Abstract
Background
Major depressive disorder (MDD) is a debilitating mental illness and a major cause of lost productivity worldwide. MDD patients often suffer from life-long recurring episodes of increasing severity, reduced therapeutic response and shorter remission periods, suggesting the presence of a persistent and potentially progressive pathology.
Methods
Subgenual anterior cingulate cortex postmortem samples from four MDD cohorts (single episode, n=20; single episode in remission, n=15; recurrent episode, n=20 and recurrent episode in remission, n=15), and one control cohort (n=20) were analyzed by mass spectrometry (MS)-based proteomics (n=3630 proteins) combined with statistical analyses. The data was investigated for trait and state progressive neuropathologies in MDD using both unbiased approaches and tests of a priori hypotheses.
Results
The data provided weak evidence for proteomic differences as a function of state (depressed/remitted) or number of prior episodes. Instead it suggested the presence of persistent MDD effects, regardless of episodes or remitted state, namely on proteomic measures related to presynaptic neurotransmission, synaptic function, cytoskeletal re-arrangements, energy metabolism, phospholipid biosynthesis/metabolism and calcium ion homeostasis. Selected proteins (DRP-1, SNAP-29, GAD-67, mGluR1 and EAAT3) were validated by Western blot analysis. The findings were independent of technical, demographic (sex, age) or other clinical parameters (death by suicide and drug treatment).
Conclusion
Collectively, the results provide evidence for persistent MDD effects across current episodes or remission, in the absence of detectable progressive neuropathology.
Keywords: Major depressive disorder, subgenual cingulate cortex, progressive neuropathologies, remission, mass spectrometry, bioinformatics
INTRODUCTION
Major depressive disorder (MDD) is a severe mental disorder with heterogeneous clinical symptoms, including prominent emotion dysregulation, low mood, poor cognition, and co-morbid anxiety (1, 2). Globally, the World Health Organization cites MDD as the leading cause of years lost due to disability (3), reflecting the fact that for most patients MDD is a lifelong illness characterized by recurring episodes, often of increasing symptom severity, longer duration, with shorter and/or partial remission periods and increasing resistance to antidepressants (4, 5). An increase in frequency and duration of depressive episodes is suggested to enhance vulnerability to further relapses and accelerate disease progression, leading to worsening functional deficits (6). This disease trajectory suggests the presence of progressive neuropathologies where outcomes (7) and treatment efficacy (8, 9), are inversely correlated with disease severity, as measured by number and/or length of depressive episodes (10).
The subgenual anterior cingulate cortex (sgACC) has previously been implicated in both acute sadness and antidepressant treatment effects, which is suggestive of a critical role in modulating negative mood states (11, 12). Patients with treatment resistant depression were observed to have a metabolically overactive sgACC whose elevated activity could be modulated by deep brain stimulation that resulted in sustained remission of depression (13). Previous transcriptome analyses of postmortem hippocampus, temporal and prefrontal cortices showed dysregulation of mRNA transcripts involved in presynaptic neurotransmission, synaptic function and cytoskeletal re-arrangement of neuronal processes (14–18). Particularly, the alterations in presynaptic neurotransmission were associated with dysregulated GABA and glutamatergic receptor signaling (15–18). Reduced expression of somatostatin (SST) and other dendritic targeting GABA-neurons markers were observed in postmortem dorsal lateral prefrontal cortex (DLPFC), sgACC and amygdala from MDD subjects, in correlation with reduced BDNF/TrkB signaling (19–22). These latter molecular studies are consistent with proton magnetic spectroscopy (23–25) and transcranial magnetic stimulation (26) analyses demonstrating decreased inhibitory GABA levels and functions in MDD subjects (27, 28). Together these findings suggest an altered excitation/inhibition balance in MDD that is mediated by both GABA and glutamate dysregulations (15, 29).
Although transcriptomic studies have advanced our knowledge of the neurobiology of MDD, they provide only one facet of molecular changes associated with complex neuropsychiatric disorders. Mass spectrometry (MS)-based proteomics (which assays protein concentrations at lower dynamic range than the protein abundances in a cell compared to measures of mRNA levels by transcriptomic studies) (30, 31), is increasingly used to survey variations in protein levels during health and disease states (32–34). It is therefore well suited for unbiased discovery of alterations in protein levels associated with neuropsychiatric disorders. Previous proteomic studies performed in postmortem brains of MDD subjects have indicated enrichment of proteins involved in energy metabolism (35, 36), synaptic function (35), myelination (37) and presynaptic glutamatergic neurotransmission (36). These studies were limited by the lack of more specific distinction between MDD disease states and traits, relatively few proteins being investigated (i.e. 1422, 56 and 1310) or by small sample size (n= 36, 44 and 46) (35–37). Western blot analysis provided evidence of reduced GAD-67 protein levels, a GABA synthesizing enzyme (38).
Here, we applied MS-based proteomics supported by bioinformatics and statistics to perform the first large scale protein investigation (n=3630) of biological changes in MDD, more specifically in the sgACC of four MDD cohorts at various stages of disease and in one control cohort (Figure 1a). We focused on the sgACC based on robust clinical and imaging evidence of deregulated function in the area (11, 13), and due to prior findings from our group suggesting a more severe molecular phenotype in this brain region in comparison to other investigated areas (21). We tested for the presence of persistent pathological findings across all MDD patients, regardless of disease state (hypothesis 1), for distinct neuropathologies corresponding to current episodes or remission states (hypotheses 2 and 3), and for evidence of progressive neuropathology in association with recurrent episodes (hypothesis 4).
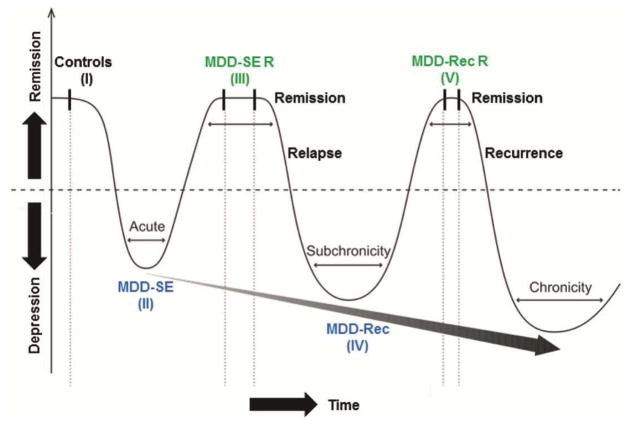
Figure 1. Overview of MDD cohorts and control subjects used in the study.
Hypothesized progressive model of major depressive disorder (MDD) showing recurring episodes of increasing severity, reduced therapeutic response and shorter remission periods. MDD episodes and treatment phases are indicated by valleys and crests, respectively. A gray arrow depicts the predicted trajectory of MDD pathology across various diseases stages. Abbreviations used: MDD-SE, MDD single episode; MDD-SE R, MDD single episode remission; MDD-Rec, MDD-recurrent and MDD-Rec R, MDD-recurrent remission. Groups are indicated by I–V (adapted from Sibille 2013 (53)).
MATERIALS AND METHODS
Detailed methods are available online
Human postmortem brain samples
Postmortem subgenual anterior cingulate cortex (sgACC; Brodmann’s area 25) samples were obtained through the University of Pittsburgh Brain Tissue Donation Program with consent from the next of kin. Sample collection was done during routine autopsies performed at the Allegheny County Medical Examiner’s Office (Pittsburgh) with procedures approved by the University of Pittsburgh Institutional Review Board and the Committee for Oversight of Research and Clinical Training Involving the Dead. Consensus DSM-IV diagnoses were made by an independent committee of experienced clinicians using information from structured interviews with family members, clinical records, toxicology results, and standardized psychological autopsies (39). The same approach was used to confirm the absence of a psychiatric diagnosis in comparison subjects. The DSM-IV diagnosis is at time of death whereas psychosis history is lifetime. Ninety samples including control subjects and four MDD cohorts were analyzed (Figure 1, Supplementary Table S1). The MDD cohorts were closely matched with controls to ensure that they did not differ in mean age, postmortem interval (PMI), brain pH or RNA integrity number (RIN) (Figure 1b). Samples comprising all six cortical layers were harvested from coronal sections, as previously described (40).
Sample preparation and Liquid Chromatography tandem Mass Spectrometry (LC-MS/MS) analysis (Supplementary Figure S1)
Total sample homogenates (approx. 20 μg) were subjected to a modified filter-aided sample preparation (FASP) protocol as previously described (41), with additional precipitation using an equal volume of 2M KCL for depletion of residual detergents. Lys-C and tryptic peptides were combined and processed on Pierce C18 Tips reversed phase resin (Thermo Scientific) for desalting and concentration. Peptides were applied to a UPLC system (EASY-nLC 1000, Thermo Scientific) and separated on a 50 cm column (75 μm inner diameter) packed with PepMap®RSLC C18 resin at 60°C, using a flow rate of 250 nl/min on a 5%–30% acetonitrile in 0.1% formic acid gradient over 224 min. Column washes were performed on a 30% to 90% acetonitrile in 0.1% formic acid gradient for 2 min, then 12 min in 90% acetonitrile in 0.1% formic acid. Peptides were introduced into an Orbitrap Elite Mass Spectrometer (Thermo Scientific) using a nano-electrospray ion source (EASY-SPRAY, Thermo Scientific). Mass spectra were acquired in a 400–1200 m/z range with a resolution of 240,000 at 400 m/z in the Orbitrap (automatic gain control target: 100,000), followed by 10 data-dependent MS/MS scans (automatic gain control target: 10,000). The “top-10” most intensive ions were selected and fragmented by collision induced dissociation with normalized collision energies of 30 in the ion trap. Dynamic exclusion duration was set at 50 s and the maximum exclusion list size at 500.
MS data analysis
Raw MS data was processed by MaxQuant software v1.5.3.8 and searched against the human UniProt database (released November, 2015; 20, 193 entries), for label free quantitation of peptides and proteins. The following settings were used: fragment ion mass tolerance of 20ppm, maximum two missed cleavages (Trypsin and Lys-C), fixed modification as carbamidomethylation of cysteine, and variable modification as oxidation of methionine and acetylation of protein N-terminal. False discovery rates were set at 1% for both peptide and protein levels in target/decoy to minimize false positives. The match between runs feature was utilized. Reverse sequences, potential contaminants and those only identified by site (Supplementary Table S2–1) were removed from the MaxQuant data prior to statistical analysis. This processing yielded 4301 proteins (Supplementary Table S2–2) for further analysis.
Statistical analysis
Data points at zero were added a value of 1, before log2 transformation of processed MS intensity data from MaxQuant. Data imputation was performed to address the observed zero values (real zeros or missing data), a common procedure in proteomics data (42). Zero values were imputed based on three criteria: proportion of samples with non-zero intensity in each group, the molecular weight of the protein, and the mean intensity over all samples. Consequently, we classified all proteins into five categories: (I) complete data (proportion of non-zero equal to 1 in both control and MDD-ALL groups); (II) mild missing but imputable data (proportion of non-zero greater than or equal to 0.7 in both groups); (III) moderate missing and non-imputable data (proportion of non-zero less than 0.7 in exactly one group); (IVA) severe missing and non-interpretable data (proportion of non-zero less than 0.7 in both groups, both its molecular weight and mean intensity are above 33th percentile in all proteins); and (IVB) severe missing but interpretable data (proportion of non-zero less than 0.7 in both groups, and its molecular weight or mean intensity is below 33th percentile in all proteins). Only the zeros in category II were imputed using the K-nearest neighbor method (“KNN”, K=10 in our case). We accepted the zeros in all the other categories. Downstream analyses were based on proteins from categories I (2421 proteins) and II (1209), in order to ensure high confidence in our data. Proteins from categories III (144), IVA (0) and IVB (527), which were of low confidence were not analyzed further (Table S2–3). Differentially-expressed label free quantified proteins were identified using the random intercept model (RIM) with parameter selection employing the smallest Bayesian Information Criterion (43) to account for potential covariates (adjusting for up to 2 cofactors among age, pH, PMI and RIN). To correct the potential bias of the variable selection procedure, we performed a permutation analysis that randomly shuffled the disease labels within each pair to generate a null distribution for p-value assessment (B=500). For the exploratory purpose of this study, we used p≤0.05 and ≥ 20% fold change (RIM coefficient ≥±0.26) as thresholds for differentially expressed proteins. Post hoc analysis to correct for the potential confound effects of psychosis, alcohol dependence, antidepressant drug use and death by suicide on differential protein expression was performed using analysis of variance (ANOVA). To test the potential of progressive effects, we hypothesized a linear relationship between MDD groups and the expression level of some proteins, across all five groups, in current episodes or in remission only. In comparison to the use of categorical or nominal variable where each category is relatively independent, the ordinal approach used in this study assumes that the categories can be ordered. The five groups in our data reflect progressive stages of the MDD disease, which we assumed are ordered categories and therefore hypothesized that a linear (stage) effect on protein expression might exist.
Western blot analysis
Details on sample processing, protein detection and antibodies are described in Supplementary Methods.
Functional and network analysis
Gene ontology (GO) analysis was performed using the PANTHER database v11.1 (Protein ANalysis THrough Evolutionary Relationships, http://pantherdb.org) (44). The following settings were used: analysis type-PANTHER Overrepresentation Test (release 20170413); Annotation version and release date- GO database released 2017-05-25; Reference List-MDD background list of all identified proteins in ≥ 70% samples (n=3630); Annotation Data Set-GO biological process complete and no correction for multiple testing, due to the limitations in sample availability of postmortem tissues. We also probed for enrichmnet in PANTHER pathways. Differentially expressed proteins were subjected to Ingenuity Pathway Analysis (IPA, www.ingenuity.com) for canonical pathways and disease categories. The following parameters were used for core analysis: Ingenuity Knowledge Base (Genes only) as a reference set, direct and indirect relationships, interaction networks, all data sources, confidence of experimentally observed data only, the species set to humans and including data from only brain tissues. The scoring method was based on Fisher’s exact p value.
RESULTS
Do MDD patients exhibit a persistent disease effect, regardless of current episode or remission state (MDD-ALL), in comparison to controls?
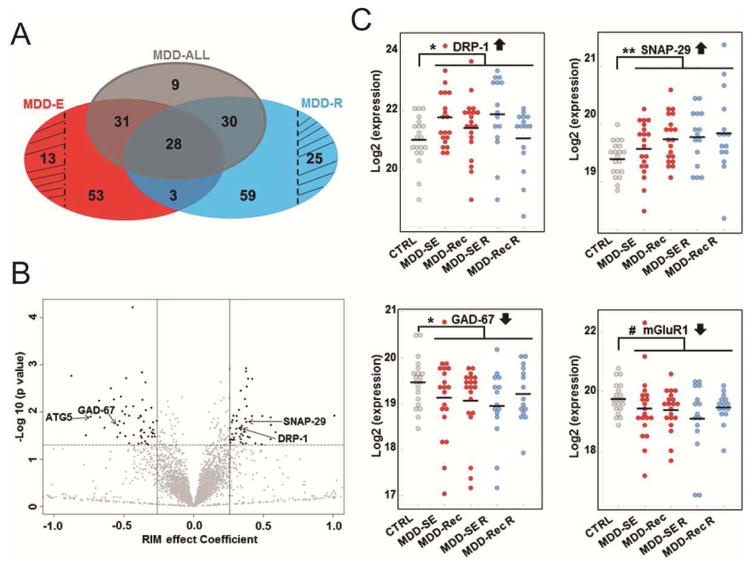
Characteristics of the MDD cohorts corresponding to current episodes, remission states and control subjects are summarized in Figure 1 and Table 1. sgACC gray matter samples from postmortem brain tissues were used in LC-MS/MS-based proteomic analyses (Supplementary Figure S1), followed by label free quantification of proteins using MaxQuant software. We detected 4301 proteins (Supplementary Table S2–2), of which 3630 were identified in at least 70% of all samples (Supplementary Table S2–3) and subsequently used for downstream analyses. A comparison of control subjects to all MDD patients using a random intercept model (RIM) to correct for potential confounders yielded 98 differentially expressed proteins, at p≤0.05 with fold change ≥20% (Log2-ratio ≥±0.26) (Figure 2a, gray shading; Table 2 and Supplementary Table S2–4). Examples of upregulated proteins (Figure 2b) within this group include: synaptosomal-associated protein 29 (SNAP-29) which is involved in autophagy/vesicle exocytosis; dihydropyrimidinase-related protein 1 (DRP-1), an axonal guidance signaling protein; and rho GTPase-activating protein 32 (p250GAP), a protein involved in NMDA receptor activity-dependent actin reorganization in dendritic spines. Examples of downregulated proteins (Figure 2b) include: glutamate decarboxylase 1 (GAD-67) which catalyzes the production of gamma-aminobutyric acid (GABA); calcium/calmodulin dependent protein kinase type 1G (CAMK1G) and autophagy related protein 5 (ATG5) (Supplementary Table S2–4). Selected scatter (gg) plots of two upregulated (DRP-1, SNAP-29) and downregulated (GAD-67, mGluR1) proteins in the various subgroups of all MDD subjects are shown in Figure 2c.
Table 1. Characteristic features of MDD cohorts and control subjects used in the study.
Postmortem brain samples from all cortical layers of the subgenual cingulate cortex included: control subjects and four major MDD cohorts as described above. Most subjects in the groups are White: controls (75%), single episode (80%), single episode remission (93%), recurrent (85%) and recurrent remission (93%). There were no significant differences between the groups on mean: Age, PMI, pH, RIN and RNA ratio. However, the groups varied in sex ratios, suicide rates and antidepressant use ATOD.
| Variables | Controls (I) n=20 | MDD-Single episode (II) n=20 | MDD-Single episode in Remission (III) n=15 | MDD- recurrent episode (IV) n=20 | MDD-recurrent episode in Remission (V) n=15 |
|---|---|---|---|---|---|
| Mean Age | 47.9 | 42.3 | 47.6 | 40.9 | 48.1 |
| Sex (% Male) | 90% | 80% | 53% | 65% | 67% |
| Mean PMI | 15.35 | 15.99 | 12.17 | 16.8 | 16.4 |
| Mean pH | 6.76 | 6.62 | 6.63 | 6.62 | 6.61 |
| RNA ratio | 1.55 | 1.62 | 1.69 | 1.57 | 1.42 |
| Mean RIN | 8.09 | 8.08 | 8.45 | 8.03 | 8.03 |
| Suicide (%) | 0% | 50% | 0% | 40% | 0% |
| Antidepressant use ATOD (%) | 5% | 60% | 20% | 75% | 60% |
Abbreviations: PMI, postmortem interval; RIN, RNA integrity number; ATOD, at time of death.
Figure 2. Summary of identified proteins across MDD episodes and remission phases.
A. Venn diagram of the differentially expressed proteins associated with all MDD patients (MDD-ALL, gray shading) and those in current episodes (MDD-E, red shading) or remission phases (MDD-R, blue shading) at p≤0.05 and RIM coefficient effect (log 2 fold change) ≥±0.26 thresholds. Differentially expressed proteins associated only with patients in current episodes (n=13) or remission (n=25) are indicated with black line shading. B. Volcano plot indicating distribution of identified proteins based on their RIM effect Coefficient (log 2 fold change) and −log 10 (p-value). Upregulated and downregulated proteins are highlighted in black (see right and left panels, respectively). Upregulated (DRP-1 and SNAP-29) and downregulated (GAD-67, mGluR1 and EAAT3) proteins selected for Western Blot confirmation (Fig. 3) are indicated in the plot. C. Scatter (gg) plots of selected differentially expressed proteins across in subgroups of all MDD patients. #, * and ** denote trend, statistical significance of p≤0.05 and 0.01, respectively.
Table 2. Biological processes and pathways associated with all MDD patients.
Differentially expressed proteins were determined by statistical analysis using the random intercept model, based on (p≤0.05 and RIM effect Coefficient ≥ ±0.26) thresholds. Gene ontology (GO) biological process analysis was performed using the PANTHER database v11.1 (http://pantherdb.org). Ingenuity Pathway Analysis (IPA) was utilized to identify the top 3 canonical pathways and significantly associated disease categories linked to the differentially expressed proteins.
| GO Biological Process Term | Overlap | Fold enrichment | p-value | Associated Proteins |
|---|---|---|---|---|
| Ganglioside metabolic process (GO:0001573) | 66.67% | 24.13 | 3.22E-3 | HEXB; ITGB8 |
| Sequestering of metal ion (GO:0051238) | 50.00% | 18.10 | 5.63E-3 | FTH1; S100A8 |
| PERK-mediated unfolded protein response (GO:0036499) | 50.00% | 18.10 | 5.63E-3 | ASNS; HEBP1 |
| Phosphatidylglycerol biosynthetic process (GO:0006655) | 33.33% | 12.07 | 1.22E-2 | TAM41; PTPMT1 |
| Positive regulation of cytokine-mediated signaling pathway (GO:0001961) | 28.57% | 10.34 | 1.63E-2 | 1-AGPAT 1; CNTFR |
| CDP-diacylglycerol biosynthetic process (GO:0016024) | 28.57% | 10.34 | 1.63E-2 | TAM41; 1-AGPAT 1 |
| IPA Canonical pathways | Overlap | p-value | Associated Proteins |
|---|---|---|---|
| Phosphatidylglycerol Biosynthesis II (Non-plastidic) | 3/22 (13.6%) | 3.47E-4 | TAM41, PTPMT1, 1-AGPAT 1 |
| Calcium Signaling | 5/144 (3.5%) | 2.19E-3 | ara CALC, CAMK1G, TPM3, TPM1, SLC8A1 |
| Retinoate Biosynthesis II | 1/1 (100%) | 6.35E-3 | RBP1 |
The differentially expressed proteins were most significantly associated with neurological diseases(Psychological disorders) in the disease category of Ingenuity Pathway Analysis.
Abbreviations: GO, Gene ontology.
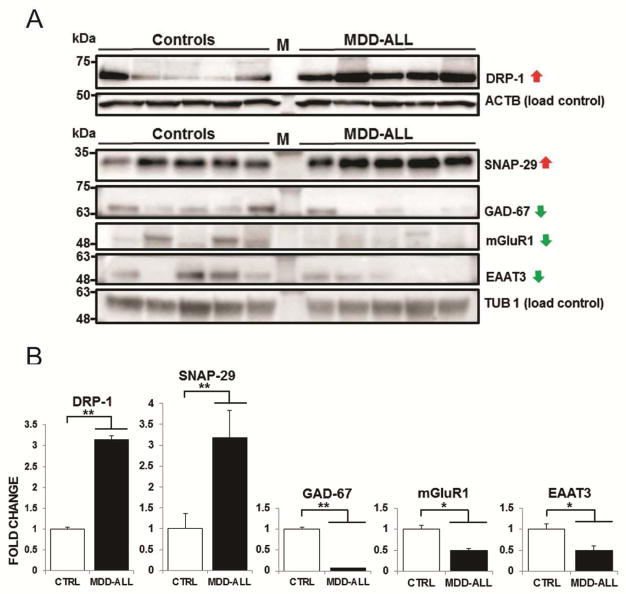
A few differentially expressed proteins were selected for independent evaluation by Western blot analysis, on the basis of their known involvement in MDD or other psychiatric disorders (DRP-1, GAD-67) and autophagy/vesicle exocytosis (SNAP-29). We also included proteins identified at p<0.1 and ≥20% fold change based on prior association with psychiatric disorders (mGluR1 and EAAT3). Glutamate decarboxylase 1 (mGluR1) which is reduced in MDD patients and was also downregulated in our proteomic analyses at the trend level, served as an internal validation of MDD effects (38). We anticipated that proteins which were upregulated (DRP-1 and SNAP-29) or downregulated (GAD-67, mGluR1 and EAAT3) in our proteomic analyses would show increased/decreased expression in immunoblots of MDD cohorts in comparison to control subjects. Western blot analysis of samples from a randomly selected subset of 5 MDD patients and control subjects that were equally matched for mean age, postmortem interval (PMI), brain pH or RNA integrity number (RIN), indicated immunoreactive bands corresponding to endogenously expressed DRP-1, SNAP-29, GAD-67, mGluR1 and EAAT3, respectively (Figure 3a). We determined significantly increased protein expression for DRP-1 (p=0.0081); SNAP-29 (p=0.0029), and reduced protein expression for GAD-67 (p=0.0004); mGluR1 (p=0.0296) and EAAT3 (p=0.0471) (Figure 3b) in the subset of the MDD patients we tested. Results of the Western blot validation are summarized in Supplementary Table S3.
Figure 3. Western blot analysis of selected differentially expressed proteins in MDD cohorts.
(A) Immunoblotting with DRP-1, SNAP-29, GAD-67, mGluR1 and EAAT3 antibodies showed increased protein expression of DRP-1 (62 kDa) and SNAP-29 (29 kDa), but reduced protein expression of GAD-67, mGluR1, EAAT3 (67, 62 and 57 kDa, respectively) for MDD patients in current episodes compared to control subjects. Anti-beta actin (ACTB, 42 kDa) and anti-beta tubulin (TUB1, 56 kDa) were used to assess equal loading of protein lysates. The two protein standards (labelled as M) used were: All Blue Prestained Protein Standards (#1610373, BioRad Canada) and BLUeye Prestained Protein Ladder (ThermoScientific). (B) Quantitation of the bands using Image Lab software (Biorad) indicated statistical significance, as denoted by * and ** for p≤0.05 and 0.01, respectively.
As a group, differentially expressed proteins associated with a persistent disease effect (at p<0.05 and >20% fold change) were previously linked in the literature to neurological diseases (Ingenuity Pathway Analysis, IPA; Table 2) and most significantly implicated in ganglioside metabolic process (66.67% overlap; fold enrichment: 24.13), sequestering of metal ion (50.00% overlap; fold enrichment: 18.10), PERK-mediated unfolded protein response (50.00% overlap; fold enrichment: 18.10) and phosphatidylglycerol biosynthetic process (33.33% overlap; fold enrichment: 12.07) (Table 2). These findings were independent of technical, demographic or other clinical parameters (death by suicide and drug treatment) (Supplementary Table S2–4 – S2–11). Moreover, IPA indicated phosphatidylglycerol biosynthesis II (non-plastidic; 13.6% overlap) and calcium signaling (3.5% overlap), as the two most significantly affected canonical pathways (Table 2 and Supplementary Table S4). Additionally, we detected proteins of previous interest based on literature knowledge, including: metabotropic glutamate receptor 1 (mGluR1), and excitatory amino acid transporter 3(EAAT3) at the less stringent p<0.1 threshold (Supplementatry Table S2–4). Together the data supports the presence of a persistent disease effect amongst all MDD patients that involves dysregulation of phospholipid biosynthesis/metabolism, calcium ion homeostasis, axon guidance and GABA or glutamate receptor signaling related proteins.
Are current episodes of depression associated with a unique profile of differentially expressed proteins?
We next probed for proteins that were differentially expressed exclusively in MDD patients in current episodes. For this, protein expression had to be significantly different from controls and from the group of MDD patients in remission (Figure 2a). The analysis yielded 13 proteins that are restricted to MDD patients in current episodes (Figure 2a, red with black line shading and Supplementary Table S2–7). Examples of the downregulated proteins in this group include: 26S proteasome non-ATPase regulatory subunit 5 (PSMD5) and Serine/threonine-protein phosphatase PP1-alpha catalytic subunit (PP-1A). The few number of differentially expressed proteins in this category precluded meaningful functional analysis. Together, the data provides weak evidence for the presence of a distinct molecular disease state for patients in current episodes compared to those in remission and healthy controls.
Are MDD patients in remission associated with a unique profile of differentially expressed proteins?
Similarly, we probed for proteins that were differentially expressed exclusively in MDD patients in remission. For this, protein expression had to be significantly different from controls and from the group of MDD patients who had a current episode of MDD at the time of death (Figure 2a). The analysis yielded 25 proteins that are restricted to MDD patients in remission (Figure 2a, blue with black line shading and Supplementary Table S2–8). Examples of the upregulated proteins in this group include: voltage-dependent L-type calcium channel subunit beta-1 (CAB1) and BTB/POZ domain-containing protein KCTD8 (KCTD8). Downregulated proteins constituted: synaptophysin (SYP) and D-2-hydroxyglutarate dehydrogenase, mitochondrial (D2HGDH). Differentially expressed proteins specific to MDD patients in remission were too few to allow for meaningful functional analysis. Overall, the data provide weak evidence in support of a distinct molecular state for the MDD patients in remission, compared to those who experienced a current episode of depression at time of death and healthy controls.
Additional proteins were identified that fell into an intermediate category. These proteins show significant expression changes between MDD patients in current episodes (MDD-E; n=53) or in remission (MDD-R; n=59) compared to control subjects without being either differentially expressed between MDD cohorts (i.e. MDD-E versus MDD-R) or identified in the first analysis combining all MDD subjects (Grey shading in Figure 2a). The functional groups with these proteins demonstrated a considerable overlap with those corresponding to differentially expressed proteins identified in the combined cohort (i.e. either in current episodes or in remission; Supplementary Tables: S2–5, S2–6 and S5). Hence, although these proteins had not been identified in the combined cohort, they may be considered as belonging to the general MDD category, rather than representing distinct pathologies between remission and current episodes.
Do MDD patients exhibit progressive neuropathology across MDD episodes and remission phases in comparison to controls?
Statistical analysis was performed by recoding the group variable as an ordinal variable (groups: I–V) and fitting a random intercept model (RIM) by adjusting for up to 2 cofactors amongst age, PMI, pH and RIN), to detect a possible linear effect across groups. Only 3 proteins fit a progressive neuropathology profile across MDD episodes and remission phases, at the less stringent p≤0.1 and ≥±20% fold change (Supplementary Table S2–9). Given this very low signal, we tested for the possibility of a progressive neuropathology restricted to patients in current episodes, i.e. from controls to early (single episode) and late (recurrent) MDD stages. Similar analysis was performed for MDD patients in remission. 39 and 41 differentially expressed proteins were associated with progressive MDD effect for patients in current episodes and remission, respectively, based on p≤0.05 and fold change ≥±20% (Supplementary Tables: S2–10 and S2–11). The differentially expressed proteins associated with the progressive MDD effect for patients in current episodes were mostly implicated in mitochondrial calcium ion homeostasis, whereas regulation of cytokine production/phosphatidylinositol 3-kinase signaling, were the top functional groups linked to a similar disease effect for patients in remission (Supplementary Table S5). Together these findings provide no supporting evidence for a progressive neuropathology across episodes and remission, and only weak evidence for progressive MDD effects restricted to patients in either current episodes or remission.
DISCUSSION
Previous large-scale human postmortem studies on depression focused on the depressive state and used analysis of the transcriptome (14–18) as a proxy measure for biological function. Here, we investigated for the first time biological changes associated with various disease states (current episode, remission and recurrence) in the sgACC in MDD and opted for a large-scale proteomic analysis using mass-spectrometry (MS)-based approaches in a large postmortem cohort.
The results provide novel evidence in support of persistent disease effects in all MDD patients (regardless of depressive episodes or remission). In contrast, the data provided weak evidence in support of distinct pathological states in patients who experienced current episodes of MDD or who were in remission from the illness at time of death. Interestingly we found even weaker evidence for a progressive disease effect, whether the subjects were analyzed together or in subgroups based on current episode or remission. These latter findings are in contrast to clinical evidence showing increased disease severity with recurrence. They suggest that the increasing disease severity and treatment resistance observed in patients with recurrent depression may be mediated by other factors (e.g. sex differences, regional connectivity, neural network changes) rather than by greater molecular pathology, at least within the limits of detection of our assays. These results provide insight into disease mechanisms and progression that have implications for treatment strategies.
Analysis of differentially expressed proteins provided some insight into biological pathways associated with presynaptic neurotransmission, energy metabolism, synaptic function and cytoskeletal re-organization (Supplementary Table S2–4), in agreement with previous findings using combined cohorts (14–18, 35, 36, 45). Moreover, we also determined novel enrichment in phospholipid biosynthesis and calcium signaling functional groups/pathways in these patients (Table 1 and Supplementary Table S4). These findings were not attributable to the effect of sex, antipsychotic medications or other potential confounders and therefore likely reflect the underlying disease process (Supplementary Tables: S2–4, S6 and S7; Supplementary Figures: S2 and S3). Phospholipids are the main constituent of biological membranes and in neurons are critical for subcellular compartmentalization of integral membrane proteins involved in neuronal communication (46). A major role of Ca2+ in neurons is to regulate activity dependent signaling by controlling neuronal excitability. Loss of neuronal Ca2+ homeostasis during aging has been associated with alterations in neuronal excitability and consequently changes in neuronal networks and metabolism (47).
Downregulated proteins associated with the persistent MDD pathology included GABA/glutamate receptor signaling proteins (GAD-67, mGluR1 and EAAT3) (Supplementary Tables: S2–4, S2–5 and S2–6), consistent with prior reports suggesting impaired GABAergic and glutamatergic neurotransmission in MDD (15–18). Moreover, dysregulation of cytoskeletal organization by Rho GTPase is associated with altered dendritic spine morphogenesis, which has been suggested to contribute to deficits in synaptic function in MDD (48). Glutamate decarboxylase 1 (GAD-67) was shown to be significantly reduced in the frontal cortex of depressed subjects at the protein (38) and mRNA levels (19–22), whereas reduced somatostatin (SST) mRNA expression in the DLPFC, sgACC and amygdala of MDD patients has previously been demonstrated (19–22). Here we also observed a downregulation of SST, although at the trend level (p≤0.1), as one of the few proteins potentially associated with a progressive MDD effect for patients in remission (Supplementary Table S2–11). These GABA/Glutamate/synaptic results are noteworthy as they provide internal control validity since we used exploratory statistical criteria for the identification and functional annotation of differentially expressed proteins. We detected neuronal proteins, including DRP-1 and SNAP-29 (upregulated), mGluR1 and EAAT3 (downregulated) which supports the sensitivity of our MS analyses for probing differential protein expression in human postmortem brain tissues. A subset of the neuronal proteins was further validated using Western blot analyses (Figure 3). Interestingly, upregulated DRP-1, as well as downregulated mGluR1 and SLC1A1 mRNA transcripts have previously been associated with Schizophrenia (49–51). Future targeted MS experiments with selected reaction monitoring (SRM) or parallel reaction monitoring (PRM) based on a priori knowledge from this study and known MDD hypotheses, should allow for more accurate and sensitive detection of lowly expressed proteins that are putatively altered at different stages of depression. SRM has already been successfully utilized to probe for changes in protein levels of selected candidate markers for neuropsychiatric disorders (37).
Although our study provided novel and interesting insights, there were several limitations: first, MS-based analysis has a relatively lower dynamic range in comparison to the complexity of the cellular proteome (30). Second, proteomic analysis of human postmortem tissues is also challenging due to the variation in postmortem interval before autopsy, during which protein degradation may occur (52). Third, although our assay exceeded prior proteomic studies (35, 36, 45) with over 4300 proteins identified, it was limited by sample availability and likely under representation of membrane proteins, which may have precluded the identification of receptors associated with the monoamine hypothesis of MDD. Finally, we did not measure post-translational modifications.
Supplementary Material
Supplementary Figure S1: MS-based proteomics workflow for analysis of sgACC gray matter. Human postmortem subgenual cingulate cortex samples from control subjects and MDD cohorts were processed using our MS-based proteomics approach. The strategy for analysis of the brain samples comprised of 4 key steps: tissue extraction, sample preparation, LS-MS/MS analysis and bioinformatics.
Supplementary Figure S2: Histogram of p-value distribution for identified proteins associated with all MDD patients in comparison to controls, with sex as a cofactor. The p-values for all the identified proteins with sex as a cofactor are evenly distributed from 0 through 1, which is very close to the distribution of p-value under null.
Supplementary Figure S3: Histogram of p-value distribution for identified proteins associated with MDD patients in recurrence in comparison to controls, with antipsychotic medications as a cofactor. The p-values for all the identified proteins with antipsychotic medications as a cofactor are evenly distributed from 0 through 1, which is very close to the distribution of p-value under null.
Supplementary Figure S4: Plot of batch effect on protein expression distribution in controls and MDD cohorts. The log2 intensity and batches are shown on the y and x-axes, respectively. The plot indicates that there is no batch effect on protein expression distribution in the samples analyzed.
Supplementary Table S1: Description of MDD cohorts and control subjects used in the study.
Postmortem brain samples were collected from control subjects (group I, n=20) and four major MDD cohorts corresponding to single episode (group II, n=20), single episode remission (group III, n=15), recurrent (group IV, n=20), and recurrent-partial/full remission (group V, n=15). DSM-IV diagnoses, mode of death (MOD), cause of death (COD) and demographic features (including: sex, age, race, PMI, pH, RNA ratio, RIN) of each subject are identified. A few control subjects with a DSM diagnosis were used and their duration of illness (DOI) is entered in parentheses (). The DOI is determined from the first depressive episode until remission or death, whichever comes first. The DSM-IV diagnosis is at time of death whereas psychosis history is lifetime. For all others subjects DOI is only for the MDD diagnosis. Abbreviations: ASCVD, atherosclerotic cardiovascular disease; ADC, Alcohol Dependence, current at time of death; ADR, Alcohol Dependence, in remission at time of death; AAC, Alcohol Abuse, current at time of death; AAR, Alcohol Abuse, in remission at time of death; ODC, Other Substance Dependence, current at time of death; ODR, Other Substance Dependence, in remission at time of death; OAC, Other Substance Abuse, current at time of death and OAR, Other Substance Abuse, in remission at time of death; Meds/Tob/Dep ATOD, Prescribed medications, tobacco use and depression at time of death; B, Benzodiazepines; C, Anticonvulsants; D, Antidepressants; N, No medications; O, Other medication(s); P, Antipsychotic; T, Tobacco; U, Unknown; V, Valproic Acid; X, Medications unknown; N/A, not applicable; 0, absence of trait; 1, presence of trait and 0/1, partial trait.
Supplementary Table S2: Analysis of MS data from MDD cohorts and control subjects by MaxQuant.
S2-1: Label free quantitation of MS data from MDD cohorts and control subjects by MaxQuant analysis-unfiltered. S2-2: Label free quantitation of MS data from MDD cohorts and control subjects by MaxQuant analysis-filtered. S2-3: Testing of hypotheses in support or not of MDD episodes and remission phases. S2-4: Differentially expressed proteins associated with a persistent disease effect in all MDD patients (hypothesis 1). S2-5: Differentially expressed proteins associated with the MDD state of patients in current episodes (hypothesis 1a). S2-6: Differentially expressed proteins associated with the residual MDD state of patients in remission (hypothesis 1b). S2-7: Differentially expressed proteins associated with distinct MDD state for patients in current episodes in comparison to those in remission (hypothesis 2). S2-8: Differentially expressed proteins associated with distinct MDD state for patients in remission in comparison to those in current episodes (hypothesis 3). S2-9: Differentially expressed proteins associated with neuro-progressive pathology across MDD episodes and remission phases (hypothesis 4). S2-10: Differentially expressed proteins associated with a progressive MDD effect for patients in current episodes (hypothesis 4a). S2-11: Differentially expressed proteins associated with a progressive MDD effect for patients in remission (hypothesis 4b). Post hoc analysis to test for the effects of psychosis, alcohol dependence, antidepressant drug use and death by suicide on differential protein expression was performed using analysis of variance (ANOVA).
Supplementary Table S3: Summary of Western blot validation of selected differentially expressed proteins.
Selected differentially expressed proteins (DRP-1, SNAP-29, GAD-67, mGluR1 and EAAT3) in all MDD patients based on MS analysis, were validated by Western blot analysis. The directionality of protein changes observed by Western blot analysis was consistent with findings from the MS data. DRP-1 and SNAP-29 were upregulated, whereas GAD-67, mGluR1 and EAAT3 were observed to be downregulated in both analyses.
Supplementary Table S4: Canonical pathways associated with a persistent disease effect in all MDD patients (hypothesis 1).
S4-1: The pathways associated with differentially expressed (DE) proteins associated with a persistent disease effect in all MDD patients (hypothesis 1) at p ≤ 0.05 and RIM Coefficient effect ≥ ± 0.26 thresholds, were identified using Ingenuity Pathway Analysis (IPA). S4-2: Pathways associated with the DE proteins in all MDD patients at less stringent p ≤ 0.1 and RIM Coefficient effect ≥ ± 0.26 thresholds.
Supplementary Table S5: Overview of differentially expressed proteins associated with MDD episodes and remission phases.
Differentially expressed proteins were identified based on p ≤ 0.1 and RIM Coefficient effect ≥ ± 0.26 thresholds. The biological significance of differentially expressed proteins in the MDD cohorts and association with disease categories were explored by GO biological process using PANTHER and Ingenuity Pathway Analysis (IPA), respectively.
Supplementary Table S6: Post hoc analysis of identified proteins associated with all MDD patients in comparison to controls, with adjustment of one cofactor at a time.
Generated p-values associated with individual cofactors served as a measure of their respective importance. This analysis indicated that sex had little effect on the p-value of identified proteins in comparison to the other cofactors, e.g. pH and Age, which we selected and adjusted through the random intercept model (RIM).
Supplementary Table S7: Post hoc analysis of identified proteins associated with MDD patients in recurrence in comparison to controls, with adjustment of one cofactor at a time.
This analysis indicated that antipsychotic medications had little effect on the p-value of identified proteins in comparison to the other cofactors, e.g. pH and Age, which we accounted for in the random intercept model (RIM).
Acknowledgments
This work was supported by National Institute of Mental Health (NIMH) R01 MH077159-09 and by the Campbell Family Mental Health Research Institute of CAMH (to Etienne Sibille). The funding agencies had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Mental Health (NIMH). We thank Drs. Paul Taylor and Jonathan Krieger (SPARC Biocentre, SickKids Toronto) for mass spectrometry analysis. Dr. Mounira Banasr (Campbell Family Mental Health Research Institute of CAMH) is acknowledged for critical review of the manuscript.
Footnotes
Supplementary information is available at Biological Psychiatry’s website (https://www.elsevier.com/journals/biological-psychiatry).
Financial Disclosures/Conflicts of Interest:
The authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Belmaker RH, Agam G. Major depressive disorder. The New England journal of medicine. 2008;358(1):55–68. doi: 10.1056/NEJMra073096. [DOI] [PubMed] [Google Scholar]
- 2.Kendler KS, Prescott CA, Myers J, Neale MC. The structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Archives of general psychiatry. 2003;60(9):929–37. doi: 10.1001/archpsyc.60.9.929. [DOI] [PubMed] [Google Scholar]
- 3.WHO. World Health Organization - The Global Burden of Disease - 2004 update. WHO Library; 2008. [Google Scholar]
- 4.Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Archives of general psychiatry. 2005;62(6):593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- 5.Moylan S, Maes M, Wray NR, Berk M. The neuroprogressive nature of major depressive disorder: pathways to disease evolution and resistance, and therapeutic implications. Molecular psychiatry. 2013;18(5):595–606. doi: 10.1038/mp.2012.33. [DOI] [PubMed] [Google Scholar]
- 6.Kendler KS, Thornton LM, Gardner CO. Genetic risk, number of previous depressive episodes, and stressful life events in predicting onset of major depression. The American journal of psychiatry. 2001;158(4):582–6. doi: 10.1176/appi.ajp.158.4.582. [DOI] [PubMed] [Google Scholar]
- 7.Gorwood P, Corruble E, Falissard B, Goodwin GM. Toxic effects of depression on brain function: impairment of delayed recall and the cumulative length of depressive disorder in a large sample of depressed outpatients. The American journal of psychiatry. 2008;165(6):731–9. doi: 10.1176/appi.ajp.2008.07040574. [DOI] [PubMed] [Google Scholar]
- 8.Tominaga K, Okazaki M, Higuchi H, Utagawa I, Nakamura E, Yamaguchi N. Symptom predictors of response to electroconvulsive therapy in older patients with treatment-resistant depression. International journal of general medicine. 2011;4:515–9. doi: 10.2147/IJGM.S21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okuda A, Suzuki T, Kishi T, Yamanouchi Y, Umeda K, Haitoh H, et al. Duration of untreated illness and antidepressant fluvoxamine response in major depressive disorder. Psychiatry and clinical neurosciences. 2010;64(3):268–73. doi: 10.1111/j.1440-1819.2010.02091.x. [DOI] [PubMed] [Google Scholar]
- 10.Kendler KS, Thornton LM, Gardner CO. Stressful life events and previous episodes in the etiology of major depression in women: an evaluation of the “kindling” hypothesis. The American journal of psychiatry. 2000;157(8):1243–51. doi: 10.1176/appi.ajp.157.8.1243. [DOI] [PubMed] [Google Scholar]
- 11.Mayberg HS, Liotti M, Brannan SK, McGinnis S, Mahurin RK, Jerabek PA, et al. Reciprocal limbic-cortical function and negative mood: converging PET findings in depression and normal sadness. The American journal of psychiatry. 1999;156(5):675–82. doi: 10.1176/ajp.156.5.675. [DOI] [PubMed] [Google Scholar]
- 12.Seminowicz DA, Mayberg HS, McIntosh AR, Goldapple K, Kennedy S, Segal Z, et al. Limbic-frontal circuitry in major depression: a path modeling metanalysis. NeuroImage. 2004;22(1):409–18. doi: 10.1016/j.neuroimage.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Mayberg HS, Lozano AM, Voon V, McNeely HE, Seminowicz D, Hamani C, et al. Deep brain stimulation for treatment-resistant depression. Neuron. 2005;45(5):651–60. doi: 10.1016/j.neuron.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 14.Aston C, Jiang L, Sokolov BP. Transcriptional profiling reveals evidence for signaling and oligodendroglial abnormalities in the temporal cortex from patients with major depressive disorder. Molecular psychiatry. 2005;10(3):309–22. doi: 10.1038/sj.mp.4001565. [DOI] [PubMed] [Google Scholar]
- 15.Sequeira A, Mamdani F, Ernst C, Vawter MP, Bunney WE, Lebel V, et al. Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PloS one. 2009;4(8):e6585. doi: 10.1371/journal.pone.0006585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duric V, Banasr M, Stockmeier CA, Simen AA, Newton SS, Overholser JC, et al. Altered expression of synapse and glutamate related genes in post-mortem hippocampus of depressed subjects. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum. 2013;16(1):69–82. doi: 10.1017/S1461145712000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medina A, Watson SJ, Bunney W, Jr, Myers RM, Schatzberg A, Barchas J, et al. Evidence for alterations of the glial syncytial function in major depressive disorder. Journal of psychiatric research. 2016;72:15–21. doi: 10.1016/j.jpsychires.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray AL, Hyde TM, Deep-Soboslay A, Kleinman JE, Sodhi MS. Sex differences in glutamate receptor gene expression in major depression and suicide. Molecular psychiatry. 2015;20(9):1139. doi: 10.1038/mp.2015.114. [DOI] [PubMed] [Google Scholar]
- 19.Sibille E, Morris HM, Kota RS, Lewis DA. GABA-related transcripts in the dorsolateral prefrontal cortex in mood disorders. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum. 2011;14(6):721–34. doi: 10.1017/S1461145710001616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tripp A, Kota RS, Lewis DA, Sibille E. Reduced somatostatin in subgenual anterior cingulate cortex in major depression. Neurobiology of disease. 2011;42(1):116–24. doi: 10.1016/j.nbd.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tripp A, Oh H, Guilloux JP, Martinowich K, Lewis DA, Sibille E. Brain-derived neurotrophic factor signaling and subgenual anterior cingulate cortex dysfunction in major depressive disorder. The American journal of psychiatry. 2012;169(11):1194–202. doi: 10.1176/appi.ajp.2012.12020248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guilloux JP, Douillard-Guilloux G, Kota R, Wang X, Gardier AM, Martinowich K, et al. Molecular evidence for BDNF- and GABA-related dysfunctions in the amygdala of female subjects with major depression. Molecular psychiatry. 2012;17(11):1130–42. doi: 10.1038/mp.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanacora G, Mason GF, Rothman DL, Behar KL, Hyder F, Petroff OA, et al. Reduced cortical gamma-aminobutyric acid levels in depressed patients determined by proton magnetic resonance spectroscopy. Archives of general psychiatry. 1999;56(11):1043–7. doi: 10.1001/archpsyc.56.11.1043. [DOI] [PubMed] [Google Scholar]
- 24.Hasler G, van der Veen JW, Tumonis T, Meyers N, Shen J, Drevets WC. Reduced prefrontal glutamate/glutamine and gamma-aminobutyric acid levels in major depression determined using proton magnetic resonance spectroscopy. Archives of general psychiatry. 2007;64(2):193–200. doi: 10.1001/archpsyc.64.2.193. [DOI] [PubMed] [Google Scholar]
- 25.Sanacora G, Gueorguieva R, Epperson CN, Wu YT, Appel M, Rothman DL, et al. Subtype-specific alterations of gamma-aminobutyric acid and glutamate in patients with major depression. Archives of general psychiatry. 2004;61(7):705–13. doi: 10.1001/archpsyc.61.7.705. [DOI] [PubMed] [Google Scholar]
- 26.Levinson AJ, Fitzgerald PB, Favalli G, Blumberger DM, Daigle M, Daskalakis ZJ. Evidence of cortical inhibitory deficits in major depressive disorder. Biological psychiatry. 2010;67(5):458–64. doi: 10.1016/j.biopsych.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 27.Sanacora G, Mason GF, Rothman DL, Krystal JH. Increased occipital cortex GABA concentrations in depressed patients after therapy with selective serotonin reuptake inhibitors. The American journal of psychiatry. 2002;159(4):663–5. doi: 10.1176/appi.ajp.159.4.663. [DOI] [PubMed] [Google Scholar]
- 28.Sanacora G, Mason GF, Rothman DL, Hyder F, Ciarcia JJ, Ostroff RB, et al. Increased cortical GABA concentrations in depressed patients receiving ECT. The American journal of psychiatry. 2003;160(3):577–9. doi: 10.1176/appi.ajp.160.3.577. [DOI] [PubMed] [Google Scholar]
- 29.Choudary PV, Molnar M, Evans SJ, Tomita H, Li JZ, Vawter MP, et al. Altered cortical glutamatergic and GABAergic signal transmission with glial involvement in depression. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15653–8. doi: 10.1073/pnas.0507901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zubarev RA. The challenge of the proteome dynamic range and its implications for in-depth proteomics. Proteomics. 2013;13(5):723–6. doi: 10.1002/pmic.201200451. [DOI] [PubMed] [Google Scholar]
- 31.Lundberg E, Fagerberg L, Klevebring D, Matic I, Geiger T, Cox J, et al. Defining the transcriptome and proteome in three functionally different human cell lines. Molecular systems biology. 2010;6:450. doi: 10.1038/msb.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wisniewski JR, Dus-Szachniewicz K, Ostasiewicz P, Ziolkowski P, Rakus D, Mann M. Absolute Proteome Analysis of Colorectal Mucosa, Adenoma, and Cancer Reveals Drastic Changes in Fatty Acid Metabolism and Plasma Membrane Transporters. Journal of proteome research. 2015;14(9):4005–18. doi: 10.1021/acs.jproteome.5b00523. [DOI] [PubMed] [Google Scholar]
- 33.Andreev VP, Petyuk VA, Brewer HM, Karpievitch YV, Xie F, Clarke J, et al. Label-free quantitative LC-MS proteomics of Alzheimer’s disease and normally aged human brains. Journal of proteome research. 2012;11(6):3053–67. doi: 10.1021/pr3001546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C, Qu B, Wang Z, Ju J, Wang Y, Wang Z, et al. Proteomic identification of differentially expressed proteins in vascular wall of patients with ruptured intracranial aneurysms. Atherosclerosis. 2015;238(2):201–6. doi: 10.1016/j.atherosclerosis.2014.11.027. [DOI] [PubMed] [Google Scholar]
- 35.Martins-de-Souza D, Guest PC, Harris LW, Vanattou-Saifoudine N, Webster MJ, Rahmoune H, et al. Identification of proteomic signatures associated with depression and psychotic depression in post-mortem brains from major depression patients. Translational psychiatry. 2012;2:e87. doi: 10.1038/tp.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gottschalk MG, Wesseling H, Guest PC, Bahn S. Proteomic enrichment analysis of psychotic and affective disorders reveals common signatures in presynaptic glutamatergic signaling and energy metabolism. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum. 2015;18(2) doi: 10.1093/ijnp/pyu019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wesseling H, Gottschalk MG, Bahn S. Targeted multiplexed selected reaction monitoring analysis evaluates protein expression changes of molecular risk factors for major psychiatric disorders. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum. 2014;18(1) doi: 10.1093/ijnp/pyu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karolewicz B, Maciag D, O’Dwyer G, Stockmeier CA, Feyissa AM, Rajkowska G. Reduced level of glutamic acid decarboxylase-67 kDa in the prefrontal cortex in major depression. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum. 2010;13(4):411–20. doi: 10.1017/S1461145709990587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glantz LA, Austin MC, Lewis DA. Normal cellular levels of synaptophysin mRNA expression in the prefrontal cortex of subjects with schizophrenia. Biological psychiatry. 2000;48(5):389–97. doi: 10.1016/s0006-3223(00)00923-9. [DOI] [PubMed] [Google Scholar]
- 40.Sibille E, Wang Y, Joeyen-Waldorf J, Gaiteri C, Surget A, Oh S, et al. A molecular signature of depression in the amygdala. The American journal of psychiatry. 2009;166(9):1011–24. doi: 10.1176/appi.ajp.2009.08121760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scifo E, Szwajda A, Soliymani R, Pezzini F, Bianchi M, Dapkunas A, et al. Proteomic analysis of the palmitoyl protein thioesterase 1 interactome in SH-SY5Y human neuroblastoma cells. Journal of proteomics. 2015;123:42–53. doi: 10.1016/j.jprot.2015.03.038. [DOI] [PubMed] [Google Scholar]
- 42.Karpievitch YV, Dabney AR, Smith RD. Normalization and missing value imputation for label-free LC-MS analysis. BMC bioinformatics. 2012;13(Suppl 16):S5. doi: 10.1186/1471-2105-13-S16-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding Y, Chang LC, Wang X, Guilloux JP, Parrish J, Oh H, et al. Molecular and Genetic Characterization of Depression: Overlap with other Psychiatric Disorders and Aging. Molecular neuropsychiatry. 2015;1(1):1–12. doi: 10.1159/000369974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic acids research. 2017;45(D1):D183–D9. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wesseling H, Gottschalk MG, Bahn S. Targeted multiplexed selected reaction monitoring analysis evaluates protein expression changes of molecular risk factors for major psychiatric disorders. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum. 2015;18(1) doi: 10.1093/ijnp/pyu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuge H, Akahori K, Yagyu K, Honke K. Functional compartmentalization of the plasma membrane of neurons by a unique acyl chain composition of phospholipids. The Journal of biological chemistry. 2014;289(39):26783–93. doi: 10.1074/jbc.M114.571075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foster TC. Calcium homeostasis and modulation of synaptic plasticity in the aged brain. Aging cell. 2007;6(3):319–25. doi: 10.1111/j.1474-9726.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- 48.Kasai H, Fukuda M, Watanabe S, Hayashi-Takagi A, Noguchi J. Structural dynamics of dendritic spines in memory and cognition. Trends in neurosciences. 2010;33(3):121–9. doi: 10.1016/j.tins.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 49.Bader V, Tomppo L, Trossbach SV, Bradshaw NJ, Prikulis I, Leliveld SR, et al. Proteomic, genomic and translational approaches identify CRMP1 for a role in schizophrenia and its underlying traits. Human molecular genetics. 2012;21(20):4406–18. doi: 10.1093/hmg/dds273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta DS, McCullumsmith RE, Beneyto M, Haroutunian V, Davis KL, Meador-Woodruff JH. Metabotropic glutamate receptor protein expression in the prefrontal cortex and striatum in schizophrenia. Synapse. 2005;57(3):123–31. doi: 10.1002/syn.20164. [DOI] [PubMed] [Google Scholar]
- 51.Afshari P, Myles-Worsley M, Cohen OS, Tiobech J, Faraone SV, Byerley W, et al. Characterization of a novel mutation in SLC1A1 associated with schizophrenia. Molecular neuropsychiatry. 2015;1(3):125–44. doi: 10.1159/000433599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lewis DA. The human brain revisited: opportunities and challenges in postmortem studies of psychiatric disorders. Neuropsychopharmacology: official publication of the American College of Neuropsychopharmacology. 2002;26(2):143–54. doi: 10.1016/S0893-133X(01)00393-1. [DOI] [PubMed] [Google Scholar]
- 53.Sibille E, French B. Biological substrates underpinning diagnosis of major depression. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum. 2013;16(8):1893–909. doi: 10.1017/S1461145713000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: MS-based proteomics workflow for analysis of sgACC gray matter. Human postmortem subgenual cingulate cortex samples from control subjects and MDD cohorts were processed using our MS-based proteomics approach. The strategy for analysis of the brain samples comprised of 4 key steps: tissue extraction, sample preparation, LS-MS/MS analysis and bioinformatics.
Supplementary Figure S2: Histogram of p-value distribution for identified proteins associated with all MDD patients in comparison to controls, with sex as a cofactor. The p-values for all the identified proteins with sex as a cofactor are evenly distributed from 0 through 1, which is very close to the distribution of p-value under null.
Supplementary Figure S3: Histogram of p-value distribution for identified proteins associated with MDD patients in recurrence in comparison to controls, with antipsychotic medications as a cofactor. The p-values for all the identified proteins with antipsychotic medications as a cofactor are evenly distributed from 0 through 1, which is very close to the distribution of p-value under null.
Supplementary Figure S4: Plot of batch effect on protein expression distribution in controls and MDD cohorts. The log2 intensity and batches are shown on the y and x-axes, respectively. The plot indicates that there is no batch effect on protein expression distribution in the samples analyzed.
Supplementary Table S1: Description of MDD cohorts and control subjects used in the study.
Postmortem brain samples were collected from control subjects (group I, n=20) and four major MDD cohorts corresponding to single episode (group II, n=20), single episode remission (group III, n=15), recurrent (group IV, n=20), and recurrent-partial/full remission (group V, n=15). DSM-IV diagnoses, mode of death (MOD), cause of death (COD) and demographic features (including: sex, age, race, PMI, pH, RNA ratio, RIN) of each subject are identified. A few control subjects with a DSM diagnosis were used and their duration of illness (DOI) is entered in parentheses (). The DOI is determined from the first depressive episode until remission or death, whichever comes first. The DSM-IV diagnosis is at time of death whereas psychosis history is lifetime. For all others subjects DOI is only for the MDD diagnosis. Abbreviations: ASCVD, atherosclerotic cardiovascular disease; ADC, Alcohol Dependence, current at time of death; ADR, Alcohol Dependence, in remission at time of death; AAC, Alcohol Abuse, current at time of death; AAR, Alcohol Abuse, in remission at time of death; ODC, Other Substance Dependence, current at time of death; ODR, Other Substance Dependence, in remission at time of death; OAC, Other Substance Abuse, current at time of death and OAR, Other Substance Abuse, in remission at time of death; Meds/Tob/Dep ATOD, Prescribed medications, tobacco use and depression at time of death; B, Benzodiazepines; C, Anticonvulsants; D, Antidepressants; N, No medications; O, Other medication(s); P, Antipsychotic; T, Tobacco; U, Unknown; V, Valproic Acid; X, Medications unknown; N/A, not applicable; 0, absence of trait; 1, presence of trait and 0/1, partial trait.
Supplementary Table S2: Analysis of MS data from MDD cohorts and control subjects by MaxQuant.
S2-1: Label free quantitation of MS data from MDD cohorts and control subjects by MaxQuant analysis-unfiltered. S2-2: Label free quantitation of MS data from MDD cohorts and control subjects by MaxQuant analysis-filtered. S2-3: Testing of hypotheses in support or not of MDD episodes and remission phases. S2-4: Differentially expressed proteins associated with a persistent disease effect in all MDD patients (hypothesis 1). S2-5: Differentially expressed proteins associated with the MDD state of patients in current episodes (hypothesis 1a). S2-6: Differentially expressed proteins associated with the residual MDD state of patients in remission (hypothesis 1b). S2-7: Differentially expressed proteins associated with distinct MDD state for patients in current episodes in comparison to those in remission (hypothesis 2). S2-8: Differentially expressed proteins associated with distinct MDD state for patients in remission in comparison to those in current episodes (hypothesis 3). S2-9: Differentially expressed proteins associated with neuro-progressive pathology across MDD episodes and remission phases (hypothesis 4). S2-10: Differentially expressed proteins associated with a progressive MDD effect for patients in current episodes (hypothesis 4a). S2-11: Differentially expressed proteins associated with a progressive MDD effect for patients in remission (hypothesis 4b). Post hoc analysis to test for the effects of psychosis, alcohol dependence, antidepressant drug use and death by suicide on differential protein expression was performed using analysis of variance (ANOVA).
Supplementary Table S3: Summary of Western blot validation of selected differentially expressed proteins.
Selected differentially expressed proteins (DRP-1, SNAP-29, GAD-67, mGluR1 and EAAT3) in all MDD patients based on MS analysis, were validated by Western blot analysis. The directionality of protein changes observed by Western blot analysis was consistent with findings from the MS data. DRP-1 and SNAP-29 were upregulated, whereas GAD-67, mGluR1 and EAAT3 were observed to be downregulated in both analyses.
Supplementary Table S4: Canonical pathways associated with a persistent disease effect in all MDD patients (hypothesis 1).
S4-1: The pathways associated with differentially expressed (DE) proteins associated with a persistent disease effect in all MDD patients (hypothesis 1) at p ≤ 0.05 and RIM Coefficient effect ≥ ± 0.26 thresholds, were identified using Ingenuity Pathway Analysis (IPA). S4-2: Pathways associated with the DE proteins in all MDD patients at less stringent p ≤ 0.1 and RIM Coefficient effect ≥ ± 0.26 thresholds.
Supplementary Table S5: Overview of differentially expressed proteins associated with MDD episodes and remission phases.
Differentially expressed proteins were identified based on p ≤ 0.1 and RIM Coefficient effect ≥ ± 0.26 thresholds. The biological significance of differentially expressed proteins in the MDD cohorts and association with disease categories were explored by GO biological process using PANTHER and Ingenuity Pathway Analysis (IPA), respectively.
Supplementary Table S6: Post hoc analysis of identified proteins associated with all MDD patients in comparison to controls, with adjustment of one cofactor at a time.
Generated p-values associated with individual cofactors served as a measure of their respective importance. This analysis indicated that sex had little effect on the p-value of identified proteins in comparison to the other cofactors, e.g. pH and Age, which we selected and adjusted through the random intercept model (RIM).
Supplementary Table S7: Post hoc analysis of identified proteins associated with MDD patients in recurrence in comparison to controls, with adjustment of one cofactor at a time.
This analysis indicated that antipsychotic medications had little effect on the p-value of identified proteins in comparison to the other cofactors, e.g. pH and Age, which we accounted for in the random intercept model (RIM).