Abstract
BACKGROUND & AIMS
Obesity-induced non-alcoholic fatty liver disease (NAFLD) develops, in part, via excess insulin-stimulated hepatic de novo lipogenesis, which increases, paradoxically, in patients with obesity-induced insulin resistance. Pleckstrin homology domain leucine-rich repeat protein phosphatase 2 (PHLPP2) terminates insulin signaling by dephosphorylating Akt; levels of PHLPP2 are reduced in livers from obese mice. We investigated whether loss of hepatic PHLPP2 is sufficient to induce fatty liver in mice, mechanisms of PHLPP2 degradation in fatty liver, and expression of genes that regulate PHLPP2 in livers of patients with NAFLD.
METHODS
C57BL/6J mice (controls), obese db/db mice and mice with liver-specific deletion of PHLPP2 (L-PHLPP2) fed either normal chow or high-fat diet (HFD) were analyzed for metabolic phenotypes including glucose tolerance and hepatic steatosis. PHLPP2-deficient primary hepatocytes or CRISPR/Cas9-mediated PHLPP2-knockout hepatoma cells were analyzed for insulin signaling and gene expression. We performed mass spectrometry analyses of livers tissues from C57BL/6J mice transduced with Ad-HA-FLAG-PHLPP2 to identify post-translational modifications to PHLPP2 and proteins that interact with PHLPP2. We measured levels of mRNAs by quantitative reverse transcription PCR in liver biopsies from patients with varying degrees of hepatic steatosis.
RESULTS
PHLPP2-knockout hepatoma cells and hepatocytes from L-PHLPP2 mice showed normal initiation of insulin signaling, but prolonged insulin action. Chow-fed L-PHLPP2 mice had normal glucose tolerance but hepatic steatosis. In HFD-fed C57BL/6J or db/db obese mice, endogenous PHLPP2 was degraded by glucagon and PKA-dependent phosphorylation of PHLPP2 (at Ser1119 and Ser1210), which led to PHLPP2 binding to potassium channel tetramerization domain containing 17 (KCTD17), a substrate-adaptor for Cul3-RING ubiquitin ligases. Levels of KCTD17 mRNA were increased in livers of HFD-fed C57BL/6J or db/db obese mice and in liver biopsies patients with NAFLD, compared with liver tissues from healthy control mice or patients without steatosis. Knockdown of KCTD17 with small hairpin RNA in primary hepatocytes increased PHLPP2 protein but not Phlpp2 mRNA, indicating that KCTD17 mediates PHLPP2 degradation. KCTD17 knockdown in obese mice prevented PHLPP2 degradation and decreased expression of lipogenic genes.
CONCLUSIONS
In mouse models of obesity, we found that PHLPP2 degradation induced lipogenesis without affecting gluconeogenesis. KCTD17, which is upregulated in liver tissues of obese mice and patients with NAFLD, binds to phosphorylated PHLPP2 to target it for ubiquitin-mediated degradation; this increases expression of genes that regulate lipogenesis to promote hepatic steatosis. Inhibitors of this pathway might be developed for treatment of patients with NAFLD.
Keywords: triglyceride, insulin signaling, gene regulation, signal transduction
Introduction
The liver integrates nutrient state with the need to preserve glucose and lipids in a narrow physiologic range, with assistance from hormonal inputs.1 In normal physiology, fasting triggers the release of glucagon from pancreatic α-cells, which stimulates gluconeogenesis and glycogenolysis to maintain normoglycemia.2 In the postprandial state, pancreatic β-cells dial up insulin levels to block hepatic glucose production3–5 and utilize excess glucose for long-term energy storage in the liver in the form of glycogen, and indirectly, triglycerides.6
The elegance of this system is undermined by the chronic overnutrition associated with obesity, which leads to insulin resistance and an inability to restrain hepatic glucose production and the fasting hyperglycemia of Type 2 Diabetes (T2D). Nonetheless, compensatory hyperinsulinemia is apparently sufficient to activate sterol regulatory element-binding protein 1c (Srebp1c)-mediated lipogenesis,7, 8 which contributes to the excess hepatic triglyceride (TG) accumulation that defines Non-Alcoholic Fatty Liver Disease (NAFLD).9 The mechanism underlying this “selective” insulin resistance remains under debate, but we and others have shown that insulin signaling diverges at the critical molecular node, Akt, to activate mTORC1-dependent and -independent lipogenic pathways.10–12 But insulin doesn’t act alone – the lipogenic program is under complex control by nutrient state and other hormones. Of note in this cadre is glucagon,13 postprandial plasma levels of which remain inappropriately high in T2D patients.14–16 The interplay between insulin and glucagon pathways is further highlighted by the unique shared phenotype of reduced hepatic and plasma TG in liver-specific insulin receptor knockout17 and glucagon receptor knockout mice,18 but the potential role of glucagon in the selective insulin resistance seen in obesity has not been explored.
Pleckstrin homology (PH) domain leucine-rich repeat protein phosphatases (PHLPPs) have been shown to dephosphorylate the key regulatory Ser473 residue of Akt, thereby terminating growth factor-stimulated Akt activity and suppress tumor growth in cancer models.19–21 Our recent work has demonstrated that PHLPPs, specifically the PHLPP2 isoform, similarly terminates insulin action in obesity.12 We observed that diet-induced (HFD-feeding) or genetic (leptin-signaling deficient ob/ob or db/db mice) mouse models of obesity have lower PHLPP2 protein levels, which renders these mice unable to terminate insulin signaling, ultimately resulting in increased DNL and fatty liver.12 Consistently, exogenous “rescue” of hepatic PHLPP2 levels in obese mice reduced inappropriately elevated Akt Ser473 phosphorylation in the “late” refed state, and reversed fatty liver, but surprisingly, did not affect glucose tolerance. These data suggested that PHLPP2 is necessary to prevent insulin-potentiated DNL, but does not affect early post-prandial events such as insulin/Akt repression of FoxO-dependent hepatic gluconeogenesis in the bifurcation model of hepatic insulin signaling.10
Several important questions remained from these studies, however – 1) is chronic reduction of hepatic PHLPP2 levels sufficient to induce hepatic steatosis; 2) how is hepatic PHLPP2 lost in obesity, as Phlpp2 mRNA is unaffected in HFD-fed and genetic mouse models; and, 3) whether these data may have clinical and/or therapeutic implications. Here, we generate and characterize hepatocyte-specific PHLPP2 knockout (L-PHLPP2) mice, which show prolonged insulin-stimulated Akt Ser473 phosphorylation and excess DNL and fatty liver. To understand the endogenous regulatory machinery that mediates PHLPP2 degradation in obese liver, we performed sequential LC/MS-MS-based screens, which identified novel glucagon/PKA-mediated PHLPP2 phosphorylation at Ser1119 and Ser1210, which augments PHLPP2 interaction with KCTD17, an adaptor protein for Cul3-RING ubiquitin ligases.22 Interestingly, KCTD17 expression is increased in livers from obese mice and patients with NASH, linking glucagon-induced PHLPP2 phosphorylation with degradation. Thus, knockdown of KCTD17 in obese mice increases endogenous PHLPP2 and ameliorates obesity-induced fatty liver, revealing a novel strategy in the treatment of obesity-mediated NAFLD/NASH.
Methods
Bioinformatics
Hepatic Phlpp2 was assessed in BXD mouse genetic reference populations fed normal chow diet (GeneNetwork Accession No: GN432), and transcriptomes corresponding to the top and bottom sextiles subjected to GSEA. All raw transcriptomic data related to human NASH samples are publicly available on Gene Expression Omnibus under accession numbers (GSE60149 and GSE48452).23, 24 Heat maps were built using GENE-E; depth of shading at the correlation matrices (correlogram) indicates the magnitude of the correlation (Spearman's Rho). Correlogram and interaction network were generated using RStudio, with positive and negative correlations represented in blue and red, respectively. Only correlations with Spearman's Rho > 0.5 or < −0.5 are displayed in the interaction network.
Constructs
AAV8-TBG-GFP or AAV8-TBG-Cre were generated by Penn Vector Core. Site-directed mutagenesis was performed using Q5 site-directed mutagenesis (NEB) to generate pcDNA3/HA/Flag/PHLPP2-S1119A, -S1210A or -S1119/1210A (2A). Ad-GFP, Ad-shControl, and Ad-HA/Flag/PHLPP2 adenoviruses have been described.12 Ad-HA/Flag/PHLPP2 (2A) and Ad-shKctd17 were generated by Welgen, Inc. (Worcester, MA). 20 nucleotide single-guide RNA (sgRNA) sequences for PHLPP2 were designed using a CRISPR design tool, cloned into LentiCRISPRv2 (Addgene #52961).
Animals
Male C57BL/6J (#664) and leptin receptor-deficient db/db (#642) mice were purchased from Jackson Labs. PHLPP2 floxed mice were generated with a targeting vector (EUCOMM) that was modified, linearized and electroporated into embryonic stem (ES) cells. Antibiotic-resistant ES cell clones were microinjected into C57BL/6 blastocysts to obtain chimeric mice. Founders that exhibited germline transmission were bred with C57BL/6J mice expressing flp recombinase (Jackson #016226) to remove the neo cassette (PHLPP2flox/+), backcrossed >5 generations with C57BL/6J mice, then intercrossed to generate homozygous PHLPP2flox/flox mice. Male PHLPP2flox/flox mice were transduced at 7 weeks of life with AAV8-TBG-GFP or AAV8-TBG-Cre (Penn Vector Core) to generate Cre- and L-PHLPP2 mice, respectively – all mice were transduced with AAV – then continued on standard chow (Purina Mills 5053) or switched to HFD (Harlan Laboratories TD.06414) as indicated. For adenovirus experiments, we injected 2.5–5×108 purified viral particles per gram body weight of control (Ad-GFP or Ad-shControl) or experimental viruses, continued HFD-feeding, performed metabolic analysis on days 3–7 and killed the mice at day 7 or 10 post-injection. We sacrificed mice after a 18 h fast followed by refeeding for 8 h. Mice were housed 3–5 animals per cage, with a 12 h light/dark cycle, in a temperature-controlled environment. All animal experiments were approved by the Columbia University Institutional Animal Care and Utilization Committee.
CRISPR generation of PHLPP2-knockout (KO) cells and cell culture studies
Lentivirus encoding PHLPP2 sgRNAs was generated by co-transfection of 293T cells with the lentiviral vectors psPAX2 and pMD2.G. Viral supernatant was applied to HepG2 or Hepa1c1c7 cells with polybrene (Sigma), which were selected with puromycin (ThermoFisher) prior to expansion of single clones, screening by western blot and genomic sequencing. In some experiments, PHLPP2 KO cells were transfected with PHLPP2-WT plasmid (Lipofectamine 3000, ThermoFisher) to “reconstitute” PHLPP2. Primary hepatocytes were transfected with plasmid expressing PHLPP2-WT or mutants using Lipofectamine 3000 (ThermoFisher) as per the manufacturer’s protocol.
Hepatocyte isolation and glucose production
Primary hepatocytes12, 25 isolated from PHLPP2flox/flox mice were transduced with Ad-GFP or Ad-Cre adenoviruses at MOI 10 two hours after plating on collagen-coated plates; 28 h later, cells were incubated for 4 h in glucose-free Krebs-Ringer buffer containing 20 mM sodium lactate and 2 mM sodium pyruvate, with or without dexamethasone, forskolin, or insulin. Glucose released was measured using Glucose (HK) assay kit (Sigma) and normalized to cellular protein concentration (BCA assay, Pierce).
Antibodies, western blots and immunoprecipitation
Immunoblots were conducted on 3–7 samples randomly chosen within each experimental cohort with antibodies against Akt (#9272), p-Akt (S473, #4060), p-Akt (T308, #13038), p-S6K1 (T389, #9205), p-S6 (S240/244, #2215), S6 (#2217), p-(Ser/Thr) PKA substrate (#9621), PKA (#4782), DYKDDDDK-tag (#14793), HA-tag (#3724), myc-tag (#2276), and β-actin (#4970) from Cell Signaling; S6K1 (sc-8418) from Santa Cruz; and PHLPP1 (A300-660A) and PHLPP2 (A300-661A) from Bethyl Laboratories, Inc. Antibodies directed to human p-PHLPP2 (S1119 or S1210, identical to S1116 or S1207 in mouse PHLPP2) were generated by immunizing rabbits with synthetic phospho-specific peptides, verified by ELISA and affinity purified (Lifetein).
Liquid chromatography-tandem mass spectrometry (LC-MS/MS)
PHLPP2 was purified from livers of male C57BL/6J mice transduced with Ad-HA-FLAG-PHLPP2. To identify PHLPP2 PTMs, liver lysates were prepared in stringent lysis buffer [25 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% NP-40, 0.05% SDS, 1% sodium deoxycholate, 10 mM β-glycerophosphate, 10 mM sodium pyrophosphate, 50 mM sodium fluoride, 1.5 mM sodium orthovanadate, 30 μM Trichostatin A (Sigma), 20 mM sodium butyrate, 20 mM nicotinamide, 25 mM N-ethylmaleimide and protease inhibitor cocktail (Pierce)], immunoprecipitated with anti-FLAG-conjugated M2 agarose (Sigma) and separated by SDS-PAGE. After Coomassie blue staining, the band corresponding to PHLPP2 was excised, extracted and trypsin digested.26 To examine PHLPP2 interacting proteins, liver lysates were extracted in less stringent lysis buffer [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1mM EDTA, 10% glycerol, 1% NP-40, 10 mM β-glycerophosphate, 10 mM sodium pyrophosphate, 50 mM sodium fluoride, 1.5 mM sodium orthovanadate, and protease inhibitor cocktail], immunoprecipitated with anti-Flag M2 agarose, and bound polypeptides eluted with FLAG peptide (Sigma). Aliquots of each sample were loaded onto an Acclaim PepMap 100 precolumn (75 μm × 2 cm, C18, 3 μm, 100Å, Thermo Scientific) in-line with an EASY-Spray, PepMap column (75 μm × 50 cm, C18, 2 μm, 100Å Thermo Scientific) using the autosampler of an EASY-nLC 1000 (Thermo Scientific). Peptides were gradient eluted into a Q Exactive mass spectrometer (Thermo Scientific) using a 60 min gradient from 2% solvent B to 40% solvent B. Solvent A was 2% acetonitrile in 0.5% acetic acid and solvent B was 90% acetonitrile in 0.5% acetic acid. The Q Exactive mass spectrometer was set up to acquire high resolution full MS spectra with a resolution of 70,000 at m/z 200, an AGC target of 1e6, with a maximum ion time of 120ms, and scan range of 400 to 1500 m/z. Following each full MS twenty data-dependent high resolution HCD MS/MS spectra were acquired using the following instrument parameters: resolution of 17,500 at m/z 200, AGC target of 5e4, maximum ion time of 250 ms, one microscan, 2 m/z isolation window, fixed first mass of 150 m/z, and NCE of 27, dynamic exclusion 30 seconds. For the phosphorylation analysis the MS/MS spectra were searched against a PHLPP2 fasta file using Byonic™27 and the site of phosphorylation verified by manual inspection of the spectrum. For the binding partner analysis, MS/MS spectra were searched against the uniprot mouse database and filtered using a <1% False Discovery Rate searched against a decoy database and excluding proteins with less than two unique peptides.
Metabolic analyses
Blood tail vein glucose was measured using a glucose meter (Bayer). Glucose tolerance tests or pyruvate tolerance tests were performed by intraperitoneal injection of 1 or 2 g per kg body weight glucose or 2 g per kg body weight sodium pyruvate after a 16 h fast. Hepatic lipids were extracted by the Folch method,28 and plasma and hepatic triglyceride (Thermo), Cholesterol E, or NEFA (Wako) measured using colorimetric assays according to the manufacturer’s protocol. Plasma insulin concentrations were measured using a mouse insulin ELISA kit (Mercodia).
Cross-sectional analysis of KCTD17 in human liver biopsy specimens
We analyzed liver gene expression in individuals who underwent liver biopsy for suspected NASH, due to persistent elevations in liver enzymes or severe obesity.29 The protocol was approved by the Ethical Committee of the Fondazione IRCCS of Milan, and each patient signed a written informed consent.
RNA/quantitative PCR
RNA was isolated by TRIzol (Invitrogen) or NucleoSpin RNA (Clontech), and cDNA synthesized with the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems) or SuperScript VILO cDNA synthesis kit (Life Technologies), followed by quantitative RT-PCR with Power SYBR Green PCR master mix (Applied Biosystems) in a CFX96 Real-Time PCR detection system (Bio-Rad). Primer sequences are available in Supplementary Table 1.
Statistical analysis
All data shown as mean ± s.e.m. ANOVA was used for comparison of means among multiple groups. For the liver biopsy cross-sectional cohort, comparisons were made by fitting data to generalized linear models, unadjusted (univariate analyses), or considering as independent variables: age, sex, BMI, T2D, PNPLA3 I148M alleles, histological steatosis grade, and activity. Hepatic KCTD17 mRNA levels were normalized for β-actin and log transformed before analyses to ensure a normal distribution. A P value < .05 was considered significant.
Results
PHLPP2 loss-of-function induces fatty liver but does not affect glucose tolerance
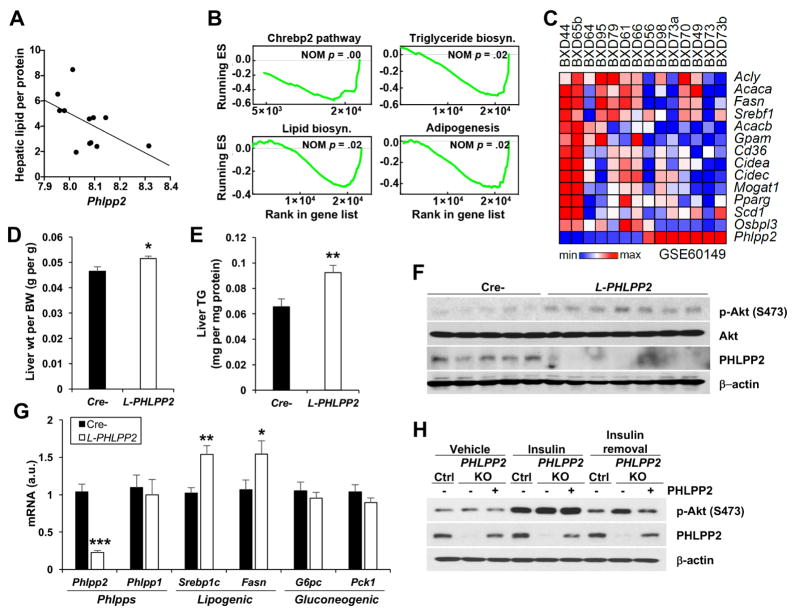
To explore endogenous differences in hepatic PHLPP2 levels and relationship to metabolism, we examined 41 strains of genetically diverse chow-fed BXD mice descendant from sequential C57BL/6J and DBA/2J crosses – BXD mice have ~5 million diverged sequence variants and represents, at present, the largest and best-characterized mouse genetic reference population.23 We found a significant, inverse correlation between Phlpp2 expression and liver TG across these chow-fed strains (Figure 1A). Next, we performed gene set enrichment analysis (GSEA) using hepatic transcriptomes of 14 BXD strains showing highest and lowest expression of Phlpp2 (Supplementary Figure 1A). In this analysis, we found that Phlpp2 is negatively correlated with lipogenic gene sets [Chrebp2 (NOM P < .001), adipogenesis (NOM P < .02), triglyceride biosynthesis (NOM P < .02) and lipid biosynthesis (NOM P < .02)] which contain Srebp1 and its target lipogenic genes (Figure 1B and C, Supplementary Figure 1B). Of note, we observed no correlation between Phlpp2 and gene sets associated with hepatic glucose production, or plasma levels of glucose (Spearman rho = .11, P = .567), insulin (Spearman rho = −.11, P = .502) and HOMA-IR (Spearman rho = .10, P = .553). These data show that across a genetically diverse mouse population where mRNA and protein levels are highly correlated,30 higher hepatic Phlpp2 expression is associated with lower lipid biosynthesis and may distinguish insulin-mediated functions in hepatic glucose and lipid metabolism, even in chow-fed lean mice.
Figure 1. Loss of Phlpp2 causes fatty liver.
(A) Hepatic Phlpp2 expression negatively correlates with hepatic lipid across different BXD populations (Spearman rho = −0.60, p = .04). (B) Normalized enrichment score (ES) of GSEA indicating gene sets that show the most significant negative correlation with hepatic Phlpp2 expression. (C) Heat map showing co-regulation of Phlpp2 and lipogenic genes in BXD strains. (D) Liver weight and (E) triglycerides, (F) western blots of liver lysates and (G) hepatic gene expression in chow-fed liver-specific PHLPP2 knockout (L-PHLPP2) and Cre- control mice (n=9–10/group). (H) Western blots from control, PHLPP2 KO, or PHLPP2-reconstituted PHLPP2 KO cells “pulsed” with 10 nM insulin for 30 min, with or without a 2 h “chase” in insulin-free medium. *P < .05, **P < .01, ***P < .001 as compared to the indicated control by two-way ANOVA. All data are shown as the means ± s.e.m.
To mimic the pathologic loss of hepatic PHLPP2 seen in obese mice,12 which may mirror endogenous differences in BXD strains, we generated homozygous PHLPP2flox/flox mice (Supplementary Figure 2A and B), and transduced with AAV8-TBG-GFP or AAV8-TBG-Cre to produce Cre- and hepatocyte-specific PHLPP2 knockout (L-PHLPP2) mice. Livers and primary hepatocytes showed >80% PHLPP2 deletion, without compensatory increase of the related isoform, PHLPP1 (Supplementary Figure 2C and D). L-PHLPP2 mice had normal body weight, adiposity and cholesterol/non-esterified fatty acid (NEFA) (Supplementary Figure 3A–E), but at sacrifice, showed increased liver weight and TG, with associated mild hypertriglyceridemia (Figure 1D and E, Supplementary Figure 3F). L-PHLPP2 mice had higher hepatic Akt Ser473 phosphorylation in “late” refeeding, similar to Ad-Cre-transduced PHLPP2flox/flox hepatocytes after extended insulin stimulation in vitro (Figure 1F, Supplementary Figure 3G). Prolonged activation of hepatic insulin signaling through Akt induced higher expression of Srebp1c and its canonical targets (i.e., Fasn), but did not affect gluconeogenic gene expression (Figure 1G). Consistently, we observed no differences in fasting or refed blood glucose, plasma insulin, glucose tolerance, pyruvate tolerance or hepatic glucose production (Supplementary Figure 3H–L). Similarly, loss of PHLPP2 did not affect key fatty acid oxidation genes (Supplementary Figure 3M).
As endogenous PHLPP2 levels are reduced in livers of WT mice fed a high-fat diet (HFD) 12, we hypothesized that HFD feeding to lower ΔPHLPP2 between L-PHLPP2 and Cre-control mice would negate differences in lipogenesis and fatty liver. Indeed, hepatic PHLPP2 levels were >70% reduced in HFD-fed as compared to chow-fed Cre- mice (Supplementary Figure 4A and B); Thus, in contrast to chow-fed cohorts, HFD-fed L-PHLPP2 mice showed unchanged liver weight, TG and lipogenic gene expression, and really no discernible metabolic phenotype, as compared to Cre- controls (Supplementary Figure 4C–L).
To confirm that PHLPP2 terminates insulin-Akt signaling in a cell-autonomous manner, we generated PHLPP2 knockout (KO) hepatoma cells using CRISPR/Cas9 (Supplementary Figure 5A–C). PHLPP2 KO cells showed similar insulin-induced Akt phosphorylation as compared to control cells, but delayed Akt dephosphorylation in the “chase” period after the insulin pulse (Supplementary Figure 5D), which was normalized by PHLPP2 reconstitution by transfection of WT PHLPP2 (Figure 1H). Similar to data from L-PHLPP2 mice, PHLPP2 KO cells had augmented insulin-stimulated Srebp1c and Fasn as compared with control cells (Supplementary Figure 5E). In combination, these data suggest that early postprandial insulin action, necessary for nuclear FoxO exclusion and cessation of gluconeogenesis31 is unimpeded, but that late postprandial insulin action on lipogenesis is heightened by either endogenous (HFD feeding) or artificial (L-PHLPP2 mice, PHLPP2 KO cells) loss of hepatocyte PHLPP2.
PHLPP2 is phosphorylated on Ser1119 and Ser1210 by glucagon/PKA action
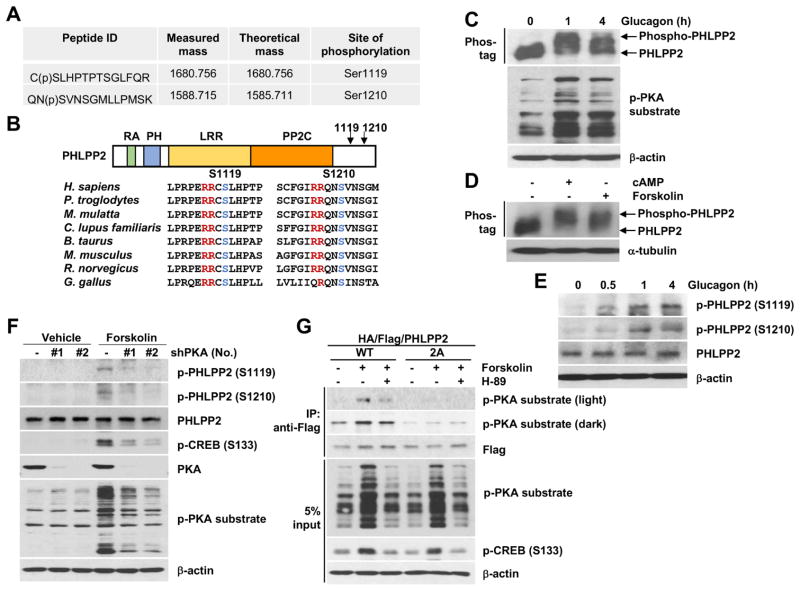
Obese mice show lower hepatic PHLPP2 levels, but unchanged Phlpp2 gene expression12. Based on this finding, we hypothesized that PHLPP2 levels may be affected by hormone or nutrient-regulated post-translational modifications (PTMs). To test this hypothesis, we performed LC-MS/MS analysis in immunoprecipitated PHLPP2 from livers of C57BL/6 mice transduced with HA/Flag-tagged PHLPP2, and found 2 phosphopeptides – RCpSLHPTPTSGLFQR (Ser1119) and RQNpSVNSGMLLPMSK (Ser1210) – at evolutionarily-conserved PKA consensus [RRX(S/T)] sites (Figure 2A and B). As glucagon is the major driver of PKA activity in liver,32 we next tested whether glucagon can induce PHLPP2 phosphorylation. Indeed, using Phos-tag-based electrophoresis, we found that PHLPP2 is rapidly phosphorylated in primary hepatocytes exposed to glucagon (Figure 2C), similar to treatment with potent PKA activators such as cAMP or forskolin (Figure 2D), but not after activation of other hormone/nutrient signaling (i.e. insulin/PI3K, MEK, GSK or mTOR) pathways (Supplementary Figure 6A). We next used site-directed mutagenesis to create Ser→Ala mutations at Ser1119 and Ser1210. Although mutation of Ser1119 or Ser1210 did not fully abolish forskolin-induced PHLPP2 phosphorylation (Supplementary Figure 6B), mutation of both serine residues (S1119/S1210A; 2A) prevented forskolin-induced PHLPP2 phosphorylation (Supplementary Figure 6C), suggesting that both sites are phosphorylated by PKA. We next generated polyclonal antibodies to p-PHLPP2 (Ser1119) and p-PHLPP2 (Ser1210), and confirmed specificity of these antibodies in primary hepatocytes transfected with WT, single (S1119A or S1210A) or double (2A) PHLPP2 Ser→Ala mutants (Supplementary Figure 6D and E). Using these novel reagents, we found that endogenous PHLPP2 phosphorylation at Ser1119 and Ser1210 is increased with acute glucagon (Figure 2E) or forskolin (Supplementary Figure 6F), but not with serum, insulin, or amino acid (AA) stimulation (Supplementary Figure 6F). As such, acute forskolin-induced phosphorylation at both Ser1119 and Ser1210 was reduced by knockdown of PKA signaling (Figure 2F). Thus, mutation of both Serine sites (2A) led to near-absent PHLPP2 phosphorylation in primary hepatocytes treated with glucagon or forskolin (Supplementary Figure 6G), recapitulating treatment with the PKA inhibitor H-89 (Figure 2G). In sum, these data prove that glucagon-induced PKA activity phosphorylates hepatocyte PHLPP2 at Ser1119 and Ser1210.
Figure 2. PHLPP2 is phosphorylated on Ser1119 and Ser1210 by glucagon/PKA action.
(A) Measured and theoretical mass (singly charged) of phosphopeptides of PHLPP2 isolated from Ad-PHLPP-transduced liver via immunoprecipitation and analyzed by LC-MS/MS, with the phosphorylated serine residue indicated with pS. (B) PHLPP2 contains Ras-association (RA), pleckstrin homology (PH), hydrophobic leucine-rich repeat (LRR), protein phosphatase 2C (PP2C) domains; amino acid sequence comparison from selected mammalian species with PKA targeting sequence (red) and phosphorylated serine (blue) residues highlighted. (C and D) Western blots from primary hepatocytes exposed to (C) 100 nM glucagon or (D) 100 μM cAMP or 10 μM forskolin for 1 h, using Phos-tag to separate phosphorylated from unphosphorylated PHLPP2. (E) Western blots from primary hepatocytes exposed to glucagon for the indicated times, or (F) HepG2 cells stably expressing two different shRNA directed at PKA or shControl (−) with or without forskolin. (G) Co-immunoprecipitation (CoIP) of PHLPP2 (WT) but not nonphosphorylatable mutant PHLPP2 (2A) with phospho-PKA substrate in response to forskolin, sensitive to the PKA inhibitor H-89, in primary hepatocytes.
Glucagon-mediated PHLPP2 phosphorylation reduces its stability in obesity
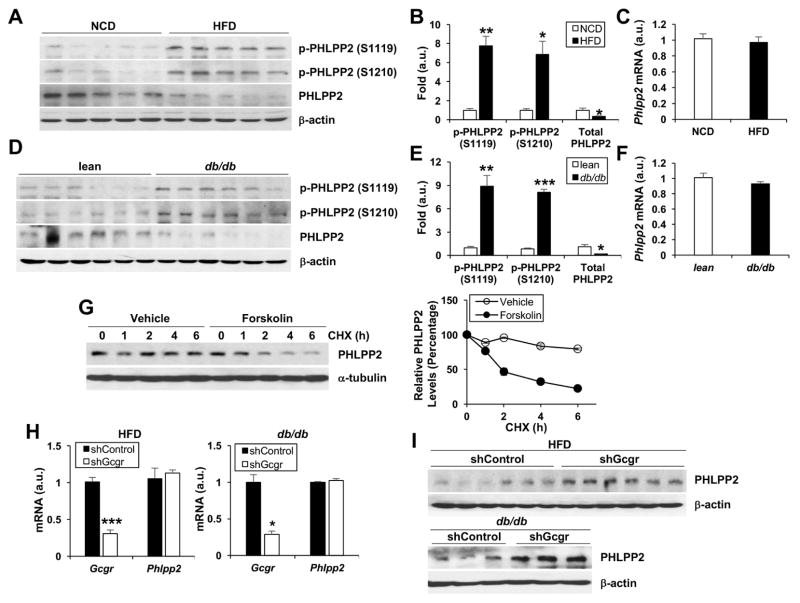
Glucagon is acutely increased in the fasted animal2. Thus, we find that hepatic PHLPP2 phosphorylation at both Ser1119 and Ser1210 is dynamically regulated by the fasting-refeeding transition in vivo (Supplementary Figure 7A and B). Obesity and T2D are associated with persistent hyperglucagonemia, which may regulate hepatic lipogenesis.33–35 We hypothesized that obesity may provoke “chronic” PHLPP2 phosphorylation. Indeed, we observed markedly increased hepatic PHLPP2 phosphorylation and a simultaneous reduction in total PHLPP2 protein in HFD-fed (Figure 3A–C) and in leptin-receptor (db/db) deficient (Figure 3D–F) mice, but strikingly, no change in Phlpp2 mRNA. These results suggested that PHLLP2 phosphorylation may directly affect its stability; to test this, we treated cells with cycloheximide to inhibit new protein translation, and found markedly decreased PHLPP2 protein in the presence of forskolin (Figure 3G). Next, we examined whether inhibition of hepatic glucagon signaling, by means of AAV8 encoding shRNA to the glucagon receptor (AAV8-shGcgr), could prevent obesity-induced loss of PHLPP2. Indeed, as compared to AAV8-shControl-transduced mice, AAV8-shGcgr transduction robustly increase hepatic PHLPP2 in HFD-fed and db/db mice, without affecting Phlpp2 mRNA (Figure 3H and I). Similar data were obtained with use of glucagon receptor antagonists (GRAs) in primary hepatocytes (not shown). We conclude that glucagon-induced hepatic PHLPP2 phosphorylation affects PHLPP2 stability.
Figure 3. Glucagon-mediated PHLPP2 phosphorylation and destabilization in obesity.
(A–F) Western blots of liver lysate with quantification of p-PHLPP2/total PHLPP2 and PHLPP2/β-actin, and corresponding liver Phlpp2 expression in (A–C) C57BL/6 WT mice fed normal chow or HFD for 16 weeks, and (D–F) 8-week-old C57BL/6J WT as compared to db/db mice. (G) Western blot of PHLPP2 in cycloheximide (CHX, 50 mg ml−1)-treated primary hepatocytes, with or without forskolin, with quantification of PHLPP2/α-tubulin relative to time 0. (H) qPCR and (I) western blots from AAV8-shControl- or shGcgr-transduced, HFD-fed C57BL/6 WT or db/db mice. *P < .05, **P < .01, ***P < .001 as compared to the indicated control by two- way ANOVA. All data are shown as the means ± s.e.m.
KCTD17 binds PHLPP2 to induce its degradation in obese liver
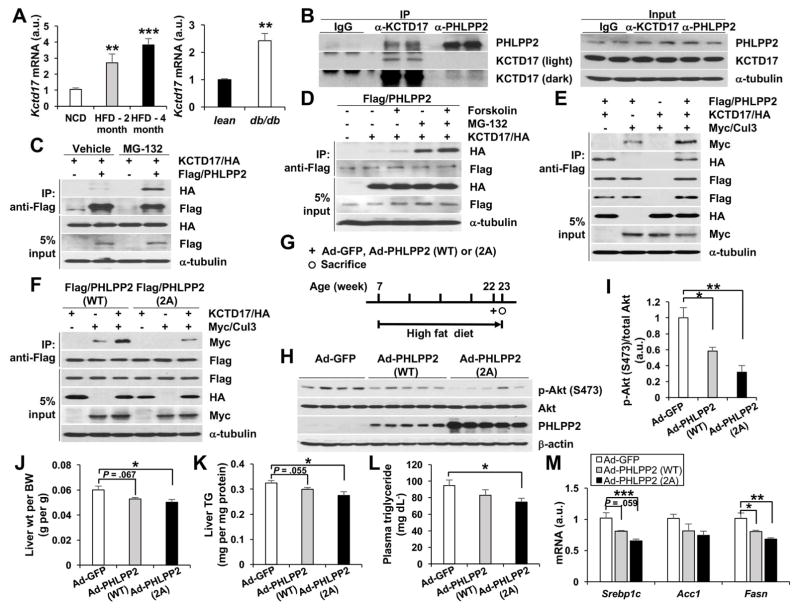
We were intrigued that although glucagon induces PHLPP2 phosphorylation in both fasting and obesity, only obesity reduced PHLPP2 protein levels. These data imply that PHLPP2 phosphorylation may be necessary but cannot be sufficient to reduce PHLPP2 stability, and that a “second hit” beyond hyperglucagonemia is necessary for the post-transcriptional loss of PHLPP2 seen in obese liver. To identify the regulatory mechanisms of glucagon-induced PHLPP2 degradation, we again performed LC/MS-MS but adjusted our purification conditions to isolate PHLPP2-binding proteins. We next sequentially queried the resultant list of PHLPP2 interactors for potential regulatory roles in protein stability and/or hepatic metabolism, and for altered expression in obese liver. From this analysis, we identified KCTD17 (potassium channel tetramerization domain containing 17) as a novel PHLPP2-associated protein. Despite its name, KCTD17 has been shown to act as a substrate-adaptor for Cul3-RING ubiquitin ligases (CRL3s) to regulate proteosomal degradation of specific targets.22, 36, 37 We observed that hepatic Kctd17 expression is robustly increased in HFD-fed and db/db mice (Figure 4A). Next, we confirmed our LC-MS/MS results by immunoprecipitation of endogenous KCTD17 in livers from HFD-fed mice with anti-PHLPP2 antibody, and vice versa (Figure 4B). KCTD17-PHLPP2 interaction was increased by proteosomal inhibitors (Figure 4C) and synergistically with forskolin co-treatment (Figure 4D), suggesting that glucagon-mediated PHLPP2 phosphorylation and degradation are linked by KCTD17. Indeed, KCTD17 increased WT PHLPP2-Cul3 interaction (Figure 4E), but far less efficiently with the nonphosphorylatable PHLPP2-2A mutant (Figure 4F). This suggested that the PHLPP2-2A mutant may be relatively resistant to degradation – to test this, we transduced HFD-fed mice with adenovirus encoding GFP, PHLPP2-WT or PHLPP2-2A (Figure 4G). Despite similar Phlpp2 mRNA induction as compared to Ad-GFP-transduced mice (Supplementary Figure 8A), PHLPP2-2A induced far greater increase in hepatic PHLPP2 levels than PHLPP2-WT, and thus greater decrease in refed hepatic Akt (Ser473) phosphorylation (Figure 4H and I). Thus, we found a gradated reduction in liver weight, TG and lipogenic gene expression, and plasma TG from control→PHLPP2-WT→PHLPP2-2A-transduced mice (Figure 4J–M), despite similar body weight/adiposity and measures of glucose homeostasis among the groups (Supplementary Figure 8B–E). In sum, these data show that glucagon-induced phosphorylation and KCTD17-recruitment of Cul3 synergistically target PHLPP2 for degradation, to increase DNL and provoke hepatic steatosis.
Figure 4. KCTD17 binds phosphorylated PHLPP2 to induce its degradation in obese liver.
(A) Liver Kctd17 expression in C57BL/6 WT mice fed normal chow or HFD for either 2 or 4 months (left) or 8-week-old C57BL/6J WT as compared to db/db mice (right). (B) Endogenous CoIP of KCTD17 and PHLPP2 in liver lysate from HFD-fed mice. (C) CoIP of KCTD17 by PHLPP2 in primary hepatocytes is increased by MG-132 and (D) synergistically by forskolin. (E) KCTD17 facilitates the interaction of PHLPP2 (WT), (F) but not the nonphosphorylatable mutant PHLPP2 (2A), with the E3-ligase, Cullin3. (G-I) Experimental outline (G) and western blots of liver lysate (H and I) from C57BL/6 mice fed HFD for 15 weeks, then transduced with Ad-GFP, Ad-PHLPP2 (WT), or Ad-PHLPP2 (2A) prior to sacrifice at a total of 16 weeks HFD feeding. (J) Liver weight and (K) triglycerides, (L) plasma triglycerides and (M) hepatic gene expression from HFD-fed C57BL/6 mice transduced with Ad-GFP, Ad-PHLPP2 (WT), or Ad-PHLPP2 (2A). (n=6–7/group). *P < .05, **P < .01 as compared to the indicated control by two-way ANOVA. All data are shown as the means ± s.e.m.
Knockdown of KCTD17 prevents diet-induced hepatic steatosis
We next predicted that KCTD17 knockdown may be sufficient to protect PHLPP2 from degradation. We produced shRNA directed against Kctd17, and found that Kctd17 knockdown increased PHLPP2 protein but not Phlpp2 mRNA expression in primary hepatocytes (Figure 5A and B) and in hepatoma cell lines (Supplementary Figure 9A and B). As hypothesized, acute Kctd17 knockdown strongly reduced PHLPP2 ubiquitination (Figure 5C), to completely block insulin-mediated induction of lipogenic genes (Supplementary Figure 9C).
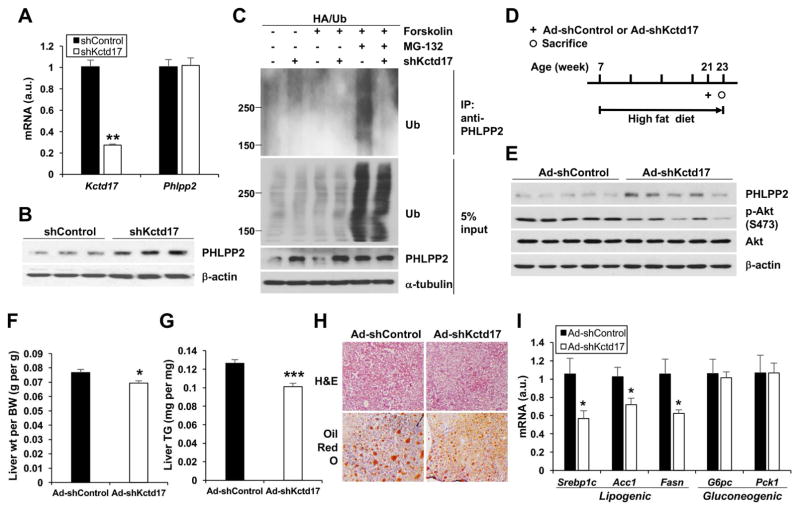
Figure 5. Knockdown of hepatic KCTD17 prevents diet-induced hepatic steatosis.
(A) qPCR and (B) western blots from primary hepatocytes transduced with Ad-shControl or Ad-shKctd17. (C) Knockdown of Kctd17 reduces endogenous PHLPP2 ubiquitination in HA/Ub-transfected primary hepatocytes. (D) Experimental outline, (E) western blots from liver lysate, (F) liver weight and (G) triglycerides, (H) liver H&E and Oil-Red-O staining, and (I) hepatic gene expression in C57BL/6 WT mice fed HFD for 14 weeks, then transduced with Ad-shControl or Ad-shKctd17 (n=6–7/group) prior to sacrifice at a total of 16 weeks HFD feeding. *P < .05, **P < .01, ***P < .001 as compared to the indicated control by two-way ANOVA. All data are shown as the means ± s.e.m.
Based on these findings, we predicted that reducing obesity-mediated increase of hepatic Kctd17 would reproduce the lower liver TG seen with Ad-PHLPP2 or -2A-transduction, by “rescuing” lower PHLPP2 levels in obese mice. Consistent with in vitro data, HFD-fed mice transduced with Ad-shKctd17 (Figure 5D) showed increased hepatic PHLPP2 protein levels and concomitantly reduced late refed Akt Ser473 phosphorylation (Figure 5E), without change in Phlpp2 expression (Supplementary Figure 9D). Remarkably, this acute reduction in Kctd17 was sufficient to lower liver weight and TG relative to Ad-shControl mice (Figure 5F–H), without confounding changes in body weight/adiposity (Supplementary Figure 9E and F). Further, similar to Ad-PHLPP2 or -2A-transduced mice, we observed lower lipogenic but unchanged gluconeogenic gene expression in Ad-shKctd17-transduced livers (Figure 5I).
KCTD17 correlates with lipogenic gene expression and hepatic steatosis in NASH
These data show that KCTD17 regulates hepatic lipogenesis, at least in part by mediating PHLPP2 degradation. To begin to understand the implication of these findings for human disease, we analyzed liver KCTD17 expression in patients undergoing liver biopsy for suspected NASH (Supplementary Table 2), and found a significant positive correlation between KCTD17 and hepatic steatosis (Figure 6A). This association with steatosis was the primary driver for a similar positive correlation between KCTD17 and NAFLD Activity Score (NAS) (Figure 6B), as higher hepatic KCTD17 expression was no longer associated with increased NAS once adjusted for steatosis in multivariate regression analysis (Supplementary Table 3). Interestingly, KCTD17 expression was not associated with the PNPLA3 I148M variant, the predominant genetic determinant of hepatic fat accumulation in humans, consistent with our mouse data showing increased Kctd17 is causative and not a simple consequence of hepatic steatosis.
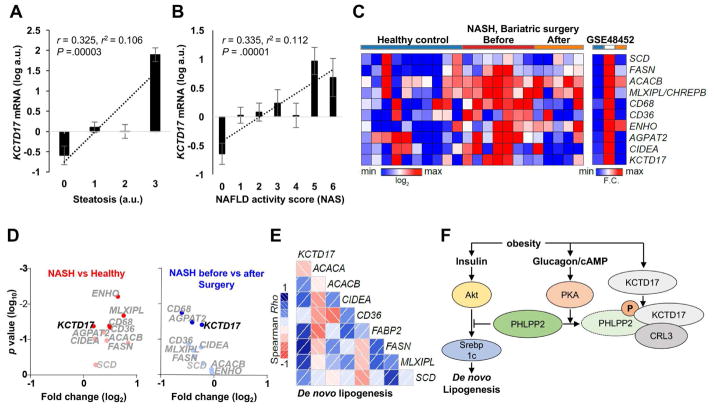
Figure 6. KCTD17 correlates with lipogenic gene expression and hepatic steatosis in NASH.
(A and B) KCTD17 expression in liver biopsy specimens (n=158) as related to (A) steatosis subscore or (B) total NAFLD activity score (NAS). (C) Heat map, (D) volcano plot and (E) correlogram of hepatic KCTD17 expression and de novo lipogenesis (DNL) gene set in hepatic transcriptomes of healthy controls and NASH patients before and after bariatric surgery (n=5–10/group), with the depth of shading of correlogram according to the magnitude of the correlation and positive and negative correlations represented in blue and red, respectively. (F) Model representing the parallel effects of obesity to increase glucagon-mediated PHLPP2 phosphorylation and KCTD17 expression/activity, which synergistically induce PHLPP2 degradation and prolongation of Akt-induced Srebp1c-mediated DNL, and fatty liver.
These data derived from a cross-sectional cohort, which may limit its generalizability. To determine if KCTD17 expression co-varies with NAFLD/NASH severity, we analyzed liver transcriptomes in patients with NASH before and after bariatric surgery, as well as normal controls,24 and found a striking co-regulation of KCTD17 and lipogenic gene sets in patients with NASH (Figure 6C). In paired analysis, KCTD17 expression was significantly higher in patients with NASH as compared to normal controls, and significantly reduced after bariatric surgery (Figure 6D). Thus, KCTD17 showed strong positive correlation with lipogenic and inflammatory gene expression (Figure 6E, Supplementary Figure 10A), which closely clustered as a gene network (Supplementary Figure 10B), but weaker/indirect associations with other pathways associated with NASH, such as β-oxidation, ER stress and fatty acid esterification (Supplementary Figure 10B). In combination with data from L-PHLPP2 and the similar phenotypes of Ad-PHLPP2-2A and Ad-shKctd17 mice, these results establish the functional significance of glucagon-mediated KCTD17/Cul3 degradation of PHLPP2 in regulation of hepatic lipid accumulation.
Discussion
Obesity-induced fatty liver is the most common chronic liver disease, and even its most severe manifestations of NASH and fibrosis/cirrhosis have no approved pharmacotherapy.38 This therapeutic need is due in large part to the complex hormonal regulation of DNL, a relatively minor contributor to liver TG content in the lean individual but a pathway that is ramped up in the obese patient.9, 39 For instance, obesity causes insulin resistance and compensatory hyperinsulinemia, which then activates both mTORC1-dependent and -independent pathways to induce DNL.10–12 Less appreciated is the possible effects of obesity-induced hyperglucagonemia,40 traditionally thought to antagonize insulin’s beneficial effects on glucose homeostasis.18 Our work has revealed the first clear intersection between insulin and glucagon signaling pathways on DNL, through PHLPP2 (Figure 6F). PHLPP2, previously shown by our group and others to dephosphorylate Akt at Ser473 to terminate insulin action, is rapidly phosphorylated by glucagon/PKA action to trigger PHLPP2 degradation. In fact, expression of a glucagon-resistant nonphosphorylatable PHLPP2 mutant prevents obesity-induced hyperinsulinemia from inducing DNL, and thus reverses fatty liver, echoing results from various glucagon antagonists in preclinical development.41–43 These results may explain the molecular mechanism underlying these observations, but also imply that other unrecognized molecular bridges between two critical hormone-regulated pathways to regulate DNL may exist. This may have implications even beyond metabolism, as DNL has emerged as a promising pathway for antineoplastic therapy.
We find that L-PHLPP2 mice show fatty liver, but normal glucose homeostasis. While this may appear paradoxical on face, this finding is actually quite consistent with normal signal transduction through InsR → PI3K → Akt observed in response to feeding in L-PHLPP2 mice, or following an insulin “pulse” in PHLPP2 KO cells. In the absence of PHLPP2, however, we observe prolonged Akt phosphorylation during the “chase” period, and downstream increase in DNL due to a failure to terminate the insulin action. Conversely, adenovirus-mediated overexpression of PHLPP2 reduces liver TG, but also does not affect glucose homeostasis.12 These data suggest the bifurcation model of insulin signaling10 may still explain the “selective” insulin resistance seen in the obese liver, but show that the kinetics of downstream signaling is as important as overall insulin levels – early post-prandial Akt activation is necessary to reduce hepatic glucose output, but prolonged Akt activity only increases DNL. These data suggest that this smoldering, chronic hepatic insulin signaling is partially responsible for increased lipogenesis, which contributes to higher liver TG in obesity, without accompanying glycemic benefit.
An unbiased LC/MS-MS screen identified PHLPP2 phosphorylations at both Ser1119 and Ser1210 in a previously undefined but evolutionarily-conserved domain of PHLPP2. We cannot as yet disentangle these phosphorylation events, nor do we have compelling evidence that these phosphorylations affect PHLPP2 phosphatase activity independent of lowering protein levels. Integrating observations from fasted and obese mice, however, glucagon/PKA-mediated PHLPP2 phosphorylation at these sites is necessary but not sufficient to induce its degradation. The second LC/MS-MS screen pointed to one mechanism for this by identifying KCTD17 as a novel PHLPP2-interacting protein. We show that the obesity-mediated increase in hepatic Kctd17 expression is necessary to link PHLPP2 phosphorylation with degradation, as Kctd17 knockdown in obese mice prevented PHLPP2 degradation, normalized Akt signaling and reduced liver TG. Studies to determine the molecular mechanism underlying increased Kctd17 expression in obese liver are ongoing. In addition, KCTD17 expression seems to preferentially predict steatosis, but given the additional correlation between KCTD17 and inflammatory gene sets, we cannot exclude the possibility that KCTD17 may mediate “multiple hits” in NAFLD/NASH pathogenesis44–46 and are in the process of testing KCTD17 effects in dietary mouse models of NASH.47
In summary, we find that obesity induces glucagon-mediated phosphorylation of PHLPP2, which targets its degradation by KCTD17/Cul3, which in turn increases insulin-mediated DNL. Although DNL is only one of several pathways for liver TG accumulation in NAFLD, the relative contribution of this pathway is higher in insulin resistance.9 Thus, with ablation of KCTD17 in HFD-fed obese mice, we find higher PHLPP2 levels and lower liver TG, but normal glucose tolerance – an uncoupling of two major insulin-regulated processes in liver, inhibition of gluconeogenesis and increased fatty acid synthesis. These data further suggest that while glucagon receptor antagonists may have adverse effects,48, 49 inhibition of PHLPP2 degradation, perhaps by dint of KCTD17 antagonism, may ameliorate obesity-induced fatty liver without affecting glucose homeostasis and stem the tide of liver disease in an increasingly obese population.
Supplementary Material
Acknowledgments
We thank A. Flete, T. Kolar, and V. Lin (Columbia University Transgenic Core Facility) for excellent technical support, S.B. Lee for providing the shRNA encoding PKA, F. Zhang for the LentiCRISPRv2 plasmid, as well as members of the Pajvani laboratory for insightful discussion.
Funding
This work was supported by NIH DK103818 (UBP) and AG043930 (JA), an Edward Mallinckrodt, Jr. Foundation Grant (UBP), the École Polytechnique Fédérale de Lausanne (JA), the Swiss National Science Foundation (31003A-140780; JA), the AgingX program of the Swiss Initiative for Systems Biology (51RTP0-151019; JA), MyFirst AIRC Grant (16888; LV), and a Paul Marks Scholarship (UBP).
Footnotes
Conflicts of interest
The authors disclose no conflicts.
Author contributions
K.K. designed, performed and interpreted experiments, and wrote the manuscript. D.R., P.D., L.O., S.N., B.U., L.V. and J.A. performed and interpreted experiments. U.P. designed and interpreted experiments, and wrote the manuscript. All authors critically read the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pajvani UB, Accili D. The new biology of diabetes. Diabetologia. 2015;58:2459–68. doi: 10.1007/s00125-015-3722-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jelinek LJ, Lok S, Rosenberg GB, et al. Expression cloning and signaling properties of the rat glucagon receptor. Science. 1993;259:1614–6. doi: 10.1126/science.8384375. [DOI] [PubMed] [Google Scholar]
- 3.Summers SA, Birnbaum MJ. A role for the serine/threonine kinase, Akt, in insulin-stimulated glucose uptake. Biochem Soc Trans. 1997;25:981–8. doi: 10.1042/bst0250981. [DOI] [PubMed] [Google Scholar]
- 4.Puigserver P, Rhee J, Donovan J, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–5. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 5.Petersen KF, Laurent D, Rothman DL, et al. Mechanism by which glucose and insulin inhibit net hepatic glycogenolysis in humans. J Clin Invest. 1998;101:1203–9. doi: 10.1172/JCI579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 7.Shimomura I, Matsuda M, Hammer RE, et al. Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol Cell. 2000;6:77–86. [PubMed] [Google Scholar]
- 8.Matsumoto M, Han S, Kitamura T, et al. Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. J Clin Invest. 2006;116:2464–72. doi: 10.1172/JCI27047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donnelly KL, Smith CI, Schwarzenberg SJ, et al. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J Clin Invest. 2005;115:1343–51. doi: 10.1172/JCI23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S, Brown MS, Goldstein JL. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc Natl Acad Sci U S A. 2010;107:3441–6. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yecies JL, Zhang HH, Menon S, et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim K, Qiang L, Hayden MS, et al. mTORC1-independent Raptor prevents hepatic steatosis by stabilizing PHLPP2. Nat Commun. 2016;7:10255. doi: 10.1038/ncomms10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kersten S. Mechanisms of nutritional and hormonal regulation of lipogenesis. EMBO Rep. 2001;2:282–6. doi: 10.1093/embo-reports/kve071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butler PC, Rizza RA. Contribution to postprandial hyperglycemia and effect on initial splanchnic glucose clearance of hepatic glucose cycling in glucose-intolerant or NIDDM patients. Diabetes. 1991;40:73–81. [PubMed] [Google Scholar]
- 15.Dunning BE, Gerich JE. The role of alpha-cell dysregulation in fasting and postprandial hyperglycemia in type 2 diabetes and therapeutic implications. Endocr Rev. 2007;28:253–83. doi: 10.1210/er.2006-0026. [DOI] [PubMed] [Google Scholar]
- 16.Ali S, Drucker DJ. Benefits and limitations of reducing glucagon action for the treatment of type 2 diabetes. Am J Physiol Endocrinol Metab. 2009;296:E415–21. doi: 10.1152/ajpendo.90887.2008. [DOI] [PubMed] [Google Scholar]
- 17.Biddinger SB, Hernandez-Ono A, Rask-Madsen C, et al. Hepatic insulin resistance is sufficient to produce dyslipidemia and susceptibility to atherosclerosis. Cell Metab. 2008;7:125–34. doi: 10.1016/j.cmet.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conarello SL, Jiang G, Mu J, et al. Glucagon receptor knockout mice are resistant to diet-induced obesity and streptozotocin-mediated beta cell loss and hyperglycaemia. Diabetologia. 2007;50:142–50. doi: 10.1007/s00125-006-0481-3. [DOI] [PubMed] [Google Scholar]
- 19.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Grzechnik AT, Newton AC. PHLPPing through history: a decade in the life of PHLPP phosphatases. Biochem Soc Trans. 2016;44:1675–1682. doi: 10.1042/BST20160170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newton AC, Trotman LC. Turning off AKT: PHLPP as a drug target. Annu Rev Pharmacol Toxicol. 2014;54:537–58. doi: 10.1146/annurev-pharmtox-011112-140338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasahara K, Kawakami Y, Kiyono T, et al. Ubiquitin-proteasome system controls ciliogenesis at the initial step of axoneme extension. Nat Commun. 2014;5:5081. doi: 10.1038/ncomms6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams EG, Wu Y, Jha P, et al. Systems proteomics of liver mitochondria function. Science. 2016;352:aad0189. doi: 10.1126/science.aad0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahrens M, Ammerpohl O, von Schonfels W, et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18:296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Pajvani UB, Qiang L, Kangsamaksin T, et al. Inhibition of Notch uncouples Akt activation from hepatic lipid accumulation by decreasing mTorc1 stability. Nat Med. 2013;19:1054–60. doi: 10.1038/nm.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balasubramanian D, Ohneck EA, Chapman J, et al. Staphylococcus aureus Coordinates Leukocidin Expression and Pathogenesis by Sensing Metabolic Fluxes via RpiRc. MBio. 2016:7. doi: 10.1128/mBio.00818-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bern M, Kil YJ, Becker C. Byonic: advanced peptide and protein identification software. Curr Protoc Bioinformatics. 2012;Chapter 13(Unit13.20) doi: 10.1002/0471250953.bi1320s40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 29.Mancina RM, Dongiovanni P, Petta S, et al. The MBOAT7-TMC4 Variant rs641738 Increases Risk of Nonalcoholic Fatty Liver Disease in Individuals of European Descent. Gastroenterology. 2016;150:1219–1230. e6. doi: 10.1053/j.gastro.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andreux PA, Williams EG, Koutnikova H, et al. Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits. Cell. 2012;150:1287–99. doi: 10.1016/j.cell.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haeusler RA, Hartil K, Vaitheesvaran B, et al. Integrated control of hepatic lipogenesis versus glucose production requires FoxO transcription factors. Nat Commun. 2014;5:5190. doi: 10.1038/ncomms6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKnight GS. Cyclic AMP second messenger systems. Curr Opin Cell Biol. 1991;3:213–7. doi: 10.1016/0955-0674(91)90141-k. [DOI] [PubMed] [Google Scholar]
- 33.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–31. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strable MS, Ntambi JM. Genetic control of de novo lipogenesis: role in diet-induced obesity. Crit Rev Biochem Mol Biol. 2010;45:199–214. doi: 10.3109/10409231003667500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perry RJ, Samuel VT, Petersen KF, et al. The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature. 2014;510:84–91. doi: 10.1038/nature13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inaba H, Goto H, Kasahara K, et al. Ndel1 suppresses ciliogenesis in proliferating cells by regulating the trichoplein-Aurora A pathway. J Cell Biol. 2016;212:409–23. doi: 10.1083/jcb.201507046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji AX, Chu A, Nielsen TK, et al. Structural Insights into KCTD Protein Assembly and Cullin3 Recognition. J Mol Biol. 2016;428:92–107. doi: 10.1016/j.jmb.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 38.Valenti L, Bugianesi E, Pajvani U, et al. Nonalcoholic fatty liver disease: cause or consequence of type 2 diabetes? Liver Int. 2016;36:1563–1579. doi: 10.1111/liv.13185. [DOI] [PubMed] [Google Scholar]
- 39.Leavens KF, Birnbaum MJ. Insulin signaling to hepatic lipid metabolism in health and disease. Crit Rev Biochem Mol Biol. 2011;46:200–15. doi: 10.3109/10409238.2011.562481. [DOI] [PubMed] [Google Scholar]
- 40.Lefebvre PJ. Glucagon and its family revisited. Diabetes Care. 1995;18:715–30. doi: 10.2337/diacare.18.5.715. [DOI] [PubMed] [Google Scholar]
- 41.Gelling RW, Du XQ, Dichmann DS, et al. Lower blood glucose, hyperglucagonemia, and pancreatic alpha cell hyperplasia in glucagon receptor knockout mice. Proc Natl Acad Sci U S A. 2003;100:1438–43. doi: 10.1073/pnas.0237106100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sloop KW, Cao JX, Siesky AM, et al. Hepatic and glucagon-like peptide-1-mediated reversal of diabetes by glucagon receptor antisense oligonucleotide inhibitors. J Clin Invest. 2004;113:1571–81. doi: 10.1172/JCI20911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sorensen H, Brand CL, Neschen S, et al. Immunoneutralization of endogenous glucagon reduces hepatic glucose output and improves long-term glycemic control in diabetic ob/ob mice. Diabetes. 2006;55:2843–8. doi: 10.2337/db06-0222. [DOI] [PubMed] [Google Scholar]
- 44.Tilg H, Moschen AR. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology. 2010;52:1836–46. doi: 10.1002/hep.24001. [DOI] [PubMed] [Google Scholar]
- 45.Peverill W, Powell LW, Skoien R. Evolving concepts in the pathogenesis of NASH: beyond steatosis and inflammation. Int J Mol Sci. 2014;15:8591–638. doi: 10.3390/ijms15058591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buzzetti E, Pinzani M, Tsochatzis EA. The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD) Metabolism. 2016;65:1038–48. doi: 10.1016/j.metabol.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 47.Wang X, Zheng Z, Caviglia JM, et al. Hepatocyte TAZ/WWTR1 Promotes Inflammation and Fibrosis in Nonalcoholic Steatohepatitis. Cell Metab. 2016;24:848–862. doi: 10.1016/j.cmet.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kelly RP, Garhyan P, Raddad E, et al. Short-term administration of the glucagon receptor antagonist LY2409021 lowers blood glucose in healthy people and in those with type 2 diabetes. Diabetes Obes Metab. 2015;17:414–22. doi: 10.1111/dom.12446. [DOI] [PubMed] [Google Scholar]
- 49.Kazierad DJ, Bergman A, Tan B, et al. Effects of multiple ascending doses of the glucagon receptor antagonist PF-06291874 in patients with type 2 diabetes mellitus. Diabetes Obes Metab. 2016;18:795–802. doi: 10.1111/dom.12672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.