Abstract
Objective
The relationship between inflammation, obesity and adverse metabolic conditions is associated with adipose tissue macrophages (ATM). We compared the measurements of human ATM using flow cytometry, immunohistochemistry (IHC) and RT-PCR of ATM markers.
Methods
We evaluated a new software program (AMCounter) to help measure ATM using IHC and compared this to flow cytometry and RT-PCR.
Results
IHC had good intra-individual reproducibility for total (CD68), pro-inflammatory (CD14) and anti-inflammatory (CD206) ATM. The AMCounter improved inter-reader agreement and was more time efficient. Flow cytometry had acceptable intra-individual reproducibility for the percent of CD68+ cells that were CD14+ or CD206+, but not for ATM/g tissue. ATM/g tissue was much greater using IHC than flow cytometry. The flow cytometry and IHC measures of ATM from the same biopsies were not correlated. There were statistically significant correlations between RT-PCR CD68 and IHC CD68, CD14 and CD206 ATM’s per 100 adipocytes. Of interest, were also statistically significant correlations between RT-PCR CD68 and IHC CD68, CD14 and adipose flow cytometry measures of CD68+, CD68+/CD14+ and CD68+/CD206+ ATM’s per g tissue.
Conclusions
The AMCounter software helps reproducibly and efficiency measures of IHC ATM’s. Flow cytometry, immunohistochemistry and RT-PCR measures of adipose inflammation provide somewhat different information.
Keywords: Adipose tissue, inflammation, macrophages, obesity, quantitation
Introduction
Obesity is linked to adipose tissue inflammation (reviewed in 1–3) with an increase in M1 or pro-inflammatory adipose tissue macrophages (ATM) and a decrease in M2 or anti-inflammatory ATM 4. Animal studies suggest that increased adipose tissue inflammation is a mechanism for the insulin resistance associated with obesity 5–7,8–10. We investigated methods to quantify ATM’s and, as part of our laboratory protocol, we evaluated the reproducibility and comparability of the different approaches.
Immunohistochemistry (IHC) and flow cytometry analyses of ATMs have been used for animal 11, 12 and human studies 13, 14. The advantages of immunohistochemistry include the preservation of all cells in the tissue and the ability to store the paraffin-embedded tissue for long periods, allowing for reuse with additional antibodies. However, quantifying ATM using IHC is time consuming and requires considerable amounts of tissue. Flow cytometry requires less tissue, can count thousands of cells in a few minutes, and multiple antibodies can be used simultaneously on a single tissue sample. However, flow cytometry requires digestion and extensive handling (potentially resulting in loss of cells) and the sample cannot be saved for future use. Additional problems include macrophage autofluorescence that complicate the analysis of flow cytometry data. We found no reports defining the agreement between IHC and flow cytometry quantification of human ATM and therefore tested how well these methods compare.
In attempts to reduce the time and resource burden of the IHC method we collaborated with Biomedical Imaging Resources to develop software to automate portions of the IHC analysis. We found that software-assisted IHC quantification of ATM burden offers some advantages and may improve the some aspects of the procedure.
Materials and Methods
Tissue collection and fat cell size
All protocols were approved by the Mayo Clinic Institutional Review Board and the volunteers provided written, informed consent. Subcutaneous adipose tissue biopsies were acquired by needle liposuction under sterile conditions using local anesthesia. Biopsies were collected from the abdominal (lateral to the umbilicus) and/or femoral (on the anterior-lateral aspect of the mid-thigh) regions. Fat cell size was measured as previously described 15. The portion of the sample destined for mRNA measurements was immediately frozen in liquid nitrogen and then stored at −80°C. The adipose tissue samples that were collected to analyze inter- and intra-individual variations for both the IHC and flow cytometry methods were obtained on two separate occasions approximately two weeks apart. To evaluate the intra-individual variability for IHC we analyzed 38 tissue samples (abdominal and femoral) from 15 individuals collected in duplicate. The analysis of intra-individual variability of flow cytometry was completed using 12 tissue samples from 6 individuals collected in duplicate. Forty samples were used to analyze reader agreement as well as agreement between manual IHC counting data and AMCounter software data. We used 47 tissues from 27 individuals to compare ATM content using IHC and flow cytometry methods.
Antibody Selection
After careful review of the literature and testing of numerous different antibodies, we selected CD68 for total macrophages using IHC. We chose the CD14 receptor as our pro-inflammatory macrophage marker for both the IHC and flow cytometry experiments because in vitro studies have shown that activation of the CD14 receptor results in a pro-inflammatory cell signaling cascade 16–18. We selected CD206 for the anti-inflammatory macrophage marker for IHC and flow cytometry. CD206 is a mannose-receptor that induces an anti-inflammatory cell signaling cascade following activation17, 19. We initially tested a FITC conjugated CD11b antibody for total macrophages by flow cytometry 20, but experienced unacceptable variability, consistent with descriptions on the Purdue List 21 (an online forum for flow cytometry experts) that adipose tissue samples gave excessive auto-fluorescence in the FITC channel. We changed to the allophycocyanin (APC) conjugated CD68 receptor antibody for flow cytometry to avoid FITC channel auto-fluorescence and to allow direct comparison with the antigen we selected for IHC. Our strategy for developing the optimal antibody concentration to ensure adequate capture of positive events and to reduce non-specific staining is provided in the supplemental online material and Figure S1.
Immunohistochemistry (IHC)
Details of the immunohistochemistry techniques for sample processing are provided in the Supplemental Online Material. The stained tissue sections were visualized using an Olympus BX43 light microscope. Ten to twelve randomly selected images per slide were taken at 40x magnification and two independent observers counted positively stained macrophages, crown-like structures (CLS) and total adipocytes for each field of view. We counted positively stained cells as ATM’s if they displayed the known morphological characteristics of macrophages. From this we derived the number of positively stained cells per 100 adipocytes. All slides were marked with a code rather than the sample identity to assure the independent observers were blinded to the other reader’s data, to the participant, the research protocol, and biopsy site. Similarly coded quality control slide photographs were included every 30–40 samples in the experimental data set to assess whether the readers’ interpretation changed over time.
IHC Calculations
Because the data is expressed per 100 adipocytes, but adipocyte size may vary between adipose regions and individuals, we also report ATM relative to tissue mass. To estimate the number of macrophages per gram of tissue we performed a quantitative lipid extraction of adipose and calculated the number of adipocytes per gram of tissue by dividing the lipid content by the average adipocyte size. For the few samples with insufficient tissue to perform a separate lipid extraction we used the average lipid content of adipose tissue (0.76 g lipid/g tissue) because there was minimal sample to sample variability in this value.
Automated IHC Software Development
The AMCounter automated image analysis program was developed using the Analyze/AVW imaging platform (Biomedical Imaging Resource, Mayo Clinic, Rochester, MN). Adipocytes and macrophages are automatically segmented and the results presented to the user for approval/modification. The automatic segmentation process takes less than 15 seconds per image on a standard PC workstation. Details of the AMCounter automated image analysis program are provided in the Supplemental Online Material.
IHC Automated Software Program Use and Statistical Analysis
The software places circles to mark and hence count adipocytes and macrophages. By moving the cursor over a cell, the user can insert new circles at missing sites or delete incorrectly-placed circles. The program saves the results and images, including the inserted circles.
Two readers manually counted the number of adipocytes and macrophages from 40 subcutaneous adipose tissue samples (ten images each) and separately using the AMCounter software. Both readers analyzed the same set of images to allow us to assess inter-reader agreement.
The between-method agreement was calculated with Lin’s concordance correlation coefficient (CCC), which is the product of precision (“how far each observation deviates from the best-fit line”) and accuracy (“how far the best-fit deviates from the 45 degree line”). The degree of concordance was defined as excellent (0.81–1.00), substantial (0.61–0.80), moderate (0.41–0.60), fair (0.21–0.40), slight (0.00–0.20), and poor (<0.00). For visual assessment, the paired readings from both methods were plotted (for each reader separately) in a scatter plot and a 45° line through the origin, the concordance line, was drawn.
Altman and Bland 22 approaches were used to assess disagreements between methods and the contribution of bias. For graphical presentation of the agreement, the differences between measurements by the two methods were plotted against their mean (for each reader separately). The graph allows visual assessment of method agreement and reveals outlying observations.
The intra-class correlation coefficient (ICC) was used to quantify measurement reliability of each method. It was calculated to assess inter-observer variability for each method: reader 1 versus reader 2 for manual counting and reader 1 versus reader 2 for software counting.
Flow Cytometry and Data Analysis
Details of the flow cytometry measures are provided in the Supplemental Online Material and Figure S2. All samples were analyzed using FlowJo Version 10 software (Ashland, OR). Forward scatter-area (FSC-A) and side scatter-area (SSC-A) gates were initially set based on size and density using control sample tissues. Within this gated population, a second gate was set using live/dead viability stained cells (Apc-Cy7+) versus FSC-A to exclude apoptotic cells. Both gates were then back-gated to our sample of interest. Within the gated populations on the sample of interest, the singlet cell population was selected using FSC-area vs. FSC-height. We further gated this population of single cells (as identified above) to CD68+ cells (APC-CD68 vs. FSC-Area). Using the CD68+ population, quadrant gating methods were applied to plot for the selected antigens (APC-CD68 vs. PE-CD14, or APC-CD68 vs. PE-Cy7-CD206). The pro-inflammatory or M1 macrophage population was defined as cells dual-stained with the CD68-APC conjugated antibody and CD14-PE conjugated antibody (CD68+/CD14+). The CD206+ macrophages, or M2 population was measured as dual-stained CD68-APC and CD206-PE-Cy7 (CD68+/CD206+). To measure the total CD68+ macrophages we used the number of cells within quadrants which contained the single stained CD68+ cells and the dual stained for either CD14 or CD206.
All of the flow cytometry results are expressed per gram of tissue, taking into account the number of aliquots utilized per sample run. We also report the percentage of ATM’s that are CD14+ and CD206+ and the ratio of CD14+ to CD206+ macrophages.
Real Time PCR
We used the RNeasy Lipid Tissue mini kit (Qiagen # 74804) to isolate RNA from adipose tissue samples. The isolated RNA was then reverse transcribed using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems #4368813) into cDNA as described by the manufacturer. RT-PCR was performed using Taqman Gene Expression assays (Applied Biosystems (CD68=Hs02836816_g1, CD206=Hs00267207_m1, CD14=Hs02621496_s1, p16=Hs00923894_m1, and CYCA=Hs99999904_m1) and TaqMan Fast Advanced Master Mix (Applied Biosystems #4444964) on an ABI Quant thermocycler using “Fast” settings in duplicate. The ΔΔCt method was used to analyze the data and cyclophilin A was used to normalize samples.
Statistics
Values are provided as average ± SEM. Paired Student t test was used for comparison of results between depots within the same individuals. Univariate regression analyses were used to test for correlations between visit one and visit two for IHC and flow cytometry as well as correlations between the IHC and flow cytometry methods.
Results
Participant Characteristics
The participant characteristics are outlined in Table 1. The study cohort included 49 participants, 13 males and 36 females.
Table 1.
Subject characteristics.
| Male (n=13) | Female (n=36) | |
|---|---|---|
| Age (years) | 41 ± 10 | 37 ± 10 |
| BMI (kg/m2) | 32 ± 4.5 | 32.1 ± 4.2 |
| Abdominal adipocyte size (μg lipid/cell) | 0.80 ± 0.37 | 0.76 ± 0.29 |
| Femoral adipocyte size (μg lipid/cell) | 0.78 ± 0.32 | 1.02 ± 0.34 |
Data are shown as means ± SD
n = no. of subjects
BMI = body mass index.
Inter- and Intra-individual variance in IHC Measurements
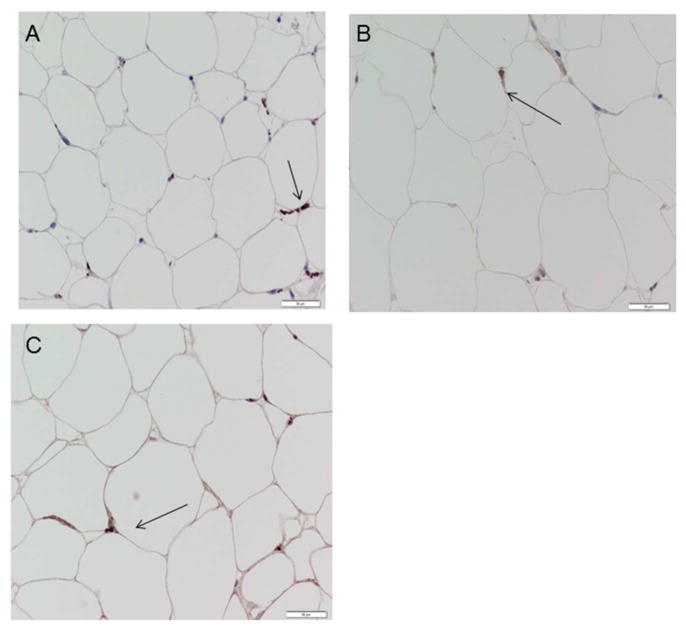
Subcutaneous abdominal and/or femoral adipose tissue biopsies were collected from fifteen participants 2–3 weeks apart in order to assess intra-individual variance in macrophage population measured using IHC. The agreements between the total monocyte/macrophage (CD68 +), CD14+ and CD206+ cells for visit one and visit two were judged as good whether expressed per 100 adipocytes or relative to tissue mass (Table 2, representative images shown in Figure 1).
Table 2.
Adipose tissue macrophage content reproducibility
| First biopsy | Second biopsy | Per gram tissue | Per 100 adipocytes | Per gram tissue | Per 100 adipocytes | |||
|---|---|---|---|---|---|---|---|---|
| IHC | /g tissue | /100 adipocytes | /g tissue | /100 adipocytes | p value | p value | r, p | r, p |
| CD68 | 120,408 ± 10,384 | 14.1 ± 1.5 | 130,349 ± 10,098 | 15.4 ± 1.6 | 0.51 | 0.61 | r = 0.63, p= 0.001 | r = 0.73, p= 0.001 |
| CD14 | 43,388 ± 4,939 | 5.5 ± 0.8 | 45,412 ± 4,837 | 5.5 ± 0.7 | 0.92 | 0.93 | r = 0.63, p= 0.001 | r = 0.76, p= 0.0001 |
| CD206 | 119,975 ± 7,223 | 14.0 ± 1.2 | 120,903 ± 7,939 | 13.8 ± 1.0 | 0.93 | 0.87 | r = 0.54, p= 0.006 | r = 0.80, p= 0.0003 |
| Flow cytometry | /g tissue | % of CD68+ | /g tissue | % of CD68+ | r, p per g tissue | r, p % of CD68+ | ||
| CD68+ | 3182 ± 2772 | 2930 ± 2474 | 0.77 | r = 0.39, p = 0.21 | ||||
| CD14+/CD68+ | 2514 ± 2319 | 75 ± 11 % | 2314 ± 1985 | 76 ± 10 % | 0.78 | r = 0.40, p = 0.20 | r = 0.58, p = 0.049 | |
| CD206+/CD68+ | 2629 ± 2423 | 78 ± 10 % | 2384 ± 2,154 | 73 ± 24 % | 0.73 | r = 0.44, p = 0.16 | 0.87, p = 0.0003 | |
For the immunohistochemistry (IHC) reproducibility there were 30 samples (abdominal and femoral) from 15 volunteers collected in duplicate 2–3 weeks apart. For flow cytometry there are 12 samples (6 abdominal and 6 femoral) from 6 volunteers collected 2–3 weeks apart. Values are given as mean ± SD.
Figure 1.
Representative images of positive macrophage staining. Positively stained cells are shown with an arrow. A.) CD68 or total macrophage. B.) CD14+ macrophage. C.) CD206+ macrophage.
For the total macrophage (CD68) marker, the absolute difference between reader one and two was 8 ± 5 macrophages/100 adipocytes (mean ± SD, range 2.4 – 13.9). The absolute difference in CD14+ macrophages between reader one and two was 3 ± 1 macrophages/100 adipocytes (mean ± SD, range 1 – 6 macrophages/100 adipocytes) and the absolute difference in CD206+ macrophages between reader one and reader two was 6 ± 3 macrophages/100 adipocytes (mean ± SD, range 0.4 – 11 macrophages/100 adipocytes).
We also counted the crown-like structures (CLS); there was minimal variation between the two readers and replicate measures, with an average of 1 CLS per 10 fields of view present in CD68 stained slides, 0.07 CLS present in CD14 stained slides, and 0.44 CLS present in CD206 slides. Parenthetically, in our population there were many samples with no CLS in 10 fields of view. The distribution of the number of CLS/10 fields of view paired with the number of CD68+ macrophages/100 adipocytes for 51 separate biopsies is provided in Figure S3.
Between Method Agreement: AMCounter Software and Manual Counting
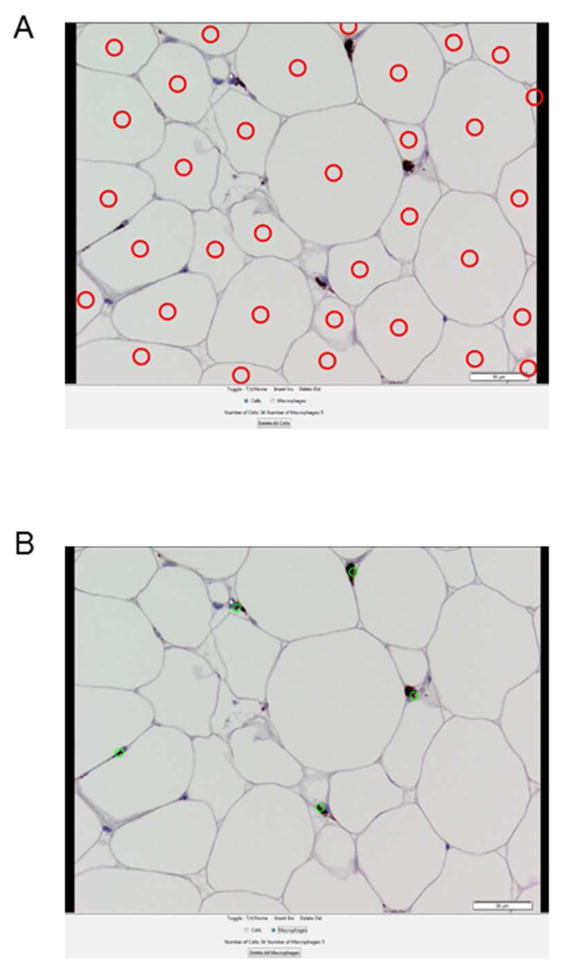
Between method agreement was determined using 40 different adipose tissue samples (representative image - Figure 2). For each reader and each antibody the concordance line plot was constructed separately. The number of macrophages and adipocytes from the same samples by two methods clustered around the line of identity at 45° (Figure 3A–F).
Figure 2.
Representative images from the AMCounter software program. A) Adipocyte counting image. B) Macrophage counting image.
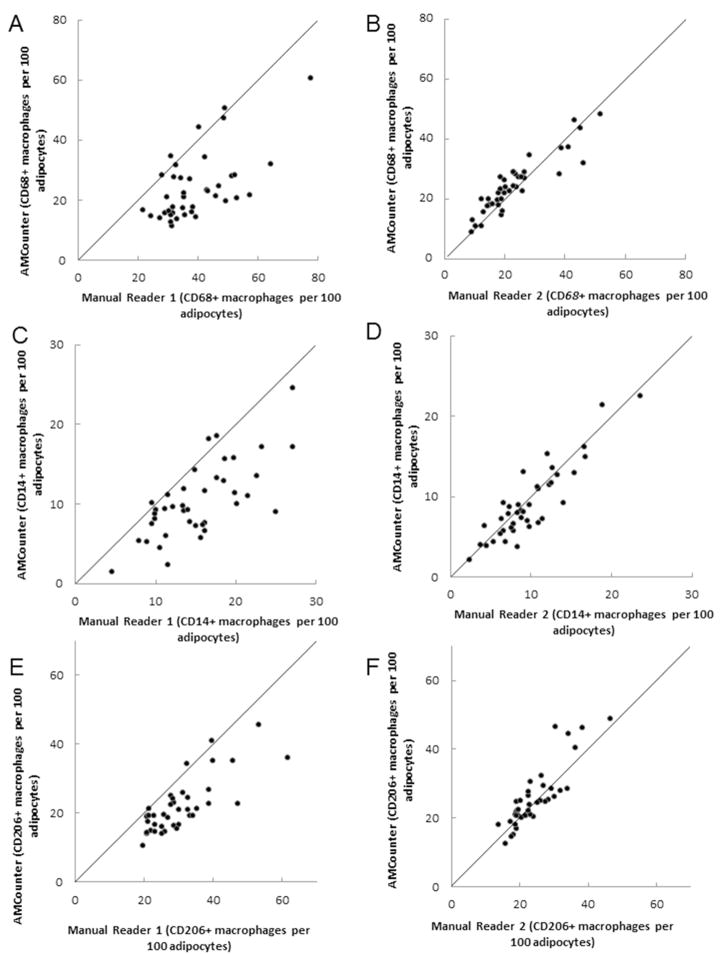
Figure 3.
Agreement between macrophages per 100 adipocytes quantified by manual reading and automated software program reading of immunohistochemistry slides. A.,C., E.: Reader 1, B.,D.,F: Reader 2, A+B: CD68+ macrophages, C., D: CD14+ macrophages, E., F: CD206+ macrophages. Drawn 45° line through the origin indicates perfect agreement.
The between-method comparisons for reader 1 had CCCs of 0.32 for CD68, 0.47 for CD14, and 0.52 for CD206, indicating fair to moderate agreements, respectively. All of the between-method comparisons for reader 2 had CCCs > 0.83 (CD68: 0.90; CD14: 0.88; CD206: 0.83) indicating excellent agreement.
The differences between the methods were plotted against their mean (Figure 3A, C and E). For reader 1 more differences are above the “0” line (4:36, 3:37 and 2:38 for CD68, CD14, and CD206 respectively), implying a tendency of reader 1 to count more cells when counting manually than when using the AMCounter. The patterns of reader 2 indicated that manual counts were very similar to the AMCounter software approach.
The inter-rater ICCs for manual counting were 0.32 for CD68, 0.41 for CD14, and 0.62 for CD206, consistent with the definition of poor between-reader agreement using the manual method (Figure 3A–F). In contrast, the inter-rater ICCs calculated for the AMCounter software were 0.88 for CD68, 0.83 for CD14, and 0.84 for CD206, consistent with the definition of excellent between-reader agreement of the AMCounter approach.
Flow Cytometry - Intra-individual Variance of Total Macrophage Analysis
We tested the reproducibility of total macrophage burden using duplicate abdominal and femoral adipose tissue samples collected from six participants approximately two weeks apart. There were no significant differences in the number of APC-conjugated CD68+ (total ATM’s), CD14+/CD68+ (M1 ATM’s) or CD206+/CD68+ (M2 ATM’s) per g tissue between visit 1 and visit 2 (Table 2).
There were no significant correlations between the duplicates of these measures per gram tissue. However, for replicate data between the first and second biopsies there were statistically significant correlations between the percent of CD68+ cells that stained positive for both CD68 and CD14 and that stained positive for both CD68 and CD206 (Table 2). There was not a significant correlation between the ratio of CD14+/CD68+ (M1 ATM’s) to CD206+/CD68+ (M2 ATM’s) for the first and second biopsies (r = 0.29, P = 0.35)
IHC and Flow Cytometry Comparison
Forty-seven adipose tissue samples (abdominal and femoral) from 24 participants were used to analyze the agreement between the IHC and flow cytometry methods. As presented in Table 2, the estimated numbers of ATM’s were markedly greater with IHC than flow cytometry. There was not a statistically significant association between the number of IHC CD68+ ATM’s/g tissue and the number of CD68+ cells/g tissue measured by flow cytometry (r= 0.20, p = 0.18), or between the IHC CD14+ (r=0.18, p = 0.24) or CD206+ (r=0.04, p = 0.77) ATM’s/g tissue and the number of CD68+/CD14+ and CD68+/CD14+ cells/g tissue, respectively, measured by flow cytometry. Likewise, there were no statistically significant correlations between the IHC and flow cytometry methods for CD14+ percentage of CD68+ ATM’s or for CD206+ percentage of CD68+ ATM’s.
RT-PCR Comparison to IHC and Flow Cytometry
Forty-three adipose tissue samples (abdominal and femoral) from 21 participants were used to analyze the agreement between the RT-PCR, IHC and flow cytometry methods. There were significant correlations between RT-PCR CD68 and IHC CD68 (r= 0.36, p = 0.017), CD14 (r= 0.58, p < 0.0001) and CD206 ATM’s (r= 0.56, p = 0.0001) per 100 adipocytes. These relationships between RT-PCR data and IHC were less strong when IHC data was expressed per gram tissue; CD14 – r = 0.46, p = 0.002, CD206 – r = 0.29, p = 0.05, CD68 – r = 0.13, p = 0.40.
Adipose total macrophage burden per g tissue as assessed by flow cytometry CD68 was also significantly related to RT-PCR CD68+ (r = 0.43, p = 0.004). The macrophage content per gram tissue by flow cytometry of CD68+/CD14+ (r= 0.43, p = 0.004) and CD68+/CD206+ (r= 0.48, p = 0.001) were likewise correlated with RT-PCR CD14 and CD206mRNA.
Discussion
An important aspect of adipose inflammation is the burden of pro- and anti-inflammatory macrophages23-26. In the process of establishing our laboratory standards for the measurement of ATM’s we evaluated the reproducibility and comparability IHC and flow cytometry methods for human tissue. Several issues arose that seem not to have been addressed in the literature. We also tested an image analysis software program that we commissioned in hopes it would reduce the burden of manual image analysis. We found that 1) IHC is reproducible in measuring ATM populations within the same individual; 2) the AMCounter software allows better agreement between readers than the traditional manual read method and saves time; 3) there is not good agreement between the IHC and flow cytometry methods; and 4) the expression of CD68, CD14 and CD206 mRNA in adipose tissue can predict some of the inter-individual variability in CD68+, CD14+ and CD206+ ATM’s measured using either IHC or flow cytometry.
The test-retest differences in IHC ATM measurements for the same individuals biopsied twice with no intervention suggests that IHC quantification of ATM’s is reproducible, albeit tedious and time intensive. The Biomedical Imaging Resources staff developed a semi-automated software approach to quantify the ATM’s that improves the between-reader agreement and reduces time needed from 8–10 min for one slide with 10 pictures for purely manual reading to 5–8 min using AMCounter.
The agreement (CCC) between relatively novice readers was variable using the manual counting method, but much better when using the software program (Figure 3). This difference may relate to the need to manually track the number of adipocytes and positively stained cells on each slide and enter this data into spreadsheets – both tasks are automatically tracked with AMCounter. Another factor may be that different users do not always agree on what constitutes a positively stained, nucleated macrophage when counting manually. This selection bias is unavoidable because different readers count according to their judgment of whether the cell has the morphology of a macrophage, which may also change over time. Thus, the importance of double reading and quality control slides for longitudinal data integrity. The software selects macrophages and adipocytes based on the intensity of color and contrast to the surrounding tissue, which should allow better consistency over time. To examine how many CD68+ cells did not have macrophage morphology we re-reviewed a subset of 20 IHC samples and counted all CD68+ cells, sorting them into macrophage and non-macrophage appearing cells. We found 16.6 ± 5.3 CD68+ macrophages per 100 adipocytes and 9.3 ± 2.7 CD68+ cells that did not have the appearance of macrophages per 100 adipocytes. Providing this is a representative sample, it suggests that the well-known expression of CD68 in other immune cells in adipose tissue can be significant. Finally, the AMCounter program saves a permanent photographic record with inserted circles and a corresponding spreadsheet, allowing investigators to retrieve images and determine the reasons for discrepancies between users.
Our goal was to develop a reliable and reproducible flow cytometry protocol for quantifying ATMs because we didn’t know whether IHC or flow cytometry would better suit our needs. To our surprise, we found poor agreement between the two methods in terms of ATM per gram tissue, although we had only 12 replicate samples and we may have missed a modest correlation. We hypothesize that this lack of agreement is due to the variability in macrophage recovery from tissue during the digestion and processing for flow cytometry. In support of this hypothesis, there was good reproducibility of flow cytometry for measuring the percent of CD68+ cells that were also positive for CD14 or CD206 even with only 12 replicate samples. Another possible explanation for the lack of agreement between IHC and flow cytometry is that the two methods use different antibody clones to detect the CD68, CD14 and CD206 antigens. Unfortunately, the antibodies used for flow cytometry cannot be used for IHC, which prevented us from determining how much of the disagreement was due to this factor. This can’t explain the suboptimal reproducibility of some aspects of flow cytometry, however. An advantage of flow cytometry is the opportunity to quickly analyze a more comprehensive list of immune cells of adipose tissue. It is possible that different gating strategies would improve the agreement between flow cytometry and immunohistochemistry measures of ATM’s. To that end, we provide as supplemental data the .fcs data files and immunohistochemistry data from 20 of the samples we used for comparison so that interested investigators can apply their own flow cytometry approaches to our dataset.
We found positive correlations between RT-PCR measures of CD68, CD14 and CD206 expression and direct ATM measures from both IHC and flow cytometry. This is reassuring in that it suggests all three measures provide some common information. However, even the best RT-PCR association could explain only 34% of the variance in the direct measure of ATM burden. This is not surprising given that cells other than macrophages can express these three receptors. The flow cytometry gating strategy we use to sort for macrophage size, live, single cells excludes other CD68 positive cells. When we re-analyzed some of our data counting all CD68+ cells irrespective of the FSC-A and SSC-A there were ~ 5 times more CD68+ cells than if we counted only live, singlet cells without gating on FSC and SSC. Thus, while RT-PCR may be adequate to distinguish group differences in adipose inflammatory burden, it is probably insufficiently specific to be valuable as a measure of individual differences.
We suggest there is a population of macrophages stain positively for both pro- (CD14+) and anti-inflammatory (CD206+) markers, as indicated by the ratio of CD14+:CD206+ macrophage burden quantified via the flow cytometry. These findings are in agreement with recent studies using immunofluorescent techniques to show that both CD14+ and CD206+ receptors can be present within the same cell 27. In addition, recent studies have also described that both CD14+ and CD206+ macrophages are present within CLS 28, indicating greater inflammation in animal models 29. These findings suggest that the inflammatory story is not as simple as pro- versus anti-inflammatory macrophage burden. It may be that total macrophage burden, regardless of specific receptor make-up, is a good measure of adipose inflammation. Further studies of human samples should be conducted to determine if indeed CD14+ and CD206+ macrophage receptors co-exist within the same cell and/or CLS.
Although our data suggests that IHC is a reliable way to measure ATM’s, and that the AMCounter may be more reproducible and efficient, there are some remaining questions. For example, we don’t know whether our results apply only to adipose tissue that has been obtained via needle aspirate, or whether it would also apply to samples collected via surgical excision. Our data also suggests that flow cytometry measures of ATM’s are better suited to understanding the relative distribution of different kinds of macrophages rather than the total macrophage burden per se. These observations have the potential to help investigators better select their measures of human adipose inflammation depending upon their goals.
Supplementary Material
Study Importance Questions.
What is already known about this subject?
Adipose tissue macrophages represent chronic, sterile inflammation
Adipose tissue inflammation is associated with metabolic disease
Immunohistochemistry, flow cytometry and mRNA of CD68, CD14 and CD206 have been used to measure adipose inflammation
What does your study add?
A software program to assist in measuring adipose macrophages by immunohistochemistry compares well with the manual method and is time-saving
Immunohistochemistry, but not flow cytometry measures of adipose tissue macrophages/g tissue were reproducible.
Flow cytometry, RT-PCR and immunohistochemistry measures of adipose macrophages provide different information.
Acknowledgments
Funding: These studies were supported by National Center for Research Resources Grant1UL1RR024150, National Institutes of Health Grants, DK45343, DK40484, DK50456 and 5T32 DK007352. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We would like to thank the personnel of the Mayo Clinic Clinical Research and Trials Unit, the Mayo Clinic Rochester Pathology Research Cores and the Flow Cytometry and Cell Sorting Microscopy team of the Cell Analysis Core, as well as Ms. Monica Davis for editorial assistance and our volunteers.
Footnotes
Clinical Trials Registration: N/A
Disclosure: Mr. Karwoski is a salaried employee of Biomedical Imaging Resources, Mayo Clinic, Rochester, Minnesota, who developed the AMCounter program. Potential sales of the AMCounter program could reduce the annual license fee Dr. Jensen pays to use AMCounter.
References
- 1.Masoodi M, Kuda O, Rossmeisl M, Flachs P, Kopecky J. Lipid signaling in adipose tissue: Connecting inflammation & metabolism. Biochim Biophys Acta. 2014;1851:503–18. doi: 10.1016/j.bbalip.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 2.Bai S, Sun Q. Macrophage recruitement in obese adipose tissue. Obesity. 2015;16:127–36. doi: 10.1111/obr.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel PS, Buras ED, Balasubramanyam A. The role of the immune system in obesity and insulin resistance. J Obes. 2013;2013:1–9. doi: 10.1155/2013/616193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraakman MJ, Murphy AK, Jandeleit-Dahm K, Kammoun HL. Macrophage polarization in obesity and type 2 diabetes: weighing down our understanding of macrophage function? Front Immunol. 2014;5:470. doi: 10.3389/fimmu.2014.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bargut TC, Mandarim-de-Lacerda CA, Aguila MB. A high-fish-oil diet prevents adiposity and modulates white adipose tissue inflammation pathways in mice. J Nutr Biochem. 2015;26:960–9. doi: 10.1016/j.jnutbio.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Hameed I, Masoodi SR, Mir SA, Nabi M, Ghazanfar K, Ganai BA. Type 2 diabetes mellitus: From a metabolic disorder to an inflammatory condition. World J Diabetes. 2015;6:598–612. doi: 10.4239/wjd.v6.i4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buras E, Yang L, Saha P, et al. Proinsulin-producing, hyperglycemia-induced adipose tissue macrophages underlie insulin resistance in high fat-fed diabetic mice. FASEB J. 2015;29:3537–48. doi: 10.1096/fj.15-271452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q, Cheng X, Zhang D, et al. Tectorigenin Attenuates Palmitate-Induced Endothelial Insulin Resistance via Targeting ROS-Associated Inflammation and IRS-1 Pathway. PloS One. 2013;8:e66417. doi: 10.1371/journal.pone.0066417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng X, Zhu S, Chang S, et al. Protective effects of chronic resveratrol treatment on vascular inflammatory injury in streptozotocin-induced type 2 diabetic rats: Role of NF-kappa B signaling. Euro J Pharmacol. 2013 doi: 10.1016/j.ejphar.2013.10.034. [DOI] [PubMed] [Google Scholar]
- 10.Dou L, Zhao T, Wang L, et al. miR-200s contribute to interleukin-6 (IL-6)-induced insulin resistance in hepatocytes. J Biol Chem. 2013;288:22596–606. doi: 10.1074/jbc.M112.423145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S, Khalyfa A, Wang Y, et al. Sleep fragmentation promotes NADPH oxidase 2-mediated adipose tissue inflammation leading to insulin resistance in mice. Int J Obes. 2014;38:619–24. doi: 10.1038/ijo.2013.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao B, Chen Y, Zhang L, et al. Momordica charantia (Bitter Melon) reduces obesity-associated macrophage and mast cell infiltration as well as inflammatory cytokine expression in adipose tissues. PloS One. 2013;8:e84075. doi: 10.1371/journal.pone.0084075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Beek L, Lips MA, Visser A, et al. Increased systemic and adipose tissue inflammation differentiates obese women with T2DM from obese women with normal glucose tolerance. Metabolism. 2014;63:492–501. doi: 10.1016/j.metabol.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Heilbronn L, Campbell L. Adipose tissue macrophages, low grade inflammation and insulin resistance in human obesity. Curr Pharm Des. 2008;14:1225–30. doi: 10.2174/138161208784246153. [DOI] [PubMed] [Google Scholar]
- 15.Tchoukalova YD, Koutsari C, Karpyak MV, Votruba SB, Wendland E, Jensen MD. Subcutaneous adipocyte size and body fat distribution. Am J Clin Nutr. 2008;87:56–63. doi: 10.1093/ajcn/87.1.56. [DOI] [PubMed] [Google Scholar]
- 16.Aron-Wisnewsky J, Tordjman J, Poitou C, et al. Human adipose tissue macrophages: m1 and m2 cell surface markers in subcutaneous and omental depots and after weight loss. J Clin Endocrinol Metab. 2009;94:4619–23. doi: 10.1210/jc.2009-0925. [DOI] [PubMed] [Google Scholar]
- 17.Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–46. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- 18.Płóciennikowska A, Hromada-Judycka A, Borzęcka K, Kwiatkowska K. Co-operation of TLR4 and raft proteins in LPS-induced pro-inflammatory signaling. Cell Mol Life Sci. 2015;72:557–81. doi: 10.1007/s00018-014-1762-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eto H, Ishimine H, Kinoshita K, et al. Characterization of human adipose tissue-resident hematopoietic cell populations reveals a novel macrophage subpopulation with CD34 expression and mesenchymal multipotency. Stem Cells Dev. 2013;22:985–97. doi: 10.1089/scd.2012.0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michaud A, Pelletier M, Noël S, Bouchard C, Tchernof A. Markers of macrophage infiltration and measures of lipolysis in human abdominal adipose tissues. Obesity. 2013;21:2342–9. doi: 10.1002/oby.20341. [DOI] [PubMed] [Google Scholar]
- 21.Purdue Cytometry List. 2016 http://www.cyto.purdue.edu/hmarchiv/index.htm.
- 22.Altman DG, Bland JM. Measurement in medicine: the analysis of method comparison studies. The Statistician. 1971;221:850–92. [Google Scholar]
- 23.Bigornia SJ, Farb MG, Mott MM, et al. Relation of depot-specific adipose inflammation to insulin resistance in human obesity. Nutr Diabetes. 2012;2:e30. doi: 10.1038/nutd.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harman-Boehm I, Bluher M, Redel H, et al. Macrophage infiltration into omental versus subcutaneous fat across different populations: effect of regional adiposity and the comorbidities of obesity. J Clin Endocrinol Metab. 2007;92:2240–7. doi: 10.1210/jc.2006-1811. [DOI] [PubMed] [Google Scholar]
- 25.Lumeng CN, Deyoung SM, Bodzin JL, Saltiel AR. Increased inflammatory properties of adipose tissue macrophages recruited during diet-induced obesity. Diabetes. 2007;56:16–23. doi: 10.2337/db06-1076. [DOI] [PubMed] [Google Scholar]
- 26.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prieur X, Mok CYL, Velagapudi VR, et al. Differential lipid partitioning between adipocytes and tissue macrophages modulates macrophage lipotoxicity and M2/M1 polarization in obese mice. Diabetes. 2011;60:797–809. doi: 10.2337/db10-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haase J, Weyer U, Immig K, et al. Local proliferation of macrophages in adipose tissue during obesity-induced inflammation. Diabetologia. 2014;57:562–71. doi: 10.1007/s00125-013-3139-y. [DOI] [PubMed] [Google Scholar]
- 29.Eguchi A, Feldstein AE. Adipocyte cell death, fatty liver disease and associated metabolic disorders. Dig Dis. 2014;32:579–85. doi: 10.1159/000360509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.