Abstract
Understanding how and why animals regenerate complex tissues has potential to transform regenerative medicine. Here we present an overview of genetic approaches that have recently been applied to dissect mechanisms of regeneration. We describe new advances that relate to central objectives of regeneration biolsts researching different tissues and species. These objectives include defining the cellular sources and key cell behaviors in regenerating tissue; elucidating molecular triggers and brakes for regeneration; and defining the earliest events that control the presence of these molecular factors.
Keywords: Regeneration, genetics, blastema, imaging, zebrafish, salamanders
1. Introduction
The ability of animals to replace injured body parts has been a subject of fascination since ancient times (25). In Greek mythology, the second labor of Hercules included a famous battle with Hydra, a mythical serpent that could regenerate any of its many heads after amputation. Millennia later in the mid-1700s, the Swiss scientist Abraham Trembley described the remarkable capacities of invertebrate polyps to regenerate their heads or feet in a laboratory setting, fittingly referring to these animals as “Hydra” (25). In this same era of scientific curiosity, the Italian scientist Lazzaro Spallanzani reported for the first time that certain vertebrates such as salamanders can regenerate complex tissues like limbs and tails. He also documented how regeneration can be affected by the severity or type of injury, or by environmental factors. For instance, regeneration of a near-whole salamander hindlimb occurred in a similar timeframe as regeneration of an amputated digit of the same animal (116). This principle holds true during regeneration of goldfish fins, identified by Pierre Broussonet at the end of the 18th century, and elaborated upon by Thomas Hunt Morgan at the beginning of the 20th century (10; 77; 88; 116). Observations like these were made centuries ago, yet it is intriguing that many questions Spallanzani and Morgan identified in their time remain unanswered and are under pursuit in this modern era of regeneration research.
Though widespread and presumably advantageous (8), high regenerative capacity is not universal, and, diversity in this trait can exist even among species that otherwise have many similarities. For instance, whereas planarians are famous for their ability to regenerate whole animals from tiny fragments, certain platyhelminthes cannot regenerate their heads after amputation and die from the sequelae (68; 109; 128). Similarly, the capacity for skin regeneration has evolved differently between Mus musculus (representing mouse strains common in laboratories) and African spiny mouse species, the latter able to regenerate large areas of skin shed in escape mechanisms (108). One possible explanation for these disparities is that regenerative capacity is an adaptive trait, but it might be less associated than other traits with overall reproductive fitness. For instance, rapid scarring mechanisms and custom regulation of tumor suppressor genes in certain tissues might contribute greatly to overall fitness, whereas optimized mechanisms for generating a tissue replicate might not (90). Interestingly, regenerative capacity changes during development and progression through life stages. For instance, fetal and newborn mice are better able than adults to regenerate complex tissues like the heart (91). An intriguing notion is that most or all species have maintained the genetic machinery that effects tissue regeneration, but not the mechanisms to retain developmental competence and positional information, or to activate expression of key regulatory factors, after major injury to certain tissues (54; 68).
In recent years, most attention in the domain of regenerative medicine was directed toward the therapeutic potential of transplanted stem and progenitor cells. However, it is becoming clear that transplanted cells have limitations in what they can provide, and they are not applicable for many tissues. Moreover, as most scientists feel that the most effective therapies of the future will be molecular - stimulating regeneration de novo from spared tissue – not cellular, animals and tissues with high regenerative capacities provide blueprints for successful innate tissue renewal. Therefore, elucidating the mechanisms of successful (and also failed) innate regenerative events in multiple contexts should inspire new clinical strategies.
An onslaught of recent studies in a variety of laboratory animals has provided exciting mechanistic insights into regeneration. It is evident that the accessibility of genetic tools has been a primary driver for these advances. For instance, new transgenic mice, axolotls, and zebrafish have been employed to determine the sources of new cells in regenerating tissues (122). In addition, genome-wide profiling, which can be combined with new genome-editing technologies, has uncovered novel factors and concepts during tissue regeneration (13; 29; 79; 94). In this review, we focus on some of the central questions in tissue regeneration research, what has been learned recently to address these questions, and how genetic strategies have enabled these studies. We discuss a handful of animal model systems and tissue types sampled from a broad and growing encyclopedia of discoveries.
2. Genetic approaches to monitor cell behavior during regeneration
Over the past decade, major advances have been achieved in our understanding of which cellular sources are activated upon injury to give rise to new tissues during regeneration (121; 124). This line of research is highly mechanistic, as cell-level resolution is necessary to interpret possible molecular players, and clear answers yield the target cells for possible therapies.
Cell labeling strategies and source determination
Before contemporary genetic tools became accessible in model systems employed for regeneration, key experiments involved attempts to transiently label cells in situ with fluorescent dyes or electroporated DNA constructs, or to transplant exogenously labeled cells or tissues. For instance, these approaches generated a model for formation of the blastema, a mass of proliferating cells, during salamander limb regeneration. The results indicated that skeletal myofibers fragment into mononuclear cells that are progenitors for multiple new tissues (27; 70). With the onset of transgenesis in the axolotl species, models for regeneration were refined. By transplantation of green fluorescent protein (GFP)-expressing cells from transgenic donors into unlabeled hosts, Kragl et al. identified that the blastema is a heterogeneous collection of proliferating cells with restricted cell fates. For example, new, regenerated skeletal muscle derives from spared skeletal muscle, which makes little or no contribution to other tissue types during regeneration (59).
To avoid the potential artifacts that transplantation can cause, intricate genetic approaches now enable direct tagging and observation of specific cell types in their natural habitat during regeneration. The most commonly used strategy employs a cell type-restricted Cre recombinase in a transgenic line, with activity that can be controlled by the estrogen analog tamoxifen. When paired with a transgene that cages a fluorescent reporter cassette downstream of a transcriptional stop sequence flanked by loxP recognition sites for Cre, one can induce permanent labeling of a specific cell type and then trace its progeny during regeneration. Cre recombinases were originally discovered in bacteriophages (2) and have provided a windfall for a variety of genetic manipulations in vertebrate systems, such as conditional gene knockout and conditional gene expression (80). Genetic fate-mapping studies using tools like this have together supported a theme in which appendage regeneration in the salamander limb, fish fin, or mouse digit tip occurs through proliferation of lineage-restricted cells (57; 63; 97; 110; 115; 118). Intriguingly, species that are considered closely related may have alternative routes to regenerate a tissue like skeletal muscle during limb regeneration. Sandoval-Guzman et al. found recently that, whereas newts appear to utilize dedifferentiation and fragmentation of myofibers to create a progenitor pool, axolotls likely mobilize a muscle stem cell population that is analogous to mammalian satellite cells (104).
There are now many molecular genetic tools available for precise genetic ablation of a specific cell type. These include strategies for spatiotemporal release of the cytotoxin diphtheria toxin A (9; 131), genetically triggering apoptotic mechanisms, or targeted expression of a bacterial nitroreductase that can convert the non-toxic, external substrate Metronidazole to a potent toxin (22; 89). For instance, in regenerating zebrafish fins, which go through a blastema stage like planarian heads or salamander limbs, several Cre-based fate-mapping studies found that new bone-depositing osteoblasts are restored by the proliferation of spared osteoblasts (57; 110; 115; 118). However, Singh et al. ablated the osteoblast population animal-wide using the nitroreductase methodology, and found that new osteoblasts in zebrafish could rise from cryptic sources to support bone regeneration at a normal pace (110). Other analogous examples of source plasticity have been demonstrated by experiments like these across numerous species and tissues. For instance, mice are able to restore insulin production after extreme β-cell ablation, not by β-cell division as typical (26), but by differentiation from other sources like glucagon-producing α-cells (123). The age of the animal has also been reported to affect choices of cellular source after β-cell ablation. In juvenile animals, pancreatic δ-cells can proliferate and later become insulin producers like β-cells (17). Thus, genetic ablation and fate-mapping strategies can reveal alternative cellular mechanisms for tissue regeneration if the typical source mechanism is disrupted.
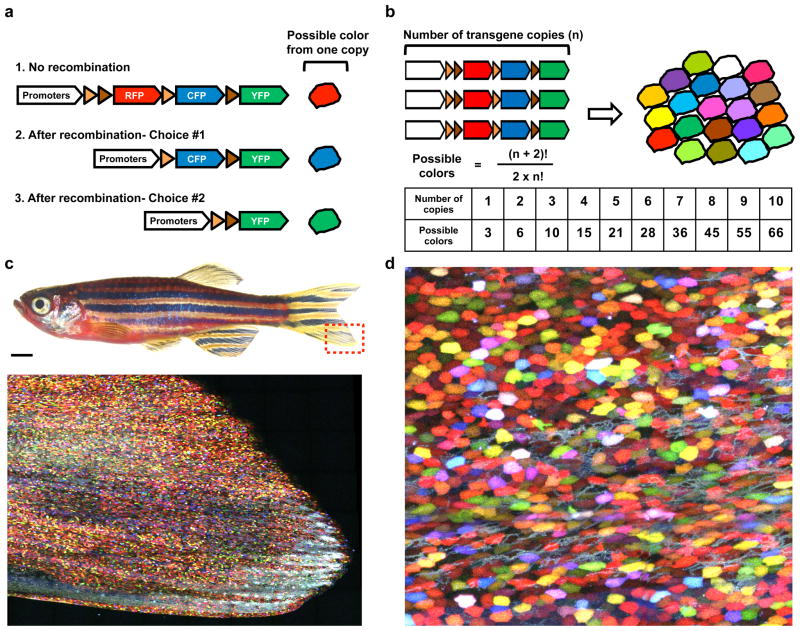
Clonal analysis
In addition to genetically labeling entire populations of specific cell types with a single color, strategies exist to examine the contributions of single cells in regenerating tissues. Limiting cell tagging to rare events can enable striking inferences that are impossible to assay in entire labeled populations. These include the expansion capacity of single stem cells, whether determination of contributions by cells in a population is stochastic or hierarchical, and how the spatial characters of a tissue are collectively established through the proliferation of individual cells (24; 101). Multicolor cell tagging strategies can be even more powerful. Livet et al. developed Cre-loxP-based genetic tool methodology, called Brainbow, to simultaneously assign unique color barcodes to many individual cells in a population (69). Brainbow uses distinct, paired loxP sites, to enable a random choice of expression of 3 different fluorescent protein cassettes upon induced Cre recombination. Transgene concatemerization can lead to many copies integrated at a single locus, which greatly expands the color selection possibilities of each cell in a population (Figure. 1). Brainbow was first exploited to distinguish nerve cells from each other in complex brain tissues. More recent studies have used Brainbow-based systems to retrospectively assess the contributions of several cells together during tissue regeneration. For instance, heart regeneration in zebrafish occurs by proliferation of cardiac muscle cells (51; 56), and a priZm reporter found that there does not appear to be a hierarchy in contributions to regeneration by these source muscle cells (39). Multicolor cell fate-mapping was also applied to study Lgr5-positive stem cells in the intestinal epithelium (5). A Brainbow-based Confetti mouse line was used to visualize that stem cell progeny at the crypt base drift toward clonality, that is, derivation from a single stem cell, over long periods (113). Thus, multicolor cell labeling enables the experimenter to elucidate mechanistic insights that are technically challenging to gain from single color cell-tagging strategies.
Figure 1.
Transgenic multicolor approach to visualize entire cell populations in live animals. (a) Schematic drawing of a Brainbow cassette. Each copy of a Brainbow cassette can result in one of three distinct colors after limited Cre-mediated recombination. In principle, the color choice is stochastically determined. (b) A high-copy number of Brainbow cassette can provide more color choices. For instance, transgenic animals with five Brainbow cassettes can theoretically generate twenty-one different colors to barcode cells of interest. (c) Brightfield view of an adult skinbow transgenic zebrafish. A red dashed-box indicates areas where the z-stacked confocal image shown below was captured. An entire population of skin epithelial cells is multicolor-barcoded. Scale bar, 1 mm. (d) A high-magnification view of skin surface cells in a skinbow zebrafish.
Live cell imaging during regeneration
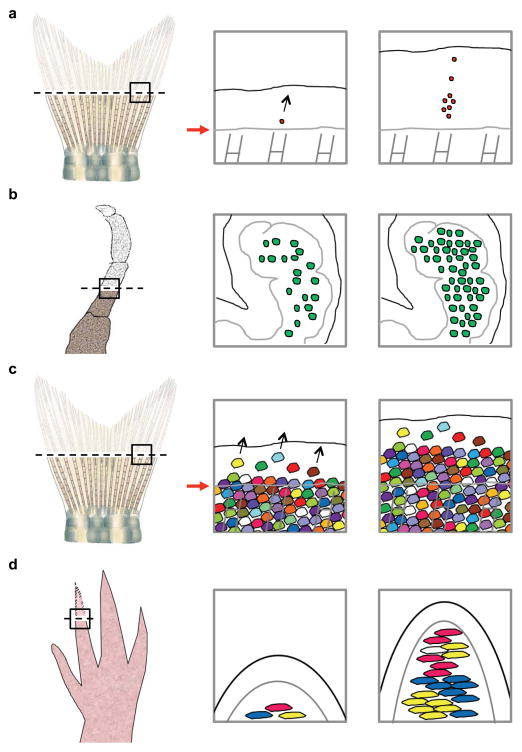
In vivo imaging platforms enable the direct visualization of intricate cell activities in live regenerating tissues (Figure 2) (4; 16; 23; 125). This has been effectively employed to visualize the behaviors of labeled cells. For instance, Tornini et al. recently performed longitudinal tracking of Cre-labeled single blastemal cells and their progeny sets in hundreds of regenerating fin rays, mapping out the contributions of each blastemal cell over entire regeneration events (Figure 2a). One observation from this study was that the numbers of progeny derived from individual blastema cells are surprisingly variable, representing a spectrum from no division to dozens of division events. The distribution of progeny within the regenerates was also heterogeneous, although data analysis indicated that the position within the blastema is predictive of the proximodistal (PD) contributions made to regeneration (125). Images were acquired every 12 hours to 3 days for this study, yet in contexts where cell behaviors are highly dynamic, continuous live imaging over long-time periods is preferred over repetitive snapshot imaging. However, keeping anaesthetized animals alive during the entire course of regeneration is technically challenging as the process can take days to weeks to complete. Notably, using a crustacean model Parhyale hawaiensis, Alwes et al. achieved continuous live imaging of individual epidermal cells over the first 96 hours after leg amputation (4) (Figure 2b). To cover the surface of a newly grown leg, continuous tracking of ~54 cells reveals that most epidermal cells on the blastema are progenitors capable of dividing on a small scale. One interesting observation was that there appears to be a sharp transition from a quiescent phase to an active phase where many cells start to divide, an unexpected finding that might not have been captured by repetitive snapshot imaging.
Figure 2.
Live cell imaging during regeneration. (a) Live imaging of single blastemal cells in regenerating fin tissue. Through tracking hundreds of permanently labeled single blastemal cells over entire regeneration events, the progeny sizes and distributions from these cells were found to be highly variable, ranging from no division to populating the entire distal-proximal axis of the regenerate. (b) Live imaging of nuclear-tagged epidermal cells in a regenerating crustacean leg. Through continuous live imaging of many cells over several days, the behaviors of epidermal cells covering the blastema were found to be highly coordinated. After a quiescent phase, many cells simultaneously start to divide on a small scale. (c) Live imaging of skin epithelial cells in regenerating fin tissue. Through direct tracking of hundreds of skin epithelial cells during fin regeneration, pre-existing, post-mitotic skin cells were found to travel long distances across the amputation plane. (d) Live imaging of connective tissue cells in a regenerating axolotl digit tip. Through tracking several cell types in the connective tissue, cells sources that migrate and contribute to the blastema were unambiguously identified. Black-dashed lines indicate anatomic sites of amputation in each system. Black arrows indicate direction of cell migration. Red arrows indicate plane of amputation
Multicolor platforms have the potential to be highly informative for live imaging during regeneration. Instead of tracking one cell and its progeny, the ability to assign many color tags enables monitoring of many cells in a population and their collective behaviors. With Brainbow-based genetic technology, Chen et al. recently developed a transgenic line, referred to as skinbow, to simultaneously monitor individual superficial epithelial cells (SECs) in a large population during tissue homeostasis and regeneration (16) (Figures 1 and 2c). Live imaging of skinbow surface tissue allows tracking of hundreds of cells over long periods of time, permitting acquisition of epithelial cell size, mobility, and cell-cell interactions during cell turnover and injury-induced regeneration. Moreover, pre-existing SECs and de novo emerging SECs could be identified. For example, major injuries like fin amputation induced massive new cell creation, whereas minor injuries such as exfoliation recovered through accelerated differentiation (16). In an analogous study, transgenic “Limbow” axolotls were employed to define cell behaviors in the connective tissue of amputated digit tips (23) (Figure 2d). Currie et al. found that cells from different connective tissue compartments behave in distinct manners. For instance, cells from chondrocyte compartments proliferate but do not migrate into the regenerate, whereas fibroblasts residing within 50–500 μm below the amputation plane can migrate above the plane. Periskeletal cells and fibroblasts in connective tissues are major contributors to form the blastema, a dynamic process influenced by Platelet-derived growth factor signaling (23). Not surprisingly, similar to axolotl digit regeneration, blastema formation during zebrafish tailfin regeneration also occurs by a definable zone of cell recruitment (125). Only cells that are located within 200– 300 μm below the amputation plane are able to contribute to the blastema. Thus, tracking at single cell resolution during regeneration can yield quantitative information about regeneration that is of great biological importance.
3. Identification of molecular factors required for regeneration
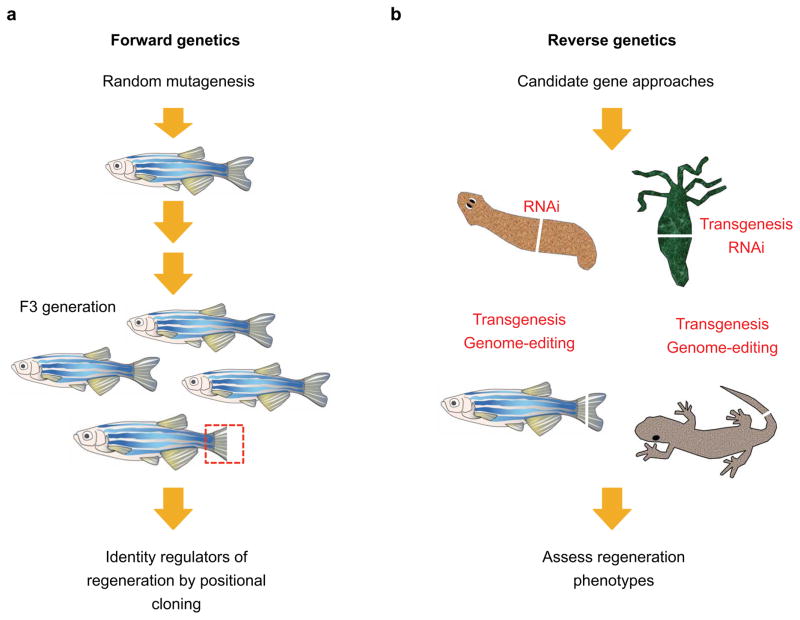
Embryologists are accustomed to studying molecular factors and their mechanisms of action in great detail, owing to vast experimental toolboxes. By contrast, regeneration biologists have traditionally lacked this level of precision. One of the main challenges for regeneration research is that the familiar model systems, such as planarians, salamanders, and fish, have been late in acquiring genetic approaches compared with others like fruit flies, nematodes, and mice. Much progress has been made in recent years, and the field will continue to be propelled by the increasing accessibility of genetic tools in model systems from the genome editing revolution. We discuss recent studies that have identified molecular regulators of regeneration and highlight the responsible genetic tools (Figure 3).
Figure 3.
Forward and reverse genetic approaches to identify regulators of regeneration. (a) Forward genetic screens in the zebrafish system. Random genetic mutations are induced by N-ethyl-N-nitrosourea (ENU) treatment. Homozygous recessive mutations that affect tailfin regeneration can be screen in the F3 generation. Notably, screens in zebrafish can be carried out to identify temperature-sensitive mutant alleles. (b) Candidate gene approaches in highly-regenerative animal models. Familiar model systems in regeneration like such as planarians, hydra, salamanders, and zebrafish have relatively recently received access from genetic approaches. Much progress has been made in recent years, and the field is propelled by the increasing accessibility of genetic tools in each model systems.
Forward genetics in regeneration research
Forward genetics is an unbiased approach to identify genes based on phenotypes. Once robust methods to perform random mutagenesis and phenotypic screening are established, the search for key regulatory factors is unbiased and requires no prior knowledge or hypothesis about the molecular nature of the phenotype. A foundation for genetic approaches to tissue regeneration was pioneered by Hadorn, Schubiger, Bryant, and others, who studied imaginal disc regeneration in Drosophila larva (11; 40; 105). Regenerative growth of imaginal discs is driven by a defined zone of cell proliferation (3; 55), reminiscent of the proliferative blastema that is a landmark feature of appendage regeneration in vertebrate model organisms. This line of study is still bearing fruit today with new injury models and mutagenesis strategies (7; 74; 106; 112). In vertebrate species, the first forward genetic screen for tissue regeneration was carried out in 1995 by Johnson and Weston using zebrafish, which have relatively short generation times and robust chemical (ENU) mutagenesis protocols (50; 78; 114). This screening strategy attempted to find mutations that block regeneration of amputated tail fins. As mutations that affect regeneration are likely to have additional roles during animal development, the investigators screened for temperature-sensitive alleles that would conceivably bypass the lethal effect of the mutation during early development at a permissive temperature (e.g. 25 degrees) but allow a regeneration phenotype to manifest a restrictive temperature (e.g. 33 degrees) (50). These screens have the additional benefit of generating conditional mutations that could allow for functional tests at any stage in development.
Remarkably, there have been only five genes from such screens identified by positional cloning of the responsible mutation. The first gene, mps1, a mediator of centrosome duplication and the spindle checkpoint and that is upregulated in proliferating blastemal cells, was identified in 2002 (93), and other screens have identified the fibroblast growth factor ligand gene fgf20a (133), the protein-folding chaperon hsp60 (73), the protein-trafficking gene sly1 (81), and the extracellular matrix component gene lamb1a (15). Among these genes, identification of fgf20a is exciting for several reasons. First, the regeneration phenotype found in fgf20a mutants is not entirely temperature-sensitive. The mutants display no embryonic phenotypes despite a predicted null allele. These findings allow one to speculate that a primary function of fgf20a is regeneration, and that its regulation and function may have in part been selected during evolution to support regeneration (see below). Also, as a signaling ligand that is induced early upon amputation injures, fgf20a appears to act very early in the regenerative process. Interestingly, one of the roles of fgf20a appears to be in establishing an epithelial-mesenchymal signaling interface, which includes induction of lamb1a to establish the basal epithelial layer (15).
To improve the productivity of forward genetics screens as discovery tools for fin regeneration, several technical challenges must be addressed. First, maintaining hundreds to thousands of aquarium tanks for screening and mapping over several years is a barrier for many labs. Successful implementation of a three-generation mutagenesis screen for adult phenotypes requires infrastructural support and careful planning. Second, positional cloning of causative mutations at the adult stage is another rate-limiting step - a process that is both labor-intensive and time-consuming. Clearly, efficiency leaps are needed to increase the discovery rate of regeneration genes from isolated mutant families. We expect recent advances in high-throughput sequencing-based mapping, zebrafish genome assembly, web-based bioinformatic tools for SNP analysis, and efficient genome-editing tools will contribute significantly to augment this classic genetic approach (41; 42; 45; 65).
Reverse genetics in regeneration research
With the advent of sequenced genomes and genome editing tools, potential mediators of regeneration can be examined by targeted perturbation of specific genes. Gene inhibition via RNA interference (RNAi) is a routine approach in planarians and hydra to deplete transcripts (71; 82; 103). By feeding planarians with bacteria that express double-stranded RNA targeting specific genes, multiple screens have been conducted for molecular factors involved in regeneration, tissue maintenance, or cell fate determination (31; 96; 98; 132). Planarians have great potential to answer questions in stem cell biology, as a single multipotent stem cell called a neoblast can replenish all necessary cell types upon transplantation to a lethally irradiated host planarian (130). Recent single-cell transcriptome analyses have revealed that neoblasts comprise a heterogeneous population with distinct molecular profiles and functions (107; 129). Additionally, through characterization of a panel of genes implicated in positional control, a subepidermal layer of cells with muscle cell markers was found to affect the process by which pluripotent neoblasts become specified as eye progenitors during head regeneration (134). Transgenesis and clustered regularly interspaced short palindromic repeats (CRISPRs) technology have proven more difficult to establish in common flatworm species used for regeneration studies. This has delayed exciting discoveries; for instance, experiments to visualize by live imaging the contributions of a single transplanted transgenic neoblast to whole-animal regeneration.
In adult salamanders and zebrafish, transient inhibition of gene activity can be achieved by electroporation of antisense morpholinos into adult tissues. For instance, this approach recently indicated functions for a novel MARCKS-like protein sufficient to activate cell proliferation during axolotl limb and tail regeneration (120). Although accessing gene function in vivo using antisense morpholinos is quick and inexpensive, their off-target effects have drawn high-profile concern (58; 100; 117). As an alternative method to study gene function during regeneration, many groups achieve precise, spatiotemporal control of gene activity through several transgenesis strategies. For instance, using the Cre-loxP system, effects of gene overexpression can be assayed in specific cell types like cardiomyocytes at adult stages (35; 41). To block gene activity, one can choose to manipulate expression of dominant-negative cassettes during regeneration using either the Cre-loxP system or a heat shock-inducible promoter (39; 62). In one recent study, Ablain et al. combined transgenesis and CRISPRs technology in zebrafish to achieve tissue-specific gene disruption. Using a transgenic cassette that co-expresses sgRNA and Cas9 in specific cell types, gene knockouts were obtained for loss-of-function studies (1).
CRISPR/Cas9 technology allows the generation of targeted mutations to study gene function in zebrafish or salamanders during regeneration. For instance, through a candidate gene approach, Fei et al. reported that genomic deletion of Sox2, a SRY-related high-mobility group box transcription factor, has no impact on the development of axolotls (29). Interestingly, upon tail amputation in adult animals, spinal cord regeneration was affected, supporting a preferential role during regeneration. Similarly, mutations in the extracellular ctgfa allow zebrafish to develop grossly normally to adulthood, but block regeneration of the spinal cord after it is severed (76). Mutant phenotypes for fgf20a, Sox2, and ctgfa are consistent with the idea that some genes might be evolutionally preserved in certain species for their roles in adult tissue regeneration (29; 133). However, it is equally or more possible that adult contexts like regeneration lack buffering by genetic compensatory mechanisms that are present in during early development (100). Rapid expansion in the CRISPRs/Cas9 toolkit and their ease of use in a wide range of traditionally “non-genetic” model systems are likely to increase the catalog of mutant phenotypes that preferentially affect regeneration versus embryogenesis (37).
Applying reverse genetics to study regeneration has some limitations. First, as alluded to earlier, animals harboring null mutations in candidate genes with essential roles during early development would never survive to adulthood to enable regeneration assays. More sophisticated genetic strategies, such as generating conditional knockouts or inducible dominant-negative transgenes in adult animals, are thus essential to access tests of function in regeneration. Second, reverse genetics often relies on an educated guess of the gene function based on prior knowledge or its known expression domains within a tissue. As a candidate-driven approach, reverse genetics can overlook essential, unsuspected regulators of regeneration.
4. Early signals for regeneration
Tissue damage like amputation somehow activates regeneration programs to produce a replicate. Answering the question of “What is the earliest signal for regeneration?” is technically challenging, but recent studies have identified new concepts and mechanisms to begin to address this deficit.
Injury signals and initiation of regeneration
Within hours of limb or fin amputation, the stump is covered by a thin layer of epithelium that soon synthesizes growth factors (92). In zebrafish fins, epithelial cell sheets migrate at different velocities corresponding to ray and interray tissue, covering the fin stump within 30–60 minutes (16). Mechanical properties of epithelial cells are likely to mobilize epithelial cell sheets (16), but the molecular basis of their differential motility and its significance are unclear. Studies of wound healing responses in zebrafish larvae and Drosophila embryos indicate clues to early molecular responses (83; 95). At the onset of wounding, initial tissue damage is thought to be detected by osmotic surveillance (28; 33). Instant activation of calcium signaling in spared cells may mediate activation of an NADPH oxidase to produce reactive oxygen species (ROS) (95), providing additional injury signals to reinforce wound detection (84; 136).
Injury causes clotting and inflammation, and there are many recent studies that report evidence for pro-regenerative roles of immune cells like macrophages and T-cells (12; 67; 87; 135). Among several known early wound-released signals (83), the production of hydrogen peroxide along the wound edge can recruit leukocytes to sites of tissue damage by activation of the redox sensor Lyn, a Src family kinase (84; 137). In addition to transient activation at the early phase of wounding (< 2 hours) for recruiting immune cells, ROS remain elevated at the wound edge for days in both zebrafish and Xenopus models (34; 72). This suggests that local tissue-derived ROS, including hydrogen peroxide, might have additional roles during tissue regeneration. Indeed, prolonged perturbation of ROS production through pharmacological approaches can impair tissue regeneration in amputated finfolds of zebrafish larvae, adult zebrafish tailfins, Xenopus tadpole tails, and gecko tails (34; 72; 136; 138). Reported targets of ROS include apoptotic events and activation of JNK signaling, which activate several pathways such as Wnt/β-catenin and Fgf signaling for tissue regeneration (34).
Cell death contributes to a vigorous regenerative response in multiple species and contexts, including salamander retinae, Xenopus tails, planarian heads, and Hydra heads (18; 34; 44; 53; 85; 126). How do dying cells trigger regeneration? Multiple studies provide evidence that apoptotic cells can release mitogens that directly activate cell proliferation in spared tissues and facilitate tissue regeneration (32; 86; 102). For instance, apoptotic cells at the amputation site can release Wnt3 ligand to activate cell proliferation during Hydra head regeneration (18). These apoptotic events are thought to be triggered by the MAPK/CREB pathway (19), consistent with the known role of ROS as a potential trigger of cell death through MAPK/JNK signaling (52). Notably, for tissues with a high demand for regeneration, apoptotic cells are integral parts of normal tissue homeostasis. In the Drosophila midgut, apoptotic enterocytes can release cytokines to activate proliferation of local stem cells, which differentiate to replenish dying enterocytes. Both Jak/Stat and EGFR/Ras/MAPK signaling pathways are key to regulate this delicate feedback system (47–49).
In planarians, pro-regenerative apoptotic events can also be regulated by bioelectric signals (6). In any living cells, not just neuronal and muscle cells, endogenous bioelectricity (i.e. transmembrane voltage potential) is generated and maintained by specific ion channels and pumps within cell membranes. Manipulating ion current across cell membranes is sufficient to trigger tail regeneration in Xenopus tadpoles during the so-called, non-regenerative refractory period (127). Although bioelectricity has long been implicated in regeneration (66), the molecular mechanism by which endogenous bioelectrical signals are triggered upon injuries remain elusive. A recent study by Ferreira et al. suggests that ROS may act upstream to modulate early bioelectrical activities in amputated tails of Xenopus tadpoles (30). Upon pharmacological inhibition of the ROS source (i.e. NADPH oxidase), multiple bioelectrical features like trans-epithelial membrane potential and electrical current densities are disrupted, coinciding with defects in regeneration. Intriguingly, short-term supplement of hydrogen peroxide can consistently modulate bioelectrical activities of the tissue through activation of sodium channels, and it can trigger tail regeneration to occur during the refractory period (30). The downstream targets that bioelectrical signals might regulate remain to be identified (30; 127). Thus, early injury signals like hydrogen peroxide may have multiple roles during regeneration, e.g. as a chemical attractant to leukocytes and as a molecular trigger to activate regeneration by modulating bioelectrical activities.
Activation of gene regulatory elements for regeneration
To interpret regenerative mechanisms, it is crucial to consider how regeneration programs are triggered at the level of DNA sequences by early injury signals. Two decades of research into gene regulatory elements has highlighted the involvement of enhancers, short DNA sequences that engage with transcription factors and gene promoters to control gene expression (61). Recent studies indicate that there are enhancer elements that preferentially or specifically activate gene expression in the contexts of injury and tissue regeneration. For instance, activation of embryonic gene expression in the epicardial cell layer of the heart is a hallmark of the cardiac injury response in zebrafish or mouse (64; 111). To identify responsible enhancer elements, Huang et al developed a mouse heart explant culture system for screening conserved putative enhancer elements linked to epicardial marker genes (43). Two short enhancer elements were identified that helped pinpoint C/EBP transcription factors as important for epicardial gene expression in developing mouse embryos and in the injured adult heart. This study provides a molecular basis to explain how some epicardial genes that are transcriptionally activated during embryonic development can be re-induced after injuries; that is, by sharing regulatory sequences. Similarly, by deleting different genomic regions surrounding mouse Bmp5, essential for skeletal development and bone repair, Guenther et al identified separable genomic regions responsible for Bmp5 expression in discrete anatomic domains during normal development or following injuries (38). Intriguingly, the 18-kb injury-responsive region is sufficient to trigger gene expression in mesenchymal or epithelial cells in multiple tissues, suggesting it might contain an injury-responsive enhancer element.
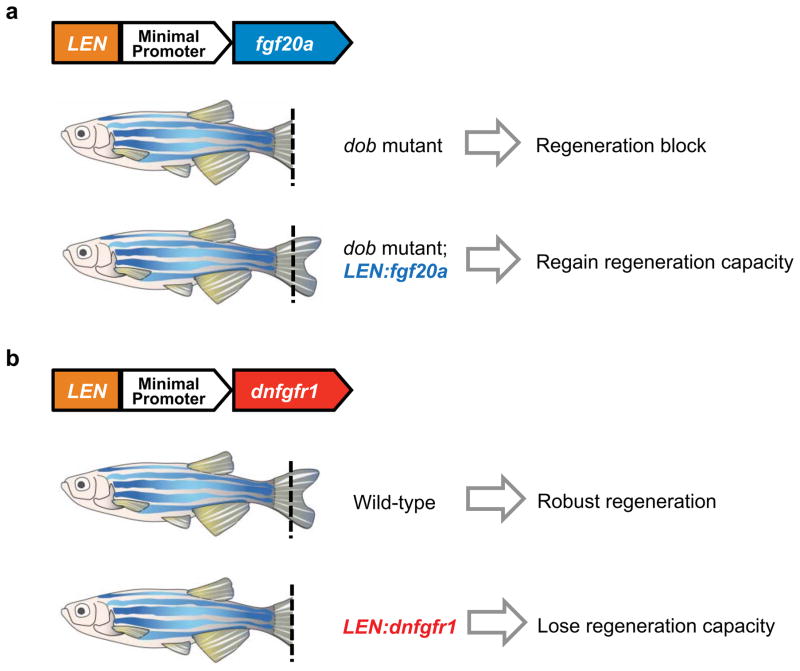
Recent reports indicate that enhancer elements preferential or specific to regeneration may be widespread, and that several genes with induced expression during regeneration have nearby DNA elements with features expected of regeneration-activated enhancers (Figure 4) (36; 54). These elements, coined “tissue regeneration enhancer elements”, or TREEs, were identifiable by chromatin profiling of uninjured and regenerating zebrafish heart tissue. For example, short sequences upstream of leptin b (lepb), a zebrafish ortholog of mammalian Leptin, acquire a mark of open chromatin typically found at enhancers, the acetylated lysine 27 of Histone H3, specifically in regenerating tissue. This sequence, combined with a minimal promoter, was sufficient to direct expression of reporter transgenes to injured fins and hearts (54). Interestingly, the lepb-linked enhance sequence could also be employed to control expression of a gene encoding a dominant-negative of fgfr1 (dnfgfr1), the fgf20a ligand gene, or the cardiomyocyte mitogen gene neuregulin1 in stable transgenic lines during tissue regeneration. These animals undergo normal development, but show a loss or gain of regenerative capacity after injury. These findings, along with the studies mentioned above, indicate that there exist cis-regulatory elements that activate gene expression after tissue injures and/or during regeneration, and that there may be thousands of TREEs that are active in each context of regeneration. Evidence indicates that some of these elements respond to the injury component of regeneration, whereas others are active during cell proliferation (36).
Figure 4.
Tissue regeneration enhancer elements (TREEs) control regeneration capacity in zebrafish. (a) After amputation injuries, a TREE linked to the leptin b gene (LEN) can rescue regeneration defects in adult fgf20a mutants (dob) when paired with an fgf20a expression cassette. (b) Conversely, the same TREE can effect a block in tailfin regeneration in the wild-type background when paired with the expression of a dominant negative Fgfr1. Intriguingly, these transgenic animals undergo normal development, whereas their regeneration capacity is modified. Thus, the LEN element appears to be specifically activated after injury and during regeneration.
A key task ahead is determination of upstream factors that bind to TREEs, guided by bioinformatical assessment of possible transcription factor binding sites. Identification of these transcription factors can provide the missing link between early injury signals and the activation of genetic programs upon injury. Also, the Kang et al. study found that the zebrafish lepb-linked element can be recognized and activate expression in injured neonatal mouse digits and hearts (54). Thus, it will be interesting to determine whether a TREE strategy can be adopted to target pro-regenerative factors and instruct regeneration in mammalian tissues. Finally, it is interesting to speculate that TREE sequences are different among species, and that the capacity to regenerate a given tissue might be impacted by these sequence differences. Addressing these questions can improve our understanding of regeneration mechanisms and suggest potential therapeutic strategies to control tissue regenerative capacity with surgical precision.
5. Concluding remarks and future perspectives
Access to new genetic tools is empowering researchers to better address central questions in regeneration, using a range of model systems with distinct technical advantages. To visualize how regeneration occurs, new genetic tools and imaging platforms are enabling a high-resolution view of intricate cell behaviors. Studies to date have focused on tracking one or two specific cell types in two-dimensional space. For instance, through spatial visualization of both hair follicle cells and adjacent dermal papilla cells, recent live imaging combined with cell ablation found that dermal papilla cells in the mesenchyme are essential to regulate follicular stem cell division during growth and regression phases (75; 99). The finding provides a direct support for the niche function of dermal papilla cells in initiating and sustaining hair regeneration (20). Extending this approach, regeneration of complex tissue involves many diverse cell types like epithelium, fibroblasts, neural and vascular tissues, and parenchymal cells that are tissue-specific, viewed in a 3-D space. An in toto view of cell behavior during regeneration, by employing the color spectrum to label various cell types, has the potential to be transformative.
In addition to cell behaviors, live imaging studies will enable concurrent imaging of signaling pathways and/or subcellular features during tissue regeneration. Dynamic regulation of molecular signals and subcellular structures (e.g. microtubules) instruct cell behaviors, for instance as visualized for the maintenance of germline stem cells in fruitflies (46). Similarly, spatiotemporal activation of signaling pathways can be quantitatively monitored in individual cells at large-scale during regeneration with genetic biosensors or reporters. Concurrent imaging of cell behaviors and cell signaling events at high-resolution should illuminate how wound healing, regenerative outgrowth, and pattern formation are achieved through dynamic regulation of morphogenetic factors in spared tissue after injuries.
To address how regeneration occurs, technical advances in genome-editing tools have enabled straightforward tests of potential regeneration factors. Established model systems either recent (e.g. zebrafish and crustaceans) or centuries-old (e.g. salamanders and planarians) will continue to offer unique technical advantages, bolstered by availability of these tools. Parallel advances in different model systems can identify not only evolutionarily conserved regeneration strategies, but also alternative strategies that might be applied across different phyla. In contrast to what we know about positive regulators of regeneration, only a handful of negative regulators have been described (60; 119); these factors may be rare, or they may be more difficult to predict. Incorporation of sensitive reporters (e.g. FUCCI or luciferase (14; 21)) into forward and reverse genetic screens, as a means to quantify proliferating cells in the regenerate, will facilitate identification of both positive and negative regulators of regeneration.
Summary points.
Three central questions in regeneration are discussed: What are the cellular sources and key cell behaviors in regenerating tissue? What molecular factors mediate regeneration by these cell sources? What are the earliest signals that control the presence of these molecular factors?
A combination of sophisticated genetic technologies, live imaging platforms, and quantitative analyses is enabling deconstruction of complex tissue regeneration at single-cell resolution.
Forward and reverse genetic approaches, evolving rapidly in the genome editing era, remain essential to uncover molecular and cellular bases of regeneration in different model systems.
Studies of early injury signals and regeneration-activated enhancer elements are exciting frontiers that may yield clues to jump-start regeneration in poorly regenerative contexts.
Acknowledgments
We apologize to the many scientists whose work we omitted or could only cover briefly due to space and bandwidth limitations. We thank Junsu Kang for comments on the manuscript. We acknowledge support from Institute of Cellular and Organismic Biology, Academia Sinica to C.H.C., and from NIH (R01-GM074057, R01-HL0810674, R01-HL131319, R01-HL136182, and R21-NS096617), Fondation Leducq, and American Heart Association to K.D.P.
Footnotes
Disclosure statement
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
References
- 1.Ablain J, Durand EM, Yang S, Zhou Y, Zon LI. A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev Cell. 2015;32:756–64. doi: 10.1016/j.devcel.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abremski K, Hoess R. Bacteriophage P1 site-specific recombination. Purification and properties of the Cre recombinase protein. J Biol Chem. 1984;259:1509–14. [PubMed] [Google Scholar]
- 3.Adler PN, MacQueen M. Cell proliferation and DNA replication in the imaginal wing disc of Drosophila melanogaster. Dev Biol. 1984;103:28–37. doi: 10.1016/0012-1606(84)90004-6. [DOI] [PubMed] [Google Scholar]
- 4.Alwes F, Enjolras C, Averof M. Live imaging reveals the progenitors and cell dynamics of limb regeneration. Elife. 2016:5. doi: 10.7554/eLife.19766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 6.Beane WS, Morokuma J, Lemire JM, Levin M. Bioelectric signaling regulates head and organ size during planarian regeneration. Development. 2013;140:313–22. doi: 10.1242/dev.086900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergantinos C, Corominas M, Serras F. Cell death-induced regeneration in wing imaginal discs requires JNK signalling. Development. 2010;137:1169–79. doi: 10.1242/dev.045559. [DOI] [PubMed] [Google Scholar]
- 8.Brockes JP, Kumar A. Comparative aspects of animal regeneration. Annu Rev Cell Dev Biol. 2008;24:525–49. doi: 10.1146/annurev.cellbio.24.110707.175336. [DOI] [PubMed] [Google Scholar]
- 9.Brockschnieder D, Lappe-Siefke C, Goebbels S, Boesl MR, Nave KA, Riethmacher D. Cell depletion due to diphtheria toxin fragment A after Cre-mediated recombination. Mol Cell Biol. 2004;24:7636–42. doi: 10.1128/MCB.24.17.7636-7642.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broussonet PMA. Observations sur la regeneration de quelques parties du corps des Poissons. Hist de l’Acad Roy des Sciences 1786 [Google Scholar]
- 11.Bryant PJ. Regeneration and duplication following operations in situ on the imaginal discs of Drosophila melanogaster. Dev Biol. 1971;26:637–51. doi: 10.1016/0012-1606(71)90146-1. [DOI] [PubMed] [Google Scholar]
- 12.Burzyn D, Kuswanto W, Kolodin D, Shadrach JL, Cerletti M, et al. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–95. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao J, Navis A, Cox BD, Dickson AL, Gemberling M, et al. Single epicardial cell transcriptome sequencing identifies Caveolin 1 as an essential factor in zebrafish heart regeneration. Development. 2016;143:232–43. doi: 10.1242/dev.130534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen CH, Durand E, Wang J, Zon LI, Poss KD. zebraflash transgenic lines for in vivo bioluminescence imaging of stem cells and regeneration in adult zebrafish. Development. 2013;140:4988–97. doi: 10.1242/dev.102053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CH, Merriman AF, Savage J, Willer J, Wahlig T, et al. Transient laminin beta 1a Induction Defines the Wound Epidermis during Zebrafish Fin Regeneration. PLoS Genet. 2015;11:e1005437. doi: 10.1371/journal.pgen.1005437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen CH, Puliafito A, Cox BD, Primo L, Fang Y, et al. Multicolor Cell Barcoding Technology for Long-Term Surveillance of Epithelial Regeneration in Zebrafish. Dev Cell. 2016;36:668–80. doi: 10.1016/j.devcel.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chera S, Baronnier D, Ghila L, Cigliola V, Jensen JN, et al. Diabetes recovery by age-dependent conversion of pancreatic delta-cells into insulin producers. Nature. 2014;514:503–7. doi: 10.1038/nature13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chera S, Ghila L, Dobretz K, Wenger Y, Bauer C, et al. Apoptotic cells provide an unexpected source of Wnt3 signaling to drive hydra head regeneration. Dev Cell. 2009;17:279–89. doi: 10.1016/j.devcel.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 19.Chera S, Ghila L, Wenger Y, Galliot B. Injury-induced activation of the MAPK/CREB pathway triggers apoptosis-induced compensatory proliferation in hydra head regeneration. Dev Growth Differ. 2011;53:186–201. doi: 10.1111/j.1440-169X.2011.01250.x. [DOI] [PubMed] [Google Scholar]
- 20.Chi W, Wu E, Morgan BA. Dermal papilla cell number specifies hair size, shape and cycling and its reduction causes follicular decline. Development. 2013;140:1676–83. doi: 10.1242/dev.090662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi WY, Gemberling M, Wang J, Holdway JE, Shen MC, et al. In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development. 2013;140:660–6. doi: 10.1242/dev.088526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curado S, Stainier DY, Anderson RM. Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat Protoc. 2008;3:948–54. doi: 10.1038/nprot.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Currie JD, Kawaguchi A, Traspas RM, Schuez M, Chara O, Tanaka EM. Live Imaging of Axolotl Digit Regeneration Reveals Spatiotemporal Choreography of Diverse Connective Tissue Progenitor Pools. Dev Cell. 2016;39:411–23. doi: 10.1016/j.devcel.2016.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Talia S, Poss KD. Monitoring Tissue Regeneration at Single-Cell Resolution. Cell Stem Cell. 2016;19:428–31. doi: 10.1016/j.stem.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dinsmore CE. A History of Regeneration Research: Milestones in the Evolution of a Science. Cambridge University Press; 1991. [Google Scholar]
- 26.Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–6. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 27.Echeverri K, Clarke JD, Tanaka EM. In vivo imaging indicates muscle fiber dedifferentiation is a major contributor to the regenerating tail blastema. Dev Biol. 2001;236:151–64. doi: 10.1006/dbio.2001.0312. [DOI] [PubMed] [Google Scholar]
- 28.Enyedi B, Kala S, Nikolich-Zugich T, Niethammer P. Tissue damage detection by osmotic surveillance. Nat Cell Biol. 2013;15:1123–30. doi: 10.1038/ncb2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fei JF, Schuez M, Tazaki A, Taniguchi Y, Roensch K, Tanaka EM. CRISPR-mediated genomic deletion of Sox2 in the axolotl shows a requirement in spinal cord neural stem cell amplification during tail regeneration. Stem Cell Reports. 2014;3:444–59. doi: 10.1016/j.stemcr.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferreira F, Luxardi G, Reid B, Zhao M. Early bioelectric activities mediate redox-modulated regeneration. Development. 2016;143:4582–94. doi: 10.1242/dev.142034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forsthoefel DJ, James NP, Escobar DJ, Stary JM, Vieira AP, et al. An RNAi screen reveals intestinal regulators of branching morphogenesis, differentiation, and stem cell proliferation in planarians. Dev Cell. 2012;23:691–704. doi: 10.1016/j.devcel.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuchs Y, Steller H. Live to die another way: modes of programmed cell death and the signals emanating from dying cells. Nat Rev Mol Cell Biol. 2015;16:329–44. doi: 10.1038/nrm3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gault WJ, Enyedi B, Niethammer P. Osmotic surveillance mediates rapid wound closure through nucleotide release. J Cell Biol. 2014;207:767–82. doi: 10.1083/jcb.201408049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gauron C, Rampon C, Bouzaffour M, Ipendey E, Teillon J, et al. Sustained production of ROS triggers compensatory proliferation and is required for regeneration to proceed. Sci Rep. 2013;3:2084. doi: 10.1038/srep02084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gemberling M, Karra R, Dickson AL, Poss KD. Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. Elife. 2015:4. doi: 10.7554/eLife.05871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldman JA, Kuzu G, Lee N, Karasik J, Gemberling M, et al. Resolving Heart Regeneration by Replacement Histone Profiling. Dev Cell. 2017 doi: 10.1016/j.devcel.2017.01.013. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grillo M, Konstantinides N, Averof M. Old questions, new models: unraveling complex organ regeneration with new experimental approaches. Curr Opin Genet Dev. 2016;40:23–31. doi: 10.1016/j.gde.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 38.Guenther CA, Wang Z, Li E, Tran MC, Logan CY, et al. A distinct regulatory region of the Bmp5 locus activates gene expression following adult bone fracture or soft tissue injury. Bone. 2015;77:31–41. doi: 10.1016/j.bone.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta V, Gemberling M, Karra R, Rosenfeld GE, Evans T, Poss KD. An injury-responsive gata4 program shapes the zebrafish cardiac ventricle. Curr Biol. 2013;23:1221–7. doi: 10.1016/j.cub.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hadorn E. Differenzierungsleistungen Wiederholt Fragmentierter Teilstucke Mannlicher Genitalscheiben Von Drosophila Melanogaster Nach Kultur in Vivo. Developmental Biology. 1963;7:617–29. [Google Scholar]
- 41.Hoshijima K, Jurynec MJ, Grunwald DJ. Precise Editing of the Zebrafish Genome Made Simple and Efficient. Dev Cell. 2016;36:654–67. doi: 10.1016/j.devcel.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang GN, Thatcher JE, McAnally J, Kong Y, Qi X, et al. C/EBP transcription factors mediate epicardial activation during heart development and injury. Science. 2012;338:1599–603. doi: 10.1126/science.1229765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hwang JS, Kobayashi C, Agata K, Ikeo K, Gojobori T. Detection of apoptosis during planarian regeneration by the expression of apoptosis-related genes and TUNEL assay. Gene. 2004;333:15–25. doi: 10.1016/j.gene.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 45.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–9. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Inaba M, Buszczak M, Yamashita YM. Nanotubes mediate niche-stem-cell signalling in the Drosophila testis. Nature. 2015;523:329–32. doi: 10.1038/nature14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang H, Grenley MO, Bravo MJ, Blumhagen RZ, Edgar BA. EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell. 2011;8:84–95. doi: 10.1016/j.stem.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–55. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin Y, Ha N, Fores M, Xiang J, Glasser C, et al. EGFR/Ras Signaling Controls Drosophila Intestinal Stem Cell Proliferation via Capicua-Regulated Genes. PLoS Genet. 2015;11:e1005634. doi: 10.1371/journal.pgen.1005634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson SL, Weston JA. Temperature-sensitive mutations that cause stage-specific defects in Zebrafish fin regeneration. Genetics. 1995;141:1583–95. doi: 10.1093/genetics/141.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–9. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamata H, Honda S, Maeda S, Chang L, Hirata H, Karin M. Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 2005;120:649–61. doi: 10.1016/j.cell.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 53.Kaneko Y, Matsumoto G, Hanyu Y. The occurrence of apoptosis during retinal regeneration in adult newts. Brain Res Dev Brain Res. 1999;117:225–8. doi: 10.1016/s0165-3806(99)00124-8. [DOI] [PubMed] [Google Scholar]
- 54.Kang J, Hu J, Karra R, Dickson AL, Tornini VA, et al. Modulation of tissue repair by regeneration enhancer elements. Nature. 2016;532:201–6. doi: 10.1038/nature17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kiehle CP, Schubiger G. Cell proliferation changes during pattern regulation in imaginal leg discs of Drosophila melanogaster. Dev Biol. 1985;109:336–46. doi: 10.1016/0012-1606(85)90460-9. [DOI] [PubMed] [Google Scholar]
- 56.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–5. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knopf F, Hammond C, Chekuru A, Kurth T, Hans S, et al. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev Cell. 2011;20:713–24. doi: 10.1016/j.devcel.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 58.Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell. 2015;32:97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kragl M, Knapp D, Nacu E, Khattak S, Maden M, et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–5. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- 60.Kujawski S, Lin W, Kitte F, Bormel M, Fuchs S, et al. Calcineurin regulates coordinated outgrowth of zebrafish regenerating fins. Dev Cell. 2014;28:573–87. doi: 10.1016/j.devcel.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 61.Lagha M, Bothma JP, Levine M. Mechanisms of transcriptional precision in animal development. Trends Genet. 2012;28:409–16. doi: 10.1016/j.tig.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132:5173–83. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- 63.Lehoczky JA, Robert B, Tabin CJ. Mouse digit tip regeneration is mediated by fate-restricted progenitor cells. Proc Natl Acad Sci U S A. 2011;108:20609–14. doi: 10.1073/pnas.1118017108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–19. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 65.Leshchiner I, Alexa K, Kelsey P, Adzhubei I, Austin-Tse CA, et al. Mutation mapping and identification by whole-genome sequencing. Genome Res. 2012;22:1541–8. doi: 10.1101/gr.135541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Levin M. Bioelectric mechanisms in regeneration: Unique aspects and future perspectives. Semin Cell Dev Biol. 2009;20:543–56. doi: 10.1016/j.semcdb.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li L, Yan B, Shi YQ, Zhang WQ, Wen ZL. Live imaging reveals differing roles of macrophages and neutrophils during zebrafish tail fin regeneration. J Biol Chem. 2012;287:25353–60. doi: 10.1074/jbc.M112.349126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu SY, Selck C, Friedrich B, Lutz R, Vila-Farre M, et al. Reactivating head regrowth in a regeneration-deficient planarian species. Nature. 2013;500:81–4. doi: 10.1038/nature12414. [DOI] [PubMed] [Google Scholar]
- 69.Livet J, Weissman TA, Kang H, Draft RW, Lu J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 70.Lo DC, Allen F, Brockes JP. Reversal of muscle differentiation during urodele limb regeneration. Proc Natl Acad Sci U S A. 1993;90:7230–4. doi: 10.1073/pnas.90.15.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lohmann JU, Endl I, Bosch TC. Silencing of developmental genes in Hydra. Dev Biol. 1999;214:211–4. doi: 10.1006/dbio.1999.9407. [DOI] [PubMed] [Google Scholar]
- 72.Love NR, Chen Y, Ishibashi S, Kritsiligkou P, Lea R, et al. Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nat Cell Biol. 2013;15:222–8. doi: 10.1038/ncb2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Makino S, Whitehead GG, Lien CL, Kim S, Jhawar P, et al. Heat-shock protein 60 is required for blastema formation and maintenance during regeneration. Proc Natl Acad Sci U S A. 2005;102:14599–604. doi: 10.1073/pnas.0507408102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McClure KD, Sustar A, Schubiger G. Three genes control the timing, the site and the size of blastema formation in Drosophila. Dev Biol. 2008;319:68–77. doi: 10.1016/j.ydbio.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mesa KR, Rompolas P, Zito G, Myung P, Sun TY, et al. Niche-induced cell death and epithelial phagocytosis regulate hair follicle stem cell pool. Nature. 2015;522:94–7. doi: 10.1038/nature14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mokalled MH, Patra C, Dickson AL, Endo T, Stainier DY, Poss KD. Injury-induced ctgfa directs glial bridging and spinal cord regeneration in zebrafish. Science. 2016;354:630–4. doi: 10.1126/science.aaf2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morgan TH. Regeneration. Macmillan; 1901. [Google Scholar]
- 78.Mullins MC, Hammerschmidt M, Haffter P, Nusslein-Volhard C. Large-scale mutagenesis in the zebrafish: in search of genes controlling development in a vertebrate. Curr Biol. 1994;4:189–202. doi: 10.1016/s0960-9822(00)00048-8. [DOI] [PubMed] [Google Scholar]
- 79.Nachtrab G, Kikuchi K, Tornini VA, Poss KD. Transcriptional components of anteroposterior positional information during zebrafish fin regeneration. Development. 2013;140:3754–64. doi: 10.1242/dev.098798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- 81.Nechiporuk A, Poss KD, Johnson SL, Keating MT. Positional cloning of a temperature-sensitive mutant emmental reveals a role for sly1 during cell proliferation in zebrafish fin regeneration. Dev Biol. 2003;258:291–306. doi: 10.1016/s0012-1606(03)00129-5. [DOI] [PubMed] [Google Scholar]
- 82.Newmark PA, Reddien PW, Cebria F, Sanchez Alvarado A. Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc Natl Acad Sci U S A. 2003;100(Suppl 1):11861–5. doi: 10.1073/pnas.1834205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Niethammer P. The early wound signals. Curr Opin Genet Dev. 2016;40:17–22. doi: 10.1016/j.gde.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Niethammer P, Grabher C, Look AT, Mitchison TJ. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459:996–9. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pellettieri J, Fitzgerald P, Watanabe S, Mancuso J, Green DR, Sanchez Alvarado A. Cell death and tissue remodeling in planarian regeneration. Dev Biol. 2010;338:76–85. doi: 10.1016/j.ydbio.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Perez-Garijo A, Martin FA, Morata G. Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila. Development. 2004;131:5591–8. doi: 10.1242/dev.01432. [DOI] [PubMed] [Google Scholar]
- 87.Petrie TA, Strand NS, Yang CT, Rabinowitz JS, Moon RT. Macrophages modulate adult zebrafish tail fin regeneration. Development. 2014;141:2581–91. doi: 10.1242/dev.098459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pfefferli C, Jazwinska A. The art of fin regeneration in zebrafish. Regeneration (Oxf) 2015;2:72–83. doi: 10.1002/reg2.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech Dev. 2007;124:218–29. doi: 10.1016/j.mod.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pomerantz JH, Blau HM. Tumor suppressors: enhancers or suppressors of regeneration? Development. 2013;140:2502–12. doi: 10.1242/dev.084210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–80. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Poss KD. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat Rev Genet. 2010;11:710–22. doi: 10.1038/nrg2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Poss KD, Nechiporuk A, Hillam AM, Johnson SL, Keating MT. Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development. 2002;129:5141–9. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- 94.Rabinowitz JS, Robitaille AM, Wang Y, Ray CA, Thummel R, et al. Transcriptomic, proteomic, and metabolomic landscape of positional memory in the caudal fin of zebrafish. Proc Natl Acad Sci U S A. 2017;114:E717–E26. doi: 10.1073/pnas.1620755114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Razzell W, Evans IR, Martin P, Wood W. Calcium flashes orchestrate the wound inflammatory response through DUOX activation and hydrogen peroxide release. Curr Biol. 2013;23:424–9. doi: 10.1016/j.cub.2013.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sanchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell. 2005;8:635–49. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rinkevich Y, Lindau P, Ueno H, Longaker MT, Weissman IL. Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature. 2011;476:409–13. doi: 10.1038/nature10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roberts-Galbraith RH, Brubacher JL, Newmark PA. A functional genomics screen in planarians reveals regulators of whole-brain regeneration. Elife. 2016:5. doi: 10.7554/eLife.17002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rompolas P, Deschene ER, Zito G, Gonzalez DG, Saotome I, et al. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature. 2012;487:496–9. doi: 10.1038/nature11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–3. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- 101.Rulands S, Simons BD. Tracing cellular dynamics in tissue development, maintenance and disease. Curr Opin Cell Biol. 2016;43:38–45. doi: 10.1016/j.ceb.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 102.Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev Cell. 2004;7:491–501. doi: 10.1016/j.devcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 103.Sanchez Alvarado A, Newmark PA. Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc Natl Acad Sci U S A. 1999;96:5049–54. doi: 10.1073/pnas.96.9.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sandoval-Guzman T, Wang H, Khattak S, Schuez M, Roensch K, et al. Fundamental differences in dedifferentiation and stem cell recruitment during skeletal muscle regeneration in two salamander species. Cell Stem Cell. 2014;14:174–87. doi: 10.1016/j.stem.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 105.Schubiger G. Regeneration, duplication and transdetermination in fragments of the leg disc of Drosophila melanogaster. Dev Biol. 1971;26:277–95. doi: 10.1016/0012-1606(71)90127-8. [DOI] [PubMed] [Google Scholar]
- 106.Schuster KJ, Smith-Bolton RK. Taranis Protects Regenerating Tissue from Fate Changes Induced by the Wound Response in Drosophila. Dev Cell. 2015;34:119–28. doi: 10.1016/j.devcel.2015.04.017. [DOI] [PubMed] [Google Scholar]
- 107.Scimone ML, Kravarik KM, Lapan SW, Reddien PW. Neoblast specialization in regeneration of the planarian Schmidtea mediterranea. Stem Cell Reports. 2014;3:339–52. doi: 10.1016/j.stemcr.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Seifert AW, Kiama SG, Seifert MG, Goheen JR, Palmer TM, Maden M. Skin shedding and tissue regeneration in African spiny mice (Acomys) Nature. 2012;489:561–5. doi: 10.1038/nature11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sikes JM, Newmark PA. Restoration of anterior regeneration in a planarian with limited regenerative ability. Nature. 2013;500:77–80. doi: 10.1038/nature12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Singh SP, Holdway JE, Poss KD. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev Cell. 2012;22:879–86. doi: 10.1016/j.devcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Smart N, Risebro CA, Melville AA, Moses K, Schwartz RJ, et al. Thymosin beta4 induces adult epicardial progenitor mobilization and neovascularization. Nature. 2007;445:177–82. doi: 10.1038/nature05383. [DOI] [PubMed] [Google Scholar]
- 112.Smith-Bolton RK, Worley MI, Kanda H, Hariharan IK. Regenerative growth in Drosophila imaginal discs is regulated by Wingless and Myc. Dev Cell. 2009;16:797–809. doi: 10.1016/j.devcel.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–44. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 114.Solnica-Krezel L, Schier AF, Driever W. Efficient recovery of ENU-induced mutations from the zebrafish germline. Genetics. 1994;136:1401–20. doi: 10.1093/genetics/136.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sousa S, Afonso N, Bensimon-Brito A, Fonseca M, Simoes M, et al. Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development. 2011;138:3897–905. doi: 10.1242/dev.064717. [DOI] [PubMed] [Google Scholar]
- 116.Spallanzani L. An essay on animal reproductions. London: 1769. Printed for T. Becket, and P.A. de Hondt. [Google Scholar]
- 117.Stainier DY, Kontarakis Z, Rossi A. Making sense of anti-sense data. Dev Cell. 2015;32:7–8. doi: 10.1016/j.devcel.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 118.Stewart S, Stankunas K. Limited dedifferentiation provides replacement tissue during zebrafish fin regeneration. Dev Biol. 2012;365:339–49. doi: 10.1016/j.ydbio.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, et al. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–89. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- 120.Sugiura T, Wang H, Barsacchi R, Simon A, Tanaka EM. MARCKS-like protein is an initiating molecule in axolotl appendage regeneration. Nature. 2016;531:237–40. doi: 10.1038/nature16974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tanaka EM. The Molecular and Cellular Choreography of Appendage Regeneration. Cell. 2016;165:1598–608. doi: 10.1016/j.cell.2016.05.038. [DOI] [PubMed] [Google Scholar]
- 122.Tanaka EM, Reddien PW. The cellular basis for animal regeneration. Dev Cell. 2011;21:172–85. doi: 10.1016/j.devcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Thorel F, Nepote V, Avril I, Kohno K, Desgraz R, et al. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464:1149–54. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tornini VA, Poss KD. Keeping at arm’s length during regeneration. Dev Cell. 2014;29:139–45. doi: 10.1016/j.devcel.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tornini VA, Puliafito A, Slota LA, Thompson JD, Nachtrab G, et al. Live Monitoring of Blastemal Cell Contributions during Appendage Regeneration. Curr Biol. 2016;26:2981–91. doi: 10.1016/j.cub.2016.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tseng AS, Adams DS, Qiu D, Koustubhan P, Levin M. Apoptosis is required during early stages of tail regeneration in Xenopus laevis. Dev Biol. 2007;301:62–9. doi: 10.1016/j.ydbio.2006.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tseng AS, Beane WS, Lemire JM, Masi A, Levin M. Induction of vertebrate regeneration by a transient sodium current. J Neurosci. 2010;30:13192–200. doi: 10.1523/JNEUROSCI.3315-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Umesono Y, Tasaki J, Nishimura Y, Hrouda M, Kawaguchi E, et al. The molecular logic for planarian regeneration along the anterior-posterior axis. Nature. 2013;500:73–6. doi: 10.1038/nature12359. [DOI] [PubMed] [Google Scholar]
- 129.van Wolfswinkel JC, Wagner DE, Reddien PW. Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell. 2014;15:326–39. doi: 10.1016/j.stem.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332:811–6. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang J, Panakova D, Kikuchi K, Holdway JE, Gemberling M, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–30. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang Y, Stary JM, Wilhelm JE, Newmark PA. A functional genomic screen in planarians identifies novel regulators of germ cell development. Genes Dev. 2010;24:2081–92. doi: 10.1101/gad.1951010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Whitehead GG, Makino S, Lien CL, Keating MT. fgf20 is essential for initiating zebrafish fin regeneration. Science. 2005;310:1957–60. doi: 10.1126/science.1117637. [DOI] [PubMed] [Google Scholar]
- 134.Witchley JN, Mayer M, Wagner DE, Owen JH, Reddien PW. Muscle cells provide instructions for planarian regeneration. Cell Rep. 2013;4:633–41. doi: 10.1016/j.celrep.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wynn TA, Vannella KM. Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity. 2016;44:450–62. doi: 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yoo SK, Freisinger CM, LeBert DC, Huttenlocher A. Early redox, Src family kinase, and calcium signaling integrate wound responses and tissue regeneration in zebrafish. J Cell Biol. 2012;199:225–34. doi: 10.1083/jcb.201203154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yoo SK, Starnes TW, Deng Q, Huttenlocher A. Lyn is a redox sensor that mediates leukocyte wound attraction in vivo. Nature. 2011;480:109–12. doi: 10.1038/nature10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang Q, Wang Y, Man L, Zhu Z, Bai X, et al. Reactive oxygen species generated from skeletal muscles are required for gecko tail regeneration. Sci Rep. 2016;6:20752. doi: 10.1038/srep20752. [DOI] [PMC free article] [PubMed] [Google Scholar]