Abstract
De-identification, or identifying and removing protected health information (PHI) from clinical data, is a critical step in making clinical data available for clinical applications and research. This paper presents a natural language processing system for automatic de-identification of psychiatric notes, which was designed to participate in the 2016 CEGS N-GRID shared task Track 1. The system has a hybrid structure that combines machine leaning techniques and rule-based approaches. The rule-based components exploit the structure of the psychiatric notes as well as characteristic surface patterns of PHI mentions. The machine learning components utilize supervised learning with rich features. In addition, the system performance was boosted with integration of additional data to the training set through domain adaptation. The hybrid system showed overall micro-averaged F-score 90.74 on the test set, second-best among all the participants of the CEGS N-GRID task.
Keywords: de-identification, psychiatric notes, natural language processing
Graphical abstract
1. Introduction
Clinical narratives of patient medical records contain rich information such as medication history and treatment information. Thus, the narratives have gained much attention from health care providers and researchers as an important resource for medical applications and clinical studies. Before using narrative resources, researchers must acquire both the informed consent from the patients and the approval from the Institutional Review Board (IRB). However, when the narratives are de-identified, i.e., when the information that may reveal patient’s identification is identified and removed from the narratives, they can be utilized without patient content. The Health Insurance Portability and Accountability Act (HIPAA) defines 18 categories of protected health information (PHI) that are required to be removed in order for a record to be considered de-identified.
Due to the costs associated with manual de-identification of large clinical corpora, significant research has focused on automatic de-identification methods. Researchers have utilized natural language processing (NLP) techniques to build systems that automatically recognize PHI from various types of clinical notes [1]–[12]. Community challenges have also been organized under the i2b2 project [13], [14] to facilitate empirical system comparison and thus expedite the development of automatic de-identification methods.
The 2016 CEGS N-GRID shared task Track 1 [15] is the most recent community challenge to address de-identification of clinical notes. Following 2014 i2b2/UTHealth shared task Track 1 [14], the 2016 task defines a wider set of PHI categories than the categories required by HIPPA; the task defines seven main categories (Name, Profession, Location, Contact, ID, Age, and Date) and 30 sub-categories. Furthermore, the 2016 task focused on psychiatric notes, as opposed to discharge summaries or progress notes studied in previous challenges[13], [14]. Psychiatric notes are an important but understudied type of clinical data in terms of de-identification research, and not included in previous community challenges. In addition, the 2016 challenge evaluated both the performance of de-identification systems without providing labeled data (Track 1A) as well as the more traditional evaluation of performance when labeled data is provided (Track 1B). Track 1A was designed to test a common issue in clinical NLP: how well do de-identification systems perform on data that is different in nature than the system’s original training data? Given the wide variety of clinical note types and the difficulty involved in manually annotating all of these possible types, it is important to determine how robust a de-identification system is when operating in this non-ideal, but common use case.
In this paper, we describe our approach to PHI de-identification and its performance on the 2016 CEGS N-GRID data. For Track 1A, we utilized an existing, traditional supervised learning-based de-identification system based on conditional random fields (CRF). For Track 1B, the system was first modified to better suit the structure and form of psychiatric notes, including pre- and post-processing, a rule-based tagger for certain PHI elements, and splitting the supervised learning system into two separate CRF models. Then, we significantly improved the system’s performance by adding rich features and employing domain adaptation to incorporate an external de-identification corpus during CRF training. Our approach yielded excellent results in both sub-tracks, achieving the second-best ranking participant in each, demonstrating the quality and robustness of the approach.
2. Related work
PHI de-identification methods fall into one of three categories: rule-based, machine learning-based, and hybrid methods that combine rules and machine learning.
Rule-based de-identification systems appeared as early as 1996, when Sweeney [1] developed rules to identify 25 categories of PHIs in pediatric EHR notes. Since then, a number of rule-based systems introduced extensive hand-coded rules and specialized dictionaries [2]–[7]. Friedlin and McDonald [6] developed the Medical De-identification System (MeDS), which is designed to scrub Health Level Seven (HL7) observation messages. In addition to extensive regular expressions and dictionaries, the system has a word sense disambiguation module based on part-of-speech (POS) information. Neamatullah et al. [7] developed a system that uses lexical look-up tables, regular expressions, and other heuristics, and tested the system on nursing notes. Incorporating context information in order to deal with misspelled PHIs, the system achieved high recall (96.7) with moderate precision (74.9). While rule-based systems do not require a large amount of training data, curating rules can require significant manual work by domain experts, and yet even human-curated rules often make assumptions about the data that limit their robustness on unseen note types.
Machine learning (ML) based systems usually cast de-identification as a token classification or sequence classification problem. Various supervised machine learning algorithms, including CRFs [16]-[18], Support Vector Machines (SVM) [19], and Decision Trees [20], [21], have been employed. Chen et al. [8] proposed a non-parametric Bayesian Hidden Markov Model (HMM) that learns a potentially infinite number of latent variables by using a Dirichlet process prior. More recently, Dernoncourt et al. [9] proposed a bidirectional Long Short-Term Memory (LSTM) based Recurrent Neural Network (RNN) that utilizes both token and character embeddings for de-identification, showing higher F-measure results than state-of-the-art CRF-based systems. The advantage of the character-based representation is that it is robust to small changes in spelling. In general, ML-based systems outperform rule-based systems when given sufficient annotated data from the dataset of interest (it is difficult, however, to generalize the performance of ML- and rule-based methods when evaluated data that differs substantially from the annotated data). On the other hand, ML-based systems typically perform worse than rules on rare types of PHI due to a dearth of training data.
Hybrid systems attempt to combine the benefits of both rules and machine learning. Rare types of PHIs or PHIs with predictable lexical patterns are better-suited for rules, while frequently occurring PHI types, especially those with unpredictable lexical variation are better-suited for machine learning [10]–[12]. Liu et al. [10] proposed a hybrid system combining a token-level CRF, a character-level CRF, and a rule-based classifier. The output of the three component classifiers is considered in a cascaded manner: the rule-based classifier is given the highest preference, then the character-level classifier, and then finally the token-level classifier. Yang and Garibaldi [11] proposed a hybrid system consisting of a CRF and rules with dictionaries and regular expressions. In addition, they include a post-processing step, in which trusted PHI mentions are utilized to uncover more potential terms.
Our approach follows the general structure of the hybrid systems (i.e., rules for rare and regularized PHIs and ML for others), but is different from the previous systems in two aspects. First, the system uses an extensive and rich feature set (e.g., word embeddings from both medical and open domains, token-shape N-gram, and information derived from the structured parts of the clinical notes). Second, the system performance is further improved by utilizing an external de-identification corpus through domain adaptation.
3. Methods
In this section, we first introduce the de-identification corpus used in our experiments. Then, we describe the pre-existing de-identification system utilized in Task 1A which also acts as our baseline system in Task 1B. After that, we introduce our hybrid de-identification method. Finally, we explain the evaluation metrics used.
3.1. Data
The 2016 CEGS N-GRID Task 1 data consists of 1,000 psychiatric notes: a training set of 600 notes with 20,845 PHIs, and a test set of 400 notes with 13,519 PHIs. All notes are annotated with the 7 main PHI categories and 30 sub-categories. The distribution of the PHI categories in the corpus is shown in Table 1 of the Supplementary Document.
Initially, the 600 notes of the training set were released for Task 1A without any gold standard PHI annotations. After submission for Task 1A, the gold standard PHI annotations for the training set were released for Task 1B, along with the unlabeled test set. Task 1A performance was thus measured only on the training set, whereas Task 1B performance was measured on the test set, whose gold standard annotations were released after the challenge.
3.2. Baseline system (Task 1A)
Our baseline system, which is used for Task 1A, is based on an existing de-identification method in the CLAMP toolkit [22]. The system consists of pre-processing steps, a CRF tagger, and post-processing rules (Figure 1(a)). In the pre-processing step, psychiatric notes are tokenized using the default CLAMP tokenizer, POS tagged using OpenNLP [23], and section parsed (identifying sections of the notes) using a dictionary-matching algorithm and a dictionary of standard section names in clinical notes provided by CLAMP (e.g., history of past illness, chief complaint, mediations).
Figure 1.
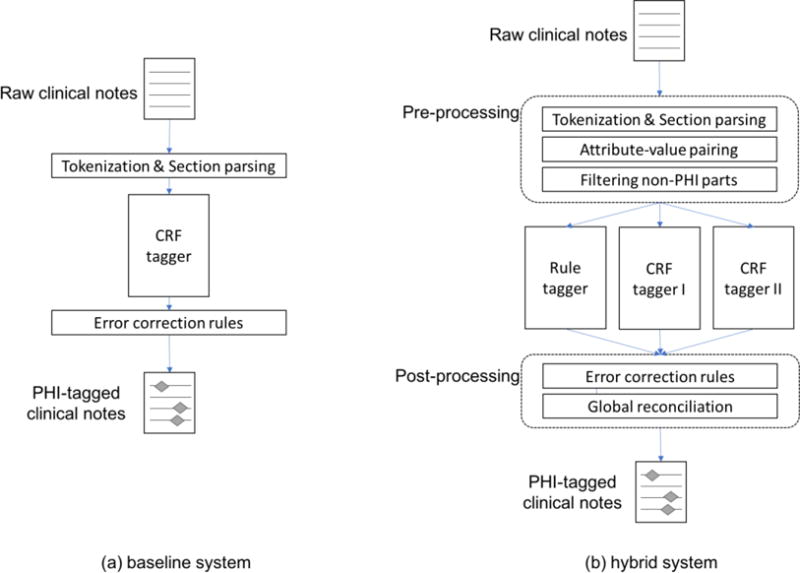
Overview of the de-identification systems for Task 1A (the baseline system) and Task 1B (the hybrid system)
After pre-processing, a token-based CRF tagger identifies all 30 types of PHI mentions using an IOB tagging scheme; the tagger classifies each token as either O (outside of any PHI mention), B-type (beginning of a PHI mention of type), or I-type (inside of a PHI mention of type), where type can be any of the 30 PHI types. The CRFsuite [24] implementation of CRF is used. Without labeled psychiatric notes for Task 1A, the CRF was trained on the 2014 i2b2/UTHealth shared task Track 1 data [14], which is annotated with the same PHI categories as the 2016 data.
The CRF uses lexical, syntactic, semantic, and discourse level feature types:
-
(1)
Orthographic token shape: orthographic forms of the token produced by substituting numbers, uppercase letters, and lowercase letters with ‘#’, ‘A’, and ‘a’, respectively
-
(2)
Regex token shape: token-level regular expressions for special token shape categories (e.g., years, phone numbers, init-caps, all caps)
-
(3)
Prefix/suffix: prefix and suffix of the token (up to three characters)
-
(4)
Token n-gram: unigrams, bigrams and trigrams of the tokens within a window of +/− 2
-
(5)
POS n-gram: unigrams, bigrams and trigrams of POS tags of tokens within a window of +/− 2
-
(6)
Sentence length: length of the sentence containing the target token (“6+” when the sentence is longer than five tokens, which is to distinguish short phrases from full sentences)
-
(7)
Sentence shape: whether the sentence ends with a colon or starts with an enumeration indicator such as ‘a)’ or ‘2.’1
-
(8)
Section header: type of the section containing the target token (e.g., Chief Complaint, Medication)
-
(9)
Word representations: Brown clusters [25], random indexing [26], and clusters [27] based on word2vec embeddings [28]. All three word representations were built from MIMIC II [29].
-
(10)
Dictionary-matching: based on frequent PHI terms such as country names, city names, popular first and last names
Finally, a post-processing step corrects common ML errors observed on the training data.
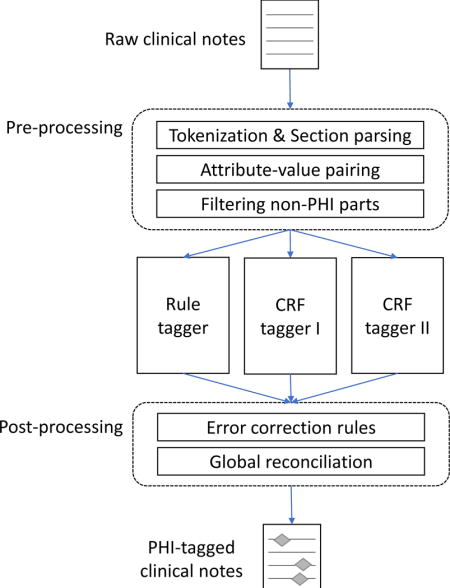
3.3. Hybrid system (Task 1B)
The baseline system was heavily modified to add extensive pre-processing and post-processing rules that are customized to the psychiatric notes, transforming the ML-based system into a hybrid system. Our hybrid approach consists of a pre-processing component, two CRF taggers, one rule-based tagger, and a post-processing component. Supplementary Table 3 summarizes which of the taggers in the hybrid system is applied to which of the PHI categories.
3.3.1. Pre-processing
Pre-processing consists of tokenization, POS tagging, and section parsing. The pre-processing modules of the baseline system were modified based on the structure and form of the psychiatric notes. For example, the notes contain a number of conjoined words (e.g., “rangeImpression”), which are likely the result of faulty EHR data extraction as opposed to human error in the original composition. But these errors can result in tokens that contain part of a PHI mention (e.g., “winterHx” where “winter” is PHI type DATE; “stantonPsychiatric” where “stanton” is PHI type NAME). To handle this, an overly aggressive tokenization strategy is used: tokens are split when a number follows a letter, or vice versa, and when an uppercase letter follows a lowercase letter. In addition, the section header dictionary for the dictionary-based section parser was also extended to increase the coverage of section headers in the psychiatric notes.
Additionally, the hybrid system employs two new pre-processing steps that specifically target the structure of the particular psychiatric notes in this data. First, all section headers were considered non-PHI and removed from training. Second, the notes contain many attribute-value pairs such as “cocaine: no”. 244 regular expressions were compiled to identify such attribute-value pairs and transform these parts of the notes into semi-structured data (intended for Track 2). Then, the attributes that do not contain PHI were removed from training. These two filtering steps were employed under the hypothesis that removing non-PHI text from further processing would benefit the CRF training, creating a more balanced training set and focusing model learning on more difficult cases.
3.3.2. Rule-based tagger
The hybrid system employs a rule-based tagger for PHI categories that show distinct surface patterns or that occur infrequently in the training data. A number of regular expressions are used to identify CONTACT-URL, CONTACT-EMAIL, CONTACT-PHONE, NAME-USERNAME, ID-MEDICALRECORD, ID-IDNUM, ID-LICENSE, LOCATION-STATE, LOCATION-STREET, AGE-AGE and DATE-DATE type PHI mentions. Table 1 shows a sample of the regular expressions for each PHI category. The number of regular expressions for each PHI category is shown in Table 2 of the Supplementary Document.
Table 1.
Example regular expressions for rule-based tagger. In the examples, italicized letters indicate the context that is matched to the look-behind or look-ahead parts of the regular expressions, not the predicted PHI mentions.
| PHI category | Regular expression | Example PHI mention |
|---|---|---|
| URL | (http://)?(www\.)?[a-zA–Z0-9]+\.(com|net|org|gov) | www.womensmentalhealth.org |
| [a-zA–Z0-9]+@[a-zA–Z0–9]+\.(com|net|org|gov) | akilj@hospital.org | |
| PHONE | \(\d{3}\)[\-\. ]?\d{3}[\-\. ]?\d{4} | 447-742-0756 |
| (?<=pager)\d{5} | pager 07516 | |
| USERNAME | (?<=\[\s{0,1}][a-z]+[0-9]+(?=\s{0,1}\)) | [hpp2] |
| MEDICALRECORD | (?<=MRN:\s{0,2})\d{7} | MRN: 2418195 |
| NUM | (?<=Devon Pharm ID:\s{0,2})\d{11} | Devon Pharm ID: 50970046433 |
| LICENSE | (?<!member)[A-Z][A-Z]-? ?\d{7}(-? ?\d{4})? | XF-1747210-9837 |
| STATE | Texas(?=?([^A-Z\-]|$)) | Texas |
| (?<=(in|to|from))FL(?=?([^A-Z\-]|$)) | FL | |
| STREET | (?<!-)\d+([A-Z][a-z]*)+Drive(?![A-Za-z]) | 82 Brook Drive |
| AGE | (?<=\b[Aa]ged?)\d+(?=? -?years) | Aged 82 years |
| DATE | [Ss]undays? | Sunday |
| [Cc]hristmas | Christmas |
3.3.3. CRF taggers
Two CRF taggers predict PHI mentions of different types. The first CRF is for quantitative types: DATE, AGE, and CONTACT. The second CRF is for name types: NAME, LOCATION, and PROFESSION. Both CRFs use a common feature base as well as some additional classifier-specific features. The base feature set includes the 10 features for the baseline system (Section 3.2), plus the following:
-
(11)
Open-domain word embeddings: binarized [30] pre-built GloVe word embeddings [31]
-
(12)
Token shape N-gram: unigram, bigram and trigram of word shapes of nearby tokens in window of +/−2 (using orthographic token shapes)
-
(13)
Attribute name: the attribute name when the token is part of the value in an attribute-value pair
Features that are specific to the quantitative CRF are as follows:
-
(1)
Context token shape: regex-based token shapes of nearby tokens in widow of +/−3
Features that are specific to the name CRF are as follows:
-
(1)
Profession suffix: whether the token ends with a suffix that is common to PROFESSION mentions, i.e., -ist, -ian, -man.
-
(2)
General domain NER: output of Stanford NER [32]
-
(3)
Semantic role labeling: output of SENNA semantic role labeling [33], combining both role and predicate (e.g., “A1-see” for a token which is part of the direct object of the predicate “see”.).
Additionally, some base features were also updated (e.g., new dictionary entries, new token shape regular expressions). The features that are used for each of the CRF taggers are summarized in Table 4 of the Supplementary Document.
Finally, for both CRFs, a domain adaptation method is used to incorporate the 2014 de-identification challenge corpus [14] into the CRF training data. Domain adaptation is a set of techniques that enables learning from a dataset that is annotated with the same set of labels as the task at hand, but has different distribution [34]–[36]. While the 2014 corpus is annotated with the same PHI types as the target psychiatric notes for CEGS N-GRID challenge, the data distribution is different due to different clinical note types. The feature augmentation method EasyAdapt [37] is used for domain adaptation, as the algorithm was shown to be effective in previous work [38] and is easy to implement.
EasyAdapt works by mapping feature vectors into higher dimensions. Given features from the target data (2016 corpus in our case) and the source data (2014 corpus) the algorithm generates three versions of feature sets: general, source-specific, and target-specific versions. As a result, the augmented feature vectors become three times longer than the originals. Formally, given a source feature vector Xs and a target feature vector Xt, the EasyAdapt feature augmentation function EA can be described as follows:
where 0 is a zero vector of length |X|. In the tuples above, the first elements represent the general version features, the second elements represent the source-specific version features, and the third are the target-specific version features. The intuition behind this algorithm is to leverage three versions of features to find best feature representations for the target domain. The general-version features will get higher weights for common instances of both target and source, whereas that the target-specific or the source-specific versions of the features will gain weights for instances unique for target or source, respectively.
3.3.4. Post-processing
Post-processing is composed of three steps. In the first step, the results from the three taggers (one rule-based tagger and two CRF taggers) are merged. PHI mentions from one of the CRF taggers that overlap with a mention from the rule-based tagger are removed, under an assumption that the rules would produce results with higher precision than the CRFs. Overlapping mentions from the CRF taggers are both kept, which occur very rarely.
In the second step, error correction is performed. Common CRF tagger errors (based on a cross-validation of the training data) are fixed. For instance, FAX numbers, which are often misclassified as PHONE numbers, are corrected using context information. STATE names not in the US state name dictionary are removed. ORGANIZATION names misclassified as HOSPITAL are also corrected when nearby tokens (+/−3) contain school-related keywords such as ‘study’, ‘degree’, and ‘senior’.
In the third step, global reconciliation is applied to force type agreement between entities in the same document with the same string value. PHI mentions—especially patient, doctor, and hospital names— often appear several times in a document, thus errors in a subset of identical-string mentions can be fixed by a combination of high-precision rules and a type-counting strategy. The high-precision rules consist of regular expressions built using the training data. For instance, if a DOCTOR PHI mention has preceding ‘Dr’ in its immediate context, other mentions of the name can be assumed to be a DOCTOR as well. The attribute-value structures provide another source for rules, e.g. if a doctor name is found in the value part of a “referral source” attribute. For type-counting, if the same name is predicted as being the same type at least two times in a document, then all names are altered to that frequent type.
3.4. Evaluation
De-identification performance is measured using precision, recall, and F1-measure at the entity, token, and binary level. At the entity level (the traditional NER evaluation), system outputs are compared to the gold standard using both the type and character offsets. A ‘strict’ entity match requires the offsets to be exactly correct, whereas a ‘relaxed’ match allows for up to two characters of difference in the ending offset. At the token level, both type and position must also match, but the evaluation is done on a per-token basis (thus assigning partial credit if a predicted PHI is off by a word). Finally, at the binary level, only the positions of the predicted PHI mentions are evaluated, and the type information is ignored (i.e., any type of PHI). Binary level evaluation is done as both a ‘strict’ entity level match and as a token level match. For statistical significance testing, the approximate randomization test [39] with N=9999 and α=0.1 is used.
Two sets of PHI types are evaluated. One is the CEGS N-GRID PHI types (i.e., 7 main categories and 30 sub-categories) defined by the challenge organizers. The other is the HIPAA PHI types, a subset of PHI types defined by the Health Insurance Portability and Accountability Act (HIPAA). The CEGS N-GRID challenge defined the micro-averaged F1-measure of the ‘strict’ entity level for the full set of types as the primary evaluation measure.
4. Results and Discussion
In this section, we report the performance of our systems and discuss the results. For both Task 1A and 1B, the participants were allowed to submit up to three runs. Here, we report the results from our official runs as well as a few additional experiments.
4.1. Task 1A
Table 2 shows the Task 1A (no labeled data) performance of the baseline system evaluated on the 600-document set (the training set for Task 1B). With strict entity level evaluation, the system F1 is 74.5. While this is the second best performance achieved by the challenge participants [15], the performance is much lower than the performance of the top-ranked systems on the 2014 i2b2 challenge [14]. The top system on the 2014 data achieved an F1 of 93.60, while the median system had an F1 of 81.19. Table 3 shows the performance of the baseline system with different combinations of training and test data. Without new data, our baseline CRF model trained on 2014 training set showed drops in F1 from 91.97 (2014 test set) to 74.5 (Track 1A). The CRF model trained on 2016 showed a lower performance of 87.55. This demonstrates the importance of incorporating appropriately similar data for machine learning-based systems. It also indicates that either de-identification on psychiatric notes is more challenging than on the 2014 notes, or that more annotated data is necessary to achieve similar results to other datasets.
Table 2.
The overall performance of the baseline system. Entity-Strict level evaluation results, which are the primary results used by the N-GRID challenge [15], are shown in bold face.
| PHI type set | Evaluation level | Precision | Recall | F1-measure |
|---|---|---|---|---|
| N-GRID PHI types | Token | 89.01 | 71.03 | 79.01 |
| Entity-Strict | 85.54 | 65.98 | 74.5 | |
| Entity-Relaxed | 85.84 | 66.2 | 74.75 | |
| Binary-Token | 95.64 | 76.32 | 84.9 | |
| Binary-Strict | 90.61 | 69.89 | 78.91 | |
| HIPAA PHI types | Token | 92.09 | 73.69 | 81.87 |
| Entity-Strict | 88.05 | 67.91 | 76.68 | |
| Entity-Relaxed | 88.46 | 68.23 | 77.04 | |
| Binary-Token | 93.59 | 74.89 | 83.2 | |
| Binary-Strict | 89.02 | 68.66 | 77.52 |
Table 3.
The performance of baseline system with different combinations of training and test data (micro-averaged, at strict entity level). 2014: i2b2/UTHealth Challenge[14], 2016: CEGS N-GRID Challenge [15]
| Training data | Test data | Precision | Recall | F-measure |
|---|---|---|---|---|
| 2014 training | 2014 test | 94.95 | 89.17 | 91.97 |
| 2014 training + test | 2016 training | 85.54 | 65.98 | 74.50 |
| 2014 training + test | 2016 test | 83.94 | 66.57 | 74.25 |
| 2016 training | 2016 test | 92.34 | 83.23 | 87.55 |
4.2. Task 1B
4.2.1. Overall performance
Table 4 shows the overall performance of the hybrid system evaluated on the test set. The performance at binary-level is generally the highest, followed by token-level, with entity-level evaluation having the lowest scores. Also, the system performed better with HIPAA categories than with N-GRID categories, as difficult categories such as PROFESSION are excluded in the HIPAA categories. Based on the primary measure used by the challenge (i.e., F1 by entity-level strict evaluation with N-GRID categories), our hybrid system achieved second best performance among all the participants [15].
Table 4.
The overall performance of the hybrid system. Entity-Strict level evaluation results, which are the primary results used by the N-GRID challenge [15], are shown in bold face.
| PHI category set | Evaluation level | Precision | Recall | F1-measure |
|---|---|---|---|---|
| N-GRID categories | Token | 95.23 | 90.15 | 92.62 |
| Entity-Strict | 93.39 | 88.23 | 90.74 | |
| Entity-Relaxed | 93.5 | 88.33 | 90.84 | |
| Binary-Token | 97.84 | 92.62 | 95.16 | |
| Binary-Strict | 95.66 | 90.38 | 92.94 | |
| HIPAA categories | Token level | 96.3 | 92.24 | 94.23 |
| Entity-Strict | 94.56 | 90.47 | 92.47 | |
| Entity-Relaxed | 94.67 | 90.57 | 92.58 | |
| Binary-Token | 97.18 | 93.09 | 95.09 | |
| Binary-Strict | 95.34 | 91.21 | 93.23 |
Table 5 shows the performance for each main PHI category. While DATE and AGE performed the best among the main categories, PROFESSION, LOCATION and ID had the lowest F1 scores. PROFESSION and LOCATION are known to be difficult from the previous challenge [14]; those categories show surface patterns with lower regularity and higher variety. The low performance of ID category is due to previously unseen types of ID PHI mentions that only appear in the test set, not in the training set. Thus, they are not covered by the regular expressions of the rule-based tagger.
Table 5.
The performance of i2b2 main PHI categories (micro-averaged on 2016 test set, at strict entity level). Number of training instances for each main category is also shown.
| PHI category | Precision | Recall | F1-measure | # training instances (%) |
|---|---|---|---|---|
| CONTACT | 93.5 | 91.27 | 92.37 | 154 (0.74) |
| NAME | 94.48 | 92.47 | 93.46 | 3,691 (17.71) |
| DATE | 97.04 | 95.32 | 96.17 | 5,723 (27.46) |
| AGE | 96.0 | 93.88 | 94.93 | 3,637 (17.45) |
| PROFESSION | 86.44 | 64.36 | 73.78 | 1,471 (7.06) |
| ID | 95.24 | 60.61 | 74.07 | 44 (0.21) |
| LOCATION | 88.47 | 81.36 | 84.76 | 7,213 (34.60) |
Table 6 shows the performance for each PHI subcategory. Subcategories that show regular surface patterns showed high F1 scores (i.e., PHONE, DATE, LICENSE, STATE, COUNTRY, ZIP). DOCTOR and AGE also showed high performance, probably due to the existence of strong context cues (e.g., ‘Dr.’ for doctor name, ‘age’ or ‘yo’ for AGE) in addition to the abundance of training examples. Subcategories with a small number of examples such as FAX, EMAIL, URL and LOCATION-OTHER had the lowest F1 scores. Finally, subcategories that have highly varied surface forms such as PROFESSION, STREET and ORGANIZATION had especially poor recall.
Table 6.
The performance of i2b2 PHI subcategories (micro-averaged on 2016 test set, at strict entity level). Only the categories that appear in the test set are shown in the table. Number of training instances for each sub category is also shown.
| Main category | Subcategory | Precision | Recall | F1-measure | # training instances (%) |
|---|---|---|---|---|---|
| CONTACT | PHONE | 98 | 96 | 97 | 143 (0.69) |
| FAX | 50 | 60 | 55 | 4 (0.02) | |
| 100 | 60 | 75 | 2 (0.01) | ||
| URL | 25 | 33 | 29 | 5 (0.02) | |
| NAME | DOCTOR | 95 | 96 | 96 | 2,396 (11.49) |
| PATIENT | 93 | 85 | 89 | 1,270 (6.09) | |
| DATE | DATE | 97 | 95 | 96 | 5,723 (27.46) |
| AGE | AGE | 96 | 94 | 95 | 3,637 (17.45) |
| PROFESSION | PROFESSION | 86 | 64 | 74 | 1,471 (7.06) |
| ID | HEALTHPLAN | 0 | 0 | 0 | 0 (0.00) |
| LICENSE | 95 | 95 | 95 | 38 (0.18) | |
| MEDICALRECORD | 0 | 0 | 0 | 4 (9.09) | |
| IDNUM | 0 | 0 | 0 | 2 (0.01) | |
| LOCATION | HOSPITAL | 87 | 82 | 84 | 2,196 (10.53) |
| STREET | 92 | 68 | 78 | 46 (0.22) | |
| ORGANIZATION | 82 | 61 | 70 | 1,113 (5.34) | |
| CITY | 88 | 89 | 88 | 1,394 (6.69) | |
| STATE | 94 | 95 | 94 | 662 (3.18) | |
| COUNTRY | 95 | 88 | 92 | 666 (3.20) | |
| ZIP | 100 | 88 | 94 | 23 (0.11) | |
| OTHER | 67 | 11 | 18 | 25 (0.12) |
4.2.2. Component impact analysis
Table 7 shows the performance of the hybrid system at each processing step. The addition of the rule-tagger and the two post-processing steps (i.e. the error correction and global reconciliation steps) increased the system performance. In particular, the rule-tagger and error correction steps show statistically significant performance improvement over previous steps. However, the system performance is shown to be heavily dependent on the performance of the CRF taggers.
Table 7.
The performance of the hybrid system measured at each step. “CRF taggers” stands for combination of the two CRFs (one for quantitative types, and another for name types). Statistically significant performance improvement over previous step is marked with*.
| Precision | Recall | F1-measure | |
|---|---|---|---|
| CRF taggers | 93.22 | 87.16 | 90.09 |
| + rule tagger | 93.09 | 87.54 | 90.23* |
| + error correction | 93.55 | 87.75 | 90.56* |
| + global reconciliation | 93.39 | 88.23 | 90.74 |
In order to further investigate the performance of the CRF taggers, we performed additional experiments. Table 8 shows the performance of the two CRF taggers with and without 2014 data. Note that we report system performance without the rule tagger and post-processing in order to emphasize the effect of additional training data on the CRF taggers. The addition of the i2b2 2014 de-identification corpus increased both precision and recall, resulting in statistically significant F1-measure improvement. EasyAdapt showed better recall than simply merging the two corpora. In contrast, simple merge showed better precision. In terms of F1-measure, the use of EasyAdapt did not show statistically significant performance improvement compared to simple merge.
Table 8.
The performances of CRF taggers with/without 2014 data. +DA stands for the use of domain adaptation algorithm, while −DA stands for simply merging 2016 training and 2014 data. Statistically significant performance improvement over using only 2016 training data is marked with*.
| Precision | Recall | F1-measure | |
|---|---|---|---|
| 2016 training | 92.87 | 86.46 | 89.55 |
| + 2014 data (+DA) | 93.22 | 87.16 | 90.09* |
| + 2014 data (−DA) | 93.59 | 86.74 | 90.03* |
Table 9 shows the ablation test results on features that are newly introduced to the hybrid system. Only the features used for both CRF taggers are shown. The test result shows that the new features are all useful in improving the performance, showing statistically significant performance increases. Additional ablation test results on more features are shown in Table 4 of the Supplementary Documents.
Table 9.
Ablation test for additional features of hybrid system. Statistically significant performance decrease over using all features is marked with *.
| Precision | Recall | F1-measure | ΔF | |
|---|---|---|---|---|
| All | 93.22 | 87.16 | 90.09 | |
| - open domain word embeddings | 93.51 | 86.23 | 89.72* | −0.37 |
| - attribute name | 92.98 | 86.89 | 89.83* | −0.26 |
| - token shape n-gram | 93.05 | 86.65 | 89.73* | −0.36 |
The hybrid system uses both word embeddings trained on medical corpora and word embeddings trained on open domain corpora. Table 10 shows the result of experiments with various sub-sets of word embeddings. Open domain word embedding show slightly better performance than medical domain word embeddings. However, the best performance is achieved with both sets of embeddings. This is consistent with the finding by Roberts [40] of the importance of utilizing a variety of embeddings for clinical NLP.
Table 10.
The performance of CRF taggers with different word embedding (WE) features. Statistically significant performance increase over using no embeddings is marked with*. Statistically significant performance decrease over using both embeddings is marked with ◊.
| Precision | Recall | F1-measure | |
|---|---|---|---|
| No WE feature | 93.25 | 85.86 | 89.41 ◊ |
| Medical domain WE only | 93.51 | 86.23 | 89.72*◊ |
| Open domain WE only | 93.03 | 86.84 | 89.83*◊ |
| Medical WE + Open WE | 93.22 | 87.16 | 90.09* |
Finally, we tested the effect of different rules for global reconciliation, shown in Table 11. The global reconciliation rules resulted in a minimal gain, largely through improved recall. Overall, the performance increase by the global reconciliation rules was not shown to be statistically significant.
Table 11.
System performance with additional global reconciliation (GR) modules
| Precision | Recall | F1-measure | |
|---|---|---|---|
| Before GR | 93.55 | 87.75 | 90.56 |
| + GR with high-precision rules | 93.58 | 87.94 | 90.67 |
| + GR with type-counting | 93.39 | 88.23 | 90.74 |
4.2.3. Error analysis
We performed detailed error analysis based on the confusion matrix shown in Table 12. The errors fall into three categories: type errors, missing errors, and spurious errors.
Table 12.
Confusion matrix of the hybrid system’s output (on the test set at strict entity level)
| predicted | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PHO | FAX | EMA | URL | DOC | PAT | DAT | AGE | PRO | HEA | LIC | MED | IDN | HOS | STR | ORG | CIT | STA | COU | ZIP | OTH | missing | total | ||
| gold | PHO | 108 | 2 | 3 | 113 | |||||||||||||||||||
| FAX | 1 | 3 | 1 | 5 | ||||||||||||||||||||
| EMA | 3 | 2 | 5 | |||||||||||||||||||||
| URL | 1 | 2 | 3 | |||||||||||||||||||||
| DOC | 1510 | 10 | 2 | 1 | 44 | 1567 | ||||||||||||||||||
| PAT | 19 | 713 | 31 | 8 | 66 | 837 | ||||||||||||||||||
| DAT | 3642 | 13 | 1 | 165 | 3821 | |||||||||||||||||||
| AGE | 1 | 2210 | 143 | 2354 | ||||||||||||||||||||
| PRO | 1 | 1 | 650 | 5 | 1 | 352 | 1010 | |||||||||||||||||
| HEA | 2 | 2 | ||||||||||||||||||||||
| LIC | 20 | 1 | 21 | |||||||||||||||||||||
| MED | 2 | 2 | ||||||||||||||||||||||
| IDN | 8 | 8 | ||||||||||||||||||||||
| HOS | 12 | 6 | 1082 | 13 | 22 | 190 | 1327 | |||||||||||||||||
| STR | 1 | 1 | 23 | 1 | 2 | 2 | 6 | 34 | ||||||||||||||||
| STA | 1 | 3 | 4 | 8 | 23 | 428 | 18 | 1 | 211 | 697 | ||||||||||||||
| ORG | 2 | 5 | 12 | 9 | 731 | 6 | 3 | 1 | 51 | 820 | ||||||||||||||
| CIT | 1 | 1 | 455 | 24 | 481 | |||||||||||||||||||
| COU | 1 | 1 | 2 | 11 | 9 | 332 | 20 | 376 | ||||||||||||||||
| ZIP | 15 | 2 | 17 | |||||||||||||||||||||
| OT | 1 | 5 | 5 | 2 | 6 | 19 | ||||||||||||||||||
| spurious | 1 | 1 | 2 | 42 | 23 | 110 | 79 | 93 | 1 | 82 | 2 | 61 | 32 | 12 | 13 | 554 | ||||||||
| total | 110 | 6 | 3 | 4 | 1590 | 763 | 3753 | 2302 | 752 | 0 | 21 | 0 | 0 | 1240 | 25 | 519 | 833 | 484 | 349 | 15 | 3 | 1301 | ||
A type error occurs when a PHI mention’s offsets are correct, but the type is incorrect. Type errors occurred the most between HOSPITAL names and other name categories such as PATIENT, ORGANIZATION and CITY. When PATIENT names were written as initials not in full names, they were frequently mis-identified as HOSPITAL names. For instance, in “yet KC denies feelings of depression”, KC is the PATIENT name initials, but is mis-identified as HOSPITAL. Confusion between HOSPITAL and ORGANIZATION, and between HOSPITAL and CITY are due to similar context around the PHI terms. For instance, “psychotherapy in Gallatin Valley at age 17 – 18” contains HOSPITAL type PHI ‘Gallatin Valley’, which was misclassified as a CITY name.
A missing error occurs when a PHI mention is completely missed (i.e., a false negative). The PROFESSION category produced the most missing errors. We believe that this is due to the great variety of PROFESSION mentions. For instance, PROFESSION mentions occur with various syntactic forms. E.g., “dancing”, “danced”, and “dancer” can be a PROFESSION. Moreover, verb phrases such as “leads a development team” can be a PROFESSION. Even professions in a medical field, such as “Doctor” and “RN” were frequently missed. The ORGANIZATION category produced the second most missing errors, similarly due to the great variety of the surface forms, especially abbreviations.
Finally, a spurious error occurs when a non-PHI mention is wrongly identified as a PHI mention (i.e., a false positive). The DATE category produced the most spurious errors, mostly due to partial or overlapping matches for multiword DATE mentions. For instance, while “Wednesday, 4/17/94” is the gold standard DATE mention, the system predicted “Wednesday” and “4/17/94” separately as DATE mentions. PROFESSION produced the second most spurious errors. Again, multiword PROFESSION mentions were the major source of the errors. For instance, for “Senior Manager of Education”, only “Senior Manager” was predicted. Conversely, the system wrongly predicted “professional fashion designer” as a PROFESSION mention, but only “fashion designer” was annotated in the gold standard annotation.
The three types of errors have different consequences from each other. While missing errors induce the risk of revealing patient’s identity, type errors and spurious errors are free of such risk. On the other hand, type errors and spurious errors may bring difficulties for downstream utilization of the notes. Type errors would leave false placeholders (e.g., placeholder for CITY after removing a hospital name), confusing the readers of de-identified notes. Spurious errors falsely remove non-PHI information, which may be important for clinical or research applications that use the de-identified notes. This observation raises the issue of how to optimize the trade-off of recall and precision. Higher recall ensures better protection of the patient’s identity, and higher precision implies losing less non-PHI information that may be of medical importance. While F1-measure is primarily used to compare de-identification systems, it hides the trade-off between precision and recall, which might be important when choosing which de-identification system to use. Other variants of F-measure would be used to weight precision or recall more highly, unlike the balanced F1.
Also, it is interesting to see that some errors can be considered as artifacts that result from the particular evaluation method being used. For instance, a spurious error produced by splitting “Wednesday, 4/17/94” into “Wednesday” and “4/17/94” neither reveals any PHI nor loses any non-PHI information. In fact, such a case will be considered as an error only under entity-level evaluation scheme. Using a token-level evaluation, both “Wednesday” and “4/17/94” will be considered as true positives.
5. Conclusion
In this paper, we described a hybrid de-identification system that is developed for psychiatric notes, as part of the CEGS N-GRID challenge. The system is composed of a rule tagger and two CRF taggers. The rule tagger is optimized through customization to the psychiatric notes. The performance of the CRF taggers is boosted with both a rich feature set as well as the integration of additional training data through domain adaptation. Our system achieved F-score 90.74, second-best among all the participants.
The CEGS N-GRID 2016 challenge uniquely emphasizes the importance of de-identification system robustness across different note types. Our baseline system showed an F1-measure of 74.5 for Task 1A without utilizing labeled psychiatric notes, but the performance was significantly increased to 87.55 when labeled training data was incorporated. This indicates that being able to customize a de-identification system to the data is critical. While a sufficient amount of target data for maximum performance of ML component is ideal, having even a limited amount of labeled data can also help (i.e., through simple error analysis and developing rule-based system). Beyond this, when the training data in the domain is not enough to maximize ML performance, one can utilize another existing dataset to bridge the gap, as shown by our experiment with CRF taggers (F-score 90.09 with 2014 data through domain adaptation, and 89.55 without 2014 data).
Supplementary Material
Highlights.
A hybrid approach for automatic de-identification of psychiatric notes is proposed.
The rule-based components exploit the structure of the psychiatric notes.
The machine learning components integrate additional data to training set.
The system showed second-best performance at the CEGS N-GRID task 1.
Acknowledgments
The authors were supported by NIH grants 5R01LM010681-05, 1R01GM102282-01A1, 1U24CA194215-01A1, and 4R00LM012104. The CEGS N-GRID challenge was supported by NIH grants P50MH106933, 4R13LM011411. We would additionally like to thank the reviewers for their valuable comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
As is commonly done in clinical NLP, we consider a colon followed by a newline as a sentence separator.
Conflict of interest
None declared.
References
- 1.Sweeney L. Replacing personally-identifying information in medical records, the Scrub system. Proc AMIA Annu Fall Symp. 1996:333–337. [PMC free article] [PubMed] [Google Scholar]
- 2.Ruch P, Baud RH, Rassinoux AM, Bouillon P, Robert G. Medical document anonymization with a semantic lexicon. Proc AMIA Symp. 2000:729–733. [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas SM, Mamlin B, Schadow G, McDonald C. A successful technique for removing names in pathology reports using an augmented search and replace method. presented at the AMIA Symposium. 2002:777–781. [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta D, Saul M, Gilbertson J. Evaluation of a deidentification (De-Id) software engine to share pathology reports and clinical documents for research. Am J Clin Pathol. 2004 Feb;1212:176–186. doi: 10.1309/E6K3-3GBP-E5C2-7FYU. [DOI] [PubMed] [Google Scholar]
- 5.Beckwith BA, Mahaadevan R, Balis UJ, Kuo F. Development and evaluation of an open source software tool for deidentification of pathology reports. BMC Medical Informatics and Decision Making. 2006 Mar;6(1):12. doi: 10.1186/1472-6947-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedlin FJ, McDonald CJ. A software tool for removing patient identifying information from clinical documents. J Am Med Inform Assoc. 2008 Sep;155:601–610. doi: 10.1197/jamia.M2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neamatullah I, Douglass MM, Lehman L-WH, Reisner A, Villarroel M, Long WJ, Szolovits P, Moody GB, Mark RG, Clifford GD. Automated de-identification of free-text medical records. BMC Medical Informatics and Decision Making. 2008 Jul;8(1):641–17. doi: 10.1186/1472-6947-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen T, Cullen RM, Godwin M. Hidden Markov model using Dirichlet process for de-identification. Journal of Biomedical Informatics. 2015 Dec;58:S60–6. doi: 10.1016/j.jbi.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dernoncourt F, Lee JY, Szolovits P, Uzuner OZ. De-identification of Patient Notes with Recurrent Neural Networks. 2016 Jun; doi: 10.1093/jamia/ocw156. p. arXiv:1606.03475 [cs.CL] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Chen Y, Tang B, Wang X, Chen Q, Li H, Wang J, Deng Q, Zhu S. Automatic de-identification of electronic medical records using token-level and character-level conditional random fields. Journal of Biomedical Informatics. 2015 Dec;58:S47–S52. doi: 10.1016/j.jbi.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang H, Garibaldi JM. Automatic detection of protected health information from clinic narratives. Journal of Biomedical Informatics. 2015 Dec;58:S30–S38. doi: 10.1016/j.jbi.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dehghan A, Kovacevic A, Karystianis G, Keane JA, Nenadic G. Combining knowledge- and data-driven methods for de-identification of clinical narratives. Journal of Biomedical Informatics. 2015 Dec;58:S53–S59. doi: 10.1016/j.jbi.2015.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uzuner O, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc. 2007 Sep;14(5):550–563. doi: 10.1197/jamia.M2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stubbs A, Kotfila C, Uzuner O. Automated systems for the de-identification of longitudinal clinical narratives: Overview of 2014 i2b2/UTHealth shared task Track 1. Journal of Biomedical Informatics. 2015 Dec;58:S11–S19. doi: 10.1016/j.jbi.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stubbs A, Filannino M, Uzuner O. De-identification of psychiatric intake records: Overview of 2016 CEGS N-GRID Shared Tasks Track 1. Journal of Biomedical Informatics. 2017 doi: 10.1016/j.jbi.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardner J, Xiong L. HIDE: An Integrated System for Health Information DE-identification. presented at the 21st International Symposium on Computer-Based Medical Systems (CBMS) 2008:254–259. [Google Scholar]
- 17.Aberdeen J, Bayer S, Yeniterzi R, Wellner Ben, Clark C, Hanauer D, Malin B, Hirschman L. The MITRE Identification Scrubber Toolkit: Design, training, and assessment. International Journal of Medical Informatics. 2010 Dec;79(12):849–859. doi: 10.1016/j.ijmedinf.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Benton A, Hill S, Ungar L, Chung A, Leonard C, Freeman C, Holmes JH. A system for de-identifying medical message board text. BMC Bioinformatics. 2011 Jun;12(3):S2. doi: 10.1186/1471-2105-12-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uzuner O, Sibanda TC, Luo Y, Szolovits P. A de-identifier for medical discharge summaries. Artificial Intelligence in Medicine. 2008 Jan;42(1):13–35. doi: 10.1016/j.artmed.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMurry AJ, Fitch B, Savova G, Kohane IS, Reis BY. Improved de-identification of physician notes through integrative modeling of both public and private medical text. BMC Medical Informatics and Decision Making. 2013 Oct;13(1):112. doi: 10.1186/1472-6947-13-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szarvas G, Farkas R, Busa-Fekete R. State-of-the-art Anonymization of Medical Records Using an Iterative Machine Learning Framework. J Am Med Inform Assoc. 2007 Sep;14(5):574–580. doi: 10.1197/j.jamia.M2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu H. CLAMP: Clinical Language Annotation, Modeling, and Processingb Toolkit. 2017 [Online]. Available: http://clamp.uth.edu. [Accessed: 13-Mar-2017]
- 23.Baldridge J. The opennlp project. 2005 [Online]. Available: http://opennlp.apache.org/index.html. [Accessed: 13-Mar-2017]
- 24.Okazaki N. CRFsuite: a fast implementation of conditional random fields (CRFs) http://www.chokkan.org/software/crfsuite. 2007 [Online]. Available: http://www.chokkan.org/software/crfsuite/. [Accessed: 13-Mar-2017]
- 25.Brown PF, deSouza PV, Mercer RL, Pietra VJD, Lai JC. Class-based n -gram models of natural language. Computational Linguistics. 1992 Dec;18(4):467–479. [Google Scholar]
- 26.Lund K, Burgess C. Producing high-dimensional semantic spaces from lexical co-occurrence. Behavior Research Methods, Instruments, & Computers. 1996;28(2):203–208. [Google Scholar]
- 27.Nikfarjam A, Sarker A, OConnor K, Ginn R, Gonzalez G. Pharmacovigilance from social media: mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. J Am Med Inform Assoc. 2015 May;22(3):671–681. doi: 10.1093/jamia/ocu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mikolov T, Sutskever I, C K, Corrado GS, Dean J. Distributed Representations of Words and Phrases and their Compositionality. presented at the NIPS. 2013:3111–3119. [Google Scholar]
- 29.Saeed M, Villarroel M, Reisner AT, Clifford G, Lehman L-W, Moody G, Heldt T, Kyaw TH, Moody B, Mark RG. Multiparameter Intelligent Monitoring in Intensive Care II (MIMIC-II): A public-access intensive care unit database. Critical care medicine. 2011 May;39(5):952–960. doi: 10.1097/CCM.0b013e31820a92c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo J, Che W, Wang H, Liu T. Revisiting Embedding Features for Simple Semi-supervised Learning. EMNLP. 2014 [Google Scholar]
- 31.Pennington J, Socher R, Christopher M. GloVe: Global Vectors for Word Representation. presented at the EMNLP 2014. 2014:1–12. [Google Scholar]
- 32.Finkel JR, Grenager T, Manning C. Incorporating non-local information into information extraction systems by Gibbs sampling. presented at the the 43rd Annual Meeting; Morristown, NJ, USA. 2005. pp. 363–370. [Google Scholar]
- 33.Collobert R, Weston J, Bottou L, Karlen M, Kavukcuoglu K, Kuksa P. Natural Language Processing (Almost) from Scratch. JMLR. 2011 Aug;12:2493–2537. [Google Scholar]
- 34.Xu Q, Yang Q. A Survey of Transfer and Multitask Learning in Bioinformatics. Journal of Computing Science and Engineering. 2011 Sep;5(3):257–268. [Google Scholar]
- 35.Weiss K, Khoshgoftaar TM, Wang D. A survey of transfer learning. J Big Data. 2016;3(1):9. [Google Scholar]
- 36.Pan SJ, Yang Q. A Survey on Transfer Learning. IEEE Trans Knowl Data Eng. 2010 Oct;22(10):1345–1359. [Google Scholar]
- 37.Daumé H., III Frustratingly Easy Domain Adaptation. arXiv.org. 2009 Jul 10; [Google Scholar]
- 38.Zhang Y, Tang B, Jiang M, Wang J, Xu H. Domain adaptation for semantic role labeling of clinical text. J Am Med Inform Assoc. 2015 Sep;22(5):967–979. doi: 10.1093/jamia/ocu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chinchor N. THE STATISTICAL SIGNIFICANCE OF THE MUC-4 RESULTS. presented at the Proceedings of the th conference on Message understanding. Association for Computational Linguistics. 1992:30–50. [Google Scholar]
- 40.Roberts K. Assessing the Corpus Size vs. Similarity Trade-off for Word Embeddings in Clinical NLP. presented at the Proceedings of the Clinical Natural Language Processing Workshop. 2016:54–63. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


