Microorganisms can catabolize a wide range of organic compounds and therefore have the potential to perform many industrially relevant bioconversions. One barrier to realizing the potential of bio-refining strategies lies in our incomplete knowledge of metabolic pathways, including those that can be used to assimilate naturally abundant or easily-generated feedstocks. For instance, levulinic acid (LA) is a carbon source that is readily obtainable as a dehydration product of lignocellulosic biomass and can serve as the sole carbon source for some bacteria. Yet, the genetics and structure of LA catabolism have remained unknown. Here, we report the identification and characterization of a seven-gene operon that enables LA catabolism in Pseudomonas putida KT2440. When the pathway was reconstituted with purified proteins, we observed the formation of four acyl-CoA intermediates including a unique 4-phosphovaleryl-CoA and the previously observed 3-hydroxyvaleryl-CoA product. Using adaptive evolution, we obtained a mutant of Escherichia coli LS5218 with functional deletions of fadE and atoC that was capable of robust growth on LA when it expressed the five enzymes from the P. putida operon. This discovery will enable more efficient utilization of biomass hydrolysates and metabolic engineering to develop bioconversions using LA as a feedstock.
Levulinic acid (LA) is a five carbon γ-keto acid that can be readily obtained from biomass through non-enzymatic, acid hydrolysis of a wide range of feedstocks1,2. LA was named one of the US Department of Energy’s “Top 12 value-added chemicals from biomass”3 because it can be used as a renewable feedstock for generating a variety of molecules, such as fuel additives4–6, flavors, fragrances7,8 and polymers9,10, through chemical catalysis. In addition, microbes can use LA as a sole carbon source and have been shown to convert LA into polyhydroxyalkanoates11–13, short chain organic acids14–16, and trehalose14. All of these bioconversion studies were conducted with natural bacterial isolates because the enzymes comprising a LA assimilation pathway were unknown14. This knowledge gap limited metabolic engineering and the potential of creating LA-based bioconversions.
While enzymes involved with LA assimilation were unknown at the time of these bioconversion demonstrations, other studies identified putative intermediates and suggested pathways for LA catabolism. In a study where crude cell lysates of Cupriavidus necator were fed LA, the concentration of LA and free CoA decreased over time while acetyl-CoA and propionyl-CoA concentrations increased, suggesting that LA is catabolized via CoA thioesters like other short-chain organic acids17. In a second study, cultures of Pseudomonas putida KT2440 expressing a heterologous TesB thioesterase were fed LA. Here, 4-hydroxyvalerate (4HV) and 3-hydroxyvalerate (3HV) transiently accumulated extracellularly before ultimately disappearing18. This observation strongly suggested that 4HV and 3HV (or their CoA thioesters) were pathway intermediates. Lastly, a metabolomic study of rat livers suggested that LA is catabolized to acetyl-CoA and propionyl-CoA via a unique phosphorylated acyl-CoA19,20. In sum, these observations suggested a relatively direct route from LA to beta-oxidation intermediates, but the enzymes comprising such a pathway remained unknown. In this work, we investigated the genetic and biochemical factors that allow P. putida KT2440 to catabolize LA and demonstrated that the pathway could be reconstituted in vitro and in E. coli.
Results
Identification of Genes Involved in Levulinic Acid Metabolism
P. putida KT2440 can metabolize LA as a sole carbon source and demonstrates diauxic growth in the presence of glucose and LA (Supplementary Figure 1a). Therefore, we initiated a genetic study to identify genes involved in LA catabolism. We constructed a mutant library with a Tn5 mini transposase21 and screened for P. putida mutants lacking the ability to grow on LA as the sole carbon source. Thirteen out of 7,000 colonies screened demonstrated LA growth deficiencies. The location of each transposon insertion was determined by sequencing PCR products created with a primer nested in the transposon paired with a degenerate random primer. Table 1 shows the ten unique isolates from these thirteen hits and the putative function of the disrupted genes. Two mutants had disruptions in genes involved in propionate metabolism, supporting the hypothesis that LA is catabolized to the central metabolites, acetyl-CoA and propionyl-CoA. Three transposon mutants had disruptions in a putative operon that had not been previously characterized (disrupting genes PP_2791, PP_2793, and PP_2794). Other mutants had disruptions in genes with no obvious connection to LA catabolism (bioH, gcvP, a hypothetical zinc protease, mrdA, and fpvA). To confirm that we had screened a sufficient number of clones, we performed random bar code transposon-site sequencing (RB-TnSeq) for cultures enriched by growth on LA and 4HV relative to growth on glucose. RB-TnSeq is an efficient method for determining gene essentiality under different conditions with high genomic coverage22. This analysis identified additional genes involved in LA metabolism including an acetoacetyl-CoA transferase important for growth on LA, genes functioning in β-oxidation and propionyl-CoA metabolism, and 14 transcriptional regulators potentially involved in LA metabolism. The RB-TnSeq dataset also revealed that 3-hydroxybutyryl-CoA dehydrogenase (FadB) and β-ketothiolase (FadA) are also necessary for growth on LA and 4HV, supporting our hypothesis that LA metabolism terminates through β-oxidation. For a more complete summary and analysis of the fitness data, please see Supplementary Table 1 and Supplementary Note.
Table 1.
P. putida Levulinic Acid Transposon Insertion Sites
| Locus | Insertion Point* | Gene Name | Description/Homology |
|---|---|---|---|
| PP_0364 | 442685 | bioH | pimeloyl-ACP methyl ester esterase |
| PP_0988 | 1128706 | gcvP-1 | glycine dehydrogenase |
| PP_2332 | 2660666 | N/A | ATP-dependent zinc protease family |
| PP_2336 | 2666405 | acnA-II | aconitate hydratase |
| PP_2337 | 2666944 | prpF | aconitate isomerase |
| PP_2791 | 3181098 | N/A | Phosphotransferase family |
| PP_2793 | 3182533 | N/A | acyl-CoA dehydrogenase family protein |
| PP_2794 | 3183601 | N/A | short chain dehydrogenase/reductase family |
| PP_3741 | 4271628 | mrdA-I | transpeptidase |
| PP_4217 | 4765953 | fpvA | TonB-dependent outer membrane ferripyoverdine receptor |
Insertion point based on location from P. putida KT2440 origin
Operon Characterization and Induction
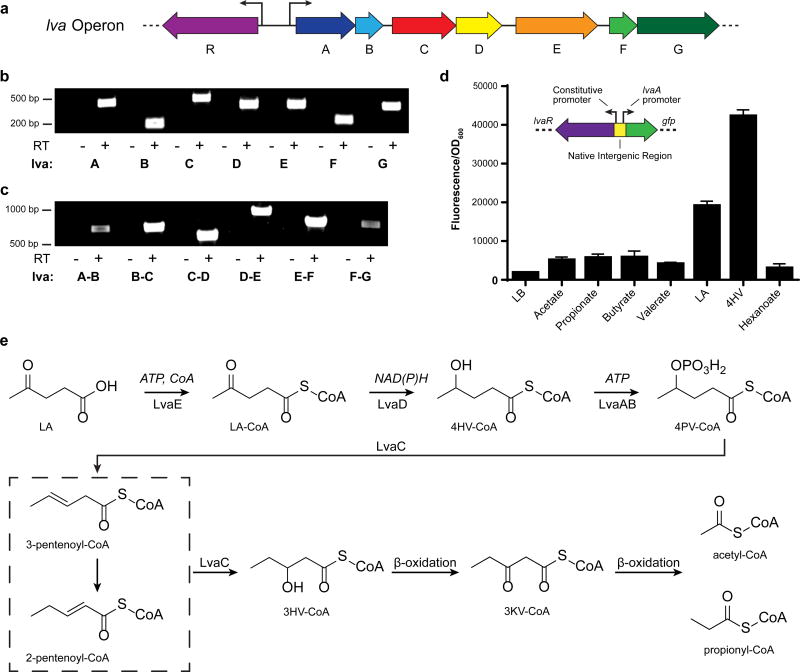
Given the propensity of bacteria to cluster related genes into operons, we examined the putative seven-gene operon, PP_2791-PP_2797, which contained three of our transposon hits (PP_2791, PP_2793 and PP_2794). We analyzed the sequence homology of the seven genes in the operon using the basic local alignment search tool (BLAST23) and assigned predicted functions, listed in Table 2. We were unable to find any published studies about these genes beyond the automated sequence annotations. Therefore, we investigated the expression and function of these genes involved in LA catabolism. First, we isolated RNA from wild type P. putida grown in minimal media with LA as the carbon source and demonstrated that we could locate all seven genes by PCR amplification of cDNA created with a reverse primer specific to PP_2797 (Figure 1a–c). The transcription start site (TSS) of the operon was isolated by 5’ RACE24 (Supplementary Figure 1b) and implicated a different start codon for PP_2791, 72 bp downstream of the one originally reported25. A σ54 promoter sequence located upstream of PP_2791 was identified by comparing sequence upstream of the new TSS with published σ54 promoter consensus sequences26 (Supplementary Figure 1c). The data presented below suggests the proteins encoded by this operon are important in LA catabolism and we propose that the polycistronic genes be designated as lvaABCDEFG.
Table 2.
P. putida LA Operon Knockout and Complementation
| Growth on LA | Growth on 4HV | ||||
|---|---|---|---|---|---|
| Genotype | Predicted Function | EV | Complement | EV | Complement |
| WT | ++ | N/A | ++ | N/A | |
| ΔlvaR | σ54 dependent sensory box protein | - | ++ | - | ++ |
| ΔlvaA | Phosphotransferase family | - | ++ | - | ++ |
| ΔlvaB | Hypothetical protein | - | ++ | - | ++ |
| ΔlvaC | acyl-CoA dehydrogenase family protein | - | ++ | + | ++ |
| ΔlvaD | Short chain dehydrogenase reductase family | - | ++ | ++ | ++ |
| ΔlvaE | Acyl-CoA synthetase | ++ | ++ | - | + |
(EV) empty vector plasmid; (N/A) not applicable; (-) No growth; (+) Visible growth; (++) Robust growth
Figure 1. Genetic characterization and proposed catabolic activity of the P. putida lva operon.
a, Organization of the lvaRABCDEFG (9,323 bp) operon. b, Reverse Transcriptase (RT) PCR demonstrates that each gene is expressed in cells grown on LA. Samples were compared with the negative control (-RT) where reverse transcriptase was omitted from the reaction (n=1). c, RT-PCR of cDNA created with primer JMR237 demonstrates that the operon is polycistronic. Note that a product spanning each intergenic region was observed (n=1). d, lva operon induction assay. GFP fluorescence was measured from LB-cultures supplemented with various organic acids (20 mM) (n=3, biological). Error bars represent s.d. Insert shows the schematic of transcriptional GFP fusion used to test induction of the lva operon. lvaR was cloned onto a plasmid containing its native constitutive promoter and the native promoter region for lvaA. The fluorescent protein sfGFP was cloned in place of lvaA. e, Proposed pathway for LA metabolism. LA, levulinic acid; 4HV, 4-hydroxyvalerate; 3HV, 3-hydroxyvalerate; LA-CoA, levulinyl-CoA; 4HV-CoA, 4-hydroxyvaleryl-CoA; CoA, coenzyme-A; ATP, adenosine triphosphate; 4PV-CoA, 4-phosphovaleryl-CoA; 3KV-CoA, 3-ketovaleryl-CoA; NAD(P)H, Nicotinamide adenine dinucleotide (phosphate) reduced; GFP, green fluorescent protein.
Upstream of lvaABCDEFG, we identified a gene oriented divergently from the operon (PP_2790) and predicted to encode a transcription factor with a σ54 interaction domain and homology to the propionate metabolism activator, prpR. The genomic organization strongly suggested that the gene encoded a regulator for the lva operon. Consequently, we deleted PP_2790 and evaluated growth of P. putida strains on both LA and a likely intermediate, 4HV. The ΔPP_2790 mutant was unable to grow on LA and 4HV suggesting that it acts as an activator. Expression of PP_2790 on a plasmid restored growth of the deletion strain on LA and 4HV. To identify compounds that activate lvaABCDEFG expression, we built a transcriptional reporter system that linked sfGFP to the σ54 promoter sequence located upstream of lvaA. The reporter cassette was cloned onto a broad host range vector (Figure 1d) and the resulting construct was transformed into wild type P. putida. We tested a variety of short and medium chain length acids by adding them to rich media and evaluating the corresponding sfGFP expression levels. We observed strong sfGFP fluorescence only when LA or 4HV was added to the system (Figure 1d). For these reasons, we suggest that PP_2790 encodes a transcriptional regulator responsive to the LA pathway and should be designated lvaR.
Genetic and Biochemical Studies of lvaABCDEFG Operon
To confirm the involvement of the lva operon in LA catabolism, we created a deletion mutant of each lva gene predicted to encode an enzymatic protein and a corresponding complementation plasmid using the ParaBAD promoter. We tested the ability of the resulting strains to grow on LA and 4HV (Table 2, Supplementary Figure 2). In addition, we purified the five enzymes from cultures of E. coli BL21 (DE3), reconstituted the enzymatic reactions in vitro, and used liquid chromatography/mass spectrometry (LC/MS) to identify reaction products. We used selective ion scanning to monitor the masses for likely intermediates based on prior studies17,18,20 and developed the following hypothesized pathway (Figure 1e, Supplementary Figure 3). First, LA is activated as a coenzyme A-thioester, levulinyl-CoA (LA-CoA). Second, LA-CoA is reduced to 4-hydroxyvaleryl-CoA (4HV-CoA). Third, 4HV-CoA is phosphorylated at the γ-position to yield 4-phosphovaleryl-CoA (4PV-CoA). Fourth, 4PV-CoA is dephosphorylated to yield a pentenoyl-CoA species (likely 3-pentenoyl-CoA). Last, pentenoyl-CoA is hydrated to yield 3-hydroxyvaleryl-CoA (3HV-CoA) which can be further oxidized via β-oxidation to yield acetyl-CoA and propionyl-CoA or incorporated into PHA polymers. The remainder of this manuscript will provide evidence supporting our hypothesized metabolic pathway for converting LA to 3-hydroxyvaleryl-CoA (3HV-CoA) and assignment of enzymes to each reaction.
LvaE
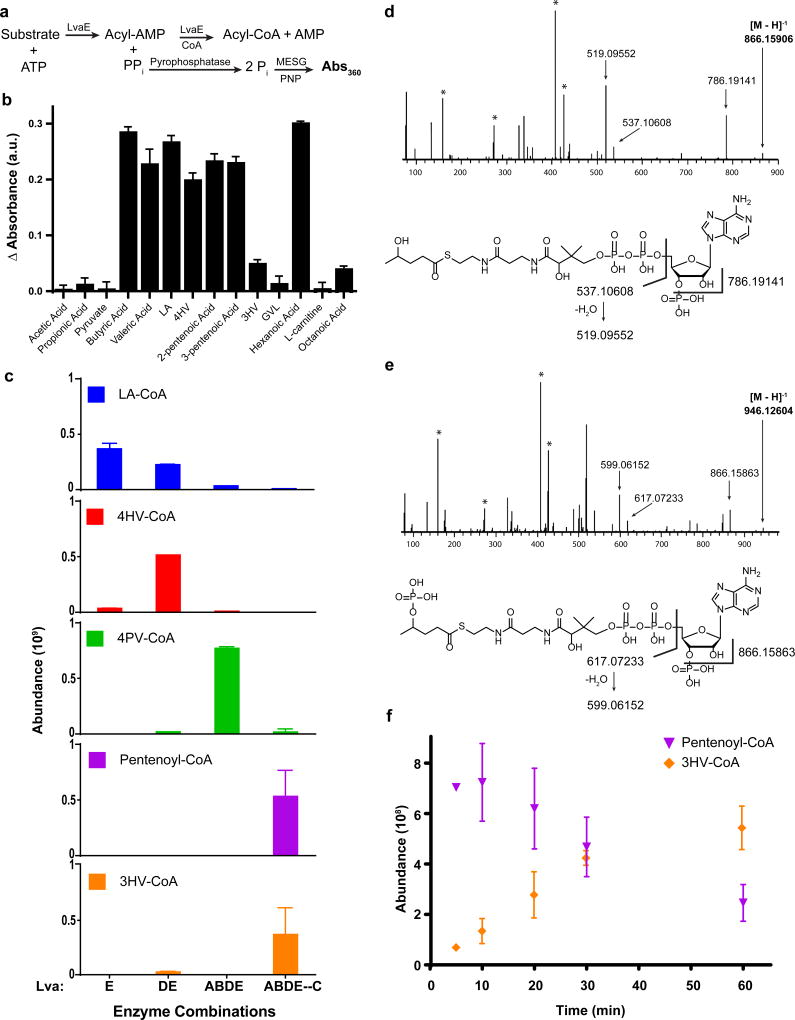
The presence of an enzyme (encoded by lvaE) with homology to an acyl-CoA synthetase (including a putative CoA binding region and an AMP binding site) suggested that the degradation pathway acts on CoA thioesters and begins with the activation of acids to acyl-CoA’s. The ΔlvaE strain grew on LA but not on 4HV, indicating that LA may also be activated by other CoA-synthetases in P. putida. We quantified the activity of purified LvaE (6x-His N-terminal fusion) on a variety of organic acid substrates using the EnzChek® Pyrophosphate Assay Kit which detects pyrophosphate released in the first half of the reaction that creates an acyl-AMP intermediate (Figure 2a). LvaE demonstrated activity on C4-C6 carboxylic acids, including LA and 4HV (Figure 2b), but showed minimal activity on other organic acids (pyruvate, acetate, propionate, octanoate). Using LC/MS to detect reaction products (Figure 2c), we demonstrated that LvaE was necessary and sufficient to catalyze the ligation of CoA to LA, generating levulinyl-CoA (LA-CoA). None of the other enzymes from the operon catalyzed this or any other reaction using LA as a substrate (Supplementary Figure 4), confirming that the pathway proceeds via acyl-CoA intermediates.
Figure 2. Enzymatic activity and pathway characterization for lva operon.
a, CoA-ligase activity assay schematic. Using the Enzchek® Pyrophosphatase Assay kit, the amount of pyrophosphate released during the CoA ligase reaction was measured as an increase of absorbance at 360 nm. b, Activity of LvaE towards short and medium chain acids (n=3, technical). Baseline subtraction was performed on all samples with a control reaction containing no substrate, indicated by Δ absorbance. c, CoA species abundance in LC/MS analysis of in vitro enzyme combinations following 30 min incubation. Reactions contained LA, CoA, ATP, and NAD(P)H with varying enzyme combinations (n=3, technical). ABDE—C indicates that the LvaABDE reaction was performed first, metabolites were separated from LvaABDE, and the resulting solution was supplemented with LvaC. The reaction confirms that LvaC is capable of converting 4PV-CoA to 3HV-CoA. d, MS/MS spectra for 4HV-CoA. Assignment of selected fragments from 4HV-CoA below. e, MS/MS spectra for 4PV-CoA. Assignments of selected fragments from 4PV-CoA below. The masses between the selected fragments of 4PV-CoA and 4HV-CoA differ by the mass of PO3H (79.967), indicating 4PV-CoA contains a phosphate group not found in 4HV-COA. Bold values indicate the mass of the parent ion. Peaks identified with the symbol (*) are fragments resulting from coenzyme A. See Supplementary Figure 6 and Supplementary Table 2 for additional fragmentation information. f, Abundance of pentenoyl-CoA and 3HV-CoA over a 60 min timecourse for a mixture of LvaABCDE, LA, CoA, ATP, and NAD(P)H (n=3, technical). AMP, adenosine monophosphate; PPi, pyrophosphate; Pi, phosphate; MESG, 2-amino-6-mercapto-7-methylpurine ribonucleoside; PNP, purine nucleoside phosphorylase; Abs, absorbance; a.u., arbitrary units. Error bars represent s.d.
LvaD
The second step in our proposed pathway is the reduction of LA-CoA to 4HV-CoA which we predicted to be catalyzed by lvaD. LvaD is annotated as an oxidoreductase containing an NADH binding domain and was found to be required for growth on LA but not necessary for growth on 4HV (Table 2). We purified LvaD in a similar manner to LvaE but used an N-terminal maltose binding protein (MBP) tag to increase the solubility of the enzyme27. The in vitro reaction containing LvaD and LvaE verified that LvaD is involved in the production of 4HV-CoA (Figure 2c). Furthermore, LvaDE was the only enzyme combination capable of generating 4HV-CoA in vitro (Supplementary Figure 4). LvaD can catalyze the reduction of LA-CoA with either NADH or NADPH (Supplementary Figure 4).
LvaAB
We hypothesized that the third intermediate would be 4-phospho-valeryl-CoA (4PV-CoA) based off its observation in LA degradation in rat livers19,20. The first enzyme encoded in the operon, LvaA, has putative homology, including an ATP binding site, that associated it with the kinase superfamily and phosphotransferase family of enzymes. The second protein in the operon (LvaB) has no listed function and is predicted to be only 12 kDa in size. Orthologous sequence alignments of lvaB reveal that in all other organisms the gene encoding this hypothetical protein is located immediately downstream of an lvaA ortholog. Therefore, a pull down experiment was used to determine if the two proteins interact28,29. LvaA was N-terminally tagged with MBP and cloned into a pET expression vector. LvaB was cloned directly downstream of LvaA as it is found in P. putida’s genome. The recombinant proteins were expressed in E. coli BL21(DE3) and purified using the MBP tag. An SDS-page gel of the eluent contained two bands at 85 kDa and 12 kDa, closely matching the predicted sizes of MBP-LvaA and untagged LvaB respectively (Supplementary Figure 5a,b). We performed liquid chromatography tandem mass spectrometry on a trypsin digest of the 12 kDa band and identified the protein sequence to be LvaB (Supplementary Figure 5c).
Growth studies of deletion mutants revealed that lvaA and lvaB are both essential genes for growth on either LA or 4-HV. This supports the hypothesis that they catalyze a reaction after the conversion of LA-CoA to 4HV-CoA. To confirm that the association between LvaA and LvaB was important for enzymatic activity, we tested the following enzymatic combinations: i) LvaA, LvaD and LvaE, ii) LvaB, LvaD, and LvaE, iii) LvaAB, LvaD and LvaE. We observed a decrease of 4HV-CoA and an increase of the predicted 4PV-CoA intermediate only when all four of the enzymes were present (Figure 2c, Supplementary Figure 4).
To verify the identity of 4PV-CoA, we performed tandem mass spectrometry (Figure 2d–e, Supplementary Figure 6, Supplementary Table 2). We compared the MS/MS spectra of 4HV-CoA and 4PV-CoA and detected major ion fragments at m/z 786.191, 537.106 and 519.095 (4HV-CoA) and 866.158, 617.072 and 599.061 (4PV-CoA). For each compound, these fragments can be assigned to the cleavage of a P-O bond, an O-C bond and the dehydration of O-C cleaved product, respectively. Both compounds are fragmenting at the same bonds, but the resulting m/z values for the daughter ions differ by 79.967. This mass corresponds to the m/z of PO3H−, supporting the existence of the phosphorylated 4HV-CoA species, 4PV-CoA.
LvaC
The final step in the hypothesized pathway is the formation of 3HV-CoA. Given that the combination of LvaABDE was responsible for generating 4PV-CoA and no 3HV-CoA was detected in these reactions, we postulated that LvaC was responsible for the final conversion steps. LvaC has homology to the dehydrogenase family of enzymes and 30% amino acid sequence identity to the E. coli acyl-CoA dehydrogenase protein. The ΔlvaC strain was unable to grow on LA, but grew weakly on 4HV. LvaC was purified as an MBP fusion and the resulting protein pellet displayed a yellow hue. This is often indicative of a co-purified flavin and an absorbance scan of the protein revealed absorbance maxima that are consistent with a flavin co-factor (Supplementary Figure 5d,e). When the LvaC sample was treated with trichloroacetic acid and centrifuged30, a white protein pellet and a yellow hued supernatant were observed. This indicates that the co-factor was not covalently bound to LvaC.
When LvaC was added to the in vitro reaction mixture, the concentrations of intermediates (LA-CoA, 4HV-CoA, 4PV-CoA) were reduced while the abundance of 3HV-CoA and a pentenoyl-CoA species increased (Figure 2c). This species is likely either 2-pentenoyl-CoA and/or 3-pentenoyl-CoA, which could not be resolved with our methods. Both compounds eluted at the same retention time with the same molecular mass. To test if LvaC was solely responsible for the conversion of 4PV-CoA to 3HV-CoA, we ran a two-step reaction. First, we performed the LvaABDE reaction with LA, CoA, ATP, NAD(P)H and separated the CoA products from the enzymes. To the enzyme-free mixture, we added LvaC without additional co-factors. After 30 min, we observed signals for both pentenoyl-CoA and 3HV-CoA. This indicated that the putative oxidoreductase, LvaC, is responsible for both the removal of the phosphate group to produce the enoyl-CoA and the hydration of the double bond at the 3-position. To reconstitute the whole pathway, we set-up a time course reaction with all five Lva enzymes and LA as the starting substrate. Over time, we observed a rapid increase in pentenoyl-CoA followed by a slow disappearance that mirrored the increase in the 3HV-CoA signal (Figure 2f). This suggested that the hydration reaction may be the limiting step in the overall pathway.
LvaFG
Based on homology alignments, lvaG is predicted to encode a protein with 95% amino acid sequence identity to a Pseudomonas aeruginosa cation acetate symporter and LvaF shares 33% amino acid sequence identity with the E. coli inner membrane protein Yhjb23. Sequence alignments of lvaF orthologs indicate that lvaF and lvaG are found with the same spatial relationship to each other in many organisms. These proteins are likely involved in organic acid transport but are unlikely to be involved in the catabolism of LA given that they were not necessary for the enzymatic conversion of LA to 3HV-CoA in vitro.
Conferring Growth on Levulinic Acid to E. coli
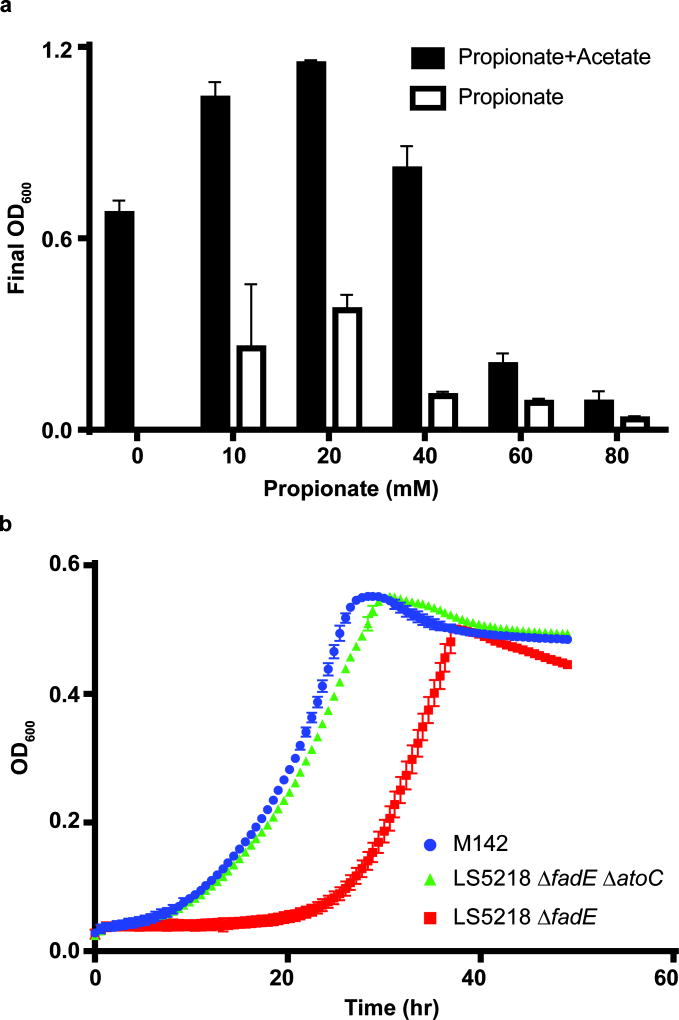
To demonstrate the ability of the lvaABCDE operon to enable LA catabolism, we augmented the metabolism of Escherichia coli LS5218 by transforming a plasmid linking LvaABCDE expression to an anhydrotetracycline inducible promoter (pJMR5). E. coli LS5218 carries two known mutations, fadR and atoC(Con) that enhance its ability to metabolize organic acids. The disruption in fadR is particularly useful because it deregulates expression of β-oxidation enzymes31, including FadB and FadA that are needed to convert 3HV-CoA to acetyl-CoA and propionyl-CoA. While E. coli contains the necessary genes for metabolism of propionyl-CoA32, elevated propionyl-CoA concentrations are known to be inhibitory33. Therefore, we fed 20 mM LA (~0.2% by weight) to minimize the impact of propionate toxicity on growth (Figure 3a). Initial cultures of E. coli LS5218 pJMR5 failed to grow on LA as a sole carbon source, so we performed adaptive evolution in effort to obtain mutants that could. The first three rounds were conducted in media containing both LA and acetate as available carbon to stimulate growth and allow cells to adapt to the presence of LA. In these experiments, we observed an increase in final cell density when both carbon sources were present relative to parallel cultures that were fed only acetate. Subsequent rounds of evolution were conducted with LA as the sole carbon source. After 14 rounds of sub-culturing on LA, we isolated two mutant strains, M141 and M142, capable of robust LA catabolism.
Figure 3. E. coli growth on propionate and LA.
a, E. coli utilization of propionate (n=3, biological). We performed a growth study to evaluate E. coli growth on various concentrations of propionate, with and without acetate as a secondary carbon source. The maximum allowable concentration that stimulated growth was 20 mM propionate, both in the presence and absence of acetate. Using this information, the LA concentration was limited to 20 mM for growth and induction studies. b, Growth curve of E. coli strains on LA. All strains harbor lvaABCDE on pJMR32 (n=3, biological). Error bars represent s.d.
We purified the LvaABCDE expression plasmid from each mutant and discovered a mutation in the ribosome binding sequence (RBS) linked to lvaA (Supplementary Table 3). The new RBS sequence was predicted to increase the translation initiation rate relative to the original sequence25. Therefore, we retransformed the isolated plasmid, designated p2, back into unevolved LS5218. The resulting strain failed to grow on LA, indicating genomic mutations were also necessary. Therefore, we submitted M141 and M142 for whole genome sequencing and identified two conserved mutations (Supplementary Table 3) relative to the LS5218 genome (GCA_002007165.1). The common mutations were a point mutation in fadE and insertion of transposons into atoC that resulted in premature stop codons and functional deletion of both genes. To validate these mutations, we generated knockouts of fadE and/or atoC using CRISPR-Cas9 mediated genome engineering34–36 and transformed each strain with pJMR32, a redesigned pJMR5 with a stronger lvaA RBS. Wild type LS5218 and LS5218 ΔatoC were unable to grow on LA whereas LS5218 ΔfadE and LS5218 ΔatoC ΔfadE grew robustly on LA as the sole carbon source (Supplementary Table 4). In liquid media, LS5218 ΔfadE demonstrated a significant lag relative to LS5218 ΔatoC ΔfadE and M142 (Figure 3b). This indicated that in E. coli LS5218, a fadE deletion is necessary, but an atoC deletion is beneficial for growth on LA.
Discussion
The work described herein identified an operon that was essential for assimilating LA into the β-oxidation pathway of P. putida. Through an integrated genetic and in vitro biochemistry study, we demonstrated that the genes lvaABCDE were upregulated in the presence of LA and were sufficient for the conversion of LA to 3HV-CoA, an intermediate of native β-oxidation. Removing any enzyme from the reaction mixture abolished 3HV-CoA production, indicating all five enzymes were necessary for this pathway. The biochemical assays confirmed the presence of 4PV-CoA, an intermediate previously observed in the metabolism of LA in rat livers. In sum, the pathway consumed at least 2 ATP and one reducing equivalent to produce 3HV-CoA (Figure 1e). β-oxidation of 3HV-CoA to acetyl-CoA and propionyl-CoA would recover the reducing equivalent. Given the energy demands of the pathway, growth on LA must be performed aerobically or in the presence of an alternative electron acceptor to enable ATP synthesis via respiration.
Like many catabolic pathways, expression of the lva operon is regulated by the presence of the pathway substrates. Using a transcriptional reporter assay, we demonstrated that the lva operon is upregulated by a transcriptional activator encoded by the divergent lvaR gene. Additionally, we suspect that the lva operon is also regulated by Crc, a global carbon catabolite repressor. Crc is an mRNA binding protein that prevents protein translation when bound to a specific mRNA sequence in P. putida, AAnAAnAA37,38. This sequence pattern is found immediately upstream of lvaE (Supplementary Figure 1d), which encodes an acyl-CoA synthetase that initiates the pathway. The presence of the Crc target sequence suggests that the operon is also subject to P. putida’s carbon catabolite repression system which may explain the diauxic growth curves observed for mixtures of glucose and LA.
The lva operon is highly conserved among the various Pseudomonas species (Supplementary Table 5). Gene clusters comprised of the main enzymatic proteins can also be found in a variety of alpha-, beta- and gamma-proteobacteria, graphically represented in Figure 4. The alpha-proteobacteria species (Azospirillum, Bradyrhizobium, Rhodopseudomonas, Sphingobium) are primarily isolated from soil environments, similar to Pseudomonas putida. The beta-proteobacteria species (Azoarcus, Limnobacter) and the gamma-proteobacteria species (Acinetobacter, Marinobacter) are isolated from both soil and ocean environments. Supplementary Table 5 lists all species that were found to contain individual homologs to LvaABCD positioned throughout the genome. Supplementary Table 6 lists all species that contain LvaACD homologs. Further investigation into the utilization of LA by these species could help determine whether spatial relationship of the lva operon genes is important.
Figure 4. Predicted LA catabolism gene clusters in other genomes.
a, Representation of lva operon enzymatic genes. b, Comparison of lva operon from P. putida KT2440 with homologous, predicted LA degradation gene clusters found in other organisms. *Cupriavidus necator had less than 30% homology to LvaA, below the homology cutoff set for species isolation listed in Supplementary Tables 5 and 6
While LvaB was shown to be essential for LA catabolism, its exact role remains unclear. LvaB is a small protein (~100 aa) that is unlikely to contain enzymatic activity by-itself. Furthermore, LvaB co-purifies with LvaA, is essential for the phosphorylation of 4HV-CoA, and its orthologs are consistently found adjacent to orthologs of lvaA in the genomes of other organisms. Similar examples where small proteins provide critical support to enzymatic function have been indentified39,40. For example, nonribosomal peptide synthetase gene clusters often contain a small protein that belongs to the MbtH-like protein family, a family of proteins that are known to bind adenylation domains and enable catalytic activity. MbtH-like proteins form the necessary complexes required for domain activation but are not predicted to interact directly with the catalytic site41,42. Although LvaB does not share significant sequence homology with known MbtH-like proteins, we speculate that it could be playing a similar role with LvaA, where the presence of LvaB is required to form an active LvaAB complex. Without a crystal structure, the specific interaction between LvaA and LvaB and its role in catalysis will be difficult to unlock.
Interestingly, the isomerization of 4HV-CoA to 3HV-CoA in P. putida proceeds through a phosphorylated intermediate, 4PV-CoA, a compound also observed in a study of LA metabolism in rat livers20. This study suggested the 3HV-CoA was generated via a pathway comprised of complex phosphorylated intermediates. We did not detect MS peaks corresponding to any of these compounds in our in vitro reaction mixtures. Instead, based on changes we observed in total ion abundance over time, we propose that 4PV-CoA is dephosphorylated to an enoyl-CoA and subsequently rehydrated to 3HV-CoA. We suspect that the phosphorylation of 4HV-CoA by LvaAB generates a better leaving group and makes the subsequent dehydration more thermodynamically favorable. However, the mechanism for these last steps remains unclear. Previous groups studying the nonmevalonate pathway have identified phosphate elimination steps for the formation of a double bond that is reminiscent of the intermediates we observed43,44, but these reactions do not include a rehydration step. The timecourse measurements collected for the full reaction indicate that the formation of the pentenoyl-CoA happens quickly, but the transition from the pentenoyl-CoA to the 3HV-CoA is a much slower reaction (Figure 2f). Our tests indicate that LvaC is capable of converting 4PV-CoA to 3HV-CoA, but those reactions still contain a higher abundance of pentenoyl-CoA compared to 3HV-CoA. A more detailed mechanistic study of the final steps may clarify the specific role of LvaC.
We selected the E. coli strain LS5218 for our complementation studies because it constitutively expresses enzymes involved in beta-oxidation and catabolism of organic acids. Interestingly, this choice of host led to the requirement of additional mutations in fadE and atoC to permit robust growth on LA. These deletions likely prevent detrimental side reactions catalyzed by FadE and AtoDA (activated by AtoC(con)45,46) that would compete with the desired catabolic flux to central metabolism. FadE is an acyl-CoA dehydrogenase that catalyzes the formation of a trans-2-enoyl-CoA from an acyl-CoA47. We hypothesize that FadE, which is upregulated in FadR-deletions, may act on LA-CoA, adding a double bond between the 2- and 3-positions of the γ-ketovaleryl-CoA species and sequestering the molecule from further metabolism. Unfortunately, FadE is an inner membrane protein48,49 that has not been purified or characterized in vitro. While a deletion of atoC was not a necessary mutation, it did confer a growth benefit. We suspect that this mutation was isolated during through the directed evolution process because we were screening for mutants with rapid initial growth. Constitutive activation of the ato regulon by the atoC(Con) mutation in LS5218 causes overexpression of an acetoacetyl-CoA transferase (encoded by atoDA), an acetyl-CoA acetyltransferase (encoded by atoB) and a short chain fatty acid transporter (encoded by atoE)46,50. We suspect that 3-ketovaleryl-CoA, the product of FadB acting on 3HV-CoA, was diverted away from the desired FadA reaction by increased AtoDA activity that released 3-ketovalerate. This sequestration of LA as 3-ketovalerate would reduce overall carbon flow to central metabolites and stunt growth until cells adapt to consume 3-ketovalerate. Reducing expression of AtoDA through the deletion of atoC would prevent the shunt pathway and allow direct flux of LA to central metabolites. Further investigation of the competing metabolic pathways will be critical to developing LA-based bioconversions.
Methods
Chemicals, Strains, and Media
All chemicals were obtained from Sigma-Aldrich or Fisher Scientific. 4-hydroxyvalerate was made through the saponification of γ-valerolactone (GVL)(Martin and Prather, 2009). The pH of 2M GVL was increased to a pH of 12, using 10 M sodium hydroxide (NaOH), and incubated for 1 h. For use in bacterial growth conditions, 4HV stocks were adjusted to a pH of 8 using 5 M HCl.
Bacterial strains and plasmids used in this study are summarized in Supplementary Table 7. Plasmid sequences are listed in the Supplementary Files – Plasmids and Jupyter Notebook. E. coli strains were grown at 37°C and P. putida strains were grown at 30°C unless otherwise noted.
Plasmid construction was completed using Phusion® High Fidelity DNA Polymerase (NEB) for the PCR reactions and Gibson assembly(Gibson et al., 2009). P. putida genomic DNA sequences retrieved from NCBI database, with the following designations: PP_2791, lvaA; PP_2792, lvaB; PP_2793, lvaC; PP_2794, lvaD; PP_2795, lvaE; PP_2790, lvaR. 2 µL of the Gibson reaction mixture was transformed into chemically competent E. coli DH5α cells and plated on appropriate media. Minimal media was prepared from the following references: M9 minimal media was made according to Sambrook and Russell(Sambrook and Russell, 2001) and MOPS minimal media was made according to Neidhardt et al(Neidhardt et al., 1974). Kanamycin was used at final concentration of 50 µg/ml. Ampicillin was used at a final concentration of 100 µg/ml. Anhydrotetracycline (aTc) was used at a final concentration of 200 µg/ml. 5-Fluorouracil was used at a final concentration of 20 µg/mL.
Transposon Library and Screening
The transposon library was created following a protocol adapted from Martinez-Garcia et al(Martínez-García et al., 2011). Suicide vector delivery was achieved through bi-parental mating. Overnights of P. putida KT2440 and E. coli CC118λpir with pBAM1 were grown with appropriate antibiotics. From overnight cultures, 1 mL of cells was pelleted by centrifugation, washed with 10 mM MgSO4, and resuspended in 1 mL of 10 mM MgSO4. Cells were mixed in a 1:1 ratio into a final volume of 1 mL 10 mM MgSO4, with the final concentration of each strain at an OD600 of 0.03 (3×107 cells). The mixture was concentrated down to 30 µL and plated on 0.22 µm filter paper. The filter paper was incubated for 16 hrs on LB agar plates at 30°C. After incubation, the filter paper was removed from the plate and transferred into a 1.5 mL microfuge tube with 1 mL of 10 mM MgSO4. The cells were resuspended through vortexing and plated onto kanamycin selective M9 citrate plates, to isolate P. putida cells with transposon insertions. P. putida transposon library was screened by replica plating colonies from the M9 citrate plates onto LB, M9 glucose and M9 LA plates supplemented with kanamycin. Positive hits were identified as colonies that exhibited growth on LB and glucose plates but not on LA plates.
P. putida Barcoded Transposon Library Preparation, Enrichment, and Analysis
We generated a DNA-barcoded transposon mutant library of P. putida KT2440 using previously described methods and resources(Wetmore et al., 2015). Briefly, we conjugated wild-type P. putida KT2440 with an E. coli strain (WM3064) carrying the transposon vector library pKMW3(Wetmore et al., 2015). pKMW3 is a mariner class transposon vector library containing a kanamycin resistance marker and millions of random 20mer DNA barcodes. Conjugations were performed at 1:1 donor:recipient ratio on LB + diaminopimelic acid (DAP) plates for 6 h and finally plated on LB plates supplemented with 100 ug/mL kanamycin. The E. coli conjugation strain WM3064 is auxotrophic for DAP and does not grow on media that is not supplemented with this compound. We combined thousands of kanamycin-resistant P. putida colonies into a single tube, made multiple aliquots, and stored these samples at −80°C for future use. We also extracted genomic DNA and mapped the transposon insertion locations and their associated DNA barcodes via a TnSeq-like Illumina sequencing protocol, as previously described(Wetmore et al., 2015). We named the final, sequenced mapped transposon mutant library Putida_ML5.
An aliquot of the P. putida RB-TnSeq library (Putida_ML5) was grown for 5 h in a shake flask containing 25mL of LB media with 50 ug/mL Kanamycin Sulfate to late log phase (30°C, 250 RPM). 1 OD600*mL of cells were pelleted, decanted, and frozen at −20°C for barcode sequencing as the time zero inoculum control. 1 OD600*mL of cells per treatment were washed with three volumes of minimal media with no carbon source and then resuspended in 2× minimal media with no carbon source for a new OD600 measurement. These cells were diluted into 2× minimal media to an OD600 of 0.04. This culture was then diluted in half with 2× solutions of each carbon source of interest to a final volume of 10mL in a culture tube for 4HV and 1.2mL total volume in the well of a 24-well microplate for LA. The carbon sources tested were 40mM 4HV (pH adjusted to 7 with NaOH), 40mM LA (pH adjusted to 7 with NaOH), 20mM potassium acetate, and 40mM glucose, each with two replicates. The 4HV and acetate experiments were performed one day and the LA experiments were performed on a different day, each day with its own 40 mM Glucose control. The culture tubes were placed in a shaker incubator (30°C, 250 RPM) until they achieved and OD600 of ~3 for 40 mM Glucose (~20 h), ~0.25 for 20 mM potassium acetate (~44 h), or ~0.3–0.5 for 40mM 4HV (~68 h). For LA, the samples were grown in a 24-well microplate in a Multitron shaker set to 30°C and 700 rpm. We monitored the OD of the microplate in a Tecan M1000 microplate reader. A 1 mL sample from each culture tube was pelleted and frozen at −20°C for barcode sequencing.
We performed DNA barcode sequencing (BarSeq) as previously described(Wetmore et al., 2015), with a slight variation in our common P1 oligo design. In this study, we used a mixture of P1 oligos (Supplementary Table 10) with variable length N space regions (2–5 nt) to “phase” our BarSeq PCR products for sequencing on the Illumina HiSeq4000.
Both the TnSeq data and the BarSeq data were processed using analysis scripts as described previously(Wetmore et al., 2015). Briefly, the fitness of a strain in the normalized log2 ratio of barcode reads in the experimental sample to barcode reads in the time zero sample. The fitness of a gene is the weighted average of the strain fitness for insertions in the central 10–90% of the gene. The gene fitness values are normalized so that the typical gene has a fitness of zero. The primary statistic t-value is of the form of fitness divided by the estimated variance across different mutants of the same gene. All experiments described herein pass the quality metrics described previously unless noted otherwise(Wetmore et al., 2015).
The fitness values reported in Supplementary Table 1 are the average of 2 replicates. Fitness scores for LA and 4HV relative to glucose were calculated using the following equation:
Annotations in Supplementary Table 1 and discussed in the text were adapted from www.MicrobesOnline.org(Dehal et al., 2009).
RNA Extraction
Wild type P. putida KT2440 cells were grown in MOPS minimal media supplemented with 20 mM LA to OD600 0.8. 10 OD-mL were collected by centrifugation at 5000 × g for 10 min at 4°C in Beckman Coulter Allegra X-15R. The supernatant was decanted and the pellet frozen at −80°C for 24 hrs. The RNA extraction protocol is adapted from Pinto et al(Pinto et al., 2009). The frozen pellet was thawed, resuspended in 1.5 mL Trizol and transferred to a 2.0 mL microfuge tube. The suspension was incubated for 5 min at 95°C and then for 5 min on ice. After the incubation, 300 µL chloroform was added and the tube shaken vigorously for 15 sec. The Trizol-chloroform mixture was incubated at room temperature for 15 min and then centrifuged for 15 min at 12000 × g and 4°C. The upper phase was transferred to a fresh tube and an equal volume of isopropanol was added. This mixture was incubated for 10 min at room temperature and then centrifuged for 10 min at 12,000 × g and 4°C. The supernatant was discarded and the pellet resuspended in 1 mL of 75% ethanol. This was centrifuged for 5 min at 8,000 × g and 4°C. The supernatant was discarded, the pellet air dried for 3 min and then resuspended in 100 µL RNase-free water and stored at −80°C.
Transcription Start Site Isolation
The transcription start site for genes lvaR and lvaA were isolated using an adapted 5’ Race protocol from Schramm et al(Schramm et al., 2000). The RNA isolated from P. putida KT2440 was treated with the TURBO DNA-free™ Kit from Ambion® to remove any contaminating DNA. The Promega GoScript RT PCR kit was used to generate cDNA using 1 µL of a 10 µM gene specific oligo (JMR2 for lvaR and JMR287 for lvaA) instead of the random oligo mixture. Following the inactivation of the reverse transcriptase, the cDNA was purified using Qiagen PCR Purification kit. Tailing of the cDNA was achieved using the terminal deoxynucleotidyl transferase (TdT) enzyme from Thermo Scientific. The final reaction mixture contained 1× reaction buffer, 1 pmol cDNA fragments, 60 pmol dGTP or dCTP and 30 U TdT. The reaction was incubated at 37°C for 15 min and then quenched by heating to 70°C for 10 min and the tailed cDNA fragments cleaned up using a Qiagen PCR Purification kit. The tailed cDNA was amplified using GoTaq Green Master Mix with an annealing temperature of 55°C and an extension time of 30 sec. Primer GG318 was used for dGTP tailing and ALM244 was used for dCTP tailing. The reverse primer for lvaR was JMR150 and for lvaA was JMR296. The resulting PCR product was submitted for sequencing.
Polycistronic Verification
Using the DNAse treated RNA isolated from LA grown P. putida KT2440, cDNA for the operon was generated with the Promega GoScript RT PCR kit using 1 µL of a 10 µM gene specific oligo (JMR237). The cDNA was then used as the template for PCR reactions using GoTaq Green Master Mix with an annealing temperature of 55°C and an extension time of 0:30 sec. Primers used for each gene are given in Supplementary Table 8.
P. putida Knockouts
The genetic knockout of lvaD was performed following the protocol from Schafer et al(Schäfer et al., 1994). Knockouts of the remaining genes in P. putida were performed following the protocol from Graf et al(Graf and Altenbuchner, 2011). Knockout constructs were designed with 500 bp of homology up and down stream of the deletion site. This region was cloned into the pJOE vector backbone. This suicide vector was transformed into P. putida KT2440 Δupp (P. putida KTU) through electroporation and colonies that successfully integrated the plasmid into the chromosome were selected on LBkan plates. A colony was then grown in LB media overnight to cure the counter-selection cassette. Various dilutions of the overnight culture were plated on LB5-FU plates to isolate colonies that had successfully excised the plasmid insertion. Colonies were then screened by colony PCR to isolate deletion strains.
Transcriptional Reporter Assay
P. putida KT2440 was transformed with pJMR74 through electroporation. pJMR74 is a broad host range plasmid containing a kan resistance marker and the predicted regulator for the lva operon, lvaR. Expressed divergent of lvaR is sfGFP cloned under the native promoter for lvaA. P. putida KT2440 containing empty vector pBAD35 was used as the no fluorescence control. Overnights of P. putida + pJMR74 or pBAD35 were inoculated at an OD600 of 0.05 in LB + kan50 + 20 mM of the appropriate carboxylic acid (acetate, propionate, butyrate, valerate, LA, 4HV, or hexanoate). Final timepoints were taken at 24 h in a Tecan infinite m1000, with OD absorbance measured at 600 nm and fluorescence measured with an excitation of 485 nm and emission of 510 nm. Standard deviation error propagation was performed for the normalization of fluorescence and optical density measurements.
Protein Production and Purification
Vectors were constructed using the pET28b backbone and individually cloned genes from the P. putida genome. The plasmid containing lvaAB was constructed using the pET28b backbone and the lvaAB genes cloned as an operon directly out of P. putida’s genome. E. coli BL21 (DE3) strains with sequenced verified plasmids were grown at 37°C in LB. Cultures were induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at an OD600 of 0.4. The cultures were then chilled on ice for 10 min before incubation at 16°C for 18 h in New Brunswick Incubator I-26. Then the cultures were centrifuged for 20 min at 5,000 × g in a Beckman Coulter Avanti J-E centrifuge. The supernatant was decanted and the cells resuspended in 30 mL of LB before another centrifugation at 5,000 × g for 20 min. The supernatant was removed and pellets stored at −80°C for at least 24 h.
Purification of His6- (lvaE) and Maltose Binding Protein (MBP)-tagged proteins (lvaABCD)
Frozen cell pellets were thawed on ice and resuspended in His6-lysis buffer (50mM Na2HPO4, 300 mM NaCl, 10mM imidazole, 2 mM DTT, pH 8.0) supplemented with 2 µL of benzonase or MBP-lysis buffer (20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA, 1mM DTT, pH 7.4) supplemented with 2 µL of benzonase. Cell suspensions were sonicated 3 times using the program: 1.5 sec pulse, 1.5 sec pause, 40% duty, for a total of 30 sec. Between each sonication cycle, the solution was stored on ice for 5 min. Lysed cells were centrifuged at 25,000 × g at 4°C for 30 min and the supernatant filtered through a 0.45 µm filter.
For the purification of His6-tagged proteins, a GE Äkta Start System with a 1 mL HisTrap HP column and a constant flow rate of 1 mL/min was used. 5 column volume (CV) of wash buffer (50 mM Na2HPO4, 300 mM NaCl, 40 mM imidazole, 2 mM DTT, pH 8.0) was used to equilibrate the column. The sample was loaded and washed with 15 CV wash buffer. The protein was eluted with 5 CV elution buffer (50 mM Na2HPO4, 300 mM NaCl, 250 mM imidazole, 2 mM DTT, pH 7.8). 1 mL fractions of eluted protein were collected. A GE PD-10 desalting column was used to buffer exchange the protein into the desalting buffer (100 mM Tris, 4.1 M glycerol and 2 mM DTT). An Amicon® Ultra 4 mL Centrifugal Filter with a 10 kDa cut-off size was used to concentrate the protein. Each protein was stored at −80°C until use.
For the purification of MBP-tagged proteins, a GE Äkta Start System with a 1 mL MBPTrap HP column and a constant flow rate of 1 mL/min was used. 5 column volume (CV) of wash buffer was used to equilibrate the column. The sample was loaded and washed with 15 CV wash buffer. The protein was eluted with 5 CV elution buffer (20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA, 1mM DTT, 10 mM maltose, pH 7.4). 1 mL fractions of eluted protein were collected. A GE PD-10 desalting column was used to buffer exchange the protein into the desalting buffer (100 mM Tris, 4.1 M glycerol, 2 mM DTT, pH 8.2). An Amicon® Ultra 4 mL Centrifugal Filter with a 10 kDa cut-off size was used to concentrate the protein. The protein was stored at −80°C until use.
LvaAB Pulldown Experiment
All proteins for the pulldown experiment were purified on a 1 mL MBPTrap HP column, as previously described, regardless of the protein tag. LvaA was tagged with an N-terminal MBP tag. LvaAB was designed with LvaA tagged with an N-terminal MBP tag and LvaB untagged. Both proteins were expressed from the same construct as they appear in a native operon. For controls, LvaA contained an N-terminal His tag and was expressed with the native LvaB and the last control was an N-terminal MBP tagged LvaA containing a frameshift stop codon expressed with native LvaB. The purified proteins were analyzed on a 15% SDS-page gel to determine the major protein products.
CoA Ligase Assay
A CoA ligase activity assay was performed with the EnzChek® Pyrophosphate Assay Kit. The maximum allowable concentration of LvaE that did not affect assay sensitivity was determined to be 0.2 µM through an enzymatic dilution test. The final reaction volume of 100 µL contained 0.1 mM ATP, 0.1 mM CoA, 0.2 µM LvaE, 0.2 mM MESG, 1 U purine nucleoside phosphorylase, 0.01 U pyrophosphatase, 50 mM Tris-HCL, 1 mM MgCl2 and 0.1 mM substrate (sodium acetate, sodium propionate, butyric acid, valeric acid, levulinic acid, hexanoic acid, octanoic acid, 4HV, 2-pentenoic acid, 3-pentenoic acid, 3HV, γ-valerolactone, pyruvate, L-carnitine). All substrate stocks were adjusted to pH 7 before use. Reactions were incubated at 25°C for 30 min before measuring the absorbance at 360 nm in a Tecan m1000. A control reaction that did not contain a substrate was used for performing an absorbance baseline subtraction.
Enzyme Assays and Metabolite Purification
All in vitro enzyme assays were performed in a 30° water bath at a pH of 7.5 and contained 50 mM Tris-HCL, 1 mM MgCl2, and 2 mM DTT. Final reaction concentrations included the following components, depending on enzymes added: 0.5 mM LA, 0.55 mM CoA, 0.55 mM ATP (1.05 mM ATP when lvaAB were present), 0 mM NAD(P)H (0.55 mM NAD(P)H when lvaD was present). Final protein concentrations were: LvaA (0.2 µM), LvaB (0.8 µM), LvaAB (0.4 µM), LvaC (0.4 µM), LvaD (0.2 µM), and LvaE (0.2 µM) (Supplementary Figure 5). The in vitro enzyme assays were incubated for 30 min, excluding the timecourse which was incubated for various intervals up to 60 min. Reaction metabolites were purified following a modified protocol from Zhang et. al. 2009(Zhang et al., 2009). Reactions were quenched by adding methanol/water 1:1 containing 5% acetic acid in a 1:1 volume ratio (extraction buffer). Quenched reactions were run on a 1 mL ion exchange column prepacked with 100 mg 2-2(pyridyl)ethyl silica gel from Sigma. The column had been preconditioned with 1 mL methanol followed by 1 mL of extraction buffer. Metabolites loaded on the column were washed with 750 µL extraction buffer before being eluted with 1 mL of 4:1 methanol/250 mM ammonium formate, pH 6.3 and 1 mL methanol. Samples were dried using Thermo Scientific Savant SC250EXP Speedvac Concentrator and stored at −80°C until LC/MS analysis. Samples for LC/MS analysis were resuspended in 100 µL 50 mM ammonium formate.
Liquid Chromatography Mass spectrometry (LC/MS, LC/MS/MS)
Samples were analyzed using an HPLC-MS/MS system consisting of a VanquishTM UHPLC system (Thermo Scientific) coupled by electrospray ionization (ESI; negative polarity) to a hybrid quadrupole - high-resolution mass spectrometer (Q Exactive orbitrap, Thermo Scientific) operated in full scan mode for detection of targeted compounds based on their accurate masses. Properties of Full MS – SIM included resolution of 140,000, AGC target of 1E6, maximum IT of 40 ms, and scan range from 70–1000 m/z. Liquid chromatography (LC) separation was achieved using an ACQUITY UPLC® BEH C18 (2.1 × 100 mm column, 1.7µm particle size; Part No. 186002352; Serial No. 02623521115711; Waters, Milford, MA). Solvent A was 97:3 water:methanol with 10 mM tributylamine (TBA) adjusted to pH 8.1–8.2 with 9 mM acetic acid. Solvent B was 100% methanol. Total run time was 25 min with the following gradient: 0 min, 5% B; 2.5 min, 5% B; 5 min, 20% B; 7.5 min, 20% B; 13 min, 55% B; 15.5 min, 95% B; 18.5 min, 95% B; 19 min, 5% B; 25 min, 5% B. Flow rate was 200 µL/min. The autosampler and the column temperatures were 4°C and 25°C, respectively. Fragmentation of CoA, 4HV-CoA, and phosphorylated 4HV-CoA was achieved using parameters indicated in Supplementary Table 9.
Enzymatic “In Gel” Digestion and Indentification of Lva Proteins
“In gel” digestion and mass spectrometric analysis was done at the Mass Spectrometry Facility (Biotechnology Center, University of Wisconsin-Madison). The digestion was performed as outlined on the website: http://www.biotech.wisc.edu/ServicesResearch/MassSpec/ingel.htm. In short, Coomassie Blue R-250 stained gel pieces were de-stained twice for 5 min in MeOH/H20/NH4HCO3 (50%:50%:100mM), dehydrated for 5 min in ACN/H20/NH4HCO3 (50%:50%:25mM) then once more for 1 min. in 100% ACN, dried in a Speed-Vac for 2 min., reduced in 25mM DTT (Dithiotreitol in 25mM NH4HCO3) for 30 min. at 56°C, alkylated with 55mM IAA (Iodoacetamide in 25mM NH4HCO3) in darkness at room temperature for 30 min., washed twice in H20 for 30 sec., equilibrated in 25mM NH4HCO3 for 1 min., dehydrated for 5 min. in ACN/H20/NH4HCO3 (50%:50%:25mM) then once more for 30sec in 100% ACN, dried again and rehydrated with 20µl of trypsin solution (10ng/µl trypsin Gold (PROMEGA) in 25mM NH4HCO3 / 0.01% ProteaseMAX w/v (PROMEGA)). Additional 30µl of digestion solution (25mM NH4HCO3 / 0.01% ProteaseMAX w/v (PROMEGA)) was added to facilitate complete rehydration and excess overlay needed for peptide extraction. The digestion was conducted for 3hrs at 42°C. Peptides generated from digestion were transferred to a new tube and acidified with 2.5% TFA (Trifluoroacetic Acid) to 0.3% final. Degraded ProteaseMAX was removed via centrifugation (max speed, 10 min) and the peptides solid phase extracted (ZipTip® C18 pipette tips Millipore, Billerica, MA).
Peptides were analyzed by nanoLC-MS/MS using the Agilent 1100 nanoflow system (Agilent) connected to a new generation hybrid linear ion trap-orbitrap mass spectrometer (LTQ-Orbitrap Elite™, Thermo Fisher Scientific) equipped with an EASY-Spray™ electrospray source. Chromatography of peptides prior to mass spectral analysis was accomplished using capillary emitter column (PepMap® C18, 3µM, 100Å, 150×0.075mm, Thermo Fisher Scientific) onto which 2µl of extracted peptides was automatically loaded. NanoHPLC system delivered solvents A: 0.1% (v/v) formic acid, and B: 99.9% (v/v) acetonitrile, 0.1% (v/v) formic acid at 0.50 µL/min to load the peptides (over a 30 min period) and 0.3µl/min to elute peptides directly into the nano-electrospray with gradual gradient from 3% (v/v) B to 30% (v/v) B over 77 min and concluded with 5 min fast gradient from 30% (v/v) B to 50% (v/v) B at which time a 5 min flash-out from 50–95% (v/v) B took place. As peptides eluted from the HPLC-column/electrospray source survey MS scans were acquired in the Orbitrap with a resolution of 120,000 followed by MS2 fragmentation of 20 most intense peptides detected in the MS1 scan from 300 to 2000 m/z; redundancy was limited by dynamic exclusion.
Raw MS/MS data were converted to mgf file format using MSConvert (ProteoWizard: Open Source Software for Rapid Proteomics Tools Development). Resulting mgf files were used to search against Pseudomonas putida amino acid sequence database containing a list of common contaminants (5,388 total entries) using in-house Mascot search engine 2.2.07 (Matrix Science) with variable Methionine oxidation with Asparagine and Glutamine deamidation plus fixed cysteine Carbamidomethylation. Peptide mass tolerance was set at 15 ppm and fragment mass at 0.6 Da.
Identification of Organisms with Potential Homologous LA Catabolism Pathways
Possible LvaABCD homologs were identified by performing a BLAST search of each protein sequence against the NCBI non-redundant protein sequence database using the BioPython library (python code provided as Juptyer Notebook in Supplementary Files- Plasmids and Jupyter Notebook) (Altschul et al., 1990; Cock et al., 2009). From the search results, the organism name was extracted from the sequence title and added to a set for each protein. The list of organisms containing the full set of LvaABCD enzymes was found by determining the intersection of the four sets of organism names from the BLAST results from each protein. A similar list was found for those organisms containing only LvaACD homologs. These lists were then used to query the original search results and find the lists of proteins that have homology to the proteins in the Lva pathway.
Genome sequencing and analysis
DNA was isolated from E. coli strains using the Wizard® Genomic DNA Purification Kit (Promega) and sequenced by the University of Wisconsin Biotechnology Center. A paired end library (2×250) was run on an Illumina Hi-Seq. Sequencing reads (as FASTQ files) of E. coli mutants were mapped to the sequenced reference genome E. coli LS5218 (GCA_002007165.1) using Bowtie2 using the “fast-local” setting (Langmead and Salzberg, 2012). The output sequence alignment map (SAM) file was converted to a binary alignment map (BAM) file and sorted using SAMtools (Li et al., 2009). Variants were then called using Naïve Variant Caller (Galaxy open source bioinformatics tool) (Goto et al., 2011).
Directed evolution of E. coli LS5218
Sub-culturing experiments were performed with a volume of 5 ml in glass test tubes (20 × 150mm, Fisher Scientific) with 250 rpm agitation in a I26 shaker (New Brunswick Scientific). Starting media contained 20 mM LA and 40 mM acetate or 40 mM acetate only for negative control. Cultures were grown for 72 h and optical density measurements taken with a Spectronic 20 (Milton Roy Company), then culture were diluted 1:100 into fresh media. Once the OD600 in the LA and acetate cultures exceeded the OD600 of the acetate only cultures, further growth media was 20 mM LA only. These cultures were incubated until turbidity was observed visually, then diluted 1:100 into fresh media. This occurred for a total of 14 dilutions steps in LA media, spanning two weeks.
Plasmids were prepped (QIAprep® Miniprep Kits, Qiagen) and sequenced (Functional Biosciences) to find mutations. Plasmids were cured out of mutate strains through serial culturing in rich media (LB broth) and patch plated on LB and LBkan50.
Genome engineering with CRISPR-Cas9
CRISPR/Cas recombineering was performed following an adapted protocol from(Li et al., 2015; Qi et al., 2013).
Data Availability
All data from the P. putida transposon sequencing experiments is available through the fitness browser at http://fit.genomics.lbl.gov/cgi-bin/exps.cgi?orgId=Putida&expGroup=carbon%20source.
All plasmid sequences are included in the Supplementary Files. P. putida genomic DNA sequences retrieved from the NCBI database, with the following designations: PP_2791, lvaA; PP_2792, lvaB; PP_2793, lvaC; PP_2794, lvaD; PP_2795, lvaE; PP_2790, lvaR. The E. coli LS5218 reference genome can be retrieved from the NCBI database under the following accession number: GCA_002007165.1.
The python code used to identify potential lva homologs is provided in the Supplementary Files.
Any additional data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
Work in the Pfleger lab was funded by the National Science Foundation (CBET-114678) and the William F. Vilas Trust. Work in the Deutschbauer and Arkin labs was funded by ENIGMA, a Scientific Focus Area Program, supported by the U. S. Department of Energy, Office of Science, Office of Biological and Environmental Research and Genomics:GTLFoundational Science through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy. Work in the Amador-Noguez lab was funded by the HHMI International Student Research Fellowship. R.L.C. was supported by the NIH NHGRI Genomic Sciences Training Program (T32 HG002760). A.L.M. was supported by NSF SEES fellowship (GEO-1215871). J.M.R. was supported by a NSF Graduate Research Fellowship (DGE-1256259).
We want to thank J. Escalante for providing the plasmid pK18mobsacB and J. Altenbuchner for providing the strain P. putida KTU and the plasmid pJOE6261.2. We would like to thank the Mass Spectrometry/Proteomics Facility at the UW-Madison Biotechnology Center for performing the in-gel digest and providing the LC-MS/MS results and the UW-Madison Biotechnology Center DNA Sequencing Facility for providing genomic sequencing services. We could also like to thank G. Gordon for helping analyze the E. coli genomic sequencing SNPs.
Footnotes
Author Contribution
J.M.R., D.E.A., and B.F.P. conceived the study. J.M.R. designed and performed the experiments and analyzed the data with the following exceptions. T.P. and D.A.M. designed the LC/MS/MS experiments and T.P. performed the LC/MS and LC/MS/MS experiments. D.E.A. and J.M.T. performed the transposon library screen. C.E.C. assisted with the promoter and CoA ligase assay. A.L.M. proposed, and helped design and perform, the pull-down experiment. Y.S. and J.R. prepared the RB-TnSeq mutant library of P. putida KT2440 (Putida_ML5). K.M.W., R.L.C, J.R. and A.M.D. performed the fitness assays with the Putida_ML5 library. M.N.P. performed the data analysis to determine fitness values. R.L.C. prepared the supplementary analysis of the Putida_ML5 fitness experiments. A.M.D. and A.P.A. managed the Bar-Seq experiments. C.R.M helped design and analyze the prevalence of the lva operon in other organisms. J.M.R. and B.F.P wrote the manuscript.
References
- 1.Pileidis FD, Titirici MM. Levulinic Acid Biorefineries: New Challenges for Efficient Utilization of Biomass. ChemSusChem. 2016;9:562–582. doi: 10.1002/cssc.201501405. [DOI] [PubMed] [Google Scholar]
- 2.Bozell JJ, et al. Production of levulinic acid and use as a platform chemical for derived products. Resour. Conserv. Recycl. 2000;28:227–239. [Google Scholar]
- 3.Werpy T, Petersen G. Top Value Added Chemicals from Biomass. Program. 2004;76 doi: 10.2172/926125. [DOI] [Google Scholar]
- 4.Lange JP, et al. Valeric biofuels: A platform of cellulosic transportation fuels. Angew. Chemie - Int. Ed. 2010;49:4479–4483. doi: 10.1002/anie.201000655. [DOI] [PubMed] [Google Scholar]
- 5.Alonso DM, Wettstein SG, Dumesic JA. Gamma-valerolactone, a sustainable platform molecule derived from lignocellulosic biomass. Green Chem. 2013;15:584. [Google Scholar]
- 6.Joshi H, Moser BR, Toler J, Smith WF, Walker T. Ethyl levulinate: A potential bio-based diluent for biodiesel which improves cold flow properties. Biomass and Bioenergy. 2011;35:3262–3266. [Google Scholar]
- 7.Zhang Z, Dong K, Zhao Z. Efficient conversion of furfuryl alcohol into alkyl levulinates catalyzed by an organic-inorganic hybrid solid acid catalyst. ChemSusChem. 2011;4:112–118. doi: 10.1002/cssc.201000231. [DOI] [PubMed] [Google Scholar]
- 8.Demolis A, Essayem N, Rataboul F. Synthesis and Applications of Alkyl Levulinates. ACS Sustain. Chem. Eng. 2014;2:1338–1352. [Google Scholar]
- 9.Moore JA, Tannahill T. Homo- and co-polycarbonates and blends derived from diphenolic acid. High Perform. Polym. 2001;13:305–316. [Google Scholar]
- 10.Guo Y, Li K, Yu X, Clark JH. Mesoporous H3PW12O40-silica composite: Efficient and reusable solid acid catalyst for the synthesis of diphenolic acid from levulinic acid. Appl. Catal. B Environ. 2008;81:182–191. [Google Scholar]
- 11.Chung SH, Choi GG, Kim HW, Rhee YH. Effect of Levulinic Acid on the Production of Poly ( 3-hydroxybutyrate-co-3-hydroxyvalerate) by Ralstonia eutropha KHB-8862. Society. 2001;39:79–82. [Google Scholar]
- 12.Berezina N, Yada B. Improvement of the poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV) production by dual feeding with levulinic acid and sodium propionate in Cupriavidus necator. N. Biotechnol. 2016;33:231–236. doi: 10.1016/j.nbt.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Jang JH, Rogers PL. Effect of levulinic acid on cell growth and poly-beta-hydroxyalkanoate production by Alcaligenes sp SH-69. J. Chem. Inf. Model. 1996;18:219–224. [Google Scholar]
- 14.Habe H, et al. Bacterial production of short-chain organic acids and trehalose from levulinic acid: A potential cellulose-derived building block as a feedstock for microbial production. Bioresour. Technol. 2015;177:381–386. doi: 10.1016/j.biortech.2014.11.048. [DOI] [PubMed] [Google Scholar]
- 15.Martin CH, Wu D, Prather KLJ, Jones Prather KL. Integrated bioprocessing for the pH-dependent production of 4-valerolactone from levulinate in pseudomonas putida KT2440. Appl. Environ. Microbiol. 2010;76:417–424. doi: 10.1128/AEM.01769-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeon YJ, Park HY, Yoo YJ. Enzymatic reduction of levulinic acid by engineering the substrate specificity of 3-hydroxybutyrate dehydrogenase. Bioresour. Technol. 2013;134:377–380. doi: 10.1016/j.biortech.2013.01.078. [DOI] [PubMed] [Google Scholar]
- 17.Jaremko M, Yu J. The initial metabolic conversion of levulinic acid in Cupriavidus necator. J. Biotechnol. 2011;155:293–298. doi: 10.1016/j.jbiotec.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 18.Martin CH, Prather KLJ. High-titer production of monomeric hydroxyvalerates from levulinic acid in Pseudomonas putida. J. Biotechnol. 2009;139:61–67. doi: 10.1016/j.jbiotec.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Zhang GF, et al. Catabolism of 4-hydroxyacids and 4-hydroxynonenal via 4-hydroxy-4-phosphoacyl-CoAs. J. Biol. Chem. 2009;284:33521–33534. doi: 10.1074/jbc.M109.055665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris SR, et al. Metabolism of levulinate in perfused rat livers and live rats: Conversion to the drug of abuse 4-hydroxypentanoate. J. Biol. Chem. 2011;286:5895–5904. doi: 10.1074/jbc.M110.196808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez-García E, Calles B, Arévalo-Rodríguez M, de Lorenzo V. pBAM1: an all-synthetic genetic tool for analysis and construction of complex bacterial phenotypes. BMC Microbiol. 2011;11:38. doi: 10.1186/1471-2180-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wetmore KMM, et al. Rapid Quantification of Mutant Fitness in Diverse Bacteria by Sequencing Randomly Bar-Coded Transposons. MBio. 2015;6:1–15. doi: 10.1128/mBio.00306-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Schramm G, Bruchhaus I, Roeder T. A simple and reliable 5’-RACE approach. Nucleic Acids Res. 2000;28:E96. doi: 10.1093/nar/28.22.e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Espah Borujeni A, Channarasappa ASS, Salis HMM. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 2014;42:2646–2659. doi: 10.1093/nar/gkt1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrios H, Valderrama B, Morett E. Compilation and analysis of sigma(54)-dependent promoter sequences. Nucleic Acids Res. 1999;27:4305–4313. doi: 10.1093/nar/27.22.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fox JD, Routzahn KM, Bucher MH, Waugh DS. Maltodextrin-binding proteins from diverse bacteria and archaea are potent solubility enhancers. FEBS Lett. 2003;537:53–57. doi: 10.1016/s0014-5793(03)00070-x. [DOI] [PubMed] [Google Scholar]
- 28.Striebel F, et al. Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat. Struct. Mol. Biol. 2009;16:647–651. doi: 10.1038/nsmb.1597. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto S, Kutsukake K. FliT acts as an anti-FlhD2C2 factor in the transcriptional control of the flagellar regulon in Salmonella enterica serovar typhimurium. J. Bacteriol. 2006;188:6703–8. doi: 10.1128/JB.00799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dijkman WP, Fraaije MW. Discovery and characterization of a 5-hydroxymethylfurfural oxidase from Methylovorus sp. strain MP688. Appl. Environ. Microbiol. 2014;80:1082–1090. doi: 10.1128/AEM.03740-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simons RW, Egan PA, Chute HT, Nunn W. D. Regulation of Fatty Acid Degradation in Escherichia coli : Isolation and Characterization of Strains Bearing Insertion and Temperature-Sensitive Mutations in gene fadR. 1980;142:621–632. doi: 10.1128/jb.142.2.621-632.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brock M, Maerker C, Schutz A, Volker U, Buckel W. Oxidation of propionate to pyruvate in Escherichia coli - Involvement of methylcitrate dehydratase and aconitase. Eur. J. Biochem. 2002;269:6184–6194. doi: 10.1046/j.1432-1033.2002.03336.x. [DOI] [PubMed] [Google Scholar]
- 33.Man WJ, Li Y, O’Connor CD, Wilton DC. The Binding of Propionyl-Coa and Carboxymethyl-Coa to Escherichia-Coli Citrate Synthase. Biochim. Biophys. Acta. 1995;1250:69–75. doi: 10.1016/0167-4838(95)00044-u. [DOI] [PubMed] [Google Scholar]
- 34.Jiang W, Bikard D, Cox D, Zhang F, Marraffini La. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Y, et al. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015;81:2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, et al. Metabolic engineering of Escherichia coli using CRISPR-Cas9 meditated genome editing. Metab. Eng. 2015;31:13–21. doi: 10.1016/j.ymben.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Moreno R, Marzi S, Romby P, Rojo F. The Crc global regulator binds to an unpaired A-rich motif at the Pseudomonas putida alkS mRNA coding sequence and inhibits translation initiation. Nucleic Acids Res. 2009;37:7678–7690. doi: 10.1093/nar/gkp825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreno R, Martínez-Gomariz M, Yuste L, Gil C, Rojo F. The Pseudomonas putida Crc global regulator controls the hierarchical assimilation of amino acids in a complete medium: Evidence from proteomic and genomic analyses. Proteomics. 2009;9:2910–2928. doi: 10.1002/pmic.200800918. [DOI] [PubMed] [Google Scholar]
- 39.Storz G, Wolf YI, Ramamurthi KS. Small proteins can no longer be ignored. Annu. Rev. Biochem. 2014;83:753–77. doi: 10.1146/annurev-biochem-070611-102400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su M, Ling Y, Yu J, Wu J, Xiao J. Small proteins: Untapped area of potential biological importance. Front. Genet. 2013;4:1–9. doi: 10.3389/fgene.2013.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Felnagle EA, et al. MbtH-like proteins as integral components of bacterial nonribosomal peptide synthetases. Biochemistry. 2010;49:8815–8817. doi: 10.1021/bi1012854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baltz RH. Function of MbtH homologs in nonribosomal peptide biosynthesis and applications in secondary metabolite discovery. J. Ind. Microbiol. Biotechnol. 2011;38:1747–1760. doi: 10.1007/s10295-011-1022-8. [DOI] [PubMed] [Google Scholar]
- 43.Gräwert T, et al. Structure of active IspH enzyme from escherichia coli provides mechanistic insights into substrate reduction. Angew. Chemie - Int. Ed. 2009;48:5756–5759. doi: 10.1002/anie.200900548. [DOI] [PubMed] [Google Scholar]
- 44.Hecht S, et al. Studies on the nonmevalonate pathway to terpenes: the role of the GcpE (IspG) protein. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14837–14842. doi: 10.1073/pnas.201399298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jenkins LS, Nunn WD. Genetic and molecular characterization of the genes involved in short-chain fatty acid degradation in Escherichia coli: the ato system. J Bacteriol. 1987;169:42–52. doi: 10.1128/jb.169.1.42-52.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matta MK, Lioliou EE, Panagiotidis CH, Kyriakidis DA, Panagiotidis CA. Interactions of the antizyme AtoC with regulatory elements of the Escherichia coli atoDAEB operon. J. Bacteriol. 2007;189:6324–32. doi: 10.1128/JB.00214-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Campbell JW, Cronan JEJ. The Enigmatic Escherichia coli fadE Gene Is yafH. J. Bacteriol. 2002;184:3759–3764. doi: 10.1128/JB.184.13.3759-3764.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daley DO, et al. Global topology analysis of the Escherichia coli inner membrane proteome. Science (80-.) 2005;308:1321–1323. doi: 10.1126/science.1109730. [DOI] [PubMed] [Google Scholar]
- 49.Díaz-Mejía JJ, Babu M, Emili A. Computational and experimental approaches to chart the Escherichia coli cell-envelope-associated proteome and interactome. FEMS Microbiol. Rev. 2009;33:66–97. doi: 10.1111/j.1574-6976.2008.00141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jenkins LS, Nunn WD. Regulation of the ato operon by the atoC gene in Escherichia coli. J. Bacteriol. 1987;169:2096–2102. doi: 10.1128/jb.169.5.2096-2102.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJL. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–3. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, Friedland GD, Huang KH, Keller K, Novichkov PS, Dubchak IL, Alm EJ, Arkin AP. MicrobesOnline: An integrated portal for comparative and functional genomics. Nucleic Acids Res. 2009;38:396–400. doi: 10.1093/nar/gkp919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–5. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Goto H, Dickins B, Afgan E, Paul IM, Taylor J, Makova KD, Nekrutenko A, Nekrutenko A, Goto H, Dickins B, Afgan E, Paul I, Taylor J, Makova K, Nekrutenko A, Chinnery P, Thorburn D, Samuels D, White S, Dahl H, Turnbull D, Lightowlers R, Howell N, Jacobs H, DiMauro S, Mercer T, Neph S, Dinger M, Crawford J, Smith M, Shearwood A-M, Haugen E, Bracken C, Rackham O, Stamatoyannopoulos J, Filipovska A, Mattick J, Li M, Wang I, Li Y, Bruzel A, Richards A, Toung J, Cheung V, Chen R, Mias G, Li-Pook-Than J, Jiang L, Lam H, Chen R, Miriami E, Karczewski K, Hariharan M, Dewey F, Cheng Y, Clark M, Im H, Habegger L, Balasubramanian S, O’Huallachain M, Dudley J, Hillenmeyer S, Haraksingh R, Sharon D, Euskirchen G, Lacroute P, Bettinger K, Boyle A, Kasowski M, Grubert F, Seki S, Garcia M, Whirl-Carrillo M, Gallardo M, Blankenberg D, Taylor J, Schenck I, He J, Zhang Y, Ghent M, Veeraraghavan N, Albert I, Miller W, Makova K, Hardison R, Nekrutenko A, Goecks J, Nekrutenko A, Taylor J, Afgan E, Baker D, Coraor N, Goto H, Paul I, Makova K, Nekrutenko A, Taylor J, Marth G, Korf I, Yandell M, Yeh R, Gu Z, Zakeri H, Stitziel N, Hillier L, Kwok P, Gish W, Li M, Schonberg A, Schaefer M, Schroeder R, Nasidze I, Stoneking M, Bar-Yaacov D, Avital G, Levin L, Richards A, Hachen N, Jaramillo BR, Nekrutenko A, Zarivach R, Mishmar D, Nekrutenko A, Taylor J, Danecek P, Auton A, Abecasis G, Albers C, Banks E, Depristo M, Handsaker R, Lunter G, Marth G, Sherry S, McVean G, Durbin R. Dynamics of mitochondrial heteroplasmy in three families investigated via a repeatable re-sequencing study. Genome Biol. 2011;12:R59. doi: 10.1186/gb-2011-12-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf N, Altenbuchner J. Development of a method for markerless gene deletion in Pseudomonas putida. Appl. Environ. Microbiol. 2011;77:5549–52. doi: 10.1128/AEM.05055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lin Z, Huang C, Zhang Y, Wang Z, Tang Y, Chen T, Zhao X. Metabolic engineering of Escherichia coli using CRISPR-Cas9 meditated genome editing. Metab. Eng. 2015;31:13–21. doi: 10.1016/j.ymben.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Martin CH, Prather KLJ. High-titer production of monomeric hydroxyvalerates from levulinic acid in Pseudomonas putida. J. Biotechnol. 2009;139:61–67. doi: 10.1016/j.jbiotec.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Martínez-García E, Calles B, Arévalo-Rodríguez M, de Lorenzo V. pBAM1: an all-synthetic genetic tool for analysis and construction of complex bacterial phenotypes. BMC Microbiol. 2011;11:38. doi: 10.1186/1471-2180-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC, Bloch PL, Smith DF. Culture Medium for Enterobacteria. J. Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto FL, Thapper A, Sontheim W, Lindblad P. Analysis of current and alternative phenol based RNA extraction methodologies for cyanobacteria. BMC Mol. Biol. 2009;10:79. doi: 10.1186/1471-2199-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-γuided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW, David W. Molecular cloning : a laboratory manual. Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Schramm G, Bruchhaus I, Roeder T. A simple and reliable 5’-RACE approach. Nucleic Acids Res. 2000;28:E96. doi: 10.1093/nar/28.22.e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore KM, Price MN, Waters RJ, Lamson JS, He J, Hoover CA, Blow MJ, Bristow J, Butland G, Arkin AP, Deutschbauer A. Rapid Quantification of Mutant Fitness in Diverse Bacteria by Sequencing Randomly Bar-Coded Transposons. MBio. 2015;6:e00306–15. doi: 10.1128/mBio.00306-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GF, Kombu RS, Kasumov T, Han Y, Sadhukhan S, Zhang J, Sayre LM, Ray D, Gibson KM, Anderson VA, Tochtrop GP, Brunengraber H. Catabolism of 4-hydroxyacids and 4-hydroxynonenal via 4-hydroxy-4-phosphoacyl-CoAs. J. Biol. Chem. 2009;284:33521–33534. doi: 10.1074/jbc.M109.055665. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data from the P. putida transposon sequencing experiments is available through the fitness browser at http://fit.genomics.lbl.gov/cgi-bin/exps.cgi?orgId=Putida&expGroup=carbon%20source.
All plasmid sequences are included in the Supplementary Files. P. putida genomic DNA sequences retrieved from the NCBI database, with the following designations: PP_2791, lvaA; PP_2792, lvaB; PP_2793, lvaC; PP_2794, lvaD; PP_2795, lvaE; PP_2790, lvaR. The E. coli LS5218 reference genome can be retrieved from the NCBI database under the following accession number: GCA_002007165.1.
The python code used to identify potential lva homologs is provided in the Supplementary Files.
Any additional data that support the findings of this study are available from the corresponding author upon request.