Abstract
As a de-ubiquitin enzyme, ubiquitin C-terminal hydrolase (UCH)-L1 has been shown to be overexpressed in several human cancers. However, the function of UCH-L1 in invasion of breast cancers is still unclear. Here we report that the expression of UCH-L1 is significantly higher in cancer cells with higher invasive ability. While ectopic UCH-L1 expression failed to alter cell proliferation in MCF-7 cells, it caused a significant upregulation of cellular invasion. Furthermore, siRNA mediated knockdown of UCH-L1 led to suppression of invasion in UCH-L1 overexpressing MCF-7 cells. In order to identify molecular mechanisms underlying these observations, a novel in vitro proximity-dependent biotin identification method was developed by fusing UCH-L1 protein with a bacterial biotin ligase (E. coli BirA R118G, BioID). Streptavidin magnetic beads pulldown assay revealed that UCH-L1 can interact with Akt in MCF-7 cells. Pulldown assay with His tagged recombinant UCH-L1 protein and cell lysate from MCF-7 cells further demonstrated that UCH-L1 preferentially binds to Akt2 for Akt activation. Finally, we demonstrated that overexpression of UCH-L1 led to activation of Akt as evidenced by upregulation of phosphorylated Akt. Thus, these findings demonstrated that UCH-L1 promotes invasion of breast cancer cells and might serve as a potential therapeutic target for treatment of human patients with breast cancers.
Keywords: UCH-L1, AKT, breast cancer, invasion
Introduction
Ubiquitin(Ub)-proteasome system is an essential component of eukaryotic cells for protein quality control and homeostasis[Etlinger and Goldberg, 1977]. Alteration of ubiquitin-proteasome system has been linked to many human diseases including cancers[Hoeller et al., 2006], cardiovascular diseases[Calise and Powell, 2013; Powell et al., 2012], and neurodegenerative diseases[Ciechanover and Brundin, 2003]. Mono- or poly- ubiquitin can be conjugated to its target proteins by ubiquitin ligases for protein degradation, relocation and functional alteration. Conversely, ubiquitin can be detached from its targets by de-ubiquitination enzymes (DUBs)[Nijman et al., 2005]. There are totally 5 sub-families of DUBs, including the Ub C-terminal hydrolases (UCHs), the Ub-specific proteases (USPs), the ovarian tumor domain containing (OTU), the Josephin-domain containing and the jab1/MPN domain-associated metalloiso-peptidase class[Nijman et al., 2005]. Similar as ubiquitin ligases, dysfunctions or mutations of DUBs have been found in many human diseases[Hanpude et al., 2015; Wilkinson, 2000].
As indicated by its name, Ubiquitin C-terminal hydrolase (UCH)-L1 belongs to the Ub C-terminal hydrolases (UCHs) sub-family. Tissue distribution analysis of UCH-L1 revealed that it is predominantly expressed in brain tissue[Day and Thompson, 2010], indicating it plays a critical role in central nerve system. Indeed, genetic ablation of UCH-L1 leads to degeneration of presynaptic terminals at the neuromuscular junction, a loss of synaptic vesicles[Chen et al., 2010]. Furthermore, several UCH-L1 mutations have been identified as the causes of neurodegenerative diseases[Bilguvar et al., 2013; Leroy et al., 1998]. Extensive studies have also demonstrated that activity and solubility alterations of UCH-L1 can cause neurodegenerative diseases[Butterfield et al., 2006; Choi et al., 2004; Gong et al., 2006].
Despite its expression level is low in other tissues, overexpression of UCH-L1 has been found in some cancers, including pancreatic cancer[Saettler and Temple, 2000; Tezel et al., 2000], myeloma[Otsuki et al., 2004], prostate cancer[Leiblich et al., 2007], neuroblastoma[Ootsuka et al., 2008], and osteosarcoma[Liu et al., 2009]. Furthermore, expression of UCH-L1 has been shown to be positively correlated with cancer cell chemotherapy resistance[Jin et al., 2015; Wang et al., 2016], metastasis[Kim et al., 2015], and negatively correlated with prognosis of cancer patients[Schroder et al., 2013; Yang et al., 2015].
Although a potential role for UCH-L1 as an oncogene has been observed, the molecular mechanisms underlying these observations have not been extensively studied. Here we show that expression of UCH-L1 is positively correlated with invasive ability of breast cancer cells. A novel live cell protein proximal biotination assay and magnetic beads pull down assay have identified that UCH-L1 can interact with endogenous Akt2 and in turn, to activate Akt signaling for promoting cell invasiveness. Thus, our study suggests that UCH-L1 promotes breast cancer cell invasion and might serve as a potential therapeutic target for treatment of human patients with breast cancers.
Materials and methods
Cell cultures
Human breast cancer cell lines, MCF-7, MDA-MB-468, MDA-MB-436, T-47D and MCF-7/Adr were obtained from American Type Culture Collection (ATCC, Manassas, USA). MCF-7, T-47D, MCF-7/Adr were cultured in RPMI-1640 medium containing 10% fetal bovine serum (Gibco) and 100 I.U./ml penicillin and 100 μg/ml streptomycin at 37 °C in a humidified atmosphere with 5% CO2. MDA-MB-468 and MDA-MB-436 were cultured in DMEM medium containing 10% fetal bovine serum (Gibco) and 100 I.U./ml penicillin and 100 μg/ml streptomycin at 37 °C in a 5% CO2 air atmosphere as mentioned before.
Western blotting
Protein of tissue samples or cell lines was extracted by using Total Protein Extraction Kit (Millipore, Darmstadt, Germany). The protein concentration was determined using Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-rad, USA). Equivalent proteins were denatured in protein loading buffer, loaded onto 10% SDS-PAGE gels, and subsequently transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA) by electroblotting. The PVDF membranes were blocked with 5% nonfat milk in PBST buffer for 1 h and incubated overnight at 4 °C with antibody against UCH-L1 (Abcam), pan-Akt (Cell Signaling), Akt1 (Cell Signaling), Akt2 (Cell Signaling), Akt3 (Cell Signaling), pAkt (Ser473) (Cell Signaling), ERK (Cell Signaling), pERK (Cell Signaling), GAPDH (Santa Cruz) and α-tubulin (Cell Signaling). Signals were detected using ECL detection reagent (Pierce) following the manufacturer’s instructions.
Establishment of stably transfected cells
The cDNA for UCH-L1 (NCBI gene symbol UCHL1) was amplified by PCR from HEK293 cells, and the product was subcloned into either pcDNA3.1 (Invitrogen, CA, USA) or pcDNA3.1 mycBioID (Addgene, MA, USA) vectors to generate UCH-L1 overexpression plasmids. Different breast cancer cells were transfected with pcDNA3.1-UCH-L1, pcDNA3.1 mycBioID-UCH-L1(BirA-UCH-L1) recombinant plasmid or empty vectors using Lipofectamine 3000 according to the manufacturer’s instructions (Invitrogen, USA) and retained in medium containing 10% FBS and 1 mg/ml G418 to select the stable transfected cells.
siRNA mediated UCH-L1 knockdown
2×105 MCF-7/Adr cells in 3 ml antibiotic-free medium were plated in 6-well plate. 24 h later, after cells reach 80% confluence, siRNA against UCH-L1 and control siRNA (GE Dharmacon, CO, USA) were mixed at equivalent ratio and then co-transfected into MCF-7/Adr cells using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions. Twelve hours after transfection, the cells were plated to Boyden chamber to test their invasive ability.
Cell proliferation assay
Cell proliferation assays were carried out with a MTT assay. Cells were plated in 96-well plates at approximately 1×103 cells per well. The numbers of cells per well were detected by the absorbance (570 nm) of MTT at the indicated time points. The absorbance (570 nm) was measured by using Flex Station 3 microplate reader (Molecular Devices, Sunnyvale, CA).
Cell Invasion assay
Transwell inserts that precoated with Matrigel (8 μm pore; Corning Costar) were placed into the 24-well plate, a total of 5×104 cells in 0.5% FBS-medium were seeded in the upper chamber and 10% FBS-medium were added in the lower chamber. After 24 h incubation, cells on the top surface of the insert were removed by wiping with a cotton swab. The cells that had invaded the bottom side of the membrane were fixed with 4% formaldehyde and stained with Crystal violet (Sigma-Aldrich). The stained cells were photographed and quantified. In addition, BD BioCoat™ FluoroBlok™ Invasion System (Becton Dikinson Labware) was used to quantify invasive ability of each cell line. Briefly, a total of 5 × 104 cells in 0.5% FBS-medium were seeded to the apical chambers and 10% FBS medium were added to the basal chambers. After 24 hours incubation in cell culture condition, the apical chambers were carefully removed and inserted into a second 24-well plate containing 500 μL/well of 4 μg/mL Calcein AM in HBSS. The plates were incubated for 1 hour at 37°C, 5% CO2. During incubation, Calcein AM was converted into green fluorescent calcein by cytosolic esterases. The invaded cells were quantified by the fluorescence at wavelengths of 494/517 nm (Ex/Em) on a bottom-reading fluorescent plate reader (Flex Station 3, Molecular Devices, Sunnyvale, CA).
Affinity pulldown of biotinylated proteins
A promiscuous biotin ligase fusion protein (BirA) was used to identify potential UCH-L1 interaction proteins [Roux et al., 2012]. Briefly, cDNA of UCH-L1 was subcloned into pcDNA3.1 mycBioID plasmid (pcDNA3.1 mycBioID was a gift from Kyle Roux (Addgene plasmid # 35700). MCF-7 cells were transfected with either mycBioID-UCH-L1 or empty mycBioID plasmids using Lipofectamine 3000 transfection reagent. Twenty-four hours after transfection, the transfected cells were cultured for 16 hours in RPMI-1640 culture medium containing 50 μM biotin. After washing three times with DPBS to wash out extracellular biotin, cells were lysed on ice in RIPA buffer containing complete protease and phosphatase inhibitors (Roche). Then sonicated using the Branson Sonifier 250 at 15% output level for 1 min (5 second on and 5 second off) on ice. Then the lysates were centrifuged at 14,000 g for 15 mins at 4°C. Supernatants were incubated with Dynabeads (MyOne Streptavidin C1, Invitrogen) overnight at 4°C with a rotator. Beads were collected and washed four times (5 mins/time) at room temperature in PBS. After wash, the samples were mixed with equal volume of loading buffer (Bio-Rad) and boiled for immunoblot analysis.
Recombinant human UCH-L1 expression and in vitro Ni-NTA pulldown assay
Human UCHL1 DNA sequence was subcloned into the pET28a vector (EMD Biosciences) with T4 DNA ligase (NEB, Ipswich, USA) to generate pET28a-rhUCH-L1 plasmid. The insertion accuracy was verified by DNA sequencing. The pET28a-rhUCH-L1 plasmid (with 6-His-Tag) was transformed into competent E.coli strain BL21 (DE3) cells (Invitrogen, USA). Then, E. coli cells were maintained at 37°C in Luria–Bertani medium with vigorous shaking (250 rpm). Isopropyl-β-D-thiogalactopyranoside (Amresco, OH, USA) was added at a concentration of 1 mM when the OD600 of the E. coli reached 0.4. After further incubation at 24 °C for 6 h, the cells were harvested for further use.
Rapid screening of expression cultures was operated according to the manual for high-level expression and purification of 6xHis-tagged proteins (Qiagen, USA). The roughly 24.8–kDa rhUCH-L1 protein was purified and afterward it was verified by SDS-PAGE analysis. The purified His-rhUCH-L1 protein was further established by western blot, probed with anti-His and UCH-L1 antibodies. The obtained purified protein were harvested for further use.
His-rhUCH-L1 protein was used as a bait to pulldown its interaction proteins from different cell lysates. The pulldown protocol was modified from previous study [Rahmeh et al., 2012]. Briefly, purified His-rhUCH-L1 protein or equal volume of saline was first incubated with Ni-NTA spin column, then cell lysates derived from either MDA-MB-231 or MCF-7 was loaded to Ni-NTA column and incubated for 1, 2 and 4 hours. The columns were washed with wash buffer for four times (5 mins/wash) and eluted with elution buffer. The elution fraction was collected and subjected to immunoblotting analysis for proteins of interest (pan-Akt, Akt1, Akt2 and Akt3).
Statistical analysis
All statistical analyses were performed using Graphpad Prism V.5.00 software (GraphPad Software, San Diego CA, USA). Statistical significance was determined at p<0.05, and tests were two sided. Comparison between two groups for statistical significance were performed with unpaired Student’s t test. For more groups, one-way ANOVA followed by Neuman-Keuls post hoc test was used.
Results
UCH-L1 expression is positively correlated with cancer invasive ability
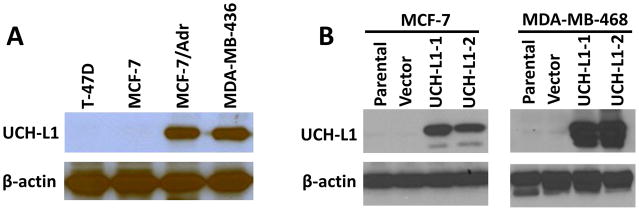
To determine the potential role of UCH-L1 in regulation of cancer invasion, we firstly examined the expression of UCH-L1 in different breast cancer cell lines with different invasive ability. The cell lines we chose were divided into two groups based on previous reports [Gumireddy et al., 2007; Li et al., 2007]: MDA-MB-468, MCF-7 and T-47D are the cell lines with low invasive ability, MDA-MB-436 and MCF-7/Adr are the cell lines with high invasive ability. Western blotting analysis showed that protein expression of UCH-L1 was significantly increased in cell lines with high invasive ability (Fig. 1A), indicating high level of UCH-L1 might contribute to high invasiveness of cancer cells. To further test the role of UCH-L1 on cancer cell invasiveness, we transfected UCH-L1 plasmid into both MCF-7 and MDA-MB-468 cells to generate stable cell lines that overexpress UCH-L1. As it is shown in Fig. 1B, we successfully obtained single clones that overexpress UCH-L1 from parental MCF-7 and MDA-MB-468 cells.
Figure 1. UCH-L1 expression is increased in the cancer cell lines with high invasive ability.
(A) UCH-L1 expression in different cancer cell lines was detected by Western blotting, β-actin was used as the internal control. (B) Western blotting analysis showed that UCH-L1 was successfully overexpressed in both MCF-7 and MDA-MB-468 cells. UCH-L1-1 and UCH-L1-2 indicated two independent clones generated from antibiotic selection.
Overexpression of UCH-L1 enhances invasive ability of cancer cells
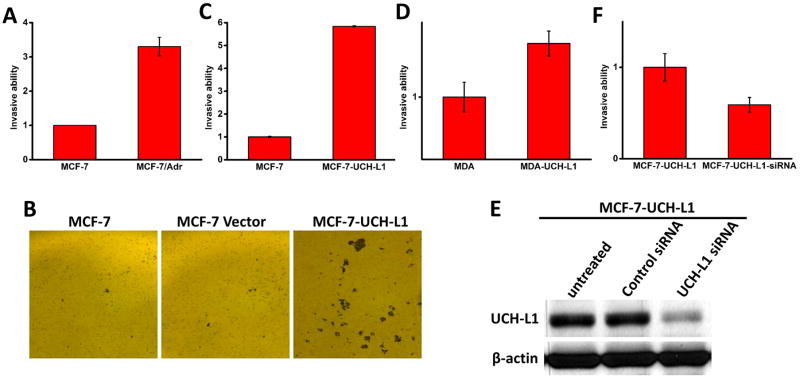
In order to further establish the correlation of UCH-L1 expression with cell invasiveness, we first compared invasive ability of MCF-7 and MCF-7/Adr cells. As it is shown in Fig. 2A, MCF-7/Adr displayed significantly higher ability of invasiveness, which was consistent with our previous studies [Li et al., 2007]. To further test our hypothesis that expression of UCH-L1 is positively correlated with invasiveness of the cells, we used UCH-L1 overexpressing cell lines we generated from MCF-7 and MDA-MB-468 cells, thus we could compare invasive ability of cells with same genetic background (MCF-7 vs MCF-7-UCH-L1; MDA-MB-468 vs MDA-MB-468-UCH-L1). With matrigel invasion chamber, we can stain invaded cells with crystal violet and found that MCF-7-UCH-L1 cells invaded more through matrigel than both parental MCF-7 cells and MCF-7 cells transfected with empty vector (Fig. 2B). To further quantify invaded cells among different cell lines, a florescence based method was used[Partridge and Flaherty, 2009]. Since calcein AM is used to fluorescently label the invaded cells and the number of invaded cells and calcein florescence reading are in linear correlation, this method is an ideal way to quantify invaded cells accurately and objectively. As it is shown in Fig. 2C and 2D, overexpression of UCH-L1 led to enhanced invasiveness in both MCF-7 (5.8 fold increase, p<0.01) and MDA-MB-468 (1.4 fold increase, p<0.05) cells. Since the overexpression of UCH-L1 required relatively long-time selection for single clones, there might be other alterations happened during selection process, which affected invasive ability of the cells. To test this possibility, we used siRNA to acutely knockdown UCH-L1 in MCF-7-UCH-L1 cells. As it is shown in Fig. 2E, siRNA against UCH-L1 could significantly reduce its expression level, while scramble siRNA had no effect as a control. We have also tested the time window of the siRNAs we used and found that the knockdown effect of siRNA could last for 96 hours which is within the period of our invasion experiments. As it shown in Fig. 2F, acute UCH-L1 siRNA treatment significantly reduced expression of UCH-L1 as well as cell invasiveness. Thus, our results suggested that UCH-L1 indeed regulates cancer cell invasion.
Figure 2. Expression of UCH-L1 is positively correlated with cancer cell invasion.
Matrigel invasion assay was performed and invaded cells were fluorescently labelled with Calcein AM dye. Normalized fluorescent intensity showed that MCF-7/Adr cells displayed higher invasiveness than MCF-7 cells (A). Similarly, overexpression of UCH-L1 led to enhancement of invasion in both MCF-7 (B, C) and MDA-MB-468 (D) cells. Since there is possibility that long term antibiotic selection might cause alterations of other genes that potentially contribute to cell invasion, MCF-7-UCH-L1 cells were acutely transfected siRNA against UCH-L1. Western blotting analysis showed that siRNA treatment can effectively knockdown expression of UCH-L1 (E) and inhibit cell invasion as compared to the cells treated with scramble siRNA as a control. Data presented as Mean ± SD, n = 3 independent experiments. * p<0.05, ** p<0.01.
UCH-L1 activates Akt pathway through directly binding to Akt2
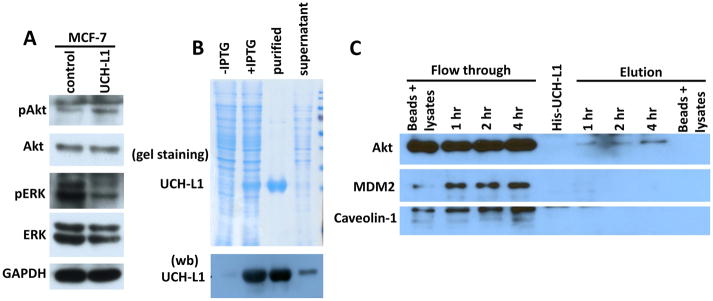
Activation of Akt and/or ERK signaling pathways have been reported to promote cell invasion in multiple cancers [Dhillon et al., 2007; Kim et al., 2001; Schuierer et al., 2004; Vasko et al., 2004]. To test whether Akt and/or ERK pathways are involved in UCH-L1 mediated cell invasion, we performed immunoblot analysis to detect phosphorylated Akt and ERK in MCF-7 and MCF-7-UCH-L1 cells. As shown in Figure 3A, UCH-L1 over-expression led to upregulation of p-Akt and downregulation of p-ERK as compared with that in empty vector-transfected cells, indicating UCH-L1 might activate Akt pathway to promote cancer cell invasion.
Figure 3. UCH-L1 activates Akt signaling pathway.
(A) MCF-7 cells were transfected with plasmid to generate stable UCH-L1 overexpression cell line. Overexpression of UCH-L1 led to activation of Akt as evidenced by upregulation of p-Akt level. (B) A His tagged recombinant human UCH-L1 expression plasmid was generated and transformed into E. Coli. Coomassie blue staining of SDS-PAGE (upper panel of B) and Western blotting (lower panel of B) show that His-rhUCH-L1 was successfully purified by Ni-NTA column. (C) His-rhUCH-L1 was used as a bait to pulldown interacting proteins from cell lysates derived from MCF-7 cells. Western blot analysis show that Akt can be pulled down by His-rhUCH-L1 protein, while other proteins, such as MDM2 and Cavin-3 cannot be found in elution fraction of the experiments. Empty Ni-NTA beads incubated with cell lysates and purified His-rhUCH-L1 were also included as controls. n = 3 independent experiments.
To test the molecular mechanisms underlying these observation, we purified His tagged recombinant human UCH-L1 protein (His-rhUCH-L1) from E. Coli fermentation and used it as a bait to pulldown its interaction proteins from cell lysates from MCF-7 cells. As it shown in Fig. 3B, we can generate His-rhUCH-L1 with high purity. When incubating with MCF-7 cell lysates with indicated time points, we observed that AKT protein can be pulled down by His-rhUCH-L1, and the amount of binding is time dependent (Fig. 3C).
To further confirm our biochemical observation in vitro, a more physiological assay was utilized to examine UCH-L1 and AKT interaction in live cancer cells. This approach took advantage of the biotin-ligase (BioID) system[Roux et al., 2012], a modified promiscuous biotin ligase was fused to N-terminal of UCH-L1 protein to form fused protein mycBioID-UCH-L1 (Fig. 4A). We then expressed mycBioID-UCH-L1 in MCF-7 cells and it will biotinylates proteins that come in close proximity, which can then be pulled down by streptavidin magnetic beads. As all of the reaction happens in live cells, the potential interaction obtained from this method will considered to be more physiological as compared to conventional co-immunoprecipitation and in vitro pulldown assays showing in Fig. 3C. As it is shown in Fig. 4B, mycBioID-UCH-L1 and mycBioID have been successfully generated and overexpressed in MCF-7 cells. More importantly, Akt can be biotinylated by mycBioID-UCH-L1, but not by mycBioID as a control (Fig. 4C).
Figure 4. UCH-L1 interacts with Akt in live cancer cells.
A novel protein/protein interaction approach was used to confirm interaction between UCH-L1 and Akt in live cancer cells. (A) A schematic figure shows the working flow of BioID system to identify interacting partners of protein of interest. Specifically, a biotin ligase, BioID, was fused with UCH-L1 and the fusion protein was expressed in MCF-7 cells. BioID-UCH-L1 can biotinylate proteins in close proximity, which can be pulled down by streptavidin magnetic beads for analysis. Both BirID and BirID-UCH-L1 plasmids were successfully expressed in MCF-7 cells (B) and streptavidin beads pulldown assay suggested that BirID-UCH-L1 can biotinylate Akt, while neither BirID alone not BirID-UCH-L1 can biotinylate MDM2, GAPDH or caveolin-1 as negative controls (C). n = 3 independent experiments.
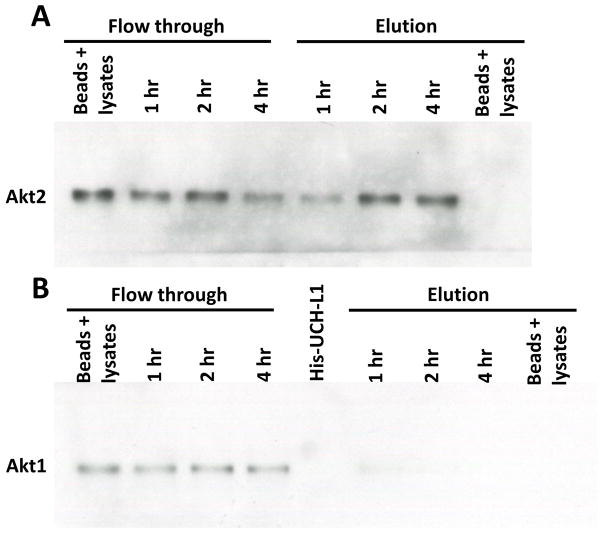
Finally, we asked the question that which isoform of Akt protein family is the target for UCH-L1. As it is well known, Akt protein family has at least 3 isoforms, Akt1, Akt2 and Akt3 (summarized in review article [Chin and Toker, 2009]). Interestingly, Akt1 and Akt2 play opposite role on controlling invasion in breast cancer cells, while Akt2 promotes cell invasion[Arboleda et al., 2003; Cheng et al., 2007], Akt1 actually inhibits cell invasion [Irie et al., 2005; Veeriah, 2012; Yoeli-Lerner et al., 2005]. To further test which Akt isoform(s) interacts with UCH-L1, we performed His-rhUCH-L1 pulldown assay as described in Fig. 3. As it is shown in Fig. 5, while His-rhUCH-L1 protein can pulldown Akt2 from MCF-7 cell lysate (Fig. 5A), the signal of Akt1 in pulldown sample is almost non-detectable and not time dependent as Akt1 (Fig. 5B) (only showed a faint band in sample at binding of 1 hour). Consistent with previous studies[Faridi et al., 2003; Nakatani et al., 1999], we didn’t detect Akt3 signal due to the fact that MCF-7 cells do not expression endogenous Akt3 (data not shown).
Figure 5. UCH-L1 preferentially binds to Akt2 in MCF-7 cells.
His-rhUCH-L1 was used as a bait to pulldown interacting proteins from cell lysates derived from MCF-7 cells. Western blot analysis show that Akt2 can be pulled down by His-rhUCH-L1 protein (A), while Akt1 signal is almost non-detectable (B).
Thus, we demonstrated that UCH-L1 positively regulates cancer cell invasion through directly binding to Akt2 and activating Akt signaling pathway. This functional interaction might be a potential target for treatment of invasive cancers.
Discussion
As a de-ubiquitination enzyme, UCH-L1 is considered as a tumor promoting protein in many human tumors [Leiblich et al., 2007; Liu et al., 2009; Ootsuka et al., 2008; Otsuki et al., 2004; Saettler and Temple, 2000; Tezel et al., 2000]. However, the function of UCH-L1 in tumor initiation, progression and invasion is still controversial, as UCH-L1 has also been found to be downregulated in other tumor types including prostate cancer [Ummanni et al., 2011], ovarian cancer [Jin et al., 2013], and nasopharyngeal cancer [Li et al., 2010]. Therefore, the potential role of UCH-L1 as an oncogene or a tumor suppressor may be cell context or tissue specific. In this study, we firstly reported that overexpression of UCH-L1 led to enhancement of cell invasion in multiple breast cancer cell lines. Cell biology and biochemical experiments show that the molecular actions of UCH-L1 for promotion of cell invasion is through directly binding to Akt2 and activating Akt signaling pathway.
There have been several studies reporting the roles of UCH-L1 in breast cancers. A clinical study has reported that overexpression of UCH-L1 is statistically correlated with metaplastic carcinomas of the breast and associated with poorer overall survival. By pathological analysis, the authors attributed these clinical observations to the potential role of UCH-L1 on promoting epithelial-mesenchymal transition (EMT) in breast cancer [Lien et al., 2013]. Similarly, an elegant study demonstrated that overexpression of UCH-L1 promotes metastasis of breast and lung cancers through its de-ubiquitin activity for stabilizing Hypoxia-inducible factor 1 (HIF-1) [Goto et al., 2015]. Interestingly, a study found that UCH-L1 inhibits breast cancer cell proliferation by stabilizing p53 and blocking G0/G1 cell cycle [Xiang et al., 2012]. Thus, the current studies about the roles of UCH-L1 on breast cancers have been focused on cell proliferation and metastasis. We have also tested the role of UCH-L1 on proliferation of MCF-7 cells and found the proliferation rate of MCF-7-UCH-L1 cells is same as MCF-7 transfected with empty vector (data not shown), which is different from the observations in previous study, where they found that overexpression of UCH-L1 inhibits cell proliferation in MDA-MB-231 cells[Xiang et al., 2012]. Given there the different genetic background of MCF-7 and MDA-MB-231, for example, MCF-7 cells express wild type p53 [Lu et al., 2001] but MDA-MB-231 cells express a mutant gain-of-function p53 which actually protects cancer cells against different stresses and promotes cell survival [Hui et al., 2006]. Thus, our studies again suggested the role of UCH-L1 is cell type and genetic background specific. Thus, the role of UCH-L1 on cell proliferation definitely requires further investigation. For its role of cell invasion, our biochemical and live cell approaches have suggested a novel target of the action of UCH-L1, which is Akt. However, there are several aspects that need more studies, first, mutagenesis studies are required to dissect the domains in UCH-L1 and Akt for their functional interaction; second, from our observation in Fig. 3B, we only observed p-Akt was increased upon overexpression of UCH-L1, while the total Akt remained the same. Our observation is consistent with previous studies on lung cancer cells and melanoma cells [Kim et al., 2009; Wulfanger et al., 2013], where overexpression of UCH-L1 didn’t alter total Akt levels. In addition, many studies have demonstrated that regulation of Akt is mainly through caspase-mediated degradation and heat shock protein-mediated protein stabilization, while only few studies suggested Akt might be degraded by ubiquitin-proteasom system (UPS) (summarized in the review article [Liao and Hung, 2010]). Thus, it is unlikely that UCH-L1 activates Akt signaling pathway by de-ubiquitinating and stabilizing Akt. So the detailed molecular mechanism underlying UCH-L1 mediated activation of Akt signaling remains to be elucidated; third, we believe that there are more UCH-L1 interacting proteins in cancer cells that might also contribute to UCH-L1 mediated promotion of cell invasion. For this purpose, we have performed BioID assay to identify other two proteins that could be labeled by biotin, they are MDM2 and Caveolin-1. The reason we performed this additional experiments is to identify a third partner interacting with Akt-UCH-L1. Given that both MDM2 and Caveolin-1 have been identified as a partner for Akt[Mayo and Donner, 2001; Wu et al., 2014] and more interestingly, MDM2 has been identified to interact with UCH-L1 as well[Li et al., 2010]. Our initial idea was to test whether UCH-L1 can form complex with Akt and MDM2 and/or Caveolin-1 to regulate invasion of breast cancer cells. However, our results couldn’t support this hypothesis. We think these interactions could be tissue specific, or our BioID assay is not sensitive enough to reveal the interactions between UCHL1 with MDM2/Caveolin-1 other than Akt, which might further suggest that the interaction between UCHL1 and AKT is more direct than other interactions. Nevertheless, our established mycBioID-UCH-L1 system will be utilized to identify more UCH-L1 binding partners for regulation of invasion in breast cancer cells. In addition to its advantages mentioned in previous text, this system is also ideal for identification of interacting partners in different treatment conditions. For example, UCH-L1 has been linked to cancer cell chemotherapy resistance [Jin et al., 2015; Wang et al., 2016], thus we can compare biotinylated protein profile derived from mycBioID-UCH-L1 overexpressing cells treated with or without chemotherapy drugs with the hope to identify specific proteins that responsible for UCH-L1 mediated chemotherapy resistance. Finally, the role of UCH-L1 on Akt2 activation for regulation of cell invasion could be tested in more details. Theoretically, specific inhibitors for specific isoform of Akt should be useful to determine whether the role of UCH-L1 on cell invasion is through activating Akt2. However, the specificity of chemical Akt inhibitors are not ideal to differentiate different isoforms of Akt [Barnett et al., 2005; Huck and Mochalkin, 2017], moreover, they usually have a high cytotoxicity [Barnett et al., 2005], which greatly obstructs our experiments by using different Akt inhibitors. Thus, some molecular approaches, such as CRISPR mediated gene editing might be useful in this situation.
In our study, we have noticed that UCH-L1 promotes invasion of breast cancer cells, future studies need to delineate whether UCH-L1 regulate chemo-resistance in breast cancer cells, as well as the role of UCH-L1 in cancer stem cells of breast cancers. Collectively, our findings implicate that UCH-L1 is an essential promotor during breast cancer metastasis and may serve as a potential biomarker to predict prognosis of the human patients with breast cancers.
Acknowledgments
This work was supported by U.S. National Institute of Health (NIH) Grant #CA109371 to J.Y., NIH grants #HL124122, #AR067766 and American Heart Association Grant 12SDG12070174 to H.Z., and the National Natural Science Foundation of China (Grant No. 81401155) to T.T.
Footnotes
Conflicts of Interest
The authors declare that there are no conflict of interest.
References
- Arboleda MJ, Lyons JF, Kabbinavar FF, Bray MR, Snow BE, Ayala R, Danino M, Karlan BY, Slamon DJ. Overexpression of AKT2/protein kinase Bbeta leads to up-regulation of beta1 integrins, increased invasion, and metastasis of human breast and ovarian cancer cells. Cancer Res. 2003;63:196–206. [PubMed] [Google Scholar]
- Barnett SF, Defeo-Jones D, Fu S, Hancock PJ, Haskell KM, Jones RE, Kahana JA, Kral AM, Leander K, Lee LL, Malinowski J, McAvoy EM, Nahas DD, Robinson RG, Huber HE. Identification and characterization of pleckstrin-homology-domain-dependent and isoenzyme-specific Akt inhibitors. Biochem J. 2005;385:399–408. doi: 10.1042/BJ20041140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilguvar K, Tyagi NK, Ozkara C, Tuysuz B, Bakircioglu M, Choi M, Delil S, Caglayan AO, Baranoski JF, Erturk O, Yalcinkaya C, Karacorlu M, Dincer A, Johnson MH, Mane S, Chandra SS, Louvi A, Boggon TJ, Lifton RP, Horwich AL, Gunel M. Recessive loss of function of the neuronal ubiquitin hydrolase UCHL1 leads to early-onset progressive neurodegeneration. Proc Natl Acad Sci U S A. 2013;110:3489–94. doi: 10.1073/pnas.1222732110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield DA, Gnjec A, Poon HF, Castegna A, Pierce WM, Klein JB, Martins RN. Redox proteomics identification of oxidatively modified brain proteins in inherited Alzheimer’s disease: an initial assessment. J Alzheimers Dis. 2006;10:391–7. doi: 10.3233/jad-2006-10407. [DOI] [PubMed] [Google Scholar]
- Calise J, Powell SR. The ubiquitin proteasome system and myocardial ischemia. Am J Physiol Heart Circ Physiol. 2013;304:H337–49. doi: 10.1152/ajpheart.00604.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Sugiura Y, Myers KG, Liu Y, Lin W. Ubiquitin carboxyl-terminal hydrolase L1 is required for maintaining the structure and function of the neuromuscular junction. Proc Natl Acad Sci U S A. 2010;107:1636–41. doi: 10.1073/pnas.0911516107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng GZ, Chan J, Wang Q, Zhang W, Sun CD, Wang LH. Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel. Cancer Res. 2007;67:1979–87. doi: 10.1158/0008-5472.CAN-06-1479. [DOI] [PubMed] [Google Scholar]
- Chin YR, Toker A. Function of Akt/PKB signaling to cell motility, invasion and the tumor stroma in cancer. Cell Signal. 2009;21:470–6. doi: 10.1016/j.cellsig.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, Li L. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson’s and Alzheimer’s diseases. J Biol Chem. 2004;279:13256–64. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–46. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- Day IN, Thompson RJ. UCHL1 (PGP 9.5): neuronal biomarker and ubiquitin system protein. Prog Neurobiol. 2010;90:327–62. doi: 10.1016/j.pneurobio.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–90. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- Etlinger JD, Goldberg AL. A soluble ATP-dependent proteolytic system responsible for the degradation of abnormal proteins in reticulocytes. Proc Natl Acad Sci U S A. 1977;74:54–8. doi: 10.1073/pnas.74.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faridi J, Wang L, Endemann G, Roth RA. Expression of constitutively active Akt-3 in MCF-7 breast cancer cells reverses the estrogen and tamoxifen responsivity of these cells in vivo. Clin Cancer Res. 2003;9:2933–9. [PubMed] [Google Scholar]
- Gong B, Cao Z, Zheng P, Vitolo OV, Liu S, Staniszewski A, Moolman D, Zhang H, Shelanski M, Arancio O. Ubiquitin hydrolase Uch-L1 rescues beta-amyloid-induced decreases in synaptic function and contextual memory. Cell. 2006;126:775–88. doi: 10.1016/j.cell.2006.06.046. [DOI] [PubMed] [Google Scholar]
- Goto Y, Zeng L, Yeom CJ, Zhu Y, Morinibu A, Shinomiya K, Kobayashi M, Hirota K, Itasaka S, Yoshimura M, Tanimoto K, Torii M, Sowa T, Menju T, Sonobe M, Kakeya H, Toi M, Date H, Hammond EM, Hiraoka M, Harada H. UCHL1 provides diagnostic and antimetastatic strategies due to its deubiquitinating effect on HIF-1alpha. Nat Commun. 2015;6:6153. doi: 10.1038/ncomms7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumireddy K, Sun F, Klein-Szanto AJ, Gibbins JM, Gimotty PA, Saunders AJ, Schultz PG, Huang Q. In vivo selection for metastasis promoting genes in the mouse. Proc Natl Acad Sci U S A. 2007;104:6696–701. doi: 10.1073/pnas.0701145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanpude P, Bhattacharya S, Dey AK, Maiti TK. Deubiquitinating enzymes in cellular signaling and disease regulation. IUBMB Life. 2015;67:544–55. doi: 10.1002/iub.1402. [DOI] [PubMed] [Google Scholar]
- Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6:776–88. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- Huck BR, Mochalkin I. Recent progress towards clinically relevant ATP-competitive Akt inhibitors. Bioorg Med Chem Lett. 2017;27:2838–2848. doi: 10.1016/j.bmcl.2017.04.090. [DOI] [PubMed] [Google Scholar]
- Hui L, Zheng Y, Yan Y, Bargonetti J, Foster DA. Mutant p53 in MDA-MB-231 breast cancer cells is stabilized by elevated phospholipase D activity and contributes to survival signals generated by phospholipase D. Oncogene. 2006;25:7305–10. doi: 10.1038/sj.onc.1209735. [DOI] [PubMed] [Google Scholar]
- Irie HY, Pearline RV, Grueneberg D, Hsia M, Ravichandran P, Kothari N, Natesan S, Brugge JS. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial-mesenchymal transition. J Cell Biol. 2005;171:1023–34. doi: 10.1083/jcb.200505087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Yu W, Lou X, Zhou F, Han X, Zhao N, Lin B. UCHL1 Is a Putative Tumor Suppressor in Ovarian Cancer Cells and Contributes to Cisplatin Resistance. J Cancer. 2013;4:662–70. doi: 10.7150/jca.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Zhang W, Xu J, Wang H, Zhang Z, Chu C, Liu X, Zou Q. UCH-L1 involved in regulating the degradation of EGFR and promoting malignant properties in drug-resistant breast cancer. Int J Clin Exp Pathol. 2015;8:12500–8. [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim S, Koh H, Yoon SO, Chung AS, Cho KS, Chung J. Akt/PKB promotes cancer cell invasion via increased motility and metalloproteinase production. FASEB J. 2001;15:1953–62. doi: 10.1096/fj.01-0198com. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim YM, Lim S, Nam YK, Jeong J, Lee KJ. Ubiquitin C-terminal hydrolase-L1 is a key regulator of tumor cell invasion and metastasis. Oncogene. 2009;28:117–27. doi: 10.1038/onc.2008.364. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Magesh V, Lee JJ, Kim S, Knaus UG, Lee KJ. Ubiquitin C-terminal hydrolase-L1 increases cancer cell invasion by modulating hydrogen peroxide generated via NADPH oxidase 4. Oncotarget. 2015;6:16287–303. doi: 10.18632/oncotarget.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiblich A, Cross SS, Catto JW, Pesce G, Hamdy FC, Rehman I. Human prostate cancer cells express neuroendocrine cell markers PGP 9.5 and chromogranin A. Prostate. 2007;67:1761–9. doi: 10.1002/pros.20654. [DOI] [PubMed] [Google Scholar]
- Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein MJ, Jonnalagada S, Chernova T, Dehejia A, Lavedan C, Gasser T, Steinbach PJ, Wilkinson KD, Polymeropoulos MH. The ubiquitin pathway in Parkinson’s disease. Nature. 1998;395:451–2. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- Li L, Tao Q, Jin H, van Hasselt A, Poon FF, Wang X, Zeng MS, Jia WH, Zeng YX, Chan AT, Cao Y. The tumor suppressor UCHL1 forms a complex with p53/MDM2/ARF to promote p53 signaling and is frequently silenced in nasopharyngeal carcinoma. Clin Cancer Res. 2010;16:2949–58. doi: 10.1158/1078-0432.CCR-09-3178. [DOI] [PubMed] [Google Scholar]
- Li QQ, Wang WJ, Xu JD, Cao XX, Chen Q, Yang JM, Xu ZD. Involvement of CD147 in regulation of multidrug resistance to P-gp substrate drugs and in vitro invasion in breast cancer cells. Cancer Sci. 2007;98:1064–9. doi: 10.1111/j.1349-7006.2007.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Hung MC. Physiological regulation of Akt activity and stability. Am J Transl Res. 2010;2:19–42. [PMC free article] [PubMed] [Google Scholar]
- Lien HC, Wang CC, Lin CH, Lu YS, Huang CS, Hsiao LP, Yao YT. Differential expression of ubiquitin carboxy-terminal hydrolase L1 in breast carcinoma and its biological significance. Hum Pathol. 2013;44:1838–48. doi: 10.1016/j.humpath.2013.02.006. [DOI] [PubMed] [Google Scholar]
- Liu X, Zeng B, Ma J, Wan C. Comparative proteomic analysis of osteosarcoma cell and human primary cultured osteoblastic cell. Cancer Invest. 2009;27:345–52. doi: 10.1080/07357900802438577. [DOI] [PubMed] [Google Scholar]
- Lu X, Errington J, Curtin NJ, Lunec J, Newell DR. The impact of p53 status on cellular sensitivity to antifolate drugs. Clin Cancer Res. 2001;7:2114–23. [PubMed] [Google Scholar]
- Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci U S A. 2001;98:11598–603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani K, Thompson DA, Barthel A, Sakaue H, Liu W, Weigel RJ, Roth RA. Up-regulation of Akt3 in estrogen receptor-deficient breast cancers and androgen-independent prostate cancer lines. J Biol Chem. 1999;274:21528–32. doi: 10.1074/jbc.274.31.21528. [DOI] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–86. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Ootsuka S, Asami S, Sasaki T, Yoshida Y, Nemoto N, Shichino H, Chin M, Mugishima H, Suzuki T. Useful markers for detecting minimal residual disease in cases of neuroblastoma. Biol Pharm Bull. 2008;31:1071–4. doi: 10.1248/bpb.31.1071. [DOI] [PubMed] [Google Scholar]
- Otsuki T, Yata K, Takata-Tomokuni A, Hyodoh F, Miura Y, Sakaguchi H, Hatayama T, Hatada S, Tsujioka T, Sato Y, Murakami H, Sadahira Y, Sugihara T. Expression of protein gene product 9.5 (PGP9.5)/ubiquitin-C-terminal hydrolase 1 (UCHL-1) in human myeloma cells. Br J Haematol. 2004;127:292–8. doi: 10.1111/j.1365-2141.2004.05205.x. [DOI] [PubMed] [Google Scholar]
- Partridge J, Flaherty P. An in vitro FluoroBlok tumor invasion assay. J Vis Exp. 2009 doi: 10.3791/1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell SR, Herrmann J, Lerman A, Patterson C, Wang X. The ubiquitin-proteasome system and cardiovascular disease. Prog Mol Biol Transl Sci. 2012;109:295–346. doi: 10.1016/B978-0-12-397863-9.00009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmeh AA, Morin B, Schenk AD, Liang B, Heinrich BS, Brusic V, Walz T, Whelan SP. Critical phosphoprotein elements that regulate polymerase architecture and function in vesicular stomatitis virus. Proc Natl Acad Sci U S A. 2012;109:14628–33. doi: 10.1073/pnas.1209147109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–10. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saettler EB, Temple WJ. The surgeon as a prognostic factor in the management of pancreatic cancer. Surg Oncol Clin N Am. 2000;9:133–42. viii. [PubMed] [Google Scholar]
- Schroder C, Milde-Langosch K, Gebauer F, Schmid K, Mueller V, Wirtz RM, Meyer-Schwesinger C, Schluter H, Sauter G, Schumacher U. Prognostic relevance of ubiquitin C-terminal hydrolase L1 (UCH-L1) mRNA and protein expression in breast cancer patients. J Cancer Res Clin Oncol. 2013;139:1745–55. doi: 10.1007/s00432-013-1496-z. [DOI] [PubMed] [Google Scholar]
- Schuierer MM, Bataille F, Hagan S, Kolch W, Bosserhoff AK. Reduction in Raf kinase inhibitor protein expression is associated with increased Ras-extracellular signal-regulated kinase signaling in melanoma cell lines. Cancer Res. 2004;64:5186–92. doi: 10.1158/0008-5472.CAN-03-3861. [DOI] [PubMed] [Google Scholar]
- Tezel E, Hibi K, Nagasaka T, Nakao A. PGP9.5 as a prognostic factor in pancreatic cancer. Clin Cancer Res. 2000;6:4764–7. [PubMed] [Google Scholar]
- Ummanni R, Jost E, Braig M, Lohmann F, Mundt F, Barett C, Schlomm T, Sauter G, Senff T, Bokemeyer C, Sultmann H, Meyer-Schwesinger C, Brummendorf TH, Balabanov S. Ubiquitin carboxyl-terminal hydrolase 1 (UCHL1) is a potential tumour suppressor in prostate cancer and is frequently silenced by promoter methylation. Mol Cancer. 2011;10:129. doi: 10.1186/1476-4598-10-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasko V, Saji M, Hardy E, Kruhlak M, Larin A, Savchenko V, Miyakawa M, Isozaki O, Murakami H, Tsushima T, Burman KD, De Micco C, Ringel MD. Akt activation and localisation correlate with tumour invasion and oncogene expression in thyroid cancer. J Med Genet. 2004;41:161–70. doi: 10.1136/jmg.2003.015339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeriah S. Opposing roles of the oncogene Akt isoforms in tumour progression: is there a dark side to Akt pathway inhibition? J Chem Biol. 2012;5:115–7. doi: 10.1007/s12154-012-0076-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zou L, Zhou D, Zhou Z, Tang F, Xu Z, Liu X. Overexpression of ubiquitin carboxyl terminal hydrolase-L1 enhances multidrug resistance and invasion/metastasis in breast cancer by activating the MAPK/Erk signaling pathway. Mol Carcinog. 2016;55:1329–42. doi: 10.1002/mc.22376. [DOI] [PubMed] [Google Scholar]
- Wilkinson KD. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol. 2000;11:141–8. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- Wu SZ, Peng FF, Li JL, Ye F, Lei SQ, Zhang BF. Akt and RhoA activation in response to high glucose require caveolin-1 phosphorylation in mesangial cells. Am J Physiol Renal Physiol. 2014;306:F1308–17. doi: 10.1152/ajprenal.00447.2013. [DOI] [PubMed] [Google Scholar]
- Wulfanger J, Biehl K, Tetzner A, Wild P, Ikenberg K, Meyer S, Seliger B. Heterogeneous expression and functional relevance of the ubiquitin carboxyl-terminal hydrolase L1 in melanoma. Int J Cancer. 2013;133:2522–32. doi: 10.1002/ijc.28278. [DOI] [PubMed] [Google Scholar]
- Xiang T, Li L, Yin X, Yuan C, Tan C, Su X, Xiong L, Putti TC, Oberst M, Kelly K, Ren G, Tao Q. The ubiquitin peptidase UCHL1 induces G0/G1 cell cycle arrest and apoptosis through stabilizing p53 and is frequently silenced in breast cancer. PLoS One. 2012;7:e29783. doi: 10.1371/journal.pone.0029783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Zhang C, Fang S, Ou R, Li W, Xu Y. UCH-LI acts as a novel prognostic biomarker in gastric cardiac adenocarcinoma. Int J Clin Exp Pathol. 2015;8:13957–67. [PMC free article] [PubMed] [Google Scholar]
- Yoeli-Lerner M, Yiu GK, Rabinovitz I, Erhardt P, Jauliac S, Toker A. Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol Cell. 2005;20:539–50. doi: 10.1016/j.molcel.2005.10.033. [DOI] [PubMed] [Google Scholar]