Abstract
CD4+ T cells have been long known to play an important role in the pathogenesis of rheumatoid arthritis (RA), but the specific cell populations and states that drive the disease have been challenging to identify with low dimensional single cell data and bulk assays. The advent of high dimensional single cell technologies – like single cell RNA-seq or mass cytometry – has offered promise to defining key populations, but brings new methodological and statistical challenges. Recent single cell profiling studies have revealed a broad diversity of cell types among CD4+ T cells, identifying novel populations that are expanded or altered in RA. Here we will review recent findings on CD4+ T cell heterogeneity and RA that have come from single cell profiling studies and discuss the best practices for conducting these studies.
Introduction
Rheumatoid arthritis (RA) is a common autoimmune disorder characterized by chronic inflammation of the synovial tissues, leading to joint damage, disability, and increased mortality [1,2]. The pathophysiology of RA involves a complex interplay between multiple cell types, including leukocyte populations, synovial fibroblasts, chondrocytes, osteoclasts and others [3]. Multiple lines of evidence drawn from genetic, histologic, and clinical observations point to key role for CD4+ T cells in directing the autoimmune response in RA. Genome-wide association studies (GWAS) have highlighted the major histocompatibility complex (MHC) as by far the strongest contributor to disease heritability, driven by variants in HLA-DRB1, HLA-DPB1, and HLA-B [4,5]. HLA-DRB1 and HLA-DPB1 are components of the MHC class II molecule, which antigen presenting cells use to present antigens to CD4+ T cells. We have further demonstrated that genetic risk alleles outside of the MHC locus also point to a role for CD4+ T cells, as genes associated with these loci are preferentially expressed in effector memory CD4+ T cells [6–8]. In addition, CD4+ T cells are frequently found infiltrating the synovium in RA, often in dense lymphocyte aggregates [9,10]. Importantly, interfering with T cell activation by blocking costimulatory signals with abatacept (CTLA4-Ig) is an effective therapy for clinical RA [3].
While it is clear that T cells play an important role in promoting RA pathology, pinpointing the specific T cell phenotypes or functions that are most relevant in this disease has been challenging. CD4+ T cells are typically categorized by the level of expression of surface and intracellular proteins that reflect functionally distinct cell types [11,12]. However, T cells are highly heterogeneous, displaying diverse combinations of surface markers and effector functions. This heterogeneity makes it difficult to describe T cell infiltrates as bulk populations and has highlighted the value of single cell analyses to resolve this heterogeneity.
Single cell analyses by flow cytometry have contributed major insights into T cell abnormalities in RA [13,14], yet flow cytometry analyses have been hampered the limited number of parameters that can be detected simultaneously, which are often insufficient to adequately assess a diverse T cell population. The recent rapid expansion of single cell technologies has led to a dramatic advance in the ability to study complex populations in large-scale with high dimensionality (Figure 1). This high-dimensional single cell profiling may lead to the identification of specific T cell populations or states that are mechanistically linked to disease and ideal for therapeutic targeting. In this review, we discuss recent advances in single cell immunoprofiling and describe their early application in RA. We will then discuss methodological and bioinformatic considerations to maximize the potential of single cell technologies in its application to define mechanisms of immune-mediated diseases.
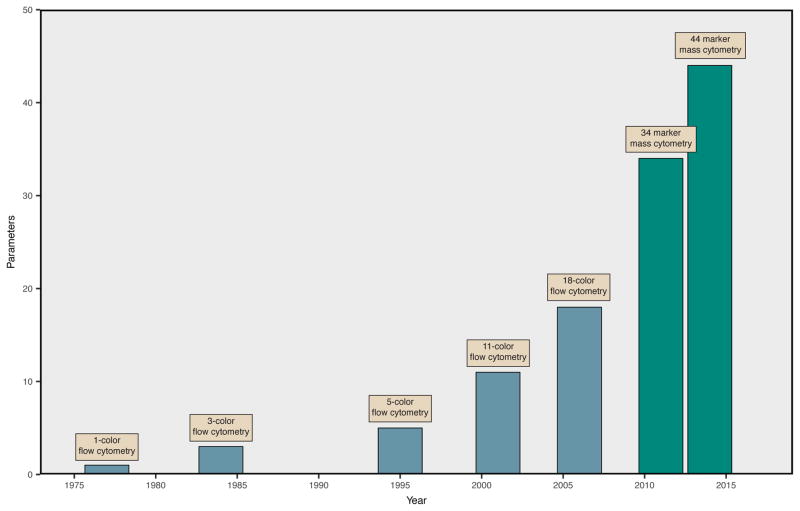
Figure 1. Advances in Single Cell Cytometry.
The number of unique molecules that can be simultaneously characterized for a single cell has progressively increased. The introduction of new fluorchromes has improved polychromatic flow cytometry and enabled the development of 18-color assays. Mass cytometry, which uses stable isotopes of non-biological rare earth metals linked to antibodies to detect protein epitopes, is currently capable of acquiring 44 markers simultaneously. Current equipment for mass cytometry supports the acquisition of over 100 markers, but experiments are limited by the availability of isotopically pure reagents.
Low-dimensional single cell analysis of T cells in RA
Single cell assays have a long history in the field of autoimmunity, beginning in 1969 with the initial use of fluorescent assays to label and sort immune cell populations [15–18]. Cytometry has been thoroughly exploited in the exploration of lymphocyte heterogeneity in RA [19–23]. Subsequent improvements in flow cytometry technology have steadily increased the number of parameters that can be measured for each cell, provided access to cytoplasmic and nuclear protein expression through intracellular staining, and facilitated measurement of cell signaling using antibodies specific for the phosphorylation state of signaling molecules [24]. Flow cytometric analyses of T cells from RA synovial tissue and fluid have highlighted the dramatic ‘activated’ phenotype of T cells within the RA joint, consistent with an ongoing autoimmune response directed at the synovium [25,26]. Synovial T cells frequently express CXCR3, suggesting Th1 differentiation, and loss of CD27, suggesting a chronically activated state [27–29].
Immunophenotyping of peripheral blood CD4+ T cells from RA patients has also identified characteristic changes, including expansion of Th17 cells relative to Tregs [30,31], and an expansion of CD28− T cells [22,32]. Unfortunately, studies of peripheral blood T cells in RA have often yielded inconsistent results. For example, the abundance of regulatory T cells (TREG) in RA peripheral blood has been observed to be reduced or expanded compared to healthy controls in different studies [33–37]; in addition, conflicting results have been reported concerning the suppressive capability of Treg cells in RA [38–42]. Inconsistent results can be partially attributed to variation in markers used across different studies or the difficulty of resolving highly heterogeneous populations with bulk cell assays – which advancing single cell technologies might help to obviate. However, some of this inconsistency is rooted in methodological issues that will need to be addressed as investigators begin to apply single cell technologies to autoimmune diseases. Specific issues have included the use of small sample sizes, variability in cohorts, technical noise resulting in batch effects, publication bias, and the lack of principled statistical methodology and criteria.
High-dimensional analyses reveal an expanded view of CD4+ T cell heterogeneity
The recent development of mass cytometry - a fusion of mass spectrometry and flow cytometry that is capable of the simultaneous acquisition of over 40 parameters on a single cell level – has further extended the dimensionality of single cell cytometric assays [43]. Mass cytometry relies upon staining cells with the same target-specific antibodies that are commonly used in flow cytometry to tag markers of interest; however, in mass cytometry antibodies are labeled with pure, non-radioactive rare earth isotopes instead of fluorescent proteins. After staining, single cells are analyzed by a time-of-flight mass spectrometer by integrating the detection of heavy metal reporter ions to determine expression levels for each labeled antibody [44–46].
Single cell immunoprofiling by mass cytometry has already been used to reveal remarkable heterogeneity within conventional T cell subsets. Wong et al. used mass cytometry to profile CD4+ T cells across eight human tissue types and described 75 different populations, including multiple Th1 populations for each TH subset. Many cell populations were tissue-specific and differed based the expression of trafficking receptors and cytokine production [47••]. They observed that certain populations co-expressed “key” cytokines like IFN-γ, IL-4, and IL-17A that are typically restricted to a single CD4+ TH subset, in line with previous findings highlighting the phenotypic plasticity between CD4+ TH lineages [48–51], reviewed in [52]. Other studies have taken advantage of high-dimensional single cell mass cytometry analysis to describe multiple populations of TREG and TFH cells [53,54].
While advances in flow cytometry and mass cytometry enable users to define single cells across many parameters, the set of proteins to be measured must be decided a priori, limiting the use of these technologies in unbiased discovery studies. In contrast, single cell transcriptomic analysis presents an opportunity to define single cell expression profiles without relying on prior knowledge. Several different single cell RNA-seq (scRNA-seq) methods have been developed over the past decade [55–59] and successfully applied in various immunological studies, such as identifying differentiation pathways in immune cell lineages [60,61], establishing novel transcriptional regulatory networks [62], and revealing functional diversity among lymphoid cell populations [63,64].
Single cell RNA-seq technologies provide an orthogonal approach to cytometry-based methods for establishing CD4+ T cell heterogeneity. As CD4+ T cell subsets are differentiated by their putative functionality, quantifying of transcript expression on the single cell level can be used to identify gene expression programs that underlie those functional divisions. Single cell sequencing of T cells isolated from patients with liver cancer identified 11 distinct CD4 and CD8 T cell populations, some of which were expanded in hepatocellular carcinoma and marked by specific gene signatures [65••]. The functional diversity of natural killer T (NKT) cells is difficult to characterize using cytometry alone; however, single cell RNA-seq analysis revealed differential patterns of gene expression that resolve NKT subsets and indicate potential functions [66]. Single cell transcriptomic profiling is also particularly useful for understanding T cell differentiation and proliferation, as the expression of key transcription factors and other regulatory genes can be easily ascertained and used to assign cells to differentiation trajectories [67,68].
Early high-dimensional analyses of T cells in RA
These same technologies are already being used in RA tissue and blood to define key features of pathogenic CD4+ T cell populations in RA. We have recently applied mass cytometry to evaluate the heterogeneity of CD4+ T cells that infiltrate RA synovium [69]. With this high-dimensional analysis, we identified a T ‘peripheral helper’ (TPH) cell population that is markedly expanded in RA synovium, constituting ~25% of synovial CD4+ T cells. TPH cells, characterized as PD-1hi CXCR5− CD4+, display a unique capacity to infiltrate inflamed tissues and enhance local B cell antibody production and differentiation into plasma cells. A preliminary single-cell RNA-seq analysis of a single RA synovial sample has also demonstrated multiple T cell subsets, including a population of TPH cells, in the RA T cell infiltrate [70].
In a distinct approach, Ishigaki and colleagues used parallel single cell transcriptomics and T cell receptor (TCR) sequencing to identify and analyze expanded CD4+ T cell clones in RA patients [71••]. Expanded memory CD4+ T cells in both the synovium and periphery are phenotypically similar in expression to senescent T cells, upregulating Granzyme B and downregulating CD28. Intriguingly, the majority of expanded memory T cell clones did not belong to the well-defined TH1 or TH17 subsets despite their established association with RA [31,72–74]. Although the findings are limited by the small number of donors studied, this study suggests that as yet undefined CD4+ T cell populations may undergo expansion in RA and may be relevant to RA pathology.
One potential benefit of characterizing the extent of CD4+ T cell diversity with high-dimensional analyses is that it may provide a means to differentiate between pathogenic and non-pathogenic variants of known T cell subsets. For example, single cell RNA-seq was used to define a spectrum of pathogenicity for TH17 cells isolated from mice with experimental autoimmune encephalomyelitis (EAE) and identify key genes involved in the process [75]. Similarly, immunoprofiling of TREG cells in RA described the discovery of a novel senescent-like TREG cell population characterized by the loss of CD28 expression and increased numbers of double stranded DNA breaks. Compared to standard TREG cells, CD28− TREG cells had impaired suppressive function and produced higher amounts of proinflammatory cytokines IFN-γ and TNF [76•].
Identifying biomarkers through cell phenotyping
As the diversity, precision, and cost of therapeutics in RA has increased, the importance of being able to determine the option best-suited for a given patient up front has become increasingly clear. There is now a major need for biomarkers to predict response to therapies with distinct mechanisms of action; however, efforts using multiplexed cytokine profiling and genetic variation have not yet led to clinically applicable tools [77,78]. The increased resolution of single cell assays is an asset for revealing disease biomarkers, as the ability to characterize the diversity of lymphocyte populations can be leveraged to monitor the abundances of multiple populations longitudinally or in a case-control context. Changes in the frequency of disease-associated populations that can be easily measured in peripheral blood can be used as a powerful readout of disease state in less accessible compartments.
Several studies have suggested the potential ability to identify specific lymphocyte populations whose peripheral frequencies are predictive of treatment response in order to guide therapeutic decisions. Tracking CD4+ T cell populations by flow cytometry in patients with early RA receiving methotrexate and healthy controls revealed that higher abundances of naïve CD4+ T cells are significantly associated with increased chances of remission [79]. Response to treatment with tocilizumab, an IL-6 receptor inhibitor, is associated with higher baseline frequencies of natural killer (CD3− CD56+) cells [80] and higher increases in the frequencies of TREG cells in the periphery [81]. A case-control study of RA patients and healthy controls demonstrated that IL-10+ producing LAG3+ TREG cells are specifically increased after treatment with abatacept, and that the magnitude of this increase is correlated with the strength of response [82]. Immunoprofiling studies have also revealed changes in the function of lymphocyte populations in response to therapy: for example, RA patients who respond well to anti-TNF treatment have higher production of GM-CSF from T cells [83]. Response to TNF inhibition therapy is also associated with a higher abundance of CD8+ T cells that are specifically reactive to apoptotic epitopes [84]. Studies such as these fuel hope for the development of predictive cellular biomarkers, though none have been prospectively validated and adopted for use clinically to date.
The Future of Single Cell Immunoprofiling
Recent advances in availability and throughput have made single cell technologies a practical choice for conducting immunoprofiling studies to understand mechanisms of disease and define predictive biomarkers. The application of these methods in RA include the profiling of blood, as many studies that we refer to above already have done, but also performing immunoprofiling in human tissue. We and others are pursuing these goals in Accelerating Medical Partnerships Rheumatoid Arthritis/Systemic Lupus Erythematosus (AMP RA/SLE network; URL: https://www.niams.nih.gov/Funding/Funded_Research/AMP_RA_Lupus/), which involves obtaining, disaggregating, and performing single cell profiling on synovial tissue from cases and controls to query both immune infiltration and stromal adaptions. For human immunology to successfully leverage the large quantities of observational data that emerge from single cell queries of the immune system, we will need to develop and reliably apply robust statistical methods and study design principles in single cell studies. Taking full advantage of the power of single cell analysis will require overcoming technical, methodological, and bioinformatic challenges.
Among the many considerations that must be taken into account when designing single cell immunophenotyping experiments, one of the most prominent is determining how to handle batch effects. Here we use the term ‘batch’ to refer to a set of samples processed together in a single experimental run, and the term ‘batch effect’ to refer to variation in a dataset caused by technical variation in the processing of different batches of samples. Large-scale microarray assays powerfully illustrated the dramatic effects that differences in machine sensitivity, preparation or handling of samples, or protocol variations can have on the results of transcriptomic analyses [85–87]. Single-cell technologies such as mass cytometry and scRNA-seq are even more vulnerable to confounding from batch effects due to extensive intra-individual and inter-individual heterogeneity of expression among single cells. Application of single cell profiling to human tissues, where cases and controls may respond differently to sample processing and manipulation, could provide an additional source of batch effects.
Indeed, Hicks et al. has demonstrated that variable detection rate and other technical effects account for much of the “biological” variation that has been presented in some of the early single cell transcriptomic studies [88••]. Careful experimental design can partially alleviate the influence of batch effects in single cell profiling studies; however, we note that Tung et al. have shown that common normalization methods for scRNA-seq like spike-in controls and the use of unique molecular identifiers (UMIs) are insufficient for fully removing technical variation [89]. For single cell transcriptomic studies, critical steps include applying quality control methods to remove poorly captured cells and quantifying transcripts to determine cell expression levels. In single cell cytometry studies, quality control is effectively performed by selecting cells for analysis based upon forward and side scatter parameters (flow cytometry) or DNA content (mass cytometry) and inclusion of a live/dead marker, while marker expression quantification is normally provided by onboard software.
However, since batch variability is difficult to completely eliminate post hoc, careful experimental design is essential. First, the importance of minimizing variation in experimental procedure cannot be overstated. Best practices include ensuring that samples are collected from the same source, handled in the same fashion, and assayed using the same protocols to the extent that it is possible. Ideally, samples would be prepared using the same lot of reagents; however, this can be difficult to achieve, and steps such as RNA preparation or antibody staining should be performed in a limited number of batches. Second, as large-scale studies typically require performing assays in batches, sample randomization is crucial. Interspersing cases and controls within each batch guards against the possibility of discovering biological associations that are perfectly confounded with batch. Finally, ensuring that sample processing is done in a short window of time and that samples are assayed using the same equipment also minimizes technical variation. For example, the AMP RA/SLE network significantly reduced batch effects by processing and assaying samples in a single location, as opposed to trying to analyze data obtained at different sites.
The choice of tools for computational analysis of high-dimensional data is another important consideration in conducting single cell immunoprofiling studies. Although produced using very different technologies, both transcriptomic and cytometric single cell data can be analyzed similarly by treating the data as matrices where rows represent single cells and columns represent expression measurements for transcripts or proteins. In the context of studying disease association, analysis of single cell immunoprofiling data can be split into two steps: clustering, where the goal is to identify groups of cells that are related by similarity of expression, and association testing, where the goal is to determine significant changes in the abundance or character of immune cell populations in disease.
While many different algorithms have been applied to the analysis of single cell data, we believe that the following methods represent some of the best tools for use with single cell immunophenotyping. Seurat is an R package that contains multiple methods for clustering and visualizing single cell sequencing data, as well as performing differential expression testing between groups and finding associations [51]. One particularly intriguing application of Seurat is to use single cell transcriptomic data to reconstruct the spatial organization of cells, which has been demonstrated in zebrafish embryos [90••]. Multiple clustering methods have been developed for the analysis of flow cytometry [91,92] and mass cytometry [46,93–97]; a recent comparison of these methods identified FlowSOM [96] and PhenoGraph [97] as the best performers [98].
While the set of algorithms available for clustering single cell data is rapidly expanding, there is a relative paucity of methods designed to perform association testing with cytometry data [99,100]. We have recently presented MASC (mixed-effect Modeling of Associations of Single Cells), which accepts user-identified populations regardless of clustering method, directly reports the significance of case-control associations for each cluster, provides an estimate of the effect size of the association itself, and incorporates both technical covariates (e.g. batch) and clinical covariates when modeling associations, a key feature when analyzing high-dimensional datasets of large disease cohorts[113••]. In comparison, the association testing method Citrus uses nested hierarchical clustering and penalized regression models to identify features (defined here as clusters of single cells or median expression levels of markers within a cluster) that are predictive of clinical endpoints; however, Citrus requires down-sampling cells from each sample and does not retain single cell resolution, which impedes the interpretation of clusters found to be predictive. Tools such as these now empower investigators to efficiently identify novel cell phenotypes associated with a disease state.
Given the high levels of inter-individual variability in the human immune system, the ability to aggregate data across multiple studies is an attractive goal for conducting well-powered analyses. Currently, data aggregation is challenging due to the high dimensionality of single cell data and the difficulty of overcoming different datasets for analysis which include differences in the use of specific sequencing protocols, technical batch effects, and differences in sample handling. Standardization of normalization and quality control methods will be key, as small differences in data processing can overpower biological signals in the noisy context of immunoprofiling; for example, the use of different software pipelines for processing single cell RNA-seq data will impede combined analysis. One important question is whether the use of imputation-based techniques will be effective to fill in missing data and meta-analyze across multiple studies in single cell analyses. These approaches have been critical in allowing human genetic studies to scale rapidly, and have supported meta-analysis of different data sets obtained on different platforms. While methods for single cell RNA-seq data have been recently described, the effectiveness of imputation is an active question in the field [101–103]. Finally, given the identifiable nature of single-cell transcriptomic sequencing data in particular, a framework to support data sharing while protecting patient privacy is essential.
For immunological applications, a key initial step should be to better characterize human lymphocytes using single cell data. Building a reference map of the human immune system is a difficult and complicated task; however, the dendritic cell atlas or the work of Wong et al. characterizing T cells across tissues provide examples of the power of this approach [57••, 104•]. Incorporating data on from multiple assays to define lymphocyte profiles will be essential for understanding their functional impact, as shown by multiple studies that utilize repertoire sequences or expression data in combination with single cell cytometry to identify disease-relevant populations [71••,105•,106,107]. The development of new peptide-MHC multimeric complexes supports the detection and isolation of antigen-specific lymphocytes at much lower frequencies [108] than was previously feasible. New methods have been recently developed to provide high-throughput single cell repertoire sequencing of B and T lymphocytes [109•,110].
Beyond integrating data across studies and across assays, the next stage of advancement for single cell technologies will be the simultaneous acquisition of transcriptomic and proteomic data from a single cell. Multiple methods for conducting such analyses have been described [111, ••112] but have yet to be applied in any large-scale immunoprofiling efforts. The ability to obtain this type of data would allow research into the temporal dynamics of transcription and protein expression as well as provide higher-resolution definition of single cells.
Conclusions
The advent of single cell technologies has the potential to revolutionize the study of RA by offering an unbiased approach to detecting and characterizing cell heterogeneity in blood and tissue. High-dimensional single cell analyses of RA synovium have revealed novel lymphocyte and stromal cell populations that are pathologically expanded in the joints of RA patients. These cell populations may now be evaluated as potential therapeutic targets. Single cell transcriptomics and TCR repertoire sequencing have enabled detailed characterization of the specific clones of CD4+ T cells that are expanded in RA and may highlight new cell phenotypes to pursue as therapeutic targets or biomarkers.
However, the current absence of rigorous standards for experimental design and analysis significantly limits the value of single cell assays. The increased resolution of single cell analyses will be wasted without defining a set of standards for experiments that enable combining experimental data across batches, assays, and studies. This will be particularly important for studying CD4+ T cells in RA, where heterogeneity among both cell types and patients has yielded conflicting and contradictory results. As the magnitude of data that is produced by single cell immunoprofiling increases and reveals unprecedented levels of diversity among immune cell, methodological rigor will be critical for deciphering mechanisms of disease.
Highlights.
Single cell immunoprofiling reveals extensive heterogeneity among CD4+ T cells.
Multidimensional analyses identify novel CD4+ T cell populations associated with rheumatoid arthritis.
Single cell disease association studies require careful attention to study design to avoid confounding technical effects.
New analysis methods are emerging to take full advantage of complex single cell datasets.
Acknowledgments
This work is supported in part by funding from the National Institutes of Health (UH2AR067677, 1U01HG009088, and 1R01AR063759 (SR)), and the Doris Duke Charitable Foundation Grant #2013097. D.A.R. is supported by Rheumatology Research Foundation Tobe and Stephen Malawista, MD Endowment in Academic Rheumatology.
Footnotes
Conflicts of interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alamanos Y, Drosos AA. Epidemiology of adult rheumatoid arthritis. Autoimmunity Reviews. 2005;4:130–136. doi: 10.1016/j.autrev.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Scott DL, Wolfe F, Huizinga TWJ. Rheumatoid arthritis. Lancet. 2010;376:1094–1108. doi: 10.1016/S0140-6736(10)60826-4. [DOI] [PubMed] [Google Scholar]
- 3.McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. 2011;365:2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 4.Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee H-S, Jia X, Alfredsson L, Padyukov L, Klareskog L, Worthington J, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nature Publishing Group. 2012;44:291–296. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boissier M-C, Semerano L, Challal S, Saidenberg-Kermanac’h N, Falgarone G. Rheumatoid arthritis: from autoimmunity to synovitis and joint destruction. Journal of Autoimmunity. 2012;39:222–228. doi: 10.1016/j.jaut.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 6.Hu X, Kim H, Stahl E, Plenge R, Daly M, Raychaudhuri S. Integrating Autoimmune Risk Loci with Gene-Expression Data Identifies Specific Pathogenic Immune Cell Subsets. The American Journal of Human Genetics. 2011;89:496–506. doi: 10.1016/j.ajhg.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diogo D, Okada Y, Plenge RM. Genome-wide association studies to advance our understanding of critical cell types and pathways in rheumatoid arthritis: recent findings and challenges. Current Opinion in Rheumatology. 2014;26:85–92. doi: 10.1097/BOR.0000000000000012. [DOI] [PubMed] [Google Scholar]
- 8.Vahedi G, Kanno Y, Furumoto Y, Jiang K, Parker SCJ, Erdos MR, Davis SR, Roychoudhuri R, Restifo NP, Gadina M, et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature. 2015;520:558–562. doi: 10.1038/nature14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young CL, Adamson TC, Vaughan JH, Fox RI. Immunohistologic characterization of synovial membrane lymphocytes in rheumatoid arthritis. Arthritis Rheum. 1984;27:32–39. doi: 10.1002/art.1780270106. [DOI] [PubMed] [Google Scholar]
- 10.Takemura S, Braun A, Crowson C. Lymphoid neogenesis in rheumatoid synovitis. The Journal of …. 2001 doi: 10.4049/jimmunol.167.2.1072. [DOI] [PubMed] [Google Scholar]
- 11.Geginat J, Paroni M, Facciotti F, Gruarin P, Kastirr I, Caprioli F, Pagani M, Abrignani S. The CD4-centered universe of human T cell subsets. Semin Immunol. 2013;25:252–262. doi: 10.1016/j.smim.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the Human Immunology Project [Internet] Nature Reviews Immunology. 2012 doi: 10.1038/nri3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gizinski AM, Fox DA. T cell subsets and their role in the pathogenesis of rheumatic disease. Current Opinion in Rheumatology. 2014;26:204–210. doi: 10.1097/BOR.0000000000000036. [DOI] [PubMed] [Google Scholar]
- 14.Ermann J, Rao DA, Teslovich NC, Brenner MB, Raychaudhuri S. Immune cell profiling to guide therapeutic decisions in rheumatic diseases. Nat Rev Rheumatol. 2015;11:541–551. doi: 10.1038/nrrheum.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hulett HR, Bonner WA, Barrett J, Herzenberg LA. Cell sorting: automated separation of mammalian cells as a function of intracellular fluorescence. Science. 1969;166:747–749. doi: 10.1126/science.166.3906.747. [DOI] [PubMed] [Google Scholar]
- 16.Loken MR, Herzenberg LA. ANALYSIS OF CELL POPULATIONS WITH A FLUORESCENCE-ACTIVATED CELL SORTER. Ann N Y Acad Sci. 1975;254:163–171. doi: 10.1111/j.1749-6632.1975.tb29166.x. [DOI] [PubMed] [Google Scholar]
- 17.Dean PN, Pinkel D. High resolution dual laser flow cytometry. J Histochem Cytochem. 1978;26:622–627. doi: 10.1177/26.8.357646. [DOI] [PubMed] [Google Scholar]
- 18.Wilder ME, Cram LS. Differential fluorochromasia of human lymphocytes as measured by flow cytometry. J Histochem Cytochem. 1977;25:888–891. doi: 10.1177/25.7.70458. [DOI] [PubMed] [Google Scholar]
- 19.Fox RI, Fong S, Sabharwal N, Carstens SA, Kung PC, Vaughan JH. Synovial fluid lymphocytes differ from peripheral blood lymphocytes in patients with rheumatoid arthritis. The Journal of Immunology. 1982;128:351–354. [PubMed] [Google Scholar]
- 20.Pitzalis C, Kingsley G, Murphy J, Panayi G. Abnormal distribution of the helper-inducer and suppressor-inducer T-lymphocyte subsets in the rheumatoid joint. Clin Immunol Immunopathol. 1987;45:252–258. doi: 10.1016/0090-1229(87)90040-7. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt D, Goronzy JJ, Weyand CM. CD4+ CD7− CD28− T cells are expanded in rheumatoid arthritis and are characterized by autoreactivity. J Clin Invest. 1996;97:2027–2037. doi: 10.1172/JCI118638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martens PB, Goronzy JJ, Schaid D, Weyand CM. Expansion of unusual CD4+ T cells in severe rheumatoid arthritis. Arthritis Rheum. 1997;40:1106–1114. doi: 10.1002/art.1780400615. [DOI] [PubMed] [Google Scholar]
- 23.Berner B, Wolf G, Hummel KM, Müller GA, Reuss-Borst MA. Increased expression of CD40 ligand (CD154) on CD4+ T cells as a marker of disease activity in rheumatoid arthritis. Annals of the Rheumatic Diseases. 2000;59:190–195. doi: 10.1136/ard.59.3.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler’s guide to cytometry. Trends Immunol. 2012;33:323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brennan FM, Hayes AL, Ciesielski CJ, Green P, Foxwell BMJ, Feldmann M. Evidence that rheumatoid arthritis synovial T cells are similar to cytokine-activated T cells: involvement of phosphatidylinositol 3-kinase and nuclear factor kappaB pathways in tumor necrosis factor alpha production in rheumatoid arthritis. Arthritis Rheum. 2002;46:31–41. doi: 10.1002/1529-0131(200201)46:1<31::AID-ART10029>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 26.Steiner G, Tohidast-Akrad M, Witzmann G, Vesely M, Studnicka-Benke A, Gal A, Kunaver M, Zenz P, Smolen JS. Cytokine production by synovial T cells in rheumatoid arthritis. Rheumatology (Oxford) 1999;38:202–213. doi: 10.1093/rheumatology/38.3.202. [DOI] [PubMed] [Google Scholar]
- 27.Qin S, Rottman JB, Myers P, Kassam N, Weinblatt M, Loetscher M, Koch AE, Moser B, Mackay CR. The chemokine receptors CXCR3 and CCR5 mark subsets of T cells associated with certain inflammatory reactions. J Clin Invest. 1998;101:746–754. doi: 10.1172/JCI1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isomaki P, Luukkainen R, Lassila O, Toivanen P, Punnonen J. Synovial fluid T cells from patients with rheumatoid arthritis are refractory to the T helper type 2 differentiation-inducing effects of interleukin-4. Immunology. 1999;96:358–364. doi: 10.1046/j.1365-2567.1999.00712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohem CL, Brezinschek RI, Wisbey H, Tortorella C, Lipsky PE, Oppenheimer-Marks N. Enrichment of differentiated CD45RBdim,CD27- memory T cells in the peripheral blood, synovial fluid, and synovial tissue of patients with rheumatoid arthritis. Arthritis Rheum. 1996;39:844–854. doi: 10.1002/art.1780390518. [DOI] [PubMed] [Google Scholar]
- 30.Niu Q, Cai B, Huang Z-C, Shi Y-Y, Wang L-L. Disturbed Th17/Treg balance in patients with rheumatoid arthritis. Rheumatol Int. 2012;32:2731–2736. doi: 10.1007/s00296-011-1984-x. [DOI] [PubMed] [Google Scholar]
- 31.Wang W, Shao S, Jiao Z, Guo M, Xu H, Wang S. The Th17/Treg imbalance and cytokine environment in peripheral blood of patients with rheumatoid arthritis. Rheumatol Int. 2012;32:887–893. doi: 10.1007/s00296-010-1710-0. [DOI] [PubMed] [Google Scholar]
- 32.Pawlik A, Ostanek L, Brzosko I, Brzosko M, Masiuk M, Machalinski B, Gawronska-Szklarz B. The expansion of CD4+CD28− T cells in patients with rheumatoid arthritis. Arthritis Res Ther. 2003;5:R210–3. doi: 10.1186/ar766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawson CA, Brown AK, Bejarano V, Douglas SH, Burgoyne CH, Greenstein AS, Boylston AW, Emery P, Ponchel F, Isaacs JD. Early rheumatoid arthritis is associated with a deficit in the CD4+CD25high regulatory T cell population in peripheral blood. Rheumatology (Oxford) 2006;45:1210–1217. doi: 10.1093/rheumatology/kel089. [DOI] [PubMed] [Google Scholar]
- 34.Han GM, O’Neil-Andersen NJ, Zurier RB, Lawrence DA. CD4+CD25high T cell numbers are enriched in the peripheral blood of patients with rheumatoid arthritis. Cell Immunol. 2008;253:92–101. doi: 10.1016/j.cellimm.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moradi B, Schnatzer P, Hagmann S, Rosshirt N, Gotterbarm T, Kretzer JP, Thomsen M, Lorenz H-M, Zeifang F, Tretter T. CD4+CD25+/highCD127low/− regulatory T cells are enriched in rheumatoid arthritis and osteoarthritis joints--analysis of frequency and phenotype in synovial membrane, synovial fluid and peripheral blood. Arthritis Res Ther. 2014;16:R97. doi: 10.1186/ar4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuki F, Saegusa J, Miyamoto Y, Misaki K, Kumagai S, Morinobu A. CD45RA-Foxp3(high) activated/effector regulatory T cells in the CCR7 + CD45RA-CD27 + CD28+central memory subset are decreased in peripheral blood from patients with rheumatoid arthritis. Biochem Biophys Res Commun. 2013;438:778–783. doi: 10.1016/j.bbrc.2013.05.120. [DOI] [PubMed] [Google Scholar]
- 37.Walter GJ, Fleskens V, Frederiksen KS, Rajasekhar M, Menon B, Gerwien JG, Evans HG, Taams LS. Phenotypic, Functional, and Gene Expression Profiling of Peripheral CD45RA+ and CD45RO+ CD4+CD25+CD127(low) Treg Cells in Patients With Chronic Rheumatoid Arthritis. Arthritis Rheumatol. 2016;68:103–116. doi: 10.1002/art.39408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao D, Malmström V, Baecher-Allan C, Hafler D, Klareskog L, Trollmo C. Isolation and functional characterization of regulatory CD25brightCD4+ T cells from the target organ of patients with rheumatoid arthritis. Eur J Immunol. 2003;33:215–223. doi: 10.1002/immu.200390024. [DOI] [PubMed] [Google Scholar]
- 39.Flores-Borja F, Jury EC, Mauri C, Ehrenstein MR. Defects in CTLA-4 are associated with abnormal regulatory T cell function in rheumatoid arthritis. Proc Natl Acad Sci US A. 2008;105:19396–19401. doi: 10.1073/pnas.0806855105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mottonen M, Heikkinen J, Mustonen L, Isomaki P, Luukkainen R, Lassila O. CD4+ CD25+ T cells with the phenotypic and functional characteristics of regulatory T cells are enriched in the synovial fluid of patients with rheumatoid arthritis. Clin Exp Immunol. 2005;140:360–367. doi: 10.1111/j.1365-2249.2005.02754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ehrenstein MR, Evans JG, Singh A, Moore S, Warnes G, Isenberg DA, Mauri C. Compromised function of regulatory T cells in rheumatoid arthritis and reversal by anti-TNFalpha therapy. Journal of Experimental Medicine. 2004;200:277–285. doi: 10.1084/jem.20040165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alunno A, Manetti M, Caterbi S, Ibba-Manneschi L, Bistoni O, Bartoloni E, Valentini V, Terenzi R, Gerli R. Altered immunoregulation in rheumatoid arthritis: the role of regulatory T cells and proinflammatory th17 cells and therapeutic implications. Mediators Inflamm. 2015;2015:751793–12. doi: 10.1155/2015/751793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spitzer MH, Nolan GP. Mass Cytometry: Single Cells, Many Features. Cell. 2016;165:780–791. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 45.Ornatsky O, Bandura D, Baranov V, Nitz M, Winnik MA, Tanner S. Highly multiparametric analysis by mass cytometry. Journal of Immunological Methods. 2010;361:1–20. doi: 10.1016/j.jim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 46.Bendall SC, Simonds EF, Qiu P, Amir E-AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••47.Wong MT, Ong DEH, Lim FSH, Teng KWW, McGovern N, Narayanan S, Ho WQ, Cerny D, Tan HKK, Anicete R, et al. A High-Dimensional Atlas of Human T Cell Diversity Reveals Tissue-Specific Trafficking and Cytokine Signatures. Immunity. 2016;45:442–456. doi: 10.1016/j.immuni.2016.07.007. This study uses mass cytometry to profile the expression of over 40 cellular markers in human lymphocytes and provides an excellent example of the breadth of human CD4+ T cell heterogeneity. The authors identify an abundance of lymphocyte populations that differ across tissues, including 75 different subtypes of CD4+ T cells. The high dimensionality of this dataset provdes better definition of T cell plasticity, as the authors find that only a restricted combination of key cytokines are expressed by non-canonical T helper subsets. [DOI] [PubMed] [Google Scholar]
- 48.Gagliani N, Vesely MCA, Iseppon A, Brockmann L, Xu H, Palm NW, de Zoete MR, Licona-Limón P, Paiva RS, Ching T, et al. Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature. 2015 doi: 10.1038/nature14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komatsu N, Okamoto K, Sawa S, Nakashima T, Oh-hora M, Kodama T, Tanaka S, Bluestone JA, Takayanagi H. Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nat Med. 2014;20:62–68. doi: 10.1038/nm.3432. [DOI] [PubMed] [Google Scholar]
- 50.Reiner SL, Adams WC. Lymphocyte fate specification as a deterministic but highly plastic process. Nature Reviews Immunology. 2014;14:699–704. doi: 10.1038/nri3734. [DOI] [PubMed] [Google Scholar]
- 51.Nakayamada S, Takahashi H, Kanno Y, O’Shea JJ. Helper T cell diversity and plasticity. Current Opinion in Immunology. 2012;24:297–302. doi: 10.1016/j.coi.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu J, Yamane H, Paul WE. Differentiation of Effector CD4 T Cell Population. Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mason GM, Lowe K, Melchiotti R, Ellis R, de Rinaldis E, Peakman M, Heck S, Lombardi G, Tree TIM. Phenotypic Complexity of the Human Regulatory T Cell Compartment Revealed by Mass Cytometry. J Immunol. 2015;195:2030–2037. doi: 10.4049/jimmunol.1500703. [DOI] [PubMed] [Google Scholar]
- 54.Wong MT, Chen J, Narayanan S, Lin W, Anicete R, Kiaang HTK, De Lafaille MAC, Poidinger M, Newell EW. Mapping the Diversity of Follicular Helper T Cells in Human Blood and Tonsils Using High-Dimensional Mass Cytometry Analysis. Cell Rep. 2015;11:1822–1833. doi: 10.1016/j.celrep.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 55.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Meth. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 56.Islam S, Kjällquist U, Moliner A, Zajac P, Fan J-B, Lönnerberg P, Linnarsson S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Research. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 58.Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Meth. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 59.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, Grimes HL. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature. 2016;537:698–702. doi: 10.1038/nature19348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ishizuka IE, Chea S, Gudjonson H, Constantinides MG, Dinner AR, Bendelac A, Golub R. Single-cell analysis defines the divergence between the innate lymphoid cell lineage and lymphoid tissue-inducer cell lineage. Nat Immunol. 2016;17:269–276. doi: 10.1038/ni.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Björklund ÅK, Forkel M, Picelli S, Konya V, Theorell J, Friberg D, Sandberg R, Mjösberg J. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol. 2016;17:451–460. doi: 10.1038/ni.3368. [DOI] [PubMed] [Google Scholar]
- 64.Mahata B, Zhang X, Kolodziejczyk AA, Proserpio V, Haim-Vilmovsky L, Taylor AE, Hebenstreit D, Dingler FA, Moignard V, Göttgens B, et al. Single-cell RNA sequencing reveals T helper cells synthesizing steroids de novo to contribute to immune homeostasis. Cell Rep. 2014;7:1130–1142. doi: 10.1016/j.celrep.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••65.Zheng C, Zheng L, Yoo J-K, Guo H, Zhang Y, Guo X, Kang B, Hu R, Huang JY, Zhang Q, et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell. 2017;169:1342–1356. e16. doi: 10.1016/j.cell.2017.05.035. This work, one of the largest single cell sequencing studies published to date, reports enrichment of CD4+ TREG cells and exhausted CD8+ T cells in hepatocellular carcinoma (HCC) patients. Single cell transcriptome analysis of these populations identified specific overexpression of the LAYN gene, potentially highlighting a new therapeutic target. Through the use of TCR sequencing and trajectory modeling, this study provides an atlas of HCC-infiltrating T cells characterized by unique expression profiles, representing the power of single cell immunoprofiling. [DOI] [PubMed] [Google Scholar]
- 66.Engel I, Seumois G, Chavez L, Samaniego-Castruita D, White B, Chawla A, Mock D, Vijayanand P, Kronenberg M. Innate-like functions of natural killer T cell subsets result from highly divergent gene programs. Nat Immunol. 2016;17:728–739. doi: 10.1038/ni.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Proserpio V, Piccolo A, Haim-Vilmovsky L, Kar G, Lönnberg T, Svensson V, Pramanik J, Natarajan KN, Zhai W, Zhang X, et al. Single-cell analysis of CD4+ T-cell differentiation reveals three major cell states and progressive acceleration of proliferation. Genome Biol. 2016;17:103. doi: 10.1186/s13059-016-0957-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kakaradov B, Arsenio J, Widjaja CE, He Z, Aigner S, Metz PJ, Yu B, Wehrens EJ, Lopez J, Kim SH, et al. Early transcriptional and epigenetic regulation of CD8(+) T cell differentiation revealed by single-cell RNA sequencing. Nat Immunol. 2017;18:422–432. doi: 10.1038/ni.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •69.Rao DA, Gurish MF, Marshall JL, Slowikowski K, Fonseka CY, Liu Y, Donlin LT, Henderson LA, Wei K, Mizoguchi F, et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature. 2017;542:110–114. doi: 10.1038/nature20810. This study presents the first description of peripheral T helper (TPH) cells, a PD-1hi CXCR5− CD4+ effector population that interacts with B cells and is expanded in the RA synovium. TPH cells express unique trafficking and migratory molecules, allowing TPH cells to accumulate at sites of inflammation, and are positively correlated with disease activity in RA patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stephenson W, Donlin LT, Butler A, Rozo C. Single-Cell RNA-Seq Of Rheumatoid Arthritis Synovial Tissue Using Low Cost Microfluidic Instrumentation. bioRxiv. 2017 doi: 10.1101/140848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••71.Ishigaki K, Shoda H, Kochi Y, Yasui T, Kadono Y, Tanaka S, Fujio K, Yamamoto K. Quantitative and qualitative characterization of expanded CD4+ T cell clones in rheumatoid arthritis patients. Nature Publishing Group. 2015;5:12937. doi: 10.1038/srep12937. This study uses single-cell transcriptomics and TCR sequencing to identify gene expression profiles of expanded memory CD4 T cells in the peripheral blood and synovium of RA patients. Here, the combination of single cell sequencing and TCR repertoire analysis permits the identification of expanded T cell clones and reveals that these cells do not resemble canonical CD4 T helper subsets. However, the small sample size of this study significantly limits the broader applicability of these results. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mellado M, Martínez-Muñoz L, Cascio G, Lucas P, Pablos JL, Rodríguez-Frade JM. T Cell Migration in Rheumatoid Arthritis. Front Immunol. 2015;6:384. doi: 10.3389/fimmu.2015.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Noack M, Miossec P. Th17 and regulatory T cell balance in autoimmune and inflammatory diseases. Autoimmunity Reviews. 2014;13:668–677. doi: 10.1016/j.autrev.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 74.Alzabin S, Williams RO. Effector T cells in rheumatoid arthritis: Lessons from animal models. FEBS Lett. 2011;585:3649–3659. doi: 10.1016/j.febslet.2011.04.034. [DOI] [PubMed] [Google Scholar]
- 75.Gaublomme JT, Yosef N, Lee Y, Gertner RS, Yang LV, Wu C, Pandolfi PP, Mak T, Satija R, Shalek AK, et al. Single-Cell Genomics Unveils Critical Regulators of Th17 Cell Pathogenicity. Cell. 2015;163:1400–1412. doi: 10.1016/j.cell.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •76.Fessler J, Raicht A, Husic R, Ficjan A, Schwarz C, Duftner C, Schwinger W, Graninger WB, Stradner MH, Dejaco C. Novel Senescent Regulatory T-Cell Subset with Impaired Suppressive Function in Rheumatoid Arthritis. Front Immunol. 2017;8:300. doi: 10.3389/fimmu.2017.00300. This study is the first to report the discovery of CD4+ CD28− FoxP3+ T cell subset specific to the periphery of RA patients. The TREG-like population shows hallmarks of senescence and impaired function as detected by TCR sequencing and suppression assays. These findings may help resolve conflicting reports on the functional capacity of TREG cells in RA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cui J, Saevarsdottir S, Thomson B, Padyukov L, van der Helm-van Mil AHM, Nititham J, Hughes LB, de Vries N, Raychaudhuri S, Alfredsson L, et al. Rheumatoid arthritis risk allele PTPRC is also associated with response to anti-tumor necrosis factor alpha therapy. Arthritis Rheum. 2010;62:1849–1861. doi: 10.1002/art.27457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cui J, Stahl EA, Saevarsdottir S, Miceli C, Diogo D, Trynka G, Raj T, Mirkov MU, Canhao H, Ikari K, et al. Genome-wide association study and gene expression analysis identifies CD84 as a predictor of response to etanercept therapy in rheumatoid arthritis. PLoS Genet. 2013;9:e1003394. doi: 10.1371/journal.pgen.1003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ponchel F, Goëb V, Parmar R, El-Sherbiny Y, Boissinot M, Jawhari El J, Burska A, Vital EM, Harrison S, Conaghan PG, et al. An immunological biomarker to predict MTX response in early RA. Annals of the Rheumatic Diseases. 2014;73:2047–2053. doi: 10.1136/annrheumdis-2013-203566. [DOI] [PubMed] [Google Scholar]
- 80.Daïen CI, Gailhac S, Audo R, Mura T, Hahne M, Combe B, Morel J. High levels of natural killer cells are associated with response to tocilizumab in patients with severe rheumatoid arthritis. Rheumatology (Oxford) 2015;54:601–608. doi: 10.1093/rheumatology/keu363. [DOI] [PubMed] [Google Scholar]
- 81.Kikuchi J, Hashizume M, Kaneko Y, Yoshimoto K, Nishina N, Takeuchi T. Peripheral blood CD4(+)CD25(+)CD127(low) regulatory T cells are significantly increased by tocilizumab treatment in patients with rheumatoid arthritis: increase in regulatory T cells correlates with clinical response. Arthritis Res Ther. 2015;17:10. doi: 10.1186/s13075-015-0526-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakachi S, Sumitomo S, Tsuchida Y, Tsuchiya H, Kono M, Kato R, Sakurai K, Hanata N, Nagafuchi Y, Tateishi S, et al. Interleukin-10-producing LAG3(+) regulatory T cells are associated with disease activity and abatacept treatment in rheumatoid arthritis. Arthritis Res Ther. 2017;19:97. doi: 10.1186/s13075-017-1309-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bystrom J, Clanchy FI, Taher TE, Al-Bogami MM, Muhammad HA, Alzabin S, Mangat P, Jawad AS, Williams RO, Mageed RA. Response to Treatment with TNFα Inhibitors in Rheumatoid Arthritis Is Associated with High Levels of GM-CSF and GM-CSF(+) T Lymphocytes. Clinic Rev Allerg Immunol. 2017;365:2205. doi: 10.1007/s12016-017-8610-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Citro A, Scrivo R, Martini H, Martire C, De Marzio P, Vestri AR, Sidney J, Sette A, Barnaba V, Valesini G. CD8+ T Cells Specific to Apoptosis-Associated Antigens Predict the Response to Tumor Necrosis Factor Inhibitor Therapy in Rheumatoid Arthritis. PLoS ONE. 2015;10:e0128607. doi: 10.1371/journal.pone.0128607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 87.Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, Geman D, Baggerly K, Irizarry RA. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••88.Hicks SC, Teng M, Irizarry RA. On the widespread and critical impact of systematic bias and batch effects in single-cell RNA-Seq data. bioRxiv. 2015 doi: 10.1101/025528. This study lays out the challenges of analyzing single cell sequencing data due to batch effects. Wide variety in gene detection rate provides a major source of cell-to-cell variation, masking biological signatures and producing false discoveries. Differences in detection rate can inflate distance calculations between similar cells and influence unbiased clustering methods. Importantly, the explicitly modeling batch effects in a regression framework is not sufficient to overcome the confounding effects of technical variation. [DOI] [Google Scholar]
- 89.Tung P-Y, Blischak JD, Hsiao CJ, Knowles DA, Burnett JE, Pritchard JK, Gilad Y. Batch effects and the effective design of single-cell gene expression studies. Nature Publishing Group. 2017;7:39921. doi: 10.1038/srep39921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••90.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nature Publishing Group. 2015;33:495–502. doi: 10.1038/nbt.3192. This report presents a new tool, Seraut, for analyzing single cell RNA-seq data. Seraut uses an imputation-like process to compensate for technical variation in single cell gene expression measurements. Here, in situ hybridization data for the expression of a small set of landmark genes was used to reconsctruct the spatial position of single cells by comparing their expression profiles to three-dimensional hybridization expression patterns. This work showcases one approach to integrating single cell transcriptomic data with other datasets to better describe cellular populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aghaeepour N, Finak G, Dougall D, Khodabakhshi AH, Mah P, Obermoser G, Spidlen J, Taylor I, Wuensch SA, Bramson J, et al. Critical assessment of automated flow cytometry data analysis techniques. Nat Meth. 2013;10:228–238. doi: 10.1038/nmeth.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hu X, Kim H, Brennan PJ, Han B, Baecher-Allan CM, De Jager PL, Brenner MB, Raychaudhuri S. Application of user-guided automated cytometric data analysis to large-scale immunoprofiling of invariant natural killer T cells. Proc Natl Acad Sci US A. 2013;110:19030–19035. doi: 10.1073/pnas.1318322110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shekhar K, Brodin P, Davis MM, Chakraborty AK. Automatic Classification of Cellular Expression by Nonlinear Stochastic Embedding (ACCENSE) Proc Natl Acad Sci US A. 2014;111:202–207. doi: 10.1073/pnas.1321405111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Becher B, Schlitzer A, Chen J, Mair F, Sumatoh HR, Teng KWW, Low D, Ruedl C, Riccardi-Castagnoli P, Poidinger M, et al. High-dimensional analysis of the murine myeloid cell system. Nat Immunol. 2014;15:1181–1189. doi: 10.1038/ni.3006. [DOI] [PubMed] [Google Scholar]
- 95.Bendall SC, Davis KL, Amir E-AD, Tadmor MD, Simonds EF, Chen TJ, Shenfeld DK, Nolan GP, Pe’er D. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157:714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, Saeys Y. FlowSOM. Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015;87:636–645. doi: 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
- 97.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir E-AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Weber LM, Robinson MD. Comparison of clustering methods for high-dimensional single-cell flow and mass cytometry data. Cytometry A. 2016;89:1084–1096. doi: 10.1002/cyto.a.23030. [DOI] [PubMed] [Google Scholar]
- 99.Lun ATL, Richard AC, Marioni JC. Testing for differential abundance in mass cytometry data. Nat Meth. 2017;14:707–709. doi: 10.1038/nmeth.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ, Nolan GP. Automated identification of stratifying signatures in cellular subpopulations. Proc Natl Acad Sci US A. 2014;111:E2770–7. doi: 10.1073/pnas.1408792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin P, Troup M, Ho JWK. CIDR: Ultrafast and accurate clustering through imputation for single-cell RNA-seq data. Genome Biol. 2017;18:59. doi: 10.1186/s13059-017-1188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Azizi E, Prabhakaran S, Carr A. Bayesian Inference for Single-cell Clustering and Imputing. Genomics and Computational Biology. 2017:3. [Google Scholar]
- 103.Li WV, Li JJ. scImpute: accurate and robust imputation for single cell RNA-seq data. bioRxiv. 2017 doi: 10.1101/141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •104.Villani A-C, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017:356. doi: 10.1126/science.aah4573. This study defined new subtypes of human dendritic cells and monocytes using single-cell RNA sequencing. Functional and transcriptomic assays identified a common progenitor of conventional CD1c+ dendritic cells alongside other novel dendritic and monocyte populations. This work demonstrates the power of single cell profiling to better define human immunology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •105.Kinslow JD, Blum LK, Deane KD, Demoruelle MK, Okamoto Y, Parish MC, Kongpachith S, Lahey LJ, Norris JM, Robinson WH, et al. Elevated IgA Plasmablast Levels in Subjects at Risk of Developing Rheumatoid Arthritis. Arthritis Rheumatol. 2016;68:2372–2383. doi: 10.1002/art.39771. This study takes advantage of single cell repertoire sequencing to find an expansion of IgA+ plasmablasts in seropositive first-degree relatives of RA probands. This discovery has the potential to improve detection of preclinical RA and further implicates mucosal immunity in early RA pathogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, Dekker CL, Mackey S, Maecker H, Swan GE, et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med. 2013;5:208ra145. doi: 10.1126/scitranslmed.3006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Strauss-Albee DM, Fukuyama J, Liang EC, Yao Y, Jarrell JA, Drake AL, Kinuthia J, Montgomery RR, John-Stewart G, Holmes S, et al. Human NK cell repertoire diversity reflects immune experience and correlates with viral susceptibility. Sci Transl Med. 2015;7:297ra115–297ra115. doi: 10.1126/scitranslmed.aac5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang J, Zeng X, Sigal N, Lund PJ, Su LF, Huang H, Chien Y-H, Davis MM. Detection, phenotyping, and quantification of antigen-specific T cells using a peptide-MHC dodecamer. Proc Natl Acad Sci US A. 2016;113:E1890–7. doi: 10.1073/pnas.1602488113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •109.Esfandiary L, Gupta N, Voigt A, Wanchoo A, Chan EKL, Sukumaran S, Nguyen CQ. Single-cell antibody nanowells: a novel technology in detecting anti-SSA/Ro60- and anti-SSB/La autoantibody-producing cells in peripheral blood of rheumatic disease patients. Arthritis Res Ther. 2016;18:107. doi: 10.1186/s13075-016-1010-5. This work describes a new, high-throughput method of detecting autoantibody production on a single cell level using nanowells (SCAN). While this study focused on autoantibodies specific to systemic lupus erythematosus (SLE), the technology presented here could be easily extended for use in studying RA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bentzen AK, Marquard AM, Lyngaa R, Saini SK, Ramskov S, Donia M, Such L, Furness AJS, McGranahan N, Rosenthal R, et al. Large-scale detection of antigen-specific T cells using peptide-MHC-I multimers labeled with DNA barcodes. Nat Biotechnol. 2016;34:1037–1045. doi: 10.1038/nbt.3662. [DOI] [PubMed] [Google Scholar]
- 111.Genshaft AS, Li S, Gallant CJ, Darmanis S, Prakadan SM, Ziegler CGK, Lundberg M, Fredriksson S, Hong J, Regev A, et al. Multiplexed, targeted profiling of single-cell proteomes and transcriptomes in a single reaction. Genome Biol. 2016;17:188. doi: 10.1186/s13059-016-1045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••112.Frei AP, Bava F-A, Zunder ER, Hsieh EWY, Chen S-Y, Nolan GP, Gherardini PF. Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat Meth. 2016;13:269–275. doi: 10.1038/nmeth.3742. This study is the first to present a method of acquiring transcriptomic and cytometric data for single cells in one assay. Named PLAYR, this approach uses proximity ligation of DNA oligonucleotide probes to detect individual mRNA transcripts, which are then amplified through rolling circle amplification and detected by complimentary oligonucleotide probe that can be detected by flow or mass cytometry. The ability of PLAYR to perform simultaneous detection of 8 transcripts and 18 protein epitopes is demonstrated, as well as the ability to uncover differences in temporal regulation of transcriptional activity and proteomic expression. This method has great potential for characterizing single cells and identifying disease-relevant populations by clustering cells on a combination of surface markers and transcript abundance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••113.Fonseka CY, Rao DA, Teslovich NC, Hannes SK, Slowikowski K, Gurish MF, Donlin LT, Weinblatt ME, Massarotti E, Coblyn JS, et al. Reverse Association Of Single Cells To Rheumatoid Arthritis Accounting For Mixed Effects Identifies An Expanded CD27- HLA-DR+ Effector Memory CD4+ T Cell Population. bioRxiv. 2017 doi: 10.1101/172403. Here we present a mixed modeling single cell association strategy, MASC (Mixed Effects Modeling of Associations of Single Cell Populations) that identifies populations associated with clinical endpoints while controlling for technical variability in single cell measurements and inter-individual biologic variation. Using MASC, we identified an HLA-DR+ CD27- population of effector memory T cells expanded in RA patients compared to non-inflammatory controls. We believe MASC is a broadly applicable method to identify disease-associated cell populations in high-dimensional single cell data. [DOI] [Google Scholar]