Abstract
A large amount of evidence has demonstrated the power of exercise to support cognitive function, the effects of which can last for considerable time. An emerging line of scientific evidence indicates that the effects of exercise are longer lasting than previously thought up to the point to affect future generations. The action of exercise on epigenetic regulation of gene expression seem central to building an “epigenetic memory” to influence long-term brain function and behavior. In this review article, we discuss new developments in the epigenetic field connecting exercise with changes in cognitive function, including DNA methylation, histone modifications and microRNAs (miRNAs). The understanding of how exercise promotes long-term cognitive effects is crucial for directing the power of exercise to reduce the burden of neurological and psychiatric disorders.
Keywords: Exercise, Memory, Epigenetics, Brain, DNA methylation, Histones, miRNAs
1. Introduction
Exercise is perceived as a indispensable aspect of our daily routine to maintain overall health of body and brain. In particular, abundant evidence supports the action of exercise in sharpening cognitive abilities throughout lifespan such that the lack of exercise is considered a risk for the incidence of several neurological disorders (Gomez-Pinilla and Hillman, 2013, Cotman and Berchtold, 2002, Cotman et al., 2007, Intlekofer and Cotman, 2013, Lista and Sorrentino, 2010). New studies indicate that the effects of exercise on brain function go beyond those previously thought, and indeed, the brain has the potential to save the effects of exercise for considerable time. There has been tremendous progress in the last decade about the molecular mechanisms through which exercise and other environmental challenges modify the program of genes with functional consequences. The action of epigenetics has come to play as it refers to alterations in chromatin that modify the transcription of genes which are saved as “an epigenetic memory” that influences long-term brain plasticity and function. An increasing body of research indicates that epigenetic modifications elicited by exercise seem to confer individuals throughout lifespan and even their progeny with the capacity to resist diseases (Denham et al., 2015, Laker et al., 2014, McPherson et al., 2015). Here we discuss recent research revealing main epigenetic mechanisms by which exercise affects brain function, particularly focusing on cognitive abilities.
1.1. Effects of exercise on cognitive abilities
The positive actions of exercise on learning and memory in humans and animals have received abundant support (Lista and Sorrentino, 2010, Cotman and Berchtold, 2002, Erickson et al., 2011, van Praag et al., 1999a, Gomez-Pinilla and Hillman, 2013). In older adults, exercise has been shown to improve cognitive performance (Colcombe et al., 2004, Cassilhas et al., 2007) and to counteract the mental decline associated with ageing (Yaffe et al., 2009, Heyn et al., 2008), and these effects have been associated with modifications in hippocampal size (Erickson et al., 2011). In schoolchildren, exercise has been found to be associated with cognitive performance: children who engaged in greater amounts of aerobic exercise generally performed better on verbal, perceptual, and mathematical tests (Sibley and Etnier, 2003). Recently, a meta-analysis study reported that a single bout of moderate aerobic exercise improves inhibitory control, cognitive flexibility, and working memory in preadolescent children and older adults (Ludyga et al., 2016), indicating that beyond the well know effects of long-term exercise on the brain, acute exercise also can be used as a tool for situations demanding a high executive control. Interestingly, we and others have found that a single bout of both aerobic (Siette et al., 2014) and resistance (Fernandes et al., 2016) exercise is able to enhance memory consolidation in rats.
Exercise is also perceived as one of the most effective therapies to reduce depression (Rethorst and Trivedi, 2013,Kvam et al., 2016) and to improve several aspects of other brain-related diseases such as Parkinson and Alzheimer's, Epilepsy, anxiety and traumatic brain injury (Grealy et al., 1999, Chin et al., 2015, Intlekofer and Cotman, 2013, Matura et al., 2016, de Almeida et al., 2017, Peixinho-Pena et al., 2012, Shu et al., 2014, Reynolds et al., 2016, Jayakody et al., 2014). Concerning to depression, a randomized controlled trial showed a dose-response relationship between exercise and depression score (reduction of 47% of high-dose aerobic exercise and 30% in the low-dose exercise) evaluated by Hamilton Rating Scale for Depression (HAM-D) (Dunn et al., 2005). Furthermore, Blumenthal et al. (1999) reported that aerobic exercise by itself or in combination with sertraline (a selective serotonin reuptake inhibitor) lowered depressive score and relapse rate at 6-month of follow-up (Blumenthal et al., 1999).
In rodents, the memory improvements induced by physical exercise are accompanied by increases in cell proliferation, neurogenesis, dendritic complexity and spine density (van Praag, 2008, Lista and Sorrentino, 2010, van Praag et al., 1999b). Moreover, these studies showed that the positive effects of exercise on learning and memory are rooted to molecular regulatory mechanisms involved in neurotransmission, metabolism and synaptic plasticity (Vaynman et al., 2004, Cotman et al., 2007, van Praag, 2008, Lista and Sorrentino, 2010). As discussed below, exercise activates the transcriptional machinery inside the nucleus to modulate the expression of genes associated with regulation of synaptic plasticity, learning and memory using epigenetic mechanisms.
2. Epigenetics mechanisms and brain
It is well accepted that adaptations to an ever-changing environment involve long-lasting physiological modifications that cannot be explained by mutations. This belief has led to the search of alternative mechanisms to account for how environmental influences can be saved in the genome. Conrad Hal Waddington coined the term "epigenetics" based on a conceptual model to account for how genes might interact with their surroundings to produce the phenotype (Waddington, 1939). Nowadays, epigenetics refers to changes in gene expression through modulation of chromatin without alterations in the DNA sequence (Jaenisch and Bird, 2003, Goldberg et al., 2007). These concepts also opened up to the possibility that epigenetic modifications could be inherited and provide a source for individual variability.
The epigenetic research has been centered on the analysis of changes on top of the genome that do not involve alterations in the nucleotide sequence. The two most studied epigenetic mechanisms are covalent modifications of DNA (methylation) or of histone proteins (i.e. acetylation and methylation), and their resulting effects on altering gene expression (Figure 1). These changes in gene expression are generally associated with the intermediate action of proteins that act as transcription activators or repressors by binding to regulatory regions of the DNA (Jaenisch and Bird, 2003, Goldberg et al., 2007). Original studies showed that exercise regulates the transcription of brain-derived neurotrophic factor (Bdnf) gene by engaging changes in histone acetylation and DNA methylation (Gomez-Pinilla et al., 2011, Ieraci et al., 2015, Sleiman et al., 2016). Thereafter, an increasing number of studies indicate the involvement of epigenetic mechanisms on the action of exercise on the brain (Table 1).It appears that exercise-induced changes in acetylation and methylation are instrumental to regulate synaptic plasticity and learning and memory (Abel and Rissman, 2013, Gomez-Pinilla et al., 2011, Intlekofer et al., 2013, Ieraci et al., 2015). Indeed, a growing body of evidence indicates that physical exercise activates signaling cascades that trigger a wave of phosphorylation and other post-translational modifications that reach the nucleus, and engage epigenetic mechanisms to alter chromatin conformation and gene expression. As discussed below, a group of small non-coding RNAs, particularly microRNAs (miRNAs), have emerged as potent epigenetic regulators of brain plasticity and memory function (Saab and Mansuy, 2014, Konopka et al., 2011, Wang et al., 2012), and this information has paved the road to a better understanding of the epigenetic mechanism underlying the action of exercise on the brain.
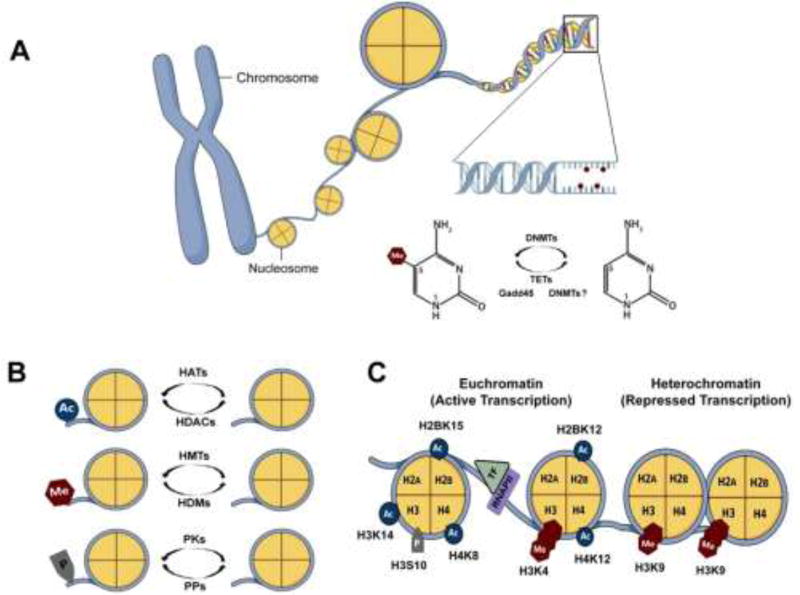
Figure 1.
Dynamic regulation of DNA methylation and histone modifications. (A) Nucleosome, the basic repeat element of chromatin, consists of 147bp of DNA wrapped around an octamer of four core histone proteins (H2A, H2B, H3 and H4) compacted by a linker histone. DNA methylation is related to repressed transcription and involves the covalent addition of a methyl group to the 5-carbon of cytosine bases [5-methylcytosine (5mC)] located in CpG dinucleotides by DNMTs. Conversely, active DNA demethylation can occur through mechanisms involving enzymes from Gadd45, TET and even DNMT families. (B) Chromatin-modifying enzymes regulate the addition and removal of histone modifications to establish an open or a silent transcriptional state. While HATs catalyze the addition of acetyl group, HDACs catalyze its removal. The histone tail is methylated by HMTs and demethylated by HDMs. PKs and PPs are responsible for the addition and removal of phosphate groups. (C) Chromatin can be found at an open (euchromatin) or a closed (heterochromatin) state according to the pattern of histone modifications. In general, euchromatin is characterized by hyperacetylated, hyperphosphorylated and specific histone methylation patterns related to active transcription whereas heterochromatin is featured by opposite post-translational modifications. Ac, acetyl group; CpG, cytosine-phosphate-guanine dinucleotides; DNMTs, DNA methyltransferases; Gadd45, growth arrest and DNA damage 45 proteins; HATs, histone acetyltransferases; HDACs, histone deacetylases; HDMs, histone demethylases; HMTs, histone methyltransferases; Me, methyl group; P, phosphate group; PKs, protein kinases; PPs, protein phosphatases; RNAPII, RNA polymerase II; TET, ten-eleven translocation proteins; TF, transcription factor.
Table 1.
Summary of studies on brain epigenetic changes induced by physical exercise.
| Study (year) |
Species | Exercise model |
Brain region | Epigenetic changes |
|---|---|---|---|---|
|
| ||||
| Collins et al. (2009) | Rats | Running wheel | Dentate gyrus | ↑ histone H3 phospho-acetylation |
|
| ||||
| Elsner et al. (2011) | Rats | Treadmill exercise | Hippocampus | ↓ HDAC activity; ↑ HAT activity; ↑ HAT/HDAC balance |
|
| ||||
| Gomez-Pinilla et al. (2011) | Rats | Running wheel | Hippocampus | ↓ DNA demethylation at Bdnf promoter IV; ↑ pMeCP2 levels; ↑ histone H3 acetylation; ↓ HDAC5 expression |
|
| ||||
| Elsner et al. (2013) | Rats | Treadmill exercise | Hippocampus | Adult rats: ↓ DNMT1 and DNMT3b; ↓ H3K9 methylation |
| Aged rats: ↑ H3K9 methylation | ||||
|
| ||||
| Intlekofer et al. (2013) | Mice | Running wheel | Hippocampus | ↑ histone H4K8 acetylation at Bdnf promoters I and IV |
|
| ||||
| Abel et al. (2013) | Mice | Running wheel | Hippocampus and cerebellum | ↑ histone H3 acetylation in both regions; |
| Cerebellum: ↑ HDAC2 and ↓ MeCP2, HDAC8 and DNMT1; | ||||
| Hippocampus: ↓ HDAC5, HDAC7, HDAC 8, DNMT1, DNMT3a, DNMT3b | ||||
|
| ||||
| Lovatel et al. (2013) | Rats | Treadmill exercise | Hippocampus | ↑ histone H4 acetylation in aged rats |
|
| ||||
| Patki et al. (2014) | Rats | Treadmill exercise | Hippocampus | Exercise normalized stress-induced changes in histone H3 acetylation, HDAC5 and MeCP2 |
|
| ||||
| Spindler et al. (2014) | Rats | Treadmill exercise | Frontal cortex | ↑ HAT activity; ↓ HDAC activity. |
|
| ||||
| Cosín-Tomás et al. (2014) | Mice | Running wheel | Hippocampus | ↑ miR-28a-5p, miR-98a-5p, miR-148b-3p, miR-7a-5p and miR-15b-5p; ↓ miR-105, and miR-133b-3p |
|
| ||||
| Bao et al. (2014) | Mice | Running wheel | Hippocampus | ↑ 20 miRNAs and ↓ 12 miRNAs |
|
| ||||
| Hu et al. (2015) | Mice | Running wheel | Hippocampus | Exercise restored traumatic brain injury (TBI)-induced changes in miR-21 |
|
| ||||
| Pan-Vazquez et al. (2015) | Mice | Running wheel | Hippocampus | Exercise attenuated the increased expression of miR-124 in a stress model |
|
| ||||
| Ieraci et al. (2015) | Mice | Running wheel | Hippocampus | ↑ histone H3 acetylation at Bdnf promoters I, II, III, IV, VI and VII; ↓ HDAC5 expression in stressed mice |
|
| ||||
| Zhong et al. (2016) | Mice | Swimming exercise | Hippocampus | ↑ H3K9, H3K14, H4K5, H4K8 and H4K12 acetylation; ↑ CBP expression |
|
| ||||
| Kim et al. (2016) | Mice | Running wheel | Basolateral amygdala | Exercise prevented the reduction in G9a histone methyltransferase expression induced by chronic stress; ↑ histone H3K9 dimethylation at oxytocin and vasopressin gene promoters |
|
| ||||
| Sleiman et al. (2016) | Mice | Running wheel | Hippocampus | ↓ HDAC2 and HDAC3 expression; ↓ HDAC2 and HDAC3 occupancy at Bdnf promoters |
2.1. DNA methylation
DNA methylation is a post-replication modification in which a methyl group is covalently added to the 5-carbon of cytosine bases (Guo et al., 2011a) located in cytosine-phosphate-guanine (CpG) dinucleotides. The CpG notation is used to distinguish a cytosine followed by a guanine in one strand from a cytosine base paired to a guanine in double-stranded sequences. Although underrepresented throughout the mammalian genome, these sites are occasionally clustered in CpG islands located close to, and in, approximately 40% of gene promoters (Jaenisch and Bird, 2003, Goldberg et al., 2007). Interestingly, DNA methylation of CpG islands is usually associated with silencing of genes with dramatic importance for cell function under homeostatic and disease conditions (Goll and Bestor, 2005). DNA methylation is catalyzed by two groups of enzymes called DNA methyltransferases (DNMTs), which transfer methyl groups from S-adenosyl-L-methionine (SAM) to the cytosine residue (Bird, 1992).As discussed below, much of what is known about the heritability of methylation depends on the action of DNMTs. While de novo DNMTs (DNMT3a and DNMT3b) target unmethylated cytosines on both DNA strands (Okano et al., 1999), the maintenance DNMT1 recognizes hemimethylated DNA and transfers a methyl group to the complementary cytosine base (Okano et al., 1999, Pradhan et al., 1999).
The methylated DNA can repress transcription by interfering with the binding of transcription factors and by assembling the transcriptional machinery to regulatory sites of genes (Takizawa et al., 2001). Moreover, the methylation of CpG dinucleotides alters gene transcription by serving as a docking site for proteins containing the methyl-binding domain (MBD) (Nan et al., 1998). The methyl-CpG-binding protein 2 (MeCP2) is the best characterized member of MDB proteins in the central nervous system (CNS), and plays a role in the neurodevelopmental disorder Rett syndrome, synaptic plasticity, and memory formation (Amir et al., 1999, Chao et al., 2007, Moretti et al., 2006). Mechanistically, MeCP2 modulates gene expression by promoting a closed chromatin, via the recruitment of transcriptional repressors (i.e. HDACs and mSin3) (Drewell et al., 2002, Fuks et al., 2003), or by activating transcription through interaction with the transcriptional activator cAMP responsive element binding protein 1 (CREB1) (Chahrour et al., 2008).
Evidence shows that DNA methylation is dynamically modulated byactivity-dependent events (Kangaspeska et al., 2008,Métivier et al., 2008, Guo et al., 2011b, Guo et al., 2011a). The growing interest in this area of research has contributed to discover putative mechanisms regulating DNA demethylation. One of these mechanisms is represented by the action of ten-eleven translocation (TET) proteins on converting the 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC), which involves base excision repair by thymine DNA glycosylase (TDG) (Guo et al., 2011b, Kohli and Zhang, 2013). Alternatively, it has been suggested that growth arrest and DNA damage 45 (Gadd45) proteins can promote DNA demethylation through recruitment of nucleotide and/or base excision repair machinery (Niehrs and Schäfer, 2012). Métivier et al., (2008) have shown that DNMTs are implicated in DNA demethylation through deamination of 5mCs, which served to demonstrate the dual role of these enzymes in maintaining the cyclical changes in the methylation status of promoter CpGs.
2.1.1. Exercise, DNA methylation, and memory
Over the last decade, several studies have been conducted to understand the role of DNA methylation on neuronal function and long-term memory (LTM). Miller and Sweatt (2007) reported the participation of de novo DNMTs in the consolidation of LTM. In this study, the authors found increased hippocampal levels of DNMT3a and DNMT3b after the acquisition of contextual fear memory (CFM), with concomitant increased DNA methylation at the promoter of protein phosphatase 1 (PP1; a memory-suppressor gene) and decreased methylation at the promoter of reelin (a plasticity-associated gene) (Miller and Sweatt, 2007). Additional studies showed that learning induces changes in CpG methylation at promoters of genes with essential roles in synaptic plasticity and memory formation such as Bdnf, arc, and calcineurin (Lubin et al., 2008, Miller et al., 2010, Penner et al., 2011).For example, the effects of CFM on increasing the expression of Bdnf was shown to be accompanied by a significant demethylation of its promoter region (Lubin et al., 2008); these changes were reversed by intrahippocampal infusions of DNMT inhibitors, drugs recognized as negative regulators of memory formation.
Unlike Bdnf, CFM triggers hypermethylation and subsequent decrease in mRNA levels of the memory suppressor gene calcineurin in the prefrontal cortex (Miller et al., 2010). Importantly, this pattern of methylation can persist for at least 30 days following the acquisition of CFM and can be abolished by anterior cingulate cortex infusions of DNMTs inhibitors, which also prevent memory retrieval. Beyond memory deficits and deregulation of plasticity-related genes, pharmacological and transgenic inhibition of DNMTs in hippocampal and forebrain neurons promotes defects in dendritic branching, action potential and long-term potentiation (LTP) (Golshani et al., 2005, Levenson et al., 2006, Feng et al., 2010). Taken together, these findings suggest the existence of an intricate balance between DNA methylation and demethylation that is necessary for proper neuronal function underlying memory processing. In this case scenario, DNA methylation relieves the repressive effects of memory-suppressor genes to favor the expression of plasticity-promoting genes and memory consolidation.
In agreement with its well-described role in cognition, physical exercise can coordinate the action of genes involved in synaptic plasticity with resulting effects on memory preservation. For example, while exercise enhances the expression of genes (i.e. Bdnf, igf-1 and creb) that positively regulate memory consolidation (Ding et al., 2006a, Vaynman et al., 2004, Molteni et al., 2002), it downregulates genes (i.e.PP1 and calcineurin) with a repressive role in these events (Chen et al., 2007, Dao et al., 2015, Zagaar et al., 2012). Evidence shows that DNA methylation is an important mechanism by which exercise affects gene expression. It is known that exercise differentially modulates the methylation pattern of specific CpG islands located at Bdnf gene (Zajac et al., 2010), decreases hippocampal expression of DNMTs (DNMT1, DNMT3a and DNMT3b) (Abel and Rissman, 2013, Elsner et al., 2013, Kashimoto et al., 2016), attenuates the global methylation changes induced by stress (Rodrigues et al., 2015, Kashimoto et al., 2016), and increases Bdnf transcription through demethylation of its promoter IV (Gomez-Pinilla et al., 2011).
Similarly to Bdnf, it has been recently demonstrated that 2 weeks of physical exercise increases the hippocampal expression of Tet1 while promotes demethylation of CpG islands located at the gene promoter of VegfA (Sølvsten et al., 2016), a growth factor involved in the beneficial effects of physical exercise on the brain (Jin et al., 2002, Zacchigna et al., 2008, Fabel et al., 2003, Kiuchi et al., 2012). Interestingly, a previous study showed that aged mice exposed to environmental enrichment - a procedure known to elicit positive brain changes based on its physical activity component - had an improvement in learning and memory as well as a reduction in 5hmC abundance in the hippocampus (Irier et al., 2014). Altogether, these findings indicate that an enhancement of physical activity levels can reprogram the brain's methylation pattern to modulate the transcription of genes necessary for the maintenance of brain health and cognitive function.
2.2. Chromatin and histone modifications
Nucleosome is the basic repeat element of chromatin and consists of 147bp of DNA wrapped around an octamer of four core histone proteins (two pairs of H2A, H2B, H3 and H4) compacted by association with the linker histone H1. The histone proteins are structurally characterized by a globular core domain and their N-terminal tails which are susceptible to a wide range of post-translational modifications, including acetylation, methylation, phosphorylation, ubiquitination, sumoylation, ADP-ribosylation, deamination and proline isomerization (Bernstein et al., 2007). A substantial progress has been made in understanding the role of acetylation and methylation in determining the chromatin conformation and accessibility to transcriptional machinery (Kouzarides, 2007). Whereas acetylation of lysine residues on histone tails is thought to activate transcription, lysine methylation can have an activating or inactivating effect depending on the target residue and the state of methylation (i.e., mono-, di- or trimethylated). For example, while the methylation of histone (H) 3 lysine (K) 4 (H3K4) and H3K36 is related with a transcriptionally active chromatin (euchromatin), the methylation of H3K9, H3K27, and H4K20 is commonly associated with a silent state of chromatin (heterochromatin) and transcriptional repression (Kouzarides, 2007, Bernstein et al., 2007).
The histone modifications are bidirectionally regulated by a specific group of enzymes (Strahl and Allis, 2000). Histone acetyltransferases (HATs) catalyze the transfer of acetyl groups from Acetyl-Coenzyme A to the lysine residue of histones, whereas histone deacetylases (HDACs) act by removing the acetyl groups. The phosphorylation pattern of histone tails, which is tightly associated with histone acetylation, is regulated by nuclear kinases and protein phosphatases (Brami-Cherrier et al., 2009, Koshibu et al., 2009). In a similar fashion, histone methylation is catalyzed by histone methyltransferases (HMTs) such as G9a and SUV39H2, whereas demethylation of specific histone residues is dependent on histone demethylases (HDMs) such as lysine-specific demethylase 1A (LSD1) and jumonji domain-containing protein 2A (JMJD2) (Shin and Janknecht, 2007, Benevento et al., 2015). This array of histone modifications gives enormous potential for epigenetic responses, especially when considering that each of these changes can impact each other and may also interact with DNA methylation through recruitment of chromatin remodeling complexes.
There are two ways by which histone modifications can alter the chromatin organization and gene expression. While the first is related to the acetylation-induced neutralization of the charge interaction between the histones and DNA, the second requires the binding of larger multi-molecular complexes containing enzymatic activities (i.e., remodeling ATPases) to specific histone modifications marks. Acetylated histone residues are recognized by bromodomains within chromatin remodeling complexes, whereas methylation and phosphorylation are recognized by chromodomain and 14-3-3 domain containing proteins, respectively (Kouzarides, 2007, Bernstein et al., 2007, Campos and Reinberg, 2009). CHD1 (chromo-ATPase/helicase-DNA binding domain 1), a component of the SAGA chromatin-remodeling complex, recognizes di- and trimethyl H3K4 (H3K4me2 and H3K4me3) via one of its two chromodomains to establish an epigenetic environment favoring gene transcription (Pray-Grant et al., 2005, Sims et al., 2005). Conversely, chromatin condensation can be mediated through the binding of HP1 (Heterochromatin protein 1) and Polycomb proteins to methylated H3K9 and H3K27, respectively (Margueron et al., 2005). This pattern of recognition is complex and depends on the methylation state of lysine residues. For instance, while HP1 shows similar affinities for H3K9me2 and H3K9me3, Polycomb proteins bind preferentially to H3K27m3 (Fischle et al., 2003). These modifications altogether raise the possibility that histone proteins act as platforms for convergence and integration of upstream signaling pathways to direct the chromatin configuration to an open or a closed state.
2.2.1. Exercise, histone modifications and memory
It has been shown that the acetylation of histone proteins is a requisite for LTM (Day and Sweatt, 2011, Levenson et al., 2004, Guan et al., 2015, Vecsey et al., 2007, Barrett and Wood, 2008). For example, intrahippocampal injection of global HDAC inhibitors (HDACis) such as sodium butyrate (NaB) and trichostatin A (TSA) enhances LTP at Schaffer collaterals in CA1 area of the hippocampus, and results in improved consolidation of CFM (Vecsey et al., 2007, Levenson et al., 2004). The pro-cognitive function of HDACis is partially attributed to their ability to increase histone acetylation and consequently the establishment of an open chromatin state. Interestingly, a previous study has shown that, like HDACis, physical exercise has the ability to transform a learning event that does not normally lead to a stable memory trace into a long-lasting form of memory (Interfolker et al., 2013). Additionally, it was found that physical exercise increases histone acetylation and reduces HDAC expression (HDAC2, HDAC3, HDAC5, HDAC7 and HDAC8) and neural activity in the hippocampus (Gomez-Pinilla et al., 2011, Sleiman et al., 2016, Abel and Rissman, 2013, Elsner et al., 2011, Intlekofer et al., 2013).
Physical exercise (Zhong et al., 2016) and the acquisition of CFM (Peleg et al., 2010) increases the acetylation of histones H3 and H4 at multiple sites including H3K9, H3K14, H4K5, H4K8, and H4K12. With regard to the role of histone H4 on cognitive function, it was demonstrated that deregulation of H4K12 acetylation is associated with memory impairments induced by aging (Peleg et al., 2010) and by neonatal exposure to isoflurane (Zhong et al., 2016) in rodents. Notably, restoration of H4K12 acetylation levels promoted by a HDACi (Peleg et al., 2010) or by physical exercise (Zhong et al., 2016) attenuated the cognitive deficits observed in these animals. Moreover, 2 weeks of treadmill exercise improved memory performance in the inhibitory avoidance task and increased hippocampal H4K12ac levels in young adult and aged rats (de Meireles et al., 2016). Using the same exercise paradigm, this research group also demonstrated the ability of physical exercise to increase the histone H4 acetylation in the prefrontal cortex of aged rats (Cechinel et al., 2016), and to restore the aversive memory-induced reduction in H3K14ac levels in the hippocampus of adult rats (de Meireles et al., 2014).
Global HAT activity has been found to be increased in the cortex (Spindler et al., 2014) and hippocampus (Elsner et al., 2011) of rodents subjected to treadmill exercise. Moreover, in a recent study, Zhong et al (2016) observed that exercise-induced memory improvements was associated with enhanced expression of cAMP response element-binding protein (CREB)-binding protein (CBP) in the hippocampus. Acting more than just a molecular scaffold for recruiting components of the transcription machinery, CBP serves this capacity by inducing chromatin remodeling via its HAT activity, which is likely necessary for activity-dependent gene expression in LTP and long-term memory formation (Day and Sweatt, 2011, Barrett and Wood, 2008).Mechanistically, the recruitment of CBP triggers histone acetylation and the formation of a transcriptional complex at the promoters of many CREB-target genes to activate transcription. Mutations in the CBP gene are responsible for the mental retardation syndrome Rubinstein–Taybi (Petrij et al., 1995) and CBP mutant mice exhibit profound deficits in synaptic plasticity and LTM (Alarcón et al., 2004, Korzus et al., 2004, Vecsey et al., 2007). Altogether, the aforementioned findings raise the idea that physical exercise promotes synaptic plasticity and memory improvements by altering the balance of HAT/HDAC enzymatic activity to favor a permissive state of chromatin, leading to the transcriptional activation of a myriad of genes with preponderant roles in cognition.
The phosphorylation and methylation of histones are also tightly associated with regulation of learning and memory. It has been demonstrated that CFM increases histone H3 phosphorylation at residue serine (S) 10 (H3S10) in the hippocampus (Chwang et al., 2006). Importantly, inhibition of histone dephosphorylation, using a transgenic mouse model in which PP1 was selectively inhibited, increased H3S10 levels and enhanced LTP and LTM in the hippocampus and amygdala (Koshibu et al., 2009, Koshibu et al., 2011). It seems that PP1 promotes chromatin condensation by dephosphorylating histone proteins and by negatively regulating the histone acetylation and methylation through association with HDACs and HDMs (Koshibu et al., 2009). Histone phosphorylation can be one of the ways by which physical exercise can enhance neurocognitive functions, i.e., increasing histone H3 phosphoacetylation (Collins et al., 2009) and reducing PP1 expression (Chen et al., 2007) in the hippocampus.
As discussed earlier, histones can be mono-, di-, or trimethylated, and depending on the residue of modification, they can repress or activate transcription. While dimethylation or trimethylation of H3K4 are associated with a transcriptionally permissive chromatin state, dimethylation at H3K9 is associated with transcriptional repression (Kouzarides, 2007). The global levels of H3K4 trimethylation and H3K9 dimethylation are transiently increased in hippocampal CA1 area 1 h after the acquisition of CFM (Gupta et al., 2010, Gupta-Agarwal et al., 2012). Indeed, mutant mice deficient in the H3K4-specific histone methyltransferase, Mll (Mixed-lineage leukemia), have shown an impairment in the consolidation of CFM (Gupta et al., 2010). Surprisingly, hippocampal inhibition of G9a/G9a-like protein (GLP) lysine complex, which control H3K9 dimethylation, has been shown to disrupt LTP at the Schaffer collateral-CA1 synapses and LTM (Gupta-Agarwal et al., 2012). These data, in conjunction with gene expression and chromatin immunoprecipitation analysis suggest that the consolidation of LTM are modulated, at least partially, by dynamic regulation of histone methylation in regulatory regions of genes with opposite roles in memory formation. While H3K4me3 induces the expression of memory-related genes, H3K9me2 suppresses the expression of memory-repressive genes (Gupta et al., 2010, Gupta-Agarwal et al., 2012). Interestingly, recent studies have demonstrated that physical exercise counteracts the aging-induced decreases in the global methylation of H3K9 in the hippocampus (Elsner et al., 2013) and reverts the stress-induced downregulation of G9a and H3K9me2 in the amygdala (Kim et al., 2016). These results suggest the capability of exercise to work at various stages of epigenetic regulation to alter brain plasticity and cognition.
2.3. The epigenetic regulatory role of miRNAs
MicroRNAs (miRNAs) are small (17 to 25 nucleotides) single stranded RNA that belong to the class of non-coding RNAs. miRNAs act as powerful silencers of gene expression by inducing translational repression and/or degradation of target transcript through partial base-pairing to its 3' untranslated region (3'-UTR) (Ambros, 2004, Bartel, 2004). Beyond the 3′ UTRs, evidence indicates that miRNAs can also bind to other sites such as 5′ UTRs and coding regions (Tay et al., 2008, Lytle et al., 2007, Eiring et al., 2010). The miRNA genes can be located in introns or exons of protein coding genes, or intergenic regions (Rodriguez et al., 2004). The number of known miRNAs has increased markedly in recent years, and it is estimated that there are more than 2500 different miRNAs in human cells. Interestingly, each miRNA has the ability to interact with a large number of mRNA (approximately 200–500 mRNA for each miRNA), suggesting that the majority of the protein-coding genes may be regulated by miRNAs (Friedman et al., 2009). Therefore, it is not surprising that miRNAs are widely expressed in the brain, and that they can participate in epigenetic mechanisms.
This group of non-coding RNAs arises from long primary microRNA (pri-miRNAs) precursors that can be transcribed by RNA polymerase II or III. Inside the nucleus, the pri-miRNA is processed by a complex consisting of the ribonuclease (RNase) III Drosha and its cofactor DGCR8 (DiGeorge syndrome critical region 8) to give rise to the precursor miRNA (pre-miRNA). Pre-miRNA has approximately 70 nucleotides (Lee et al., 2003, Han et al., 2004), and is subsequently transported to the cytoplasm by exportin-5, in a guanosine triphosphate (GTP)-dependent manner. Pre-miRNA is further cleaved by the RNase enzyme Dicer into a double-stranded molecule representative of the mature miRNA, named guide strand or 5p, and its complementary sequence miRNA*, referred as passenger strand or 3p (Chendrimada et al., 2005, Bartel, 2004). Thereafter, the miRNA:miRNA* duplex are separated by helicases, and the mature strand is incorporated into the RNA-induced silencing complex (RISC) whereas the miRNA* strand is usually degraded. Once bound to the RISC, the mature miRNA guide this complex to the target mRNA to inhibit its translation or degrade it (Ambros, 2004, Bartel, 2004) (Figure 2).
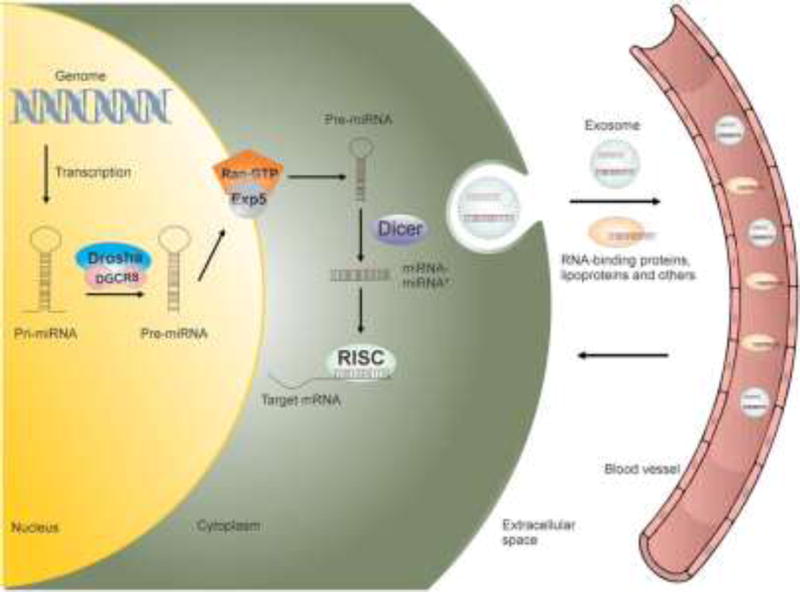
Figure 2.
miRNA biogenesis. miRNAs are initially transcribed into primary miRNA (pri-miRNA) that undergo processing by a microprocessor complex composed of Drosha and DiGeorge syndrome critical region 8 (DGCR8). The resulting precursor miRNAs (pre-miRNAs) are transported from the nucleus to the cytoplasm by exportin 5 (Exp5), in a guanosine triphosphate (GTP)-dependent manner, and further processed into its mature form by Dicer. The mature miRNA is then loaded into the RNA-induced silencing complex (RISC) to degrade or inhibit the translation of target mRNA. The miRNAs can be released in the blood inside exosomes or in association with RNA-binding proteins and lipoproteins to spread signals to cells located in the vicinity or in other parts of the organism.
2.3.1. Exercise, miRNAs and memory
Over the past decade, miRNAs have emerged as potential regulators of numerous biological processes within the brain, ranging from cell proliferation, differentiation, apoptosis, synaptic plasticity and memory formation (Saab and Mansuy, 2014). Brain miRNAs are susceptible to regulation by neuronal activity acting at the synapto-dendritic compartment resulting in control mRNA translation of several synaptic transcripts (Kye et al., 2007, McNeill and Van Vactor, 2012, Schratt, 2009). A large number of miRNAs have been identified in brain regions responsible for specific forms of memory such as hippocampus, amygdala and cortex (Schratt et al., 2006, Griggs et al., 2013, Gao et al., 2010, Konopka et al., 2010, Kye et al., 2011, Lin et al., 2011). Some miRNAs regulate memory by targeting CREB-BDNF signaling, a well know pathway activated by exercise. MicroRNA-132 (miR-132) is a CREB-regulated miRNA that is induced by neuronal activity and BDNF, and exerts a critical role in regulation of neuronal structure and excitability (Cheng et al., 2007, Wayman et al., 2008). The action of miR-132 on learning and memory is very dependent on its levels, such that very low (Wang et al., 2013) or high (Hansen et al., 2010, Scott et al., 2012) levels of miR-132 may be detrimental for memory while moderate levels can improve it (Hansen et al., 2013).In turn, the miR-134 has been shown to reduce hippocampal CREB levels and impairs LTM and LTP in CA1 hippocampal area (Gao et al., 2010).
Microarray analysis revealed that 32 miRNAs are differently expressed in the hippocampus of healthy mice subjected to physical exercise, from which 20 of them were upregulated and 12 were downregulated (Bao et al., 2014). In addition, it has been shown that exercise can mitigate the detrimental effects of traumatic brain injury and aging on cognitive function by regulating the hippocampal expression of miR-21 (Hu et al., 2015) and miR-34a (Kou et al., 2017). Physical exercise has also been shown to alter the expression of seven miRNAs (upregulated: miR-28a-5p, miR-98a-5p, miR-148b-3p, miR-7a-5p and miR-15b-5p; downregulated: miR-105, and miR-133b-3p) in the hippocampus of senescence-accelerated SAMP8 mice (Cosín-Tomás et al., 2014). Finally, physical exercise has been shown to attenuate the effects of stress-related increase in miR-124 - a miRNA known for its role in neurogenesis and memory formation (Pan-Vazquez et al., 2015).
3. Exercise and circulating miRNAs (c-miRNAs): The link between periphery and the brain
During the course of evolution, the eukaryotes have developed an elegant cell-to-cell communication system to allow efficient coordination among different cell types and tissues. Eukaryotic cells can communicate directly with each other through cell-cell contact or at distance by secreting soluble factors such as hormones, growth factors, cytokines and chemokines. In 2007, Valadi and collaborators extended this concept by showing that both RNA and miRNAs can be functionally transferred from a donor to a recipient cell via membrane-derived vesicles called exosomes. Interestingly, similarly to hormones, miRNAs are released into the circulation (Mitchell et al., 2008, Chim et al., 2008) to affect cells throughout the organism (Figure 2). The c-miRNAs are transported by exosomes (Valadi et al., 2007), high-/low-density lipoproteins (Vickers et al., 2011), apoptotic bodies (Zernecke et al., 2009), and RNA-binding proteins (Arroyo et al., 2011, Turchinovich and Burwinkel, 2012); these molecules confer c-miRNAs with stability and protection against plasma RNases. Once inside the recipient cells, the miRNAs use cellular machinery to reduce the expression of target mRNAs and to alter the cellular phenotype of the host (Cortez et al., 2011).
A growing body of evidence in human and animal has shown that exercise alters blood levels of several miRNAs (Gomes et al., 2015, Xu et al., 2015, Flowers et al., 2015), and indicates that exercise can use these epigenetic modulators to regulate communication between the brain and peripheral organs. It has been described that a single bout of acute intermittent exercise rapidly elevates circulating levels of miR-132 (Radom-Aizik et al.,2012) in young healthy men. As discussed above, miR-132 is linked to multiple functions in the brain, including neuronal development (Tognini and Pizzorusso, 2012), synaptic plasticity (Wayman et al., 2008, Impey et al., 2010) and memory formation (Wang et al., 2013, Hansen et al., 2013, Scott et al., 2012). It has been shown that miR-132 alters dendritic morphogenesis by activating the Rac1-PAK pathway (Impey et al., 2010, Wayman et al., 2008) and by regulating MeCP2 expression (Klein et al., 2007, Hansen et al., 2010). More recently, it has been shown that conditional knockout of miR-132/212 gene cluster impairs memory and promotes gross alterations in hippocampal transcriptional profile in mice (Hansen et al., 2016).
A bout of exercise reduces miR-146a levels and increases miR-223 levels in the circulation in young healthy men (Nielsen et al., 2014). The miR-146a is a NF-κB-sensitive miRNA whose enhanced expression has been associated with the proinflammatory state in neurodegenerative disorders, including Alzheimer’s disease, age-related macular degeneration, and prion disease (Su et al., 2016). This miRNA is a putative biomarker for Alzheimer’s disease and targets key inflammatory mediators such as the complement factor H (CFH), the membrane spanning beta-amyloid precursor protein (bAPP)-associated TSPAN12, and interleukin receptor-associated kinase IRAK-1 (Cui et al., 2010). It has been shown that miR-223 regulates neuronal differentiation (Jovičić et al., 2013), and can protect the brain from transient global ischemia and excitotoxic injury by targeting GluR2 and NR2B subunits of the glutamate receptor (Harraz et al., 2012). In turn, the lack of miR-223 leads to hippocampal-dependent memory deficits and neuronal cell death (Harraz et al., 2012).
Long-term physical exercise can also affect basal levels of c-miRNAs. Nielsen et al. (2014) performed global miRNA screening in young healthy men before and after 12 weeks of endurance exercise and found decreased plasma levels of miR-21, in apparent discrepancy to increased c-miR-21 levels in competitive athletes after 90 days of endurance training (Baggish et al., 2011). Given the heterogeneity of subjects involved in these studies (athletes vs non-athletes) and differences in the exercise protocols employed, it is likely that miRNAs, like hormones and others molecules, are differentially modulated by the type and intensity of physical exercise. Several studies have shown that miR-21 is one of the most commonly and dramatically up-regulated miRNA after TBI (Redell et al., 2011, Lei et al., 2009, Ge et al., 2014). This miRNA has been implicated in the control of cellular survival and apoptosis by targeting genes encoding the phosphatase and tensin homolog (PTEN), programmed cell death 4 (PDCD4), reversion-inducing-cysteine-rich protein with kazal motifs (RECK), and tissue inhibitor of metalloproteinases-3 (TIMP3) (Sayed et al., 2010, Chen et al., 2008, Gabriely et al., 2008). It has been shown that miR-21 inhibits apoptosis and improves the neurological outcome in TBI animal models by post-transcriptionally repressing PTEN expression, and by activating Akt (a downstream mediator of survival pathway) signaling in the brain (Ge et al., 2014). The overall information indicates that c-miRNAs are important conveyors of the salutary action of exercise across several tissues.
4. Can the beneficial effects of physical exercise be transmitted across generations?
The transmission of epigenetic alterations in response to environment that control gene expression and phenotypes across generations is becoming a realistic possibility (Szyf, 2015, Heard and Martienssen, 2014). Of note, it is important to differentiate parental (or intergenerational) effects, which occurs due to the exposure of embryo (F1) and its germline (F2) to an environmental factor, from truly transgenerational effects which are observed in generations not directly exposed to the environmental stimuli. Thus, transgenerational epigenetic evidence requires transmission of the epigenetic traits for at least three generations for maternal exposure and two generations for paternal exposure (Szyf, 2015, Heard and Martienssen, 2014).
Epigenetic inheritance can be triggered in response to a wide range of stimuli, including stress, infection, drugs, diet and exercise (Franklin and Mansuy, 2010, Barrès and Zierath, 2016). For example, prenatal administration of the viral mimetic poly(I:C), an experimental model for neurodevelopmental disorders, has been shown to induce significant behavioral abnormalities. Behavioral changes are accompanied by global hypoacetylation of H3K9K14 and H4K8, increased DNA methylation and MeCP2 binding in the gene promoters of glutamate decarboxylase (Gad) 1 and Gad2 in the cortex of mice offspring (Tang et al., 2013). In addition, depressive-like behavior induced by chronic and unpredictable maternal separation in F1 mice can be passed on to the F2 and F3 generations. The increased DNA methylation observed in the MeCP2 promoter of F1 sperm cells can be also found in F2 sperm and brain, suggesting that the changes in DNA methylation triggered by maternal separation survived epigenetic reprogramming during embryogenesis (Franklin et al., 2010).
Although not well explored in the CNS, current studies have provided some insights regarding the epigenetic inheritance induced by physical exercise. Denham et al. (2015) reported that 3 months of physical exercise altered global and genome wide sperm DNA methylation in young healthy men. Importantly, the authors observed that exercise increased DNA methylation at promoters of several genes associated with brain pathologies such as schizophrenia and Parkinson disease (Denham et al., 2015). Recently, it has been reported that physical exercise alters the expression of miRNAs in paternal sperm, whereas suppresses the reinstatement of fear memory and reduces anxiety in male mice offspring (Short et al., 2017).
In obese male mice, 8 wk of physical exercise restored insulin sensitivity and normalized the expression profile of X-linked sperm miRNAs that target genes playing important roles in pathways related to early embryogenesis, cell cycle and apoptosis (McPherson et al., 2015). Interestingly, paternal exercise has also been shown to restore insulin sensitivity, to reduce adiposity and to improve metabolic health in F1 female offspring. In another study, maternal exercise prevented the prenatal high-fat diet induced hypermethylation of peroxisome proliferator-activated receptor γ co-activator-1 α (PGC-1α; a transcriptional co-activator critical for mitochondrial biogenesis and oxidative metabolism) in the skeletal muscle of neonatal and 12-month-old offspring (Laker et al., 2014). Altogether, these findings provide compelling evidence suggesting that environmental stimuli can promote epigenetic modifications in germline and somatic cells to allow the transfer of parental experiences to the following generations.
Although there is a lack of experimental evidence for the existence of epigenetic alterations in the offspring brain of exercised parents, indirect studies indicate this possibility. Clinical evidence indicates that maternal exercise during pregnancy promotes a healthy intrauterine milieu for the fetus and enhances cognitive function in childhood (Clapp, 1996, Wolfe et al., 1994, Weissgerber et al., 2006). Clapp (1996) assessed the neurodevelopmental characteristics of offspring (5 years old) from mothers who exercised regularly throughout pregnancy, and found that their children had better performance on tests of general intelligence and oral language skills. Moreover, it has been shown that male offspring from physically active mothers had higher scores in academic performance (i.e. math and language) than boys from sedentary mothers (Esteban-Cornejo et al., 2016).
Rodent studies show that prenatal exercise enhances several features in the offspring's brain such as memory (Lee et al., 2006, Akhavan et al., 2008, Robinson and Bucci, 2014), antioxidant defenses (Marcelino et al., 2013), hippocampal neurogenesis (Bick-Sander et al., 2006, Lee et al., 2006), expression of neurotrophic factors (Aksu et al., 2012, Gomes da Silva et al., 2016), while prenatal exercise reduces anxiety (Aksu et al., 2012). In a recent study, we showed that maternal exercise during pregnancy enhances cognitive function (habituation behavior and spatial learning) and increases BDNF levels and cells numbers (neuronal and non-neuronal) in the hippocampus of rat offspring (Gomes da Silva et al., 2016). Furthermore, prenatal exercise was able to attenuate the progression of Alzheimer's disease-like pathology in the TgCRND8 mice (Herring et al., 2012). In this study, pups born from mothers that ran during their pregnancy time exhibited a reduction in β-amyloid (Aβ) plaque burden and amyloid precursor protein (APP), decreased expression of proinflammatory mediators, and an increased expression of genes involved in synaptic plasticity and memory formation. In addition to the positive effects of maternal exercise on the progeny, pups from physically active fathers are also privileged in terms of cognitive function (Yin et al., 2013). As demonstrated by Yin et al. (2013), 6 wk of paternal exercise enhanced the spatial learning and memory of F1 mice offspring, with a concomitant increase in hippocampal BDNF and reelin expression. Yet, it has been recently shown that paternal exercise transgenerationally alters the affective behavior and the endocrinological response to stress (Yeshurun et al., 2017). Therefore, it is reasonable to assume that physical exercise may use epigenetic mechanisms to affect future generations.
5. Exercise influences cognition engaging epigenetic mechanisms: Central action of Bdnf Transcription
It is getting recognition that exercise and BDNF can reduce depression and improve cognitive function, and that epigenetic regulation of the Bdnf gene is a biological mechanism by which exercise promote mental health and build resistance to neurological disorders (Gomez-Pinilla et al., 2011, Intlekofer et al., 2013, Sleiman et al., 2016). For example, BDNF is essential for hippocampal-dependent spatial learning (Linnarsson et al., 1997, Mizuno et al., 2000) such that elevations in hippocampal BDNF improve learning in animals (Nagahara et al., 2009, Moguel-González et al., 2008). In addition, peripheral levels of BDNF have been associated with cognitive function and hippocampal size in human subjects (Erickson et al., 2011). Data from rodent studies suggest that proteins up-regulated by exercise in the hippocampus are associated with energy metabolism and neurocognitive plasticity, in a process in which BDNF has a preponderant role (Ding et al., 2006b, Ding et al., 2006a, Vaynman et al., 2004, Gomez-Pinilla et al., 2008).
The Bdnf gene consists of nine 5’ non-coding exons and one 3’ coding exon (Aid et al., 2007). In each of these promotor regions, the methylation status of CpG islands and post-translational modifications of histones (i.e.acetylation, phosphorylation, methylation) are thought to regulate Bdnf transcription in an activity dependent manner (Martinowich et al., 2003, Lubin et al., 2008, Guo et al., 2011b). We and others have shown that exercise impacts Bdnf expression by engaging epigenetic modifications such as acetylation and methylation (Gomez-Pinilla et al., 2011, Intlekofer et al., 2013, Sleiman et al., 2016), and these mechanisms are likely associated with synaptic plasticity, and learning and memory. Exercise enhances BDNF expression and facilitates LTP through activation of N-methyl-D-aspartate receptors (NMDA-Rs) (Vaynman et al., 2003, Farmer et al., 2004). Upon activation, NMDA-Rs allow the influx of Ca2+ in the postsynaptic neuron to initiates a cascade of molecular events leading to the activation of Ca2+/Calmodulin-dependent kinase (CaMK) and mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) signaling pathways, which in turn phosphorylates the nuclear transcription factor CREB at serine 133 (S133) (Nguyen and Woo, 2003, Xia and Storm, 2012). Exercise increases the active form of CaMKII, ERK1/2 and CREB (pCaMKII, pERK1/2 and pCREB) in the hippocampus (Gomez-Pinilla et al., 2011, Muller et al., 2008), which are part of the intracellular signaling cascades through which exercise induces BDNF-mediated synaptic plasticity and cognition (Vaynman et al., 2004, Vaynman et al., 2003) (Figure 3). Blocking the function of CaMKII and MAPK in the hippocampus reduces CREB and Bdnf mRNAs and abrogates the cognitive enhancement induced by exercise (Vaynman et al., 2003, Vaynman et al., 2007).
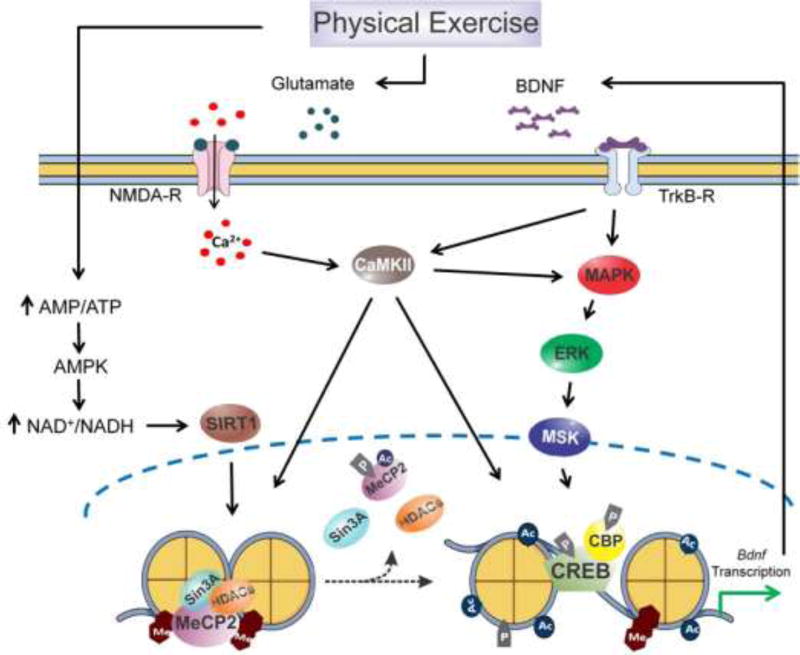
Figure 3.
Potential epigenetic mechanism through which exercise-induced Bdnf expression may enhance synaptic plasticity and cognition. Exercise-induced glutamate release stimulates NMDA-R and, consequently, the influx of Ca2+ in the postsynaptic neurons. Increases in Ca2+ levels trigger a wave of molecular events leading to the activation of CaMKII and MAPK/ERK/MSK signaling. CaMKII phosphorylates MeCP2 inducing its dissociation, along with other co-repressors (Sin3A and HDACs), from the promoter region of Bdnf gene. The effects of exercise on MeCP2 dissociation from Bdnf promoter may also involve the SIRT1-dependent deacetylation of MeCP2, given that the metabolic changes underlying SIRT1 activity and expression are directly modulated by exercise. Besides the phosphorylation of the transcription factor CREB, the activation of CAMKII and MAPK/ERK/MSK signaling induced by exercise phosphorylates some histone proteins and promotes the recruitment and activation of CBP, a HAT. Altogether, these alterations changes the configuration of chromatin to an open state allowing the transcription of Bdnf which, after translated, can bind to its receptor TrkB to engage a positive feedback loop underlying the positive effects of exercise on synaptic plasticity and cognitive abilities. Ac, acetyl group; AMP, adenosine monophosphate; AMPK, AMP-activated protein kinase; ATP, adenosine triphosphate; Bdnf, brain-derived neurotrophic factor; CaMKII, Ca2+/Calmodulin-dependent kinase II; CBP, CREB-binding protein; MAPK/ERK, mitogen-activated protein kinase/extracellular signal-regulated kinase; Me, methyl group; MeCP2, methyl-CpG-binding protein 2; NAD+, nicotinamide adenine dinucleotide (oxidized); NADH, nicotinamide adenine dinucleotide (reduced); NMDA-R, N-methyl-D-aspartate receptor; P, phosphate group; Sin3a, paired amphipathic helix protein Sin3a; HDACs, histone deacetylases; SIRT1, sirtuin 1.
The efficacy of CREB to regulate gene transcription, including Bdnf, is dependent on the activation of various transcriptional coactivators such as CBP (Vecsey et al., 2007), and various HATs (McManus and Hendzel, 2001, Perissi et al., 1999). The phosphorylation of CREB at serine-133 is a requisite to recruit CBP and to initiate subsequent transcription (Chrivia et al., 1993). It has been demonstrated that CBP acetylates lysines on histones H2B, H3, and H4, in vivo, and that although neurons lacking CBP maintain pCREB levels, fail to activate CREB/CBP-mediated gene expression. Moreover, the CBP homolog p300 is unable to compensate for the loss of CBP activity, indicating some level of specificity for CBP in hippocampal neurocognitive processes (Barrett et al., 2011). Interestingly, recent studies have shown that physical exercise increases CBP expression in the hippocampus and promotes acetylation of histones H4K8 and H3 at different Bdnf promoters (Gomez-Pinilla et al., 2011, Intlekofer et al., 2013, Ieraci et al., 2015). While acetylation of H4K8ac is enriched at Bdnf exon I and IV (Intlekofer et al., 2013), H3 acetylation is increased at exons I, II, III, IV, VI and VII (Ieraci et al., 2015).
The acetylation of histones associated with DNA can promote a permissive/active chromatin form necessary for the transcription of select genes such as Bdnf. Experience-dependent plasticity promotes acetylation of lysine amino acid residues in histones H3 and H4 in unbound, transcriptionally active chromatin (Barrett and Wood, 2008, Day and Sweatt, 2011). Neuronal activity has been shown to regulate Bdnf transcription involving epigenetic mechanisms important for regulation of synaptic plasticity, and learning and memory (Martinowich et al., 2003, Lubin et al., 2008, Levenson et al., 2006, Feng et al., 2007). Acquisition of CFM increases acetylation of histones H3 and H4 at Bdnf promoter IV (Lubin et al., 2008), while inhibition of HDACs has been shown to increase Bdnf mRNA levels in hippocampal neurons (Tian et al., 2010), LTP and LTM formation (Levenson et al., 2004). In the prefrontal cortex, the extinction of conditioned fear memory involves the acetylation of histone H4 on the Bdnf promotor region (Bredy et al., 2007). Moreover, high-frequency stimulation in the medial prefrontal cortex increases acetylation of H3 and H4 in the Bdnf gene, and local administration of an HDAC inhibitor increases LTP and LTM (Sui et al., 2012).
Beyond the acetylation, histone proteins are subjected to other post-translational modifications that impact chromatin structure, and their interplay provide an additional level of functional regulation (Latham and Dent, 2007). For example, phosphorylation of histone H3S10 is coupled to acetylation of H3K14 in the hippocampus (Crosio et al., 2003). During the acquisition of CFM, the activation of NMDA-R stimulates the MAPK/ERK pathway that, via kinase 1 (MSK1) contributes to histone H3 acetylation and phosphorylation (Levenson et al., 2004, Chwang et al., 2006, Chwang et al., 2007). Lubin et al. (2008) reported that learning-induced Bdnf transcription increases H3 phosphoacetylation at promoter IV. Recently, it has been shown that NMDA increases transcriptional activation of Bdnf through demethylation of H3K27Me3 (via JMJD3) and phosphorylation of H3K27me3 at Serine 28 (via MAPK/MSK1/2 pathway). These modifications lead to dissociation of the enzyme enhancer of zeste homologue 2 (EZH2), the catalytic subunit of polycomb repressor complex 2, allowing transcription of Bdnf (Palomer et al., 2016). Additionally, NMDA stimulation increases H3K4me3 (activation marker) levels and triggers H3K27 acetylation by recruiting the CREB/CBP complex to Bdnf promoters.
As previously discussed, physical exercise influences synaptic plasticity and memory consolidation involving alterations of histones related to transcription of the Bdnf gene. In aged rats, the effects of the environment on memory have been associated with increased levels of hippocampal H3K4me3 at Bdnf promoter IV (Morse et al., 2015), HAT activity, Pcaf (P300/CBP-associated factor) expression, and histone H3 acetylation in the Bdnf promoter I (Neidl et al., 2016). Sleiman et al. (2016) observed reduced levels of HDAC2 in the hippocampus of mice subjected to voluntary exercise, which was accompanied by decreased binding of HDAC2 to Bdnf promoter I and increased Bdnf expression. Interestingly, BDNF induces nitrosylation of HDAC2 at cysteines 262 and 274 leading to the displacement of this HDAC from chromatin, which results in increased level of histone acetylation in BDNF target genes, including Bdnf itself (Nott et al., 2008). Therefore, it can be proposed that exercise and experience-driven memory formation engage a positive feedback loop triggered by histone modifications at Bdnf promoters. This, in turn, would induce Bdnf expression that could self-sustain a transcriptional program underlying synaptic plasticity and LTM formation.
5.1. MeCP2 and SIRT1 act as transcriptional regulators of Bdnf
MeCP2 belongs to the family of methylcytosine-binding proteins that are abundantly expressed in the central nervous system and are notably recognized for their contribution to the gene silencing effect of DNA methylation (Lewis et al., 1992, Nan et al., 1998, Fuks et al., 2003). MeCP2 phosphorylation seems to be requisite for activity-dependent gene expression, dendritic patterning, spine morphogenesis, LTP and LTM (Zhou et al., 2006, Li et al., 2011). In the absence of stimulation, MeCP2 binds to methylated CpG on the Bdnf promoter IV (referred to at the time as promoter III) to silence its expression through the recruitment of a transcriptional repressor complex containing mSin3A and HDACs (Martinowich et al., 2003). The calcium influx triggered by neuronal depolarization induces phosphorylation of MeCP2 at serine 421 and, consequently, its dissociation from the Bdnf promoter IV (Chen et al., 2003, Zhou et al., 2006). This effect is mediated by a CaMKII-dependent mechanism and occurs in parallel with demethylation of promoter IV to allow Bdnf transcription.
Gomez-Pinilla et al. (2011) reported that the exercise-induced Bdnf expression was linked to reductions in promoter IV methylation and an enhancement in the levels of phospho-CaMKII and phospho-MeCP2 in the hippocampus. Importantly, blockade of CaMKII signaling in this region attenuated the increases in Bdnf expression and the cognitive improvement elicited by exercise (Vaynman et al., 2003, Vaynman et al., 2007). Furthermore, studies conducted in Mecp2 mutant mice, a mouse model of Rett syndrome, have shown that environmental enrichment restores BDNF levels (Kondo et al., 2008, Lonetti et al., 2010, Kondo et al., 2016), attenuates LTP deficits (Lonetti et al., 2010) and ameliorates several behavioral abnormalities including memory deficits, affective dysfunction, anxiety, and impairments in motor function (Lonetti et al., 2010, Kondo et al., 2008, Kondo et al., 2016, Nag et al., 2009).
It has been recently reported that Bdnf transcription can be regulated by sirtuin 1 (SIRT1)-dependent deacetylation of MeCP2 (Zocchi and Sassone-Corsi, 2012). SIRT1 is a highly conserved nicotinamide-adenine dinucleotide (NAD+)-dependent Class III HDAC (Thiagalingam et al., 2003) that, beyond its role on processes related to ageing, stress tolerance, and energy metabolism (Schwer and Verdin, 2008, Haigis and Sinclair, 2010), has emerged as a key regulator of synaptic plasticity and memory formation (Gao et al., 2010). SIRT1-dependent deacetylation of MeCP2 precedes its detachment from Bdnf promoter IV in neuronal culture and SIRT1dex4 mutant mice exhibit increased binding of MeCP2 to the Bdnf promoter IV and reduced Bdnf expression (Zocchi and Sassone-Corsi, 2012). In accordance with the positive effects of exercise on Bdnf expression and memory formation, evidence shows that physical exercise alters SIRT1 dynamics in the brain. It was demonstrated that exercise increases SIRT1 activity in the hippocampus (Marton et al., 2016), and elevates its expression in various brain regions including cortex, hippocampus and hypothalamus (Steiner et al., 2011). Furthermore, in a recent study of the effects of physical exercise on the pathophysiology of Alzheimer's disease, the downregulation of SIRT1 in the cerebral cortex of 3xTgAD mice was attenuated by exercise treatment (Revilla et al., 2014).
The mechanisms connecting SIRT1 activity to BDNF were first explored through SIRT1 loss-of-function experiments, using SIRT1Δ mutant mice lacking SIRT1 catalytic activity. SIRT1 loss-of-function results in higher levels of miR-134, the brain specific miRNA that acts as a translational block of CREB. Concomitantly, BDNF (mRNA and protein) and CREB (protein) are down regulated in the hippocampus, and LTP and memory formation are impaired (Gao et al., 2010). Notably, SIRT1 loss-of-function also abolishes the learning-related increases in Bdnf promoter (I and IV) binding by CREB (Gao et al., 2010). Mechanistically, SIRT1 normally functions in cooperation with Ying Yang 1 (YY1), a ubiquitous and highly conserved transcription factor that can act as a transcriptional repressor or activator (Shi et al., 1997). SIRT1 is recruited as a co-repressor by YY1 to inhibit the expression of the miR-134 gene, which in turn decreases the probability that miR-134 will act as a translational block of CREB and consequently facilitates Bdnf transcription via Bdnf promoter binding by phospho-CREB (Gao et al., 2010). The coordination of SIRT1/YY1 in transcriptional repression with complementary activity-dependent mechanisms may regulate Bdnf transcription in the hippocampus during exercise.
6. Concluding remarks
Exercise has played a crucial action on shaping the modern brain through thousands of years of evolution. Nowadays, the adaptive force of exercise is vitally instrumental for controlling susceptibility to a wide range of neurological and metabolic disorders with the potential to be inheritable. Accumulating scientific evidence in the last few years portray the capacity of exercise to promote profound alterations in the program of genes and their protein products in the form of epigenomic manifestations. The remarkable capacity of epigenetic mechanisms to regulate synaptic and cognitive plasticity has a fundamental value for the control of neurological and psychological disturbances. As more studies continue to reveal the details by which exercise influences a varied assortment of gene regulatory mechanisms, it is becoming clear that gene reprogramming is instrumental for transducing the effects of exercise on brain structure and function. Recent developments in the epigenetic field are leading to a paradigm shift in basic and clinical neuroscience to account for how environmental factors can impact long-term brain function with profound implications for public health of current and future generations.
Highlights.
Exercise regulates DNA methylation and histone acetylation in the hippocampus
Exercise enhances bdnf expression by engaging epigenetic mechanisms.
Exercise modulates the expression of memory-related miRNAs.
The beneficial effects of exercise are epigenetically inherited.
Acknowledgments
This work was supported by The National Institute of Health (NIH: NS50465), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP: #2013/12692-4; #2013/13725-3; #2015/09234-0) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq: #300605/2013-07).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abel JL, Rissman EF. Running-induced epigenetic and gene expression changes in the adolescent brain. Int J Dev Neurosci. 2013;31(6):382–90. doi: 10.1016/j.ijdevneu.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85(3):525–35. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhavan MM, Emami-Abarghoie M, Safari M, Sadighi-Moghaddam B, Vafaei AA, Bandegi AR, Rashidy-Pour A. Serotonergic and noradrenergic lesions suppress the enhancing effect of maternal exercise during pregnancy on learning and memory in rat pups. Neuroscience. 2008;151(4):1173–83. doi: 10.1016/j.neuroscience.2007.10.051. [DOI] [PubMed] [Google Scholar]
- Aksu I, Baykara B, Ozbal S, Cetin F, Sisman AR, Dayi A, Gencoglu C, Tas A, Büyük E, Gonenc-Arda S, Uysal N. Maternal treadmill exercise during pregnancy decreases anxiety and increases prefrontal cortex VEGF and BDNF levels of rat pups in early and late periods of life. Neurosci Lett. 2012;516(2):221–5. doi: 10.1016/j.neulet.2012.03.091. [DOI] [PubMed] [Google Scholar]
- Alarcón JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42(6):947–59. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23(2):185–8. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggish AL, Hale A, Weiner RB, Lewis GD, Systrom D, Wang F, Wang TJ, Chan SY. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training. J Physiol. 2011;589(Pt 16):3983–94. doi: 10.1113/jphysiol.2011.213363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao TH, Miao W, Han JH, Yin M, Yan Y, Wang WW, Zhu YH. Spontaneous running wheel improves cognitive functions of mouse associated with miRNA expressional alteration in hippocampus following traumatic brain injury. J Mol Neurosci. 2014;54(4):622–9. doi: 10.1007/s12031-014-0344-1. [DOI] [PubMed] [Google Scholar]
- Barrett RM, Malvaez M, Kramar E, Matheos DP, Arrizon A, Cabrera SM, Lynch G, Greene RW, Wood MA. Hippocampal focal knockout of CBP affects specific histone modifications, long-term potentiation, and long-term memory. Neuropsychopharmacology. 2011;36(8):1545–56. doi: 10.1038/npp.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RM, Wood MA. Beyond transcription factors: the role of chromatin modifying enzymes in regulating transcription required for memory. Learn Mem. 2008;15(7):460–7. doi: 10.1101/lm.917508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrès R, Zierath JR. The role of diet and exercise in the transgenerational epigenetic landscape of T2DM. Nat Rev Endocrinol. 2016;12(8):441–51. doi: 10.1038/nrendo.2016.87. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Benevento M, van de Molengraft M, van Westen R, van Bokhoven H, Kasri NN. The role of chromatin repressive marks in cognition and disease: A focus on the repressive complex GLP/G9a. Neurobiol Learn Mem. 2015;124:88–96. doi: 10.1016/j.nlm.2015.06.013. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128(4):669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bick-Sander A, Steiner B, Wolf SA, Babu H, Kempermann G. Running in pregnancy transiently increases postnatal hippocampal neurogenesis in the offspring. Proc Natl Acad Sci U S A. 2006;103(10):3852–7. doi: 10.1073/pnas.0502644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992;70(1):5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Blumenthal JA, Babyak MA, Moore KA, Craighead WE, Herman S, Khatri P, Waugh R, Napolitano MA, Forman LM, Appelbaum M, Doraiswamy PM, Krishnan KR. Effects of exercise training on older patients with major depression. Arch Intern Med. 1999;159(19):2349–56. doi: 10.1001/archinte.159.19.2349. [DOI] [PubMed] [Google Scholar]
- Brami-Cherrier K, Roze E, Girault JA, Betuing S, Caboche J. Role of the ERK/MSK1 signalling pathway in chromatin remodelling and brain responses to drugs of abuse. J Neurochem. 2009;108(6):1323–35. doi: 10.1111/j.1471-4159.2009.05879.x. [DOI] [PubMed] [Google Scholar]
- Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn Mem. 2007;14(4):268–76. doi: 10.1101/lm.500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–99. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Cassilhas RC, Viana VA, Grassmann V, Santos RT, Santos RF, Tufik S, Mello MT. The impact of resistance exercise on the cognitive function of the elderly. Med Sci Sports Exerc. 2007;39(8):1401–7. doi: 10.1249/mss.0b013e318060111f. [DOI] [PubMed] [Google Scholar]
- Cechinel LR, Basso CG, Bertoldi K, Schallenberger B, de Meireles LC, Siqueira IR. Treadmill exercise induces age and protocol-dependent epigenetic changes in prefrontal cortex of Wistar rats. Behav Brain Res. 2016;313:82–7. doi: 10.1016/j.bbr.2016.07.016. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320(5880):1224–9. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao HT, Zoghbi HY, Rosenmund C. MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron. 2007;56(1):58–65. doi: 10.1016/j.neuron.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302(5646):885–9. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Chen WQ, Viidik A, Skalicky M, Höger H, Lubec G. Hippocampal signaling cascades are modulated in voluntary and treadmill exercise rats. Electrophoresis. 2007;28(23):4392–400. doi: 10.1002/elps.200700336. [DOI] [PubMed] [Google Scholar]
- Chen Y, Liu W, Chao T, Zhang Y, Yan X, Gong Y, Qiang B, Yuan J, Sun M, Peng X. MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett. 2008;272(2):197–205. doi: 10.1016/j.canlet.2008.06.034. [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436(7051):740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54(5):813–29. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, Lo YM. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54(3):482–90. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- Chin LM, Keyser RE, Dsurney J, Chan L. Improved cognitive performance following aerobic exercise training in people with traumatic brain injury. Arch Phys Med Rehabil. 2015;96(4):754–9. doi: 10.1016/j.apmr.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365(6449):855–9. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Chwang WB, Arthur JS, Schumacher A, Sweatt JD. The nuclear kinase mitogen- and stress-activated protein kinase 1 regulates hippocampal chromatin remodeling in memory formation. J Neurosci. 2007;27(46):12732–42. doi: 10.1523/JNEUROSCI.2522-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chwang WB, O'Riordan KJ, Levenson JM, Sweatt JD. ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learn Mem. 2006;13(3):322–8. doi: 10.1101/lm.152906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapp JF. Morphometric and neurodevelopmental outcome at age five years of the offspring of women who continued to exercise regularly throughout pregnancy. J Pediatr. 1996;129(6):856–63. doi: 10.1016/s0022-3476(96)70029-x. [DOI] [PubMed] [Google Scholar]
- Colcombe SJ, Kramer AF, McAuley E, Erickson KI, Scalf P. Neurocognitive aging and cardiovascular fitness: recent findings and future directions. J Mol Neurosci. 2004;24(1):9–14. doi: 10.1385/JMN:24:1:009. [DOI] [PubMed] [Google Scholar]
- Collins A, Hill LE, Chandramohan Y, Whitcomb D, Droste SK, Reul JM. Exercise improves cognitive responses to psychological stress through enhancement of epigenetic mechanisms and gene expression in the dentate gyrus. PLoS One. 2009;4(1):e4330. doi: 10.1371/journal.pone.0004330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8(8):467–77. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosín-Tomás M, Alvarez-López MJ, Sanchez-Roige S, Lalanza JF, Bayod S, Sanfeliu C, Pallàs M, Escorihuela RM, Kaliman P. Epigenetic alterations in hippocampus of SAMP8 senescent mice and modulation by voluntary physical exercise. Front Aging Neurosci. 2014;6:51. doi: 10.3389/fnagi.2014.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotman CW, Berchtold NC. Exercise: a behavioral intervention to enhance brain health and plasticity. Trends in Neurosciences. 2002;25(6):295–301. doi: 10.1016/s0166-2236(02)02143-4. [DOI] [PubMed] [Google Scholar]
- Cotman CW, Berchtold NC, Christie LA. Exercise builds brain health: key roles of growth factor cascades and inflammation. Trends Neurosci. 2007;30(9):464–72. doi: 10.1016/j.tins.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Crosio C, Heitz E, Allis CD, Borrelli E, Sassone-Corsi P. Chromatin remodeling and neuronal response: multiple signaling pathways induce specific histone H3 modifications and early gene expression in hippocampal neurons. J Cell Sci. 2003;116(Pt 24):4905–14. doi: 10.1242/jcs.00804. [DOI] [PubMed] [Google Scholar]
- Cui JG, Li YY, Zhao Y, Bhattacharjee S, Lukiw WJ. Differential regulation of interleukin-1 receptor-associated kinase-1 (IRAK-1) and IRAK-2 by microRNA-146a and NF-kappaB in stressed human astroglial cells and in Alzheimer disease. J Biol Chem. 2010;285(50):38951–60. doi: 10.1074/jbc.M110.178848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao AT, Zagaar MA, Alkadhi KA. Moderate Treadmill Exercise Protects Synaptic Plasticity of the Dentate Gyrus and Related Signaling Cascade in a Rat Model of Alzheimer's Disease. Mol Neurobiol. 2015;52(3):1067–76. doi: 10.1007/s12035-014-8916-1. [DOI] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron. 2011;70(5):813–29. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida AA, Gomes da Silva S, Lopim GM, Vannucci Campos D, Fernandes J, Cabral FR, Arida RM. Resistance Exercise Reduces Seizure Occurrence, Attenuates Memory Deficits and Restores BDNF Signaling in Rats with Chronic Epilepsy. Neurochem Res. 2017;42(4):1230–1239. doi: 10.1007/s11064-016-2165-9. [DOI] [PubMed] [Google Scholar]
- de Meireles LC, Bertoldi K, Cechinel LR, Schallenberger BL, da Silva VK, Schröder N, Siqueira IR. Treadmill exercise induces selective changes in hippocampal histone acetylation during the aging process in rats. Neurosci Lett. 2016;634:19–24. doi: 10.1016/j.neulet.2016.10.008. [DOI] [PubMed] [Google Scholar]
- de Meireles LC, Bertoldi K, Elsner VR, Moysés FoS, Siqueira IR. Treadmill exercise alters histone acetylation differently in rats exposed or not exposed to aversive learning context. Neurobiol Learn Mem. 2014;116:193–6. doi: 10.1016/j.nlm.2014.10.008. [DOI] [PubMed] [Google Scholar]
- Denham J, O’Brien BJ, Harvey JT, Charchar FJ. Genome-wide sperm DNA methylation changes after 3 months of exercise training in humans. Epigenomics. 2015;7(5):717–31. doi: 10.2217/epi.15.29. [DOI] [PubMed] [Google Scholar]
- Ding Q, Vaynman S, Akhavan M, Ying Z, Gomez-Pinilla F. Insulin-like growth factor I interfaces with brain-derived neurotrophic factor-mediated synaptic plasticity to modulate aspects of exercise-induced cognitive function. Neuroscience. 2006a;140(3):823–833. doi: 10.1016/j.neuroscience.2006.02.084. [DOI] [PubMed] [Google Scholar]
- Ding Q, Vaynman S, Souda P, Whitelegge JP, Gomez-Pinilla F. Exercise affects energy metabolism and neural plasticity-related proteins in the hippocampus as revealed by proteomic analysis. Eur J Neurosci. 2006b;24(5):1265–76. doi: 10.1111/j.1460-9568.2006.05026.x. [DOI] [PubMed] [Google Scholar]
- Drewell RA, Goddard CJ, Thomas JO, Surani MA. Methylation-dependent silencing at the H19 imprinting control region by MeCP2. Nucleic Acids Res. 2002;30(5):1139–44. doi: 10.1093/nar/30.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AL, Trivedi MH, Kampert JB, Clark CG, Chambliss HO. Exercise treatment for depression: efficacy and dose response. Am J Prev Med. 2005;28(1):1–8. doi: 10.1016/j.amepre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, Becker H, Chandler JC, Andino R, Cortes J, Hokland P, Huettner CS, Bhatia R, Roy DC, Liebhaber SA, Caligiuri MA, Marcucci G, Garzon R, Croce CM, Calin GA, Perrotti D. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140(5):652–65. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsner VR, Lovatel GA, Bertoldi K, Vanzella C, Santos FM, Spindler C, de Almeida EF, Nardin P, Siqueira IR. Effect of different exercise protocols on histone acetyltransferases and histone deacetylases activities in rat hippocampus. Neuroscience. 2011;192:580–7. doi: 10.1016/j.neuroscience.2011.06.066. [DOI] [PubMed] [Google Scholar]
- Elsner VR, Lovatel GA, Moysés F, Bertoldi K, Spindler C, Cechinel LR, Muotri AR, Siqueira IR. Exercise induces age-dependent changes on epigenetic parameters in rat hippocampus: a preliminary study. Exp Gerontol. 2013;48(2):136–9. doi: 10.1016/j.exger.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson KI, Voss MW, Prakash RS, Basak C, Szabo A, Chaddock L, Kim JS, Heo S, Alves H, White SM, Wojcicki TR, Mailey E, Vieira VJ, Martin SA, Pence BD, Woods JA, McAuley E, Kramer AF. Exercise training increases size of hippocampus and improves memory. Proc Natl Acad Sci U S A. 2011;108(7):3017–22. doi: 10.1073/pnas.1015950108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban-Cornejo I, Martinez-Gomez D, Tejero-González CM, Izquierdo-Gomez R, Carbonell-Baeza A, Castro-Piñero J, Sallis JF, Veiga OL Group, U. D. S. Maternal physical activity before and during the prenatal period and the offspring’s academic performance in youth. The UP&DOWN study. J Matern Fetal Neonatal Med. 2016;29(9):1414–20. doi: 10.3109/14767058.2015.1049525. [DOI] [PubMed] [Google Scholar]
- Fabel K, Tam B, Kaufer D, Baiker A, Simmons N, Kuo CJ, Palmer TD. VEGF is necessary for exercise-induced adult hippocampal neurogenesis. Eur J Neurosci. 2003;18(10):2803–12. doi: 10.1111/j.1460-9568.2003.03041.x. [DOI] [PubMed] [Google Scholar]
- Farmer J, Zhao X, van Praag H, Wodtke K, Gage FH, Christie BR. Effects of voluntary exercise on synaptic plasticity and gene expression in the dentate gyrus of adult male Sprague-Dawley rats in vivo. Neuroscience. 2004;124(1):71–9. doi: 10.1016/j.neuroscience.2003.09.029. [DOI] [PubMed] [Google Scholar]
- Feng J, Fouse S, Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatr Res. 2007;61(5 Pt 2):58R–63R. doi: 10.1203/pdr.0b013e3180457635. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13(4):423–30. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J, Soares JCK, Baliego LGZdA, Arida RM. A single bout of resistance exercise improves memory consolidation and increases the expression of synaptic proteins in the hippocampus. Hippocampus. 2016;26(8):1096–1103. doi: 10.1002/hipo.22590. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17(15):1870–81. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers E, Won GY, Fukuoka Y. MicroRNAs associated with exercise and diet: a systematic review. Physiol Genomics. 2015;47(1):1–11. doi: 10.1152/physiolgenomics.00095.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin TB, Mansuy IM. Epigenetic inheritance in mammals: evidence for the impact of adverse environmental effects. Neurobiol Dis. 2010;39(1):61–5. doi: 10.1016/j.nbd.2009.11.012. [DOI] [PubMed] [Google Scholar]
- Franklin TB, Russig H, Weiss IC, Gräff J, Linder N, Michalon A, Vizi S, Mansuy IM. Epigenetic transmission of the impact of early stress across generations. Biol Psychiatry. 2010;68(5):408–15. doi: 10.1016/j.biopsych.2010.05.036. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278(6):4035–40. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28(17):5369–80. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Gräff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466(7310):1105–9. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge XT, Lei P, Wang HC, Zhang AL, Han ZL, Chen X, Li SH, Jiang RC, Kang CS, Zhang JN. miR-21 improves the neurological outcome after traumatic brain injury in rats. Sci Rep. 2014;4:6718. doi: 10.1038/srep06718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–8. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Golshani P, Hutnick L, Schweizer F, Fan G. Conditional Dnmt1 deletion in dorsal forebrain disrupts development of somatosensory barrel cortex and thalamocortical long-term potentiation. Thalamus Relat Syst. 2005;3(3):227–233. doi: 10.1017/S1472928807000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes CP, Kim TK, Wang K, He Y. The implications on clinical diagnostics of using microRNA-based biomarkers in exercise. Expert Rev Mol Diagn. 2015;15(6):761–72. doi: 10.1586/14737159.2015.1039517. [DOI] [PubMed] [Google Scholar]
- Gomes da Silva S, de Almeida AA, Fernandes J, Lopim GM, Cabral FR, Scerni DA, de Oliveira-Pinto AV, Lent R, Arida RM. Maternal Exercise during Pregnancy Increases BDNF Levels and Cell Numbers in the Hippocampal Formation but Not in the Cerebral Cortex of Adult Rat Offspring. PLoS One. 2016;11(1):0147200. doi: 10.1371/journal.pone.0147200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pinilla F, Hillman C. The influence of exercise on cognitive abilities. Compr Physiol. 2013;3(1):403–28. doi: 10.1002/cphy.c110063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pinilla F, Vaynman S, Ying Z. Brain-derived neurotrophic factor functions as a metabotrophin to mediate the effects of exercise on cognition. Eur J Neurosci. 2008;28(11):2278–87. doi: 10.1111/j.1460-9568.2008.06524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pinilla F, Zhuang Y, Feng J, Ying Z, Fan G. Exercise impacts brain-derived neurotrophic factor plasticity by engaging mechanisms of epigenetic regulation. Eur J Neurosci. 2011;33(3):383–90. doi: 10.1111/j.1460-9568.2010.07508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grealy MA, Johnson DA, Rushton SK. Improving cognitive function after brain injury: the use of exercise and virtual reality. Arch Phys Med Rehabil. 1999;80(6):661–7. doi: 10.1016/s0003-9993(99)90169-7. [DOI] [PubMed] [Google Scholar]
- Griggs EM, Young EJ, Rumbaugh G, Miller CA. MicroRNA-182 regulates amygdala-dependent memory formation. J Neurosci. 2013;33(4):1734–40. doi: 10.1523/JNEUROSCI.2873-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Xie H, Ding X. The role of epigenetic regulation in learning and memory. Exp Neurol. 2015;268:30–6. doi: 10.1016/j.expneurol.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, Zhang K, Ming GL, Gao Y, Song H. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011a;14(10):1345–51. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011b;145(3):423–34. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, Paylor RE, Lubin FD. Histone methylation regulates memory formation. J Neurosci. 2010;30(10):3589–99. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta-Agarwal S, Franklin AV, Deramus T, Wheelock M, Davis RL, McMahon LL, Lubin FD. G9a/GLP histone lysine dimethyltransferase complex activity in the hippocampus and the entorhinal cortex is required for gene activation and silencing during memory consolidation. J Neurosci. 2012;32(16):5440–53. doi: 10.1523/JNEUROSCI.0147-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–95. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KF, Karelina K, Sakamoto K, Wayman GA, Impey S, Obrietan K. miRNA-132: a dynamic regulator of cognitive capacity. Brain Struct Funct. 2013;218(3):817–31. doi: 10.1007/s00429-012-0431-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KF, Sakamoto K, Aten S, Snider KH, Loeser J, Hesse AM, Page CE, Pelz C, Arthur JS, Impey S, Obrietan K. Targeted deletion of miR-132/-212 impairs memory and alters the hippocampal transcriptome. Learn Mem. 2016;23(2):61–71. doi: 10.1101/lm.039578.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KF, Sakamoto K, Wayman GA, Impey S, Obrietan K. Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS One. 2010;5(11):e15497. doi: 10.1371/journal.pone.0015497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harraz MM, Eacker SM, Wang X, Dawson TM, Dawson VL. MicroRNA-223 is neuroprotective by targeting glutamate receptors. Proc Natl Acad Sci U S A. 2012;109(46):18962–7. doi: 10.1073/pnas.1121288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157(1):95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring A, Donath A, Yarmolenko M, Uslar E, Conzen C, Kanakis D, Bosma C, Worm K, Paulus W, Keyvani K. Exercise during pregnancy mitigates Alzheimer-like pathology in mouse offspring. FASEB J. 2012;26(1):117–28. doi: 10.1096/fj.11-193193. [DOI] [PubMed] [Google Scholar]
- Heyn PC, Johnson KE, Kramer AF. Endurance and strength training outcomes on cognitively impaired and cognitively intact older adults: a meta-analysis. J Nut Health Aging. 2008;12(6):401–9. doi: 10.1007/BF02982674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T, Zhou FJ, Chang YF, Li YS, Liu GC, Hong Y, Chen HL, Xiyang YB, Bao TH. miR21 is Associated with the Cognitive Improvement Following Voluntary Running Wheel Exercise in TBI Mice. J Mol Neurosci. 2015;57(1):114–22. doi: 10.1007/s12031-015-0584-8. [DOI] [PubMed] [Google Scholar]
- Ieraci A, Mallei A, Musazzi L, Popoli M. Physical exercise and acute restraint stress differentially modulate hippocampal brain-derived neurotrophic factor transcripts and epigenetic mechanisms in mice. Hippocampus. 2015;25(11):1380–92. doi: 10.1002/hipo.22458. [DOI] [PubMed] [Google Scholar]
- Impey S, Davare M, Lesiak A, Lasiek A, Fortin D, Ando H, Varlamova O, Obrietan K, Soderling TR, Goodman RH, Wayman GA. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Mol Cell Neurosci. 2010;43(1):146–56. doi: 10.1016/j.mcn.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer KA, Berchtold NC, Malvaez M, Carlos AJ, McQuown SC, Cunningham MJ, Wood MA, Cotman CW. Exercise and sodium butyrate transform a subthreshold learning event into long-term memory via a brain-derived neurotrophic factor-dependent mechanism. Neuropsychopharmacology. 2013;38(10):2027–34. doi: 10.1038/npp.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer KA, Cotman CW. Exercise counteracts declining hippocampal function in aging and Alzheimer’s disease. Neurobiol Dis. 2013;57:47–55. doi: 10.1016/j.nbd.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Irier H, Street RC, Dave R, Lin L, Cai C, Davis TH, Yao B, Cheng Y, Jin P. Environmental enrichment modulates 5-hydroxymethylcytosine dynamics in hippocampus. Genomics. 2014;104(5):376–82. doi: 10.1016/j.ygeno.2014.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–54. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jayakody K, Gunadasa S, Hosker C. Exercise for anxiety disorders: systematic review. Br J Sports Med. 2014;48(3):187–96. doi: 10.1136/bjsports-2012-091287. [DOI] [PubMed] [Google Scholar]
- Jin K, Zhu Y, Sun Y, Mao XO, Xie L, Greenberg DA. Vascular endothelial growth factor (VEGF) stimulates neurogenesis in vitro and in vivo. Proc Natl Acad Sci U S A. 2002;99(18):11946–50. doi: 10.1073/pnas.182296499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovičić A, Roshan R, Moisoi N, Pradervand S, Moser R, Pillai B, Luthi-Carter R. Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. J Neurosci. 2013;33(12):5127–37. doi: 10.1523/JNEUROSCI.0600-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangaspeska S, Stride B, Métivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452(7183):112–5. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- Kashimoto RK, Toffoli LV, Manfredo MH, Volpini VL, Martins-Pinge MC, Pelosi GG, Gomes MV. Physical exercise affects the epigenetic programming of rat brain and modulates the adaptive response evoked by repeated restraint stress. Behav Brain Res. 2016;296:286–9. doi: 10.1016/j.bbr.2015.08.038. [DOI] [PubMed] [Google Scholar]
- Kim TK, Lee JE, Kim JE, Park JY, Choi J, Kim H, Lee EH, Han PL. G9a-Mediated Regulation of OXT and AVP Expression in the Basolateral Amygdala Mediates Stress-Induced Lasting Behavioral Depression and Its Reversal by Exercise. Mol Neurobiol. 2016;53(5):2843–56. doi: 10.1007/s12035-015-9160-z. [DOI] [PubMed] [Google Scholar]
- Kiuchi T, Lee H, Mikami T. Regular exercise cures depression-like behavior via VEGF-Flk-1 signaling in chronically stressed mice. Neuroscience. 2012;207:208–17. doi: 10.1016/j.neuroscience.2012.01.023. [DOI] [PubMed] [Google Scholar]
- Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10(12):1513–4. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502(7472):472–9. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Gray LJ, Pelka GJ, Christodoulou J, Tam PP, Hannan AJ. Environmental enrichment ameliorates a motor coordination deficit in a mouse model of Rett syndrome--Mecp2 gene dosage effects and BDNF expression. Eur J Neurosci. 2008;27(12):3342–50. doi: 10.1111/j.1460-9568.2008.06305.x. [DOI] [PubMed] [Google Scholar]
- Kondo MA, Gray LJ, Pelka GJ, Leang SK, Christodoulou J, Tam PP, Hannan AJ. Affective dysfunction in a mouse model of Rett syndrome: Therapeutic effects of environmental stimulation and physical activity. Dev Neurobiol. 2016;76(2):209–24. doi: 10.1002/dneu.22308. [DOI] [PubMed] [Google Scholar]
- Konopka W, Kiryk A, Novak M, Herwerth M, Parkitna JR, Wawrzyniak M, Kowarsch A, Michaluk P, Dzwonek J, Arnsperger T, Wilczynski G, Merkenschlager M, Theis FJ, Köhr G, Kaczmarek L, Schütz G. MicroRNA loss enhances learning and memory in mice. J Neurosci. 2010;30(44):14835–42. doi: 10.1523/JNEUROSCI.3030-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka W, Schütz G, Kaczmarek L. The microRNA contribution to learning and memory. Neuroscientist. 2011;17(5):468–74. doi: 10.1177/1073858411411721. [DOI] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42(6):961–72. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshibu K, Gräff J, Beullens M, Heitz FD, Berchtold D, Russig H, Farinelli M, Bollen M, Mansuy IM. Protein phosphatase 1 regulates the histone code for long-term memory. J Neurosci. 2009;29(41):13079–89. doi: 10.1523/JNEUROSCI.3610-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshibu K, Gräff J, Mansuy IM. Nuclear protein phosphatase-1: an epigenetic regulator of fear memory and amygdala long-term potentiation. Neuroscience. 2011;173:30–6. doi: 10.1016/j.neuroscience.2010.11.023. [DOI] [PubMed] [Google Scholar]
- Kou X, Li J, Liu X, Chang J, Zhao Q, Jia S, Fan J, Chen N. Swimming attenuates D-galactose-induced brain aging via suppressing miR-34a-mediated autophagy impairment and abnormal mitochondrial dynamics. J Appl Physiol (1985) 2017 doi: 10.1152/japplphysiol.00018.2017. pp. jap.00018.2017. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kvam S, Kleppe CL, Nordhus IH, Hovland A. Exercise as a treatment for depression: A meta-analysis. J Affect Disord. 2016;202:67–86. doi: 10.1016/j.jad.2016.03.063. [DOI] [PubMed] [Google Scholar]
- Kye MJ, Liu T, Levy SF, Xu NL, Groves BB, Bonneau R, Lao K, Kosik KS. Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. RNA. 2007;13(8):1224–34. doi: 10.1261/rna.480407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kye MJ, Neveu P, Lee YS, Zhou M, Steen JA, Sahin M, Kosik KS, Silva AJ. NMDA mediated contextual conditioning changes miRNA expression. PLoS One. 2011;6(9):e24682. doi: 10.1371/journal.pone.0024682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laker RC, Lillard TS, Okutsu M, Zhang M, Hoehn KL, Connelly JJ, Yan Z. Exercise prevents maternal high-fat diet-induced hypermethylation of the Pgc-1α gene and age-dependent metabolic dysfunction in the offspring. Diabetes. 2014;63(5):1605–11. doi: 10.2337/db13-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham JA, Dent SY. Cross-regulation of histone modifications. Nat Struct Mol Biol. 2007;14(11):1017–24. doi: 10.1038/nsmb1307. [DOI] [PubMed] [Google Scholar]
- Lee HH, Kim H, Lee JW, Kim YS, Yang HY, Chang HK, Lee TH, Shin MC, Lee MH, Shin MS, Park S, Baek S, Kim CJ. Maternal swimming during pregnancy enhances short-term memory and neurogenesis in the hippocampus of rat pups. Brain Dev. 2006;28(3):147–54. doi: 10.1016/j.braindev.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lei P, Li Y, Chen X, Yang S, Zhang J. Microarray based analysis of microRNA expression in rat cerebral cortex after traumatic brain injury. Brain Res. 2009;1284:191–201. doi: 10.1016/j.brainres.2009.05.074. [DOI] [PubMed] [Google Scholar]
- Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279(39):40545–59. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281(23):15763–73. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69(6):905–14. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Li H, Zhong X, Chau KF, Williams EC, Chang Q. Loss of activity-induced phosphorylation of MeCP2 enhances synaptogenesis, LTP and spatial memory. Nat Neurosci. 2011;14(8):1001–8. doi: 10.1038/nn.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Wei W, Coelho CM, Li X, Baker-Andresen D, Dudley K, Ratnu VS, Boskovic Z, Kobor MS, Sun YE, Bredy TW. The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nat Neurosci. 2011;14(9):1115–7. doi: 10.1038/nn.2891. [DOI] [PubMed] [Google Scholar]
- Linnarsson S, Bjorklund A, Ernfors P. Learning deficit in BDNF mutant mice. Eur J Neurosci. 1997;9(12):2581–2587. doi: 10.1111/j.1460-9568.1997.tb01687.x. [DOI] [PubMed] [Google Scholar]
- Lista I, Sorrentino G. Biological mechanisms of physical activity in preventing cognitive decline. Cell Mol Neurobiol. 2010;30(4):493–503. doi: 10.1007/s10571-009-9488-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetti G, Angelucci A, Morando L, Boggio EM, Giustetto M, Pizzorusso T. Early environmental enrichment moderates the behavioral and synaptic phenotype of MeCP2 null mice. Biol Psychiatry. 2010;67(7):657–65. doi: 10.1016/j.biopsych.2009.12.022. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28(42):10576–86. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludyga S, Gerber M, Brand S, Holsboer-Trachsler E, Pühse U. Acute effects of moderate aerobic exercise on specific aspects of executive function in different age and fitness groups: A meta-analysis. Psychophysiology. 2016;53(11):1611–1626. doi: 10.1111/psyp.12736. [DOI] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc Natl Acad Sci U S A. 2007;104(23):9667–72. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelino TB, Longoni A, Kudo KY, Stone V, Rech A, de Assis AM, Scherer EB, da Cunha MJ, Wyse AT, Pettenuzzo LF, Leipnitz G, Matté C. Evidences that maternal swimming exercise improves antioxidant defenses and induces mitochondrial biogenesis in the brain of young Wistar rats. Neuroscience. 2013;246:28–39. doi: 10.1016/j.neuroscience.2013.04.043. [DOI] [PubMed] [Google Scholar]
- Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15(2):163–76. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302(5646):890–3. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Marton O, Koltai E, Takeda M, Mimura T, Pajk M, Abraham D, Koch LG, Britton SL, Higuchi M, Boldogh I, Radak Z. The rate of training response to aerobic exercise affects brain function of rats. Neurochem Int. 2016;99:16–23. doi: 10.1016/j.neuint.2016.05.012. [DOI] [PubMed] [Google Scholar]
- Matura S, Carvalho AF, Alves GS, Pantel J. Physical Exercise for the Treatment of Neuropsychiatric Disturbances in Alzheimer’s Dementia: Possible Mechanisms, Current Evidence and Future Directions. Curr Alzheimer Res. 2016;13(10):1112–23. doi: 10.2174/1567205013666160502123428. [DOI] [PubMed] [Google Scholar]
- McManus KJ, Hendzel MJ. CBP, a transcriptional coactivator and acetyltransferase. Biochem Cell Biol. 2001;79(3):253–66. [PubMed] [Google Scholar]
- McNeill E, Van Vactor D. MicroRNAs shape the neuronal landscape. Neuron. 2012;75(3):363–79. doi: 10.1016/j.neuron.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson NO, Owens JA, Fullston T, Lane M. Preconception diet or exercise intervention in obese fathers normalizes sperm microRNA profile and metabolic syndrome in female offspring. Am J Physiol Endocrinol Metab. 2015;308(9):E805–21. doi: 10.1152/ajpendo.00013.2015. [DOI] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13(6):664–6. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53(6):857–69. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno M, Yamada K, Olariu A, Nawa H, Nabeshima T. Involvement of brain-derived neurotrophic factor in spatial memory formation and maintenance in a radial arm maze test in rats. J Neurosci. 2000;20(18):7116–21. doi: 10.1523/JNEUROSCI.20-18-07116.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moguel-González M, Gómez-Palacio-Schjetnan A, Escobar ML. BDNF reverses the CTA memory deficits produced by inhibition of protein synthesis. Neurobiol Learn Mem. 2008;90(3):584–7. doi: 10.1016/j.nlm.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Molteni R, Ying Z, Gómez-Pinilla F. Differential effects of acute and chronic exercise on plasticity-related genes in the rat hippocampus revealed by microarray. Eur J Neurosci. 2002;16(6):1107–16. doi: 10.1046/j.1460-9568.2002.02158.x. [DOI] [PubMed] [Google Scholar]
- Moretti P, Levenson JM, Battaglia F, Atkinson R, Teague R, Antalffy B, Armstrong D, Arancio O, Sweatt JD, Zoghbi HY. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J Neurosci. 2006;26(1):319–27. doi: 10.1523/JNEUROSCI.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse SJ, Butler AA, Davis RL, Soller IJ, Lubin FD. Environmental enrichment reverses histone methylation changes in the aged hippocampus and restores age-related memory deficits. Biology (Basel) 2015;4(2):298–313. doi: 10.3390/biology4020298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller AP, Cammarota M, Dietrich MO, Rotta LN, Portela LV, Souza DO, Izquierdo I, Bevilaqua LR, Perry ML. Different effect of high fat diet and physical exercise in the hippocampal signaling. Neurochem Res. 2008;33(5):880–5. doi: 10.1007/s11064-007-9530-7. [DOI] [PubMed] [Google Scholar]
- Métivier R, Gallais R, Tiffoche C, Le Péron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452(7183):45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- Nag N, Moriuchi JM, Peitzman CG, Ward BC, Kolodny NH, Berger-Sweeney JE. Environmental enrichment alters locomotor behaviour and ventricular volumein Mecp2 1lox mice. Behav Brain Res. 2009;196(1):44–8. doi: 10.1016/j.bbr.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Nagahara AH, Merrill DA, Coppola G, Tsukada S, Schroeder BE, Shaked GM, Wang L, Blesch A, Kim A, Conner JM, Rockenstein E, Chao MV, Koo EH, Geschwind D, Masliah E, Chiba AA, Tuszynski MH. Neuroprotective effects of brain-derived neurotrophic factor in rodent and primate models of Alzheimer’s disease. Nat Med. 2009;15(3):331–7. doi: 10.1038/nm.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393(6683):386, 9. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Neidl R, Schneider A, Bousiges O, Majchrzak M, Barbelivien A, de Vasconcelos AP, Dorgans K, Doussau F, Loeffler JP, Cassel JC, Boutillier AL. Late-Life Environmental Enrichment Induces Acetylation Events and Nuclear Factor κB-Dependent Regulations in the Hippocampus of Aged Rats Showing Improved Plasticity and Learning. J Neurosci. 2016;36(15):4351–61. doi: 10.1523/JNEUROSCI.3239-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen PV, Woo NH. Regulation of hippocampal synaptic plasticity by cyclic AMP-dependent protein kinases. Prog Neurobiol. 2003;71(6):401–37. doi: 10.1016/j.pneurobio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Niehrs C, Schäfer A. Active DNA demethylation by Gadd45 and DNA repair. Trends Cell Biol. 2012;22(4):220–7. doi: 10.1016/j.tcb.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Nielsen S, Åkerström T, Rinnov A, Yfanti C, Scheele C, Pedersen BK, Laye MJ. The miRNA plasma signature in response to acute aerobic exercise and endurance training. PLoS One. 2014;9(2):e87308. doi: 10.1371/journal.pone.0087308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455(7211):411–5. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Palomer E, Carretero J, Benvegnù S, Dotti CG, Martin MG. Neuronal activity controls Bdnf expression via Polycomb de-repression and CREB/CBP/JMJD3 activation in mature neurons. Nat Commun. 2016;7:11081. doi: 10.1038/ncomms11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan-Vazquez A, Rye N, Ameri M, McSparron B, Smallwood G, Bickerdyke J, Rathbone A, Dajas-Bailador F, Toledo-Rodriguez M. Impact of voluntary exercise and housing conditions on hippocampal glucocorticoid receptor, miR-124 and anxiety. Mol Brain. 2015;8:40. doi: 10.1186/s13041-015-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixinho-Pena LF, Fernandes J, de Almeida AA, Novaes Gomes FG, Cassilhas R, Venancio DP, de Mello MT, Scorza FA, Cavalheiro EA, Arida RM. A strength exercise program in rats with epilepsy is protective against seizures. Epilepsy & Behavior. 2012;25(3):323–328. doi: 10.1016/j.yebeh.2012.08.011. [DOI] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, Salinas-Riester G, Dettenhofer M, Kang H, Farinelli L, Chen W, Fischer A. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328(5979):753–6. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- Penner MR, Roth TL, Chawla MK, Hoang LT, Roth ED, Lubin FD, Sweatt JD, Worley PF, Barnes CA. Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol Aging. 2011;32(12):2198–210. doi: 10.1016/j.neurobiolaging.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Dasen JS, Kurokawa R, Wang Z, Korzus E, Rose DW, Glass CK, Rosenfeld MG. Factor-specific modulation of CREB-binding protein acetyltransferase activity. Proc Natl Acad Sci U S A. 1999;96(7):3652–7. doi: 10.1073/pnas.96.7.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, Tommerup N, van Ommen GJ, Goodman RH, Peters DJ. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376(6538):348–51. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- Pradhan S, Bacolla A, Wells RD, Roberts RJ. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J Biol Chem. 1999;274(46):33002–10. doi: 10.1074/jbc.274.46.33002. [DOI] [PubMed] [Google Scholar]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433(7024):434–8. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- Radom-Aizik S, Zaldivar F, Leu SY, Adams GR, Oliver S, Cooper DM. Effects of exercise on microRNA expression in young males peripheral blood mononuclear cells. Clin Transl Sci. 2012;5(1):32–8. doi: 10.1111/j.1752-8062.2011.00384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redell JB, Zhao J, Dash PK. Altered expression of miRNA-21 and its targets in the hippocampus after traumatic brain injury. J Neurosci Res. 2011;89(2):212–21. doi: 10.1002/jnr.22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rethorst CD, Trivedi MH. Evidence-based recommendations for the prescription of exercise for major depressive disorder. J Psychiatr Pract. 2013;19(3):204–12. doi: 10.1097/01.pra.0000430504.16952.3e. [DOI] [PubMed] [Google Scholar]
- Revilla S, Suñol C, García-Mesa Y, Giménez-Llort L, Sanfeliu C, Cristòfol R. Physical exercise improves synaptic dysfunction and recovers the loss of survival factors in 3xTg-AD mouse brain. Neuropharmacology. 2014;81:55–63. doi: 10.1016/j.neuropharm.2014.01.037. [DOI] [PubMed] [Google Scholar]
- Reynolds GO, Otto MW, Ellis TD, Cronin-Golomb A. The Therapeutic Potential of Exercise to Improve Mood, Cognition, and Sleep in Parkinson’s Disease. Mov Disord. 2016;31(1):23–38. doi: 10.1002/mds.26484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson AM, Bucci DJ. Physical exercise during pregnancy improves object recognition memory in adult offspring. Neuroscience. 2014;256:53–60. doi: 10.1016/j.neuroscience.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues GM, Toffoli LV, Manfredo MH, Francis-Oliveira J, Silva AS, Raquel HA, Martins-Pinge MC, Moreira EG, Fernandes KB, Pelosi GG, Gomes MV. Acute stress affects the global DNA methylation profile in rat brain: modulation by physical exercise. Behav Brain Res. 2015;279:123–8. doi: 10.1016/j.bbr.2014.11.023. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902–10. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saab BJ, Mansuy IM. Neuroepigenetics of memory formation and impairment: the role of microRNAs. Neuropharmacology. 2014;80:61–9. doi: 10.1016/j.neuropharm.2014.01.026. [DOI] [PubMed] [Google Scholar]
- Sayed D, He M, Hong C, Gao S, Rane S, Yang Z, Abdellatif M. MicroRNA-21 is a downstream effector of AKT that mediates its antiapoptotic effects via suppression of Fas ligand. J Biol Chem. 2010;285(26):20281–90. doi: 10.1074/jbc.M110.109207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10(12):842–9. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439(7074):283–9. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7(2):104–12. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Scott HL, Tamagnini F, Narduzzo KE, Howarth JL, Lee YB, Wong LF, Brown MW, Warburton EC, Bashir ZI, Uney JB. MicroRNA-132 regulates recognition memory and synaptic plasticity in the perirhinal cortex. Eur J Neurosci. 2012;36(7):2941–8. doi: 10.1111/j.1460-9568.2012.08220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1….. Biochim Biophys Acta. 1997;1332(2):F49–66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- Shin S, Janknecht R. Diversity within the JMJD2 histone demethylase family. Biochem Biophys Res Commun. 2007;353(4):973–7. doi: 10.1016/j.bbrc.2006.12.147. [DOI] [PubMed] [Google Scholar]
- Short AK, Yeshurun S, Powell R, Perreau VM, Fox A, Kim JH, Pang TY, Hannan AJ. Exercise alters mouse sperm small noncoding RNAs and induces a transgenerational modification of male offspring conditioned fear and anxiety. Transl Psychiatry. 2017;7(5):e1114. doi: 10.1038/tp.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu HF, Yang T, Yu SX, Huang HD, Jiang LL, Gu JW, Kuang YQ. Aerobic exercise for Parkinson’s disease: a systematic review and meta-analysis of randomized controlled trials. PLoS One. 2014;9(7):e100503. doi: 10.1371/journal.pone.0100503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley BA, Etnier JL. The Relationship between Physical Activity and Cognition in Children: A Meta-Analysis. Pediatric Exercise Science. 2003;15(3):243–256. [Google Scholar]
- Siette J, Reichelt AC, Westbrook RF. A bout of voluntary running enhances context conditioned fear, its extinction, and its reconsolidation. Learn Mem. 2014;21(2):73–81. doi: 10.1101/lm.032557.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280(51):41789–92. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleiman SF, Henry J, Al-Haddad R, El Hayek L, Abou Haidar E, Stringer T, Ulja D, Karuppagounder SS, Holson EB, Ratan RR, Ninan I, Chao MV. Exercise promotes the expression of brain derived neurotrophic factor (BDNF) through the action of the ketone body β-hydroxybutyrate. Elife. 2016:5. doi: 10.7554/eLife.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindler C, Cechinel LR, Basso C, Moysés F, Bertoldi K, Roesler R, Lovatel GA, Rostirola Elsner V, Siqueira IR. Treadmill exercise alters histone acetyltransferases and histone deacetylases activities in frontal cortices from wistar rats. Cell Mol Neurobiol. 2014;34(8):1097–101. doi: 10.1007/s10571-014-0096-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner JL, Murphy EA, McClellan JL, Carmichael MD, Davis JM. Exercise training increases mitochondrial biogenesis in the brain. J Appl Physiol (1985) 2011;111(4):1066–71. doi: 10.1152/japplphysiol.00343.2011. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Su W, Aloi MS, Garden GA. MicroRNAs mediating CNS inflammation: Small regulators with powerful potential. Brain Behav Immun. 2016;52:1–8. doi: 10.1016/j.bbi.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui L, Wang Y, Ju LH, Chen M. Epigenetic regulation of reelin and brain-derived neurotrophic factor genes in long-term potentiation in rat medial prefrontal cortex. Neurobiol Learn Mem. 2012;97(4):425–40. doi: 10.1016/j.nlm.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Szyf M. Nongenetic inheritance and transgenerational epigenetics. Trends Mol Med. 2015;21(2):134–44. doi: 10.1016/j.molmed.2014.12.004. [DOI] [PubMed] [Google Scholar]
- Sølvsten CA, de Paoli F, Christensen JH, Nielsen AL. Voluntary Physical Exercise Induces Expression and Epigenetic Remodeling of VegfA in the Rat Hippocampus. Mol Neurobiol. 2016 doi: 10.1007/s12035-016-0344-y. [DOI] [PubMed] [Google Scholar]
- Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, Nakao M, Taga T. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1(6):749–58. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- Tang B, Jia H, Kast RJ, Thomas EA. Epigenetic changes at gene promoters in response to immune activation in utero. Brain Behav Immun. 2013;30:168–75. doi: 10.1016/j.bbi.2013.01.086. [DOI] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Thiagalingam S, Cheng KH, Lee HJ, Mineva N, Thiagalingam A, Ponte JF. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- Tian F, Marini AM, Lipsky RH. Effects of histone deacetylase inhibitor Trichostatin A on epigenetic changes and transcriptional activation of Bdnf promoter 1 by rat hippocampal neurons. Ann N Y Acad Sci. 2010;1199:186–93. doi: 10.1111/j.1749-6632.2009.05175.x. [DOI] [PubMed] [Google Scholar]
- Tognini P, Pizzorusso T. MicroRNA212/132 family: molecular transducer of neuronal function and plasticity. Int J Biochem Cell Biol. 2012;44(1):6–10. doi: 10.1016/j.biocel.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Turchinovich A, Burwinkel B. Distinct AGO1 and AGO2 associated miRNA profiles in human cells and blood plasma. RNA Biol. 2012;9(8):1066–75. doi: 10.4161/rna.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- van Praag H. Neurogenesis and exercise: past and future directions. Neuromolecular Med. 2008;10(2):128–40. doi: 10.1007/s12017-008-8028-z. [DOI] [PubMed] [Google Scholar]
- van Praag H, Christie BR, Sejnowski TJ, Gage FH. Running enhances neurogenesis, learning, and long-term potentiation in mice. Proc Natl Acad Sci U S A. 1999a;96(23):13427–31. doi: 10.1073/pnas.96.23.13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Praag H, Kempermann G, Gage FH. Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat Neurosci. 1999b;2(3):266–70. doi: 10.1038/6368. [DOI] [PubMed] [Google Scholar]
- Vaynman S, Ying Z, Gomez-Pinilla F. Interplay between brain-derived neurotrophic factor and signal transduction modulators in the regulation of the effects of exercise on synaptic-plasticity. Neuroscience. 2003;122(3):647–57. doi: 10.1016/j.neuroscience.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Vaynman S, Ying Z, Gomez-Pinilla F. Hippocampal BDNF mediates the efficacy of exercise on synaptic plasticity and cognition. Eur J Neurosci. 2004;20(10):2580–90. doi: 10.1111/j.1460-9568.2004.03720.x. [DOI] [PubMed] [Google Scholar]
- Vaynman S, Ying Z, Gomez-Pinilla F. The select action of hippocampal calcium calmodulin protein kinase II in mediating exercise-enhanced cognitive function. Neuroscience. 2007;144(3):825–33. doi: 10.1016/j.neuroscience.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, Wood MA. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J Neurosci. 2007;27(23):6128–40. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423–33. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. An introduction to modern genetics. G. Allen & Unwin ltd.; 1939. [Google Scholar]
- Wang RY, Phang RZ, Hsu PH, Wang WH, Huang HT, Liu IY. In vivo knockdown of hippocampal miR-132 expression impairs memory acquisition of trace fear conditioning. Hippocampus. 2013;23(7):625–33. doi: 10.1002/hipo.22123. [DOI] [PubMed] [Google Scholar]
- Wang W, Kwon EJ, Tsai LH. MicroRNAs in learning, memory, and neurological diseases. Learn Mem. 2012;19(9):359–68. doi: 10.1101/lm.026492.112. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105(26):9093–8. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissgerber TL, Wolfe LA, Davies GA, Mottola MF. Exercise in the prevention and treatment of maternal-fetal disease: a review of the literature. Appl Physiol Nutr Metab. 2006;31(6):661–74. doi: 10.1139/h06-060. [DOI] [PubMed] [Google Scholar]
- Wolfe LA, Brenner IK, Mottola MF. Maternal exercise, fetal well-being and pregnancy outcome. Exerc Sport Sci Rev. 1994;22:145–94. [PubMed] [Google Scholar]
- Xia Z, Storm DR. Role of signal transduction crosstalk between adenylyl cyclase and MAP kinase in hippocampus-dependent memory. Learn Mem. 2012;19(9):369–74. doi: 10.1101/lm.027128.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Liu Q, Yao J, Dai Y, Wang H, Xiao J. Circulating microRNAs in response to exercise. Scand J Med Sci Sports. 2015;25(2):e149–54. doi: 10.1111/sms.12421. [DOI] [PubMed] [Google Scholar]
- Yaffe K, Fiocco AJ, Lindquist K, Vittinghoff E, Simonsick EM, Newman AB, Satterfield S, Rosano C, Rubin SM, Ayonayon HN, Harris TB, Study HA. Predictors of maintaining cognitive function in older adults: the Health ABC study. Neurology. 2009;72(23):2029–35. doi: 10.1212/WNL.0b013e3181a92c36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeshurun S, Short AK, Bredy TW, Pang TY, Hannan AJ. Paternal environmental enrichment transgenerationally alters affective behavioral and neuroendocrine phenotypes. Psychoneuroendocrinology. 2017;77:225–235. doi: 10.1016/j.psyneuen.2016.11.013. [DOI] [PubMed] [Google Scholar]
- Yin MM, Wang W, Sun J, Liu S, Liu XL, Niu YM, Yuan HR, Yang FY, Fu L. Paternal treadmill exercise enhances spatial learning and memory related to hippocampus among male offspring. Behav Brain Res. 2013;253:297–304. doi: 10.1016/j.bbr.2013.07.040. [DOI] [PubMed] [Google Scholar]
- Zacchigna S, Lambrechts D, Carmeliet P. Neurovascular signalling defects in neurodegeneration. Nat Rev Neurosci. 2008;9(3):169–81. doi: 10.1038/nrn2336. [DOI] [PubMed] [Google Scholar]
- Zagaar M, Alhaider I, Dao A, Levine A, Alkarawi A, Alzubaidy M, Alkadhi K. The beneficial effects of regular exercise on cognition in REM sleep deprivation: behavioral, electrophysiological and molecular evidence. Neurobiol Dis. 2012;45(3):1153–62. doi: 10.1016/j.nbd.2011.12.039. [DOI] [PubMed] [Google Scholar]
- Zajac MS, Pang TY, Wong N, Weinrich B, Leang LS, Craig JM, Saffery R, Hannan AJ. Wheel running and environmental enrichment differentially modify exon-specific BDNF expression in the hippocampus of wild-type and pre-motor symptomatic male and female Huntington's disease mice. Hippocampus. 2010;20(5):621–36. doi: 10.1002/hipo.20658. [DOI] [PubMed] [Google Scholar]
- Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Köppel T, Jahantigh MN, Lutgens E, Wang S, Olson EN, Schober A, Weber C. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2(100):ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- Zhong T, Ren F, Huang CS, Zou WY, Yang Y, Pan YD, Sun B, Wang E, Guo QL. Swimming exercise ameliorates neurocognitive impairment induced by neonatal exposure to isoflurane and enhances hippocampal histone acetylation in mice. Neuroscience. 2016;316:378–88. doi: 10.1016/j.neuroscience.2015.12.049. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, Hu L, Steen JA, Weitz CJ, Greenberg ME. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52(2):255–69. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zocchi L, Sassone-Corsi P. SIRT1-mediated deacetylation of MeCP2 contributes to BDNF expression. Epigenetics. 2012;7(7):695–700. doi: 10.4161/epi.20733. [DOI] [PMC free article] [PubMed] [Google Scholar]