Abstract
Bacteria reside in externally accessible niches on and in multicellular organisms, often forming mutualistic relationships with their host. Recent studies have linked the composition of these microbial communities with alterations in the host’s health, behavior, and development, yet the causative mediators of host-microbiota interactions remain poorly understood. Advances in understanding and engineering these interactions require the development of genetic tools to probe the molecular interactions driving the structure and function of microbial communities as well as their interactions with their host. This review discusses the current challenges to rendering culturable, non-model members of microbial communities genetically tractable--including overcoming barriers to DNA delivery, achieving predictable gene expression, and applying CRISPR-based tools--and details recent efforts to create generalized pipelines that simplify and expedite the tool-development process. We use the bacteria present in the human gastrointestinal tract as representative microbiota to illustrate some of the recent achievements and future opportunities for genetic tool development.
Keywords: Cas9, genome engineering, microbiota, synthetic biology, transformation
Introduction
Humans, animals, insects, and plants are host to diverse communities of microorganisms that impact their phenotype in direct and indirect ways. Known contributions range from metabolic activities that supplement the host’s nutritional and energetic requirements to providing key cues during development and even protection from invasive pathogens. These functions are determined by the individual microbial constituents, as well as their interactions with each other, their host, and the environment. Given the ubiquity of microbiota and their expanding role in biotechnology, agriculture, medicine, and the environment, there is a pressing need to understand the molecular mechanisms underlying these interactions and how to engineer constituent microbes to alter their composition, function, and influence on the host phenotype.
Our understanding of host-associated microbial communities has largely been shaped by extensive characterization studies using ‘omics technologies. For instance, metagenomic sequencing has revealed the taxonomy and genetic makeup of microbial communities in different environments, transcriptomics and proteomics have revealed the expression levels of constituent genes, and metabolomics have revealed the chemical environment created by these communities [1]. Despite these advances, ‘omics techniques provide observations about communities and generally generate--rather than test--mechanistic hypotheses underlying community dynamics and functions.
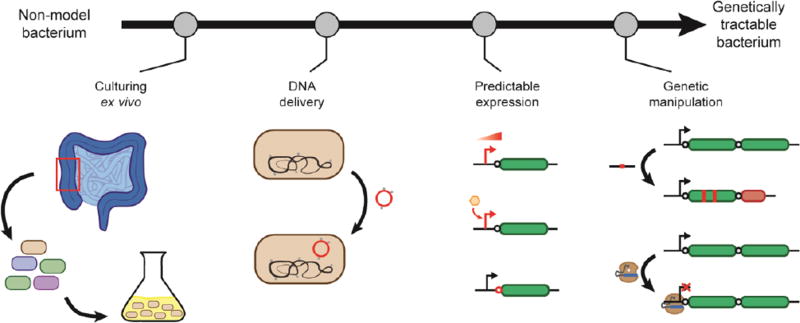
Elucidating the mechanistic underpinnings of each community and member instead comes from targeted genetic manipulation and testing. Before these experiments can occur, each bacterium must be rendered genetically tractable by determining how to culture it outside of its native environment, efficiently introduce and stably maintain synthetic DNA, and manipulate the genetic sequences and regulatory architecture (Figure 1). Each step has proven challenging, where the vast majority of host-associated species possess few if any tools. Despite these challenges, there have been recent advances in accelerating the process of genetic tool development. This review discusses these advances and the opportunities going forward using bacterial members of the human gut microbiota as case studies. As described below, we focus on these bacteria because they are critical to human health yet overwhelmingly lack genetic tools, although the techniques we describe are generally applicable to all non-model microbes.
Figure 1.
The process of developing genetic tools for a non-model bacterium. The steps include determining culturing conditions under laboratory conditions, developing methods for delivering and stably maintaining DNA in the cell, achieving tunable and predictable gene expression, and applying tools for genetic manipulation.
The largely untamed human gut microbiota
The human gastrointestinal tract is home to a diverse collection of bacteria representing roughly 1,000 species from at least four phyla [2,3]. Rather than being passive hitchhikers, these communities play dynamic and significant roles in shaping human development and health. For instance, dysbiosis of the gut has been linked to health conditions ranging from inflammatory bowel diseases and obesity to heart disease and Parkinson’s disease [3–5]. The causative nature of these links and their underlying mechanisms remain to be fully understood.
The development and application of genetic tools could provide these insights through experimental investigation of the many bacteria within the human gut microbiome. While most of these constituents remain to be cultured, the Human Microbiome Project funded through the National Institutes of Health [6] led to the isolation and sequencing of dozens of individual bacterial strains from human fecal samples that are now available through commercial stock centers. Of the strains that naturally reside in the human gut, only a handful belonging to the genera Bacteroides, Bifidobacteria, Escherichia, and Lactobacillus have sufficiently developed tools to be considered established or emerging “model” strains. To our knowledge, the remaining genera do not possess even a single genetically tractable member found in the human gut. As one example, Clostridia comprises one of the most abundant classes in human fecal samples and possess many sequenced, culturable members (e.g. Faecalibacterium prausnitzii) [2], yet no single member native to the human gut has been reported to be transformed let alone genetically manipulated. While the gut microbiome is arguably the most intensely studied host-associated microbial community, its diversity, complexity, and genetic-intractability have largely restricted the scope of research to correlational studies. Below we highlight approaches to introduce DNA, predictably express genes, and perform genome editing in culturable, but otherwise genetically intractable, bacteria. Determining methods for culturing non-model microbes is a critical aspect of genetic tool development. While these methods are not explicitly discussed in this review, we recommend recent publications on this subject [7–9].
Delivering and maintaining exogenous DNA
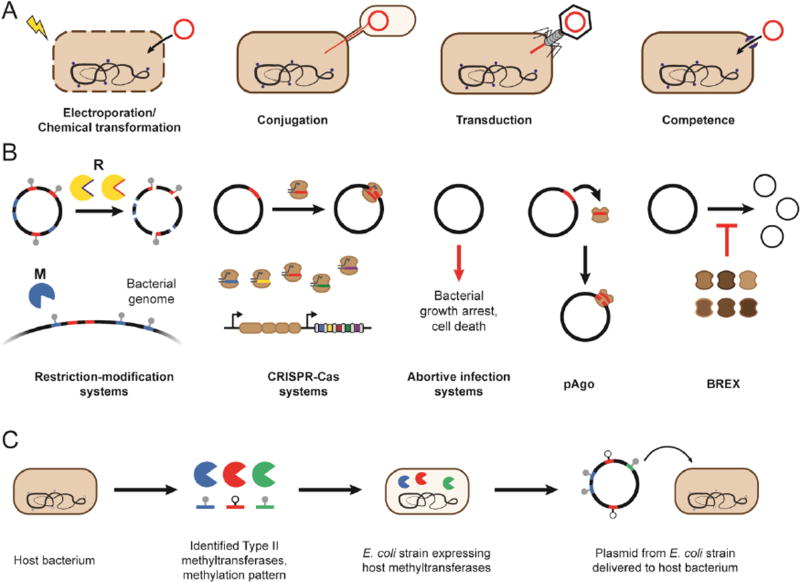
Once a bacterium can be cultured in isolation, the next critical capability is introducing and stably maintaining foreign DNA within the cell for expressing heterologous genes/pathways or serving as templates for recombination. This capability requires transporting DNA into the cytoplasm, bypassing the bacterium’s defense systems, and stably replicating the DNA. Several methods have been established for DNA transfer, including transiently disrupting the cell wall (electroporation, chemical transformation), injecting the DNA across the cell wall into the periplasm or cytoplasm (conjugation, transduction), or inducing uptake machinery already present in the cell (natural competence) (Figure 2A) [10,11]. Identifying the optimal conditions to make recipients conducive to DNA uptake can be difficult and often requires trial-and-error. However, this process can be accelerated using high-throughput methodologies [12]. Once introduced, if not degraded by the bacterial defense systems, the DNA can be stably propagated episomally by inclusion of a compatible origin-of-replication and a selectable marker. Alternatively, the DNA could be integrated into the genome through recombination. While these components may be species-specific, broad-host plasmids and transposases/recombinases have been identified and have been used extensively across bacterial taxa [13]. The middle step--bypassing the host’s defense systems--is recognized as the most challenging, and is discussed in more detail below.
Figure 2.
Overcoming barriers to DNA delivery. (A) Methods of delivering DNA into the cytoplasm. (B) Bacterial defense systems that could impede DNA delivery. (C) A systematic approach to bypass restriction-modification systems.
Bacteria possess a collection of innate and acquired defense systems that protect them from invasive genetic material. Well-known systems include restriction-modification, CRISPR-Cas, abortive infection, prokaryotic Argonaute, and the recently discovered BREX [14–17] (Figure 2B). Of these, restriction-modification systems are the most prevalent, appearing in approximately 95% of all sequenced bacterial genomes [18], and commonly are considered the greatest barrier to DNA transfer. Restriction-modification systems comprise DNA methyltransferases that methylate specific short sequences in the host DNA and restriction enzymes that cleave DNA sequences lacking the native methylation signature. If the DNA prepared for transfer encodes numerous restriction sites and its methylation pattern deviates from the bacterium, then the DNA would be targeted for immediate degradation. Furthermore, these systems can be poorly conserved among related species, resulting in restriction barriers that vary even between related strains [19].
Traditional strategies to overcome these defense systems involved modifying the methylation pattern using different intermediate hosts with varying methylation patterns or incubating the plasmid with a commercially available DNA methyltransferase. These strategies were shown to increase the transformation efficiencies in the human gut-associated Lactobacillus plantarum [20], although they are typically limited to the few types of methylation performed in E. coli or the available methyltransferases. More laborious strategies have involved deleting putative restriction-enzyme genes in the bacterium or isolating a plasmid from closely-related transformable species.
Arguably the most systematic approach to-date was first reported by Zhang and coworkers [21]. Their approach involved expressing a subset of the host bacterium’s Type II methyltransferase genes in a plasmid-propagating strain of E. coli devoid of any native restriction-modification systems (Figure 2C). The authors focused on Type II methyltransferases because they can function without additional proteins and are predicted in REBASE (http://rebase.neb.com/rebase/rebase.html), an online curated genomic database of restriction-modification systems maintained by New England Biolabs [18,21]. The approach radically boosted the transformation efficiency across a few bacterial species with some improvements as high as 106-fold.
The methodology introduced by Zhang et al. offers a promising strategy to rapidly achieve efficient DNA transformation in commensal bacteria [21], although there are opportunities to further improve this strategy. For instance, PacBio SMRT sequencing analysis could be applied to rapidly map the methylation pattern of the host bacterium under standard culturing conditions and predict which methyltransferase genes contribute to the observed methylation pattern. O’Callaghan et al. recently applied this approach to Bifidobacterium longum subs. longum NCIMB 8809 [22]. SMRT sequencing analysis of the strain’s methylome implicated three methyltransferases. Using DNA from E. coli overexpressing one of the methyltransferases boosted the transformation efficiency by 100-fold over unmodified plasmid, and within 10-fold of that of DNA isolated from the native B. longum strain. Methylome analysis has been conducted on other human gut strains, providing information necessary to begin bypassing restriction-modification systems in those bacteria [19,23,24]. Going forward, methyltransferases identified and characterized through these analyses could be catalogued and made available, allowing others to quickly recreate identified patterns, and transform hitherto non-transformable bacteria.
Achieving tunable and predictable gene expression
Transformation delivers genes that can be expressed to probe regulatory activities, interrogate functions of proteins and RNA, perturb endogenous gene expression or cellular metabolism, or introduce synthetic pathways for strain engineering. Overexpression is often sufficient for preliminary studies to elucidate gene functions. However, a range of expression levels may be desired, such as those required to match endogenous levels of gene products, express cytotoxic proteins, or optimize flux through metabolic pathways. Predictable gene expression requires identification or design of appropriate transcription and translation components; however, past work developing an extensive tool set for the model organism E. coli has yet to be fully adapted to other members of the microbiota.
Gene expression can be driven from a constitutive or inducible promoter. Inducible promoters rely on one or two-component signal transduction pathways that activate or repress transcription in response to environmental signals. These systems can be co-opted from the host bacterium, such as inducible systems that respond to the complex carbohydrates mannan, chondroitin sulfate, arabinogalactan, and rhamnose in Bacteroides thetaiotamicron [25,26]. These systems are convenient because the sensory proteins are already present, although the regulatory pathway can interface with other aspects of cellular metabolism and physiology and can result in undesirable pleiotropic effects [27]. Alternatively, heterologous inducible systems have been imported or modified from other organisms to function in the intended microbial host with some success, such as the nisin and sppIP/IP-673 induction systems in Lactobacilli, the anhydrotetracycline-responsive repressor TetR, the isopropyl β-D-1-thiogalactopyranoside (IPTG)-responsive repressor LacI, and ligand responsive riboswitches [28]. These inducible systems may be preferred because they operate orthogonally from the host’s metabolism and physiology, although factors such as compatibility and toxicity should be considered during implementation.
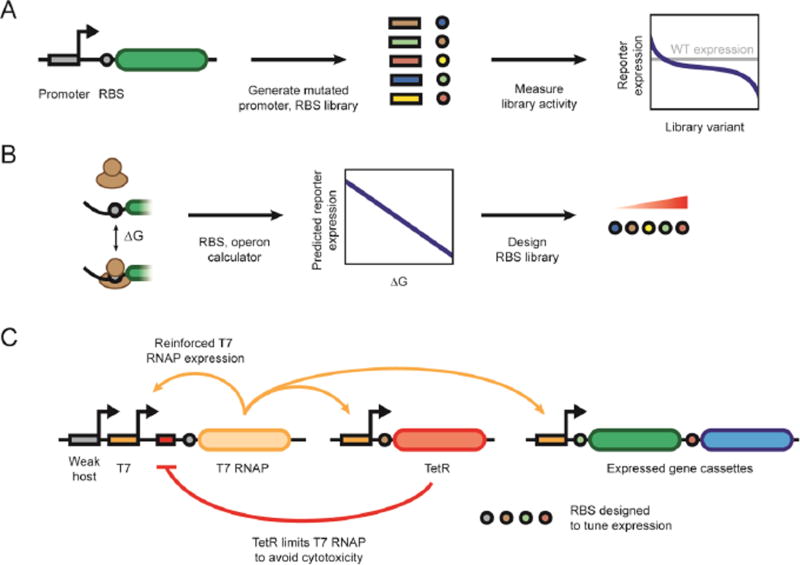
Constitutive promoters are convenient because they do not require additional accessory factors, although identified promoters might not exhibit the required expression strength. This challenge has previously been addressed by generating a diverse library of mutated promoter sequences and measuring each variant’s expression strength, yielding a library of derivative promoters covering an expansive range of transcriptional activities. For instance, Rud et al. randomized non-consensus regions within a native promoter, resulting in a library exhibiting up to 104-fold range of expression strengths in L. plantarum [29]. Similarly, alterations to the ribosomal binding site (RBS) can achieve ranges of translation, where promoter and RBS libraries have been created for B. thetaiotamicron (Figure 3A) [25].
Figure 3.
Strategies for tuning and predicting gene expression. (A) Process of generating promoter and/or RBS libraries covering a wide range of gene expression levels. (B) Use of in silico biophysical models to predict translational strength and guide the design of RBS libraries. (C) A host-independent, orthogonal gene expression system based on coordinated transcription by the T7 RNA polymerase.
While promoter and RBS libraries allow for tunable gene regulation, the exact target gene expression levels can vary depending on the downstream gene or untranslated regions. One promising solution is the use of in silico biophysical models to predict expression strengths based on sequence information (Figure 3B). The Salis lab has developed an online suite of software (https://salislab.net/software/) to predict protein expression levels based on the energetics of mRNA folding, ribosome-mRNA binding, and translational initiation as well as the impact of standby sites and translational coupling [30–33]. Aside from predicting translation strengths, these algorithms can design small libraries of RBS sequences covering a broad expression range to reduce the number of variants to be screened for pathway optimization. More complex synthetic gene regulation systems have been demonstrated in E. coli, but the lack of genetic tools has prevented these types of systems from being implemented into less characterized microbes. A separate solution is preventing interactions with the 5’ untranslated region by cleaving upstream of the RBS using a highly active ribozyme or a hairpin selectively cleaved by the CRISPR-associated protein Csy4 [34,35]. Overall, these solutions could be adopted to help ensure predictable protein expression across bacterial species.
A common caveat of designing gene expression systems based on a host’s existing transcriptional machinery is that they tend to be host-dependent. Kushwaha and coworkers reported a novel host-independent and orthogonal alternative in which expression of the T7 bacteriophage RNAP is kick-started by a weak consensus promoter and then used to drive its own expression and the expression of other genes (Figure 3C) [36]. This system was implemented in E. coli, Bacillus subtilis, and Pseudomonas putida, suggesting broad applicability across bacteria.
Applying CRISPR-based gene editing and regulation
Aside from expressing heterologous genes, DNA can be introduced into a cell to create defined edits to the bacterium’s genome. This capability often represents the pinnacle of genetic tool development and marks the transition from a non-model to a model bacterium. Traditional genome editing has relied on introduction of double-crossover events using a selectable marker. Given the low frequency of this event in most bacteria, bacteriophage-based recombinases have been introduced to enhance the frequency of homologous recombination [37]. These same recombinases also could promote recombination of single-stranded oligonucleotides. With the development of recursive editing strategies [10,38,39], recombineering could achieve efficient multiplexed genome editing without direct selection of the genomic mutations. Despite these advances, recombineering has only been demonstrated in a few organisms besides E. coli [40,41]. Instead, the most prevalent means of achieving genome editing have involved CRISPR-Cas systems, prokaryotic immune systems that use short guide RNAs to direct the system’s Cas nucleases to bind and cleave complementary DNA and/or RNA. While these systems are incredibly diverse [42,43], almost all associated CRISPR technologies however have been based on the Type II effector protein Cas9 and an engineered single-guide RNA (sgRNA).
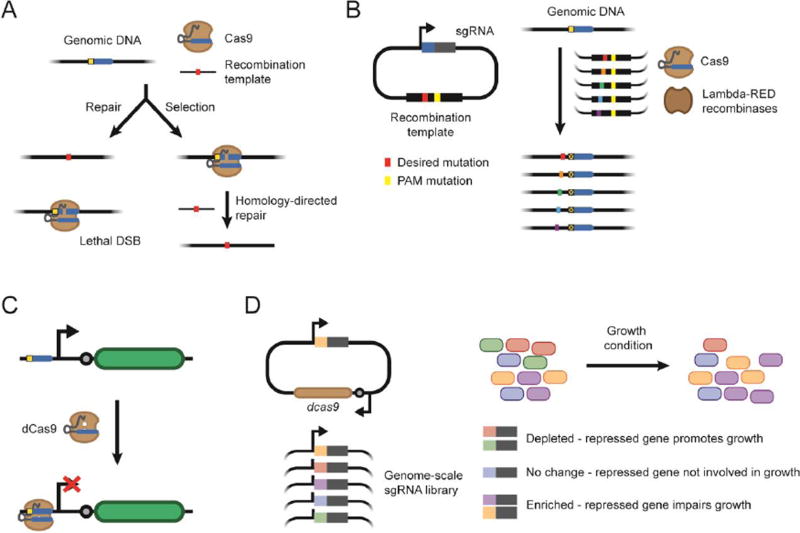
DNA cleavage by Cas9 is not sufficient to drive genome editing. In eukaryotic cells, a double strand break drives DNA repair through non-homologous end joining or homology-directed repair. However, in bacteria, evidence suggested that Cas nuclease-mediated genome cleavage cannot be readily repaired, resulting in a lethal double-stranded break that kills the bacterium [44,45] (Figure 4A). To perform genome editing, the DNA sequence is modified through recombination and the unmodified DNA is subsequently targeted for cleavage by Cas9 [45]. Cleavage therefore acts as a counter-selection to eliminate unedited cells. Recent work by Cui and Bikard challenged this paradigm by reporting that double-stranded breaks by Cas9 in the E. coli are rarely lethal [46]. Cells survived repeated cleavage by Cas9 through RecA-mediated homologous recombination, presumably by using an intact genomic copy as the template (Figure 4A). They further showed that a plasmid with point mutations flanked by homology arms resulted in efficient genome editing, potentially explaining why plasmid-encoded recombination templates are more effective than oligo-mediated recombination for Cas9-based editing [47,48]. The efficiency of Cas9-based editing with plasmid-encoded recombination templates allowed Garst and coworkers to perform high-throughput genome editing in E. coli through a methodology termed CREATE (CRISPR-enabled trackable genome engineering) [49] (Figure 4B). As part of the method, the recombination template introduced the desired modification along with a silent mutation to the required protospacer-adjacent motif (PAM). The editing efficiency was sufficiently high to allow the generation and screening of large mutant libraries. Further advances could include eliminating the recombinases or relying on endogenous CRISPR-Cas systems present in many commensal bacteria to further limit the number of required components [50,51].
Figure 4.
Applying CRISPR-based tools for genome editing and gene regulation. (A) Cas9 genome editing occurs through repair of the Cas9-mediated double-stranded break or through killing caused by an irreparable double-stranded break. (B) The CREATE method of high-throughput genome editing. (C) dCas9-mediated gene repression is achieved by introducing two point mutations to catalytically deactivate Cas9 (dCas9) and using it to block transcriptional initiation or elongation. (D) High-throughput genetic screen with dCas9-mediated gene repression.
While many advances have been made in Cas9-mediated genome engineering in E. coli, successful genome editing remains to be reported beyond only a few other bacteria [52], including limited examples of bacteria native to the human gut [44,53]. Part of the challenge is the need for efficient DNA delivery and recombineering that can be difficult to achieve in non-model bacteria. Given that Cas9 is often used to delete or disrupt genes, researchers have been pursuing an alternative: using CRISPR-Cas systems modified for programmable gene repression (Figure 4C). Targeting a catalytically dead Cas9 (dCas9) to the promoter or coding region of a gene blocks transcription initiation or elongation, a process termed CRISPR interference (CRISPRi) [54,55]. Among gut bacteria, dCas9-mediated gene repression has been implemented in B. thetaiotamicron to yield up to 45-fold downregulation of a reporter gene [25]. Other types of Cas proteins have also been harnessed for gene repression [56–59], offering a more diverse toolbox for CRISPR-mediated repression and opens the opportunity to co-opt endogenous CRISPR-Cas systems for programmable gene repression.
One attractive feature of dCas9-mediated repression is how readily it can be scaled for high-throughput genetic screens (Figure 4D). This feat was first demonstrated by Peters and coworkers in B. subtilis using dCas9-based repression [60]. They created a genome-wide library to screen for essential genes. This same strategy could be implemented in other culturable, transformable bacteria to perform genome-wide functional screens. While other procedures such as Tn-Seq are also commonly used for similar screens [61], they cannot directly identify essential genes. In addition, these approaches depend on random integration events and therefore require much larger libraries to approach full genome coverage. In contrast, CRISPR-based repression can be designed to have only a few CRISPR RNAs targeting a given gene, and the RNA sequences can be readily sequenced to determine the target locus. Genetic screens based on CRISPR-based repression therefore offer one of the most promising approaches to quickly interrogate genetic functions in diverse bacteria.
Conclusions and outlook
Our ability to progress a newly cultured host-associated bacterium to a genetically tractable host has greatly improved with advances in enhancing DNA delivery, predictable expression of genes and operons, and the use of CRISPR technologies for genome editing and gene regulation. Despite these advances, tool development remains an ad hoc process that plays out differently for each bacterial strain. Future efforts could focus on assembling a generalized tool-development pipeline that spans the identification of optimal growth conditions and efficient DNA delivery to characterizing genetic components that provide predictable gene expression and Cas9-based genomic screens and exploits information gleaned from ‘omics datasets. With further developments, a long-term outcome could be the rapid generation of genetic tools regardless of the specific bacterial host. This outcome would allow researchers and engineers to select the most application appropriate strain for research and strain engineering, rather than the ones that are currently more amenable to genetic tool development. This capability in turn could have a profound impact on our ability to understand the mechanisms underlying the dynamics and function of various host-associated microbial communities and how to engineer their members to probe and shape the community and the host.
Highlights.
Bacterial members of various microbiota are culturable but lack genetic tools
Means to boost DNA delivery by mimicking the host’s DNA methylation pattern
Predictable gene expression by designing or screening promoter, RBS libraries
Diverse CRISPR technologies for gene editing, repression, high-throughput screens
Progress toward a pipeline for expediting genetic tool development
Acknowledgments
We thank Jennie Fagen for providing critical feedback. This work was supported by funding from the National Science Foundation (MCB-1452902 to C.L.B.) and from the National Institutes of Health (1DP2HD91798-01 to N.U.N.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest:
None declared.
References
- 1.Quinn RA, Navas-Molina JA, Hyde ER, Song SJ, Vázquez-Baeza Y, Humphrey G, Gaffney J, Minich JJ, Melnik AV, Herschend J, et al. From sample to multi-omics conclusions in under 48 hours. mSystems. 2016;1:e00038–16. doi: 10.1128/mSystems.00038-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto J-M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 4.Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell. 2016;167:1469–1480. e12. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.NIH HMP Working Group. Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, Goulding D, Lawley TD. Culturing of “unculturable” human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533:543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pham VHT, Kim J. Cultivation of unculturable soil bacteria. Trends Biotechnol. 2012;30:475–484. doi: 10.1016/j.tibtech.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Schnupf P, Gaboriau-Routhiau V, Gros M, Friedman R, Moya-Nilges M, Nigro G, Cerf-Bensussan N, Sansonetti PJ. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature. 2015;520:99–103. doi: 10.1038/nature14027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalia AB, McDonough E, Camilli A. Multiplex genome editing by natural transformation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:8937–8942. doi: 10.1073/pnas.1406478111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo Scrudato M, Blokesch M. A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable. Nucleic Acids Res. 2013;41:3644–3658. doi: 10.1093/nar/gkt041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia PA, Ge Z, Moran JL, Buie CR. Microfluidic screening of electric fields for electroporation. Sci. Rep. 2016;6:21238. doi: 10.1038/srep21238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pérez-Arellano I, Zúñiga M, Pérez-Martínez G. Construction of compatible wide-host-range shuttle vectors for lactic acid bacteria and Escherichia coli. Plasmid. 2001;46:106–116. doi: 10.1006/plas.2001.1531. [DOI] [PubMed] [Google Scholar]
- 14.van Houte S, Buckling A, Westra ER. Evolutionary ecology of prokaryotic immune mechanisms. Microbiol. Mol. Biol. Rev. 2016;80:745–763. doi: 10.1128/MMBR.00011-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldfarb T, Sberro H, Weinstock E, Cohen O, Doron S, Charpak-Amikam Y, Afik S, Ofir G, Sorek R. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 2015;34:169–183. doi: 10.15252/embj.201489455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV, van der Oost J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353:aad5147. doi: 10.1126/science.aad5147. [DOI] [PubMed] [Google Scholar]
- 17.Swarts DC, Jore MM, Westra ER, Zhu Y, Janssen JH, Snijders AP, Wang Y, Patel DJ, Berenguer J, Brouns SJJ, et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014;507:258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *18.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE--a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2015;43:D298–9. doi: 10.1093/nar/gku1046. The authors develop an online database of reported and putative restriction-modification (R-M) systems in bacterial strains. The database is useful for developing methods to bypass R-M systems, allowing DNA delivery or to improve transformation efficiency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leonard MT, Davis-Richardson AG, Ardissone AN, Kemppainen KM, Drew JC, Ilonen J, Knip M, Simell O, Toppari J, Veijola R, et al. The methylome of the gut microbiome: disparate Dam methylation patterns in intestinal Bacteroides dorei. Front. Microbiol. 2014;5:361. doi: 10.3389/fmicb.2014.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spath K, Heinl S, Grabherr R. Direct cloning in Lactobacillus plantarum: electroporation with non-methylated plasmid DNA enhances transformation efficiency and makes shuttle vectors obsolete. Microb. Cell Fact. 2012;11:141. doi: 10.1186/1475-2859-11-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang G, Wang W, Deng A, Sun Z, Zhang Y, Liang Y, Che Y, Wen T. A mimicking-of-DNA-methylation-patterns pipeline for overcoming the restriction barrier of bacteria. PLoS Genet. 2012;8:e1002987. doi: 10.1371/journal.pgen.1002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *22.O’Callaghan A, Bottacini F, O’Connell Motherway M, van Sinderen D. Pangenome analysis of Bifidobacterium longum and site-directed mutagenesis through by-pass of restriction-modification systems. BMC Genomics. 2015;16:832. doi: 10.1186/s12864-015-1968-4. Using SMRT sequencing to identify the methylation pattern in B. longum, the authors cloned and overexpressed B. longum methyltransferases in E. coli to greatly increase transformation efficiency. This is the first application of the technique in B. longum and one of the few examples in human gut-associated microbes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’ Connell Motherway M, Watson D, Bottacini F, Clark TA, Roberts RJ, Korlach J, Garault P, Chervaux C, van Hylckama Vlieg JET, Smokvina T, et al. Identification of restriction-modification systems of Bifidobacterium animalis subsp. lactis CNCM I-2494 by SMRT sequencing and associated methylome analysis. PLoS One. 2014;9:e94875. doi: 10.1371/journal.pone.0094875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee WC, Anton BP, Wang S, Baybayan P, Singh S, Ashby M, Chua EG, Tay CY, Thirriot F, Loke MF, et al. The complete methylome of Helicobacter pylori UM032. BMC Genomics. 2015;16:424. doi: 10.1186/s12864-015-1585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **25.Mimee M, Tucker AC, Voigt CA, Lu TK. Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell Syst. 2015;1:62–71. doi: 10.1016/j.cels.2015.06.001. The authors greatly expanded the genetic tools for the abundant gut bacterium. B. thetaiotamicron via promoter/RBS libraries, inducible promoters, and CRISPR-mediated gene repression. This study is a comprehensive example of how genetic tool development of “non-model” organisms can lead to novel applications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horn N, Carvalho AL, Overweg K, Wegmann U, Carding SR, Stentz R. A novel tightly regulated gene expression system for the human intestinal symbiont Bacteroides thetaiotaomicron. Front. Microbiol. 2016;7:1080. doi: 10.3389/fmicb.2016.01080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Afroz T, Biliouris K, Kaznessis Y, Beisel CL. Bacterial sugar utilization gives rise to distinct single-cell behaviours. Mol. Microbiol. 2014;93:1093–1103. doi: 10.1111/mmi.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Topp S, Reynoso CMK, Seeliger JC, Goldlust IS, Desai SK, Murat D, Shen A, Puri AW, Komeili A, Bertozzi CR, et al. Synthetic riboswitches that induce gene expression in diverse bacterial species. Appl. Environ. Microbiol. 2010;76:7881–7884. doi: 10.1128/AEM.01537-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rud I, Jensen PR, Naterstad K, Axelsson L. A synthetic promoter library for constitutive gene expression in Lactobacillus plantarum. Microbiology. 2006;152:1011–1019. doi: 10.1099/mic.0.28599-0. [DOI] [PubMed] [Google Scholar]
- 30.Tian T, Salis HM. A predictive biophysical model of translational coupling to coordinate and control protein expression in bacterial operons. Nucleic Acids Res. 2015;43:7137–7151. doi: 10.1093/nar/gkv635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salis HM. The ribosome binding site calculator. Methods Enzymol. 2011;498:19–42. doi: 10.1016/B978-0-12-385120-8.00002-4. [DOI] [PubMed] [Google Scholar]
- 32.Espah Borujeni A, Salis HM. Translation initiation is controlled by RNA folding kinetics via a ribosome drafting mechanism. J. Am. Chem. Soc. 2016;138:7016–7023. doi: 10.1021/jacs.6b01453. [DOI] [PubMed] [Google Scholar]
- 33.Espah Borujeni A, Cetnar D, Farasat I, Smith A, Lundgren N, Salis HM. Precise quantification of translation inhibition by mRNA structures that overlap with the ribosomal footprint in N-terminal coding sequences. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi L, Haurwitz RE, Shao W, Doudna JA, Arkin AP. RNA processing enables predictable programming of gene expression. Nat. Biotechnol. 2012;30:1002–1006. doi: 10.1038/nbt.2355. [DOI] [PubMed] [Google Scholar]
- 35.Lou C, Stanton B, Chen Y-J, Munsky B, Voigt CA. Ribozyme-based insulator parts buffer synthetic circuits from genetic context. Nat. Biotechnol. 2012;30:1137–1142. doi: 10.1038/nbt.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *36.Kushwaha M, Salis HM. A portable expression resource for engineering cross-species genetic circuits and pathways. Nat. Commun. 2015;6:7832. doi: 10.1038/ncomms8832. Utilizing an orthogonal polymerase that drives its own expression and regulation along with the expression of other genes, the authors develop a portable expression system that can be implemented in ranging non-model microbes. This system offers a unique alternative to developing expression tools for specific bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang P, Wang J, Qi Q. Prophage recombinases-mediated genome engineering in Lactobacillus plantarum. Microb. Cell Fact. 2015;14:154. doi: 10.1186/s12934-015-0344-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA, Goodman DB, Reppas NB, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Pijkeren J-P, Britton RA. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res. 2012;40:e76. doi: 10.1093/nar/gks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krylov AA, Kolontaevsky EE, Mashko SV. Oligonucleotide recombination in corynebacteria without the expression of exogenous recombinases. J. Microbiol. Methods. 2014;105:109–115. doi: 10.1016/j.mimet.2014.07.028. [DOI] [PubMed] [Google Scholar]
- 42.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barrangou R, van Pijkeren J-P. Exploiting CRISPR-Cas immune systems for genome editing in bacteria. Curr. Opin. Biotechnol. 2016;37:61–68. doi: 10.1016/j.copbio.2015.10.003. [DOI] [PubMed] [Google Scholar]
- *46.Cui L, Bikard D. Consequences of Cas9 cleavage in the chromosome of Escherichia coli. Nucleic Acids Res. 2016;44:4243–4251. doi: 10.1093/nar/gkw223. The authors show that cleaving the E. coli genome with Cas9 is not always lethal and instead undergoes homology-directed repair. These insights impact the mechanism of Cas9-mediated genome editing in bacteria and whether editing occurs through repair or through selection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015;81:2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu T, Li Y, Shi Z, Hemme CL, Li Y, Zhu Y, Van Nostrand JD, He Z, Zhou J. Efficient genome editing in Clostridium cellulolyticum via CRISPR-Cas9 nickase. Appl. Environ. Microbiol. 2015;81:4423–4431. doi: 10.1128/AEM.00873-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **49.Garst AD, Bassalo MC, Pines G, Lynch SA, Halweg-Edwards AL, Liu R, Liang L, Wang Z, Zeitoun R, Alexander WG, et al. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat. Biotechnol. 2017;35:48–55. doi: 10.1038/nbt.3718. The authors developed a method termed CREATE for multiplexed and trackable genome editing using CRISPR and homologous recombination. This method allows rapid, high-throughput, precise editing of thousands of sites in parallel along with barcoding for edit mapping and screening. [DOI] [PubMed] [Google Scholar]
- 50.Pyne ME, Bruder MR, Moo-Young M, Chung DA, Chou CP. Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium. Sci. Rep. 2016;6:25666. doi: 10.1038/srep25666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo ML, Leenay RT, Beisel CL. Current and future prospects for CRISPR-based tools in bacteria. Biotechnol. Bioeng. 2016;113:930–943. doi: 10.1002/bit.25851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mougiakos I, Bosma EF, Weenink K, Vossen E, Goijvaerts K, van der Oost J, van Kranenburg R. Efficient genome editing of a facultative thermophile using mesophilic spCas9. ACS Synth. Biol. 2017 doi: 10.1021/acssynbio.6b00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oh J-H, van Pijkeren J-P. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 2014;42:e131. doi: 10.1093/nar/gku623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo ML, Mullis AS, Leenay RT, Beisel CL. Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res. 2015;43:674–681. doi: 10.1093/nar/gku971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rath D, Amlinger L, Hoekzema M, Devulapally PR, Lundgren M. Efficient programmable gene silencing by Cascade. Nucleic Acids Res. 2015;43:237–246. doi: 10.1093/nar/gku1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leenay RT, Maksimchuk KR, Slotkowski RA, Agrawal RN, Gomaa AA, Briner AE, Barrangou R, Beisel CL. Identifying and visualizing functional PAM diversity across CRISPR-Cas systems. Mol. Cell. 2016;62:137–147. doi: 10.1016/j.molcel.2016.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stachler A-E, Marchfelder A. Gene repression in Haloarchaea using the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-Cas I-B system. J. Biol. Chem. 2016;291:15226–15242. doi: 10.1074/jbc.M116.724062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **60.Peters JM, Colavin A, Shi H, Czarny TL, Larson MH, Wong S, Hawkins JS, Lu CHS, Koo B-M, Marta E, et al. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell. 2016;165:1493–1506. doi: 10.1016/j.cell.2016.05.003. CRISPR-mediated gene regulation is utilized as a screen for essential genes in Bacillus subtilis. This is the first example of this approach in B. subtilis and demonstrates the usefulness of CRISPR as a genetic screening tool. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]