Abstract
The CEGS N-GRID 2016 Shared Task (Filannino, Stubbs, Uzuner (2017)) in Clinical Natural Language Processing introduces the assignment of a severity score to a psychiatric symptom, based on a psychiatric intake report. We present a method that employs the inherent interview-like structure of the report to extract relevant information from the report and generate a representation. The representation consists of a restricted set of psychiatric concepts (and the context they occur in), identified using medical concepts defined in UMLS that are directly related to the psychiatric diagnoses present in the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV) ontology. Random Forests provides a generalization of the extracted, case-specific features in our representation. The best variant presented here scored an inverse mean absolute error (MAE) of 80.64%. A concise concept-based representation, paired with identification of concept certainty and scope (family, patient), shows a robust performance on the task.
Keywords: Symptom Severity Identification, Natural Language Processing, Concept Detection, Psychiatry, Positive Valence
Graphical Abstract
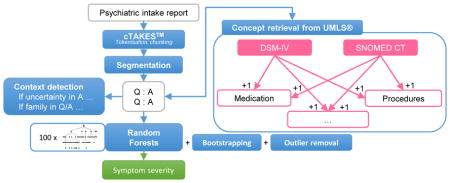
1. Introduction
A large portion of information in hospitals is stored in the form of unstructured clinical documents. Examples are doctor’s notes, daily progress reports, and intake reports. To assist decision making, relevant information in such reports needs to be extracted, for which machine learning methods are typically used. This extracted information can be used as input for several applications, such as the identification of diagnostic criteria for heart failure [1, 2] or the prediction of diagnosis and procedure codes [3]. Since the effectiveness of data-driven algorithms not only depends on data size, but also on the quality of data representation, robust information extraction techniques are required [4, 5]. These techniques need to extract qualitative concepts from clinical texts which tend to have misspellings and ungrammatical sentences.
1.1. Background
One of the basic methods of representing information in a text is as a ‘bag of words’, considering individual words as the primary source of meaning. Medical information, however, is often present in the form of a medical multi-word expression (mMWE) such as ‘alcohol abuse’. Techniques to extract these mMWEs often use knowledge resources such as the UMLS metathesaurus to map medical concepts onto texts fragments [6].
Medical concept detection has been the target of several shared tasks. The first track of the 2010 i2b2/VA shared task [7] challenged the participants to identify medical entities and label them as a ‘problem’, ‘treatment’, or ‘test’. The best-performing system used a wide range of features for this purpose, including n-grams and document-level features [8]. Their results demonstrated the beneficial impact of using semantic and syntactic features based on concepts derived from UMLS and concept mapping modules in cTAKES [9]. In the ShARe/CLEF 2013 eHealth shared task [5], the goal was to recognize mMWEs in clinical notes and normalize them to UMLS Concept Unique Identifiers (CUIs). The best-ranking system applied supervised learning techniques, representing candidates as a tf-idf weighted bag of words [10]. The detection of concepts can also be supported using distributional semantics. Concepts can then be assigned to words that have a similar semantic context to certain concepts, but are not present themselves in a lexicon [11].
Aiming for a complete and accurate representation of all concepts occurring in a text is a different focus than applying concept detection methods to a specific task (such as scoring the severity for a specific symptom). In that case, an unfiltered list of detected concepts may not be the optimal representation for a clinical text. Different methods can be suggested to zoom in on the task-specific relevant information. Often, such mechanisms improve algorithm performance, particularly in situations where training data is limited [12, 13]. This motivates us to look for relevant medical factors, yielding a more concise document representation.
The second track of the 2014 i2b2/UTHealth shared task [2] hypothesized that, to identify medical risk factors in longitudinal medical records, and in particular to assess the severity of a risk factor, identifying indicators of a disease would be more informative than identifying the actual diagnosis. If hypertension, for instance, can be managed through diet and exercise, it would be considered less severe than when it requires anticoagulants. The best performing team did not directly rely on syntactic or semantic information, but rather focused on improving the annotated training data by annotating the negation markers present in the text but absent in the annotated indicators [14]. For instance, the concept ‘diabetes’ was extended to ‘no diabetes’ if it was used in a negated context. This demonstrates that having detailed concept annotations can be useful to identify risk factors in a patient’s medical record.
The detection of negation, social context, and family history is crucial for several tasks. Garcelon et al. used unstructured information within the electronic health record (EHR) to identify rare diseases [15]. Negation detection is crucial, since mentions of rare diseases are often negated, and negated symptoms, combined with other factors, can be an indicator of a rare disease. Intake reports that have an interview style also benefit from such context detection, especially because a large proportion of the questions are answered negatively, and/or often refer to patient’s surroundings and family.
An (already sparse) concept-based representation of clinical text becomes more sparse with the addition of context to the feature representation. To adequately deal with an approach that uses sparse features, we need to either generalize these features, or use a classifier that is able to deal with sparsity without overfitting. Random Forests [16] is one algorithm that deals well with sparsity. The individual trees are designed to overfit on features (making very specific decisions that only account for part of the dataset), while the voting strategy later mitigates these effects (by generalising over the decisions of multiple trees).
1.2. CEGS N-GRID 2016 Shared Task
The Research Domain criteria (RDoc), a framework established by the National Institute of Mental Health (NIMH), describe human behavior, from normal to abnormal, by means of functional constructs. The RDoC domain in focus for the task at hand is positive valence, which refers to “Systems primarily responsible for responses to positive motivational situations or contexts, such as reward seeking, consummatory behavior, and reward/habit learning” [17]. The severity of positive valence, as a symptom, describes the ability of a person to control the processes influencing these systems (e.g., addictions, obsessive compulsive disorders).
The CEGS N-GRID 2016 Shared Task focuses on classifying symptom severity in the positive valence domain based on psychiatric intake reports [18]. In this paper, we target the task of symptom severity prediction as a multi-class classification task where the different severity scores are the labels we assign to an intake report. We have developed techniques to present the intake report to a classifier in a simple, interpretable manner. We attempt to minimize the manual collection of expert knowledge for this task specifically, to allow for scalability. The approach we present strongly builds on the questionnaire structure of the documents and on the ability for Random Forests to generalize over case-specific features generated using UMLS as a source collection of ontologies for medical concept detection.
2. Materials and methods
2.1. Dataset
The dataset available from the shared task consists of a training set of 600 psychiatric intake reports and a test set of 216 reports. The training set itself has 325 reports annotated by two or more annotators, 108 reports annotated by one annotator, and 167 unannotated reports. The annotation consists of one score per report, indicating the severity of symptoms in the positive valence domain exhibited by that person. This score corresponds to four distinct categories: absent, mild, moderate, and severe, presented as discrete classes 0, 1, 2, and 3.
A psychiatric intake report documents a first interview conducted with the patient. This interview contains case-based semi-standardized questions, which can contain a short or elaborate answer. It always contains a section in which the patient explains in his own words his or her motivation for coming to the hospital. A question is often preceded by the category on which the question applies, such as ‘OCD’ or ‘Bipolar disorder’. In Figure 1, an excerpt of an intake report is shown.
Figure 1.
An excerpt of an intake report.
2.2. Medical information extraction
Raw text needs to be processed to generate feature vectors before feeding it to a machine learning algorithm. Different levels of preprocessing are performed: text normalization, identification of crucial information like medical concepts, identification of the scope of a certain medical concept, and identification of linguistic markers - such as negation cues and family cues - that influence the factuality or context of the medical concept.
In our pipeline1, linebreaks are restored (e.g., ‘heart burnPsychiatric’) using capitalization cues. The raw texts are then processed using the cTAKES Natural Language Processing (NLP) pipeline [9] to identify sentence boundaries, token boundaries, part-of-speech tags, and syntactic chunks such as noun phrases. The resulting text is then processed further (outside of cTAKES) to extract information in the form of medical concepts with a custom concept detection algorithm.
2.3. Medical Multi-Word Expression (mMWE) detection
A weighted partial matching algorithm is used to detect medical Multi-Word Expressions (mMWE) in the psychiatric intake report. The individual words in each noun phrase are matched to definitions of concepts extracted from the UMLS metathesaurus [6]. These matches result in a weighted sum of the scores for all word matches, where each word is weighted according to its Inverse Document Frequency (IDF), for which each concept definition was considered a document [19]. Using IDF limits the contribution of frequent words while mapping concept definitions. A concept definition is mapped onto a noun phrase when a score of at least 80% is achieved. Multiple concepts can be assigned to a single noun phrase. For example, when mapping the noun phrase ‘obsessive compulsive spectrum deviation’ to the concept ‘obsessive compulsive disorder’, both ‘obsessive’ and ‘compulsive’ have an IDF of 2. The word ‘spectrum’ does not occur in the definition and as such does not contribute to the total score. ‘Disorder’ is a more common word, with an IDF of 1, but it is not mapped either, since ‘deviation’ is present in the noun phrase instead. Using the IDF scoring method, we can map the noun phrase to the concept with a certainty of 80%, since the sum of IDF of the matched words is 4, out of 5 as the total sum of IDF for all words in the definition. This score would not have been reached with a simple exact match approach, which would yield a certainty of 67% (2 out of 3 words corresponded with the definition).
2.4. Text segmentation
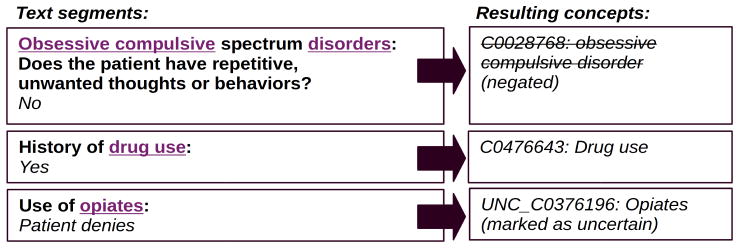
The format of the psychiatric intake report consists of headers (often in the form of a question), followed by content related to that header. A segmentation algorithm is used to convert text into header-content pairs. This allows us to distinguish between elements occurring in headers on the one hand and content on the other, and to keep track of the relation between a header and content. Individual sentences are automatically identified as either a question or an answer. Regular expressions are used to detect cues in the last character of a sentence, such as a colon or a question mark, to tag them as header or content. Typically, each question ends in a colon and a newline, whereas the content starts with a capitalized first letter, hence allowing us to use colons as sentence endings. In addition to colons and question marks, some exceptions are added to categorize certain sentences as a header. Consecutive sentences belonging to the same category are collapsed to prevent pairs with only partial information. Figure 2 shows the resulting segmentation and its effect on the concepts that were mapped.
Figure 2.
An example of segmentation and its effect on extracted concepts. A segment consists of a header and its content. For each segment, the header is marked in bold, the content is marked in italics and the context from the segment is projected onto the extracted concepts. Underlined text indicates the text that triggered a concept to be found.
2.5. Context detection
In a segment consisting of a header and content, information within a header can be confirmed or denied by analyzing the content. This largely influences the applicability of information found within the header to the patient. Contextual relations between the header and content are modelled using lexical cues. Negation is detected by retrieving lexical negation cues (e.g., ‘no’, ‘none’) at the start of the content. If negation has been detected, the corresponding header and its associated concepts are considered negated. Preliminary experiments have been conducted to extend the scope of negation to the content itself as well, but this did not yield improved performance. It is likely that follow-up content after negation (e.g., ‘No’, ‘None’) provides additional relevant information. Expressions indicating doubt from the interviewer’s point of view are also detected and marked as uncertain. This is based on our hypothesis that the choice of words by a psychiatrist indicates whether they are apprehensive about the reliability of the patient’s answer. Such uncertainty is captured by identifying usage of terms such as ‘patient denies’ as opposed to direct answers like ‘absent’ or ‘no’.
In some cases, the information does not apply to the patient, but rather to his/her environment or family. This is detected by using lexical cues in the header: if the header mentions ‘family’, concepts falling within the scope of the entire segment are marked as family-related.
The heuristics used to identify the context for the medical concepts as well as the scope of this context rely on the inherent structure of the psychiatric intake reports and may not be directly transferrable to other types of clinical notes. To summarize, in each model, unless explicitly mentioned otherwise, context is taken into account by removing concepts occurring in the header of a negated segment; by separating out concepts in the header of an uncertain segment; and by separating out concepts in the entire segment that has been tagged as family related.
2.6. Medical Information Representation
After the identification of header-content pairs, the identification of UMLS concepts, and the addition of context markers to each concept, the intake reports are used to derive several types of features which can then be used in a machine learning algorithm.
2.6.1. Baseline
Three baseline approaches were implemented: (1) a random baseline where a random severity score is assigned to an intake report, (2) a bag-of-words model which represents each psychiatric intake report as a list of individual words along with their frequency in the training vocabulary, and (3) a model using all concepts derived from UMLS as separate features, with the corresponding concept occurrences as values (we will refer to this as the UMLS model).
2.6.2. UMLS Subsets: DSM, DSM+1, DSM+2, SNOMED+1
Our UMLS metathesaurus set consists of 4,487,085 distinct concept definitions. By creating subsets of UMLS concepts, the number of concepts, hence features provided to the machine learning algorithm, can be reduced to a set of facts relevant to the psychiatric domain. A subset of UMLS concepts is defined in the DSM-IV ontology [20] as a set of psychiatric diagnoses. While this subset (which we refer to as DSM) contains diagnoses relevant for the symptom severity classification task, it provides a limited set of 8,123 purely diagnostic definitions, affecting its applicability to symptom identification in the psychiatric intake report. On expanding this list with all concepts in UMLS (a representation we will refer to as DSM+1) that can be directly related to one of the DSM-IV concepts, the resulting list of 84,563 definitions includes concept categories like symptoms and medications. A second expansion creates a list of definitions that are a maximum of two steps away from a DSM-IV code (referred to as DSM+2), with 1,252,709 definitions. Each concept in this list is thus not always directly related to a certain DSM-IV code, but one of its directly related concepts is. For example, ‘C0426582: Cannot face food’ cannot be directly related to a definition from the DSM-IV, but its related definition ‘C0029587: other eating disorders’ can be related to the DSM-IV concept ‘C0013473: eating disorders’. One concept can have multiple definitions, which results in a normalization step, as all these definitions are mapped to the same concept identifier.
As an alternative to identifying psychiatric concepts by means of the DSMIV ontology, we extracted SNOMED codes relating to ‘116367006 Psychological finding’ [21]. The SNOMED+1 variant (all concepts directly related to this set of SNOMED codes) was used for the experiments below. Preliminary results with SNOMED showed that the subset was too restrictive.
2.6.3. Question sets
We used the extracted header-content segments to discern three types of headers: closed questions, categorical headers, and elaborate response headers. A closed question starts with an affirmative, uncertain, or negative answer (or no answer at all) (e.g., ‘History of drug use: no’). Closed questions are represented with a continuous scale, where a denial yields 0 as feature value and an affirmation yields 1. Categorical headers need to have a brief, limited set of potential answers (e.g., ‘Appearance: neat’). Different answers in the training set are used as possible categories for the feature. Elaborate headers are all headers (including some questions) that are not categorical. This type of headers is represented by the concepts found in both the header and the content. Closed questions can have an elaborate answer in addition to a yes/no answer, which causes features for both a closed question and an elaborate question to be generated. By identifying header types, we enable the creation of different types of features for each header. For example, the question ‘Is the patient depressed?’ can be a feature in itself, as it can be answered with yes or no, while a question/header ‘chief complaint’ is not very meaningful as a feature in itself, because the answer is often complex and cannot be reduced to a single value.
The different types of headers are determined during the training phase. This ensures that a question is treated as the categories where it frequently has a certain type of answer. A content type needs to occur in 30% of the answers to the header and the header needs to occur in 14% of the intake reports before it is considered as a separate feature.
2.6.4. Other derived feature sets
We tried several other techniques to generate a concise text representation which we briefly discuss here, but do not include into the results section. For these approaches, we used a distributed semantic model (word2vec) [22]. Concepts were represented by the sum of vectors of the individual words within the concept [23], and differences between vectors were calculated using cosine distance. The vectors contained 300 dimensions.
One of our approaches identifies the distance between different symptoms, to check how many different types of symptoms are present in the patient. The distance can either be the distance between two symptoms in the UMLS tree, or the cosine distance between embedded vectors of the concepts representing those symptoms. A second concise representation technique grouped different related questions and answers into one. Questions were considered related when they either had several key concepts in common (e.g., two questions with ‘depressed’ in their definition), or had concepts with a similar vector representation (e.g., ‘depressed’ and ‘feeling down’, which have two similar concept vectors). Several thresholds were explored for this purpose. A third method directly used an averaged vector representation of the vectors of all occurring concepts in an intake report instead of using the concepts themselves as a representation. However, experimental results have shown that these approaches scored systematically worse than the other approaches introduced above.
We also added another set of features, which involved the identification of medication mentions in the text based on RxNorm and ATC-definitions within UMLS. These were then normalized to active compounds. We hypothesized that differences in medication prescriptions across different groups of patients could indicate symptom severity. The performance of models using this feature set was below par, since the other feature sets using concepts already included medication, rendering this approach redundant.
2.7. Bootstrapping and outlier removal
Since the training set has a limited number of documents, individual instances (assuming one instance per document) can have a large impact on the resulting model. To improve generalizability of the model on the small number of instances available, we use a bootstrapping technique on the unannotated portion of the data (which can increase the training set with up to 50% new instances). In addition, we can select instances that are more prototypical for the severity class they belong to, to avoid overfitting on outlier instances. Boot-strapping and instance selection are used to make the resulting model more robust.
Bootstrapping (also referred to as self-training) is performed by predicting the severity of positive valence symptoms on the unannotated dataset. The model is then retrained on the original training data as well as the previously unannotated instances with a prediction confidence higher than 60% (threshold determined using a grid search). This increases the number of instances the model can use to make a decision.
Outlier removal is performed by looking at the ratio of similarity of each instance to the instances in its own class and to its similarity to instances in other classes. Similarity is determined by calculating the average cosine distance between all feature vectors. Instances that are less similar to their own class than to other classes will have a lower ratio. In the presented setup of the task as a discrete classification task (as opposed to a continuous scale of severity), these instances decrease the discriminative quality of the training set, as the decision boundary will be modelled to include ambiguous instances to a certain class. For each class, the instances with the 5% lowest ratio are removed.
2.8. Experimental Setup
The features generated were first processed with a variance filter that filters out all features whose values vary by less than 5% among all instances. Subsequently, they were presented to a Random Forests classifier [16] for symptom severity classification. Random Forests was preferred over other algorithms due to its robustness to overfitting, its ability to make a good prediction on few instances, and the inherent generalization it can perform over features that are case-specific (e.g., alcohol abuse is an indicator for positive valence, but it is one of many indicators). The generalization of Random Forests expresses itself in the way trees are formed: all trees form an independent (cf. random) combination of features to assign a certain class, which means that several paths leading to a severity score can be present in several trees. Due to the training of an individual tree on a subset of the training data, patterns that are not universally present, but that are strong indicators for a few instances, can get more weight in that data subset and are thus more easily used as a decision boundary.
We also performed experiments with SVMs, but SVMs were consistently out-performed by Random Forests. The Random Forests classifier is trained using 100 trees (based on preliminary experiments). During the development phase, experiments were conducted using ten-fold cross-validation on the training set. In the testing phase, the entire training set was used to do prediction on the official test set provided by the CEGS N-GRID 2016 Shared Task [18].
We compare performance of eight models below (of which the first three are baseline models). One model used a random baseline, in which the four categories got randomly assigned to instances. The second model used a bag-of-words as features. The third and fourth model used all detected UMLS concepts, once with and once without taking context into account. The four other models applied different subsets of UMLS concepts to see whether restricting the set of medical concepts to those related to the psychiatric domain better matches the task.
Three models were selected for submission to the CEGS-NGRID shared task [18]. One model used the question-set features, while taking context into account. Two models used the DSM+1 feature set, while taking context into account. The latter of these models also used bootstrapping and instance selection, to improve quantity and quality of the training set.
2.9. Evaluation metrics
The experiments are evaluated using both inverse mean absolute error (MAE) and micro-averaged F-measure. The inverse mean absolute error is the inverse of the average error when predictions are made. This measure is weighted by the severity of the error: a predicted score deviating more from the actual class also yields a larger error. It also corrects for class imbalance: instances from a class with a lower occurrence rate will be given a higher weight than they would get based on their frequency. MAE was used as the official metric during the CEGS N-GRID 2016 Shared task [18].
Micro-averaged F-measure is the harmonic mean between precision and recall and does not apply instance weighting. It is an evaluation of the percentage of correctly assigned classes and offers a more strict evaluation - especially for determining the exact class - than inverse mean absolute error.
3. Results
Table 1 shows the results during the development phase (with cross-validation) and during the testing phase. The results on the test set consistently outperform results seen during the development phase. This can be explained by the former having 10% more training data (during the development phase, only 90% of the available annotated data is used for training). Since the algorithms have access to more training data during the testing phase, we can assume that a plateau has not been reached. An overview of results with a grid search over parameters can be found in supplementary materials. In Table 2, we see the increase/decrease for each additional parameter, averaged over all different sets of features.
Table 1.
Results for the different models, both during 10-fold cross-validation (CV) and on the test run. Models marked in bold were submitted to the challenge.
| Development phase (10-fold CV) | Testing phase | |||
|---|---|---|---|---|
| System | MAE | F-measure | MAE | F-measure |
| Random baseline | 50.96% (SD 5.32) | 24.44% (SD 5.26) | 57.28% | 30.90% |
| Bag of words (baseline) | 74.65% (SD 5.05) | 43.53% (SD 9.15) | 75.91% | 54.31% |
| UMLS concepts (baseline) | 72.76% (SD 4.42) | 42.88% (SD 7.95) | 72.88% | 47.37% |
| UMLS concepts with context | 75.49% (SD 3.73) | 51.21% (SD 8.60) | 79.41% | 62.42% |
| DSM with context | 72.82% (SD 3.22) | 47.13% (SD 6.32) | 71.91% | 49.95% |
| DSM+1 with context | 78.30% (SD 2.65) | 57.05% (SD 4.58) | 79.52% | 61.15% |
| DSM+1 with context, bootstrapping, outlier removal | 78.77% (SD 3.61) | 56.70% (SD 6.66) | 80.64% | 63.67% |
| DSM+2 with context | 76.81% (SD 3.73) | 53.62% (SD 5.52) | 79.78% | 61.09% |
| SNOMED+1 with context | 75.97% (SD 3.50) | 52.69% (SD 6.06) | 79.73% | 61.38% |
| Question sets | 78.01% (SD 2.63) | 53.57% (SD 5.24) | 79.34% | 60.46% |
Table 2.
Results over different parameter settings, averaged for all different sets of concepts (Bag of words, UMLS concepts, DSM, DSM+1, DSM+2, Snomed, Snomed+1). These results indicate the general trend seen when adding a certain parameter to the model
| Development phase (10-fold CV) | Testing phase | |||
|---|---|---|---|---|
| System | MAE | F-measure | MAE | F-measure |
| No context | 72.33% | 43.34% | 75.03% | 52.73% |
| Context added | 75.79% | 52.26% | 77.90% | 58.77% |
| Context, bootstrapping, outlier detection added | 74.94% | 50.73% | 77.85% | 58.51% |
The three models submitted to the CEGS N-GRID 2016 Shared task (DSM+1, question sets, and DSM+1 with bootstrapping and outlier removal) outperformed other models and variants during the development phase. In the testing phase, the DSM+2 and UMLS concepts+context also perform well on the test-set.
All models outperform the random baseline, but the simple bag-of-words approach seems harder to beat. The results for models that did take negated answers, family history, and uncertainty cues into account are better than models that did not have access to context cues. Restricting to DSM-only concepts has a negative impact on performance.
In Table 3, we see the confusion matrix of predictions by the DSM+1 model. We show the confusion matrix summed over the results of ten-fold cross validation during development. Cross-validation provides a more robust result than results from a single experiment during the test phase. The absent and mild severity labels are predicted best, while predictions for moderate and severe cases are more spread. The mild category seems to be the most often assigned class (due to its higher frequency). Moderate cases are often (30%) misclassified as mild and there is also a substantial amount of confusion (25%) with the severe class. Severe cases are confused with mild and moderate similarly.
Table 3.
Confusion matrix of the DSM+1 model in 10-fold cross validation setup (sum of all folds).
| Prediction | |||||
|---|---|---|---|---|---|
| Absent | Mild | Moderate | Severe | ||
| Gold | Absent | 36 | 22 | 3 | 0 |
| Mild | 9 | 142 | 10 | 5 | |
| Moderate | 11 | 33 | 38 | 28 | |
| Severe | 1 | 22 | 24 | 49 | |
3.1. Feature Analysis
In Random Forests, the importance of a feature is determined by how often that feature is used as the pivot point with the best information gain (internal scoring metric in random forests). A higher scoring feature is thus more often used as a decisive factor. Below, we interpret the results based on the features that were deemed important by Random Forests, for qualitative analysis. This analysis is performed on the results of ten-fold cross validation. We have extracted the twenty most important features for both the DSM+1 model and the question sets model, as seen in Table 4.
Table 4.
The twenty most important features according to the DSM+1 model and the question sets model. Feature importance (in %) is shown between brackets. The features have been rephrased for improved readability. ‘DSM+1’ and ‘Elaborate Header’ features consist of a UMLS concept unique identifier (CUI), while ‘Closed question’ and ‘Categorical header’ features consist of the header itself.
| Rank | DSM+1 | Question sets |
|---|---|---|
| 1 | Meta feature representing the number of DSM+1 concepts (3.0%) | (Elaborate header) C0439857; Patient dependence on (1.9%) |
| 2 | C0344315; Feeling Down (2.9%) | (Closed question) Depression: Did the patient feel little pleasure doing things for a period lasting weeks? (1.9%) |
| 3 | C0439857;Patient dependence on (2.7%) | (Closed question) Depression: Did the patient feel sad or depressed for a period lasting weeks? (1.9%) |
| 4 | C0004927;Behaviour (2.5%) | (Closed question) Psychiatric history: Has there been any inpatient treatment in the past? (1.4%) |
| 5 | C0013080;Down Syndrome (2.3%) | (Closed question) OCD: does the patient struggle with repetitive unwanted thought or behaviors? (1.4%) |
| 6 | C0013227;medication(s) (2.1%) | (Categorical header) Age (1.3%) |
| 7 | C0439861;Unspecified substance (2.0%) | (Elaborate header) C0001975; Alcohol (1.2%) |
| 8 | C0001962;Ethyl alcohol (substance) (1.8%) | (Elaborate header) C0013227;medication(s) (1.2%) |
| 9 | C0235146;Feeling high (finding) (1.6%) | (Elaborate header) C0001962; Ethyl alcohol (substance) (1.2%) |
| 10 | C0039798;treatment (1.6%) | (Elaborate header) C0039798;treatment (1.2%) |
| 11 | C0001975;Alcohol (1.6%) | (Elaborate header) C0011581;Disorder; depressive (1.2%) |
| 12 | C0003467;feeling anxious (1.5%) | (Elaborate header) C0439861;Unspecified substance (1.1%) |
| 13 | C0012634;disease (1.4%) | (Elaborate header) C0012634;disease (1.1%) |
| 14 | C0011581;Disorder; depressive (1.4%) | (Elaborate header) C0460137;Depression - motion (qualifier value) (1.0%) |
| 15 | C0011570;Disorder; depression (1.3%) | (Elaborate header) C0011570;Disorder; depression (0.9%) |
| 16 | C0460137;Depression - motion (qualifier value) (1.2%) | (Closed question) Family history: Is there any history of substance abuse? (0.9%) |
| 17 | C1457887;Symptoms (1.2%) | (Elaborate header) C0037313;Sleep (qualifier value) (0.9%) |
| 18 | C0037313;Sleep (qualifier value) (1.1%) | (Elaborate header) C0019665;Historical aspects qualifier (0.9%) |
| 19 | C0015576;Family (1.0%) | (Elaborate header) C1457887; Symptoms (0.8%) |
| 20 | C0019665;Historical aspects qualifier (0.9%) | (Closed question) Bipolar: Did the patient have periods that they felt high without the use of substances? (0.8%) |
For DSM+1, we see that concepts indicating depression (ranked 14th and 15th) and feeling down (ranked 2nd) are ranked high in the list of important features. Features indicating a dependence also score high: ‘patient dependence on’ (ranked 3rd), ‘unspecified substance’ (ranked 7th), ‘alcohol’ (ranked 8th and 11th). ‘Feeling high’ (ranked 9th) and ‘feeling anxious’ (ranked 12th) also were detected as important. Concepts with ambiguous definitions have not been disambiguated, and this is reflected in the importance of concepts such as ‘Depression (motion)’ (ranked 16th) and ‘Down syndrome (ranked 5th)’.
In the question set features, we see that some recurring questions provide interesting features, such as ‘Did the patient feel little pleasure doing things for a prolonged time?’ and ‘OCD: Does the patient struggle with repetitive unwanted thoughts or behaviors?’ Important features are found in the three different categories of question sets: closed questions, categorical headers and elaborate headers.
Overall, we see that there are no dominant features. Several features contribute to the final decision, but no single feature is important on its own.
As part of the shared task evaluation, we obtained a list of factors that were considered important to identify symptom severity in the positive valence domain [18]. Several factors were identified, including depression and its consequences, the extent of addiction to different substances, change in motivation, the extent of obsessive compulsive disorders and bipolar disorders, and the treatments prescribed for the patient.
Comparing these features with those captured by our models, we find some overlap and some deviations. While our model is able to capture features like depression, alcohol dependence, other substance dependence, bipolar disorder, and whether the patient is undergoing some treatment or is on medication, it does not capture information like change in motivation, driving under influence, and hospitalization due to positive valence symptoms. Similarly, concepts like marijuana and obsessive compulsive disorders were present in the feature set and contributed to the models but were not top-ranking features. The models consider features like ‘how does the patient feel’, and information about family history more important than ‘marijuana’.
Several features deemed important by the challenge, like OUI (Operating Under Influence), DUI (Driving Under Influence), IOP(Intensive Outpatient Program) PHP (Partial Hospitalization Program), BPAD (Bipolar Disorder), MJ (Marijuana), CBT (Cognitive Behavioral Therapy), ECT (Electro Convulsive Therapy), were not identified because they were abbreviated in text and did not match to the UMLS concept description.
4. Discussion
Error analysis showed that the detection of concepts worked best in situations where formal medical language is used (within the questions/headers and within answers that are written by the interviewer). In sections where the patient’s own words were recorded, the model fails to identify concepts that describe the patient’s situation. This indicates that by relying on rigid definitions of concepts, medically relevant informal language use cannot be identified. An effect of this can be seen in the performance drop when moving from a bag of words model to using UMLS concepts as features, while applying dimensionality reduction was expected to increase performance. Abbreviations are also not available in the DSM+1 subset and are thus also ignored. For abbreviations specifically, adding acronym and abbreviation definitions from the SPECIALIST lexicon [24] can be a solution. Techniques that do not depend on an expert-created lexicon to detect concepts can increase coverage for both abbreviations and informal language use, at the cost of more false positive concepts and losing the mapping to a structured ontology. Exploratory experiments showed that including abbreviations can boost the inverse mean absolute error with an additional 1.5%.
The scope of the definition of a medical concept plays an important role in the coverage of relevant information for document representation. However, typically, the scope is not wide enough to capture the complete meaning of text. For example, in the sentence ‘The current goal is to smoke occasionally’, concept detection methods will identify ‘smoke’ as a concept, but will lose the meaning that the person is trying to cut down on smoking. For such deep text understanding, different representation techniques to capture natural language syntax and semantics can play an important role (e.g., with the assistance of a distributed semantic model). However, it is a challenging research question in itself, and representing the text as a list of medical concepts is more robust to errors.
For disambiguating terms, relying on UMLS concept detection was insufficient. The UMLS concept representing ‘Down Syndrome’ was a high-ranking important feature, but it was erroneously assigned to sentences in which ‘down’ referred to ‘being down’. Because the challenge had a limited scope, this was not a problem, but it would be a concern when trying to generalize an approach.
Incorporating family history and negated concepts into the feature representation resulted in a substantial improvement in performance. One of the reasons is that concepts detected in standardized headers do not always apply to the patient, but are used as features nonetheless if negation cues are not identified. This confirms previous findings [15] on the influence of correctly incorporating the usage of such context cues. However, the specific technique used for detecting context heavily relies on the structure of the psychiatric intake report (e.g., negation is triggered by a negative answer), which implies limited generalizability to texts not structured as an interview.
The recurrent nature of concepts occurring in questions (as the same questions are often repeated), causes them to be present in more intake reports. This increases the usability of those concepts as a discriminative feature: the absence of a concept (e.g., in the case of negation) is often as informative as its presence. A concept that is only used occasionally in the reports is only informative when it is present. Occasional features occur more often in free text descriptions, both due to a difference in vocabulary as to the non-standardized form of these sections (e.g., ‘patient’s own words’). One can argue that the structure provided by the questions in the reports is partly responsible for the gain in performance when considering context cues.
Our best performing approaches have a simple representation of the intake reports in common. Contrary to our expectations, a psychiatry-related concept representation (DSM+1) did not yield significantly better results than an all-round representation (UMLS). Moreover, the choice of subset was less influential than we had anticipated. Restricting a subset to a limited set of concepts (i.e., diagnoses in DSM) has a negative effect on performance.
The introduction of bootstrapping and outlier removal yielded a minor improvement in classification performance. The differences in results during development and in the test phase suggest that a larger training dataset would boost performance further, suggesting the learning plateau has not been reached.
The limited success our approaches had in distinguishing between moderate and severe cases demonstrates an inability to identify the cues relevant to make this distinction. The use of concepts focused on psychiatric symptoms as identifiers may be a contributing factor. As per the organizers qualitative analysis of the task [18], the difference between these categories is the method of treatment: both severity scores required a positive valence symptom to be the reason of the main treatment, but a severe label was only assigned when the patient was in an intensive inpatient program. During the development phase, we assumed the severity class to be an indicator of the range of different positive valence symptoms that a patient inhibited.
5. Conclusion
In this paper, we presented our work in the framework of the CEGS N-GRID 2016 Shared Task in Clinical Natural Language Processing. Our approach to the identification of symptom severity in the RDoC domain of positive valence is tailored towards the questionnaire style of psychiatric intake reports. An intake report can be split up in segments, each of them containing a header (e.g., a question) and content (e.g., the answer). This segmentation is incorporated into different feature generation techniques, most notably by using context cues (e.g., negation cues, uncertainty cues, family cues) picked up in either header or content and projecting them onto information detected within the segment. Additionally, a list of concepts related to the psychiatric field was used to restrict the detected concepts in these segments.
This feature generation approach yields many concepts that are relevant but case-specific. Instead of generalising concepts to increase the number of samples covered by a general indicator (e.g., ‘addiction’), we used Random Forests, a classifier that utilizes specific features to create weak, overfitted ‘random’ trees, which are generalized by voting the trees against each other.
A model using the actual questions as features yielded similar performance to a model using a subset of concepts (DSM+1). The actual subset of concepts chosen has limited impact, provided that it is not too restrictive. Taking negation and family history into account significantly improved results, which emphasizes the need to consider the context in which detected concepts occur. Bootstrapping and outlier removal had a limited positive effect on the final results.
Supplementary Material
Acknowledgments
We would like to thank the organizers of the shared task for their initiative and efficient organization. Furthermore, we would like to extend our gratitude to the Partners Healthcare, the Neuropsychiatric Genome-Scale, and RDoC Individualized Domains (N-GRID) project [HMS PI: Kohane grant nr. NIH P50 MH106933, MGH PI: Perlis, grant nr. NIH 4R13LM011411] of Harvard Medical School for releasing the dataset for use.
This work was supported by the Agency for Innovation by Science and Technology in Flanders (IWT) [grant numbers 131137, 150056].
Footnotes
Code can be found here: https://github.com/Elyne/rdocChallenge
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Byrd RJ, Steinhubl SR, Sun J, Ebadollahi S, Stewart WF. Automatic identification of heart failure diagnostic criteria, using text analysis of clinical notes from electronic health records. International journal of medical informatics. 2014;83(12):983–992. doi: 10.1016/j.ijmedinf.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stubbs A, Kotfila C, Xu H, Uzuner Ö. Identifying risk factors for heart disease over time: Overview of 2014 i2b2/UTHealth shared task Track 2. Journal of biomedical informatics. 2015;58:S67–S77. doi: 10.1016/j.jbi.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheurwegs E, Luyckx K, Luyten L, Daelemans W, Van den Bulcke T. Data integration of structured and unstructured sources for assigning clinical codes to patient stays. Journal of the American Medical Informatics Association. 2016;23(e1):e11–e19. doi: 10.1093/jamia/ocv115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008;35(128):44. [PubMed] [Google Scholar]
- 5.Pradhan S, Elhadad N, South BR, Martinez D, Christensen L, Vogel A, Suominen H, Chapman WW, Savova G. Evaluating the state of the art in disorder recognition and normalization of the clinical narrative. Journal of the American Medical Informatics Association. 2015;22(1):143–154. doi: 10.1136/amiajnl-2013-002544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32(suppl 1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association. 2011;18(5):552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Bruijn B, Cherry C, Kiritchenko S, Martin J, Zhu X. Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. Journal of the American Medical Informatics Association. 2011;18(5):557–562. doi: 10.1136/amiajnl-2011-000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG. Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang B, Wu Y, Jiang M, Denny JC, Xu H. Recognizing and encoding disorder concepts in clinical text using machine learning and vector space model. CLEF (Working Notes) 2013 [Google Scholar]
- 11.Jonnalagadda S, Cohen T, Wu S, Gonzalez G. Enhancing clinical concept extraction with distributional semantics. Journal of biomedical informatics. 2012;45(1):129–140. doi: 10.1016/j.jbi.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vieira SM, Mendonça LF, Farinha GJ, Sousa JM. Modified binary PSO for feature selection using SVM applied to mortality prediction of septic patients. Applied Soft Computing. 2013;13(8):3494–3504. [Google Scholar]
- 13.Botsis T, Nguyen MD, Woo EJ, Markatou M, Ball R. Text mining for the vaccine adverse event reporting system: medical text classification using informative feature selection. Journal of the American Medical Informatics Association. 2011;18(5):631–638. doi: 10.1136/amiajnl-2010-000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts K, Shooshan SE, Rodriguez L, Abhyankar S, Kilicoglu H, Demner-Fushman D. The role of fine-grained annotations in supervised recognition of risk factors for heart disease from EHRs. Journal of biomedical informatics. 2015;58:S111–S119. doi: 10.1016/j.jbi.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcelon N, Neuraz A, Benoit V, Salomon R, Burgun A. Improving a full-text search engine: the importance of negation detection and family history context to identify cases in a biomedical data warehouse. Journal of the American Medical Informatics Association. 2016:ocw144. doi: 10.1093/jamia/ocw144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breiman L. Random Forests. Machine learning. 2001;45(1):5–32. [Google Scholar]
- 17.Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, Sanislow C, Wang P. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. 2010 doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- 18.Filannino M, Stubbs A, Uzuner Ö. Symptom severity prediction from neuropsychiatric clinical records: Overview of 2016 CEGS N-GRID Shared Tasks Track 2. Journal of Biomedical Informatics. 2017 doi: 10.1016/j.jbi.2017.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salton G, Buckley C. Term-weighting approaches in automatic text retrieval. Information processing & management. 1988;24(5):513–523. [Google Scholar]
- 20.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 4th Edition, Text Revision (DSM-IV-TR) 4. American Psychiatric Association; 2000. [Google Scholar]
- 21.Stearns MQ, Price C, Spackman KA, Wang AY. SNOMED clinical terms: Overview of the development process and project status. Proceedings of the AMIA Symposium; American Medical Informatics Association; 2001. p. 662. [PMC free article] [PubMed] [Google Scholar]
- 22.Mikolov T, Chen K, Corrado G, Dean J. Efficient Estimation of Word Representations in Vector Space. ArXiv e-printsarXiv:1301.3781. [Google Scholar]
- 23.Tulkens S, Šuster S, Daelemans W. Using distributed representations to disambiguate biomedical and clinical concepts. arXiv preprint arXiv:1608.05605. [Google Scholar]
- 24.McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proceedings of the Annual Symposium on Computer Application in Medical Care; American Medical Informatics Association; 1994. p. 235. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.