strain of OYE2 and OYE3 was generated (S4), in which expression of GOR and feeding of 1 led to a 99% reduction in the level of shunt product 4 (Fig. 3B). In the strain S4/pJB033, while formation of 4 was completely eliminated, we instead observed accumulation of oxidation products of 2 (Fig. S6, trace iii), indicating that endogenous aldehyde dehydrogenases can oxidize the dialdehyde in the absence of ISY. No apparent growth defects were observed in strains S2 – S4. Our results therefore show that both OYE2 and OYE3 contribute significantly to shunt product 4 formation (Fig. 2, top shunt pathway), and that a double knockout is required to prevent this mode of reduction. Several studies have shown that OYE3 expression is tightly regulated and can be induced on α,β-unsaturated alcohols (Ma and Liu, 2010; Trotter et al., 2006). Meanwhile, previous efforts to characterize monoterpene alcohol metabolism was conducted at approximately 10-fold less substrate concentration, which is potentially lower than the required threshold for OYE3 induction. The significant transcriptional changes in response to oxidative stress response, and metabolite accumulation should therefore be considered in the context of expressing redox-intensive biosynthetic pathways. Because an estimated one-sixth of all natural products contain an α,β-unsaturated carbonyl moiety (Rodrigues et al., 2016), elimination of endogenous ‘ene’-reduction may play a key role in future natural product biosynthetic efforts.
3.3. In vitro reconstitution of old yellow enzyme activity
In order to determine the exact reaction catalyzed by OYE2 and OYE3 that leads to the reduction of 8-hydroxygeraniol 1 to 4, we performed in vitro characterization of the enzymes (Fig. 3D). Both OYE as well as GOR were expressed and purified from E. coli (Fig. S4). Incubation of 8-hydroxygeraniol 1 with GOR and either OYE2 (Fig. 3D, trace vi) or OYE3 (Fig. 3D, trace vii), in the presence of cofactors NAD+ and NADPH for 1 hour resulted in the production of a compound with identical retention time and mass spectra as the saturated shunt product 4 isolated from the yeast-based bioconversion (Fig. 3F, spectra i and ii). We also observed the 8-hydroxy-2,3-dihydrogeraniol 6 that corresponds to a product that has undergone one α,β-reduction. Based on retention time and mass spectra (Fig. S8), compound 7 is likely a partially saturated aldehyde en route to the fully reduced shunt product 4. Incubation of 8-hydroxygeraniol with GOR alone produced the expected monoaldehydes 5a and 5b (Fig. 2) as well as the product dialdehyde 2, which is consistent with previous in vitro observations (Miettinen et al., 2014). Incubation of 8-hydroxygeraniol 1 directly with either OYE2 (Fig. 3D, trace iv) or OYE3 (Fig. 3D, trace v) resulted in no reaction, consistent with GOR mediated oxidation increasing shunt product 4 in yeast. Our data confirm that the ‘ene’-reductases OYE2 and OYE3 cannot act directly on the α,β-unsaturated alcohols in substrates such as 8-hydroxygeraniol 1, but can catalyze the reduction of double bonds in the activated α,β-unsaturated aldehyde 2. This observation is consistent with the canonical mechanism of ‘ene’-reductases involving a Michael-type hydride addition: NAD(P)H is not a strong enough reductant to reduce unactivated double bonds such as those present in α,β-unsaturated alcohols. From the in vitro assays, we noted that OYE2 can use either NADPH or NADH as the reducing cofactor, whereas OYE3 has a preferred cofactor of NADPH (Fig. S7). Our experience further confirms the reversible nature of GOR, and indicates the enzyme can also accept the saturated aldehydes for reduction into 4. The exact order of dehydrogenation, ‘ene’-reduction, and aldehyde reduction with respect to the different allylic alcohols in 1 have not been determined, with one possible route from 8-hydroxygeraniol 1 to 8-hydroxytetrahydrogeraniol 4 shown in Fig. 2.
3.4. Expression of ISY in yeast led to formation of the saturated shunt product even in OYE deletion strains
Having identified that both OYE2 and OYE3 are involved in the formation of 4 in the presence of GOR, we next coexpressed both GOR and ISY in the double knockout strain S4 to measure the in vivo selectivity of converting 1 to either 3 or 4. Surprisingly, whereas essentially none of substrate 1 was converted to 8-hydroxytetrahydrogeraniol 4 in S4 expressing GOR alone (Fig. S5, trace iii), over 90% of 1 was reduced to 4 (and the oxidized acid products) in S4 expressing GOR and ISY (Fig. S5, trace iv). The selectivity in the conversion of 1 to 3 increased from 0.14 to 0.27, indicating elimination of the OYEs was beneficial towards formation of iridoid. However, the coexpression of ISY unexpectedly led to the reemergence of the undesired shunt product 4. Campbell et al. were the first to report a similar observation of the involvement of ISY in monoterpenol reduction. However the current mechanistic understanding of ISY cyclization involves a Michael-type hydride addition to the unsaturated aldehyde (Figs. 1 and 2) (Lindner et al., 2014), thereby precluding direct reduction of α,β-unsaturated alcohols. ISY first catalyzes the NAD(P)H-dependent reduction of the C2-C3 α,β-unsaturated aldehyde in 2 to generate the enolate anion. This sets up an intramolecular cyclization via 1,4-addition to form the cyclopentane ring that is present in 3 (Geu-Flores et al., 2012). The enzyme was proposed to undergo a conformation change after enolate formation to accommodate folding of the substrate and subsequent cyclization (Kries et al., 2016). Studies using substrate analogs indeed showed ISY can catalyze hydride transfer to a wide variety of α,β-unsaturated carbonyls (Geu-Flores et al., 2012). We therefore rationalized that if substrate 1 is only selectively oxidized at one of the terminal alcohols to yield a mono-aldehyde (for example, compound 5a in Fig. 2) instead of the dialdehyde 2, the enone moiety in 5a can still be reduced by ISY. However, the subsequent enolate cannot perform the Michael addition to the allylic alcohol, leading to the formation of the reduced 8-hydroxy-2,3-dihydrogeranial from 5a via quenching of the enolate with a proton. Once reduced, this intermediate can be subjected to further redox modifications by both endogenous yeast enzymes and ISY for the second α,β reduction to ultimately arrive at the saturated diol 4 (Fig. 2, bottom shunt pathway). The incomplete oxidation of 1 to a monoaldehyde can therefore lead to “interrupted” cyclization by ISY, and is detrimental to the selective formation of 3 from 1.
Interestingly our in vitro assay with GOR and ISY in the presence of NAD+ and 1 led to complete conversion to 3 with no trace of shunt products (Fig. 3A, trace iv) indicating formation of the dialdehyde 2 is highly controlled to ensure no “interrupted” cyclization can occur. We therefore hypothesized that in yeast, formation of the monoaldehyde intermediates such as 5a or 5b must be catalyzed by endogenous yeast alcohol dehydrogenases (ADHs) in competition with GOR, and elimination/minimization of responsible ADH activities in S4 can lead to improvement in the selectivity towards 3. In other words, tuning the extent of in vivo oxidation of substrate 1 at the time of hydride addition by ISY will dictate the biocatalytic selectivity of cyclized versus reduced product.
3.5. Deletion of yeast dehydrogenases/reductases increases biocatalytic selectivity
The reversible oxidation of alcohols to aldehydes plays a critical role in the robust fermentative capacity of yeast. Involvement of yeast short chain alcohol dehydrogenases during both fermentative growth and ethanol respiration is well-established (de Smidt et al., 2008). Of the nearly 300 oxidoreductases annotated in the Saccharomyces Genome Database, 19 potential short-chain dehydrogenases/reductases (Kavanagh et al., 2008) and 17 potential medium-chain dehydrogenase/reductases (Jornvall et al., 1999) are predicted. A literature survey allowed us to speculate four of these ADHs with potential activity towards 1 that would lead to the formation of undesired monoaldehydes such as 5a. The candidates were selected based on expression profile of encoded genes in the presence of structurally related α,β-unsaturated aldehydes acrolein and HMF (Golla et al., 2015; Ma and Liu, 2010; Petersson et al., 2006). These included the medium-chain dehydrogenases/reductases ADH6 and ADH7 as well as the short-chain dehydrogenases/reductases ARI1 and GRE2. CRISPR/Cas9-mediated knockout of the four candidate oxidoreductases was carried out in S4, generating individual knockout strains S5–S9 (Table 1). The strains were transformed with the plasmid pJB034 expressing GOR and ISY and biocatalytic selectivity towards 3 was measured using the repeated fed-batch bioconversion assay (Fig. 3C). Knockout of ADH6 and ADH7 provided a 57% and 143% increase in selectivity towards 3, respectively, confirming our hypothesis that ADHs are directly involved in shunt product 4 formation. Knockout of ARI1 provided a modest increase in selectivity while knockout of GRE2 did not have a positive effect. To further improve selectivity, we generated a combined knockout strain (S9) in which ARI1, ADH6, and ADH7 were all deleted. The resulting strain S9 displayed biocatalytic selectivity of 0.72 ± 0.01 towards 3, which represents a 169% increase relative to S4 and 420% (5.2-fold) increase relative to S1 (Fig. 3A trace ii vs. Fig. 3A trace iii). No apparent growth defects were observed in S5 – S9.
3.6. In vitro reconstitution of second shunt pathway confirms the role of partial oxidation in non-productive reduction
In order to validate our hypothesis that endogenous dehydrogenases facilitate the non-productive reduction by ISY, we again turned to in vitro shunt pathway reconstitution (Fig. 3E). ISY, ADH6, and ADH7 were purified from E. coli and ARI1 was purified from yeast (Fig. S4). As expected, when 8-hydroxygeraniol 1 was only incubated with the C. roseus ISY, NAD+, and NADPH for 12 hours, no reaction was observed. (Fig. 3E, trace iii). We next analyzed substrate oxidation by the yeast ADHs (ARI1, ADH6, and ADH7). Incubation of ARI1, ADH6, and ADH7 with 8-hydroxygeraniol 1, NAD+, and NADPH resulted in only trace substrate oxidation (Fig. 3E, trace iv, v, vi). We also assessed cofactor preference for the NAD(P)+ dependent oxidation (Fig. S8). We again observed only trace oxidation and noted that ARI1 prefers NADP+ while ADH6 and ADH7 could accept NAD+ or NADP+. This observation is consistent with previous conclusions that ARI1, ADH6, and ADH7 perform oxidations much more inefficiently than the corresponding reductions (Jordan et al., 2011; Larroy et al., 2002a; Larroy et al., 2002b). In fact, the Keq for similar α,β-unsaturated alcohol/aldehyde equilibria has been estimated to be ~2 × 10−11 M, which matches our observation of primarily alcohol products in vitro and in vivo (Jordan et al., 2011).
We next performed a coupled in vitro assay in which 1 was incubated with each of the ADHs along with ISY, NAD+, and NADPH (Fig. 3E, trace vii, viii, ix). We observed near complete conversion of 1 to the saturated shunt products, 4 and 6. We also observed compound 8 which is likely a partially saturated aldehyde en route to the fully reduced shunt product based on retention time and mass spectra (Fig. S10). Despite the fact that the alcohols dominate the ADH-mediated equilibrium, ISY was able to drive the system towards the reduced shunt product via irreversible reduction of the aldehyde. We speculate that depleted NAD(P)H reducing equivalents can then be replenished via alcohol oxidation by ADH. The ability of ISY to readily carry out synergistic, cascaded substrate reduction in conjunction with yeast alcohol dehydrogenases suggests that control of substrate oxidation and therefore cyclization must be strictly maintained by the C. roseus GOR, as seen in the in vitro assay. A similar in vitro enzymatic “cross talk” between ISY and a GOR homologue was described previously (Krithika et al., 2015). Future studies into the mechanism of how GOR and ISY coordinate the timing of substrate oxidation and cyclization may lead to new engineering strategies to improve biocatalytic selectivity. Finally, we attempted to recapitulate the in vivo biocatalytic selectivity in vitro by incubating 1 with equimolar GOR, ISY, and the five enzymes which were previously deleted (OYE2, OYE3, ARI1, ADH6, and ADH7). This assay was performed using 500 μM NAD+, 10 μM NADH, 2 μM NADP+, and 100 μM NADPH in order to approximate physiological cofactor concentrations. After 1 hour, we observed complete conversion of 200 μM 8-hydroxygeraniol 1 to a cis-trans nepetalactol 3 and 8-hydroxytetrahydrogeraniol 4 with a selectivity of 0.25 (Fig. 3A, trace v), mirroring in vivo observations.
3.7. Bioconversion-based continuous fed-batch fermentation establishes platform scalability and yield
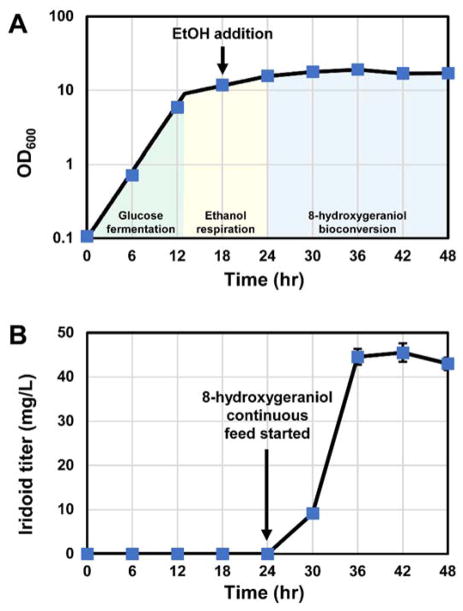
In order to further evaluate iridoid production in the final engineered strain, S9, a lab-scale fermentation was performed (Fig. 4 and Fig. S10). The fermentation process consisted of three phases: (i) glucose fermentation and biomass generation, (ii) ethanol respiration and pathway induction, and (iii) continuous 8-hydroxygeraniol addition and iridoid production. During the glucose fermentation phase, the OD600 increased rapidly with a specific growth rate of 0.35 hr−1. After 13 hours, diauxic shift occurred, as indicated by a change in the agitation rate, and the specific growth rate decreased substantially to 0.05 hr−1 (Fig. 4A). Upon addition of substrate, iridoid titer increased steadily, reaching a maximum of 45 mg/L after 42 hours (Fig. 4B). A maximum molar productivity of 3.2 μmol L−1 h−1 was observed after 36 hours of fermentation. Plasmid retention decreased over time, from 97% at the start of the fermentation to 89% after two days of growth.
Fig. 4. Bioconversion-based continuous fed-batch fermentation establishes platform scalability and yield.
(A) OD600 during fermentation. (B) Iridoid titer during fermentation.
4. CONCLUSIONS
In this study, we characterized and engineered the biocatalytic selectivity of iridoid 3 production in yeast. Bioconversion of the pathway intermediate 8-hydroxygeraniol 1 allowed for direct analysis of shunt product formation. Elimination of ‘ene’-reduction provided a clean background to study non-productive reduction by ISY. In vitro reconstitution of the two shunt pathways confirmed the distinct mechanisms of shunt product formation stemming from endogenous and exogenous reduction of α,β-unsaturated carbonyls. This critical insight enabled CRISPR/Cas9 mediated strain engineering which provided a 5.2-fold increase in the production of iridoid 3 relative to reduced shunt product 4. Our final strain was utilized for a lab-scale bioconversion-based continuous fed-batch fermentation. The strain produced 45 mg/L iridoid from a synthetically accessible precursor, with a maximum molar productivity of 3.2 μmol L−1 h−1, which correspond to the highest reported titer and productivity to date.
Our results highlight a striking example of enzymatic crosstalk between an exogenous pathway and endogenous metabolism and underscores a key challenge in engineering heterologous pathways in microbes. Many natural product biosyntheses evolved in the context of highly specialized cells that offer pathway compartmentalization and differential gene expression. Production of MIAs and other natural products in yeast will therefore rely on the continued development of tools for coordinated pathway induction (Zalatan et al., 2015) and localized pathway expression (Hammer and Avalos, 2017). Stepwise analysis around two critical steps in iridoid biosynthesis permitted a more complete understanding of the biochemical details surrounding shunt product formation, enabling rational metabolic engineering. Our results indicate the importance of understanding the biosynthetic and chemical logic that Nature employs to generate chemical diversity. Ultimately, developing a scalable platform for iridoid production sets the stage for future efforts to engineer yeast for MIA production.
Supplementary Material
Acknowledgments
This work is supported by NSF CBET1605877 and Packard Foundation. We thank Jan Schlemmer for discussions with fermentation parameters. J.M.B., J.S.B. and A.B.D are supported by NIH predoctoral training grants T32 GM067555, T32 GM008496 and T32 GM008496, respectively.
References
- Bao ZH, Xiao H, Lang J, Zhang L, Xiong X, Sun N, Si T, Zhao HM. Homology-Integrated CRISPR-Cas (HI-CRISPR) System for One-Step Multigene Disruption in Saccharomyces cerevisiae. ACS synthetic biology. 2015;4:585–594. doi: 10.1021/sb500255k. [DOI] [PubMed] [Google Scholar]
- Billingsley JM, DeNicola AB, Tang Y. Technology development for natural product biosynthesis in Saccharomyces cerevisiae. Current opinion in biotechnology. 2016;42:74–83. doi: 10.1016/j.copbio.2016.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogazkaya AM, von Buhler CJ, Kriening S, Busch A, Seifert A, Pleiss J, Laschat S, Urlacher VB. Selective allylic hydroxylation of acyclic terpenoids by CYP154E1 from Thermobifida fusca YX. Beilstein journal of organic chemistry. 2014;10:1347–1353. doi: 10.3762/bjoc.10.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–32. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brenna E, Gatti FG, Monti D, Parmeggiani F, Sacchetti A. Cascade Coupling of Ene Reductases with Alcohol Dehydrogenases: Enantioselective Reduction of Prochiral Unsaturated Aldehydes. Chemcatchem. 2012;4:653–659. [Google Scholar]
- Brown S, Clastre M, Courdavault V, O’Connor SE. De novo production of the plant-derived alkaloid strictosidine in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:3205–3210. doi: 10.1073/pnas.1423555112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A, Bauchart P, Gold ND, Zhu Y, De Luca V, Martin VJJ. Engineering of a Nepetalactol-Producing Platform Strain of Saccharomyces cerevisiae for the Production of Plant Seco-Iridoids. ACS synthetic biology. 2016;5:405–414. doi: 10.1021/acssynbio.5b00289. [DOI] [PubMed] [Google Scholar]
- Curran KA, Karim AS, Gupta A, Alper HS. Use of expression-enhancing terminators in Saccharomyces cerevisiae to increase mRNA half-life and improve gene expression control for metabolic engineering applications. Metabolic engineering. 2013;19:88–97. doi: 10.1016/j.ymben.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Srivastava PS. Variation in vinblastine production by Catharanthus roseus during in vivo and in vitro differentiation. Phytochemistry. 1997;46:135–137. [Google Scholar]
- de Smidt O, du Preez JC, Albertyn J. The alcohol dehydrogenases of Saccharomyces cerevisiae: a comprehensive review. FEMS yeast research. 2008;8:967–78. doi: 10.1111/j.1567-1364.2008.00387.x. [DOI] [PubMed] [Google Scholar]
- Fischer MJC, Meyer S, Claudel P, Bergdoll M, Karst F. Metabolic Engineering of Monoterpene Synthesis in Yeast. Biotechnology and bioengineering. 2011;108:1883–1892. doi: 10.1002/bit.23129. [DOI] [PubMed] [Google Scholar]
- Galanie S, Thodey K, Trenchard IJ, Filsinger Interrante M, Smolke CD. Complete biosynthesis of opioids in yeast. Science. 2015;349:1095–100. doi: 10.1126/science.aac9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geu-Flores F, Sherden NH, Courdavault V, Burlat V, Glenn WS, Wu C, Nims E, Cui YH, O’Connor SE. An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature. 2012;492:138-+. doi: 10.1038/nature11692. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature protocols. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- Golla U, Bandi G, Tomar RS. Molecular Cytotoxicity Mechanisms of Allyl Alcohol (Acrolein) in Budding Yeast. Chemical research in toxicology. 2015;28:1246–1264. doi: 10.1021/acs.chemrestox.5b00071. [DOI] [PubMed] [Google Scholar]
- Gramatica P, Manitto P, Ranzi BM, Delbianco A, Francavilla M. Stereospecific Reduction of Geraniol to (R)-(+)-Citronellol by Saccharomyces-Cerevisiae. Experientia. 1982;38:775–776. [Google Scholar]
- Hammer SK, Avalos JL. Harnessing yeast organelles for metabolic engineering. Nature chemical biology. 2017;13:823–832. doi: 10.1038/nchembio.2429. [DOI] [PubMed] [Google Scholar]
- Jakounas T, Sonde I, Herrgard M, Harrison SJ, Kristensen M, Pedersen LE, Jensen MK, Keasling JD. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metabolic engineering. 2015;28:213–222. doi: 10.1016/j.ymben.2015.01.008. [DOI] [PubMed] [Google Scholar]
- Jiang GZ, Yao MD, Wang Y, Zhou L, Song TQ, Liu H, Xiao WH, Yuan YJ. Manipulation of GES and ERG20 for geraniol overproduction in Saccharomyces cerevisiae. Metabolic engineering. 2017;41:57–66. doi: 10.1016/j.ymben.2017.03.005. [DOI] [PubMed] [Google Scholar]
- Jordan DB, Braker JD, Bowman MJ, Vermillion KE, Moon J, Liu ZL. Kinetic mechanism of an aldehyde reductase of Saccharomyces cerevisiae that relieves toxicity of furfural and 5-hydroxymethylfurfural. Biochimica et biophysica acta. 2011;1814:1686–94. doi: 10.1016/j.bbapap.2011.08.011. [DOI] [PubMed] [Google Scholar]
- Jornvall H, Hoog JO, Persson B. SDR and MDR: completed genome sequences show these protein families to be large, of old origin, and of complex nature. FEBS letters. 1999;445:261–264. doi: 10.1016/s0014-5793(99)00130-1. [DOI] [PubMed] [Google Scholar]
- Jung ST, Lauchli R, Arnold FH. Cytochrome P450: taming a wild type enzyme. Current opinion in biotechnology. 2011;22:809–17. doi: 10.1016/j.copbio.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh KL, Jornvall H, Persson B, Oppermann U. The SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cellular and molecular life sciences : CMLS. 2008;65:3895–906. doi: 10.1007/s00018-008-8588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A, Richard Dickinson J. Biotransformation of monoterpene alcohols by Saccharomyces cerevisiae, Torulaspora delbrueckii and Kluyveromyces lactis. Yeast. 2000;16:499–506. doi: 10.1002/(SICI)1097-0061(200004)16:6<499::AID-YEA548>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Kries H, Caputi L, Stevenson CEM, Kamileen MO, Sherden NH, Geu-Flores F, Lawson DM, O’Connor SE. Structural determinants of reductive terpene cyclization in iridoid biosynthesis. Nature chemical biology. 2016;12:6–8. doi: 10.1038/nchembio.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krithika R, Srivastava PL, Rani B, Kolet SP, Chopade M, Soniya M, Thulasiram HV. Characterization of 10-Hydroxygeraniol Dehydrogenase from Catharanthus roseus Reveals Cascaded Enzymatic Activity in Iridoid Biosynthesis. Scientific reports. 2015:5. doi: 10.1038/srep08258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivoruchko A, Nielsen J. Production of natural products through metabolic engineering of Saccharomyces cerevisiae. Current opinion in biotechnology. 2015;35:7–15. doi: 10.1016/j.copbio.2014.12.004. [DOI] [PubMed] [Google Scholar]
- Larroy C, Fernandez MR, Gonzalez E, Pares X, Biosca JA. Characterization of the Saccharomyces cerevisiae YMR318C (ADH6) gene product as a broad specificity NADPH-dependent alcohol dehydrogenase: relevance in aldehyde reduction. The Biochemical journal. 2002a;361:163–72. doi: 10.1042/0264-6021:3610163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larroy C, Pares X, Biosca JA. Characterization of a Saccharomyces cerevisiae NADP(H)-dependent alcohol dehydrogenase (ADHVII), a member of the cinnamyl alcohol dehydrogenase family. European journal of biochemistry. 2002b;269:5738–45. doi: 10.1046/j.1432-1033.2002.03296.x. [DOI] [PubMed] [Google Scholar]
- Lee KM, DaSilva NA. Evaluation of the Saccharomyces cerevisiae ADH2 promoter for protein synthesis. Yeast. 2005;22:431–440. doi: 10.1002/yea.1221. [DOI] [PubMed] [Google Scholar]
- Leonard E, Runguphan W, O’Connor S, Prather KJ. Opportunities in metabolic engineering to facilitate scalable alkaloid production. Nature chemical biology. 2009;5:292–300. doi: 10.1038/nchembio.160. [DOI] [PubMed] [Google Scholar]
- Li Y, Smolke CD. Engineering biosynthesis of the anticancer alkaloid noscapine in yeast. Nature communications. 2016;7:12137. doi: 10.1038/ncomms12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner S, Geu-Flores F, Brase S, Sherden NH, O’Connor SE. Conversion of Substrate Analogs Suggests a Michael Cyclization in Iridoid Biosynthesis. Chem Biol. 2014;21:1452–1456. doi: 10.1016/j.chembiol.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale R, Reetz MT. Reduction of alpha, beta-Unsaturated Ketones by Old Yellow Enzymes: Mechanistic Insights from Quantum Mechanics/Molecular Mechanics Calculations. Journal of the American Chemical Society. 2015;137:14733–14742. doi: 10.1021/jacs.5b08687. [DOI] [PubMed] [Google Scholar]
- Ma MG, Liu ZL. Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4, and HSF1 as key regulatory genes in genomic adaptation to the lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae. BMC genomics. 2010:11. doi: 10.1186/1471-2164-11-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen K, Dong LM, Navrot N, Schneider T, Burlat V, Pollier J, Woittiez L, van der Krol S, Lugan R, Ilc T, Verpoorte R, Oksman-Caldentey KM, Martinoia E, Bouwmeester H, Goossens A, Memelink J, Werck-Reichhart D. The seco-iridoid pathway from Catharanthus roseus. Nature communications. 2014:5. doi: 10.1038/ncomms4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J. Yeast cell factories on the horizon. Science. 2015;349:1050–1. doi: 10.1126/science.aad2081. [DOI] [PubMed] [Google Scholar]
- O’Connor SE, Maresh JJ. Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat Prod Rep. 2006;23:532–47. doi: 10.1039/b512615k. [DOI] [PubMed] [Google Scholar]
- Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, Polichuk DR, Teoh KH, Reed DW, Treynor T, Lenihan J, Fleck M, Bajad S, Dang G, Dengrove D, Diola D, Dorin G, Ellens KW, Fickes S, Galazzo J, Gaucher SP, Geistlinger T, Henry R, Hepp M, Horning T, Iqbal T, Jiang H, Kizer L, Lieu B, Melis D, Moss N, Regentin R, Secrest S, Tsuruta H, Vazquez R, Westblade LF, Xu L, Yu M, Zhang Y, Zhao L, Lievense J, Covello PS, Keasling JD, Reiling KK, Renninger NS, Newman JD. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–32. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- Peng B, Williams TC, Henry M, Nielsen LK, Vickers CE. Controlling heterologous gene expression in yeast cell factories on different carbon substrates and across the diauxic shift: a comparison of yeast promoter activities. Microbial cell factories. 2015;14:91. doi: 10.1186/s12934-015-0278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson A, Almeida JR, Modig T, Karhumaa K, Hahn-Hagerdal B, Gorwa-Grauslund MF, Liden G. A 5-hydroxymethyl furfural reducing enzyme encoded by the Saccharomyces cerevisiae ADH6 gene conveys HMF tolerance. Yeast. 2006;23:455–64. doi: 10.1002/yea.1370. [DOI] [PubMed] [Google Scholar]
- Proft M, Grzesitza D, Entian KD. Identification and characterization of regulatory elements in the phosphoenolpyruvate carboxykinase gene PCK1 of Saccharomyces cerevisiae. Molecular & general genetics : MGG. 1995;246:367–73. doi: 10.1007/BF00288610. [DOI] [PubMed] [Google Scholar]
- Renault H, Bassard JE, Hamberger B, Werck-Reichhart D. Cytochrome P450-mediated metabolic engineering: current progress and future challenges. Current opinion in plant biology. 2014;19:27–34. doi: 10.1016/j.pbi.2014.03.004. [DOI] [PubMed] [Google Scholar]
- Rodrigues T, Reker D, Schneider P, Schneider G. Counting on natural products for drug design. Nature chemistry. 2016;8:531–541. doi: 10.1038/nchem.2479. [DOI] [PubMed] [Google Scholar]
- Steyer D, Erny C, Claudel P, Riveill G, Karst F, Legras JL. Genetic analysis of geraniol metabolism during fermentation. Food microbiology. 2013;33:228–234. doi: 10.1016/j.fm.2012.09.021. [DOI] [PubMed] [Google Scholar]
- Trotter EW, Collinson EJ, Dawes IW, Grant CM. Old yellow enzymes protect against acrolein toxicity in the yeast Saccharomyces cerevisiae. Applied and environmental microbiology. 2006;72:4885–4892. doi: 10.1128/AEM.00526-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tundis R, Loizzo MR, Menichini F, Statti GA, Menichini F. Biological and pharmacological activities of iridoids: recent developments. Mini reviews in medicinal chemistry. 2008;8:399–420. doi: 10.2174/138955708783955926. [DOI] [PubMed] [Google Scholar]
- Weinhandl K, Winkler M, Glieder A, Camattari A. Carbon source dependent promoters in yeasts. Microbial cell factories. 2014;13:5. doi: 10.1186/1475-2859-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RE, Bruce NC. ‘New uses for an Old Enzyme’ - the Old Yellow Enzyme family of flavoenzymes. Microbiol-Sgm. 2002;148:1607–1614. doi: 10.1099/00221287-148-6-1607. [DOI] [PubMed] [Google Scholar]
- Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, Lim WA. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–50. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JZ, Li C, Zhang Y, Shen Y, Hou J, Bao XM. Dynamic control of ERG20 expression combined with minimized endogenous downstream metabolism contributes to the improvement of geraniol production in Saccharomyces cerevisiae. Microbial cell factories. 2017:16. doi: 10.1186/s12934-017-0641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.