Abstract
Enterococcus faecalis biofilm traits and distribution characteristics in China have not been clarified. This study aimed to determine the prevalence and characteristics of E. faecalis biofilm formation in a sample of clinical isolates and to explore the virulence factors associated with biofilm formation in those isolates. A total of 265 E. faecalis isolates were collected from patients in Shenzhen, China. Virulence genes were detected within the genomes of the microbes by polymerase chain reaction. The isolates were subjected to multilocus sequence typing (MLST) based on housekeeping genes. Biofilms were detected by crystal violet staining. The expression levels of the clinical E. faecalis isolates’ genes were determined by quantitative real-time polymerase chain reaction. The prevalence of biofilm formation among E. faecalis clinical isolates was 47.2%. MLST yielded 44 different sequence types (STs). The main STs were ST16 and ST179; the ST16 isolates were more likely to form strong or medium biofilm than the ST179 isolates (p < 0.001). Strong or medium biofilm formation was more common in linezolid-resistant isolates than in linezolid-sensitive isolates (p = 0.001). Biofilm formation was more frequently detected in enterococcal surface protein (esp+), surface aggregating protein (asa1+), cytolysin A (cylA+), or aggregation substance (agg+) positive isolates than in isolates that were negative (-) for these virulence factors. Multivariate regression analysis indicated that cylA [odds ratio (OR) 4.083, p < 0.001] was a risk factor for weak biofilm formation, and that esp (OR 8.207, p < 0.001) was a risk factor for strong or medium biofilm formation. The expression of cylA was raised (4.02 to 6.00-fold) in weak biofilm isolates compared to the biofilm-negative isolates, and the expression of esp was greatly elevated (11.39 to 134.08-fold) in strong biofilm isolates compared to biofilm-negative isolates. In conclusion, the ST16 classification and linezolid resistance were positively associated with strong/medium biofilm formation in clinical E. faecalis isolates. cylA was associated with weak biofilm formation, and esp was only associated with strong or medium biofilm formation of the clinical E. faecalis isolates.
Keywords: Enterococcus faecalis, biofilm formation, virulence genes, multilocus sequence typing, linezolid resistance
Introduction
Enterococci are normal gut microbes in humans and other animals (Sghir et al., 2000; Damborg et al., 2009). However, in the last two decades, enterococcus pathogens have emerged as a major cause of nosocomial infections affecting various tissues, including the urinary tract, respiratory tract, peritoneum, and bloodstream (Treitman et al., 2005; Bonten and Willems, 2012). The most prevalent species cultured from humans, Enterococcus faecalis and Enterococcus faecium, account for more than 90% of clinical enterococcal isolates (Fisher and Phillips, 2009). It is noteworthy that E. faecalis and E. faecium cause treatment to be increasing difficult because of their intrinsic and acquired resistance to many antibiotics. E. faecalis and E. faecium have resistance to many commonly used antimicrobial agents, such as ampicillin and vancomycin (Van Harten et al., 2017). VRE especially have emerged as a major cause of outbreaks of nosocomial infection, which with their extensive resistance to a plethora of antibiotics have attracted more and more attention in recent years (Flokas et al., 2017). Linezolid, as the first antimicrobial agent of the oxazolidinones class, was widely used to treat VRE infections. Unfortunately, growing cases of linezolid-resistant enterococci have emerged in hospitals (Gawryszewska et al., 2017).
In addition to drug resistance, enterococci usually with a high capacity of biofilm formation, which has increased the difficulty of treatment. In Italy, 86.7% of E. faecalis clinical isolates formed different levels of biofilms, whereas only 15.6% of E. faecium isolates formed biofilms (Dupre et al., 2003). Among 109 enterococcal bloodstream isolates from Britain, 100% of E. faecalis strains had the ability to form biofilm, but only 42% of E. faecium isolates formed biofilms (Sandoe et al., 2003). E. faecalis isolates from the USA also had a high capacity of biofilm formation, Mohamed et al. found 93% of 163 E. faecalis isolates from various sources classified as biofilm producers (Mohamed et al., 2004). Another study from Japan indicated 100% of 352 E. faecalis isolates in urinary tract infections with different levels of biofilm formation (Seno et al., 2005). However, only 57.2% of E. faecalis isolates from Spain had the ability to form biofilms, and the 39 E. faecium isolates in the study couldn’t produce biofilms (Toledo-Arana et al., 2001). These studies found the E. faecalis isolates to have a higher capacity of biofilm formation than the E. faecium isolates, and the prevalence of E. faecalis biofilm differs regionally. However, up to now, the prevalence and characteristics of E. faecalis biofilm in China have remained obscure.
The phylogenetic relationships of pathogenic bacteria, including Staphylococcus spp. and Enterococcus spp., have been examined with MLST (Urwin and Maiden, 2003), a methodology that has yielded STs with distinctive virulence and drug resistance strain profiles (Manning et al., 2009). Biofilm formation has also been shown to vary across isolate STs (Kozitskaya et al., 2005). ST27 of Staphylococcus epidermidis, which is prone to biofilm formation, was found to occur preferentially in hospitals and to differ from clones in the community (Kozitskaya et al., 2005). The ST17 and ST19 lineages of group B Streptococcus strains isolated from invasive disease case samples were found to form weak biofilms relative to strains recovered from individuals with asymptomatic colonization (Parker et al., 2016). The biofilm formation properties of different STs of E. faecalis have not been characterized.
Previous researchers have reported that some virulence factors play important roles in the pathogenicity of E. faecalis, and several virulence factors may be related to E. faecalis biofilm formation. Esp is a large surface protein, which has been found to support cell adherence, colonization, and persistence in the urinary tract, evasion of the immune system, and to play an important role in E. faecalis biofilm formation (Toledo-Arana et al., 2001; Paganelli et al., 2012). GelE is an extracellular metalloprotease that hydrolyzes gelatin, collagen, and hemoglobin. GelE was responsible for regulating autolysis and the release of high-molecular-weight eDNA, an important component for the development of E. faecalis biofilms, and has also been reported to contribute to the bacterial adherence and biofilm formation of E. faecalis (Kayaoglu and Ørstavik, 2004; Park et al., 2007; Thomas et al., 2008). However, some research groups have found no significant correlation between the presence of Esp or GelE and E. faecalis biofilm formation (Kristich et al., 2004; Mohamed and Murray, 2005; Anderson et al., 2016).
Another virulence factor of E. faecalis, the Asa1, mediates aggregation between bacteria and enables the transfer of plasmids, was also found to promote biofilm formation in one study (Süßmuth et al., 2000; Chuang-Smith et al., 2010). The cylA gene of E. faecalis synthesizes a protein involved in the activation of cytolysin, and the lytic action of cytolysin on various cell types has been explored, including its contribution to the virulence of E. faecalis in infections (Van Tyne et al., 2013). Another study found cylA may be associated with E. faecalis biofilm formation in urinary tract infections (Seno et al., 2005). However, evidence of the involvement of asa1 and cylA in E. faecalis biofilm formation is still deficient and needs to be confirmed by further studies. Hence, the virulence factors that enable E. faecalis biofilm formation remain ambiguous and controversial.
The aim of the present study was to explore the prevalence and characteristics of E. faecalis biofilm, and to identify virulence factors associated with E. faecalis biofilm formation. To our knowledge, this is the first study to combine data on biofilm formation, virulence genes, antibiotic resistance, and MLST of E. faecalis from China.
Materials and Methods
Bacterial Isolates
A total of 265 clinical E. faecalis isolates were obtained from patients at Shenzhen Nanshan Hospital, Shenzhen University in China between January 1 2010 and June 30 2016. Information on the antibiotics used in each case was collected from the electronic medical records database. Antibiotic use was as follows: 44 cases involved penicillins (ampicillin, piperacillin, amoxicillin), 15 cases involved glycopeptides (vancomycin, teicoplanin), 4 cases involved linezolid, and 37 cases involved other antibiotics (ciprofloxacin, levofloxacin, moxifloxacin, minocycline). The strains isolated from the patients were identified with a VITEK 2 microbial testing system (bioMérieux, Marcy l’Etoile, France). E. faecalis ATCC29212 and OG1RF (ATCC47077) strains purchased from the American Type Culture Collection were used as reference strains. All procedures involving human participants were performed in accordance with the ethical standards of Shenzhen University and with the 1964 Helsinki declaration and its later amendments, or comparable ethical standards. For this type of study, formal consent is not required.
Antibiotic-Susceptibility Testing
The susceptibilities of the isolates to clinically relevant antibiotics (i.e., penicillin G, ampicillin, imipenem, ciprofloxacin, levofloxacin, teicoplanin, vancomycin, tigecycline, cotrimoxazole, linezolid, high-level gentamicin, erythromycin, and tetracycline) were determined with the VITEK 2 system (bioMérieux, Marcy l’Etoile, France). We determined the MICs of ampicillin, vancomycin, and linezolid using the broth macrodilution method according to CLSI guidelines, with CLSI-recommended MIC breakpoints.
Isolation of DNA
DNA was extracted from isolates and purified with a DNeasy Blood and Tissue Kit (Qiagen China Co., Ltd., Zhangjiang Hi-Tech Park Pudong, Shanghai, China) according to the manufacturer’s protocol for Gram-positive bacteria. The microbial DNA was eluted with 100 μl of AE buffer (Qiagen) and stored at -20°C.
Detection of Virulence Genes by PCR
The extracted DNA served as a template for the amplification of virulence genes. All primer sequences and corresponding references are listed in Table 1 (Vankerckhoven et al., 2004; Dupre et al., 2003). PCR amplification was performed in a total volume of 50 μl, containing 2× PCR Master Mix (Tiangen Biotech Beijing Co., Ltd., Beijing, China), 0.5 mM of each primer, and 1 μl template DNA. The cycling conditions were as follows: 95°C for 3 min; followed by 30 cycles at 95°C for 30 s, 52°C for 30 s, 72°C for 60 s; and a final 10 min extension step at 72°C. Each PCR set included a no-template control and a positive control. The amplification products were analyzed by electrophoresis in 1.0% agarose gels.
Table 1.
PCR primers used for detection of Enterococcus faecalis virulence factors.
| Target | Primer | Primer sequence (5′–3′) | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| esp | esp-F | AATTGATTCTTTAGCATCTGG | 510 | Vankerckhoven et al., 2004 |
| esp-R | AGATTCATCTTTGATTCTTGG | |||
| gelE | gelE-F | TATGACAATGCTTTTTGGGAT | 213 | Vankerckhoven et al., 2004 |
| gelE-R | AGATGCACCCGAAATAATATA | |||
| asa1 | asa1-F | GCACGCTATTACGAACTATGA | 375 | Vankerckhoven et al., 2004 |
| asa1-R | TAAGAAAGAACATCACCACGA | |||
| cylA | cylA-F | ACTCGGGGATTGATAGGC | 688 | Vankerckhoven et al., 2004 |
| cylA-R | GCTGCTAAAGCTGCGCTT | |||
| hyl | hyl-F | GACTGACGTCCAAGTTTCCAA | 276 | Vankerckhoven et al., 2004 |
| hyl-R | ACAGAAGAGCTGCAGGAAATG | |||
| agg | agg-F | CCAGTAATCAGTCCAGAAACAACC | 406 | Dupre et al., 2003 |
| agg-R | TAGCTTTTTTCATTCTTGTGTTTGTT |
Multilocus Sequence Typing
Multilocus sequence typing was conducted by the reference method (Ruiz-Garbajosa et al., 2006). Briefly, seven housekeeping genes (gdh, gyd, pstS, gki, aroE, xpt, and yqiL) were PCR amplified and sequenced. All primer sequences and corresponding references are listed in Table 2. The PCR system and cycling conditions were the same as indicated above for the virulence genes. For each locus, a distinct allele number was assigned to each unique sequence, in accordance with the E. faecalis MLST database1. The seven STs were assigned in the order gdh, gyd, pstS, gki, aroE, xpt, and yqiL, corresponding to the allele numbers at the seven loci. ST names were kept consistent as much as possible for the same strains.
Table 2.
PCR primers used for E. faecalis MLST gene diversity determination.
| Target | Primer | Primer sequence (5′–3′) | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| gdh | gdh-F | GGCGCACTAAAAGATATGGT | 530 | Ruiz-Garbajosa et al., 2006 |
| gdh-R | CCAAGATTGGGCAACTTCGTCCCA | |||
| gyd | gyd-F | CAAACTGCTTAG CTCCAATGGC | 395 | Ruiz-Garbajosa et al., 2006 |
| gyd-R | CATTTCGTTGTCATACCAAGC | |||
| pstS | pstS-F | CGGAACAGGACTTTCGC | 583 | Ruiz-Garbajosa et al., 2006 |
| pstS-R | ATTTACATCACGTTCTACTTGC | |||
| gki | gki-F | GATTTTGTGGGAATTGGTATGG | 438 | Ruiz-Garbajosa et al., 2006 |
| gki-R | ACCATTAAAGCAAAATGATCGC | |||
| aroE | aroE-F | TGGAAAACTTTACGGAGACAGC | 459 | Ruiz-Garbajosa et al., 2006 |
| aroE-R | GTCCTG TCCATTGTTCAAAAGC | |||
| xpt | xpt-F | AAAATGATGGCCGTGTATTAGG, | 456 | Ruiz-Garbajosa et al., 2006 |
| xpt-R | AACGTCACCGTTCCTTCACTTA | |||
| yqiL | yqiL-F | CAGCTTAAGTCAAG TAAGTGCCG | 436 | Ruiz-Garbajosa et al., 2006 |
| yqiL-R | GAATATCCCTTCTGCTTGTGCT |
Biofilm Assay
Enterococcus faecalis biofilm formation was detected according to the reference method, with some modifications (Toledo-Arana et al., 2001; Mohamed et al., 2004). Briefly, bacteria that had been grown overnight were diluted 1:100 in 200 μl of tryptic soy broth with 0.25% glucose and inoculated onto polystyrene microtiter plates (Costar3599; Corning). After 24 h of static incubation at 37°C, the supernatant was discarded, and plates were washed thrice with deionized water to remove unattached cells, then fixed with methanol for 30 min, stained with 1% crystal violet for 30 min, and rinsed with distilled water. The optical density at 570 nm (OD570) was determined. Each assay was performed in triplicate on at least three occasions.
RNA Extraction and qRT-PCR
The expression levels of the esp and cylA genes of E. faecalis clinical isolates were determined by qRT-PCR. The primers used for qRT-PCR are listed in Table 3. The RNA extraction was carried out according to the reference method (Xu et al., 2017), with some modifications: the E. faecalis strains were cultured at 37°C until the OD600 reached 0.6. Then, the cell pellets were washed with ice-cold normal saline and then homogenized using 0.1 mm Zirconia-silica beads in a Mini-BeadBeater (Biospec, Bartlesville, OK, United States) at a speed of 4,000 rpm for 1 min, followed by cooling on ice for 1 min. This homogenization and cooling cycle was repeated five times; then, the samples were centrifuged at 15,000 rpm and the bacterial RNA in the supernatant was purified using an RNeasy Mini Kit (Qiagen) and quantified using an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States). RNA samples that had a 260/280 ratio between 2.0 and 2.2 were reverse transcribed with the PrimeScript RT Reagent Kit (TaKaRa). Finally, qRT-PCR was performed with the SYBR Premix Ex Taq II Kit (TaKaRa) on the Mastercycler ep realplex system (Eppendorf), with an initial incubation at 95°C for 2 min, followed by 40 cycles of 15 s at 95°C, and 60 s at 60°C. Each sample was analyzed in triplicate.
Table 3.
PCR primers used for determining the expression levels of esp and cylA of E. faecalis isolates by quantitative real-time polymerase chain reaction (qRT-PCR).
| Target | Primer | Primer sequence (5′–3′) | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| recA | recA-F | CGACTAATGTCTCAAGCACTAC | 106 | This study |
| recA-R | CGAACATCACGCCAACTT | |||
| esp | Resp-F | GCATCAGTATTAGTTGGT | 196 | This study |
| Resp-R | TTCCTTGTAACACATCAC | |||
| cylA | RcylA-F | GGAGGATATGGTGACAAT | 163 | This study |
| RcylA-R | TTACTTCTGGAGTTGCTAA |
For all samples, the internal control gene recA (Ruiz-Cruz et al., 2015) was used to normalize the expression of the esp and cylA genes. The threshold cycle (Ct) numbers were confirmed by the detection system software and the data were analyzed based on the 2-ΔΔCt method.
Statistical Analysis
The prevalence of E. faecalis biofilm is reported as a number (percentage) and was compared using the chi-square test or Fisher’s exact test. The virulence factors associated with biofilm formation were analyzed with a multivariate logistic regression model constructed by backward selection based on the Wald statistic. ORs are reported with 95% CIs. P-values < 0.05 were regarded as statistically significant. All data were analyzed using the statistical software SPSS (version 14.0; SPSS, Chicago, IL, United States).
Results
Biofilm Formation
OD570 microplate readings after crystal violet staining ranged from 0.05 to 3.5. The numbers and percentages of the isolates, altogether and segregated by clinical source, and the biofilm phenotype categorized based on the approach of others (Toledo-Arana et al., 2001; Mohamed et al., 2004), with a strong (OD570 > 2), medium (OD570, 1–2), or weak (OD570 > 0.5 and <1) biofilm formation phenotype, are reported in Table 4. The median OD570 values for control strains OG1RF [weak biofilm (Montealegre et al., 2015)] and ATCC29212 were 0.56 and 0.15, respectively.
Table 4.
Occurrence of E. faecalis biofilm formation by clinical source.
| Clinical source (no. isolates tested) | No. (%) of isolates with biofilm phenotype |
||||
|---|---|---|---|---|---|
| Strong | Medium | Strong or medium | Weak | All positive | |
| Blood (25) | 5 (20.0) | 1 (4.0) | 6 (24.0) | 4 (16.0) | 10 (40.0) |
| Urine (113) | 15 (13.3) | 15 (13.3) | 30 (26.6) | 27 (23.9) | 57 (50.4) |
| Pus or secretions (64) | 6 (9.4) | 1 (1.6) | 7 (11.0) | 22 (34.4) | 29 (45.3) |
| Bile (19) | 1 (5.3) | 4 (21.1) | 5 (26.3) | 2 (10.5) | 7 (36.8) |
| Othera (44) | 8 (18.2) | 3 (6.8) | 11 (25.0) | 11 (25.0) | 22 (50.0) |
| Total (265) | 35 (13.2) | 24 (9.1) | 59 (22.3) | 66 (24.9) | 125 (47.2) |
aSputum, tissue, catheters, pleural effusion, ascites fluid, amniotic fluid, puncture fluid, cerebrospinal fluid, throat swabs.
As reported in Table 4, biofilms were observed for nearly half of all of the E. faecalis isolates examined, and among these biofilm-forming strains, the majority were classified as having a weak biofilm phenotype. With respect to clinical source, isolates from urine were the most likely to exhibit biofilm formation, followed by isolates from bile; the likelihood of biofilm formation was statistically similar across the remaining sources.
Association between ST and Biofilm Formation
Multilocus sequence typing of 265 E. faecalis clinical isolates enabled us to determine the STs for 224 isolates. We found 44 different STs, 41 of which were represented by four or fewer isolates. The biofilm characteristics and numbers of isolates assigned to each ST are reported in Supplementary Table S1.
Because ST16 and ST179 were quite dominant, accounting for 65.6% (147/224) of the isolates in this study, the biofilm formation characteristics of the ST16 and ST179 isolates were subjected to further analysis. As shown in Table 5, biofilm formation, and strong or medium biofilm formation in particular, were detected significantly more frequently among ST16 isolates than among ST179 isolates. Further analysis of the clinical sources of ST16 and ST179 E. faecalis isolates indicated that the characteristics of each were consistent across clinical sources (Supplementary Table S2).
Table 5.
Biofilm-forming capacity comparison between ST16 and ST179 isolates.
| Biofilm phenotype | No. (%) with phenotype |
P | |
|---|---|---|---|
| ST16, N = 79 | ST179, N = 68 | ||
| Strong | 19 (24.1) | 1 (1.5) | <0.001 |
| Medium | 9 (11.4) | 3 (4.4) | NS |
| Strong or medium | 28 (35.4) | 4 (5.9) | <0.001 |
| Weak | 20 (25.3) | 24 (35.3) | NS |
| All positive | 48 (60.8) | 28 (41.2) | 0.018 |
NS, not significant.
Association between Biofilm Formation and Linezolid Susceptibility
Using the broth macrodilution method to determine the MICs of ampicillin, vancomycin, and linezolid, and a resistance criterion classification of MIC ≥8μg/ml, we found 10 strains with linezolid resistance (the 10 patients received no linezolid treatment), only four strains with ampicillin resistance (only 1 of the 4 patients received piperacillin-tazobactam treatment), and none with vancomycin resistance. Follow-up analysis of the relationship between E. faecalis biofilm formation and linezolid susceptibility showed that the linezolid-resistant strains were more likely to exhibit biofilm formation than the linezolid-sensitive strains (MIC ≤ 2 μg/ml), and that the biofilm formation characteristics of the isolates with a linezolid MIC in the intermediate range (2–8 μg/ml; n = 41) did not differ significantly from that of the lower or higher MIC isolate groups (Table 6). More specifically, strong or medium biofilm formation was more prevalent among linezolid-resistant isolates than among linezolid-sensitive isolates (Table 6). Two ST16 isolates and no ST179 isolates were included among the linezolid-resistant strains; biofilm prevalence did not differ significantly between linezolid-sensitive and -resistant isolates (Supplementary Table S3).
Table 6.
Correlation between biofilm-forming capacity and linezolid sensitivity.
| Biofilm phenotype | No. (%) with linezolid MIC in μg/ml |
||
|---|---|---|---|
| MIC ≤ 2, N = 214 | MIC 2–8, N = 41 | MIC ≥ 8, N = 10 | |
| Strong | 24 (11.2) | 9 (22.0) | 2 (20.0) |
| Medium | 19 (8.9) | 0 (0.0) | 5 (50.0)a |
| Strong or medium | 43 (20.1) | 9 (22.0) | 7 (70.0)b |
| Weak | 52 (24.3) | 12 (29.3) | 2 (20.0) |
| All positive | 95 (44.4) | 21 (51.2) | 9 (90.0)c |
aMedium: MIC ≥ 8 vs. MIC ≤ 2, p = 0.002. bStrong or medium: MIC ≥ 8 vs. MIC ≤ 2, p = 0.001. cPositive: MIC ≥ 8 vs. MIC ≤ 2, p = 0.007.
Correlation between Biofilm Formation and Virulence Factors
As reported in Table 7, the analysis of PCR-amplified virulence factors showed that the esp-positive (+) E. faecalis isolates had a significantly greater prevalence of biofilm formation than the esp-negative (-) isolates. Additionally, biofilm formation was detected more frequently in asa1+, cylA+ (cytolysin A-encoding gene positive), and agg+ (aggregation substance-encoding gene positive) isolates than in isolates not expressing each of these virulence factors (Table 7). Conversely, gelE- isolates were significantly more likely to have biofilm formation than gelE+ isolates, especially biofilms categorized as strong or medium (Table 7, see Footnote).
Table 7.
Relationship between biofilm-forming capacity and virulence factors.
| Virulence factors (no. isolates tested) | No. (%) of isolates with biofilm phenotype |
Pa | ||||
|---|---|---|---|---|---|---|
| Strong | Medium | Strong or medium | Weak | All positive | ||
| esp+ (163) | 30 (18.4) | 20 (12.3) | 50 (30.7) | 47 (28.8) | 97 (59.5) | <0.001 |
| esp- (102) | 5 (4.9) | 4 (3.9) | 9 (8.8) | 19 (18.6) | 28 (27.5) | |
| gelE+ (171) | 12 (7.0) | 13 (7.6) | 25 (14.6) | 46 (26.9) | 71 (41.5) | 0.013 |
| gelE- (94) | 23 (24.5)b | 11 (11.7) | 34 (36.2)c | 20 (21.3) | 54 (57.4) | |
| asa1+ (210) | 32 (19.4) | 20 (9.7) | 52 (29.1) | 57 (26.0) | 109 (55.1) | 0.003 |
| asa1- (55) | 3 (5.5) | 4 (7.3) | 7 (12.7) | 9 (16.4) | 16 (29.1) | |
| cylA+ (178) | 26 (14.6) | 19 (10.7) | 45 (25.3) | 57 (32.0) | 102 (57.3) | <0.001 |
| cylA- (87) | 9 (10.3) | 5 (5.7) | 14 (16.1) | 9 (10.3) | 23 (26.4) | |
| hyl+ (83) | 7 (8.4) | 6 (7.2) | 13 (15.7) | 20 (24.1) | 33 (39.8) | 0.103 |
| hyl- (182) | 28 (15.4) | 18 (9.9) | 46 (25.3) | 46 (25.3) | 92 (50.5) | |
| agg+ (88) | 11 (12.5) | 12 (13.6) | 23 (26.1) | 27 (30.7) | 50 (56.8) | 0.027 |
| agg- (177) | 24 (13.6) | 12 (6.8) | 36 (20.3) | 39 (22.0) | 75 (42.4) | |
aBiofilm formation: + vs. –. bStrong biofilm: gelE– vs. gelE+, p < 0.001. cStrong or medium biofilm: gelE– vs. gelE+, p < 0.001.
Virulence Factor Association with Biofilm Formation by Multiple Regression Analysis
In order to determine the independent contribution of each virulence factor to the biofilm formation of E. faecalis, multiple logistic regression analysis was performed. Thus, esp, gelE, asal, cylA, and agg were used as independent variables, and weak biofilm formation, strong or medium biofilm formation were used as a dependent variable in the multivariate model. The logistic regression model was constructed by a backward selection approach based on the Wald statistic. P-values < 0.05 were regarded as statistically significant. As shown in Table 8, only one factor, cylA, was found to be an independent risk factor for weak biofilm formation. Meanwhile, esp positivity was independently associated with strong or medium biofilm formation, and gelE emerged as an independent anti-risk factor for strong or medium biofilm formation.
Table 8.
Multivariate regression analysis of virulence factors associated with E. faecalis biofilm formation.
| Isolate biofilm phenotype | Factor | OR (95% CI) | P |
|---|---|---|---|
| Weak biofilm formation | |||
| cylA | 4.083 (1.912–8.716) | <0.001 | |
| Strong or medium biofilm formation | |||
| gelE | 0.300 (0.159–0.564) | <0.001 | |
| cylA | 0.393 (0.146–1.056) | 0.064 | |
| esp | 8.207 (2.795–24.099) | <0.001 |
Expression Levels of the esp and cylA Genes in E. faecalis Clinical Isolates
Last, in order to clarify the role of E. faecalis virulence factors during biofilm formation, the expression levels of the esp and cylA genes of E. faecalis clinical isolates were determined by qRT-PCR. For the detection of esp expression, eight clinical E. faecalis isolates that were esp positive, and with the same phenotypes of other virulence factors (asa1+; gelE-; cylA-; hyl-; agg+) were chosen. Among those 8 esp-positive isolates, the 16C383 strain with negative biofilm formation was set as the reference strain (esp expression = 1). As shown in Figure 1, the esp expression was markedly elevated (11.39 to 134.08-fold, respectively) in those strong biofilm isolates (16C1, 16C51, 16C126, and 16C353), compared with the weak biofilm producers (16C29, 4.01-fold and 16C274, 3.47-fold). These results suggest that esp was only associated with strong biofilm, not with the weak biofilm formation of E. faecalis isolates, consistent with the previous findings.
FIGURE 1.
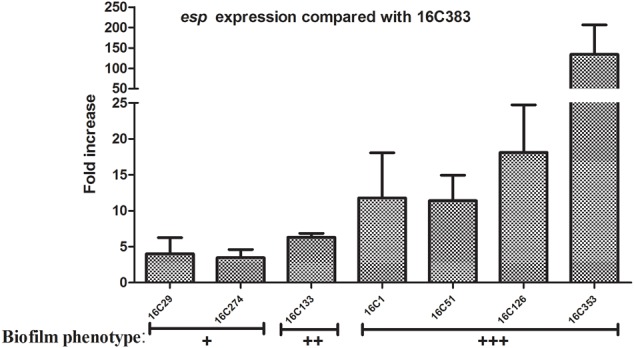
Relative fold expression of esp in seven clinical Enterococcus faecalis isolates. The expression levels of esp were determined by quantitative real-time polymerase chain reaction (qRT-PCR), with the clinical E. faecalis isolate 16C383 as the reference strain (expression = 1, biofilm negative). Biofilm phenotype: +, weak; ++, medium; +++, strong. The esp was overexpressed in the four strong biofilm strains (11.39 to 134.08-fold).
For the test of cylA expression, nine clinical E. faecalis isolates that were cylA positive, and with the same phenotypes of other virulence factors (asa1+; gelE-; esp-; hyl-; agg+) were chosen. Among those 9 cylA-positive isolates, the 16C281 strain with negative biofilm formation was set as the reference strain (cylA expression = 1). As shown in Figure 2, the cylA gene expression was higher (4.02 to 6.00-fold) in those weak biofilm isolates (16C24, 16C54, 16C305, 16C306, and 16C374), compared with in the negative biofilm isolate (16C394, 1.37-fold), or the medium (16C169, 2.82-fold) or strong (16C60, 1.70-fold) biofilm isolates. These data may confirm that the cylA gene was only associated with weak biofilm, not with the strong or medium biofilm formation of E. faecalis isolates.
FIGURE 2.
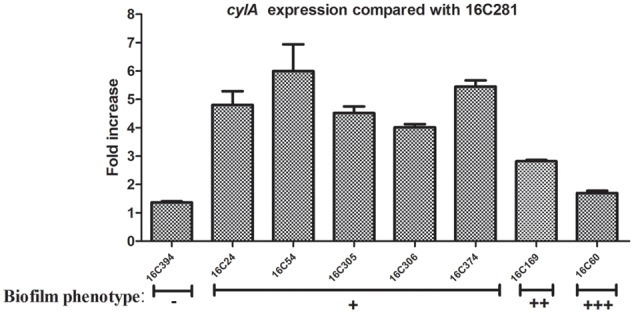
Relative fold expression of cylA in eight clinical E. faecalis isolates. The expression levels of cylA were determined by qRT-PCR, with the clinical E. faecalis isolate 16C281 as the reference strain (expression = 1, biofilm negative). Biofilm phenotype: –, negative; +, weak; ++, medium; +++, strong. The cylA was overexpressed in the five weak biofilm strains (4.02 to 6.00-fold).
Discussion
Although E. faecalis is generally less drug resistant than E. faecium, we found that biofilm formation was more prevalent for E. faecalis than has been reported previously for E. faecium (Sandoe et al., 2003). The prevalence of E. faecalis biofilm observed in this study, 47.2%, was lower than the 60–90% prevalence rates reported previously in Europe (Toledo-Arana et al., 2001; Dupre et al., 2003; Sandoe et al., 2003). In particular, the prevalence of E. faecalis biofilm in this study for isolates from urine, 50.4%, and from blood, 40.0%, were essentially half of the 100% prevalence rates reported previously for isolates from urine (Seno et al., 2005) and blood (Sandoe et al., 2003). Among the 57 E. faecalis isolates from urine with biofilm formation ability, 47.4% (27/57) of isolates were weak biofilm producers. Another study from Japan found in 352 E. faecalis strains from urinary tract infections that all of them had the capacity to form biofilm, 37.5% (132/352) of isolates exhibited weak biofilm formation (Seno et al., 2005). Our study found the cylA was associated with E. faecalis isolates with weak biofilm formation, and the prevalence of cylA in the biofilm-positive E. faecalis isolates from urine was 78.9% (45/57), which was higher than in the Seno et al. research (46.6%, 164/352). Thus, the prevalence of the weak biofilm phenotype in those E. faecalis isolates from urine may be due to the expression of cylA, based on the present study.
The lower rates of E. faecalis biofilm observed here could be due to several possible reasons. First, the E. faecalis strains isolated from different studies may have different ST frequencies. Second, because crystal violet staining analysis is a semi-quantitative method, it carries a risk of operation error. Last, there is not yet a consistent criterion for the determination of a positive biofilm, particularly with respect to a crystal violet optical density cut-off value.
The present demonstration of 44 STs provides, to the best of our knowledge, the first MLST of E. faecalis in China. The dominance of ST16 and ST179 among our samples is somewhat consistent with a prior study in Malaysia, in which ST6, ST16, ST28, ST179, and ST399 were represented (Weng et al., 2013). Our finding of greater biofilm prevalence, especially strong/medium biofilm prevalence, among ST16 isolates than among ST179 isolates is noteworthy in light of previous evidence showing that ST16 isolates may be representative of hospital-adapted strains (Ruiz-Garbajosa et al., 2006), and the observation that, relative to planktonic bacteria, biofilms enable bacteria to better survive in adverse environmental conditions, including in the presence of antibiotics and disinfectants (Lebeaux et al., 2014). E. faecalis ST16 may, like S. epidermidis ST27 (Kozitskaya et al., 2005), occur preferentially in nosocomial environments, with an atypically high prevalence of biofilm formation relative to other STs of the microbe species.
The relationship between E. faecalis biofilm formation and antibiotic resistance had not been well explored before this study. Interest in VREs has been growing in recent years (O’Driscoll and Crank, 2015). Linezolid has been used to treat VRE infection, but wider use of linezolid may be increasing the prevalence of linezolid resistance in E. faecalis (Kainer et al., 2007). Indeed, here we found ten E. faecalis isolates that were resistant to linezolid, which was with no linezolid treatment, but found no VRE isolates. Moreover, we found that the prevalence of biofilm formation, especially the strong or medium biofilm formation, was significantly higher in linezolid-resistant E. faecalis isolates than in linezolid-sensitive isolates. Although E. faecalis ST16 was found to have a high prevalence of biofilm formation, ST16 isolates were similarly represented across linezolid-sensitive and -resistant strains, indicating that biofilm formation by linezolid-resistant isolates cannot be attributed to the ST16 profile per se. Further research is needed to examine whether biofilm formation plays a role in E. faecalis resistance to linezolid.
The present finding that esp+ isolates are more likely to exhibit strong or medium biofilm formation than esp- isolates is consistent with several previous studies that have suggested a link between the virulence factor Esp and E. faecalis biofilm formation (Toledo-Arana et al., 2001; Tendolkar et al., 2004; Paganelli et al., 2012). Notwithstanding, other studies have failed to find evidence of such a link (Kristich et al., 2004; Mohamed and Murray, 2005; Di Rosa et al., 2006; Elhadidy and Elsayyad, 2013; Anderson et al., 2016). We did not observe an association between esp positivity and weak biofilm formation, suggesting the possibility that esp might enhance extant E. faecalis biofilm formation, without having a fundamental influence on the basic capacity for biofilm formation per se.
Our finding of gelE positivity being inversely associated with strong or medium biofilm formation in E. faecalis is surprising. Previous studies have suggested that the gelE-encoded protein, which can hydrolyze gelatin, collagen, and hemoglobin, may favor the development of E. faecalis biofilms (Kayaoglu and Ørstavik, 2004; Park et al., 2007; Thomas et al., 2008), while others found no significant correlation between the presence of gelE and biofilm formation in E. faecalis (Mohamed and Murray, 2005; Anderson et al., 2016). It might be that the STs of our E. faecalis strains differed from those of previous studies, or there may be as yet unidentified factors that determine E. faecalis biofilm formation.
Our negative finding with respect to asa1 affecting biofilm formation is inconsistent with a prior study showing as association between asa1 and E. faecalis biofilm formation (Chuang-Smith et al., 2010). Our finding that cylA was only a significant factor in weak biofilm formation suggests that cylA may be irrelevant to strong or medium biofilm formation in E. faecalis. The cylA gene has been reported previously to be associated with the E. faecalis biofilm formation of urinary tract isolates (Seno et al., 2005). However, thus far, there is little information regarding the potential role of cylA – which can lyse prokaryotic cells, as well as erythrocytes and other eukaryotic cells – in E. faecalis biofilm formation.
Conclusion
The present study showed that a large portion of E. faecalis isolates from clinical samples form biofilms readily. The MLST of E. faecalis in this study demonstrated substantial ST diversity, with the main STs being ST16 and ST179. In general, biofilm formation was more strongly associated with ST16 than ST179. Our findings suggest a positive association between linezolid resistance in E. faecalis and robust biofilm formation. Interestingly, we found that the cylA gene was associated with weak biofilm formation of E. faecalis isolates, while the esp gene was only associated with strong or medium biofilm formation of E. faecalis isolates.
Author Contributions
J-XZ participated in the design of the study, carried out the biofilm assay, analyzed and interpreted the data, and drafted the manuscript. YW participated in the analysis of the data and provided a critical revision of the manuscript. Z-WL conducted the RNA extraction, quantitative real-time PCR and the data analysis. Z-YP performed antibiotic-susceptibility testing, detected virulence genes by PCR, and participated in the data analysis. W-MY, ZC, and D-YL participated in the acquisition of the samples, isolated DNA, conducted MLST, and participated in the data analysis. Q-WD, DQ, and Z-JY designed the study, participated in the data analysis, and provided critical revisions of the manuscript for important intellectual content.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Xiao-fei Wang and Zhi-hui Lv (Key Laboratory of Medical Molecular Virology of Ministries of Education and Health, School of Basic Medical Science and Institutes of Biomedical Sciences, Shanghai Medical College of Fudan University) for their excellent technical support and suggestions.
Abbreviations
- Agg
aggregation substance
- Asa1
surface aggregating protein
- CI
confidence intervals
- CLSI
Clinical and Laboratory Standards Institute
- CylA
cytolysin A
- eDNA
extracellular DNA
- Esp
enterococcal surface protein
- GelE
gelatinase
- MIC
minimal inhibitory concentration
- MLST
multilocus sequence typing
- OR
Odds ratio
- qRT-PCR
quantitative real-time polymerase chain reaction
- ST
sequence type
- VRE
vancomycin-resistant enterococci
Funding. This work was supported by grants from the National Natural Science Foundation of China (no. 81170370), from a grant from Sanming Project of Medicine in Shenzhen (SMGC201705029), the Shenzhen Scientific Research Program (Nos. JCYJ20150402152130167, JCYJ20150402152130173, and JCYJ20170307153714512), the Scientific Research Project of the Shenzhen Health and Family Planning System (no. 201601058), and the Shenzhen Nanshan District Scientific Research Program of China (no. 2016010).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02338/full#supplementary-material
References
- Anderson A. C., Jonas D., Huber I., Karygianni L., Wölber J., Hellwig E., et al. (2016). Enterococcus faecalis from food, clinical specimens, and oral sites: prevalence of virulence factors in association with biofilm formation. Front. Microbiol. 6:1534 10.3389/fmicb.2015.01534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonten M. J., Willems R. J. (2012). Vancomycin-resistant Enterococcus-chronicle of a foretold problem. Ned. Tijdschr. Geneeskund. 156 A5233. [PubMed] [Google Scholar]
- Chuang-Smith O. N., Wells C. L., Henry-Stanley M. J., Dunny G. M. (2010). Acceleration of Enterococcus faecalis biofilm formation by aggregation substance expression in an ex vivo model of cardiac valve colonization. PLOS ONE 5:e15798. 10.1371/journal.pone.0015798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damborg P., Top J., Hendrickx A. P., Dawson S., Willems R. J., Guardabassi L. (2009). Dogs are a reservoir of ampicillin-resistant Enterococcus faecium lineages associated with human infections. Appl. Environ. Microbiol. 75 2360–2365. 10.1128/AEM.02035-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rosa R., Creti R., Venditti M., D’Amelio R., Arciola C. R., Montanaro L., et al. (2006). Relationship between biofilm formation, the enterococcal surface protein (Esp) and gelatinase in clinical isolates of Enterococcus faecalis and Enterococcus faecium. FEMS Microbiol. Lett. 256 145–150. 10.1111/j.1574-6968.2006.00112.x [DOI] [PubMed] [Google Scholar]
- Dupre I., Zanetti S., Schito A. M., Fadda G., Sechi L. A. (2003). Incidence of virulence determinants in clinical Enterococcus faecium and Enterococcus faecalis isolates collected in Sardinia (Italy). J. Med. Microbiol. 52 491–498. 10.1099/jmm.0.05038-0 [DOI] [PubMed] [Google Scholar]
- Elhadidy M., Elsayyad A. (2013). Uncommitted role of enterococcal surface protein, Esp, and origin of isolates on biofilm production by Enterococcus faecalis isolated from bovine mastitis. J. Microbiol. Immunol. Infect. 46 80–84. 10.1016/j.jmii.2012.02.002 [DOI] [PubMed] [Google Scholar]
- Fisher K., Phillips C. (2009). The ecology, epidemiology and virulence of Enterococcus. Microbiology 155 1749–1757. 10.1099/mic.0.026385-0 [DOI] [PubMed] [Google Scholar]
- Flokas M. E., Karageorgos S. A., Detsis M., Alevizakos M., Mylonakis E. (2017). Vancomycin-resistant enterococci colonisation, risk factors and risk for infection among hospitalised paediatric patients: a systematic review and meta-analysis. Int. J. Antimicrob. Agents 49 565–572. 10.1016/j.ijantimicag.2017.01.008 [DOI] [PubMed] [Google Scholar]
- Gawryszewska I., Żabicka D., Hryniewicz W., Sadowy E. (2017). Linezolid-resistant enterococci in Polish hospitals: species, clonality and determinants of linezolid resistance. Eur. J. Clin. Microbiol. Infect. Dis. 36 1279–1286. 10.1007/s10096-017-2934-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainer M. A., Devasia R. A., Jones T. F., Simmons B. P., Melton K., Chow S., et al. (2007). Response to emerging infection leading to outbreak of linezolid-resistant enterococci. Emerg. Infect. Dis. 13 1024–1030. 10.3201/eid1307.070019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayaoglu G., Ørstavik D. (2004). Virulence factors of Enterococcus faecalis: relationship to endodontic disease. Crit. Rev. Oral Biol. Med. 15 308–320. 10.1177/154411130401500506 [DOI] [PubMed] [Google Scholar]
- Kozitskaya S., Olson M. E., Fey P. D., Witte W., Ohlse K., Ziebuhr W. (2005). Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J. Clin. Microbiol. 43 4751–4757. 10.1128/JCM.43.9.4751-4757.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristich C. J., Li Y. H., Cvitkovitch D. G., Dunny G. M. (2004). Esp independent biofilm formation by Enterococcus faecalis. J. Bacteriol. 186 154–163. 10.1128/JB.186.1.154-163.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeaux D., Ghigo J. M., Beloina C. (2014). Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 78 510–543. 10.1128/MMBR.00013-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning S. D., Springman A. C., Lehotzky E., Lewis M. A., Whittam T. S., Davies H. D. (2009). Multilocus sequence types associated with neonatal group B streptococcal sepsis and meningitis in Canada. J. Clin. Microbiol. 47 1143–1148. 10.1128/JCM.01424-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed J. A., Huang W., Nallapareddy S. R., Teng F., Murray B. E. (2004). Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect. Immun. 72 3658–3663. 10.1128/IAI.72.6.3658-3663.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed J. A., Murray B. E. (2005). Lack of correlation of gelatinase production and biofilm formation in a large collection of Enterococcus faecalis isolates. J. Clin. Microbiol. 43 5405–5407. 10.1128/JCM.43.10.5405-5407.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montealegre M. C., La Rosa S. L., Roh J. H., Harvey B. R., Murray B. E. (2015). The Enterococcus faecalis EbpA Pilus protein: attenuation of expression, biofilm formation, and adherence to fibrinogen start with the rare initiation codon ATT. mBio. 6:e00467-15. 10.1128/mBio.00467-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll T., Crank C. W. (2015). Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infect. Drug Resist. 8 217–230. 10.2147/IDR.S54125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paganelli F. L., Willems R. J., Leavis H. L. (2012). Optimizing future treatment of enterococcal infections: attacking the biofilm? Trends Microbiol. 20 40–49. 10.1016/j.tim.2011.11.001 [DOI] [PubMed] [Google Scholar]
- Park S. Y., Kim K. M., Lee J. H., Seo S. J., Lee I. H. (2007). Extracellular gelatinase of Enterococcus faecalis destroys a defense system in insect hemolymph and human serum. Infect. Immun. 75 1861–1869. 10.1128/IAI.01473-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R. E., Laut C., Gaddy J. A., Zadoks R., Davies H. D., Manning S. (2016). Association between genotypic diversity and biofilm production in group B Streptococcus. BMC Microbiol. 16:86. 10.1186/s12866-016-0704-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Cruz S., Espinosa M., Goldmann O., Bravo A. (2015). Global regulation of gene expression by the MafR protein of Enterococcus faecalis. Front. Microbiol. 6:1521. 10.3389/fmicb.2015.01521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Garbajosa P., Bonten M., Robinson D., Robinson A., Top J., Nallapareddy S., et al. (2006). Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 44 2220–2228. 10.1128/JCM.02596-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoe J. A., Witherden I. R., Cove J. H., Heritage J., Wilcox M. H. (2003). Correlation between enterococcal biofilm formation in vitro and medical-device-related infection potential in vivo. J. Med. Microbiol. 52 547–550. 10.1099/jmm.0.05201-0 [DOI] [PubMed] [Google Scholar]
- Seno Y., Kariyama R., Mitsuhata R., Monden K., Kumon H. (2005). Clinical implications of biofilm formation by Enterococcus faecalis in the urinary tract. Acta Med. Okayama 59 79–87. [DOI] [PubMed] [Google Scholar]
- Sghir A., Gramet G., Suau A., Rochet V., Pochart P., Dore J. (2000). Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 66 2263–2266. 10.1128/AEM.66.5.2263-2266.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süßmuth S. D., Muscholl-Silberhorn A., Wirth R., Susa M., Marre R., Rozdzinski E. (2000). Aggregation substance promotes adherence, phagocytosis, and intracellular survival of Enterococcus faecalis within human macrophages and suppresses respiratory burst. Infect. Immunity 68 4900–4906. 10.1128/iai.68.9.4900-4906.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tendolkar P. M., Baghdayan A. S., Gilmore M. S., Shankar N. (2004). Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis. Infect. Immun. 72 6032–6039. 10.1128/IAI.72.10.6032-6039.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas V. C., Thurlow L. R., Boyle D., Hancock L. E. (2008). Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J. Bacteriol. 190 5690–5698. 10.1128/JB.00314-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Arana A., Valle J., Solano C., Arrizubieta M., Cucarella C., Lamata M., et al. (2001). The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 67 4538–4545. 10.1128/AEM.67.10.4538-4545.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treitman A. N., Yarnold P. R., Warren J., Noskin G. A. (2005). Emerging incidence of Enterococcus faecium among hospital isolates (1993 to 2002). J. Clin. Microbiol. 43 462–463. 10.1128/JCM.43.1.462-463.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwin R., Maiden M. C. (2003). Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11 479–487. 10.1016/j.tim.2003.08.006 [DOI] [PubMed] [Google Scholar]
- Van Harten R. M., Willems R. J. L., Martin N. I., Hendrickx A. P. A. (2017). Multidrug-resistant enterococcal infections: new compounds, novel antimicrobial therapies? Trends Microbiol. 25 467–479. 10.1016/j.tim.2017.01.004 [DOI] [PubMed] [Google Scholar]
- Van Tyne D., Martin M. J., Gilmore M. S. (2013). Structure, function, and biology of the Enterococcus faecalis cytolysin. Toxins 5 895–911. 10.3390/toxins5050895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankerckhoven V., Autgaerden T., Vael C., Lammens C., Chapelle S., Rossi R., et al. (2004). Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J. Clin. Microbiol. 42 4473–4479. 10.1128/JCM.42.10.4473-4479.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng P. L., Ramli R., Shamsudin M. N., Cheah Y. K., Hamat R. A. (2013). High genetic diversity of Enterococcus faecium and Enterococcus faecalis clinical isolates by pulsed-field gel electrophoresis and multilocus sequence typing from a hospital in Malaysia. Biomed Res. Int. 2013:938937. 10.1155/2013/938937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Wu Y., Lin Z. W., Bertram R., Götz F., Zhang Y., et al. (2017). Identification of genes controlled by the essential YycFG two-component system reveals a role for biofilm modulation in Staphylococcus epidermidis. Front. Microbiol. 8:724. 10.3389/fmicb.2017.00724 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


