Abstract
Presently three major groups of proteins from Aedes aegypti, cadherin, alkaline phosphatases (ALP) and aminopeptidases N (APN), have been identified as Cry11Aa toxin receptors. To further characterize their role on toxicity, transgenic mosquitoes with silenced Aedes cadherin expression were previously generated and the role of cadherin in mediating the toxicity of four different mosquitocidal toxins (Cry11Aa, Cry11Ba, Cry4Aa and Cry4Ba) was demonstrated. Here, we investigated the role of another reported Cry11Aa receptor, ALP1. As with Aedes cadherin, this protein is localized in the apical cell membrane of distal and proximal gastric caecae and the posterior midgut. We also successfully generated transgenic mosquitoes that knockdowned ALP1 transcript levels using an inducible Aedes heat shock promoter, Hsp70A driving dsALP1RNA. Four different mosquitocidal toxins were used for larval bioassays against this transgenic mosquito. Bioassay results show that Cry11Aa toxicity to these transgenic larvae following a heat shock decreased (4.4 fold) and Cry11Ba toxicity is slightly attenuated. But Cry4Aa and Cry4Ba toxicity to ALP1 silenced larvae is unchanged. Without heat shock, toxicity of all four toxins does not change, suggesting this heat shock promoter is heat-inducible. Notably, transgenic mosquitoes with ALP1 knockdown are about 3.7 times less resistant to Cry11Aa toxin than those with Aedes cadherin knockdown. These results demonstrate that the ALP1 is an important secondary receptor for Cry11Aa and Cry11Ba, but it might not be involved in Cry4Aa and Cry4Ba toxicity.
Keywords: Aedes aegypti, alkaline phosphatase, Bacillus thuringiensis, receptor, function, toxin
Introduction
Aedes aegypti is an important vector for transmitting several tropical fevers virus, including yellow fever, dengue and Zika. The control of this important disease vector is primarily by chemical insecticides to which increased resistance has been observed in many countries (Devillers et al. 2014). But increasing formulations of Bacillus thuringiensis subsp. Israelensis (Bti) are being used as an indispensible weapon for biological control of Aedes mosquitoes. During the sporulation phase, this bacterium produces crystalline inclusions that have a number of proteins including Cry4Aaa, Cry4Baa, Cry11Aaa and Cyt1Aa proteins (Berry et al. 2002). Among them, Cry11Aaa is one of the more active toxins against Ae. aegypti larvae (Chilcott and Ellar, 1988). Also this study evaluates another more toxic Cry toxin, Cry11Ba from B. thuringiensis subsp. jegathesan (Btj), which shares 58% amino acid identity with Cry11Aa (Delecluse et al. 1995; Kawalek et al. 1995).
In lepidopteran insects, four major Cry1A toxin protein receptors have been identified – cadherins, ABCC transporters, aminopeptidases (APNs) and alkaline phosphatases (ALPs) (Atsumi et al. 2012; Bravo et al. 2011; Gahan et al. 2010; Pardo-Lopez et al. 2012; Pigott and Ellar 2007; Soberon et al. 2009; Tay et al. 2015). APN and ALP are thought to be receptors that facilitate localization of toxin monomers to midgut microvilli before toxin interaction with cadherin. The activated Cry toxins then bind to the cadherin receptor in the microvilli, resulting in further toxin cleavage and triggering toxin oligomerization. The toxin oligomers then bind to GPI-anchored receptors, APN and/or ALP, leading to membrane insertion and pore formation (Arenas et al. 2010; Bravo et al. 2007; Soberon et al. 2007; Soberon et al. 2009). It appears that the ABCC2 transporter also binds Cry1A toxins and facilitates the formation of stable pores (Tanaka et al., 2016). This membrane insertion and pore formation by the toxin causes midgut cells lysis, which ultimately results in killing the larval insects (Soberon et al. 2007; Soberon et al. 2009). In an alternative model, the cadherin alone initiates an intracellular cascade that leads to cell toxicity (Zhang et al. 2005; Zhang et al. 2006).
ALPs from Heliothis virescens and Manduca sexta were first identified as Cry1A toxin-binding proteins (Jurat-Fuentes and Adang 2004; McNall and Adang 2003; Sangadala et al. 1994). Cry1Ac toxin binding with ALPs appears to be dependent on the presence of GalNac moieties (Jurat-Fuentes and Adang 2004; McNall and Adang 2003). More importantly, reduced levels of membrane-bound alkaline phosphatase have been noted in lepidopteran strains resistant to Cry toxins, suggesting a functional role for ALPs in toxicity (Jurat-Fuentes and Adang 2004; Jurat-Fuentes et al. 2011). Recently a midgut membrane-bound ALP from Plutella xylostella, together with ABCC2 and ABCC3, was shown to be associated with recessive Cry1Ac resistance and their down-regulation was apparently mediated by the mitogen-activated protein kinase (MAPK) signaling pathway (Guo et al. 2015). Moreover, expression of this ALP in Sf9 cells makes them susceptible to Cry1Ac toxin and RNAi knockdown of this ALP in P. xylostella highly reduces Cry1Ac toxicity, strongly suggesting the PxALP is a functional receptor in P. xylostella (Guo et al. 2015). In addition, a toxin-binding alkaline phosphatase fragment synergizes Cry1Ac toxin against susceptible and resistant Helicoverpa armigera (Chen et al. 2015).
Recent evidence suggests mosquitocidal Cry toxins also bind ALP proteins in the mosquito midguts, and toxin interaction with ALPs has been observed (Likitvivatanavong et al. 2011). For example, an ALP from Anopheles gambiae binds Cry11Baa (Hua et al. 2009) and two different ALPs from Ae. aegypti bind Cry4Aa and Cry4Ba, respectively (Dechklar et al. 2010; Stalinski et al. 2016a; Thammasittirong et al. 2011). Also, this Cry4Ba-binding ALP from Ae. aegypti, and a number of other GPI-anchored proteins, are localized in cell membrane lipid rafts (Bayyareddy et al. 2012). Moreover, Sf9 cells expressing this ALP protein become susceptible to the Cry4Ba toxin (Dechklar et al. 2010). Another ALP from Ae. Aegypti was demonstrated to bind Cry4Ba, Cry11Aaa and Cry11Ba (Fernandez et al. 2009; Jimenez et al. 2012; Likitvivatanavong et al. 2010). More significantly, ALPs play a role in Ae. aegypti mosquito resistance to Cry toxins. For example, Lee et al (2014) reported that a 40% reduction of ALPs expression in Aedes larval midgut is associated with Aedes mosquitoes resistant to Cry11Aa (Lee et al. 2014). Similarly, Bti or individual toxin (eg. Cry4Aa, Cry4Ba and Cry11Aa) resistant mosquitoes exhibited a higher level of transcription of ALP genes than the susceptible strain, even though these mosquitoes showed a lower level of total ALP activity (Stalinski et al. 2016a; Stalinski et al. 2016b). Taken together, ALPs from mosquitoes also appear to be functional toxin receptors (Likitvivatanavong et al. 2011).
Previously we reported Ae. aegypti ALP1 (AaeALP1) functions as a Cry11Aaa and Cry4Baa receptor (Fernandez et al. 2009; Jimenez et al. 2012). In this study we show its immunolocalization in larval midgut and its expression in Sf9 cells. To test its role on the toxicity for all mosquitocidal Cry toxins, we also generated a transgenic Aedes mosquito strain, which facilitates AaeALP1 transcript level knockdown.
Materials and methods
Preparation of mosquitocidal Cry toxins
B. thuringiensis strains producing Cry4Aaa, Cry4Baa, Cry11Aaa, or Cry11Baa (Chang et al. 1993; Delecluse et al. 1995), were grown in nutrient broth sporulation medium containing erythromycin (12.5 μg/ml) at 30°C for 4–5 days. For crude Cry toxin production, spores and crystal inclusions were harvested 10,000 × g for 10 min at 4 °C, washed three times with sterilized water, and stored in water at −80°C until used.
Alp1 protein antibody and peptide antibody preparation
For the ALP1 protein antibody, the N-terminally truncated ALP1 protein was expressed in the expression vector, pQE30 (Qiagen, Hilden, Germany). The truncated ALP1 protein was purified by Ni-affinity columns, then separated in SDS-PAGE gel and the gel with the purified band was excised and sent for antibody preparation. To prepare a more specific anti-ALP1 antibody, the 15 amino acids peptide, CPDRDGDDGNLNRKK, was synthesized by Genscript (Nanjing, Jiangsu, China) and then conjugated with keyhole limpet hemocyanin via the N-terminal Cys. Both antibodies were prepared in rabbits by Robert Sargeant Inc. (Pomona, CA, USA) following commonly used protocols (Sambrook et al. 1989).
Immunolocalization of AaeALP1 in larval midgut of Ae. aegypti
Late fourth instar larvae and isolated guts of Ae. aegypti were fixed overnight in 4% paraformaldehyde (PFA) at 4°C. After fixation, 8 mm thick sections were cut and analyzed as described (Chen et al. 2009). Then these sections were incubated with protein A-purified rabbit polyclonal anti-AaeALP1 antibody (1:100 dilution) in 1% bovine serum albumin (BSA)/PBST overnight at 40°C. After washing with PBST containing 2% goat serum (GS) and 0.1% BSA, sections were incubated with secondary antibodies – Cy3-conjugated goat anti-rabbit IgG (1:1000 dilution) and Alexa-488 for actin F (1:100 dilution) – both diluted in 0.1% BSA/2% GS/PBST. After incubation in the dark at room temperature for 40 min, tissues were again washed and mounted in Shur/Mount media (Electron Microscopy Sciences, Hatfield, PA). Images were obtained using a Zeiss Axioplan laser-scanning confocal microscope (LSM 510) located in the Institute of Integrative Genome Biology (IIGB) at UC, Riverside. All images were imported into Adobe Photoshop for assembly and annotation.
Expression of AaeALP1 in Sf9 cells
The full-length mCherry ORF gene was first cloned into the baculovirus expression vector, pBac4x (Merck Millipore, Darmstadt, Germany). Next the AaeALP1 ORF was subcloned into the vector, pBac4x-mCherry. These resultant transfer vectors DNA, pBac-4x-mCherry or pBac-4x-mCherry-ALP1, together with pBacMagic 3 DNA, was co-transferred into Sf9 cells seeded 1h before use. Sf9 cells were then grown and maintained in Sf-900™ II Serum Free Medium (ThermoFisher Scientific, Waltham, MA) with penicillin and streptomycin at 27°C. After 5 days incubation the recombinant baculovirus was harvested and then amplified in Sf9 cells for three times. Finally the P3 virus stocks titer was calculated and used for protein expression and cytotoxicity test.
Detection of AaeALP1 expression in Sf9 cells by Western blot
Cells expressing mCherry or AaeALP1 were harvested, washed with phosphate buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) twice and resuspended in SDS-PAGE sample buffer. The collected cells were boiled for 10 min and centrifuged at 10,000 × g for 10 min to remove insoluble material. The supernatants were loaded in SDS polyacrylamide gel (10%), and then electrotransferred to a nitrocellulose membrane. The membranes were treated with blocking solution (PBS, 5% skim milk and 0.1% Tween-20) for 1 h at room temperature, and washed with PBST (PBS and 0.1% Tween-20) three times. The blocked membranes were incubated with protein A-purified anti-AaeALP1 antibody (1: 100) overnight at 4 °C. The membranes were then washed with PBST, and subsequently incubated with anti-rabbit horseradish peroxidase (HRP, 1:5,000) secondary antibody (Sigma, St. Louis, MO) for 90 min at room temperature. After washing with PBST, HRP reactivity was revealed with a luminal substrate (ThermoFisher Scientific, Waltham, MA) and exposed to an X-ray film in a darkroom.
Construction of transformation vector and transgenic mosquitoes
The pBAC[3xP3-EGFP afm] transformation vector containing 3xP3 (eye-specific promoter), TATA box, and enhanced GFP (Horn and Wimmer 2000) and the phsp-pBac helper plasmid (Handler and Harrell 1999) were obtained from Drs. Alexander Raikhel and Vladimir Kokoza (Department of Entomology, University of California, Riverside, CA). To obtain the 3′-UTR sequences of AaeALP1 gene for making hairpin dsRNA, 3′-RACE was done and the PCR product was cloned into the TA vector, pCR2.1 Topo, and eventually sequenced. The hairpin dsRNA construct of AaeALP1 was made by using a heat-inducible Aedes heat shock promoter 70A (Gross et al. 2009), an AaeALP invert repeat with an APN intron sequence as a stem loop (140 bp, AAEL008155), and SV40 terminator (250 bp). The synthesized dsRNA construct was then cloned into pBAC[3xP3-EGFP afm] transformation vector. The mixture of pBAC[3xP3-EGFP afm, AaeALP1dsRNA] and phsp-pBac helper plasmid were injected into Ae. aegypti embryos (Insect Transformation Facility, University of Maryland, Rockville, MD). G0 eggs were hatched and reared with a mixture of dog food and yeast (3:1) in dechlorinated tap water at 29 °C, 8:12 h light:dark. Adult G0 were back crossed with wild-type females or males and G1 progeny were screened for green-fluorescent eyes using fluorescence microscopy (Nikon SMZ1500). Homozygous larvae were obtained by subsequently mating individual male and female showing green-fluorescent eyes until all progeny larvae contained green-fluorescent eyes. To express the hairpin dsRNA of AaeALP1 regulated by the Aedes heat shock promoter 70A, 3rd instar wild-type transgenic larvae were incubated at 37 °C for 1 h.
Bioassay
Bioassays were performed with crude Cry4Aaa, Cry4Baa, Cry11Aaa, and Cry11Baa toxins. In brief, after 2nd instar larvae were heat-shocked at 37°C for 1h, twenty early fourth-instar larvae were transferred to plastic cups containing 200 ml tap water, then fed Cry toxins at different concentrations and mortality was observed at 24 h. Bioassays were performed three times and the mortality values were calculated by Probit (EPA) and the data was plotted by Origin program (Origin Lab, Northampton, MA).
Inverse PCR
Inverse PCR was performed according to the previously described but slightly modified method (Handler et al. 1998; Kokoza et al. 2001). First, genomic DNA (5 μg) from transgenic and wild-type mosquitoes was digested by HpaII for either left arm or right arm inverse PCR. Then the digested fragments were circularized by ligation and the circularized fragments were used for nested PCR with primers specific to the left or right arm of the piggyBac transposon. The final PCR product was cloned into a TA cloning vector, pCR2.1-Topo and sequenced. Sequence analysis was done by Lasergene software (DNASTAR, Madison, WI) and BlastN search at NCBI.
Quantitative real-time PCR (qPCR)
Total RNA was extracted from wild type or transgenic larvae using TRIzol. cDNA was synthesized from total RNA with SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA) and 1.5 μl cDNA were used as template for qPCR. Primers specific to AaeALP1 (AAEL009077) and the reference genes actin (AAEL011197) or 40S ribosomal protein S7 (AAEL009496), were designed to have similar properties in terms of nucleotide length and %GC content. PCR conditions, including the template cDNA, primer concentrations and annealing temperatures, were adjusted for amplification efficiencies (efficiency 90–110%) for all genes. Optimized PCR master mix (25 μl) contained the following components: 12.5 μL iQSYBRGreensupermix (Bio-Rad, Irvine, CA), 2.0 μL cDNA, 0.5 μL 10 μM of each primer, and 10 μL water. The qPCR was performed using CFX Connect real-time PCR instrument (Bio-Rad). Optimized thermal program consisted of: one cycle of 95°C/2 min and 40 cycles of 95°C/15 sec, 60°C/30 sec, and 72°C/30 sec, followed by a final extension of one cycle 72°C/5 min. Homogeneity of the PCR product was confirmed by melting curve analysis. To check for gDNA contamination in RNA preparation, the melting curve of 40S ribosomal protein S7 using primers that spanned a 114-bp intron was analyzed on every run. Quantification of the transcript levels or relative copy number of the genes was conducted according to the Pfaffl method (Pfaffl 2001). Quantitative PCR was performed three times using independently prepared larval midgut cDNA.
Results
Expression of AaeALP1 in larval midgut and Sf9 cells
Previously an anti-AaeALP1 protein polyclonal antibody used for the detection of BBMV proteins separated in 2-D gel, it recognized at least four proteins with similar molecular weights (65 kDa) but with different isoelectric points (Jiménez et al., 2012). Similarly, when this antibody was used for E. coli-expressed AaeALP1, AaeALP2 and AaeALP3 detection, it could detect AaeALP1 and AaeaALP3 (data not shown). Clearly this anti-AaeALP1 protein polyclonal antibody was non-specific. Since ALP family proteins from Ae. aegypti mosquitoes are highly similar, it is a challenge to develop specific protein antibodies. Due to non-specificity of the protein antibody, we developed a new anti-AaeALP1 polyclonal antibody using the peptide, CPDRDGDDGNLNRKK, which was conjugated with keyhole limpet hemocyanin via the N-terminal Cys. This peptide fragment shows no significant similarity with the other known 11ALPs in Ae. aegypti. When this new anti-AaeALP1 peptide antibody was used for Western blotting of E. coli-expressed AaeALP1, AaeALP2 and AaeALP3, it detected only AaeALP1, but not AaeALP2 and 3 (Fig. 1A). Also when the polyclonal anti-AaeALP1 peptide antibody was used for BBMV protein western blotting, only an expected single 65-kDa protein band was observed (Fig. 1B). Therefore, compared with a previous anti-AaeALP1 protein antibody (Jiménez et al., 2012), the specificity of this peptide antibody is much improved.
Fig. 1.
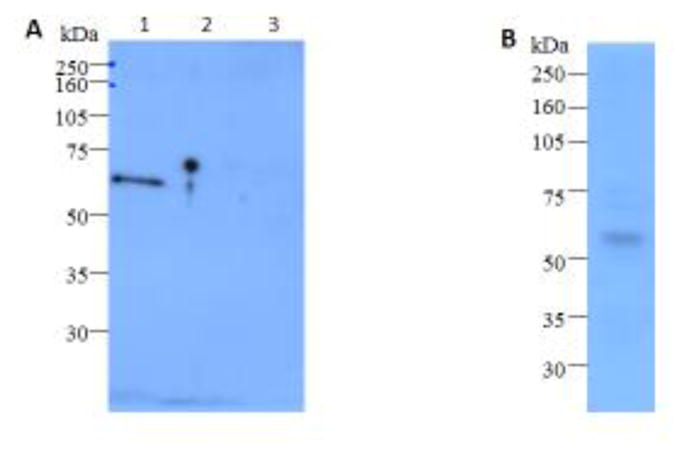
Western blot detection of AaeALP1 expression in E. coli and in BBMV.(A) AaeALP1 (lane 1), AaeALP2 (lane 2) and AaeALP3 (lane 3) were expressed in E. coli and protein-A purified peptide polyclonal anti-AaeALP1 antibody (1:100 dilution) was used for western blot assay. (B) Proteins from Ae. aegypti BBMV were separated by SDS-PAGE and electrotransferred to PVDF membrane. AaeALP1 proteins were detected using protein A-purified anti-AaeALP1 antibody (1:100).
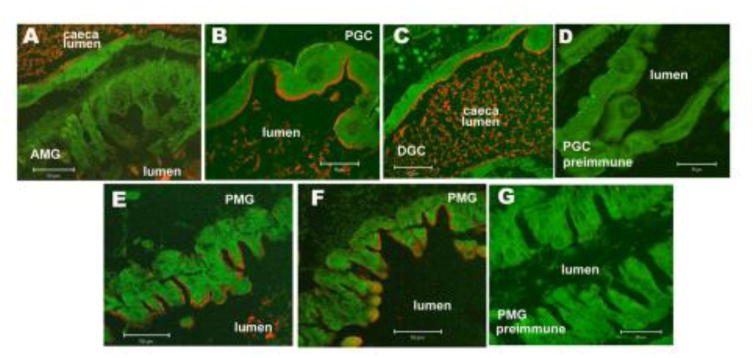
The peptide antibody mentioned above was then used for immunohistochemical detection of AaeALP1 in Aedes larval midgut. The images showed the AaeALP1 expression could be detected in the apical membrane of proximal (Fig. 2, Panel B) and distal gastric caecae (Fig. 2, Panel C), and the posterior midgut (Fig. 2, Panels E and F). No immunofluorescence was observed in the anterior midgut epithelium (Fig. 2, Panel A) or in control tissues what were incubated with preimmune serum at the same dilutions (1:100) as anti-AaeALP1 peptide antibody (Fig. 2, Panel D and G).
Fig. 2.
AaeALP1is expressed in the gastric caecae and posterior midgut of larval Ae. aegypti. Paraffin sections of fourth instar larval midgut were probed with anti-AaeALP1 peptide antibody. AaeALP1 is localized in the apical membrane of proximal (Panel B) and distal gastric caecae (Panel C) and posterior midgut (Panels E and F). No immunofluorescence was observed in the anterior midgut epithelium (Panel A). No immunoreactivity was detected in control tissues that were incubated with pre-immune serum at the same dilutions (1:100) as anti-AaeALP1 antibody (Panel D, gastric caecae; Panel G, posterior midgut). Scale bars, 50 mm.
To observe AaeALP1 function in insect cells, a mCherry gene was cloned into baculovirus expression vector, pBac4x. Red fluorescence was observed in transfected cells (Fig. 3, Panel A), suggesting mCherry successfully expresses in Sf9 cells. Then full-length AaeALP1 was subcloned into the vector, pBac4x-mCherry. AaeALP1 expression in Sf9 cells transfected with the resultant P3 virus was analyzed using the peptide antibody. Western blot results showed AaeALP1 protein was expressed in Sf9 cells containing pBac4x-mCherry-ALP1, but not in those with pBac4x-mCherry only (Fig. 3, lanes 2 and 1). However, AaeALP1 expressing Sf9 cells were not susceptible to either Cry11Aa or Cry4Ba toxin (data not shown).
Fig. 3.
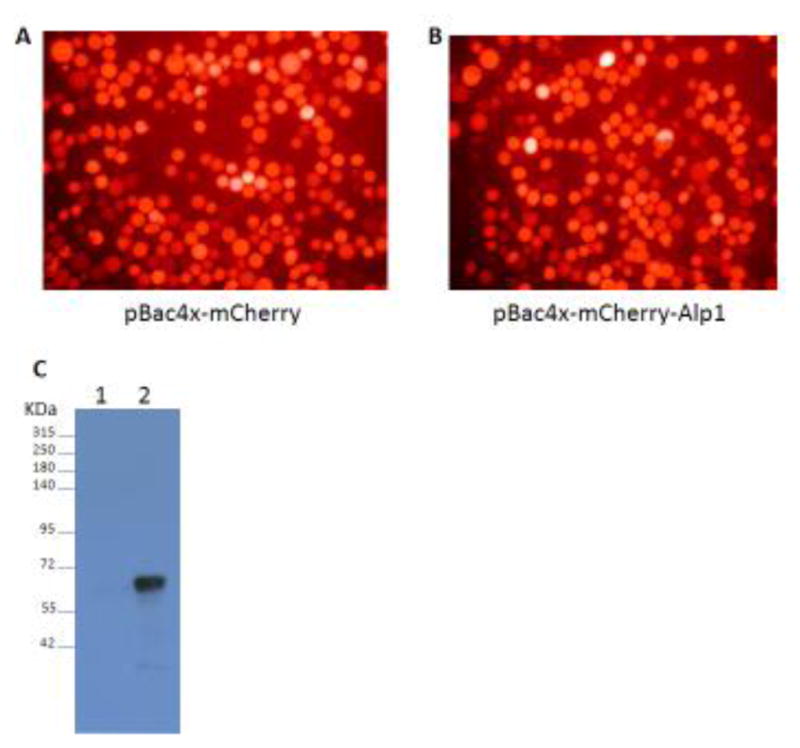
AaeALP1 expression in Sf9 cells using a baculovirus system. Sf9 cells containing the vector pBac4x with mCherry alone (A) or mCherry and AaeALP1 co-expression (B). These cells were photomicrographed at a total magnification of x 1000. Then AaeALP1 expression in Sf9 cells was detected by Western blot (C). Proteins from Sf9 cells containing expression vector pBac4x-mcherry (lane 1) and those with pBac4x-mcherry-ALP1 (lane 2) were separated by SDS-PAGE and electrotransferred to PVDF membrane. Proteins were detected using protein A-purified anti-AaeALP1 antibody (1:100).
Generation of transgenic mosquitoes with AaeALP1 gene knockdown
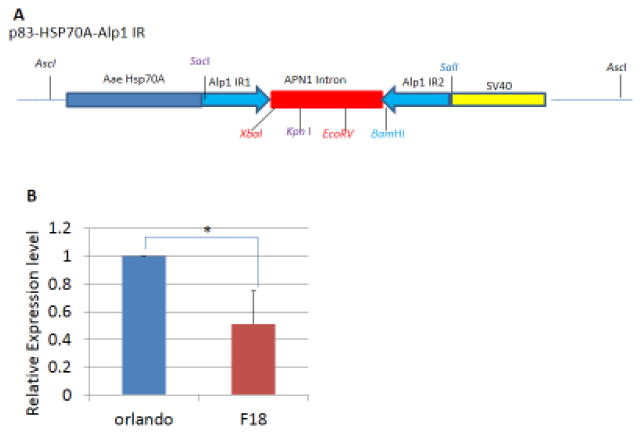
To investigate AaeALP1’s role in the toxicity of mosquitocidal Cry proteins, including Cry11Aa, Cry11Ba, Cry4Aa and Cry4Ba, we generated transgenic mosquitoes in which the AaeALP1 gene was silenced. To ensure specificity of the AaeALP1RNAi in vivo, the sequence for the hairpin dsRNA construct was obtained from the 3′ UTR of AaeALP1 gene. This sequence is quite distinct from that of the other ALPs. A highly heat-inducible heat shock 70A promoter (Gross et al. 2009) was used to drive the hairpin dsRNA expression in case Aedes mosquitoes are not tolerant to the AaeALP1 gene knockdown (Fig. 4A).
Fig. 4.
Vector construction for generating transgenic mosquitoes with AaeALP1 gene knockdown. (A) Construction of the mosquito transformation vector. The hairpin dsRNA construct of Aedes ALP1 was made with these components in order –heat-inducible Aedes heat shock 70A promoter, AaeALP1 inverted repeat (IR) left arm, APN intron (Lee et al. 2015), AaeALP1 IR right arm and SV40 terminator. This construct was cloned into pBAC[3xP3-EGFP afm] transformation vector containing 3xP3 (eye-specific promoter), TATA box, and enhanced GFP.(B) AaeALP1 gene transcript level detection by real time PCR. Orlando:a wild-type Ae. Aegypti mosquito strain used for embryo injections. F18: the stable transgenic mosquito strain with ALP1 invert repeat insert, that leads to ALP1 knockdown.
After homozygous transgenic mosquito lines were established, genomic PCR analysis was conducted to confirm stable incorporation of the transposon and the integrity of the AaeALP1 hairpin dsRNA constructs into the Ae. aegypti genome (data not shown). To analyze the AaeALP1dsRNA genomic insertion site, inverse PCR was performed with primers specific to the left and right arms of the piggyback transposon. Sequencing results showed that inverse PCR product sequence is nearly identical to the genomic contig aag2_ctg_30 (1292700-1293738). The transposon insert site is bp 1293493 of this contig. Since no physiological difference between wild-type mosquitoes and the transgenic mosquitoes was observed, we assume the dsRNA construct insertion does not cause any problem on mosquitoes physiologically. Moreover, no function has been ascribed to this gene fragment.
To ensure the AaeALP1dsRNA could successfully trigger RNAi in Aedes larval midgut we performed quantitative PCR using cDNA prepared from Aedes midgut tissues of transgenic and wild-type larvae after a heat shock. Figure 4B shows that the AaeALP1 transcript levels in transgenic mosquitoes decrease by about half compared with that of wild-type mosquitoes (Fig. 4B).
Bioassay against the transgenic mosquitoes with AaeALP1 gene knockdown
Although we previously reported (Jimenez et al. 2012) that AaeALP1 plays a role in the toxicity of Cry11Aa and Cry4Ba, its role in the toxicity of Cry4Aa and Cry11Ba was not explored. With AaeALP1dsRNA transgenic mosquitoes we were able to perform rigorous bioassays with all of these mosquitocidal Cry toxins. These bioassays, performed using heat-shocked transgenic and wild-type mosquitoes, showed that the transgenic mosquitoes are 4.4- and 1.8-fold more tolerant to Cry11Aa and Cry11Ba toxins, respectively, than wild-type mosquitoes (Fig 6). But the transgenic larvae showed no tolerance to Cry4Aa and Cry4Ba, suggesting that AaeALP1 is a functional receptor for Cry11Aa and Cry11Ba, but not for Cry4Aa and Cry4Ba toxins. In the absence of a heat shock AaeALP1 transgenic mosquitoes are almost as sensitive as wild-type mosquitoes to Cry11Aa and Cry11Ba toxins (Fig. 6), suggesting the ALP1 gene was not silenced without a heat shock. It further indicates this hsp70A promoter is heat sensitive, and therefore useful as an inducible promoter for modulating gene expression in vivo in Ae. aegypti.
Fig. 6.
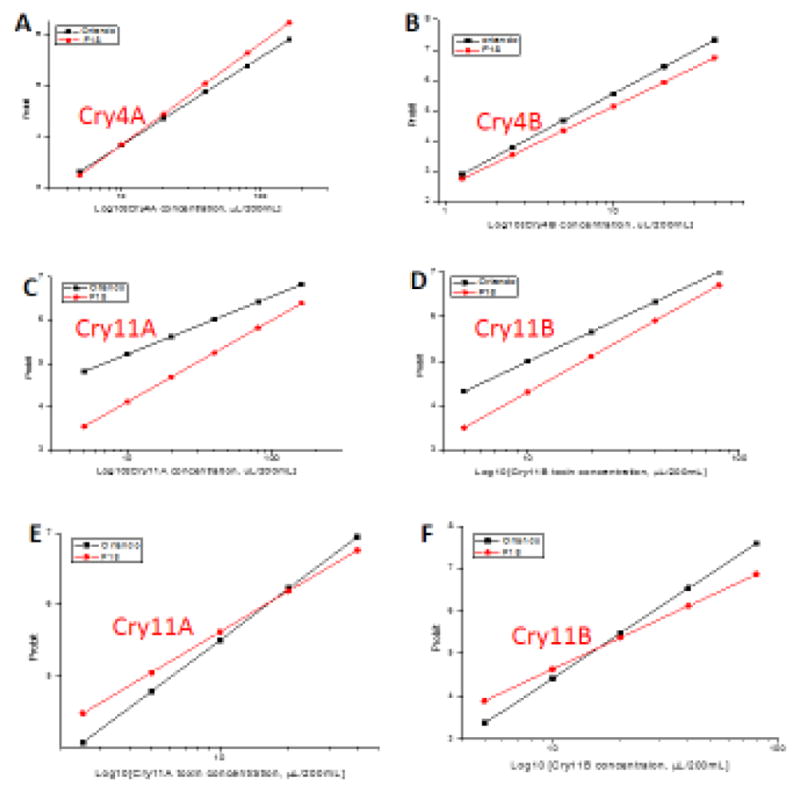
AaeALP1 knockdown mosquitoes show increased tolerance to Cry11Aa and Cry11Ba. Bioassay was performed with early 4th instar larvae after 39°C heat shock for 1 hr at the 2nd instar. Wild-type and homozygous transgenic mosquitoes larvae were exposed to Cry4Aa (A), Cry4Ba (B), Cry11Aa (C) and Cry11Ba (D) crude toxins. AaeALP1 knockdown mosquitoes without heat shock showed no difference in toxicity to Cry11Aa (E) and Cry11Ba (F) toxins. The bioassay data was analyzed by Probit and Origin software. Transgenic mosquitoes showed more tolerance to the Cry11Aa (by 4.4 fold) and Cry11Ba (by 1.8 fold) toxins, but not to Cry4Aa and Cry4Ba toxins.
Discussion
Previously we reported the Aedes cadherin, one of the Cry11Aa toxin receptors, is localized in the apical cell membrane of caecae and posterior midgut (Chen et al. 2009). Interestingly, we show here the AaeALP1 protein shares this same localization pattern (Fig. 2). Thus, co-localization of these two Cry11Aa toxin receptors is meaningful since both of them together are required for Cry11A a intoxication process according to the sequential binding model (Bravo et al. 2004). The anti-AaeALP1 peptide antibody used here, developed to a variable region, is more specific than that used previously (Jimenez et al. 2012). The antibody does not recognize other two similar ALPs (AAEL000931 and AAEL003289) (Fig. 1A). However, in Fig. 1B, there exist two additional faint bands other than a much stronger ALP1 band. Since glycosylation of membrane proteins often occurs, these bands could be glycosylated ALP1. But we cannot rule out the possibility that the peptide antibody could have weak cross-reaction with other proteins in BBMV. Importantly the expression pattern we observed is very similar to phage antibodies and Cry11Aa binding regions previously reported (Fernandez et al., 2005). Hence the expression pattern is very likely that of AaeALP1 and not that of a similar protein.
Unlike AaeALP1, however, AgALP1 in An. gambiae served as a Cry11Ba toxin receptor, and it is localized to the anterior and posterior midgut (Hua et al. 2009), although another Cry11Ba toxin receptor An. gambiae cadherin 2, is localized only in posterior midgut (Hua et al. 2012). However, the anti-AgALP1 antibody was developed against an expressed N-terminal 514 amino acid truncated AgALP1 protein (Hua et al. 2009). Since the ALP family proteins from An. gambiae also share high similarity, it is possible that the anti-AgALP1 antibody could cross-react with other ALPs in the anterior midgut of An. gambiae. Further, although Cry11A and Cry11B proteins share very high amino acid similarity, Cry11B shows little cross-resistance to Cry11A-resistant Ae. aegypti mosquitoes and hence these two toxins likely have different mechanisms of action (Lee et al. 2014).
We attempted to study the AaeALP1 function in Sf9 insect cells, but AaeALP1 expression by baculovirus system could not confer cell susceptibility to monomeric Cry11Aa toxin (data not shown). However, in the presence of another Aedes ALP (AAEL015070), the Sf9 cells undergo cell lysis after treatment with Cry4Ba toxin (Dechklar et al. 2010). It has been reported that Cry4Ba toxin, unlike Cry11Aa, can oligmerize spontaneously in the absence of the cadherin protein (Rodriguez-Almazan et al. 2012). Taken together, these results suggested as mentioned above that Cry11Aa toxin sequentially binds to cadherin and then to a GPI-anchored APN or ALP, meaning the oligomeric Cry11Aa toxin is potentially toxic to the insect cells expressed with AaeALP1.
Previously it was observed that AaeALP1dsRNA-fed Aedes mosquitoes show less than 2-fold and 4-fold more tolerance for Cry4Ba and Cry11Aa toxin, respectively, compared with wild-type mosquitoes (Jimenez et al. 2012). In this study, transgenic mosquitoes with AaeALP1 transcript knockdown showed similar 4.4 fold Cry11A a reduction in toxicity, but the transgenic mosquitoes were as sensitive as wild-type mosquitoes to Cry4Ba toxin (Fig. 6). Potentially the different bioassay methods used could contribute to this discrepancy in Cry4Ba toxicity between the two AaeALP1 knockdown mosquito lines. Here LC50 values were obtain using multiple toxin doses, but previously the mortality data was obtained from only a single toxin dose. Clearly, the bioassay method used here provides stronger statistical data. Moreover, since only marginal reduction in Cry4Ba toxicity was observed previously (Jimenez et al. 2012), it is not surprising that under more rigorous analysis we did not detect a difference in the role of AaeALP1 knockdown on Cry4Ba toxicity.
In addition to Cry4Ba and Cry11Aa, two other mosquitocidal toxins, Cry4Aa and Cry11Ba, were also used for transgenic mosquitoes bioassay. Interestingly, Cry4Aa toxicity, as with Cry4Ba, showed no correlation with AaeALP1 expression level changes, but Cry11Ba toxicity, as with Cry11Aa, was attenuated with decreased AaeALP1 expression in the larval midgut (Fig. 6). Because Cry4Aa and 4Ba and Cry11Aa and 11B belong to different Cry protein families, it is plausible that Cry4Aa and 4B and Cry11Aa and 11B highjack different ALP receptors from Aedes mosquitoes.
Compared with transgenic mosquitoes with AaeCad gene knockdown, the AaeALP1 gene knockdown mosquitoes are 3.7 times less resistant. In accordance with the sequential toxin-binding model, the Aedes cadherin works as a primary receptor, but AaeALP1 is one of many secondary receptors. Thus it is expected that knockdown of AaeALP1 affects Cry11Aa toxicity much less than knockdown of AaeCad. Apparently multiple receptors are involved in the mechanism of action for individual mosquitocidal Cry toxins. Bti resistance has not been observed in the field not only because of Cyt toxins, but also because known Cry toxins in Bti act by different mechanisms. Therefore, the mechanism of Bti action is much more complex than previously thought. Bti can be used for mosquito control for the foreseeable future. On the other hand, deep insight into mechanism of Bti toxin action could provide some novel strategies for transgenic Bt crops.
Fig. 5.
Nucleotide sequence of the inverse PCR product identifying the transposon site. The section (in yellow) represents the inserted fragment from piggyBac vector with the two arms ending with ttaa (underlined). The four nucleotides in grey show the HpaII restriction sites. This sequence is identical (99.7%) to genomic contig aag2_ctg_30 (1292700-1293738).
Highlights.
ALP1 is localized in distal and proximal gastric caecae and posterior midgut.
Transgenic mosquitoes with knockdowned ALP1 transcript levels were generated.
Cry11Aa toxicity to these transgenic larvae decreased by 4.4 fold.
ALP1 is an important secondary receptor for Cry11Aa and Cry11Ba.
Acknowledgments
This research was funded in part through grants from the National Institutes of Health, 1R01 AI066014 and the University of California Agricultural Experiment Station. The technical assistance of Maria Ramirezis gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arenas I, Bravo A, Soberon M, Gomez I. Role of alkaline phosphatase from Manduca sexta in the mechanism of action of Bacillus thuringiensis Cry1Ab toxin. J Biol Chem. 2010;285:12497–503. doi: 10.1074/jbc.M109.085266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S, Miyamoto K, Yamamoto K, Narukawa J, Kawai S, Sezutsu H, Kobayashi I, Uchino K, Tamura T, Mita K, Kadono-Okuda K, Wada S, Kanda K, Goldsmith MR, Noda H. Single amino acid mutation in an ATP-binding cassette transporter gene causes resistance to Bt toxin Cry1Ab in the silkworm, Bombyx mori. Proc Natl Acad Sci U S A. 2012;109:E1591–8. doi: 10.1073/pnas.1120698109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayyareddy K, Zhu X, Orlando R, Adang MJ. Proteome analysis of Cry4Ba toxin-interacting Aedes aegypti lipid rafts using geLC-MS/MS. J Proteome Res. 2012;11:5843–55. doi: 10.1021/pr3006167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry C, O’Neil S, Ben-Dov E, Jones AF, Murphy L, Quail MA, Holden MT, Harris D, Zaritsky A, Parkhill J. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. israelensis. Appl Environ Microbiol. 2002;68:5082–95. doi: 10.1128/AEM.68.10.5082-5095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A, Gill SS, Soberon M. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon. 2007;49:423–35. doi: 10.1016/j.toxicon.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A, Gomez I, Conde J, Munoz-Garay C, Sanchez J, Miranda R, Zhuang M, Gill SS, Soberon M. Oligomerization triggers binding of a Bacillus thuringiensis Cry1Ab pore-forming toxin to aminopeptidase N receptor leading to insertion into membrane microdomains. Biochim Biophys Acta. 2004;1667:38–46. doi: 10.1016/j.bbamem.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Bravo A, Likitvivatanavong S, Gill SS, Soberon M. Bacillus thuringiensis: A story of a successful bioinsecticide. Insect Biochem Mol Biol. 2011;41:423–31. doi: 10.1016/j.ibmb.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Yu YM, Dai SM, Law SK, Gill SS. High-level cryIVD and cytA gene expression in Bacillus thuringiensis does not require the 20-kilodalton protein, and the coexpressed gene products are synergistic in their toxicity to mosquitoes. Appl Environ Microbiol. 1993;59:815–21. doi: 10.1128/aem.59.3.815-821.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Aimanova KG, Fernandez LE, Bravo A, Soberon M, Gill SS. Aedes aegypti cadherin serves as a putative receptor of the Cry11Aa toxin from Bacillus thuringiensis subsp. israelensis. Biochem J. 2009;424:191–200. doi: 10.1042/BJ20090730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Liu C, Xiao Y, Zhang D, Zhang Y, Li X, Tabashnik BE, Wu K. A toxin-binding alkaline phosphatase fragment synergizes Bt toxin Cry1Ac against susceptible and resistant Helicoverpa armigera. PLoS One. 2015;10:e0126288. doi: 10.1371/journal.pone.0126288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechklar M, Tiewsiri K, Angsuthanasombat C, Pootanakit K. Functional expression in insect cells of glycosylphosphatidylinositol-linked alkaline phosphatase from Aedes aegypti larval midgut: a Bacillus thuringiensis Cry4Ba toxin receptor. Insect Biochem Mol Biol. 2010;41:159–66. doi: 10.1016/j.ibmb.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Delecluse A, Rosso ML, Ragni A. Cloning and expression of a novel toxin gene from Bacillus thuringiensis subsp. jegathesan encoding a highly mosquitocidal protein. Appl Environ Microbiol. 1995;61:4230–5. doi: 10.1128/aem.61.12.4230-4235.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devillers J, Lagneau C, Lattes A, Garrigues JC, Clemente MM, Yebakima A. In silico models for predicting vector control chemicals targeting Aedes aegypti. SAR QSAR Environ Res. 2014;25:805–35. doi: 10.1080/1062936X.2014.958291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez LE, Martinez-Anaya C, Lira E, Chen J, Evans A, Hernandez-Martinez S, Lanz-Mendoza H, Bravo A, Gill SS, Soberon M. Cloning and epitope mapping of Cry11Aa-binding sites in the Cry11Aa-receptor alkaline phosphatase from Aedes aegypti. Biochemistry. 2009;48:8899–907. doi: 10.1021/bi900979b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahan LJ, Pauchet Y, Vogel H, Heckel DG. An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 2010;6:e1001248. doi: 10.1371/journal.pgen.1001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross TL, Myles KM, Adelman ZN. Identification and characterization of heat shock 70 genes in Aedes aegypti (Diptera: Culicidae) J Med Entomol. 2009;46:496–504. doi: 10.1603/033.046.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Kang S, Chen D, Wu Q, Wang S, Xie W, Zhu X, Baxter SW, Zhou X, Jurat-Fuentes JL, Zhang Y. MAPK signaling pathway alters expression of midgut ALP and ABCC genes and causes resistance to Bacillus thuringiensis Cry1Ac toxin in diamondback moth. PLoS Genet. 2015;11:e1005124. doi: 10.1371/journal.pgen.1005124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler AM, Harrell RA., 2nd Germline transformation of Drosophila melanogaster with the piggyBac transposon vector. Insect Mol Biol. 1999;8:449–57. doi: 10.1046/j.1365-2583.1999.00139.x. [DOI] [PubMed] [Google Scholar]
- Handler AM, McCombs SD, Fraser MJ, Saul SH. The lepidopteran transposon vector, piggyBac, mediates germ-line transformation in the Mediterranean fruit fly. Proc Natl Acad Sci U S A. 1998;95:7520–5. doi: 10.1073/pnas.95.13.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn C, Wimmer EA. A versatile vector set for animal transgenesis. Dev Genes Evol. 2000;210:630–7. doi: 10.1007/s004270000110. [DOI] [PubMed] [Google Scholar]
- Hua G, Zhang Q, Zhang R, Abdullah AM, Linser PJ, Adang MJ. AgCad2 cadherin in Anopheles gambiae larvae is a putative receptor of Cry11Ba toxin of Bacillus thuringiensis subsp. jegathesan. Insect Biochem Mol Biol. 2012;43:153–61. doi: 10.1016/j.ibmb.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Hua G, Zhang R, Bayyareddy K, Adang MJ. Anopheles gambiae alkaline phosphatase is a functional receptor of Bacillus thuringiensis jegathesan Cry11Ba toxin. Biochemistry. 2009;48:9785–93. doi: 10.1021/bi9014538. [DOI] [PubMed] [Google Scholar]
- Jimenez AI, Reyes EZ, Cancino-Rodezno A, Bedoya-Perez LP, Caballero-Flores GG, Muriel-Millan LF, Likitvivatanavong S, Gill SS, Bravo A, Soberon M. Aedes aegypti alkaline phosphatase ALP1 is a functional receptor of Bacillus thuringiensis Cry4Ba and Cry11Aa toxins. Insect Biochem Mol Biol. 2012;42:683–9. doi: 10.1016/j.ibmb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurat-Fuentes JL, Adang MJ. Characterization of a Cry1Ac-receptor alkaline phosphatase in susceptible and resistant Heliothis virescens larvae. Eur J Biochem. 2004;271:3127–35. doi: 10.1111/j.1432-1033.2004.04238.x. [DOI] [PubMed] [Google Scholar]
- Jurat-Fuentes JL, Karumbaiah L, Jakka SR, Ning C, Liu C, Wu K, Jackson J, Gould F, Blanco C, Portilla M, Perera O, Adang M. Reduced levels of membrane-bound alkaline phosphatase are common to lepidopteran strains resistant to Cry toxins from Bacillus thuringiensis. PLoS One. 2011;6:e17606. doi: 10.1371/journal.pone.0017606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawalek MD, Benjamin S, Lee HL, Gill SS. Isolation and Identification of novel toxins from a new mosquitocidal isolate from Malaysia, Bacillus thuringiensis subsp. jegathesan. Appl Environ Microbiol. 1995;61:2965–9. doi: 10.1128/aem.61.8.2965-2969.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoza V, Ahmed A, Wimmer EA, Raikhel AS. Efficient transformation of the yellow fever mosquito Aedes aegypti using the piggyBac transposable element vector pBac[3xP3-EGFP afm] Insect Biochem Mol Biol. 2001;31:1137–43. doi: 10.1016/s0965-1748(01)00120-5. [DOI] [PubMed] [Google Scholar]
- Lee SB, Aimanova KG, Gill SS. Alkaline phosphatases and aminopeptidases are altered in a Cry11Aa resistant strain of Aedes aegypti. Insect Biochem Mol Biol. 2014;54:112–21. doi: 10.1016/j.ibmb.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Chen J, Aimanova KG, Gill SS. Aedes cadherin mediates the in vivo toxicity of the Cry11Aa toxin to Aedes aegypti. Peptides. 2015;68:140–7. doi: 10.1016/j.peptides.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likitvivatanavong S, Chen J, Bravo A, Soberon M, Gill SS. Cadherin, alkaline phosphatase, and aminopeptidase N as receptors of Cry11Ba toxin from Bacillus thuringiensis subsp. jegathesan in Aedes aegypti. Appl Environ Microbiol. 2010;77:24–31. doi: 10.1128/AEM.01852-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likitvivatanavong S, Chen J, Evans AM, Bravo A, Soberon M, Gill SS. Multiple receptors as targets of Cry toxins in mosquitoes. J Agric Food Chem. 2011;59:2829–38. doi: 10.1021/jf1036189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNall RJ, Adang MJ. Identification of novel Bacillus thuringiensis Cry1Ac binding proteins in Manduca sexta midgut through proteomic analysis. Insect Biochem Mol Biol. 2003;33:999–1010. doi: 10.1016/s0965-1748(03)00114-0. [DOI] [PubMed] [Google Scholar]
- Pardo-Lopez L, Soberon M, Bravo A. Bacillus thuringiensis insecticidal three-domain Cry toxins: mode of action, insect resistance and consequences for crop protection. FEMS Microbiol Rev. 2012;37:3–22. doi: 10.1111/j.1574-6976.2012.00341.x. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigott CR, Ellar DJ. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol Mol Biol Rev. 2007;71:255–81. doi: 10.1128/MMBR.00034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Almazan C, Reyes EZ, Zuniga-Navarrete F, Munoz-Garay C, Gomez I, Evans AM, Likitvivatanavong S, Bravo A, Gill SS, Soberon M. Cadherin binding is not a limiting step for Bacillus thuringiensis subsp. israelensis Cry4Ba toxicity to Aedes aegypti larvae. Biochem J. 2012;443:711–7. doi: 10.1042/BJ20111579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratories; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sangadala S, Walters FS, English LH, Adang MJ. A mixture of Manduca sexta aminopeptidase and phosphatase enhances Bacillus thuringiensis insecticidal CryIA(c) toxin binding and 86Rb(+)-K+ efflux in vitro. J Biol Chem. 1994;269:10088–92. [PubMed] [Google Scholar]
- Soberon M, Fernandez LE, Perez C, Gill SS, Bravo A. Mode of action of mosquitocidal Bacillus thuringiensis toxins. Toxicon. 2007;49:597–600. doi: 10.1016/j.toxicon.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Soberon M, Gill SS, Bravo A. Signaling versus punching hole: How do Bacillus thuringiensis toxins kill insect midgut cells? Cell Mol Life Sci. 2009;66:1337–49. doi: 10.1007/s00018-008-8330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalinski R, Laporte F, Despres L, Tetreau G. Alkaline phosphatases are involved in the response of Aedes aegypti larvae to intoxication with Bacillus thuringiensis subsp. israelensis Cry toxins. Environ Microbiol. 2016a;18:1022–36. doi: 10.1111/1462-2920.13186. [DOI] [PubMed] [Google Scholar]
- Stalinski R, Laporte F, Tetreau G, Despres L. Receptors are affected by selection with each Bacillus thuringiensis israelensis Cry toxin but not with the full Bti mixture in Aedes aegypti. Infect Genet Evol. 2016b;44:218–27. doi: 10.1016/j.meegid.2016.07.009. [DOI] [PubMed] [Google Scholar]
- Tay WT, Mahon RJ, Heckel DG, Walsh TK, Downes S, James WJ, Lee SF, Reineke A, Williams AK, Gordon KH. Insect Resistance to Bacillus thuringiensis Toxin Cry2Ab Is Conferred by Mutations in an ABC Transporter Subfamily A Protein. PLoS Genet. 2015;11:e1005534. doi: 10.1371/journal.pgen.1005534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thammasittirong A, Dechklar M, Leetachewa S, Pootanakit K, Angsuthanasombat C. Aedes aegypti membrane-bound alkaline phosphatase expressed in Escherichia coli retains high-affinity binding for Bacillus thuringiensis Cry4Ba toxin. Appl Environ Microbiol. 2011;77:6836–40. doi: 10.1128/AEM.05775-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Candas M, Griko NB, Rose-Young L, Bulla LA., Jr Cytotoxicity of Bacillus thuringiensis Cry1Ab toxin depends on specific binding of the toxin to the cadherin receptor BT-R1 expressed in insect cells. Cell Death Differ. 2005;12:1407–16. doi: 10.1038/sj.cdd.4401675. [DOI] [PubMed] [Google Scholar]
- Zhang X, Candas M, Griko NB, Taussig R, Bulla LA., Jr A mechanism of cell death involving an adenylyl cyclase/PKA signaling pathway is induced by the Cry1Ab toxin of Bacillus thuringiensis. Proc Natl Acad Sci U S A. 2006;103:9897–902. doi: 10.1073/pnas.0604017103. [DOI] [PMC free article] [PubMed] [Google Scholar]