Abstract
Agnoprotein (Agno) is an important regulatory protein of JC virus (JCV), BK virus (BKV) and simian virus 40 (SV40) and these viruses are unable to replicate efficiently in the absence of this protein. Recent 3D–NMR structural data revealed that Agno contains two alpha-helices (a minor and a major) while the rest of the protein adopts an unstructured conformation (Coric et al., 2017, J Cell Biochem). Previously, release of the JCV Agno from the Agno-positive cells was reported. Here, we have further mapped the regions of Agno responsible for its release by a structure-based systematic mutagenesis approach. Results revealed that amino acid residues (Lys22, Lys23, Phe31, Glu34 and Asp38) located either on or adjacent to the hydrophilic surface of the major alpha-helix domain of Agno play critical roles in release. Additionally, Agno was shown to strongly interact with unidentified components of the cell surface when cells are treated with Agno, suggesting additional novel roles for Agno during the viral infection cycle.
Keywords: Agnoprotein, viroporin, dimer/oligomer formation, polyomaviruses, JCV, BKV, SV40, Merkel cell polyomavirus, DNA replication, transcription, alpha helix, progressive multifocal leukoencephalopathy, protein release
Introduction
Viruses have evolved various strategies to modify the host cellular environment in order to successfully complete their life cycle. One of the ways to accomplish this task is to facilitate the release of some of their own proteins from infected cells to modulate the function of neighboring cells. Upon release, these viral proteins can act as cytokine inhibitors (Alcami et al., 1998; Liu et al., 2000), cytokine mimickers (Liu et al., 2004; Suzuki et al., 1995), complement inhibitors (Al-Mohanna et al., 2001; Anderson et al., 2002) and inflammatory cell inhibitors (Lucas et al., 1996) so as to evade the host immune system.
The human polyomaviruses JC (JCV), BK (BKV) and simian vacuolating virus 40 (SV40) encode a small regulatory protein from their late coding region, designated agnoprotein (Agno), which plays important regulatory roles in the viral replication cycle (Akan et al., 2006; Carswell et al., 1986; Ellis and Koralnik, 2015; Ellis et al., 2013; Hay et al., 1984; Johannessen et al., 2008; Johannessen et al., 2011; Myhre et al., 2010; Saribas et al., 2016; Saribas et al., 2014; Unterstab et al., 2010). These viruses undergo a productive life cycle in the presence of Agno. Interestingly, other human polyomaviruses, including HPyV9, HPyV10, MCV, TSV, HPyV6, HPyV7, KIPyV and WUPyV (De Gascun and Carr, 2013) do not have an Agno gene. Analysis of Agno null mutants demonstrated that it is required to sustain a successful propagation of the viral life cycle (Ellis et al., 2013; Myhre et al., 2010; Sariyer et al., 2011). Even the constitutive expression of large T antigen (LT-Ag), which is the major regulatory protein of the polyomaviruses, is unable to compensate for the loss of Agno function in the infected cells. In other words, in the absence of Agno, LT-Ag alone cannot sustain an efficient viral replication cycle (Sariyer et al., 2011). Agno is a primarily cytoplasmic protein with high concentrations accumulating in the perinuclear region of infected cells, but a small portion of the protein is also consistently detected in the nucleus, indicating a possible role for it in the nucleus (Saribas et al., 2012). An example of such a role was recently demonstrated where Agno was shown to enhance the DNA binding activity of LT-Ag to the viral origin (Ori) without directly interacting with DNA (Saribas et al., 2012). Another interesting feature of Agno is its tendency to form highly stable, SDS-resistant homodimers and oligomers (Saribas et al., 2011), which is mediated by the major alpha helical domain of the protein (Coric et al., 2014). Recent studies have also demonstrated that this region is required for the stable expression of Agno (Coric et al., 2014; Saribas et al., 2013). Furthermore, Suzuki et al (Suzuki et al., 2013; Suzuki et al., 2010) has demonstrated that Agno behaves as a viroporin indicating its possible association with the plasma membrane. It is also known that homodimer and oligomer formation is also some of the characteristics of viroporin proteins (Royle et al., 2015).
JCV establishes a persistent asymptomatic infection in most individuals during childhood and may reactivate later in life in a subset of immunocompromised patients (Saribas et al., 2016; Saribas et al., 2010) but the mechanism(s) of this reactivation is currently unknown. JCV primarily infects glial cells in the human brain, i.e., the oligodendrocytes and astrocytes, leading to a rare demyelinating white matter disease, known as the progressive multifocal leukoencephalopathy (PML), which occurs in a subset of patients with immunosuppressive conditions, such as HIV-1/AIDS, cancer and organ transplant (Berger, 2011; Berger and Concha, 1995; Major, 2010; Major et al., 1992). In recent years however, PML has also been encountered in autoimmune disorder patients, e.g., individuals with multiple sclerosis (MS), Crohn’s disease (CD) or psoriasis, who are treated with immunomodulatory antibodies such as natalizumab and efalizumab. These antibodies are known to target certain cell surface receptors on B and T cells and modulate immune function (Kleinschmidt-DeMasters and Tyler, 2005; Langer-Gould et al., 2005; Van Assche et al., 2005).
JCV Agno interacts with a number of viral and cellular proteins, including JCV large T-antigen (LT-Ag) (Safak and Khalili, 2001), JCV small t-antigen (Sm t-Ag) (Sariyer et al., 2008) and, capsid protein, VP1 (Suzuki et al., 2012), p53 (Darbinyan et al., 2002), YB-1 (Safak et al., 2002), FEZ1 and HP1-α (Okada et al., 2005), adaptor protein complex 3 (AP-3) (Suzuki et al., 2013) and has been implicated in various aspects of the JCV life cycle, including viral transcription (Safak et al., 2002), viral replication (Safak et al., 2001), functioning as a viroporin (Suzuki et al., 2013; Suzuki et al., 2010), encapsidation (Sariyer et al., 2006). BKV Agno has been implicated in interfering with exocytosis (Johannessen et al., 2011) and inhibition of the viral DNA replication (Gerits et al., 2015). In addition, JCV Agno was shown to deregulate cell cycle progression (Darbinyan et al., 2002).
A recent study by Otlu et al. (34) has unraveled another interesting feature of Agno by demonstrating its release from Agno-positive cells through an unknown mechanism without killing cells. The same investigators further showed that Agno is present in extracellular medium in free form not associated with exosomes (Otlu et al., 2014) In the current study, we have furthered these findings by mapping the region(s) of Agno responsible for its release. Our data demonstrate that the major alpha helix plays a significant role in Agno release. More specifically, charged amino acid residues located on the hydrophilic surface of the helix including Lys22, Lys23, Glu34 and Asp38, and a hydrophobic residue Phe31, which is located towards the middle portion of the helix and separates positively charged residues from the negatively charged ones, are critical for this process. In addition, we demonstrated that Agno strongly interacts with the cell surface when cells are treated with a synthetic Agno peptide, implicating additional novel roles for Agno after its release from infected cells during the viral replication cycle.
Materials and Methods
Cell lines
SVG-A is a human glial cell line established by the transformation of primary human fetal glial cells with an origin-defective SV40 mutant (Major et al. 1985), which expresses T-antigen but does not express Agno. HEK293T cells (ATCC no. CRL-1573) are a human embryonic kidney cell line. Cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) (Life Technologies, catalog no. 31600-034) supplemented with 10% heat inactivated fetal bovine serum (FBS) and antibiotics [penicillin-streptomycin (100 µg/ml), ciprofloxacin (10 µg/ml)]. Cells were maintained at 37°C in a humidified atmosphere supplemented with 7 % CO2.
Plasmids
pCGT7-Agno, an expression plasmid containing full-length JCV Agno tagged with T7 peptide (MASMTGGQQMG) at its N-terminus, was a gift from Dr. Sariyer, Temple University. We also constructed the deletion and point mutants of JCV Agno by tagging them with a T7-peptide at their N-terminus by subcloning each Agno DNA fragment into XbaI/BamHI sites of pCGT7 vector employing a PCR-based cloning strategy using appropriate primers. The wild type JCV Mad-1 Agno (1–71) and its deletion mutants; including Agno 23–71, Agno 1–39, Agno 40–71, Agno 1–22, Agno 23–71, Agno 1–56, and Agno 1–62 were subcloned into XbaI/BamHI sites of pCGT7 vector and the resulting plasmids were designated as pCGT7- JCV Agno (23–71), pCGT7-JCV Agno (1–39), pCGT7-JCV Agno (40–71), pCGT7-JCV Agno (1–22), pCGT7- JCV Agno (1–56) and pCGT7- JCV Agno (1–62). Various site-directed mutants of JCV Agno including, F31A, F35A, F39A, L33A, I30A, L29A, L32A, L36A, E34A, D38A, K22A, K23A and R27A were also subcloned into the XbaI/BamHI sites of pCGT7 vector using the QuikChange mutagenesis kit (Agilent, Santa Clara, CA, catalog no. 200521) using appropriate primers. These plasmids are designated as: pCGT7-JCV Agno (F31A), pCGT7-JCV Agno (F35A), pCGT7-JCV Agno (F39A), pCGT7- JCV Agno (L33A), pCGT7-JCV Agno (I30A), pCGT7-JCV Agno (L29A), pCGT7-JCV Agno (L32A), pCGT7-JCV Agno (L36A), pCGT7-JCV Agno (E34A), pCGT7-JCV Agno (D38A), pCGT7-JCV Agno (K22A), pCGT7-JCV Agno (K23A) and pCGT7-JCV Agno (R27A). The integrity of each construct was validated by DNA sequencing.
Transfection
Tissue culture plates (60 mm in diameter, Becton Dickinson, Franklin Lakes, NJ, catalog no. 353002) were treated with poly-D-lysine hydrobromide for 15 min (Sigma, St. Louis, MO, catalog no. p7280-5G, prepared in dH2O, 100 µg/ml final), washed with dH2O three times and air-dried under sterile conditions. HEK293T cells were then plated onto these plates (1×106 cells/plate) in triplicate the day before transfection. Cells were then washed with growth medium (DMEM), refed with fresh media, incubated for 3 h at 37°C to allow them to adopt the fresh medium conditions. Each culture plate was then transfected with each expression plasmid (15 µg/plate) by the calcium phosphate precipitation method (Graham, 1973) as described previously (Sariyer et al., 2011). Cells were incubated with DNA-calcium phosphate complexes for 4 h, washed once with 1X phosphate buffer saline (PBS) and refed with fresh medium (4 ml). Growth medium was collected from the culture plates at 24 h posttransfection, centrifuged at 3000 rpm (Effendorf 5810R) for 10 min and treated with sodium azide (0.01% final concentration) (ThermoFisher Scientific, catalog no. PB922-500) and stored at 4°C until use. In parallel, whole-cell extracts were prepared from each plate as described below.
Preparation of whole-cell extracts and Western blotting
Whole-cell extracts from transfected and untransfected cells (HEK293T cells, 1 × 106cells/plate) were prepared as described previously with modifications (Coric et al., 2014; Sariyer et al., 2011). Briefly, cells were washed with 1×PBS and directly lysed on the plate with 1 ml of the lysis buffer containing 50 mM Tris-HCl (pH7.4), 150 mM NaCl and 1.0 % NP-40 in the presence of protease inhibitors (Sigma, St. Louis, MO, catalog no. P8340) and rotated on a revolving platform at 4°C for 30 minutes. The cell lysates were then cleared by centrifugation (17,000 × g for 10 min at 4°C) and stored at −80°C until use. For Western blot, protein extracts (15 µg) were prepared in 5X SDS sample buffer [Tris-HCl (0.25 M, (pH 6.8), glycerol (50%), SDS (10%), bromophenol blue (0.25%)], denatured at 95°C for 10 min and then resolved on SDS-15%-PAGE. Protein bands on the gel were transferred onto a nitrocellulose membrane with 0.2 µm pore size (Bio-Rad, Herkules, CA, catalog no. 162-0112) for 30 minutes at 250 mA. The membranes were blocked with 5 % bovine serum albumin solution prepared in 1× TBST buffer (20 mM Tris-HCL pH 7.4 and 150 mM NaCl, 0.01% Tween-20) and incubated with a primary rabbit polyclonal anti-Agno antibody (Del Valle et al., 2002), (diluted 1:1000 in TBST) overnight at 4°C. Membranes were then washed three times with TBST and incubated with a secondary goat anti-rabbit IRDye 680LT (LI-COR, Lincoln, NE, catalog no. 926–68706) antibody for 1 hour according to the manufacturer’s recommendations. Finally, membranes were washed twice with TBST and scanned using Odyssey® CLx Infrared Imaging System (LI-COR) to detect Agno protein.
Agnoprotein release assay
Culture media from transfected and untransfected HEK293T cells were collected at 24h posttransfection and centrifuged for 5 min at 3,000 rpm (Eppendorf 5810R centrifuge). Supernatants were transferred into a new tube and stored at 4°C after the addition of sodium azide [0.02% (w/v) final]. For the analysis of the expression and release of the deletion mutants of Agno, an immunoprecipitation/Western blotting (IP/WB) method was employed as follows: Whole-cell extracts (200 µg) were incubated with monoclonal anti-T7 antibody (1 µl) (Novus biologicals, catalog no. NBP1–40393) in lysis buffer (500 µl reaction volume) for overnight on a rotating platform at 4°C. The next day, a 20 µl of protein G Sepharose 4 fast flow (FF) resin (GE Healthcare, catalog no. 17–0618-01) (50 % suspension in phosphate buffered saline, PBS) was added to the protein mix and incubated for an additional 4h +4°C. Immune complexes were subsequently washed with 1 × TBST buffer three times by centrifugation (10,000 rpm) and processed for analysis by Western blot as described above. For the analysis of the released Agno from cells, IP/WB was also performed. Supernatants from either transfected or untransfected cells (200 µl each) were first subjected to a preclearing process in order to avoid interference from the serum-born IgG during IP/WB analysis of the supernatants. To eliminate serum-born IgG, samples were initially incubated with the protein G Sepharose 4 fast flow (FF) resin (50 µl, 50 % suspension) for 5h at 4°C. The resin-bound IgG was eliminated by centrifugation (10,000 rpm) of the samples while saving the supernatants. Supernatants were then incubated for a second round of protein G resin (20 µl, 50 % suspension) plus with anti-T7 antibody (1 µl) for 24 h at + 4°C on a rotating platform. Immune complexes were then washed with 1 X TBST buffer three times and processed for analysis by Western blotting as described above. Additionally, whole-cell extracts and supernatants from each sample were also analyzed by straight Western blotting without immunoprecipitation, where 15µg extracts and 15µl supernatants were loaded on the SDS-15% PAGE for Western blot analysis. Western blot membranes were probed with primary anti-Agno polyclonal and secondary goat anti-rabbit IRDye 680LT (LI-COR) antibodies; and scanned using Odyssey® CLx Infrared Imaging System (LI-COR) in order to detect Agno protein as described above.
Indirect immunofluorescence microscopy
Indirect immunofluorescence microscopy studies were performed as previously described (Saribas et al., 2014). Briefly, SVG-A cells (1×106cells/60 mm plate) were plated onto poly-D-lysine hydrobromide-treated tissue culture plates (Becton Dickinson, Missisauga, ON, Canada, catalog no. 353002) the day before transfection. Cells were then separately transfected with either wild-type (WT) Agno or deletion mutant plasmids (5 µg/plate) using lipofectamine 3000 (Invitrogen, Carlsbad, CA, catalog no. L3000008) according to the manufacturer’s recommendations. At 24 h post transfections, cells were trypsinized (Gibco Life Technologies, catalog no. 25200-056) and seeded on poly-D-lysine hydrobromide-coated glass chamber slides (Nunc, catalog no. 154461). After 24 h incubation, the cells were washed twice with PBS, fixed in ice-cold acetone for 2 min, blocked with 5% bovine serum albumin (prepared in PBS) for 2 h, and incubated over-night with a primary monoclonal anti-T7-tag antibody (EMD Millipore, Billerica, MA, catalog no. 69522) (1:200 dilution). Cells were then washed with TBST buffer three times with 10 min intervals and incubated with a secondary FITC-conjugated goat anti-mouse antibody for 45 min. Cells were finally washed with TBST three times for 10 min each, incubated with DAPI (4',6-Diamidino-2-Phenylindole, Dihydrochloride, catalog no. D1306, ThermoFisher, prepared in PBS) for 10 min and washed in PBS three times, mounted using mounting medium (ThermoFisher, catalog no. P36934) and examined under a fluorescence microscope (Leica, DMI-6000B, objective: HCX PL APD 40×/1.25 oil, LAS AF operating software) for visualization of the expression profiles of Agno and its deletion mutants.
Treatment of cells with synthetic Agnoprotein
Full-length Agno protein was commercially synthesized by Biosynthesis Inc., (Lewisville, TX), (http://www.biosyn.com/), dissolved in dimethyl sulfoxide (DMSO, 1 µg/µl stock solution) and used to treat SVG-A cells. Briefly, SVG-A cells were grown in 2-chamber slides (ThermoFisher, catalog no, 154461) to 70–80% confluency and treated with synthetic Agno protein (10 µg/ml final) prepared in 0.5 ml DMEM plus 10% FBS supplemented with antibiotics [penicillin-streptomycin (100 µg/ml), ciprofloxacin (10 µg/ml)] and incubated for 24 h at 37°C in 7% CO2. At 24h post-treatment, cells were washed with PBS three times, fixed with ice-cold acetone and processed for immunocytochemistry using primary (anti-Agno polyclonal) and secondary FITC-conjugated goat anti-rabbit (EMD Millipore, catalog no. AP132F) antibodies; and analyzed under a fluorescence microscope (Leica, DMI-6000B, objective: HCX PL APD 40×/1.25 oil, LAS AF operating software) for visualization of Agno protein on cells.
Results
Structural features of agnoprotein
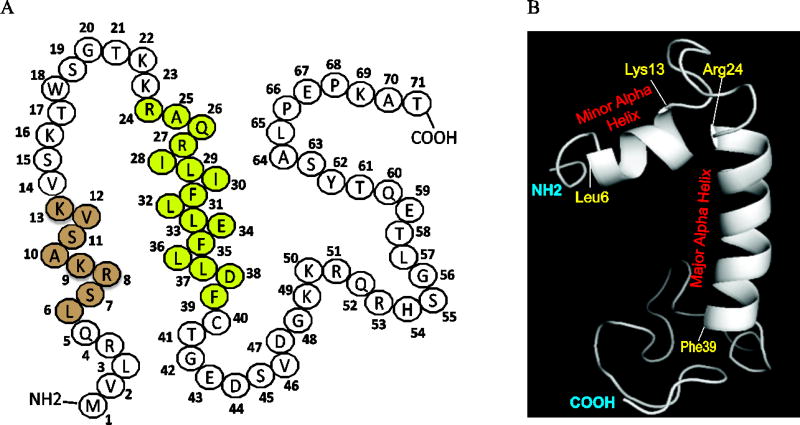
JCV agnoprotein is a small regulatory protein (71 aa long) of JCV and possesses a highly basic amino acid composition in its N-terminal and C-terminal regions, highlighted by Lys and Arg residues. However, the central portion of the protein exhibits an Ile/Leu/Phe-rich characteristic features (Fig 1A). The three dimensional (3D) structure of this protein has been recently resolved by NMR, which revealed the presence of two alpha helical regions on the protein, a minor (Leu6-Lys13) and a major (Arg24-Phe39) helix. The remaining regions of the protein adopt an intrinsically unstructured conformation (Fig. 1B). It is also interesting to note that Agno has only three Phe residues (Phe31, Phe35 and Phe39) all of which are confined to the major helix region of the protein (Fig. 1A).
Fig. 1. Structural features of JCV Agno.
(A) Primary amino acid sequence composition of JCV agnoprotein, highlighting location of the minor (leu6-Lys13, light brown colored) and major (Lys24-Phe39, light green colored) alpha helical regions. Agno is a highly basic protein in nature containing many Lys and Arg residues on its N- and C-terminus regions. The major alpha helix is however, mainly composed of hydrophobic residues. (B) A recently revealed NMR structure of the JCV full-length Agno which highlights the location of the minor (Leu6-Lys13) and major alpha helix (Lys24-Phe39) regions and the intrinsically unstructured (Met1-Gln5, Val14-Lys23 and Cyt40-Thr71) domains.
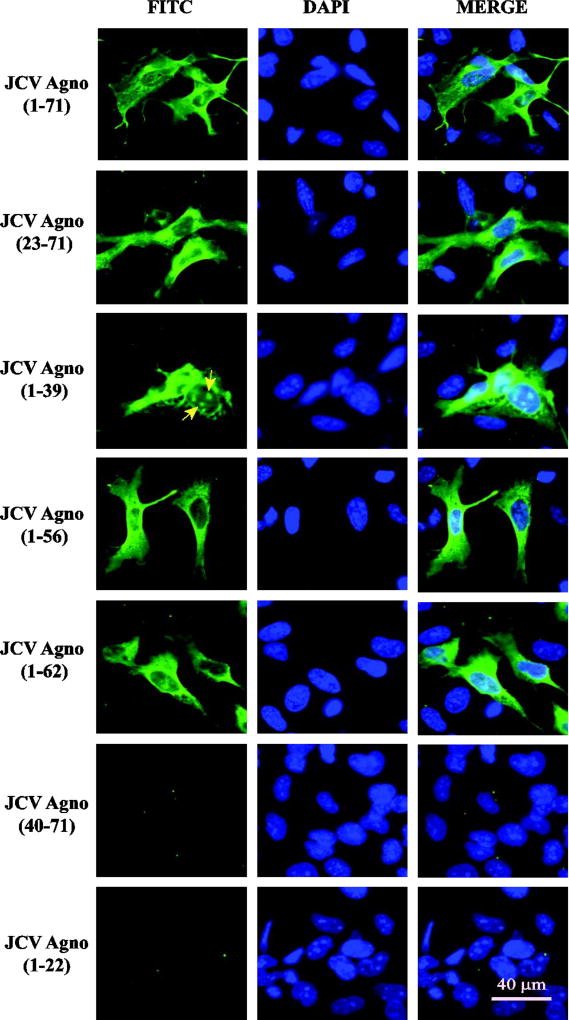
Analysis of the expression profiles of the Agno deletion mutants by immunocytochemistry
The release of JCV Agno from the infected and transfected cells was recently reported (Otlu et al., 2014). The goal of the current study was to identify and map the region(s) of Agno responsible for its release from the Agno-positive cells. To achieve this goal, a structure-based systematic mutagenesis approach has been implemented in creation of the deletion mutants of Agno and their analysis in Agno release assays. Our recent NMR structural studies revealed that this protein contains two alpha helices, a minor one located towards the amino terminus and a major one situated towards the middle portion of the protein and the remaining sequences do not form a structured conformation (Fig. 1B) (Coric et al. 2017). Thus, based on this structural data, a series of deletion mutants was created by including or excluding these two alpha helical regions in the mutants in order to analyze their contribution to Agno release. Prior to performing the release assays, we first sought to determine the stable expression and subcellular distribution profiles of each mutant by immunocytochemistry (ICC). The ICC assays were performed using SVG-A cells (Major et al., 1985). These cells were used because they are relatively large in size and therefore provide appreciable compartmentalization to examine subcellular distribution under a fluorescence microscope. It should be noted here, however, that any cell type could have been chosen for this purpose as long as the chosen cell line is relatively larger in size for the microscopic studies, since Agno does not require a cell-type specific release. These ICC studies demonstrated that the expression of any deletion mutant that retains the major alpha helix region of Agno (aa 23–71, aa 1–39, aa 1–56 and aa 1–62) is readily detectable by ICC (Fig. 2). In addition, the subcellular distribution of each mutant was similar to that observed for Agno WT except that the Agno 1–39 deletion mutant exhibited a noticeable punctate nuclear localization (Fig. 2, pointed by arrows) in addition to its WT-like cytoplasmic distribution. The remaining two mutants, one of which lacks both alpha helices from its structure (Agno 40–71) and the other one preserves the minor but lacks the major alpha helix region were both undetectable by ICC, suggesting that the major alpha helix domain of the protein is most likely required for the stable expression of both mutants, which is consistent with our previous findings, where it was demonstrated that the major helix domain of the protein plays a major role in stability of Agno (Saribas et al., 2013).
Fig. 2. Analysis of the cellular distribution properties of the various Agno deletion mutants by immunocytochemistry (ICC).
JCV Agno WT and its deletion mutants were tagged with T7-tag at their N-terminus by cloning them into pCGT7 vector at XbaI/BamHI sites and transfected into SVG-A cells as described in Materials and Methods. The cellular distribution and expression profiles of the clones were then investigated by ICC using primary monoclonal anti-T7 and secondary FITC conjugated goat anti-mouse antibodies. Finally cells were examined under a fluorescence microscope as described in Materials and Methods. Scale bar = 40 µm. Yellow arrows point to the patchy distribution of Agno 1–39 deletion mutant in the nucleus.
Investigation of the release properties of the Agno deletion mutants
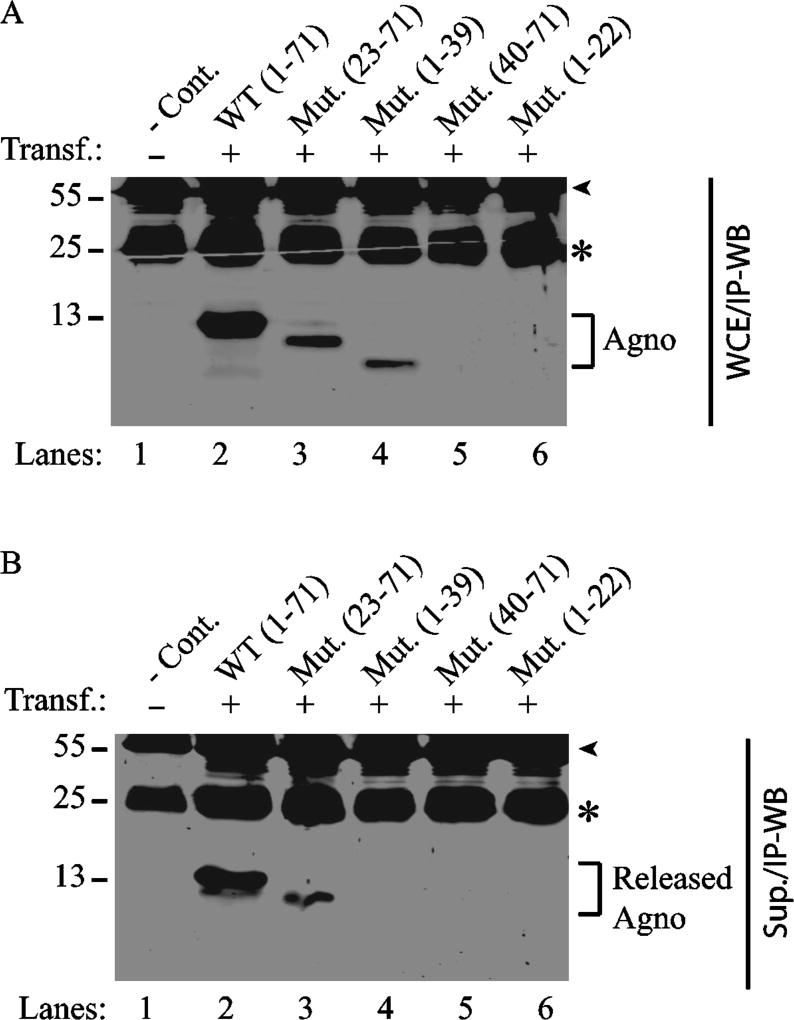
Next, the release properties of each deletion mutant were examined by immunoprecipitation followed by Western blot (IP/WB) compared to those of WT. Prior to performing the release assays, the expression dynamics of each mutant were assessed by IP/WB. Due to their remarkably high transfection efficiency, HEK293T cells were chosen to analyze the release properties of all Agno mutants in this study. As such, either Agno WT or its deletion mutant plasmids were separately transfected into HEK293T cells by the calcium phosphate precipitation method and at 24 h posttransfection, whole-cell extracts were prepared and analyzed by IP/WB to determine their expression levels as described in Materials and Methods. Consistent with the results obtained from ICC assays, results from these IP/WB assays demonstrate that again all the deletion mutants that retain the major helix domain of Agno are also detectable by IP/WB (Fig. 3A, lanes 3 and 4, Fig. 4A, lanes 3 and 4). As expected, the two deletion mutants (Agno 40–71 and Agno 1–22) that lack the major alpha helix domain were undetectable by IP/WB.
Fig. 3. Analysis of the expression and release properties of Agno deletion mutants by immunoprecipitation followed by Western blot.
(A) Analysis of the expression profiles of either WT Agno or its deletion mutants by immunoprecipitation/Western blot (IP/WB). Either Agno WT or its various deletion mutants were cloned into pCGT7 vector and transfected into 293HEKT cells as described in Materials and Methods. Whole-cells extracts (WCE) (200 µg) prepared from untransfected (lane 1) transfected (lanes 2–6) cells at 24 h posttransfection were subjected to immunoprecipitation (IP) using anti-T7 antibody followed by Western blot (WB) using the same antibody as described in Materials and Methods. (B) Analysis of the release properties of Agno WT and its deletion mutants by IP/WB. In parallel to the IP/WB analysis described for panel A, supernatants collected from each sample (200 µl) were subjected to IP/WB analysis using anti-T7 antibody as described in the Materials and Methods. Sup.: Supernatant, -Cont.: Control for the whole cells extracts prepared from the untransfected cells and for the supernatant collected from the untransfected HEK293T cells. A star sign and an arrow head for each panel point to the migration position of IgG light and IgG heavy chains respectively originating from the anti-T7 antibody that was used for IP. Transf.: Transfection., Mut.: Mutant.
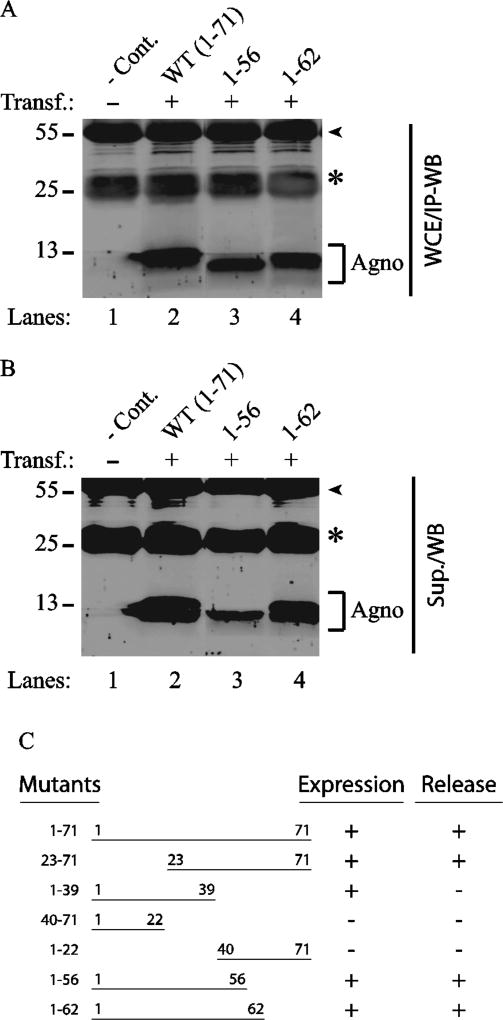
Fig. 4. Analysis of the expression and release profiles of the additional Agno C-terminal deletion mutants by IP/WB.
(A) Analysis of the expression properties of Agno 1–56 and Agno 1–63 deletion mutants. Either Agno WT or its two deletion mutants (Agno 1–56 and Agno 1–62) were cloned into pCGT7 vector and transfected into HEK293T cells. At 24 h posttransfection, WCE were prepared and analyzed by IP/WB using anti-T7 antibody as described for Fig. 3A. (B) Release properties of Agno 1–56 and Agno 1–63 deletion mutants. In parallel to the experiments described for panel A, the supernatants collected from each sample at 24h posttransfection were subjected to IP/WB analysis using anti-T7 antibody as described for Fig. 3B. (C) Summary of the expression and release properties of the Agno deletion mutants.
In parallel, the ability of each mutant’s release from the transfected cells into the cell-free culture medium was next examined by release assays as described in Materials and Methods. Results demonstrated that all the major helix-containing mutants of Agno (Fig. 3B, Agno 23–71; Fig. 4B, Agno 1–56 and Agno 1–62), except Agno 1–39 mutant, were all detectable by IP/WB in the supernatants of the transfected cells. However, we noticed that the expression and release levels of Agno 1–56 mutant were relatively lower than those of WT and the Agno 1–62 mutant, raising a critical question of whether the five amino acid residues 57–61 are important for either one or both processes. We believe that these residues are primarily important for the stability of the Agno 1–56 mutant rather than its release. In other words, in the absence of these residues (57–61), the Agno 1–56 mutant is produced in relatively low levels in cells and consequently this is reflected on its release levels as well. It should also be stressed here that “Agno release” is not associated with cell death in these assays. This important question was previously addressed by Otlu et al., who implemented various cell death detection assays with Agno positive cells including MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay) assays and found no significant effect of Agno on cell viability (Otlu et al., 2014).
It was surprising to observe that Agno 1–39 deletion mutant was undetectable in release assays although its expression was apparent in whole-cell extracts (Fig. 3B). It is possible that either it is not released from the Agno-positive cells at all or that the Agno 1–39 mutant becomes highly unstable upon its release into the culture medium. We favor the latter explanation since it is our consistent observation that this mutant protein degrades more rapidly than other deletion mutants when whole-cells extracts for all mutants are subjected to several cycles of the freeze and thaw, which is consistent with Agno 1–39 mutant degrading faster in cell culture medium after its release. One of the possible reasons for its rapid degradation is that this mutant may require Cys40 for its stability. An elegant study by Hidaka et al., showed the involvement of Cys40 in dimer/oligomer formation by a truncated Agno peptide (Lys24-Asp44) through a disulfide bridge (Hidaka et al., 2015). Thus, it is conceivable that the presence of such a disulfide bond in Agno 1–39 mutant may prevent its rapid degradation. Fig. 4C summarizes the expression and release levels of the deletion mutants.
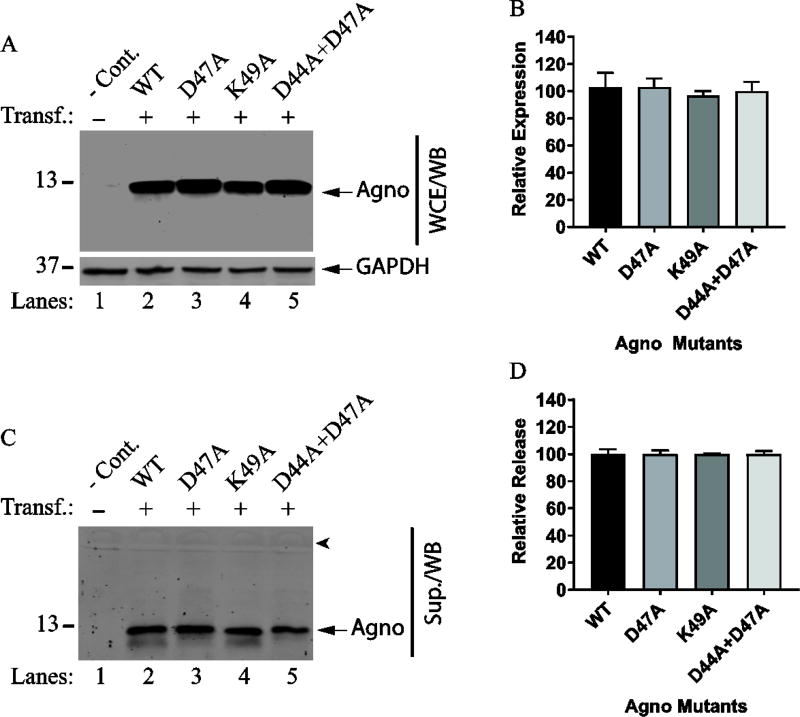
Another possibility is that Agno 1–39 mutant may require additionally charged but conserved residues beyond the major alpha helix region such as, Asp44, Asp47 and Lys49 since the release of Agno 1–56 deletion mutant is detectable (Fig. 4B). This possibility was investigated by using Agno D47A and K49A and Agno D44A+D47A substitution mutants in release assays as described in Materials and Methods. As shown in Fig. 5, all these three Agno mutants are expressed (Fig. 5A and 5B) and released (Fig. 5C and 5D) relatively well compared to Agno WT, suggesting that these residues, which are conserved among JCV, BKV and SV40 do not appear to contribute to Agno release significantly. Therefore, we concluded that the undetectable nature of the Agno 1–39 mutant in the supernatants of the transfected cells cannot be directly attributed to the lack of these residues in the mutant protein but is rather likely to be due to its unstable nature. It should also be noted here that some of the mutants were not detectable by direct Western blot due to the issues associated with their rapid degradation tendency upon cell lysis. It is our experience working with these deletion mutants that even thought their expression is readily detectable by ICC (Fig. 2), some of them showed deviations from WT Agno with respect to expression and release. This was the reason why the expression and release analysis of all the deletion mutants were all assessed by IP/WB rather than a direct Western blot.
Fig. 5. Analysis of the release properties of the several conserved residues located close proximity to the major alpha helix.
(A) Western blot of the expression levels of D47A, K49A and D44A+D47A mutants. HEK293T cells were either transfected with Agno WT or Agno Phe mutant expression plasmids separately in triplicate and WCE were prepared at 24 h posttransfection and analyzed by Western blot. Briefly, 15µg of WCE were resolved on a SDS-15% PAGE, transferred onto a nitrocellulose membrane (0.2 micron pore size) for 30 min at 250 mA. Membranes were probed with primary anti-Agno polyclonal and secondary goat anti-rabbit IRDye 680LT (LI-COR) antibodies and scanned using Odyssey® CLx Infrared Imaging System (LI-COR) to detect Agno protein as described in Materials and Methods. Blots were also probed with anti-GAPDH antibody (Santa Cruz, Dallas, TX, cat. no: sc-365062) as loading control. (B) Quantitative analysis of the results: The band intensities for Agno on Western blots on panel A were determined by using an Image J program (https://imagej.nih.gov/ij/); and subsequently statistically analyzed and graphed by using GraphPad Prism 7 program (https://www.graphpad.com/scientific-software/prism/). One way ANOVA was used to determine the statistical differences between WT Agno and mutant samples. (C) Analysis of the supernatants for Agno release by Western blotting. Briefly, 15 µl of supernatant from each transfectant or control (untransfected, -Cont.) were analyzed by WB using primary anti-Agno and secondary goat anti-rabbit IRDye 680LT (LI-COR) antibodies as described for panel A. Sup.: Supernatant. An arrowhead points to the migration position of a dominant serum protein, bovine serum albumin, transferred onto the blot. (D) Quantitative analysis of the band intensities for Agno on Western blots on panel C using an Image J program and statistical analysis of the results using GraphPad Prism 7 program.
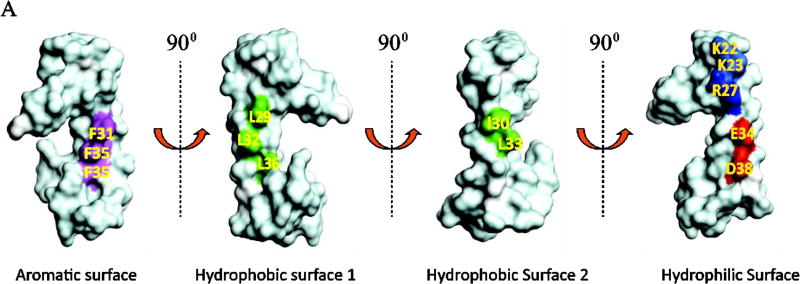
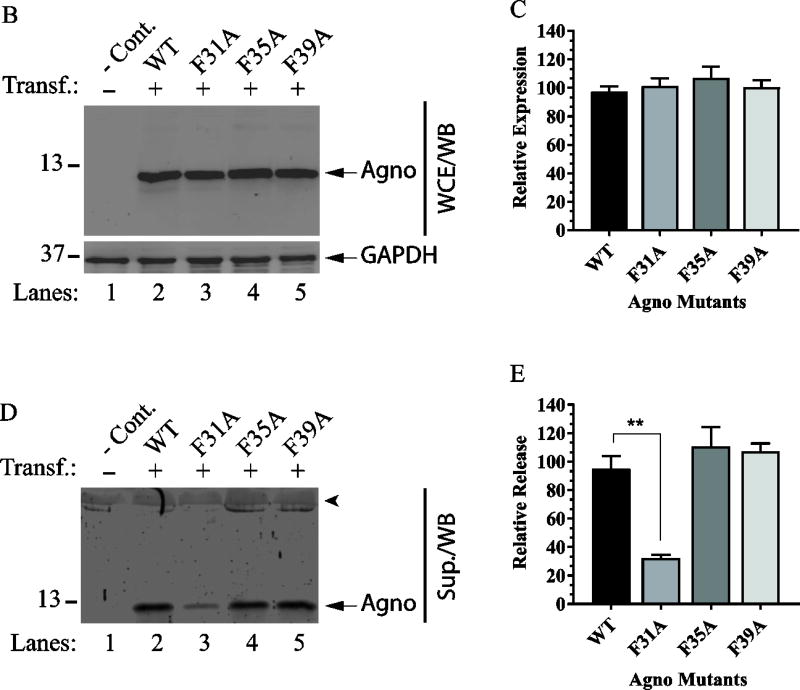
Phe31 plays a critical role in Agno release
The release analysis of the deletion mutants suggested that the major alpha helix region of Agno may play a critical role in Agno release. NMR structure-based surface representation of the major alpha helical region shows that this region contains an aromatic, a hydrophilic and two hydrophobic surfaces (Coric et al., 2017; Coric et al., 2014) (Fig. 6A). Based on this surface representation, a systematic mutational analysis of the major alpha helix region was implemented to determine the contribution of this region to Agno release. As shown in Fig. 6A, the aromatic surface contains three Phe residues, Phe31, Phe35 and Phe39. Two of these residues, Phe31 and Phe39 are highly conserved among JCV, BKV and SV40 Agno proteins (Fig. 9G). Phe35 is, however, replaced by a Leu residue in both BKV and SV40 Agno proteins. Previous mutational studies from our lab have demonstrated that these Phe residues enhance JCV large T antigen (LT-Ag) binding to the viral origin of DNA replication and thereby play roles in the viral replication cycle (Saribas et al., 2013). Another interesting observation is that the F31A, F35A and F39A mutants showed relatively even distribution of Agno in the cytoplasm and lost their ability to preferentially accumulate around the perinuclear area compared to WT Agno (Saribas et al., 2013), suggesting that these residues also play a role in a strategic distribution of Agno around the perinuclear area. The other interesting finding about these Phe mutants was that the majority of the cells infected by Phe mutant viruses showed a relatively larger cell volume compared to those of WT (Saribas et al., 2013).
Fig. 6. Release analysis of the Agno Phe mutants.
(A) NMR structure-based surface analysis of the full-length Agno (Coric et al., 2017; Coric et al., 2014). Positions of the amino acids residues mutated to Ala and used in Agno release assays are indicated. The vertical turns for each surface representation is indicated by arrows. Aromatic, hydrophilic and two hydrophobic surfaces on the major alpha helix regions are represented. (B) Analysis of the expression levels of the Phe mutants in HEK293T cells. HEK293T cells were either transfected with Agno WT or its Phe mutant expression plasmids separately and WCE were prepared at 24 h posttransfection and analyzed by Western blot as described for Fig. 5A. (C) Quantification of the band intensities of the expressed Agno on Western blots for panel B using an Image J and GraphPad Prism 7 programs as described for Fig. 5B. (D) Release analysis of Phe mutants by Western blot as described for Fig. 5C. (E) Quantification of the band intensities for the released Agno on Western blots on panel D using an Image J program and GraphPad Prism 7 program as described for Fig. 5B. ** P< 0.001: indicates statistical differences between the WT Agno and mutant samples.
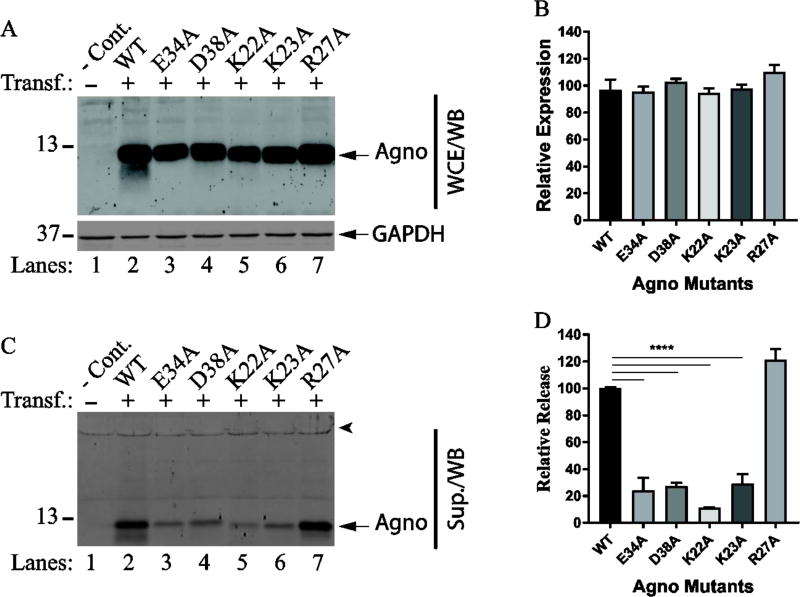
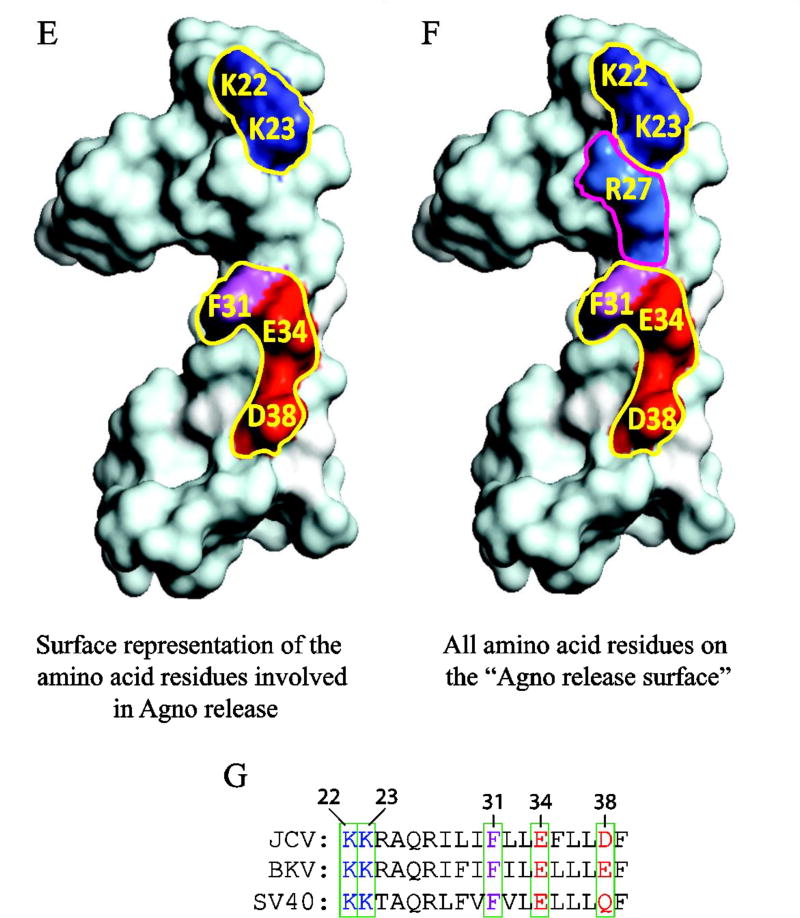
Fig. 9. The majority of the charged residues on the hydrophilic surface play critical roles Agno release.
(A) The expression analysis of the charged residue mutants located on the hydrophilic surface by Western blot as describe for Fig. 5A. (B) The intensities of the band for Agno on Western blots on panel A were quantified and graphed as described for Fig. 5B. (C) The release analysis of the mutants by Western blotting as described for Fig. 5C. (D) The intensities of Agno bands on Western blots on panel C were quantified and graphed as described for Fig. 5D. **** P< 0.001: indicates statistical differences between the Agno WT and mutant samples. (E) Locations of the amino acid residues involved in Agno release on hydrophilic surface are illustrated by a fill-in representation format of Agno structure. (F) Positions of all the amino acid residues involved or not involved in Agno release are illustrated by a fill-in representation format of Agno structure. (G) Amino acid sequence comparison for JCV, BKV and SV40 Agno primarily in the major alpha helix area. Amino acid residues involved in Agno release are encased in boxes and numbered.
Based on all these previous observations (Saribas et al., 2013) as well as the fact that all three Phe residues are structurally located on the same aromatic surface of the major alpha helix (Fig. 6A) (Coric et al., 2017; Coric et al., 2014), we reasoned that these Phe residues may individually play a role in the release of Agno from cells. For this reason, all three Phe residues were individually mutated to Ala and used for release assays. Note that Ala is known to be involved in alpha helix formation, therefore any residue within the alpha helix region substituted with Ala is assumed to preserve the structural integrity of the major alpha helical region. WT Agno or its Phe mutant expression plasmids were separately transfected into HEK293T cells. At 24 h posttransfection, whole-cell extracts were prepared and the cell-free supernatants from the transfectants were collected for analysis of Agno expression and release respectively as described in Materials and Methods. First, the relative expression levels of Phe mutants compared to that of WT Agno was assessed by Western blot. As shown in Fig. 6B, all Phe mutants exhibited a similar expression level relative to WT.
In parallel, each Phe mutant was then compared with that of Agno WT by analyzing the released Agno levels in the supernatants collected from each transfectant by Western blotting. Please note that the expression and release profiles of all mutants were investigated by Western blot without immunoprecipitation, because after analysis of the Agno deletion mutants by IP/WB, our attempts to detect the released Agno in the supernatants were successful and therefore only Western blot was employed for the rest of the Agno release assays. The quantitative analysis of the bands on the blots corresponding to each Phe mutant compared to that of WT Agno showed a statistically significant difference between the release levels of the Phe31Ala mutant and that of WT Agno (Fig. 6D, compare lane 3 with 2, Fig. 6E). The other two Phe mutants (F35A and F39A), however, behaved similar to that observed for WT (Fig. 6D, lanes 4 and 5; and Fig. 6E).
Targeted mutations within two hydrophobic surfaces of the major alpha helix do not alter Agno release
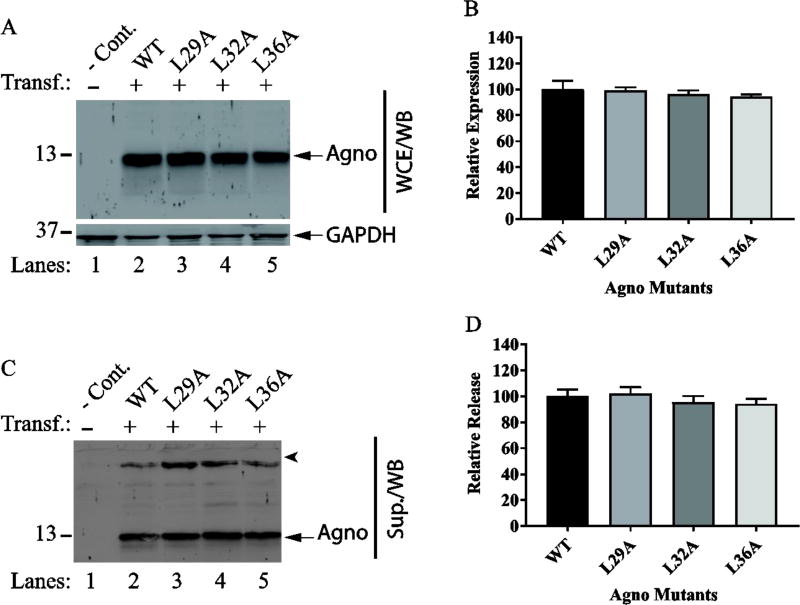
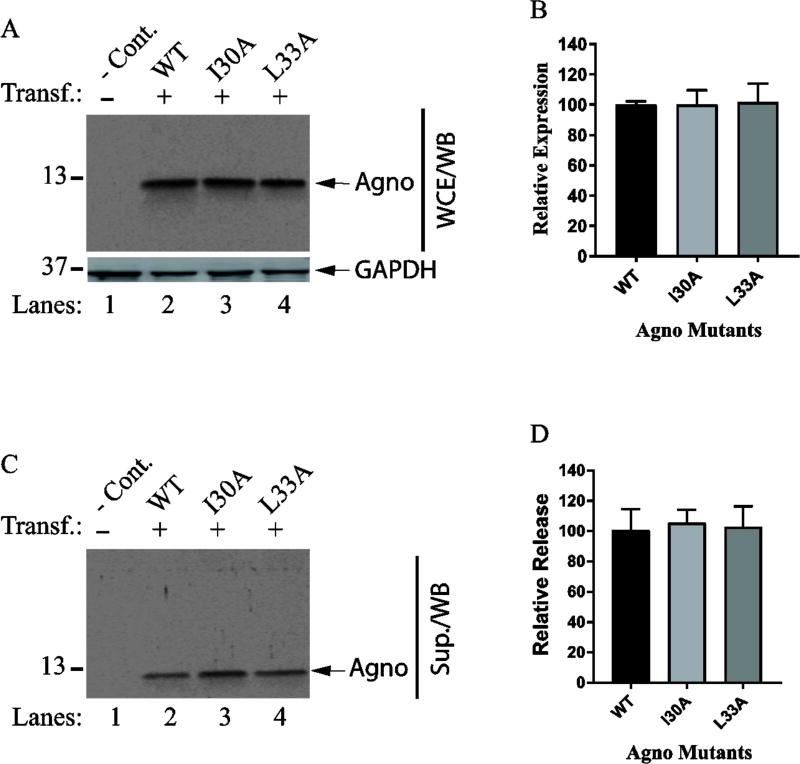
Next, we have examined the contribution of two hydrophobic surfaces of the major alpha helix to Agno release (Fig. 7 and Fig. 8). To investigate this, we have utilized a selected panel of the mutants. For the hydrophobic surface 1, we chose to analyze Leu29, Leu32 and Leu36 for their level of expression and release. In particular, special attention was devoted to two residues, Leu29 and Leu36, because our previous studies suggested that both residues play roles in stable expression and dimer formation by Agno (Coric et al., 2014). In the same studies however, Leu32 was observed not to have an effect on both processes (Coric et al., 2014). Another interesting feature of the Leu29 residue alone was that it translocates into the nucleus when mutated to Ala during the viral replication cycle (Coric et al., 2014). While Leu36 residue is conserved among JCV, BKV and SV40 Agno proteins, Leu29 is replaced by Phe in both BKV and SV40 (Fig. 9G). In addition, both Leu29 and Leu36 were shown to have a significant effect on viral DNA replication when mutated to Ala (Coric et al., 2014). For the hydrophobic surface 2, Ile30 and Leu33 were selected for analysis of Agno release, because these two residues were also previously analyzed for their contribution to protein stability and viral replication cycle (Coric et al., 2014). While Leu33 residue is conserved among JCV, BKV and SV40, Ile30 is substituted by Val in SV40 (Fig. 9G). Previous studies demonstrated that neither Ile30 nor Leu33 affect the stability of Agno, but Leu33 has an appreciable influence on viral replication (Coric et al., 2014). Finally, the expression and release features of these selected mutants of hydrophobic surface 1 and 2 by Western blot revealed no significant difference between the mutants and WT Agno, suggesting that two hydrophobic surfaces do not significantly contribute to Agno release (Fig. 7 and 8).
Fig. 7. Release analysis of the mutants on hydrophobic surface 1.
(A) Determining the expression levels of the mutants (L29A, L32A and L36A) by Western blot as described for Fig. 5A. (B) Quantification of the bands for Agno on Western blots on panel A as described for Fig. 5B. (C) In parallel to WCE preparations for panel A, the supernatants collected from the transfected and untransfected cells were analyzed for Agno release by Western blot as described for Fig. 5C. (E) Quantification of the bands on Western blots for panel C as described for Fig. 5D.
Fig. 8. Release analysis of the mutants located on the hydrophobic surface 2.
(A) Western blot analysis of the expression levels of the mutants (L33A and I30A) located on hydrophobic surface 2 as describe for Fig. 5A. (B) Band intensities on Western blots for Agno on panel A were quantified using Image J and graphed using GraphPad programs as described for Fig. 5B. (C) In parallel to WCE preparations for panel A, the released Agno levels were determined in supernatants collected from the untransfected and transfected cells by Western blotting as described for Fig. 5C. (E) Quantification of the bands for Agno on Western blots on panel C as described for Fig. 5D.
Specific charged residues on the hydrophilic surface of the major alpha helix play major roles in Agno release
We then turned our attention to charged residues on the hydrophilic surface (Fig. 6A) to examine their roles in Agno release. The hydrophilic surface exhibits several interesting features. For example, this region contains three positively (Lys22, Lys23, Arg27) and two negatively (Glu34 and Asp38) charged residues. Of these, Lys22 and Lys23 are located immediately adjacent to the major alpha helix, the other three residues are localized within the alpha helix. All but Asp38 are conserved among JCV, BKV and SV40. In addition, a close examination of 3D structure surface revealed that the Phe31 residue substantially close to the hydrophilic surface (Fig. 9F) even though its location is designated on the aromatic surface. In other word, Phe31 appears to be strategically located between the clusters of the positively charged (Lys22, Lys23, Arg27) and that of negatively charged (Glu34 and Asp38) residues; and it separates these two clusters (Fig. 9F). As for the other selected residues located on the other surfaces, all five of the charged residues were also individually mutated to Ala and analyzed for their level of expression in cells and that of release from the transfected cells into the cell culture media by Western blot. As shown in Fig. 9AB, all the mutants are expressed relatively well compared to WT Agno but the release level of each mutant drastically reduced (more than 70%) compared to that of WT (Fig. 9CD). For example, the reduction in release levels of Lys22Ala, Lys23Ala, Glu34Ala and Asp38Ala mutants was statistically significant compared to that of WT Agno (Fig. 9CD). In contrast, however, Arg27, another positively charged residue located on the same surface, did not exhibit such a drastic drop in its release levels when its charge was eliminated by a mutation to Ala, suggesting that not all the charged residues on the hydrophilic surface are involved regulating the release of Agno. Note that in the absence of Lys22 residue, a deletion mutant Agno 23–71 exhibited a relatively low level of expression and release. We believe that this differential expression and release result from the structural differences between WT Agno and the Agno 23–71 deletion mutant. Thus, the behavior of Lys22Ala mutant presented in Fig. 9C and 9D is a genuine reflection of the activity of Lys22 residue in release in the context for full-length protein. Collectively, in addition to Phe31 residue, which is also located adjacent to the hydrophilic surface (Fig. 9F), the certain charged residues on the hydrophilic surface of the major alpha helix play major roles in Agno release (Fig. 9C and Fig. 9D). Therefore, it is reasonable to designate the hydrophilic surface of the major alpha helix region as “the release surface” which is responsible for regulating Agno release from Agno-positive cells. Additionally, one can ask whether the residues involved in Agno release may play functional roles in JC virus replication cycle as well. Indeed, we have previously tested the effect of some of these mutants on the viral proliferation, including Phe31, Glu34 and Asp38. Results demonstrated that the Phe31Ala mutant showed a slight negative effect on viral proliferation (Saribas et al., 2012), but the other two mutants (Glu34Ala and Asp38Ala) significantly reduced the levels of the same process (Coric et al., 2014), suggesting that the residues involved in release may also play roles in viral proliferation.
Agno strongly interacts with cell surface
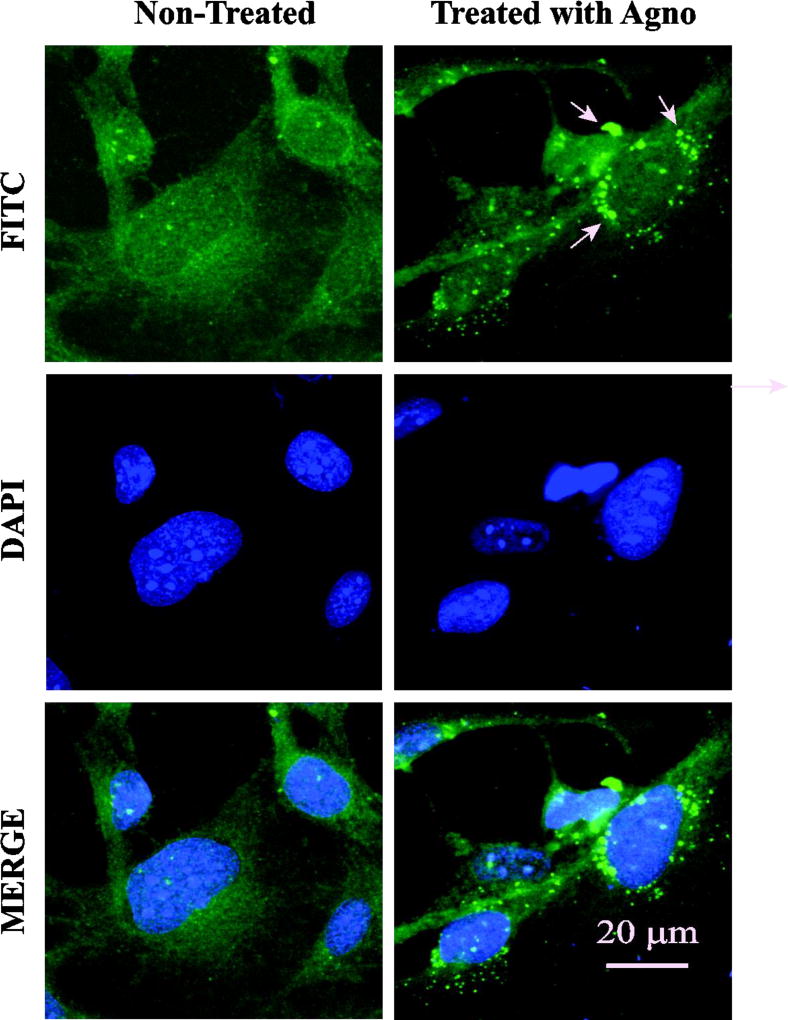
There are a number of viral proteins that were shown to be released from cells and subsequently observed to act on the neighboring cells (Ensoli et al., 1993; Luganini et al., 2016; Macdonald et al., 2005; Windheim et al., 2013). So far, our data demonstrate that Agno is released from cells, but did not address whether released Agno interacts with neighboring cells or subsequently enters them. To distinguish between these possibilities, SVG-A cells were treated with a synthetic Agno protein and monitored for the fate of Agno with respect to its interaction with the cell surface or internalization by immunocytochemistry as described in Materials and Methods. As shown in Fig. 10, Agno is readily detectable on cells by 24 h post-treatment. Treated cells were also examined under a confocal microscope to detect its internalization but these studies did not yield convincing results with respect to Agno internalization (data not shown), suggesting that Agno primarily interacts with the cell surface molecules and may exert its functional activities through them. However, there is always the possibility that various structural forms of Agno including monomers, dimers and oligomers, may enter cells in low amounts, but their level of detection is not sensitive enough to demonstrate its internalization under our experimental conditions.
Fig. 10. Treatment of SVG-A cells with synthetic Agno protein.
SVG-A cells were plated onto 2-chamber slides at the 70–80% confluency. Next day, cells were treated with DMSO only (control, non-treated) or treated with a synthetic full-length Agno protein (10 µg/ml final) prepared in 0.5 ml DMEM plus 10% FBS and incubated for 24 h at 37°C under 7% CO2. At the 24h post-treatment, cells were extensively washed with PBS, fixed with ice-cold acetone and processed for ICC using primary (anti-Agno) and secondary (goat anti-rabbit FITC-conjugated) antibodies and examined under a fluorescence microscope (Leica DM 600B) as described in Materials and Methods. Arrows point to granulated Agno deposits on cells. Scale bar = 20 µm.
Discussion
The release of JCV Agno from Agno-positive and JCV-infected cells was previously reported by Otlu et al., (Otlu et al., 2014). In this report, we aimed to further this study by mapping the regions of Agno responsible for its release and demonstrated that the major alpha-helix region of the protein plays a critical role in this process. The recent 3D NMR structural studies on JCV Agno protein demonstrated that the major alpha-helix region is composed of four different surfaces: an aromatic, a hydrophilic and two hydrophobic surfaces Fig. 6A) (Coric et al., 2017; Coric et al., 2014). The deletion mutagenesis of various Agno regions showed that integrity of the major alpha helix is required for Agno expression and its release from the Agno-positive cells suggesting that the major alpha helical region of the protein may regulate the release of Agno. We then specifically demonstrated that two positively (Lys22 and Lys23) and two negatively (Glu34 and Asp38) charged residues located on the hydrophilic surface play fundamental roles in Agno release (Fig. 9CD). In addition, an aromatic residue, Phe31, located adjacent to the hydrophilic surface was also found to significantly contribute to this process. Therefore, the hydrophilic surface of the major alpha helix region and Phe31 all together is designated as “the release surface” which controls Agno release. In addition, our studies also showed that released Agno interacts with as-yet unidentified cell surface components as evidenced by the detection of Agno on the cells treated with a synthetic Agno protein. The functional consequences of this interaction are yet to be determined.
The identified amino acids on the major alpha helix that are involved in Agno release are all but Asp38 conserved among JCV, BKV and SV40 Agno proteins. Asp38 is, however, substituted with Glu in BKV and with Gln in SV40 Agno (Fig. 9G). The high level of conservation suggests that they may also play similar roles in release of BKV and SV40 Agno from Agno-positive cells. In fact, the release of the BKV and SV40 Agno proteins were also analyzed by our group and found that they are both released from Agno-positive cells (data not shown). As mentioned above, the release of Agno from infected cells was previously reported (Otlu et al., 2014). We have also selectively investigated the impact of two mutants, Phe31 (conserved among JCV, BKV and SV40 Agno proteins) and Asp38 (not conserved) on release of Agno from the JCV-infected cells in order to justify the relevance of our infection-free release assays in HEK293T cells. In this respect, the JCV genome was individually mutated to Phe31Ala or Asp38Ala and JCV-permissive cells (SVG-A) were transfected/infected with these mutants, the consequences of which were then examined by release assays. The results showed that these mutant viruses behaved similarly with respect to Agno release during the infection cycle (data not shown), as observed for the release of the same mutants in infection-free cases in HEK293T cells (Fig. 9CD). That is, the expression levels of Agno for the both mutants in infected cells were comparatively similar to that which was observed for WT virus but their release levels were found to be significantly lower than WT (data not shown). In other words, the results obtained from the infection studies for the Phe31Ala and Asp38Ala mutants correlate with those obtained from our infection-free release assays (Fig. 9A–D).
The exact mechanism whereby Agno is released from cells is unknown. Although, the involvement of the endoplasmic reticulum (ER) to Golgi pathway was previously suggested using an ER to Golgi protein transport inhibitor, Brefeldin A, in Agno release assays (Otlu et al., 2014). In addition, it is important to address a relevant question as to whether cell death contributes significantly to the release of Agno from Agno-positive cells. This question was previously addressed by Otlu et al., by implementing various cell death detection assays including the MTT assay, which is a colorimetric assay and measures the extend of the apoptotic cell lysis (Mosmann, 1983). This group found no significant effect of Agno on cell viability in their assays. Furthermore, these investigators tested Agno release using a dual promoter expression system where Agno and green fluorescent protein (GFP) were expressed individually or together (Otlu et al., 2014). They observed no GFP release after GFP expression alone or co-expression with Agno, while Agno release occurs, as expected, indicating that Agno release was not due to cell lysis (Otlu et al., 2014).
Expression of Agno using various eukaryotic expression systems has proven to be a difficult task, since it always yields a low level of expression. In order to stabilize Agno expression, all Agno WT and its mutants were tagged at their N-terminus with a small T7-peptide (MASMTGGQQMG) as previously reported (Otlu et al., 2014). To investigate whether the tag contributes to its release from cells, another viral regulatory protein, JCV small t antigen, has been similarly tagged at its N-terminus and the contribution of this tag to protein release was analyzed as a control. Results demonstrated that small t antigen with tag was not released from cells after its expression, suggesting that the T7-tag does not play a role in the release of Agno from cells (data not shown).
Viruses have evolved a variety of strategies to evade the host anti-viral defense systems in order to successfully complete their replication cycle. Various viral proteins are reported to be secreted from infected cells as immunomodulatory proteins in the form of cytokine inhibitors, cytokine mimickers, complement inhibitors and protease inhibitors (Lucas and McFadden, 2004). Such behavior is not unique to a particular virus group. It could be a DNA or RNA virus group. For instance, DNA poxvirus-infected cells release interleukin 18 binding proteins (IL-18BPS) to modulate the host immune response (Smith et al., 2000). The human cytomegalovirus-infected cells are also known to release a number of viral proteins including UL21, UL128, UL146, UL147 and a host protein IL-10. Of which, UL21 was shown to interact with CC15/RANTES and blocks its interaction with cellular receptors. UL147, a chemokine, is believed to play a role in viral dissemination (Luganini et al., 2016). Another DNA viral protein, adenovirus E4/49K, was also found to be released and bind to leukocyte common antigen (CD45) to modulate the leukocyte function (Windheim et al., 2013).In addition, in an RNA virus group, one of the best-characterized secreted viral proteins is West Nile virus nonstructural protein, NS1, which forms stable dimers and oligomers (Muller and Young, 2013) similar to JCV Agno protein (Saribas et al., 2013). NS1 is found in different cellular locations and forms, including a cell surface attached form, as a membrane-bound form in association with the virus-induced intracellular vesicular compartments and as a secreted hexameric lipoparticle form. NS1 functions as enterotoxin and plays an essential cofactor role in viral replication (Muller and Young, 2013). It is not clear at the moment whether Agno protein has similar forms to NS1 and plays roles in evasion of the host anti-viral defense systems as some other secreted viral proteins do as mentioned above. These interesting questions are subject to further investigation but they are beyond the scope of this work.
Agno possesses a highly basic amino acid composition with multiple Lys and Arg residues, located at the amino and carboxy termini of the protein. The middle portion of the protein, however, contains highly a hydrophobic Leu/Ile/Phe-rich region, which is involved in stable dimer and oligomer formation and confers protein stability (Saribas et al., 2013). Recent NMR structural studies has shown that this region is involved in formation of a major alpha-helix. Agno also forms another short helix towards the N-terminus of the protein. The remaining regions appear to adopt an intrinsically unstructured conformation (Fig. 1B). During our initial mapping studies, we addressed the question of whether the amino acid residues in the flanking regions of the alpha helices are important for Agno release. Results demonstrated that the flanking sequences of the major alpha helix do not significantly alter Agno release (Fig. 3B and Fig. 4B). Even the elimination of the minor alpha helix region did not show an impact on this process (Fig. 3B). However, the deletion mapping studies have revealed the fact that any mutant that retains the major alpha helical region is expressed and released from Agno-positive cells (Fig. 2, Fig. 3AB and Fig. 4AB). Thus, we focused our attention to the major helix region of the protein to further examine its role in Agno release. As mentioned above, the major alpha helical region of Agno was shown to form stable dimers and oligomers (Saribas et al., 2013). In addition, the recent 3D NMR structure of the Agno (Coric et al., 2017; Coric et al., 2014) revealed the presence of four surfaces on its major alpha helix. (Fig. 6A). We have previously examined the contribution of these four surfaces to the stable dimer/oligomer formation by Agno and found out that the hydrophobic surface 1 (Fig. 6A) is highly important for dimer/oligomer formation (Coric et al., 2014). Experimental evidence by mutational analysis of the four surface areas accompanied by modeling studies suggested that particularly Leu29 and Leu36 residues on this hydrophobic surface may interact with one another in zipper-like fashion when the Agno monomers are in their anti-parallel orientation and significantly contribute to dimer formation (Coric et al., 2014). In this study, we also employed a systematic mutational approach to investigate the contribution of each surface to another interesting feature of Agno, “release from the Agno-positive cells”. To address this, we have used a selected panel of Agno mutants created on the major alpha helix region in release assays. Results demonstrated that the hydrophilic surface area of the major alpha helix plays a fundamental role in this process. Particularly, two positively, Lys22 and Lys23; and two negatively (Glu34 and Asp38) charged residues are highly critical in this process. Additionally, an aromatic amino acid, Phe31, located near these charged residues on the alpha helix turned out to contribute significantly this process as well (Fig. 6DE). It is noted that although Phe31 is located on the aromatic surface, its location is highly adjacent to the charged residues on the hydrophilic surface (Fig. 9F), which makes our experimental evidence corroborated with its location. It was also interesting to note that not every charged residue is involved in Agno release even though it is located on the hydrophilic surface. For example, the mutation of Arg27 to Ala did not restrict the Agno release but rather both its expression and release levels are relatively increased compared to those of WT Agno (Fig. 9A–D). Additionally, as previously reported, JCV Agno forms highly stable dimers and oligomers (Saribas et al., 2013; Saribas et al., 2011) and it is not known which forms of Agno protein (monomer and/or dimer and/or oligomer) are preferentially released to the cell culture media. Although it is an interesting question to address, it is beyond the scope of our current work. Additionally, a close examination of the orientation of the major alpha helical region of Agno also revealed the fact that the positions of the previously designated dimerization surface (hydrophobic surface 1) (Coric et al., 2014) and currently designated the “release surface” (hydrophilic surface) are positioned completed opposite side of the alpha helix region (Fig. 6A). As such, given the importance of Agno protein in JC virus life cycle (Sariyer et al., 2006; Sariyer et al., 2011), both surfaces can serve as the potential target regions of Agno for the drug development purposes against the JCV infection-induced white matter disease of the central nervous system, known as PML.
The functional consequences of released Agno is not known. It is conceivable that released Agno interacts with the cell surface, perhaps enters cells and exerts its effects by either one or both pathways. We have directly addressed this question by treating cells with a synthetic Agno protein and followed its behavior. Our repeated and independent experiments consistently demonstrated that Agno primarily interacts with and remains bound to the cell surface rather than visibly entering cells in significant quantities under our experimental conditions. However, there are always the possibilities that the low levels of the various forms of Agno (monomer, dimer, oligomer) (Saribas et al., 2013) may enter cells but they are not detectable under our immunocytochemistry conditions. The question then remains unanswered as to the function of released Agno. There are several possibilities in this respect: (i) The released Agno influences the neighboring cell environment by interacting with the cell surface and, directly or indirectly, modulating the host cell gene expression so that incoming virus replicates more efficiently in those cells. (ii) The released Agno may enter cells in low levels through which it influences again the host machinery in favor of efficient replication of incoming viruses. (iii) The Agno exerts its functions through either the interaction with the cell surface or by entering the cells to modulate the expression levels of various chemokines and cytokines so that the virus can evade the host immune response against JCV infection. Our future plans are to determine the functional consequences of the released extracellular Agno with respect to its effects on neighboring cells.
Acknowledgments
This work was made possible by grants awarded to M.S. (RO1NS09049) by the NIH and by the Temple University Drug Discovery Initiative (161398).
Footnotes
Conflicts of Interest:
The authors declare no conflict of interest
References
- Akan I, Sariyer IK, Biffi R, Palermo V, Woolridge S, White MK, Amini S, Khalili K, Safak M. Human polyomavirus JCV late leader peptide region contains important regulatory elements. Virology. 2006;349(1):66–78. doi: 10.1016/j.virol.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Al-Mohanna F, Parhar R, Kotwal GJ. Vaccinia virus complement control protein is capable of protecting xenoendothelial cells from antibody binding and killing by human complement and cytotoxic cells. Transplantation. 2001;71(6):796–801. doi: 10.1097/00007890-200103270-00019. [DOI] [PubMed] [Google Scholar]
- Alcami A, Symons JA, Collins PD, Williams TJ, Smith GL. Blockade of chemokine activity by a soluble chemokine binding protein from vaccinia virus. J Immunol. 1998;160(2):624–633. [PubMed] [Google Scholar]
- Anderson JB, Smith SA, Kotwal GJ. Vaccinia virus complement control protein inhibits hyperacute xenorejection. Transplant Proc. 2002;34(4):1083–1085. doi: 10.1016/s0041-1345(02)02805-1. [DOI] [PubMed] [Google Scholar]
- Berger JR. The basis for modeling progressive multifocal leukoencephalopathy pathogenesis. Curr Opin Neurol. 2011;24(3):262–267. doi: 10.1097/WCO.0b013e328346d2a3. [DOI] [PubMed] [Google Scholar]
- Berger JR, Concha M. Progressive multifocal leukoencephalopathy: the evolution of a disease once considered rare. J Neurovirol. 1995;1(1):5–18. doi: 10.3109/13550289509111006. [DOI] [PubMed] [Google Scholar]
- Carswell S, Resnick J, Alwine JC. Construction and characterization of CV-1P cell lines which constitutively express the simian virus 40 agnoprotein: alteration of plaquing phenotype of viral agnogene mutants. J Virol. 1986;60(2):415–422. doi: 10.1128/jvi.60.2.415-422.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coric P, Saribas AS, Abou-Gharbia M, Childers W, Condra JH, White MK, Safak M, Bouaziz S. Nuclear Magnetic Resonance Structure of the Human Polyoma JC Virus Agnoprotein. J Cell Biochem. 2017 doi: 10.1002/jcb.25977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coric P, Saribas AS, Abou-Gharbia M, Childers W, White MK, Bouaziz S, Safak M. Nuclear magnetic resonance structure revealed that the human polyomavirus JC virus agnoprotein contains an alpha-helix encompassing the Leu/Ile/Phe-rich domain. Journal of virology. 2014;88(12):6556–6575. doi: 10.1128/JVI.00146-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbinyan A, Darbinian N, Safak M, Radhakrishnan S, Giordano A, Khalili K. Evidence for dysregulation of cell cycle by human polyomavirus, JCV, late auxiliary protein. Oncogene. 2002;21(36):5574–5581. doi: 10.1038/sj.onc.1205744. [DOI] [PubMed] [Google Scholar]
- De Gascun CF, Carr MJ. Human polyomavirus reactivation: disease pathogenesis and treatment approaches. Clin Dev Immunol. 2013;2013:373579. doi: 10.1155/2013/373579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle L, Gordon J, Enam S, Delbue S, Croul S, Abraham S, Radhakrishnan S, Assimakopoulou M, Katsetos CD, Khalili K. Expression of human neurotropic polyomavirus JCV late gene product agnoprotein in human medulloblastoma. J Natl Cancer Inst. 2002;94(4):267–273. doi: 10.1093/jnci/94.4.267. [DOI] [PubMed] [Google Scholar]
- Ellis LC, Koralnik IJ. JC virus nucleotides 376–396 are critical for VP1 capsid protein expression. Journal of Neurovirology. 2015;21(6):671–678. doi: 10.1007/s13365-014-0278-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis LC, Norton E, Dang X, Koralnik IJ. Agnogene Deletion in a Novel Pathogenic JC Virus Isolate Impairs VP1 Expression and Virion Production. PLoS One. 2013;8(11):e80840. doi: 10.1371/journal.pone.0080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensoli B, Buonaguro L, Barillari G, Fiorelli V, Gendelman R, Morgan RA, Wingfield P, Gallo RC. Release, uptake, and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J Virol. 1993;67(1):277–287. doi: 10.1128/jvi.67.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerits N, Johannessen M, Tummler C, Walquist M, Kostenko S, Snapkov I, van Loon B, Ferrari E, Hubscher U, Moens U. Agnoprotein of polyomavirus BK interacts with proliferating cell nuclear antigen and inhibits DNA replication. Virol J. 2015;12:7. doi: 10.1186/s12985-014-0220-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hay N, Kessler M, Aloni Y. SV40 deletion mutant (d1861) with agnoprotein shortened by four amino acids. Virology. 1984;137(1):160–170. doi: 10.1016/0042-6822(84)90019-9. [DOI] [PubMed] [Google Scholar]
- Hidaka K, Hojo K, Fujioka S, Nukuzuma S, Tsuda Y. Oligomerization of neutral peptides derived from the JC virus agnoprotein through a cysteine residue. Amino Acids. 2015;47(10):2205–2213. doi: 10.1007/s00726-015-2004-3. [DOI] [PubMed] [Google Scholar]
- Johannessen M, Myhre MR, Dragset M, Tummler C, Moens U. Phosphorylation of human polyomavirus BK agnoprotein at Ser-11 is mediated by PKC and has an important regulative function. Virology. 2008;379(1):97–109. doi: 10.1016/j.virol.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Johannessen M, Walquist M, Gerits N, Dragset M, Spang A, Moens U. BKV agnoprotein interacts with alpha-soluble N-ethylmaleimide-sensitive fusion attachment protein, and negatively influences transport of VSVG-EGFP. PLoS One. 2011;6(9):e24489. doi: 10.1371/journal.pone.0024489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt-DeMasters BK, Tyler KL. Progressive multifocal leukoencephalopathy complicating treatment with natalizumab and interferon beta-1a for multiple sclerosis. N Engl J Med. 2005;353(4):369–374. doi: 10.1056/NEJMoa051782. [DOI] [PubMed] [Google Scholar]
- Langer-Gould A, Atlas SW, Green AJ, Bollen AW, Pelletier D. Progressive multifocal leukoencephalopathy in a patient treated with natalizumab. N Engl J Med. 2005;353(4):375–381. doi: 10.1056/NEJMoa051847. [DOI] [PubMed] [Google Scholar]
- Liu L, Dai E, Miller L, Seet B, Lalani A, Macauley C, Li X, Virgin HW, Bunce C, Turner P, Moyer R, McFadden G, Lucas A. Viral chemokine-binding proteins inhibit inflammatory responses and aortic allograft transplant vasculopathy in rat models. Transplantation. 2004;77(11):1652–1660. doi: 10.1097/01.tp.0000131173.52424.84. [DOI] [PubMed] [Google Scholar]
- Liu L, Lalani A, Dai E, Seet B, Macauley C, Singh R, Fan L, McFadden G, Lucas A. The viral anti-inflammatory chemokine-binding protein M-T7 reduces intimal hyperplasia after vascular injury. J Clin Invest. 2000;105(11):1613–1621. doi: 10.1172/JCI8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas A, Liu L, Macen J, Nash P, Dai E, Stewart M, Graham K, Etches W, Boshkov L, Nation PN, Humen D, Hobman ML, McFadden G. Virus-encoded serine proteinase inhibitor SERP-1 inhibits atherosclerotic plaque development after balloon angioplasty. Circulation. 1996;94(11):2890–2900. doi: 10.1161/01.cir.94.11.2890. [DOI] [PubMed] [Google Scholar]
- Lucas A, McFadden G. Secreted immunomodulatory viral proteins as novel biotherapeutics. J Immunol. 2004;173(8):4765–4774. doi: 10.4049/jimmunol.173.8.4765. [DOI] [PubMed] [Google Scholar]
- Luganini A, Terlizzi ME, Gribaudo G. Bioactive Molecules Released From Cells Infected with the Human Cytomegalovirus. Front Microbiol. 2016;7:715. doi: 10.3389/fmicb.2016.00715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald J, Tonry J, Hall RA, Williams B, Palacios G, Ashok MS, Jabado O, Clark D, Tesh RB, Briese T, Lipkin WI. NS1 protein secretion during the acute phase of West Nile virus infection. Journal of virology. 2005;79(22):13924–13933. doi: 10.1128/JVI.79.22.13924-13933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major EO. Progressive multifocal leukoencephalopathy in patients on immunomodulatory therapies. Annu Rev Med. 2010;61:35–47. doi: 10.1146/annurev.med.080708.082655. [DOI] [PubMed] [Google Scholar]
- Major EO, Amemiya K, Tornatore CS, Houff SA, Berger JR. Pathogenesis and molecular biology of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin Microbiol Rev. 1992;5(1):49–73. doi: 10.1128/cmr.5.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major EO, Miller AE, Mourrain P, Traub RG, de Widt E, Sever J. Establishment of a line of human fetal glial cells that supports JC virus multiplication. Proc Natl Acad Sci U S A. 1985;82(4):1257–1261. doi: 10.1073/pnas.82.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65(1–2):55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Muller DA, Young PR. The flavivirus NS1 protein: molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antiviral research. 2013;98(2):192–208. doi: 10.1016/j.antiviral.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Myhre MR, Olsen GH, Gosert R, Hirsch HH, Rinaldo CH. Clinical polyomavirus BK variants with agnogene deletion are non-functional but rescued by trans-complementation. Virology. 2010;398(1):12–20. doi: 10.1016/j.virol.2009.11.029. [DOI] [PubMed] [Google Scholar]
- Okada Y, Suzuki T, Sunden Y, Orba Y, Kose S, Imamoto N, Takahashi H, Tanaka S, Hall WW, Nagashima K, Sawa H. Dissociation of heterochromatin protein 1 from lamin B receptor induced by human polyomavirus agnoprotein: role in nuclear egress of viral particles. EMBO Rep. 2005;6(5):452–457. doi: 10.1038/sj.embor.7400406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otlu O, De Simone FI, Otalora YL, Khalili K, Sariyer IK. The agnoprotein of polyomavirus JC is released by infected cells: evidence for Its cellular uptake by uninfected neighboring cells. Virology. 2014;468–470:88–95. doi: 10.1016/j.virol.2014.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle J, Dobson SJ, Muller M, Macdonald A. Emerging Roles of Viroporins Encoded by DNA Viruses: Novel Targets for Antivirals? Viruses. 2015;7(10):5375–5387. doi: 10.3390/v7102880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safak M, Barrucco R, Darbinyan A, Okada Y, Nagashima K, Khalili K. Interaction of JC virus agno protein with T antigen modulates transcription and replication of the viral genome in glial cells. J Virol. 2001;75(3):1476–1486. doi: 10.1128/JVI.75.3.1476-1486.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safak M, Khalili K. Physical and functional interaction between viral and cellular proteins modulate JCV gene transcription. J Neurovirol. 2001;7(4):288–292. doi: 10.1080/13550280152537111. [DOI] [PubMed] [Google Scholar]
- Safak M, Sadowska B, Barrucco R, Khalili K. Functional interaction between JC virus late regulatory agnoprotein and cellular Y-box binding transcription factor, YB-1. J Virol. 2002;76(8):3828–3838. doi: 10.1128/JVI.76.8.3828-3838.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Abou-Gharbia M, Childers W, Sariyer IK, White MK, Safak M. Essential roles of Leu/Ile/Phe-rich domain of JC virus agnoprotein in dimer/oligomer formation, protein stability and splicing of viral transcripts. Virology. 2013;443(1):161–176. doi: 10.1016/j.virol.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Arachea BT, White MK, Viola RE, Safak M. Human polyomavirus JC small regulatory agnoprotein forms highly stable dimers and oligomers: implications for their roles in agnoprotein function. Virology. 2011;420(1):51–65. doi: 10.1016/j.virol.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Coric P, Hamazaspyan A, Davis W, Axman R, White MK, Abou-Gharbia M, Childers W, Condra JH, Bouaziz S, Safak M. Emerging From the Unknown: Structural and Functional Features of Agnoprotein of Polyomaviruses. Journal of cellular physiology. 2016;231(10):2115–2127. doi: 10.1002/jcp.25329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Mun S, Johnson J, El-Hajmoussa M, White MK, Safak M. Human polyoma JC virus minor capsid proteins, VP2 and VP3, enhance large T antigen binding to the origin of viral DNA replication: evidence for their involvement in regulation of the viral DNA replication. Virology. 2014;449:1–16. doi: 10.1016/j.virol.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Ozdemir A, Lam C, Safak M. JC virus-induced Progressive Multifocal Leukoencephalopathy. Future Virol. 2010;5(3):313–323. doi: 10.2217/fvl.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, White MK, Safak M. JC virus agnoprotein enhances large T antigen binding to the origin of viral DNA replication: Evidence for its involvement in viral DNA replication. Virology. 2012;433(1):12–26. doi: 10.1016/j.virol.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariyer IK, Akan I, Palermo V, Gordon J, Khalili K, Safak M. Phosphorylation mutants of JC virus agnoprotein are unable to sustain the viral infection cycle. J Virol. 2006;80(8):3893–3903. doi: 10.1128/JVI.80.8.3893-3903.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariyer IK, Khalili K, Safak M. Dephosphorylation of JC virus agnoprotein by protein phosphatase 2A: inhibition by small t antigen. Virology. 2008;375(2):464–479. doi: 10.1016/j.virol.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariyer IK, Saribas AS, White MK, Safak M. Infection by agnoprotein-negative mutants of polyomavirus JC and SV40 results in the release of virions that are mostly deficient in DNA content. Virol J. 2011;8:255. doi: 10.1186/1743-422X-8-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith VP, Bryant NA, Alcami A. Ectromelia, vaccinia and cowpox viruses encode secreted interleukin-18-binding proteins. The Journal of general virology. 2000;81(Pt 5):1223–1230. doi: 10.1099/0022-1317-81-5-1223. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Orba Y, Makino Y, Okada Y, Sunden Y, Hasegawa H, Hall WW, Sawa H. Viroporin activity of the JC polyomavirus is regulated by interactions with the adaptor protein complex 3. Proc Natl Acad Sci U S A. 2013;110(46):18668–18673. doi: 10.1073/pnas.1311457110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Orba Y, Okada Y, Sunden Y, Kimura T, Tanaka S, Nagashima K, Hall WW, Sawa H. The human polyoma JC virus agnoprotein acts as a viroporin. PLoS Pathog. 2010;6(3):e1000801. doi: 10.1371/journal.ppat.1000801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Semba S, Sunden Y, Orba Y, Kobayashi S, Nagashima K, Kimura T, Hasegawa H, Sawa H. Role of JC virus agnoprotein in virion formation. Microbiol Immunol. 2012;56(9):639–646. doi: 10.1111/j.1348-0421.2012.00484.x. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Tahara H, Narula S, Moore KW, Robbins PD, Lotze MT. Viral interleukin 10 (IL-10), the human herpes virus 4 cellular IL-10 homologue, induces local anergy to allogeneic and syngeneic tumors. J Exp Med. 1995;182(2):477–486. doi: 10.1084/jem.182.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterstab G, Gosert R, Leuenberger D, Lorentz P, Rinaldo CH, Hirsch HH. The polyomavirus BK agnoprotein co-localizes with lipid droplets. Virology. 2010;399(2):322–331. doi: 10.1016/j.virol.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Van Assche G, Van Ranst M, Sciot R, Dubois B, Vermeire S, Noman M, Verbeeck J, Geboes K, Robberecht W, Rutgeerts P. Progressive multifocal leukoencephalopathy after natalizumab therapy for Crohn’s disease. N Engl J Med. 2005;353(4):362–368. doi: 10.1056/NEJMoa051586. [DOI] [PubMed] [Google Scholar]
- Windheim M, Southcombe JH, Kremmer E, Chaplin L, Urlaub D, Falk CS, Claus M, Mihm J, Braithwaite M, Dennehy K, Renz H, Sester M, Watzl C, Burgert H-G. A unique secreted adenovirus E3 protein binds to the leukocyte common antigen CD45 and modulates leukocyte functions. P Natl Acad Sci USA. 2013;110(50):E4884–4893. doi: 10.1073/pnas.1312420110. [DOI] [PMC free article] [PubMed] [Google Scholar]