Abstract
Objective
Our objective was to develop a machine learning-based system to determine the severity of Positive Valance symptoms for patients, based on information included in their initial psychiatric evaluation. Severity was rated on an ordinal scale of 0–3 as follows: 0 (absent=no symptoms), 1 (mild=modest significance), 2 (moderate=requires treatment), 3 (severe=causes substantial impairment) by experts.
Materials and Methods
We treated the task of assigning Positive Valence severity as a text classification problem. During development, we experimented with regularized multinomial logistic regression classifiers, gradient boosted trees, and feedforward, fully-connected neural networks. We found both regularization and feature selection via mutual information to be very important in preventing models from overfitting the data. Our best configuration was a neural network with three fully connected hidden layers with rectified linear unit activations.
Results
Our best performing system achieved a score of 77.86%. The evaluation metric is an inverse normalization of the Mean Absolute Error presented as a percentage number between 0 and 100, where 100 means the highest performance. Error analysis showed that 90% of the system errors involved neighboring severity categories.
Conclusion
Machine learning text classification techniques with feature selection can be trained to recognize broad differences in Positive Valence symptom severity with a modest amount of training data (in this case 600 documents, 167 of which were unannotated). An increase in the amount of annotated data can increase accuracy of symptom severity classification by several percentage points. Additional features and/or a larger training corpus may further improve accuracy.
Keywords: Positive Valance, mental disorder severity, text classification, Research Domain Criteria (RDoC), machine learning
Graphical abstract
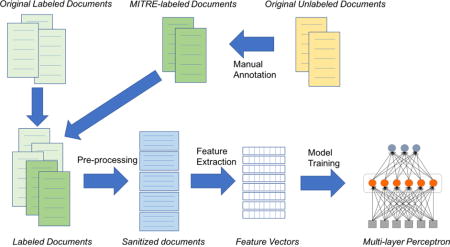
1 Introduction
We developed a neural network-based system to determine the severity of Positive Valance symptoms for patients, based on information included in their initial psychiatric evaluation. This system was developed in the context of participation in the 2016 Centers of Excellence in Genomic Science (CEGS) Neuropsychiatric Genome-Scale and RDoC Individualized Domains (N-GRID) Shared Task in Clinical Natural Language Processing [1]. The CEGS-N-GRID challenge posed the task of determining the severity of patients’ Positive Valence symptoms, based on initial psychiatric evaluation. Severity was rated on an ordinal scale of 0–3 as follows:
0 (absent=no symptoms)
1 (mild=modest significance)
2 (moderate=requires treatment)
3 (severe=causes substantial impairment)
As a participant in this challenge, we developed a machine learning-based system to determine the severity of Positive Valence symptoms of a patient.
1.1 Background
1.1.1 Research Domain Criteria
Research Domain Criteria (RDoC) constitute a research framework developed under the aegis of the National Institute of Mental Health (NIMH) to facilitate new ways of studying mental disorders. The initiative is motivated by concerns that diagnostic categories based on clinical consensus fail to align with findings emerging from clinical neuroscience and genetics; that the boundaries of these categories have not been predictive of treatment response; and that these categories, based upon presenting signs and symptoms, may not capture fundamental underlying mechanisms of dysfunction [2]. A primary goal of the initiative is to develop a classification system for mental health based on dimensions of observable behavior and neurobiological measures ([3] [4]), rather than symptom complexes based largely on clinical descriptions, which form the basis of the Diagnostic and statistical manual of mental disorders (DSM) [5] and the International Classification of Diseases (ICD) [6]. The new classification system is intended to foster diagnosis based on multiple levels of information including genetics, imaging, and cognitive science. Units of analysis include genes, molecules, cells, circuits, physiology, behavior, self-reports, and paradigms. The framework is intended to support a description of human behavior from normal to abnormal in multiple domains:
The Positive Valence Systems – events, objects or situations that signal mental disorders but are attractive to the patient to the point of active engagement. Examples are excessive alcohol or drug consumption, gambling, and compulsive behavior.
The Negative Valence Systems – responses to aversive situations or context, such as fear, anxiety, and loss
Cognitive Systems – various cognitive processes including attention, perception, learning, and memory
Systems for Social Processes – responses to interpersonal settings of various types, including perception and interpretation of others’ actions
Arousal and Regulatory Systems – activation of neural systems and providing appropriate homeostatic regulation of such systems as energy balance and sleep
1.2 Related Work
1.2.1 Classification of Patients and Clinical Notes
The ability to characterize patients or clinical records with respect to specific attributes has found practical application for patient cohort selection [7] and computerized syndrome surveillance (e.g., [8] [9]). There has been an increase in the number of studies associated with cohort identification using electronic medical records in recent years. Statistical analyses or machine learning and NLP techniques have been gaining popularity in comparison with rule-based systems [10]. Solti et al. used using Maximum Entropy with word unigram and character n-grams as features to classify chest x-ray reports as indicative of Acute Lung Injury [11]. Yetisgen-Yildiz et al. also used Maximum Entropy with bag of word and n-grams as features to classify patients with respect to Acute Lung Injury, and to each n-gram feature they added an identified assertion value (e.g., pneumonia_absent) [12]. Liao et al. developed a system that classifies subjects with rheumatoid arthritis using logistic regression and clinical named entities extracted with NLP as features [13]. Wright et al. designed a system that identifies progress notes pertaining to diabetes using a Support Vector Machine (SVM) with bag of words features [14].
More recently, classification of various types of mental health-related data has been attempted. SVMs with specific words, specific parts of speech and emotional concepts, and readability vectors as features have been applied to distinguish simulated from real suicide notes [15,16]. Cook et al. used a LIBLINEAR machine learning protocol to identify suicidal ideation in questionnaire responses associated with post hospital discharge therapeutic reminders [17]. Features included n-grams and contextual information such as negation context provided by an NLP pipeline to the text. Perlis et al. determined patient status (depressed or well) based on the information in out-patient psychiatry practice notes [18]. The researchers used logistic regression with International Classification of Diseases, Ninth Revision (ICD-9) [19] codes and NLP-extracted terms as features to classify patients as well (defined as absence or virtual absence of depressive symptoms), depressed (defined as likely to meet criteria for a current major depressive episode), or intermediate (subthreshold).
Systems designed for specific use cases such as those referenced above typically approach the task as binary classification, wherein a clinical document and (by extension a patient) is labeled or not labeled with a disorder. On the other hand, several community medical language challenges have been organized to promote development of patient classification systems, and these challenges have posed more complex tasks requiring systems to distinguish a larger number of categories.
The i2b2 Smoking Challenge invited participants to develop systems that could predict the smoking status of patients based on the narratives in medical discharge summaries [20]. The task required systems to distinguish five categories: nonsmoker, past smoker, current smoker, smoker, and unknown. Participants developed a variety of machine learning-based and hybrid systems. Only 2 out of 23 submissions were generated by systems that were entirely rule-based. The top performing system employed a hybrid of machine-leaning and rule based techniques [21].
The Obesity Challenge was a multi-class, multi-label classification task focused on obesity and its co-morbidities. Systems were challenged to classify obesity and its comorbidities into four classes based on individual discharge summaries – present, absent, questionable, or unmentioned in the documents [22]. The challenge presented two tasks with differing criteria for data annotation. For the textual judgment task, experts classified documents based on explicitly documented information. For the intuitive judgment task, experts classified documents by applying their intuition and judgment to information in the documents. It was expected that intuitive judgments would agree with textual judgments of present, absent, or questionable, but that a textual judgment of unmentioned would be given an intuitive judgment of present, absent, or questionable, depending whether there was information present that would support inference. Although most of the top performing systems were rule-based for text-based classification, machine learning approaches contributed to the top ten systems in a task to infer intuitive judgments.
The first NLP challenge that focused on applying NLP to text in the mental health domain was the Fifth i2b2/VA/Cincinnati Shared-Task and Workshop. Participants were challenged to develop systems that could classify emotions found in real suicide notes at the phrase level [23]. This challenge did not include document-level classification.
The CEGS N-GRID challenge was the first challenge to address document-level classification of mental health notes. Like other medical NLP challenges, it required distinction of multiple categories. Unlike other medical NLP challenges, it had the ambitious goal to apply categories from a research classification framework (RDoC) very different from the classification framework that underlies clinical diagnosis and description presented in the data (DSM). Also in contrast to most previous medical NLP challenges, there were no evidence annotations in the text (which, if present, could be used to support the document-level classifications).
2 Materials and Methods
2.1 Data
The training data consisted of 600 psychiatric evaluation reports. Experts reviewed and assigned Positive Valence severity scores to a subset of these reports. The number of singly annotated, doubly annotated, and unannotated documents in the training set are shown in Table 1 below. Table 1 also shows the distribution of severity ratings in the annotated data.
Table 1.
Distribution of annotation and severity ratings
| ANNOTATION | TOTAL DOCUMENTs | SEVERITY | |||
|---|---|---|---|---|---|
| Absent | Mild | Moderate | Severe | ||
| Double | 325 | 45 | 130 | 82 | 68 |
| Single | 108 | 16 | 36 | 28 | 28 |
| Total Annotated | 433 | 61 | 166 | 110 | 96 |
| Unannotated | 167 | ||||
| Total | 600 | ||||
The documents in this data set contained a variety of text formats including narrative sections that consisted of section headings, semi-structured text, and unstructured text. The semi-structured text included attribute-value pairs often occurring in lists, as well as templated text consisting of a heading followed by a question and an answer that was typically Yes, No, or occasionally Uncertain. The format was frequently obscured by a lack of space characters between words (as can happen when records are pulled from a clinical warehouse), which had an impact not only on detection of different text regions, but which also reduced the accuracy of word tokenization. In addition, almost all the documents contained DSM codes1 [5], and there were ICD-10 [24] codes in a very small number of documents.
Although the documents were annotated with a single value indicating symptom severity, the textual evidence upon which these judgements were made was not annotated.
2.2 Text Classification
We treated the task of assigning Positive Valence severity as a text classification problem. Our team did not include a clinical subject matter expert, and we did not have access to someone experienced with the RDoC criteria. Consequently, we selected a classification method that did not require annotation or other explication of the elements that might provide the basis of the document classification. One of the attributes of neural networks is their ability to detect patterns and associations in text without a priori assumptions.
2.3 Text Preprocessing
Prior to feature extraction, we applied pre-processing that included limited spelling correction for frequently misspelled words, tokenization refinement of words/headings merged together due to document formatting, and normalization of section and attribute names. Tokenization refinement included, for example, splitting apart separate words that were merged together without intervening white space. This was a common problem and non-trivial to solve, but we found certain capitalization patterns such as a token appearing as lowerUpper which should separate into lower and Upper. Other refinements included splitting punctuation.
2.4 Feature Extraction
Features that served as input to our system consisted of the terms that occurred in the document (bag-of-words features). The raw terms were used to construct features. No stemming, lemmatization or other transformations were performed; however, we did ignore case. Beyond these features, we experimented with a range of additional features that we expected would contribute to the determination of Positive Valence symptom severity.
Manually Created Lexica
Based on manual analysis of the text, we created a lexicon that included terms judged to be associated with Positive Valence symptoms, inflected forms of original terms and case variants. Terms were assigned a label that served to group semantically related terms.
Automatically Generated Term Clusters
We experimented with two other ways to reduce the dimensionality and increase the potential generalizability of lexical information in the documents. The first method involved term clustering using the Brown clustering algorithm [25]. We utilized 1000 pre-built clusters on the Gigaword corpus [26], which provides a mapping from each lexical item to a unique cluster represented as a bit vector. The bit vector encodes the location of the cluster in a dendrogram and similar clusters share bit-vector prefixes. We also obtained term clusters by K-means clustering over neural word embeddings trained using skip-gram word2vec models on a corpus of 8 billion words of PubMed articles.
Location-Based Features
The same term can carry different meanings depending on its location in a document. For example, the term culture can carry different meanings in a Social History section and the Laboratory section of a Summary note. We addressed this by first parsing documents to produce a set of template header strings. We treated the top 30 most frequent header strings as sections, and then modeled each term T that occurred in a section S by introducing a conjoined feature S_T. This ensured we had a feature for each possible section-dependent interpretation of a term.
Frequency Count of DSM Codes
For English words, we simply introduced binary features indicating whether a term was present or not. In the case of DSM codes, however, we noticed some value in capturing their frequency across the document. Accordingly, we used integer-valued features for DSM codes with their raw counts from the document.
Frequency Count of Affirmative Responses to Questions
Motivated by the distribution of affirmative answers associated with different severity levels in the data (shown in Table 2), we attempted to approximate crudely some of the information provided by Yes and No answers in templates by incorporating an aggregate feature that indicated how often the string Yes preceded by a colon occurred in the document. We excluded from the count Yes answers that indicated a neutral or positive factor, such as Does the patient feel safe in current living situation.
Table 2.
Frequency yes answers and severity
| SEVERITY | FREQUENCY PER DOCUMENT |
|---|---|
| Severe | 12.9 |
| Moderate | 11.3 |
| Mild | 9.1 |
| Absent | 4.9 |
After considerable experimentation, we used only a subset of the feature types described above in our final experiments. For example, the automatically generated and manual term clusters were excluded as they added no measurable improvements. Section 2.7.1 summarizes the final set of candidate features used for the model.
2.5 Feature Selection Via Mutual Information
Table 3 shows the twelve features with the highest overall mutual information (MI) scores where the MI was measured between each feature and the gold-standard severity category. For a given feature, a point-wise MI score is estimated for each severity category; these point-wise estimates are rolled up into a single overall MI score. Features represent terms conceptually related to substance abuse and DSM codes for drug and alcohol dependence and abuse. As noted earlier, term features ignored case, but did not involve stemming, lemmatization or other transformation. DSM [5] code 304 occurs in descriptions of drug dependence such as Opioid dependence 304.00, 304.01 opiate dependence, 304.20 Cocaine, Dependence, 304.30 Cannabis dependence, and 304.40 Amphetamine Dependence. DSM code 303 occurs in description of alcohol dependence such as EtOH dependence 303.9 and 303.90 Alcohol Dependence.
Table 3.
Terms with top twelve mutual information scores
| Feature | Topic Association | Overall MI Information Score | Count |
|---|---|---|---|
| detox | Substance Dependence | 0.105149393 | 62 |
| dependence | Substance Dependence | 0.103470645 | 141 |
| addiction | Substance Dependence | 0.086458342 | 86 |
| sober | Substance Dependence | 0.079067214 | 80 |
| sobriety | Substance Dependence | 0.069137302 | 60 |
| 304 | DSM code | 0.067738141 | 56 |
| substances | Substance | 0.058324368 | 65 |
| iop | Treatment (Intensive outpatient program) | 0.056322528 | 52 |
| suboxone | Substance | 0.053652091 | 28 |
| opioid | Substance | 0.052574696 | 31 |
| 303 | DSM code | 0.050121578 | 46 |
| opiates | Substance | 0.049796579 | 30 |
2.6 Additional Training Data
We assigned a person on our team to annotate the severity score in the available unannotated documents provided to serve as additional training data, although no member of our team had clinical expertise. The addition of 167 annotated documents improved our accuracies by 3–5% on held out data. A pilot annotation effort involving 20 documents showed that our annotator agreed with the multiple-annotated gold-standard annotations 60% of the time.
2.7 Model Development and Tuning
Most of our experimentation involved fully connected neural networks, also called multi-layered perceptrons (MLPs)2. MLPs consist of an input layer representing the input features for an example, an output layer and any number of hidden layers. For our dataset, the input layer involved the set of features for our corpus after any reductions based on feature selection; most features were binary-valued. Layers are connected to each other via a weight matrix. An MLP without any hidden layers is a linear model (a generalization of logistic regression) while the addition of hidden layers allows the model to accommodate nonlinear relationships between the inputs and outputs through non-linear activation functions. For our models, the output layer was a so-called SoftMax3 output layer that allows the network to output a probability distribution over outcomes. For this task, our output layer had four possible outcomes (0, 1, 2 and 3).
We determined the best candidate MLP configurations based on a single split of the available training data into 475 training documents and 125 test documents. All test documents were drawn randomly from the original annotated data. MITRE-annotated documents were included in the training split only. We found that MLPs with hidden layers generally outperformed linear models, though extensive measures were required to prevent overfitting. We utilized L1 and L2 regularization penalties on each layer; in addition, we found that dropout regularization was essential to prevent overfitting and co-adaptation.
Even with extensive regularization, however, we found some benefit to using the feature selection techniques described above to reduce the input dimensionality.
2.7.1 Final model
Our final models for submission to the challenge involved a neural network with three fully connected hidden layers with rectified linear unit activations. The first layer was comprised of 50 units/dimensions; the second layer was comprised of 20 units/dimensions, and the third layer was comprised of 40 units/dimensions. We used 40% dropout on each hidden layer along with an L1 penalty to prevent overfitting [27]. The output SoftMax layer used L2 regularization. All such configuration parameters were determined by selecting the best configuration via cross-fold validation on the available training data. All three submitted runs used this configuration yet varied in the dropout used on the input layer as well as the number of input features. This was done to hedge against the potential to overfit the model on our relatively small training set.
Our set of candidate input features included:
Unigrams from the entire document (including template headers) except for those found in the family history section
DSM codes and their frequency
The frequency of “Yes” responses to questions that would appear to indicate higher risk for severe psychiatric symptoms. We introduced just a single count for all “Yes” responses to all question types rather than separate counts/features for each question type.
This set resulted in over 29375 candidate features. For all documents in our training set, we computed the values for these 29375 candidate features. Given the small amount of training data, we employed feature selection, as described in Section 2.5, to reduce this the number of input features. We found reducing the number of features to anywhere from 50 to 500 features performed well, with the best performance in the range of 60 to 120.
2.8 Submissions
We submitted 3 runs which differed by number of features and/or percentage of input dropout.
Run 1 (Under) was intended to slightly underfit the training data, using only 60 input features (top-ranked features based on mutual information), and 25% dropout on the input (in addition to the hidden layer dropouts).
Run 2 (Middle) was intended to fit the training data “just right” using 20% input dropout with 120 input features.
Run 3 (Over) was intended to slightly overfit the training data using 120 input features with no dropout on the input.
We trained all three models on all 600 training documents using AdaGrad [28] for 6000 epochs using the Mandolin software package4. Our models were evaluated on 216 test documents provided by the challenge.
3 Results
Our best performing system, Run 2 (Middle), achieved an official score of 77.86% on challenge test data. The evaluation metric is an inverse normalization of the macro-averaged Mean Absolute Error presented as a percentage number between 0 and 100, where 100 indicates a perfect score. Table 4 shows the overall scores and scores for each severity category for three systems. For reference, the top scoring system in the shared task achieved a score of 86.30%, and the average score across all 65 submissions was 77.15%, with a standard deviation of 5.50 %.
Table 4.
System accuracy scores on challenge test data. Our official scores were slightly lower than our post hoc analysis scores due to one file that our system failed to process. The failure was caused by a software bug in pre-processing present in all three runs. For that file, we assigned the maximum possible misclassification penalty to compute our official score as directed by the challenge organizers.
| SEVERITY | SUBMISSION | ||
|---|---|---|---|
| Run 1 (Under) | Run 2 (Middle) | Run 3 (Over) | |
| Absent | 64.52% | 64.52% | 69.89% |
| Mild | 92.44% | 93.60% | 91.86% |
| Moderate | 71.74% | 72.83% | 70.65% |
| Severe | 79.49% | 82.05% | 74.36% |
| Score | 77.05% | 78.25% | 76.69% |
| Official Score | 76.67% | 77.86% | 76.34% |
3.1 Analysis
3.1.1 Model and Training Effects
Annotating the documents that were initially unannotated in the training data set and including that information in the training had the biggest impact on system accuracy. As can be seen in Figure 1, this effect was observable for all three configurations of our neural network model. Figure 1 also shows that Run 1, the system configured to underfit the data, performed best on held out training data. However, Run 2 (Middle), which was configured to fit the data and which performed the least well of the three on the training data, gave the best results on the evaluation data provided by the challenge.
Figure 1.
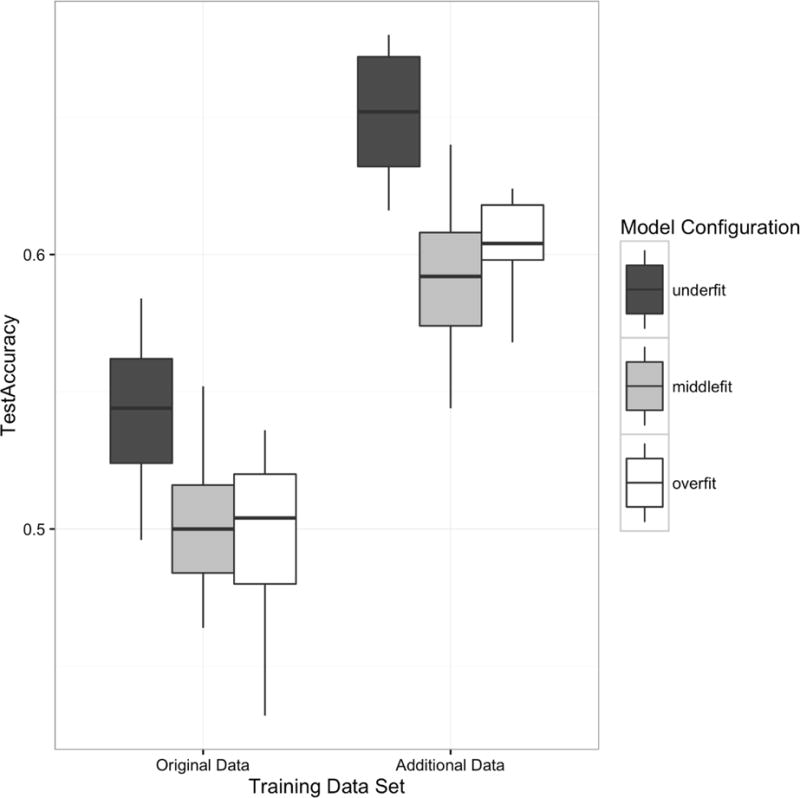
Aggregate accuracies across model types and training datasets
3.1.2 System Confusions
Table 5 presents a confusion matrix for the best performing system configuration, Middle. This configuration did not generate any absent values. 90% of the system errors involved neighboring severity categories (i.e., absent/mild, mild/moderate or moderate/severe). Only 9% of the errors involved categories that were two levels apart (i.e., absent/moderate or mild/severe). Although the system made no confusions between severe and absent, the system was unable to process one of the test documents and generated no score. This instance was assessed for the purpose of official scoring as a severe/absent confusion.
Table 5.
Confusion matrix for best system configuration, Run 2 (Middle)
| HUMAN EXPERT | ||||
|---|---|---|---|---|
| SYSTEM | Absent | Mild | Moderate | Severe |
| Absent | 0 | 0 | 0 | 0 |
| Mild | 29 | 75 | 19 | 6 |
| Moderate | 2 | 11 | 21 | 16 |
| Severe | 6 | 30 | ||
| (blank) | 1 | |||
3.1.3 Features and Mutual Information Scores
Examination of the mutual information scores of the 120 features that served as input to Run2 (Middle) yielded several observations related to system performance. Terms that ranked high in overall MI frequently ranked high in category-specific MI for severe and low for other severity categories. Most terms had positive MI scores for severe and moderate and zero or negative MI scores for mild and absent. The term with the highest overall MI score was detox. This term has a strong semantic relation to substance abuse, and its informal register meant that it never appeared in template questions or headings; it only appeared in statements by or about the patient.
We examined a few terms whose severity-specific scores for moderate and severe differed by less than 0.1 and whose relevance to symptom severity was not apparent. It was usually the case that these terms occurred in text that did not pertain to the patient. One such term was comment, which occurred in the singular primarily within the instructions of templated question: If Yes, comment on Timing, Lethality, Impulsivity, Comorbid Intoxication or Psychosis and If Yes, please comment on Branch, Dates of Service… Although the MI score for comment is high, this may be a spurious artifact of the design of the template questions rather than a true indicator of symptom severity.
3.1.4 DSM Codes
An independent manual analysis of the data revealed that although DSM [5] codes were not in themselves a predictor of Positive Valence severity scores, certain codes, shown in Table 6, never appeared in documents labeled absent. Disorders associated with most of these codes (possibly excluding schizophrenia) seem likely to be associated with of Positive Valence signs and symptoms. Several of these codes also had high MI rankings. In three cases, our tokenization grouped codes with likely different Positive Valence relevance into the same category:
296.4–296.9 (Bipolar Disorders I and II) and 296 – 296.3 (Depressive Disorder)
300.3 (Obsessive Compulsive Disorder) and 300.000–300.23 (Anxiety Disorder, Panic Disorder, Social Phobia)
307.2 (Tic Disorders), 307.5 (Eating Disorders), 307.8 (Pain Disorder), and 307.42–307.47 (Sleep Disorders)
Table 6.
Mutual information rankings for DSM codes that did not co-occur with severity absent
| DSM Code | Description | MI Rank | Count |
|---|---|---|---|
| 304.× | Drug Dependence | 6 | 56 |
| 303.× | Alcohol Dependence/Alcohol Use Disorder | 11 | 46 |
| 296.4–296.9 | Bipolar Disorders | 28 | 293 |
| 305.× | Alcohol Abuse/Substance Abuse | 55 | 46 |
| 307.× | Eating Disorders and Tic/Movement Disorders | 548 | 47 |
| 295.× | Schizophrenia | 1258 | 7 |
| 300.3 | Obsessive Compulsive Disorder | 2272 | 278 |
Bipolar Disorder and Obsessive Compulsive Disorder never appeared in documents labeled absent, but Depression Disorder and Anxiety Disorder appeared in documents of all categories. Tic Disorders and Eating Disorder would likely be associated with Positive Valence symptoms, but no such expectation applies to Pain Disorder and Sleep Disorders. Including the decimal values as part of these tokens might have yielded more informative MI scores.
3.2 Potential Sources of System Errors
The black-box nature of neural networks minimizes the ability to determine the causes of errors that the system made. However, we can speculate on likely sources of error based on our knowledge of the information the system had at its disposal, our understanding of the demands of the task, and our experience with other NLP systems.
3.2.1 Feature Representation
Positive Valence severity implied by certain disorders may have been missed by our system because behavior-related terms indicative of the disorder were not among the 120 features supplied to the system. For example, neither eating nor the DSM code for Eating Disorder was among the features used by the final system configuration. Only the top 120 features were used as input; eating had an MI rank of 190, and the DSM code for Eating Disorder ranked 548. Including bigrams such as eating disorder as features might have improved accuracy; however, this term appeared in a templated question of almost every document, so it might have been necessary to determine its context to benefit from this term as a feature. The ranking and informative value of its associated DSM code (307) may have been increased by retaining the decimal values to distinguish it (307.5) from the codes for Pain Disorder (307.8) and Sleep Disorders (307.4).
3.2.2 Polarity
Negation was often expressed in narrative and as No answers to templated questions. None of our systems addressed negation directly. We considered applying clinical information extraction technology to the records to extract clinical named entities and their assertion status (e.g., negated, uncertain). We faced two challenges with this approach. First, many concepts relevant to the evaluation of mental health status in general, and that might by relevant to determination of positive valence in particular, are not concepts that are identified by clinical information systems. Examples of such concepts include the history of inpatient psychiatric hospitalization, concepts that imply well-being (e.g., future oriented, optimistic, high functioning), and concepts that suggest social behavior problems (e.g., arrest, jail, probation). A second challenge was that in addition to negation in narrative text (e.g., no history of prior suicide attempts) which negation algorithms are generally designed to interpret, the documents contained a considerable amount of negation in semi-structured or templated text (Hx of Suicidal Behavior: No). Negation detection algorithms designed or trained to detect negation in narrative contexts will not perform well in these semi-structured contexts because the word ordering and punctuation play a crucial role in determining which words are interpreted as negated. Because of these issues, we decided to postpone incorporation of extracted concepts and assertion values to future work.
3.2.3 Gravity
Severity ratings were likely influenced by the impact of positive valence symptoms on a patient’s life. The notes expressed impact in narrative sections (e.g., in the Formulation and Risk Assessment sections), and through various assessment scores, such as DSM Axis V: Global Assessment of Functioning (GAF). The GAF is a numeric scale used by clinicians to rate the social, occupational, and psychological functioning of an individual. Scale values range from 100 (extremely high functioning) to 1 (severely impaired). While the GAF does not provide information with respect to a specific symptom category such as Positive Valence, however, the GAF score in conjunction with Positive Severity symptom information elsewhere in the psychiatric note might contribute to the severity classification. Our system did not make explicit use of this type of information.
3.2.4 Relevance
Several system errors may have resulted from the system’s failure to distinguish information about the patient from information about others. Information about significant others occurred not only within Family History sections but also in Chief Complaint and History of Present Illness sections in the context of the patient’s concerns. In one report, a patient had no problems except anxiety, but discussed a family member’s problems at length. The severity category assigned by a clinical expert was absent. Although our system explicitly disregards content of the Family History section, most of this patient’s expressed concerns about the family member were presented in the History of Present Illness and Precipitating Events section.
4 Discussion
We created a neural network-based system to determine the severity of Positive Valance symptoms for a patient, based on information included in their initial psychiatric evaluation using no manually crafted features. Despite the black-box nature of our approach, our analysis of system input features and system errors suggests several directions for future work.
4.1 Learning Curves
Our experiments showed that additional annotated data for system training improved accuracy on held out training data. We would like to experiment with varying amounts of annotation and create learning curves from which to estimate optimal training set size for this task.
4.2 Term Analysis
Our bag-of-words-based features were primarily5 individual words. Several multiple-word concepts that were likely very relevant to the severity were not represented as features. Instead, their component words were features, and the MI score for the components was likely not as high as the MI score for the phrase. (Examples include obsessive and compulsive versus obsessive compulsive, or eating and disorder versus eating disorder.) Using bigrams and possibly higher dimensional n-grams in addition to unigrams might facilitate identification of phrases with more informative MI scores.
Determining mutual information for syntactic and semantic word classes may also have resulted in additional informative features. We manually generated categories for a relatively small number of terms, and the derived features had no impact on our accuracy. However, systematic normalization on a larger scale could have a positive impact on system accuracy. This might be accomplished with term clusters derived on data more closely matching the domain than the Gigaword corpus.
4.3 Term Clustering
We experimented with a variety of methods to generate features from term clustering derived using the Brown clustering algorithm, but we found no noticeable improvements on these data, possibly due to a misalignment between the psychiatric domain and the contents of the Gigaword dataset, which is mostly news sources.
Similar to the Brown term clusters, we found no noticeable improvement with term clusters derived by K-means clustering over neural word embeddings trained using skip-gram word2vec models on a corpus of 8 billion words of PubMed articles. Due to time constraints, we were only able to train a single embedding with 50 dimensions; additional experimentation is required to determine whether embeddings with a greater dimensionality may perform better.
4.4 Assertion Analysis
Yetisgen-Yildiz et al. found that adding assertion values to their n-gram features did not improve the accuracy of their system’s ability to identify patients who are positive for Acute Lung Injury [12]. However, they concluded from their analysis that the reason was that the assertion categories they used (present, absent, conditional, hypothetical, possible) were not sufficient to capture the crucial information in many cases. As an example, the authors noted that their assertion classifier assigned the class present to the bigram bibasilar opacities in the sentence There are bibasilar opacities that are unchanged. They observed that although present was the correct assignment for bibasilar opacities, the more important piece of information was the change of state in bibasilar opacities. We would like to examine the impact of adding contextual information that occurs widely such as negation to our classification system. However, we think it may be important to go beyond the general assertion categories that have been most frequently considered up to this point to consider categories that may be important for specific domains. In addition to changed/unchanged, relevant assertion categories for the medical domain might include normal/abnormal, and WNL (within normal limits).
4.5 Text Zone Analysis
Clinical information NLP systems typically include a component that interprets the structure of a documents, identifying headings, determining boundaries and classifying sections. The type of section a concept occurs in helps determine its relevance. Available sectionizers have been developed using clinical note types such as Discharge Summaries, History and Physicals, and Progress Notes. The mental health notes provided by the challenge contained some of the same section types, but also contained many sections that do not appear in other clinical note types. Developing a sectionizer for mental health notes would provide the following benefits:
Support for more accurate assertion status detection. Clinical information extraction systems typically assign a default assertion status of positive (i.e., present), in the absence of specific assertion indicators like no or possible. However, concepts mentioned in headings are typically used to describe the topic of the subsequent text, so there is no positive assertion being made. Furthermore, distinguishing sections that describe the patient’s history from sections that describe the patient’s family history facilitates the interpretation of section content.
Facilitation of customization of NLP methods such as tokenization and sentence boundary detection to text format (free text, semi structured text, tables, etc.)
4.6 Category-Specific Models
Both manual analysis and mutual information scores indicated that the absent category was not strongly associated with any particular terms. What seemed to define the absent category was the lack of terms associated with Positive Valence symptoms or negation associated with those terms. Modeling the absent class separately from other classes, as well as creating a separate classifier for each severity category may yield a system with better performance.
4.7 Illumination of Evidence
We plan to work with a mental health subject matter expert to determine the explicit and implicit support for document-level severity ratings. Annotation of textual evidence may provide useful input features for the system, and will certainly facilitate document analysis.
4.8 Ablation Studies
We would like to run a series of ablation experiment to determine the impact of different types of features on the accuracy of the system.
5 Conclusion
We developed a neural network-based system to determine the severity of Positive Valance symptoms for a patient, based on information included in their initial psychiatric evaluation. We achieved an accuracy level of 77.05% on this classification task using a feedforward, fully-connected neural network with no manually crafted features. Regularization and feature selection via mutual information were very important to address overfitting. A modest increase in the amount of annotated data had a positive impact on classification accuracy. Our results can serve as a useful baseline for future comparisons.
Highlights.
-
■
We trained a machine learning-based system to determine psychiatric symptom severity.
-
■
Regularization and feature selection via mutual information reduced overfitting.
-
■
Increasing the amount of annotated data increased accuracy by several percent.
Acknowledgments
Funding: This work was supported by The MITRE Corporation. The 2016 Centers of Excellence in Genomic Science (CEGS) Neuropsychiatric Genome-Scale and RDoC Individualized Domains (N-GRID) Shared Task in Clinical Natural Language Processing was made possible by NIH P50 MH106933 (PI: Isaac Kohane); and NIH 4R13LM011411 (PI: Ozlem Uzuner).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The DSM codes were presented using the Multi-Axial Diagnoses/Assessment system, which indicates the version was DSM IV, because DSM-5 eliminated the multiaxial system.
We also experimented with regularized multinomial logistic regression classifiers and gradient boosted trees.
The SoftMax function is a generalization of the logistic function, which reduces a K-dimensional vector of arbitrary real values to a K-dimensional vector of real values in the range (0, 1) that add up to 1.
A small number of two-word pairings were created by replacing punctuation between words such as hyphen and slashes with underscores.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Filannino M, Stubbs A, Uzuner Ö. Symptom severity prediction from neuropsychiatric clinical records: Overview of 2016 CEGS N-GRID Shared Tasks Track 2. Journal of Biomedical Informatics. 2017 doi: 10.1016/j.jbi.2017.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- 3.Morris SE, Cuthbert BN. Research Domain Criteria: cognitive systems, neural circuits, and dimensions of behavior. Dialogues Clin Neurosci. 2012;14:29–37. doi: 10.31887/DCNS.2012.14.1/smorris. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cuthbert BN, Insel TR. Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med. 2013;11:126. doi: 10.1186/1741-7015-11-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.A.P. Association. Diagnostic and Statistical Manual of Mental Disorders. Amer Psychiatric Pub Incorporated. 2000 doi: 10.1176/appi.books.9780890420249.dsm-iv-tr. [DOI] [Google Scholar]
- 6.World Health Organization. International Statistical Classification of Diseases and Related Health Problems. World Health Organization; 2004. [Google Scholar]
- 7.Velupillai S, Mowery D, South BR, Kvist M, Dalianis H. Recent Advances in Clinical Natural Language Processing in Support of Semantic Analysis. Yearb Med Inform. 2015;10:183–193. doi: 10.15265/IY-2015-009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herasevich V, Yilmaz M, Khan H, Hubmayr RD, Gajic O. Validation of an electronic surveillance system for acute lung injury. Intensive Care Med. 2009;35:1018–1023. doi: 10.1007/s00134-009-1460-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azzam HC, Khalsa SS, Urbani R, Shah CV, Christie JD, Lanken PN, et al. Validation study of an automated electronic acute lung injury screening tool. J Am Med Inform Assoc. 2009;16:503–508. doi: 10.1197/jamia.M3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shivade C, Raghavan P, Fosler-Lussier E, Embi PJ, Elhadad N, Johnson SB, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. Journal of the American Medical Informatics Association. 2014;21:221–230. doi: 10.1136/amiajnl-2013-001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solti I, Cooke CR, Xia F, Wurfel MM. Automated Classification of Radiology Reports for Acute Lung Injury: Comparison of Keyword and Machine Learning Based Natural Language Processing Approaches. Proceedings (IEEE Int Conf Bioinformatics Biomed) 2009;2009:314–319. doi: 10.1109/BIBMW.2009.5332081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yetisgen-Yildiz M, Bejan CA, Wurfel MM. Identification of Patients with Acute Lung Injury from Free-Text Chest X-Ray Reports. Acl. 2013;2013 [Google Scholar]
- 13.Liao KP, Cai T, Gainer V, Goryachev S, Zeng-treitler Q, Raychaudhuri S, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res (Hoboken) 2010;62:1120–1127. doi: 10.1002/acr.20184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright A, McCoy AB, Henkin S, Kale A, Sittig DF. Use of a support vector machine for categorizing free-text notes: assessment of accuracy across two institutions. Journal of the American Medical Informatics Association. 2013;20:887–890. doi: 10.1136/amiajnl-2012-001576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pestian JP, Matykiewicz P, Grupp-Phelan J, Lavanier SA, Combs J, Kowatch R. Using natural language processing to classify suicide notes. AMIA Annual Symposium Proceedings / AMIA Symposium AMIA Symposium. 2008:1091. doi: 10.3115/1572306.1572327. [DOI] [PubMed] [Google Scholar]
- 16.Pestian J, Nasrallah H, Matykiewicz P, Bennett A, Leenaars A. Suicide Note Classification Using Natural Language Processing: A Content Analysis. Biomedical Informatics Insights. 2010;2010:19–28. doi: 10.4137/BII.S4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cook BL, Progovac AM, Chen P, Mullin B, Hou S, Baca-Garcia E. Novel Use of Natural Language Processing (NLP) to Predict Suicidal Ideation and Psychiatric Symptoms in a Text-Based Mental Health Intervention in Madrid. Comput Math Methods Med. 2016;2016:8708434–8. doi: 10.1155/2016/8708434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perlis RH, Iosifescu DV, Castro VM, Murphy SN, Gainer VS, Minnier J, et al. Using electronic medical records to enable large-scale studies in psychiatry: treatment resistant depression as a model. Psychol Med. 2012;42:41–50. doi: 10.1017/S0033291711000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Centers for Disease Control and Prevention. International classification of diseases, ninth revision, clinical modification (ICD-9-CM) 2013 URL: http://www.cdc.gov/nchs/about/….
- 20.Uzuner Ö, Goldstein I, Luo Y, Kohane I. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc. 2008;15:14–24. doi: 10.1197/jamia.M2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark C, Good K, Jezierny L, Macpherson M, Wilson B, Chajewska U. Identifying smokers with a medical extraction system. J Am Med Inform Assoc. 2008;15:36–39. doi: 10.1197/jamia.M2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uzuner Ö. Recognizing obesity and comorbidities in sparse data. J Am Med Inform Assoc. 2009;16:561–570. doi: 10.1197/jamia.M3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pestian JP, Matykiewicz P, Linn-Gust M, South B, Uzuner Ö, Wiebe J, et al. Sentiment Analysis of Suicide Notes: A Shared Task. Biomedical Informatics Insights. 2012;5:3–16. doi: 10.4137/BII.S9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.World Health Organization. ICD-10: International statistical classification of diseases and related health problems: tenth revision. 1989 [Google Scholar]
- 25.Brown PF, deSouza PV, Mercer RL, Pietra VJD, Lai JC. Class-based n-gram models of natural language. Comput Linguist Assoc Comput Linguist. 1992;18:467–479. [Google Scholar]
- 26.Graff D, Cieri C. English gigaword corpus. Linguistic Data Consortium. 2003 [Google Scholar]
- 27.Srivastava N, Hinton GE, Krizhevsky A. Dropout: a simple way to prevent neural networks from overfitting. Journal of Machine Learning Research. 2014;15:1929–1958. [Google Scholar]
- 28.Duchi J, Hazan E, Singer Y. Adaptive Subgradient Methods for Online Learning and Stochastic Optimization. Journal of Machine Learning Research. 2011;12:2121–2159. [Google Scholar]


