Abstract
This paper presents a novel method for automatically recognizing symptom severity by using natural language processing of psychiatric evaluation records to extract features that are processed by machine learning techniques to assign a severity score to each record evaluated in the 2016 RDoC for Psychiatry Challenge from CEGS/N-GRID. The natural language processing techniques focused on (a) discerning the discourse information expressed in questions and answers; (b) identifying medical concepts that relate to mental disorders; and (c) accounting for the role of negation. The machine learning techniques rely on the assumptions that (1) the severity of a patient’s positive valence symptoms exists on a latent continuous spectrum; and (2) all the patient’s answers and narratives documented in the psychological evaluation records are informed by the patient’s latent severity score along this spectrum. These assumptions motivated our two-step machine learning framework for automatically recognizing psychological symptom severity. In the first step, the latent continuous severity score is inferred from each record; in the second step, the severity score is mapped to one of the four discrete severity levels used in the CEGS/N-GRID challenge. We evaluated three methods for inferring the latent severity score associated with each record: (i) pointwise ridge regression; (ii) pairwise comparison-based classification; and (iii) a hybrid approach combining pointwise regression and the pairwise classifier. The second step was implemented using a tree of cascading support vector machine (SVM) classifiers. While the official evaluation results indicate that all three methods are promising, the hybrid approach not only outperformed the pairwise and pointwise methods, but also produced the second highest performance of all submissions to the CEGS/N-GRID challenge with a normalized MAE score of 84.093% (where higher numbers indicate better performance). These evaluation results enabled us to observe that, for this task, considering pairwise information can produce more accurate severity scores than pointwise regression – an approach widely used in other systems for assigning severity scores. Moreover, our analysis indicates that using a cascading SVM tree outperforms traditional SVM classification methods for the purpose of determining discrete severity levels.
1. Introduction
In 2008, the National Institute of Mental Health (NIMH) included an aim to “[d]evelop, for research purposes, new ways of classifying mental disorders based on dimensions of observable behavior and neurobiological measures” in its new strategic plan. The framework that implements this aim was named the Research Domain Criteria project, or RDoC. RDoC fostered research that integrates multiple forms of information to better understand all the dimensions of functioning underlying the full range of patient behavior, from normal to abnormal. As reported in Cuthbert [1], an NIMH workgroup was convened in early 2009 to devise an approach for RDoC. The workgroup determined five major domains of functioning: (1) negative valence systems (i.e., those that respond to aversive situations); (2) positive valence systems; (3) cognitive systems; (4) systems for social processes; and (5) arousal/regulatory systems.
While the recognition, profiling, and treatment of mental disorders benefit from the RDoC framework, the decision of whether a patient requires medical attention or hospitalization is informed by the severity of his or her symptoms. Symptoms of mental illness can vary, depending on the disorder, circumstances, and other factors. Psychological evaluation records provide insights into the severity of a patient’s symptoms based on the information documented by the psychiatrist during interviews with the patient. However, this information is documented through natural language; thus, is not readily available for automatic processing.
In this paper, we present a new method for identifying the degree of severity associated with a patient’s positive valence symptoms by processing the natural language from a set of psychological evaluation records. Natural language processing enables the extraction of features characterizing the patients’ past experiences, diagnoses, and social history, as well as behavioral interventions and mental health treatments. These features inform machine learning techniques. The machine learning techniques rely on two assumptions; namely, that (1) the severity of a patient’s positive valence symptoms exists on a latent continuous spectrum; and (2) all the patient’s answers and narratives documented in his or her psychological evaluation record(s) are informed by the patient’s latent severity score along this spectrum. These assumptions allowed us to design a two-step machine learning framework for automatically recognizing the severity of a patient’s positive valence symptoms. In the first step, we infer the latent continuous severity score which was mostly likely to have produced each psychiatric evaluation record. In the second step, we map each inferred latent severity score to a discrete severity level. We considered and evaluated three machine learning approaches for inferring the latent continuous severity score for each psychiatric evaluation record: (i) pointwise ridge regression; (ii) pairwise comparison-based classification; and (iii) a hybrid approach combining pointwise regression and the pairwise classifier. The second step of the machine learning framework was implemented using a tree of three cascading support vector machine (SVM) classifiers.
We believe that the two-step framework presented in this paper could facilitate efforts to stratify risk for adverse outcomes among psychiatric disorders as well as efforts to identify optimal treatments for patient subgroups.
2. Background
The notion of severity was considered previously when qualifying the severity of a disease, e.g. Medsger et al. [2] describes disease severity as the total effect of disease on the body while Chen et al. [3] reports that disease severity is assigned by direct observation of a patient and by pathological examination after symptoms have appeared. Severity was viewed as a “degree of illness” in Joshi and Szolovits [4], who learned a severity graph by applying the Radial Domain Folding (RDF) algorithm. The RDF algorithm is a novel multivariate clustering approach operating on the MIMIC II clinical dataset [5] without processing the language from the clinical narratives.
Disease severity was historically quantified by scores which were defined and derived in two ways: (1) by using medical expert knowledge; or (2) by predicting a score associated with the risk of experiencing an adverse event. Specifically, when a panel of experts identifies factors that are most indicative of severity of the target disease after reviewing the existing clinical literature, the severity score is produced by a weighted sum of the relative contribution of the factors to the disease severity. For example, the Acute Physiology And Chronic Health conditions score (APACHE II) [6] assesses the overall health state in an inpatient setting by using factors that are most predictive of mortality. The APACHE II disease severity classification system uses basic physiologic principles to stratify acutely ill patients by risk of death. It uses a point score based on the values of 12 routine physiologic measurements, age and previous health status to provide a general measure of severity of disease. In computing the severity score, APACHE II adds different points based on the abnormal range of the measures. Several other widely used severity scoring systems have been designed in the same way as APACHE II, including the Multiple Organ Dysfunction Score (MODS) [7] and the Medsgers scoring system [2]. More recently, the Rothman Index [8] was developed by data analysts to provide a summary score of a patient’s clinical condition based on 26 variables, including vital signs, laboratory profile data, and nursing assessments.
Alternatively, supervised machine learning methods have been used to predict disease severity scores. In Pirracchio et al. [9], the Super Learner selected the optimal regression algorithm via cross-validation to produce a severity score used to predict mortality of patients in intensive care units (ICU) on the Multiparameter Intelligent Monitoring in Intensive Care II (MIMIC-II) database including all patients admitted to an ICU at Boston Beth Israel Deaconess Medical Center from 2001 to 2008.
While no previous clinical natural language processing task has focused on discerning the symptom severity from psychiatric evaluation records, we found that we could benefit from the automatic identification of medical concepts related to mental disorders to capture the semantics of the language used in the psychological evaluation records. Nevertheless, in addition to semantics, we discovered that we also needed to uncover the discourse structure provided by the sequences of questions and answers. Thus, we developed natural language processing methods capturing semantic and discourse features, which could be used to learn the degree of psychological symptom severity. To do so, we (i) explored several machine learning techniques; and (ii) used the insight that comparisons between psychiatric evaluation records labeled with different symptom severity classes could produce better discriminators.
3. Task Description
In 2016 the Centers of Excellence in Genomic Science (CEGS) Neuropsychiatric Genome-Scale and RDoC Individualized Domains (N-GRID) organized a new challenge which aimed to extract symptom severity from neuropsychiatric clinical records [10]. The RDoC for Psychiatry Challenge of the CEGS/N-GRID focused on just one domain of functioning, namely: positive valence. The positive valence domain pertains to events or situations that signal mental disorders but are attractive to the patients, to the point that they actively engage them, e.g. alcohol or drug consumption, gambling, abuse, drinking, repetitive and/or compulsive behavior, craving, and counting. The goal of the RDoC for Psychiatry Challenge was to evaluate automatic approaches to determining symptom severity in an RDoC domain for a patient, based on information included in their initial psychiatric evaluation record. The symptom severity was measured using an ordinal scale from 0 (ABSENT) to 3 (SEVERE). Specifically:
Severity Level = 0, or ABSENT, indicating no positive valence symptoms are observed;
Severity Level = 1, or MILD, indicating while some symptoms may be present, they are not a focus of treatment;
Severity Level = 2, or MODERATE, indicating that symptoms are present and a focus of treatment;
Severity Level = 3, or SEVERE, indicating that symptoms are present and require hospitalization, an ER visit, or otherwise have a major consequence.
The participants in the RDoC for Psychiatry Challenge were provided with a training set of records in which each record was associated with the gold standard severity level for the patient’s positive valence symptoms. It should be noted that each record is associated with exactly one patient and has exactly one severity level. Systems developed for the challenge had to identify the lifetime maximum severity a patient’s symptoms in the positive valence domain, with the assumption that symptoms not documented in the record are not present. Moreover, the severity level did not need to be related to a current or recent diagnosis. As a result, even past experiences and diagnoses were considered relevant. In addition, it was assumed that predicting the severity level of the positive valence domain does not need to rely on any textual clues related to any of the other functional domains.
3.1. The Corpus
In the research reported in this paper, we considered the set of 1,000 psychiatric evaluation records provided to participants during the CEGS/N-GRID evaluation. The psychiatric evaluation records were provided by the Centers of Excellence in Genomic Science (CEGS) Neuropsychiatric Genome- Scale and RDoC Individualized Domains (N-GRID) project of Harvard Medical School, and constitute the first set of mental health records available for research. The corpus has been deidentified, and the original protected health information (PHI) has been replaced by synthetic surrogate information.
Figure 1 includes an excerpt of an initial psychiatric evaluation record. As can be seen, there are a number of typographical and punctuation anomalies. Moreover, many lines are structured as question and answer pairs where the answer may be very short (YES, NO) or may have a verbose elaboration. It should be noted that the order of questions is arbitrary and changes between records.
Figure 1.
Example psychiatric evaluation record (abridged).
A total of three expert psychiatrists were selected by the organizers of the CEGS/N-Grid evaluation task to annotate the psychiatric evaluation records used in the challenge. The corpus contained 433 human-annotated records in the training set and 216 human-annotated records in the testing set (there was no official development set). In the training set, 325 records were annotated by at least two psychiatrists, and the remaining 108 were annotated by a single psychiatrist. In the test set, all 216 annotated records were annotated by at least two psychiatrists. Records evaluated by a single psychiatrist were evaluated by the most experienced psychiatrist. In records evaluated by two psychiatrists, if there was a disagreement between the psychiatrists, the gold-standard severity level was determined by the most experienced psychiatrist. The average agreement between all pairs of psychiatrists was κ = 65.713% (computed as the weighted average of Cohen’s κ agreements).
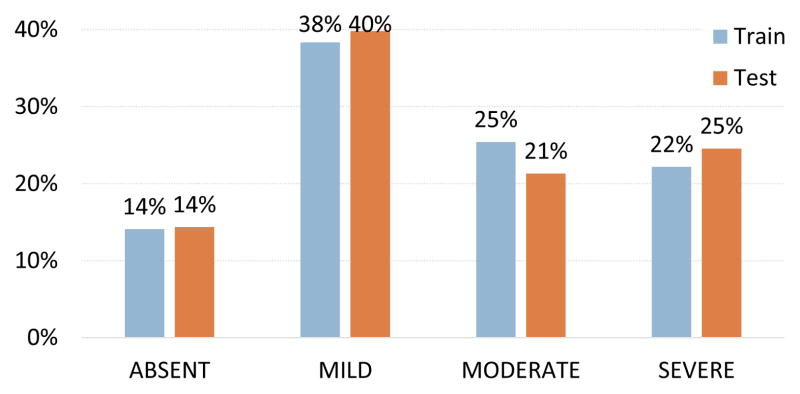
Figure 2 illustrates the distribution of each severity level within the training and testing sets. As can be the seen, the levels are reasonably balanced, with slightly more MILD than ABSENT records. Moreover, it can be seen that the distribution of severity levels is effectively the same between the training and testing sets.
Figure 2.
Distribution of severity levels in the training and testing sets.
4. Automatically Recognizing Symptom Severity Levels
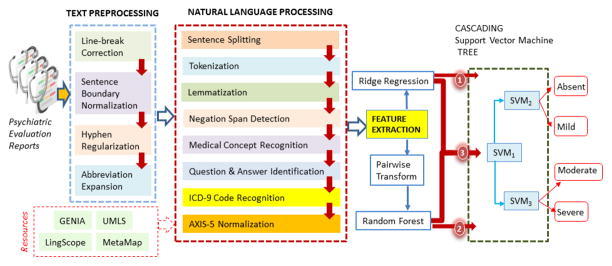
Our three approaches for the automatic recognition of symptom severity have been implemented in the system illustrated in Figure 3. First, text preprocessing was applied to each psychiatric evaluation record with methods that we detail in Section 4.1. Then, we performed natural language processing, detailed in Section 4.2, on the preprocessed records enabling us to extract a variety of features, as detailed in Section 4.3. These features were used in three different machine learning approaches described in Sections 4.4.2–4.4.5: (1) pointwise ridge regression, (2) pairwise Random Forest classification, and (3) a hybrid method which combines both approaches. Each of these machine learning approaches produced a continuous severity score which was mapped to one of the four discrete severity levels using a cascading SVM tree, as described in Section 4.4.5.
Figure 3.
Architecture of our three approaches for identifying positive valence symptom severity levels in psychiatric evaluation records.
4.1. Text Preprocessing
Clinical data is known to suffer from a myriad of idiosyncrasies [11–13]. The psychiatric evaluation records used in this challenge were no exception. For example, consider the following excerpts:
Example 1. Current living situation:
Lives with boyfriend in rented apartmentFirearms: None
Example 2. Name/Phone # of Probation/Parole Officer:
REDACTEDMilitary Service History: Hx of Military Service: No
Example 3. If Yes, comment on Timing, Lethality, Impulsivity, Weapon Use, Comorbid Intoxication or Psychosis:
Denied-Psychiatric History Hx of Inpatient Treatment: Yes Pt says she has been hospitalized three times
These three examples highlight the erroneous usage of linebreaks found throughout this corpus. In addition, it should be noted that sentence boundaries are not only obfuscated by out-of-place line-breaks but also by the omission of spaces. Moreover, most documents in the corpus contain bulleted lists which use hyphens as bullets, with no space between the hyphen and the start of the sentence; e.g. “-Psychiatric History” from Example 3. As shown in Examples 2 and 3, abbreviations such as “Hx” and “Pt” – indicating “history” and “patient”, respectively – are prevalent in the data. These types of idiosyncrasies can cause automatic tokenization tools to incorrectly conclude that “-Psychiatric” is a single token and can confound other downstream NLP tools. We address these types of errors and idiosyncrasies with four text preprocessing steps:
Line-break Correction
To address the erroneous use of line-breaks, three regular expressions were used to detect when the end of one line is concatenated to the beginning of the next line, e.g. “… apartmentFirearms … “ from Example 1. Each regular expression was designed to identify conjoined words that indicate the omission of a line-break, e.g. “43Sex:”. The pattern “ .*[a-z0-9]+[A-Z]+.+“ detects conjoined words by finding cases of a lowercase word conjoined with a capitalized word, such as “apartmentFirearms” in Example 1. Likewise the pattern “ [A-Z]2,[a-z]+“ identifies uppercase words conjoined with a capitalized word, such as “REDACTEDMilitary” in Example 2. Finally, the pattern “ [a-z]+-[A-Z].+“ discovers conjoined words in which the second word starts with a hyphen, such as “Denied-Psychiatric” in Example 3. This pattern was designed to account for the predominance of bulleted lines which begin with hyphens. When any of these patterns are detected, the line-break is removed between the line in which the pattern was detected and the previous line and a new line-break is inserted between the two conjoined words discovered by the pattern.
Sentence Boundary Normalization
In addition to the irregular use of line-breaks, another type of grammatical error present in the corpus is the omission of spaces between sentences. We used the pattern “ [a-zA-Z]+\\ .[a-zA-Z]+.*“ to find words containing a period between letters and inserted a space after the period.
Hyphen Regularization
To address the use of hyphens to denote bullets in the psychiatric evaluation records, we used the pattern “ -\\ S+“ to find words with leading hyphens (indicating bullets) and inserted a space after the hyphen.
Abbreviation Expansion
The last type of syntactic/grammatical errors we addressed concerned the use of abbreviations in the corpus. The text preprocessing system expands the following abbreviations into their full forms: “pt”, “hx”, “nml”, are expanded to “patient”, “history”, and “normal” while both “sxs” and “syx’s” and expanded to “symptoms”.
4.2. Natural Language Processing
After text preprocessing, we performed seven automatic natural language processing (NLP) steps: (1) sentence splitting, (2) tokenization, lemmatization, and part-of-speech tagging, (3) negation span detection, (4) UMLS concept identification, (5) ICD-9 code recognition, (6) question and answer identification, and (7) AXIS-V normalization. Each of these steps is detailed below.
Sentence Splitting
Despite the linebreak and sentence boundary corrections made during text preprocessing, the performance of general domain sentence splitters such as those provided by Stanford’s CoreNLP toolkit [14], and OpenNLP1 were unable to correctly process psychiatric evaluation records. For example, both of these general domain sentence splitters group the two lines from Example 1 into a single sentence along with more than 50 additional lines of text. We found that general-domain sentence splitters were unable to account for the prevalent semi-structured question-and-answer lines (as these lines do not include traditional sentence boundaries). Consequently, we relied on the GENIA sentence splitter [15], which is a maximum entropy sentence boundary detection tool trained on biomedical texts. We observed that, in most cases, the GENIA sentence splitter was able to correctly split semi-structured question-and-answer lines into individual sentences.
Tokenization, Lemmatization, & Part of Speech Tagging
After sentence splitting, we performed tokenization, lemmatization, and part of speech tagging using the GENIA tagger [16]. As with sentence splitting, we found that the GENIA tagger was more effective at processing psychiatric text than general purpose NLP toolkits.
Negation Span Detection
In the records, the psychiatrists document both the symptoms and disorders exhibited by the patient as well as any important symptoms and disorders that the patient does not currently exhibit but which may inform the patient’s psychological state. For example:
Example 4. Currently not drinking at all and no drugs.
Although the words “drinking” and “drugs” are present, the fact that the patient does not currently drink or use drugs may have a significant impact on the patient’s psychological state as both alcohol and drug use are strong indicators of positive valence symptom severity. To detect when a relevant text mention is negated, we use LingScope [17] which is a system that uses a Conditional Random Field trained to detect spans of text (not just individual words) which are negated based on syntactic information. In Example 4, LingScope is able to recognize that the entire span “drinking at all and no drugs” is negated.
UMLS Concept Identification
The psychiatric evaluation records in the corpus are rich with medical concept mentions, many of which are direct indicators of positive valence symptom severity. Consider the following example:
Example 5. Patient is a 29 y/o woman with prior diagnoses of opioid/cocaine/benzo dependence, dysthymic disorder, and PTSD who is referred for a psychopharm evaluation.
The relevant medical concepts in this excerpt are (1) “opioid … dependence”, (2) “cocaine … dependence”, (3) “benzo dependence”, (4) “dysthymic disorder”, and (5) “PTSD”. All of these medical concepts are documented within the Unified Medical Language System (UMLS) Metathesaurus [18]. In UMLS, each medical concept is associated with (1) a Concept Unique Identifier (CUI) and (2) a number of atoms, corresponding to natural language realizations of the concept (e.g. the concept Post-Traumatic Stress Disorder has the CUI C0038436 and an atom containing the text “PTSD”). UMLS also encodes hierarchical relationships between medical concepts.
We initially used MetaMap [19] to automatically identify UMLS concepts in the corpus. MetaMap was able to successfully identify four of the five relevant medical concepts in Example 5: Opiate Addiction, Cocaine Dependence, Dysthymic Disorder, and Post-Traumatic Stress Disorder (PTSD). However, it also identifies seven irrelevant concepts (Patients, 29+, Woman, Prior diagnosis, Cocaine, Referring, and Evaluation procedure) and two concepts which are not present in the source text at all (Degenerative polyarthritis and Yotta). To account for these problems, rather than using the concepts detected by MetaMap, we implemented a more focused medical concept detection approach which only considers UMLS concepts which descend from Mental Health (C0025353), Mental Disorder (C0004936), or Behavior Disorder (C0004930) in the UMLS hierarchy. Specifically, for each concept in each hierarchy, we recognized instances of that concept in each psychiatric evaluation record by detecting mentions of any of the atoms associated with that concept. Using this method, we discovered a total of 82, 634 medical concepts in the training set as opposed to the 280, 494 detected by MetaMap.
ICD-9 Code Recognition
Some of the psychiatric evaluation records contain references to diseases, symptoms, or health problems using ICD-9 (International Statistical Classification of Diseases and Related Health Problems, 9th revision) codes. Clearly, ICD-9 codes can be informative for determining positive valence symptom severity as they can indicate MILD, MODERATE, or SEVERE symptoms (or their causes). ICD-9 codes are comprised of three digits indicating a specific disease type, a decimal point, and one to three more digits indicating more specific details about the disease (e.g. a subtype or means of disease contraction). For example, the code 304.00 indicates heroin dependence, where 304 refers to drug dependence and .00 indicates that the drug is heroin. Likewise the code for cocaine dependence is 304.20. There are 19 broad categories of ICD-9 codes and the code classification is organized such that similar diseases will have similar codes. By far the most common ICD-9 code class in the records was Mental Disorders which accounted for more than 63% of the ICD-9 codes detected in the training set. We detected ICD-9 codes with the following pattern: ^(V\d{2}(\.\d{1,2})?|\d{3}(\.\d{1,2})?|E\d{3}(\.\d)?)$
Question & Answer Identification
The text in the psychiatric evaluations are largely made up of question/answer pairs that contain important information about the patient’s current and past mental state. For example:
Example 6. History of Inpatient Treatment: Yes Patient says she has been hospitalized three times
… History of Brain Injury: No. history of falls and head strikes. denies post-concussive symptoms.
Example 7. PSYCHOSIS: Has the patient had unusual experiences that are hard to explain: No
… This visit for a one-time consultation only? No
Example 8. Behavior: Cooperative
In the corpus, the majority of questions have a YES/NO answer (e.g. Examples 6 and 7) which may be accompanied with natural language elaboration (e.g. Example 6). While most of the questions are repeated between records, the elaborations contain information specific to the individual patient described in the record. Moreover, the elaborations accompanying NO answers can contain information that would not fit under any other question in the questionnaire but which is still important. In Example 6, the information regarding “falls” and “head strikes” is included as a part of the answer to a question about brain injury.
Axis V (GAF) Normalization
The Axis V Global Assessment of Functioning (GAF) [20] is a numeric scale used by mental health clinicians to rate patients’ psychological ability to function on a scale from 100 (no symptoms) to 1 (persistent danger to self or others). This metric measures the severity of overall mental health symptoms, not just positive valence symptoms. The records can contain up to three types of Axis V scores: (1) the patient’s current score, (2) the patient’s lowest recorded score to date, and (3) the patient’s highest recorded score to date. Over 90% of records in the training set contain a “current” Axis V score and over 50% of records have lowest and highest Axis V scores as well. We detected Axis V scores by taking the first contiguous string of numbers from lines that contained “axis v” or “axisv”. The type of the Axis V score was determined by searching for the strings “current”, “highest”, and “lowest” in the line containing the score. If no type was detected, we defaulted to the “current” Axis V score type. It should be noted that each Axis V score falls into one of ten ranges. Consequently, we normalize each detected score into one of these ranges. Most of the scores in the records fall into either the Mild range from 61–70 (26.4%), the Moderate range from 51–60 (44.0%), or the Serious range from 41–50 (13.5%), however at least one score from each of the other seven ranges was present in at least one record.
4.3. Feature Extraction
After text preprocessing and natural language processing, we encoded each psychiatric evaluation record as a 568-dimension feature vector. It should be noted, however, that the psychiatric evaluation records used in the CEGS/N-GRID evaluation contain a large amount of natural language content which is not directly related to positive valence symptoms (e.g. general medical history, or unrelated psychological problems) or which is negated (e.g. Example 4). Consequently, in order to ensure that the features we extract from each record are statistically meaningful, we only extracted features from the relevant portions of each record. We defined the relevant portions of a psychiatric evaluation record as (a) pseudo-structured questions answered YES as well as any elaboration, (b) non-negated narrative content, and (c) the elaboration of any question answered NO.
The features extracted from relevant portions of each psychiatric evaluation record are illustrated in Table 1. Table 1 also indicates the type or domain of each feature: (1) Boolean features, denoted by ℬ, are assigned the value ‘1’ to encode TRUE, and the value of ‘0’ to encode FALSE; (2) numeric features, denoted by 𝒩, are associated with positive integers; and (3) multivalued features, denoted by ℳ, are associated with one or more discrete values (for example, a bag-of-words feature would be classified as multivalued)2. It is important to note that the same feature vectors were processed by all three learning methods. We extract six types of features, which are described below.
Table 1.
Features used for identifying symptom severity levels.
| Name | Definition | Type |
|---|---|---|
| Question & Answer Features | ||
| Severe questions | ||
| F1 | Has the patient had inpatient treatment? | ℬ |
| F2 | Does the patient have a history of suicidal behavior? | ℬ |
| F3 | Does the patient have a history of violent behavior? | ℬ |
| F4 | Does the patient have a history of self-injurious behavior? | ℬ |
| Risk assessment questions | ||
| F5 | Has the patient experienced loss of housing? | ℬ |
| F6 | Has the patient had thoughts of harm to self? | ℬ |
| F7 | Is the patient prone to worrying excessively? | ℬ |
| Drug & substance questions | ||
| F8 | Does the patient have a history of drug use? | ℬ |
| F9 | Does the patient have a history of marijuana use? | ℬ |
| F10 | Does the patient have a history of cocaine use? | ℬ |
| F11 | Does the patient have a history of sedative/hypnotic use? | ℬ |
| F12 | Does the patient have a history of opiate use? | ℬ |
| F13 | Does the patient have a history of hallucinogens use? | ℬ |
| Psychological symptom questions | ||
| F14 | Does the patient have symptoms of depression (MDD)? | ℬ |
| F15 | Does the patient have symptoms of bipolar disorder (BP)? | ℬ |
| F16 | Does the patient have symptoms of general anxiety disorder (GAD)? | ℬ |
| F17 | Does the patient have symptoms of obsessive compulsive spectrum disorders? | ℬ |
| F18 | Does the patient have symptoms of attention deficit hyperactive disorder (ADHD)? | ℬ |
| F19 | Does the patient have symptoms of eating disorders? | ℬ |
| F20 | Does the patient have symptoms of complicated grief? | ℬ |
| F21 | Does the patient have symptoms of post-traumatic stress disorder (PTSD)? | ℬ |
| F22 | Does the patient have symptoms of dementia? | ℬ |
| Question coverage features | ||
| F23 | Were any severe questions answered yes? | ℬ |
| F24 | Number of severe questions answered yes | 𝒩 |
| F25 | Were any risk assessment questions answered yes? | ℬ |
| F26 | Number of risk assessment questions answered yes | 𝒩 |
| F27 | Were any drug & substance questions answered yes? | ℬ |
| F28 | Number of drug & substance questions answered yes | 𝒩 |
| F29 | Were any psychological symptom questions answered yes? | ℬ |
| F30 | Number of psychological symptom questions answered yes | 𝒩 |
| F31 | Were any questions answered yes? | ℬ |
| F32 | Number of questions answered yes | 𝒩 |
| Lexical & Pattern Features | ||
| F33 | Were any patterns in the ALCOHOL lexicon found? | ℬ |
| F34 | Number of patterns in the ALCOHOL lexicon that were found | 𝒩 |
| F35 | Were any patterns in the DRUG lexicon found? | ℬ |
| F36 | Number of patterns in the DRUG lexicon that were found | 𝒩 |
| F37 | Were any patterns in the EATING_DISORDER lexicon found? | ℬ |
| F38 | Number of patterns in the EATING_DISORDER lexicon that were found | 𝒩 |
| F39 | Were any patterns in the HISTORY lexicon found? | ℬ |
| F40 | Number of patterns in the HISTORY lexicon that were found | 𝒩 |
| F41 | Were any patterns in the INPATIENT lexicon found? | ℬ |
| F42 | Number of patterns in the INPATIENT lexicon that were found | 𝒩 |
| F43 | Were any patterns in the LEGAL lexicon found? | ℬ |
| F44 | Number of patterns in the LEGAL lexicon that were found | 𝒩 |
| F45 | Were any patterns in the OUTPATIENT lexicon found? | ℬ |
| F46 | Number of patterns in the OUTPATIENT lexicon that were found | 𝒩 |
| UMLS Hierarchy Features | ||
| F47 | Were any medical concepts descendants of Mental Health in UMLS? | ℬ |
| F48 | All paths from each descendant of Mental Health and Mental Health itself in UMLS | ℳ |
| F49 | Were any medical concepts descendants of Mental Disorder in UMLS? | ℬ |
| F50 | All paths from each descendant of Mental Disorder and Mental Disorder itself in UMLS | ℳ |
| F51 | Were any medical concepts descendants of Behavior Disorder in UMLS? | ℬ |
| F52 | All paths from each descendant of Behavior Disorder and Behavior Disorder itself in UMLS | ℳ |
| Patient Age Features | ||
| F53 | Was the patient at least 80 years old? | ℬ |
| F54 | Was the patient at least 70 years old? | ℬ |
| F55 | Was the patient at least 60 years old? | ℬ |
| F56 | Was the patient younger than 40 years old? | ℬ |
| F57 | The age of the patient | 𝒩 |
| Axis V GAF Features | ||
| F58 | The patient’s lowest AXIS V GAF category | 𝒩 |
| F59 | The patient’s current AXIS V GAF category | 𝒩 |
| ICD-9 Code Features | ||
| F60 | All ICD-9 codes (truncated as an integer) | ℳ |
| F61 | All ICD-9 codes (truncated to the tenths decimal place) | ℳ |
| F62 | All ICD-9 codes (truncated to the hundredths decimal place) | ℳ |
| F63 | All ICD-9 codes (truncated to as the ICD-9 code class) | ℳ |
Question & Answer Features
Thirty-two features are extracted to represent the questions and their answers that were automatically identified in each psychiatric evaluation record. The features F1-F4 correspond to individual questions concerning severe psychological symptoms, F5-F7 correspond to individual questions about social risk factors, and F8-F13 correspond to individual questions regarding the use of drugs and other substances. Eeach feature F14-F22 corresponds to a group of questions and takes the value TRUE when any question in the group is answered as TRUE, and FALSE, otherwise. Finally, features F23-F32 capture the coverage of positive answers in each psychiatric note: indicating whether at least one question in each category was answered in the affirmative, and the number of questions in each category answered as TRUE.
Lexical & Pattern Features
While the semi-structured YES/NO questions in each psychiatric evaluation record provided an abundance of high-level information for recognizing the severity of a patient’s psychiatric symptoms, we found that the elaborations as well as unstructured narratives contained an abundance of important cues. However, unlike the semi-structured YES/NO questions, the elaborations and narrative content in each psychiatric evaluation record varied substantially between patients. In order to extract information from the elaborations and narrative content, we manually created a number of lexica containing textual patterns by reviewing psychiatric evaluation records in the training set. A total of 7 lexica were created, containing lexical patterns associated with ALCOHOL, DRUGs, EATING_DISORDERs, patient HISTORY, previous INPATIENT psychiatric treatment, LEGAL consequences, and previous OUTPATIENT psychiatric treatment. Table 2 provides examples from each lexicon (the full lexica are provided in Online Supplementary Appendix A).
Table 2.
Examples of lexical patterns from seven manually crafted lexica.
| Lexicon | Examples |
|---|---|
| ALCOHOL | vodka daily, liter per day, whiskey daily, … |
| DRUG | benzo, benzos, heroin, … |
| EATING_DISORDER | bulimia, anorexia, purging, … |
| HISTORY | medical leave, raped, suicide attempt, … |
| INPATIENT | psychiatric inpatient, resident program, inpatient detox, … |
| LEGAL | incarceration, custody, arrested, … |
| OUTPATIENT | regular psychiatric, regular psychopharm, group therapy, … |
UMLS Hierarchy Features
When analyzing the content of psychiatric evaluation records, we found that although many patients had different individual symptoms, most patients within each severity level shared a number of general traits. For example, although individual drug or alcohol dependency disorders were not frequently mentioned in SEVERE psychiatric evaluation records (for example, only 7.98% contained a non-negated mention of heroin), nearly all records had at least one mention of some kind of drug-or-alcohol dependency disorder. Thus, we wanted to extract features which capture more general common behavior or mental disorders than might be explicitly stated in the content of the psychiatric evaluation records. To do this, we extracted UMLS hierarchy features. We considered the three separate UMLS hierarchies rooted at the UMLS concepts corresponding to Mental Health (C0025353), Mental Disorder (C0004936), and Behavior Disorder (C0004930) described in Section 4.2. Each hierarchy was constructed by traversing UMLS following outgoing RB relations (an outgoing RB relation in UMLS indicates “has a broader relationship”) in which any concept that would introduce a cycle into the hierarchy was ignored. Figure 4 illustrates an excerpt of the UMLS hierarchy we constructed for the concept Mental Disorder. For each UMLS hierarchy, we extracted two features from a psychiatric record encoding: (1) whether any concept in the hierarchy was mentioned within the relevant content of the record and (2) the path between every mentioned concept in the hierarchy and the root of the hierarchy. Each path was encoded as a multivalued bag-of-CUIs feature where each CUI in the hierarchy was assigned a value of ‘1’ if the CUI occurred in the path from any mentioned concept in the hierarchy to the root of the hierarchy, and a value of ‘0’, otherwise. These features correspond to F47-F52.
Figure 4.
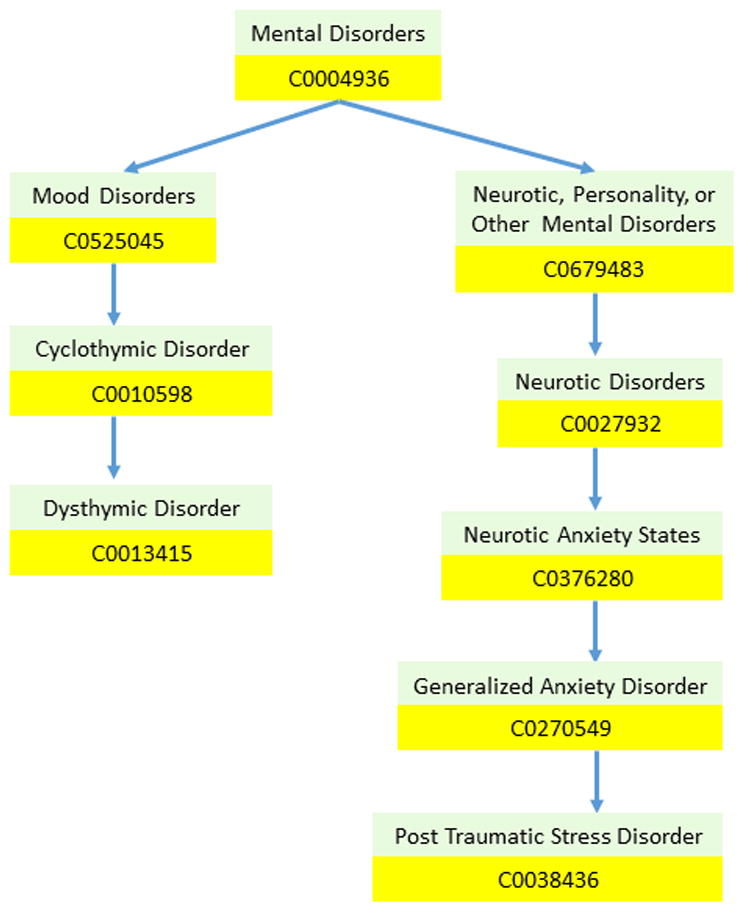
Excerpts of the UMLS hierarchy for two Mental Disorders: (1) Post Traumatic Stress Disorder (PTSD) and (2) Dysthymic Disorder.
Patient Age Features
When analyzing the content of psychiatric evaluation records, we noted that the age of a patient played a significant role. Specifically, we found that as the age of the patient increased so, too, did the number of behavioral and mental symptoms required to elevate the patient’s psychological symptoms to a more severe level. Features F53-F56 capture this phenomenon for older patients, while F56 captures this phenomenon for younger patients. F57 encodes the age of the patient directly.
Axis V GAF Features
We extracted F58 and F59 which encode the lowest, and current Axis V GAF categories as deter- mined by the Axis V GAF normalization process described in Section 4.2.
ICD-9 Features
We considered four multivalued features to encode the ICD-9 diagnostic codes associated with each psychiatric evaluation record. As with the UMLS hierarchy features, our features were designed to capture specific ICD-9 codes as well as more general information. To do this, we took advantage of the fact that ICD-9 codes are organized in a hierarchy with each digit indicating a further level of granularity. Thus, we considered four bag-of-ICD-9-code features: (1) all ICD- 9 codes truncated to integers (i.e., no decimal places), (2) all ICD-9 codes truncated to a single decimal place (i.e., the tenths digit), (3) all ICD-9 codes truncated to two decimal places (i.e., the hundredths digit), and (4) the ICD-9 code classes of all ICD- 9 codes (as described in Section 4.2).
It should be noted that, with the exception of Lexical features (F33-F46), all the features we extracted capture general psychological information with no specific emphasis on positive valence symptoms. Consequently, it is the role of the learning methods to discover which individual features – e.g., which questions or UMLS concepts – are the most indicative of positive valence symptom severity.
4.4. Learning Methods
Our approach for automatically recognizing the severity of a patient’s positive valance symptoms from psychiatric evaluation records relies on two assumptions: (1) that the severity of a patient’s positive valence symptoms exists within a latent continuous spectrum, and (2) that all of the patient’s answers and narratives documented in his or her psychological evaluation records are informed or influenced by the patient’s latent severity score along this spectrum. These two assumptions allow us to consider a two-step approach for automatically recognizing symptom severity from psychiatric evaluation record:
Step 1: Infer the latent continuous severity score that was most likely to produce a given psychiatric evaluation record; and
Step 2: Map the severity score into one of the four discrete severity levels used in the CEGS/N-GRID challenge.
We considered three methods for inferring the latent severity score from a given psychiatric evaluation record (i.e., Step 1): (1) point-wise ridge regression, (2) a pairwise random forest, and (3) a hybrid model combining pointwise ridge regression with the pairwise random forest. Step 2, associating the severity score with one of four discrete severity classes, is accomplished by a cascading SVM tree. In the remainder of this section, we present (1) a formal definition of the problem, (2) pointwise ridge regression, (3) the pairwise random forest, and (4) the hybrid model as well as (5) the cascading SVM tree used to implement Step 2.
4.4.1. Problem Definition
Formally, let 𝒯 represent the training data. Each training sample (x, y) ∈ 𝒯 corresponds to a psychiatric evaluation record where x is the feature vector representation of the record (as described in Section 4.3) and y is the gold-standard positive valence symptom severity level y ∈ {0, 1, 2, 3} (corresponding to ABSENT, MILD, MODERATE, and SEVERE, respectively). For a given training sample (x, y), our approach operates by (1) inferring the most likely latent severity score s (within the latent continuous positive valence symptom severity spectrum) to have produced x and then (2) mapping s to the severity label ŷ which is closest to y.
4.4.2. Pointwise Ridge Regression
Our first approach casts the problem of inferring latent severity score s from a given psychiatric evaluation record x as a pointwise linear regression problem. Specifically, we use ridge regression [21] (also known as Tikhonov regularization) in which we learn the set of feature weights θ which produces the severity score s with the lowest least-square error from y after L2-normalization. Formally,
| (1) |
where λ is a regularization term (in our experiments we set λ = 1). After training, we can infer the most likely severity score s for a given x as s = Σi θi · xi.
4.4.3. Pairwise Random Forest
When analyzing the behavior of the pointwise ridge regression model described above, we found that it was often easier to understand why psychiatric evaluation records were given a particular label by comparing the record to another record with a different severity level. We observed that it was often much easier for us – as non-experts without any background psychiatric knowledge – to determine which of the two records was more severe, than it was to directly determine the severity of each record independently. In light of this observation, – we wondered whether it would also be easier for the machine as well. Like us, a machine-learned model does not have access to the background psychiatric knowledge used by psychiatrists when manually assessing the severity level of a patient’s positive valence symptoms. Inspired by these observations, we considered a second model in which, rather than directly producing a severity score s from the feature vector x, we transformed the problem of recognizing patients’ symptom severity levels to a binary classification problem. To do this, we used the pairwise transform [22] which forms unique pairwise training samples by taking the difference between all pairs of feature vectors extracted from records with different gold-standard severity levels. Specifically, we created a new pairwise training sample,
| (2) |
from every pair of pointwise training samples (xi, yi), (xj, yj) ∈ 𝒯 where yi ≠ yj. In this way, each feature in the pairwise feature vector xP encodes the difference between the value of that feature in xi and its value in xj while the pairwise label yP has two possible values: +1 if yi is more severe than yj, and −1 if yi is less severe than yj. Consequently, yP indicates the ordering between xi and xj by their psychological symptom severity. Moreover, a useful property of the pairwise transform is that pairwise training data can be balanced by multiplying pointwise training samples in the minority class with −1 until both classes are balanced, improving classification performance. By applying the pairwise transform, we were able to train a binary classifier to predict yP ∈ {+1, −1} given xP. Moreover, as opposed to the pointwise regressor (described in Section 4.4.2) which weights features based on the feature’s contribution to the overall severity score, the pairwise representation allows a binary classifier to learns weights based on how well each feature can be used to discriminate between yP = +1 and yP = −1. For example, consider Feature F32 from Table 1, which indicates the number of questions in the psychological questionnaire which were answered YES. The regression model learns the weight of this feature by measuring how well the severity score can be directly computed from the value of the feature (which may not be possible). By contrast, the pairwise transform allows the model to instead learn how the difference in the number of YES-answered questions can be used to infer which of a pair of records indicates the most severe positive valence symptoms – i.e., the binary classifier can capture the intuition that if record xi has more YES-answered questions than record xj, then record xi is likely to have a higher severity score than record xj. In our approach, we considered a random forest (RF) classifier [23] for this purpose.
An RF is a meta or ensemble classifier which relies on perturb-and-combine techniques [24] to fit a diverse set of independent decision tree classifiers on various sub-samples of the training set. Formally, each tree in the ensemble or forest is built by randomly selecting (with replacement) a pairwise training sample. Unlike traditional decisions trees [25], when each decision tree in the random forest is constructed, each branch is determined using a random subset of features. By combining random samples of training data with random subsets of features, the RF has substantially reduced variance compared to many other classifiers. Unlike Breiman [23], in which the class predicted by the RF is chosen by voting, we average the probabilistic predictions across each tree. The RF was trained using Gini impurity [25] as the splitting criterion, as the number of sampled features, and 500 individual decision trees.
Given a trained RF, we inferred the latent severity score s for the feature vector x associated with a given psychiatric evaluation record by directly classifying x with the RF – that is, without applying the pairwise transform – and determined s as the average probability of assigning the class label yP = +1 (as opposed to yP = −1) from each tree in the forest. Conceptually, this corresponds to producing the score for a psychiatric evaluation record by measuring the likelihood that the record would have a higher severity level than a “blank” psychiatric evaluation record.
4.4.4. Hybrid Model
Pointwise ridge regression and pairwise random forest classification both approach the problem of inferring the severity score s from x very differently: the pointwise approach learns to infer s from a single psychiatric evaluation record directly, while the RF learns to infer s based on information learned by ordering pairs of psychiatric evaluation records. Consequently, each model learns a very different set of feature weights for determining s from x. We were interested in discovering if combining these two alternate approaches would result in more robust and accurate severity scores. Thus, our third and final method for inferring s from x uses a linear combination of the inferred severity scores predicted by both models. Formally, let s1 be the severity score inferred by ridge regression and let s2 be the severity score inferred by the random forest classifier. The hybrid model infers a new severity score s as a weighted combination of s1 and s2:
| (3) |
where α and α are weights learned by minimizing the least squares error between s and y in the training data.
4.4.5. Cascading SVM Tree
After inferring the latent continuous severity score s associated with a record, it is necessary to map s to a discrete severity level y ∈ {0, 1, 2, 3} (corresponding to ABSENT, MILD, MODERATE, and SEVERE, respectively). However, it should be noted that the severity levels are not independent (i.e. severity level 1 is closer to severity level 2 than it is to severity level 3). Consequently, standard classification techniques – which treat each class label independently – are not ideal. Specifically, there are two methods typically used to convert a binary classifier (e.g. a support vector machine, or logistic regression classifier) into a multi-class classifier: (1) one-vs-one in which, for each severity level y, three binary classifiers are learned where each classifier discriminates between y and each of the other three severity levels y′ using only the subset of the training data with severity levels y and y′; and (2) one-vs-rest in which, for each severity level y, a single binary classifier is trained to discriminate between y and the other three severity levels. While these two methods are the most commonly applied (e.g., they are used by SVM-Light [26] and scikit-learn [27]), they cannot account for the relationships between severity levels. For example, consider the severity level MILD. In the one-vs-one approach, the classifier used to distinguish between MILD and SEVERE is unable to consider any of the information from psychiatric evaluation records associated with ABSENT severity levels, despite that fact that the training examples used to distinguish between ABSENT and SEVERE may also help distinguish between MILD and SEVERE. In the one-vs-rest approach, the binary classifier for MILD would have to learn a single decision boundary (i.e., the latent continuous severity score threshold) to distinguish between MILD and SEVERE as well as between MILD and ABSENT, despite the fact that SEVERE and ABSENT correspond to opposite ends of the latent continuous severity spectrum.
To account for these problems, we used a hierarchical approach to multi-class classification inspired by Kumar et al. [28]. Specifically, we trained a tree of three cascading binary support vector machine (SVM) classifiers, as illustrated in Figure 3. This hierarchical approach allows us to (1) consider all of the training data to determine the boundaries between the severity scores associated with each discrete severity level and (2) captures the implicit relationships between severity levels. In general, each SVM is a binary classifier which is trained on pairs of severity scores and gold-standard severity levels, (s; y) and learns the optimal decision boundary to separate new, unlabeled severity scores into two buckets. The decision boundaries learned by each SVM correspond to thresholds on the latent continuous severity spectrum which classify severity scores into the four discrete severity levels. The first classifier, SVM1, learns the optimal decision boundary which separates severity scores into two buckets, one containing all the severity scores associated with y ∈ {0; 1} and the second bucket containing all severity scores associated with y ∈ {2; 3}. SVM2 learns to further separate the severity scores associated with y ∈ {0; 1} as either y = 0 or y = 1. Likewise, SVM3 learns to further separate the severity scores associated with y ∈ {2; 3} as either y = 2 or y = 3.
After training, we determine the severity level of an unlabeled psychiatric evaluation record (with severity score s) using the cascading SVM tree in two stages: (a) we use SVM1 to determine whether s is associated with a severity level ŷ ∈ {0; 1} or with ŷ ∈ {2; 3} and (b) depending on the result of stage (a), we use either SVM2 or SVM3 to determine the individual severity level. Specifically, if SVM1 predicts that ŷ is in {0; 1}, we determine the final severity level as either 0 or 1 using SVM2. Likewise, if SVM1 predicts that ŷ is in {2; 3}, we determine the final severity level as either 2 or 3 using SVM3. It should be noted that the cascading approach does not require an SVM and could be implemented using any binary classifier.
5. Results
We evaluated our approach using the official results provided by the organizers of the CEGS/N-GRID workshop [29]. A total of 65 runs were submitted by 24 teams. The official evaluation metric used by the organizers was the normalized mean absolute error (MAE):
| (4) |
where h is a system for recognizing symptom severity such that h(x) is the predicted severity level, Y is the set of severity levels (with 0 indicating ABSENT, 1 indicating MILD, 2 indicating MODERATE, and 3 indicating SEVERE), 𝒟j is the set of test documents with gold-standard score j, xi is a psychiatric evaluation record and yi is its gold-standard score. Note that in the official evaluation, the MAE was normalized and reported as a percentage where the system with the highest normalized MAE percentage obtained the highest performance. The normalized MAE evaluation has the important property that the every severity level is given the same importance, regardless of its frequency in the test collection. Table 4 presents the official evaluation results for the top ten submitted runs across all teams. As shown, our team placed second over-all, performing just below the system developed by SentiMetrix, Inc. We submitted three runs: run 1 corresponds to method 1, pointwise ridge regression; run 2 corresponds to method 2, the pairwise random forest; and run 3 corresponds to method 3, the hybrid model. From Table 4, it can be seen that run 3, our hybrid model, was the second-best performing run over all submitted runs. Moreover, it can be seen that run 2, which corresponds to the pairwise random forest classifier (method 2), was the ninth-best performing system. Table 4 also presents the median, mean, and minimum MAE scores aggregated across all runs.
Table 4.
Top performing runs across all teams.
| Rank | Team | Run | MAE Score |
|---|---|---|---|
| 1 | SentiMetrix Inc. | 3 | 0.863019 |
| 2 | University of Texas at Dallas | 3 | 0.840963 |
| 3 | University of Kentucky | 3 | 0.838615 |
| 4 | University of Kentucky | 1 | 0.837284 |
| 5 | Med Data Quest Inc. | 1 | 0.836503 |
| 6 | University of Kentucky | 2 | 0.835138 |
| 7 | SentiMetrix | 2 | 0.833281 |
| 8 | University of Pittsburgh | 3 | 0.825594 |
| 9 | University of Texas at Dallas | 2 | 0.824262 |
| 10 | University of Pittsburgh | 2 | 0.821807 |
|
| |||
| - | Median | - | 0.775880 |
| - | Mean | - | 0.771492 |
| - | Minimum | - | 0.524597 |
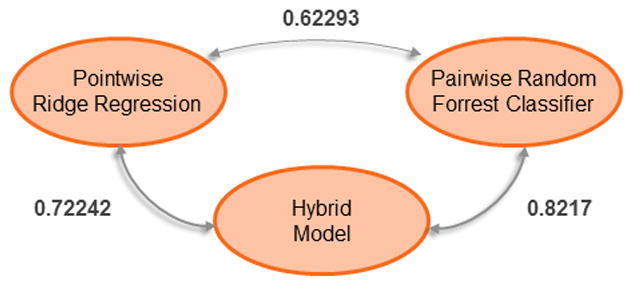
In addition to the official rankings, we were interested in comparing the performance of our three methods as well as comparing the performance between each severity level. The macro-average normalized MAE for each of our three methods are shown in Table 3 as well as the normalized MAE for each of the four individual severity levels. As shown by Tables 3 and 4, all three of our methods significantly outperformed the official median and mean MAE scores reported by the organizers. Moreover, it is also clear that the hybrid approach achieved the best performance of our three methods, while the pairwise random forest classifier obtained lower but still high performance. The pointwise ridge regressor, however, obtained the lowest performance of our three methods. We can also see that the severity labels on the extremes – ABSENT and SEVERE – were the easiest for our methods to classify, while distinguishing between MILD and MODERATE was more difficult. Moreover, the increase in performance of the hybrid model compared to the pointwise and pairwise models it combines suggests that the pointwise and pairwise models learn significantly different information from the psychiatric evaluation records. In order to determine how differently the two methods predicted severity scores, we calculated the weighted agreement between all three methods (using Cohen’s κ). Figure 5 presents κ between all pairs of methods for inferring the severity score of a psychiatric evaluation record.
Table 3.
Official performance results for each configuration of our system.
| Method | Average | Mean Absolute Error (MAE) | |||
|---|---|---|---|---|---|
| ABSENT | MILD | MODERATE | SEVERE | ||
| 1. Pointwise Ridge Regression | 0.791169 | 0.928571 | 0.778761 | 0.659091 | 0.896296 |
| 2. Pairwise Random Forest | 0.824262 | 0.884615 | 0.831579 | 0.692982 | 0.912281 |
| 3. Hybrid Model | 0.840963 | 0.920635 | 0.868750 | 0.698276 | 0.912281 |
Figure 5.
Cohen’s κ between all pairs of methods for inferring the severity score.
As shown in Figure 5, the pointwise and pairwise methods had a κ agreement of only 62:3% which clearly indicates that both methods often produced different severity scores for the same record. Moreover, we can see that the hybrid model has a higher κ agreement with the pairwise method than with the pointwise method, suggesting that, while the hybrid model often prefers the severity scores produced by the pairwise method, it is able to successfully recognize when the scores inferred by the pointwise method are more reliable.
6. Discussion
6.1. Error Analysis
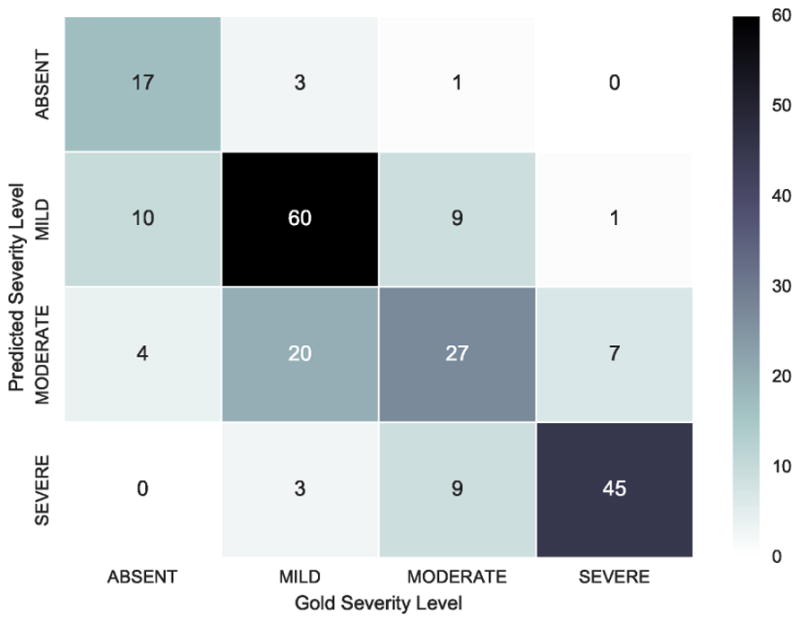
The majority of errors in predictions by our model are between adjacent severity levels, i.e. between ABSENT and MILD, MILD and MODERATE, and MODERATE and SEVERE. Figure 6 illustrates the number of records with each severity level predicted by our hybrid model against the number of records with each gold-standard severity level. Clearly, the most common misclassification errors were between the MILD and MODERATE severity levels. Recall from Section 3, that the difference between the MILD and MODERATE severity levels are that MODERATE symptoms have to be the focus of outpatient treatment, while MILD symptoms are never the focus of treatment. In the psychiatric evaluation records used for this evaluation, the focus of treatment is not explicitly stated. Moreover, the focus of treatment is difficult to infer without psychiatric expertise – for example, knowledge about what behavioral or psychological phenomena each treatment is designed to address. Future work may benefit from inferring relations between treatments and their focus in psychiatric evaluation records.
Figure 6.
Number of records with predicted severity level versus gold-standard severity level
Errors between MODERATE and SEVERE severity levels were less common, and were often due to errors in distinguishing between inpatient treatment/hospitalization for general medical conditions or for psychiatric symptoms, specifically. Consider the following two examples that indicate inpatient treatment in the past:
Example 9. History of Inpatient Treatment: Yes, Counseling center 5/15/2069-5/0021
Example 10. … Comanche Hospital 2066, complete hysterectomy
Example 9 is an excerpt from a SEVERE record our system misclassifies as MODERATE. Clearly, this patient has a history of inpatient treatment for psychological reasons since the treatment was administered in a counseling center. Example 10 is from a MODERATE record our system classifies as SEVERE and describes a history of inpatient treatment for non-psychiatric reasons. Our system is unable to reliably distinguish mentions of inpatient treatment based on the reason for hospitalization, but the inclusion of such information could benefit future work.
Finally, we observed that errors between MILD and ABSENT were often due to (1) patients recalling significantly distant (e.g. childhood) events or (2) patients describing actions of their friends or family. For example, the following excerpt is from a narrative portion of a record:
Example 11. … with psychiatric history notable for alcohol abuse in sustained remission
In Example 11, the psychiatrist notes that the patient’s past history of alcoholism is “in sustained remission” and therefore not likely to influence current positive valence symptom severity. We believe that future work may benefit from incorporating information about whether medical concepts are associated with the patient, or with someone else, i.e., incorporating the assertions investigated in the 2010 i2b2/Veterans Affairs challenge on concepts, assertions, and relations [30].
6.2. System Design
Feature extraction was implemented in Java 8 and relied on the natural language processing systems described in Section 4.2. Ridge regression and the cascading SVM trees were implemented in Python 2.7 using scikit-learn [27]. The pairwise random forest approach relied on the random forest implementation offered in RankLib3, part of the Lemur4 search engine project.
When designing our system, we considered a much larger set of features than those described in Section 4.3, however many of these features degraded performance on the training set (performance decreases are described in Section 6.3). In this section, we describe various natural language processing and feature sets that we ultimately removed based on cross validation performance during training. Note: all of these features were extracted after applying all of the text preprocessing steps described in Section 4.1.
Bag-of-Words
Given that the psychiatric evaluation records used in our experiments are text documents, a natural feature would be using a bag-of-words representation. We found that this significantly hurt performance during training. We additionally considered a bag-of-nonnegated-words features, as well as a ternary bag-of-words feature in which each word would be associated with the value 1 if it was mentioned and not negated, 0 if it was not mentioned and −1 if it was negated. None of these features improved performance.
Bag-of-Bigrams
After analyzing the failure of bag-of-words features to improve performance, we considered a bag-of-bigrams alternative. Bigrams are commonly used in language modeling and other natural language processing tasks and correspond to pairs of consecutive words in a document. As with bag-of-words, we also considered a bag-of-nonnegated-bigrams and ternary bag-of-bigrams feature. None of these three features improved performance.
Bag-of-Concepts
We experimented with a bag-of-concepts feature which was akin to bag-of-words but relied on concepts identified by MetaMap rather than individual words. As with bag-of-words, we also considered a bag-of-nonnegated-concepts and ternary bag-of-concepts feature. We observed, as with bag-of-bigrams and bag-of-words, the performance was not improved.
Doc2Vec
After observing that bag-of-words, bag-of-bigrams, and bag-of-concepts features all degraded performance, we considered modern distributional approaches to representing the content of psychiatric evaluation records. Specifically, we learned vector representations of each psychiatric evaluation record using Doc2Vec [31]. Doc2Vec relies on deep neural learning to learn an embedded representation of words, sentences, and documents. We experimented with Doc2Vec embeddings of dimensions 50, 100, and 200, and found that including the embeddings learned by Doc2Vec degraded performance.
6.3. Feature Ablation Analysis
In order to facilitate future work, we performed a feature ablation study using both the training and testing sets. Table 5 presents these results. The top half of Table 5 illustrates the loss in performance when the rejected feature sets described in Section 6.2 were incrementally incorporated. In the table, we report only the best performing representations of the bag-of-words, bag-of-bigrams, and bag-of-concepts features – namely, the ternary representation; likewise, the Doc2Vec performance was reported when using 200-dimensional embeddings. The second half of Table 5 illustrates the loss in performance when each feature sets described in Section 4.3 was incrementally removed. Interestingly, the bag-of-bigrams feature degraded performance while cross validating against the training set but improved performance on the testing set. This suggests that future work may benefit from incorporating bigram features and that bigrams features can capture information not available from UMLS concepts, ICD9 codes, or any of the lexical features. In terms of the features described in Section 4.3, the largest performance decrease in the training set (and second largest in the testing set) was observed when removing the UMLS Hierarchy features, highlighting the importance of considering relevant medical concepts especially when compared against the decrease in performance observed when all MetaMap-detected concepts were considered. Likewise, the decrease in performance after lexical features were removed, although less significant than UMLS Hierarchy features in the training set, indicates the importance of incorporating hand-tuned information characterizing positive valence symptoms. It can be seen that, overall, the importance of features was similar between the training and testing sets which suggests that the features reported in this paper are able to generalize to new psychiatric records.
Table 5.
Feature ablation analysis
| Feature(s) | Mean Absolute Error (MAE) | |
|---|---|---|
| Training | Testing | |
| + Bag-of-Words | −0.1517 | −0.0611 |
| + Bag-of-Bigrams | −0.7748 | +4.2203 |
| + Doc2Vec | −0.5167 | −5.8393 |
| + Bag-of-Concepts | −2.6129 | −2.9746 |
|
| ||
| Hybrid Model | 80.4026 | 84.0963 |
|
| ||
| −ICD-9 | −0.6078 | −0.1435 |
| −Axis V | −0.4892 | −0.0190 |
| −Age | −0.6793 | −0.4390 |
| −Lexical | −0.9280 | −1.0341 |
| −UMLS Hierarchy | −1.9248 | −0.9224 |
6.4. Cascading SVM Tree Analysis
In addition to the individual feature analyses performed above, we compared the performance when using three different methods for mapping the latent continuous severity score associated with each record to the discrete severity levels used in the CEGS/N-GRID evaluation: (1) the cascading SVM tree reported in this paper, (2) a one-vs-one SVM approach, and (3) a one-vs-rest SVM approach. For an explanation of these methods, please refer to Section 4.4.5. We report the performance of the hybrid approach when using each method (though the observations hold for the pointwise and pairwise models as well). We found that the cascading SVM tree performed 7:5142% better than the one-vs-one approach (which obtained an MAE of 78:2188) and 11:6464% better than the one-vs-rest approach (which obtained an MAE of 75:3237). This shows the importance of considering the implicit relationship between severity levels and indicates that out-of-the-box classification approaches may not be ideal.
6.5. Severity Score Analysis
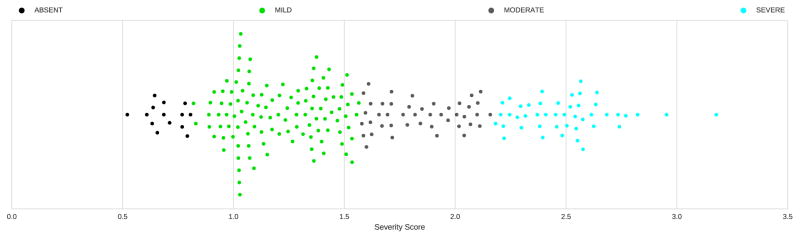
While the official evaluation results allowed us to measure the performance of our approach for recognizing discrete severity levels, we were also interested in analyzing the latent continuous severity scores inferred by our methods. Figure 7 illustrates the inferred severity scores produced by the hybrid model along with the predicted severity levels. It is interesting to note that thresholds learned by the cascading SVM tree are significantly different than what one might expect – ABSENT was associated with severity scores s ∈ [0; 0:81279), MILD was associated with severity scores s ∈ [0:81279; 1:5721065), MODERATE was associated with severity scores s ∈ [1:5721065; 2:1700965) and SEVERE was associated with severity scores s ∈ [2:1700965, ∞). Moreover, it can be seen that the severity score inferred from one psychiatric evaluation record was actually greater than 3 – the value associated with SEVERE. We analyzed the content of that record as well as the feature vector extracted from it. We found that this record had 11 active Question & Answer features, compared to an average of 6:1 active Question & Answer features for all SEVERE records (the average number of active Question & Answer features for the records associated with MODERATE, MILD, ABSENT severity levels was 4:22, 2:93 and 0:26 respectively). Moreover, we observed that the patient suffered from a substantial number of separate behavioral and mental problems. This patient’s record had 11 active Behavior Disorder UMLS Hierarchy features compared to an average of 6:5 for all SEVERE records and 30 active Mental Disorder UMLS Hierarchy features versus 26:8 on average for SEVERE records. This suggests that the symptom severity of this patient is indeed more severe than that of other patients within the SEVERE severity level. Thus, we believe that the severity scores inferred by our approach are able to provide finer grained information than the four discrete severity labels. Moreover, we also observed that the hybrid model actually performed better on the test set (0:841) than on the training set (0:804 in cross validation). The increase in performance obtained between the training and test set indicates that the features we extract (described in Section 4.3) are not only effective but robust.
Figure 7.
Inferred severity scores with predicted severity labels.
6.6. Future Work
Based on the above analyses, we believe possible avenues for future work include: (1) incorporating features capturing fine-grained information about the degree of belief associated with medical concepts such as assertions, modality and speculation; (2) considering the experiencer of a medical concept (e.g. patient, family member); and (3) modeling temporal information to differentiate between medical concepts from the patient’s present, recent past, and history. Moreover, future work may benefit from considering alternative pairwise ordering classifiers or by designing new machine learning approaches capable of jointly modeling pointwise and pairwise information.
7. Conclusion
This article has described our submissions to the 2016 CEGS/N-GRID shared tasks and workshop on Challenges in Natural Language Processing for Clinical Data. Our submissions relied on a rich set of features characterizing the patient’s answers to questions in the psychiatric evaluation report, as well as relevant medical concepts and established psychiatric metrics. We submitted three methods for recognizing positive valence symptom severity: (1) a pointwise model based on ridge regression, a (2) a pairwise model using a random forest classifier, and (3) a hybrid model combining both. Our key contributions to the task are (1) the performance comparison between pairwise and pointwise methods for predicting symptom severity as well as (2) the generation of a hybrid model which combines the strengths of both methods, and (3) the discovery of robust features which actually performed better on the test set than on the training set. Two of our submissions placed within the top ten submissions out of all participants, with the hybrid model ranking second over-all in the official evaluations. The official results of this task, as well as the post-hoc analysis we performed, demonstrate the importance of incorporating comparative (e.g. pairwise) information to recognize symptom severity.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of Mental Health (NIMH), the National Library of Medicine (NLM), and the National Human Genome Research Institute (NHGRI) of the National Institutes of Health under awards number P50MH106933, R13LM011411, and U01HG008468, respectively. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Technically, each multivalued feature actually corresponds to a set of binary features, one for each possible discrete value.
Conflicts of interest
There are no conflicts of interest.
URL: www.hlt.utdallas.edu/~travis (Travis R. Goodwin), www.hlt.utdallas.edu/~ramon (Ramon Maldonado), www.hlt.utdallas.edu/~sanda (Sanda M. Harabagiu)
References
- 1.Cuthbert BN. The rdoc framework: facilitating transition from icd/dsm to dimensional approaches that integrate neuroscience and psychopathology. World Psychiatry. 2014;13(1):28–35. doi: 10.1002/wps.20087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medsger T, Bombardieri S, Czirjak L, Scorza R, Rossa A, Bencivelli W. Assessment of disease severity and prognosis. Clinical and experimental rheumatology. 2003;21(3) SUPP/29:S42–S46. [PubMed] [Google Scholar]
- 3.Chen DP, Weber SC, Constantinou PS, Ferris TA, Lowe HJ, Butte AJ. Clinical arrays of laboratory measures, or” clinarrays”, built from an electronic health record enable disease subtyping by severity. AMIA. 2007 [PMC free article] [PubMed] [Google Scholar]
- 4.Joshi R, Szolovits P. AMIA Annual Symposium Proceedings. Vol. 2012. American Medical Informatics Association; 2012. Prognostic physiology: modeling patient severity in intensive care units using radial domain folding; p. 1276. [PMC free article] [PubMed] [Google Scholar]
- 5.Saeed M, Villarroel M, Reisner AT, Clifford G, Lehman LW, Moody G, Heldt T, Kyaw TH, Moody B, Mark RG. Multiparameter intelligent monitoring in intensive care ii (mimic-ii): a public-access intensive care unit database. Critical care medicine. 2011;39(5):952. doi: 10.1097/CCM.0b013e31820a92c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knaus WA, Draper EA, Wagner DP, Zimmerman JE. Apache ii: a severity of disease classification system. Critical care medicine. 1985;13(10):818–829. [PubMed] [Google Scholar]
- 7.Marshall JC, Cook DJ, Christou NV, Bernard GR, Sprung CL, Sibbald WJ. Multiple organ dysfunction score: a reliable descriptor of a complex clinical outcome. Critical care medicine. 1995;23(10):1638–1652. doi: 10.1097/00003246-199510000-00007. [DOI] [PubMed] [Google Scholar]
- 8.Rothman MJ, Rothman SI, Beals J. Development and validation of a continuous measure of patient condition using the electronic medical record. Journal of biomedical informatics. 2013;46(5):837–848. doi: 10.1016/j.jbi.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 9.Pirracchio R, Petersen ML, Carone M, Rigon MR, Chevret S, van der Laan MJ. Mortality prediction in intensive care units with the super icu learner algorithm (sicula): a population-based study. The Lancet Respiratory Medicine. 2015;3(1):42–52. doi: 10.1016/S2213-2600(14)70239-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filannino M, Stubbs A, Uzuner Ö. Symptom severity prediction from neuropsychiatric clinical records: Overview of 2016 CEGS N-GRID Shared Tasks Track 2. Journal of Biomedical Informatics. 2017 doi: 10.1016/j.jbi.2017.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiner M. Evidence generation using data-centric, prospective, outcomes research methodologies. San Francisco, CA. Presentation at AMIA Clinical Research Informatics Summit; 2011. [Google Scholar]
- 12.Smith PC, Araya-Guerra R, Bublitz C, Parnes B, Dickinson LM, Van Vorst R, Westfall JM, Pace WD. Missing clinical information during primary care visits. Jama. 2005;293(5):565–571. doi: 10.1001/jama.293.5.565. [DOI] [PubMed] [Google Scholar]
- 13.O’malley KJ, Cook KF, Price MD, Wildes KR, Hurdle JF, Ashton CM. Measuring diagnoses: Icd code accuracy. Health services research. 2005;40(5p2):1620–1639. doi: 10.1111/j.1475-6773.2005.00444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manning CD, Surdeanu M, Bauer J, Finkel J, Bethard SJ, Mc-Closky D. The Stanford CoreNLP natural language processing toolkit. Association for Computational Linguistics (ACL) System Demonstrations. 2014:55–60. http://www.aclweb.org/anthology/P/P14/P14-5010.
- 15.Sætre R, Yoshida K, Yakushiji A, Miyao Y, Matsubayashi Y, Ohta T. Akane system: protein-protein interaction pairs in biocreative2 challenge, ppi-ips subtask. Proceedings of the Second BioCreative Challenge Workshop; Madrid; 2007. pp. 209–212. [Google Scholar]
- 16.Tsuruoka Y, Tateishi Y, Kim JD, Ohta T, McNaught J, Ananiadou S, Tsujii J. Developing a robust part-of-speech tagger for biomedical text. Panhellenic Conference on Informatics; Springer; 2005. pp. 382–392. [Google Scholar]
- 17.Agarwal S, Yu H. Biomedical negation scope detection with conditional random fields. Journal of the American medical informatics association. 2010;17(6):696–701. doi: 10.1136/jamia.2010.003228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bodenreider O. The unified medical language system (umls): integrating biomedical terminology. Nucleic acids research. 2004;32(suppl 1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aronson AR. Effective mapping of biomedical text to the umls metathesaurus: the metamap program. Proceedings of the AMIA Symposium; American Medical Informatics Association; 2001. p. 17. [PMC free article] [PubMed] [Google Scholar]
- 20.Hall RC. Global assessment of functioning: a modified scale. Psycho-somatics. 1995;36(3):267–275. doi: 10.1016/S0033-3182(95)71666-8. [DOI] [PubMed] [Google Scholar]
- 21.Hoerl AE, Kennard RW. Ridge regression: Biased estimation for nonorthogonal problems. Technometrics. 1970;12(1):55–67. [Google Scholar]
- 22.Herbrich R, Graepel T, Obermayer K. Support vector learning for ordinal regression. Artificial Neural Networks, 1999. ICANN 99. Ninth International Conference on (Conf. Publ. No. 470); IET; 1999. pp. 97–102. [Google Scholar]
- 23.Breiman L. Random forests. Machine learning. 2001;45(1):5–32. [Google Scholar]
- 24.Breiman L. Arcing classifiers. The annals of statistics. 1998;26(3):801–849. [Google Scholar]
- 25.Breiman L, Friedman J, Stone CJ, Olshen RA. Classification and regression trees. Belmont, CA: Wadsworth; 1984. [Google Scholar]
- 26.Joachims T. LS8-Report 24. Universität Dortmund; 1998. Making large-scale svm learning practical. LS VIII-Report. [Google Scholar]
- 27.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. Scikit-learn: Machine learning in Python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- 28.Kumar S, Ghosh J, Crawford MM. Hierarchical fusion of multiple classifiers for hyperspectral data analysis. Pattern Analysis & Applications. 2002;5(2):210–220. [Google Scholar]
- 29.Filannino M, Stubbs A, Uzuner Ö. Symptom severity prediction from neuropsychiatric clinical records: Overview of 2016 CEGS N-GRID Shared Tasks Track 2. Journal of Biomedical Informatics. 2017 doi: 10.1016/j.jbi.2017.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/va challenge on concepts assertions and relations in clinical text. Journal of the American Medical Informatics Association. 2011;18(5):552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le QV, Mikolov T. Distributed representations of sentences and documents. ICML. 2014;14:1188–1196. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.